Abstract
Understanding how ethanol actions on brain signal transduction and gene expression lead to excessive consumption and addiction could identify new treatments for alcohol dependence. We previously identified glycogen synthase kinase 3-beta (Gsk3b) as a member of a highly ethanol-responsive gene network in mouse medial prefrontal cortex (mPFC). Gsk3b has been implicated in dendritic function, synaptic plasticity and behavioral responses to other drugs of abuse. Here, we investigate Gsk3b in rodent models of ethanol consumption and as a risk factor for human alcohol dependence. Stereotactic viral vector gene delivery overexpression of Gsk3b in mouse mPFC increased 2-bottle choice ethanol consumption, which was blocked by lithium, a known GSK3B inhibitor. Further, Gsk3b overexpression increased anxiety-like behavior following abstinence from ethanol. Protein or mRNA expression studies following Gsk3b over-expression identified synaptojanin 2, brain-derived neurotrophic factor and the neuropeptide Y Y5 receptor as potential downstream factors altering ethanol behaviors. Rat operant studies showed that selective pharmacologic inhibition of GSK3B with TDZD-8 dose-dependently decreased motivation to self-administer ethanol and sucrose and selectively blocked ethanol relapse-like behavior. In set-based and gene-wise genetic association analysis, a GSK3b-centric gene expression network had significant genetic associations, at a gene and network level, with risk for alcohol dependence in humans. These mutually reinforcing cross-species findings implicate GSK3B in neurobiological mechanisms controlling ethanol consumption, and as both a potential risk factor and therapeutic target for alcohol dependence.
Introduction
Alcohol use disorder (AUD), consisting of the behavioral spectrum from alcohol abuse to alcohol dependence, is a disabling disease characterized by uncontrolled consumption of ethanol, negative social and health consequences, and an enormous cost to society [1]. Alcohol dependence is uniquely dependent on environmental influences, with pathogenesis requiring repeated ethanol exposure, yet the disease develops under polygenic risk factors totaling 40–60% heritability [2]. Understanding the mechanisms leading from initial ethanol exposure to alcohol dependence is considered a requisite for development of improved therapies for the disorder. Although clearly only part of the complex neurobiology of alcohol, selective alterations in brain gene expression have been suggested as a convergent mechanism underlying transition from alcohol use to abuse and dependence [3].
Acute and chronic ethanol exposure have been shown to prominently alter brain gene expression [3, 4] and show strikingly overlapping patterns of expression [5]. Acute molecular responses to ethanol might thus predict or contribute to long-term brain adaptations corresponding to altered behaviors seen in AUD. We previously used genome-wide expression network profiling across a recombinant inbred panel of mice (BXD) following acute ethanol exposure [4]. That work identified prominent gene networks coordinately regulated by acute ethanol in medial prefrontal cortex (mPFC), a brain region implicated as ethanol-sensitive and involved in drug seeking behavior [6, 7] and chronic compulsive ethanol consumption [8]. Importantly, our prior mPFC acute ethanol expression network study identified glycogen synthase kinase-3 beta (Gsk3b) as strongly regulated by ethanol and a central member of a larger, highly interconnected ethanol-responsive gene network [4]. This finding suggests that in the mPFC, Gsk3b may serve as an important integrator of ethanol-induced signaling changes, and thus influence ethanol behavioral responses.
GSK3B is a serine/threonine kinase, first identified for its inhibition of glycogen synthase [9], and subsequently found to be multi-functional and ubiquitously expressed, with particularly high expression in the brain [10]. As part of the canonical Wnt-signaling pathway, GSK3B contributes to neuronal differentiation and growth and influences neuronal morphogenesis and plasticity in mature neurons [11, 12]. GSK3B dysregulation has been linked to Alzheimer’s disease, major depression, and bipolar disorder [13, 14]. In rodents, global reduction of GSK3B protein expression reduces behavioral responses to psychostimulants [15], while forebrain-specific gene deletion shows anxiolytic-like effects [16].
In adult animals, acute and chronic ethanol inhibit GSK3B activity in nucleus accumbens (NAC) through phosphorylation of its inhibitory Ser9 residue [17]. Ron and colleagues have suggested that ethanol-induced GSK3B inhibition in the NAC leads to elevated collapsin response mediator protein 2 (CRMP-2) microtubule-binding and contributes to increased ethanol intake [18]. Acute ethanol has also been shown to increase inhibitory phosphorylation of GSK3B in the PFC of rats [19] but the consequence on behavior is unknown. As GSK3B has been found to decrease BDNF activity [20], and decreased BDNF in PFC has been causally linked to the transition to increased ethanol consumption [21], we hypothesized that, in contrast to its role in the NAC, inhibition of GSK3B in PFC may represent a protective effect against such increased consumption.
Despite accumulating evidence of regulation of Gsk3b by ethanol, the effect of direct modulation of GSK3B on ethanol-related behaviors, particularly consumption, has not been investigated. Here we utilized genetic, pharmacologic, and behavioral studies in rodent models and humans to implicate GSK3B and a Gsk3b gene network in mechanisms affecting ethanol consumption and alcohol dependence. The initial basis for these investigations, the Gsk3b-centric network highly responsive to acute ethanol [4], was identified in microdissected mouse mPFC encapsulating the anterior cingulate, infralimbic, and prelimbic cortical subregions [22]. Thus, we began the studies described here with a Gsk3b overexpression vector targeting this brain region, specifically the anterior cingulate cortex (ACC). The ACC has been strongly implicated in alcohol craving and relapse [22, 23], features which underlie increased long-term consumption. We supplemented the behavioral and molecular findings of this gene manipulation with systemic, pharmacologic manipulation of GSK3B activity, to assess preclinical applicability of GSK3B as a drug target. Additionally, to investigate potential translational impact we studied the frequency of risk-conferring SNPs in GSK3B and other members of the ethanol-responsive network in alcohol-dependent human patients relative to healthy controls. Taken together, our findings strongly implicate GSK3B as a potential risk factor for alcohol dependence, a key signaling molecule modulating ethanol consumption and as a possible pharmacotherapeutic target for treatment of AUD.
Methods and materials
Animals
Rodent animal studies and procedures were approved by the Institutional Animal Care and Use Committee of Virginia Commonwealth University and followed the NIH Guide for the Care and Use of Laboratory Animals. Mouse and rat models, behavioral assays, viral preparation and stereotactic injections, in vivo imaging, tissue collection, and blood alcohol studies are described in detail in the Supplementary Materials. Animals were ethanol-naive at the beginning of these studies. For behavioral studies, investigators were blind to the treatment conditions.
Acute ethanol regulation of GSK3B
Adult C57BL/6J mice were habituated with daily i.p. saline injections for three days prior to the experiment. On the experimental day, mice (n = 5–6/dose) were injected with ethanol (0.5, 2.0, or 4.0 g/kg), or isotonic saline (0.028 L/kg) and returned to home cage. Mice were sacrificed 30 min post-injection via cervical dislocation and brains were rapidly dissected and stored at −80 °C. Phosphorylation levels of GSK3B-Ser9 in PFC were assessed via Western blot as described in Supplementary Materials. Data on regulation of Gsk3b mRNA by acute ethanol (1.8 g/kg i.p., 4 h post-injection) was drawn from previously published microarray analyses of PFC in BXD recombinant inbred mouse lines (GEO accession GSE28515) [4]. S-scores were used to quantify transcript-level changes from microarray data and are a measure of relative expression change across comparisons of individual oligonucleotide probe intensities, rather than probeset intensity summaries, to determine effect of a treatment on transcript expression [24]. While distinct, S-scores are generally correlated with fold-change and have a standard normal distribution as seen with a z-score transformation [25].
Mouse voluntary ethanol intake
Five weeks after viral injection, ethanol consumption by AAV-GSK3B mice compared to control AAV-IRES mice was assessed using a two-bottle choice drinking paradigm [8, 26]. Mice received increasing concentrations of ethanol for 5 days at each dose (3–15% w/v ethanol). Water was available at all times during the experiment. Ethanol and water consumption was measured every 24 h, and body weight was recorded every 5 days. Bottles containing ethanol or tap water were placed in an empty cage to account for evaporation loss and values were subtracted from the amount consumed for each mouse to calculate corrected preference ratios and ethanol intake. Ethanol consumption was calculated as grams of ethanol per kilogram body weight per 24 h. Percent ethanol preference for individual mice was obtained by dividing volume of ethanol consumption by total fluid consumption for daily drinking sessions.
Gene expression analysis of Gsk3b over-expression responses
Mice were euthanized 3 weeks after viral vector injections and mPFC brain tissue was collected for protein or RNA isolation and subsequent ELISA, microarray and quantitative real-time reverse transcription polymerase chain reaction studies (q-RT-PCR) as described in Supplementary Materials. Equal amounts of the reverse transcribed cDNA products were subjected to PCR amplification using SYBR Green as fluorescent indicator using a Bio-Rad iCycler system (Bio-Rad, Hercules, CA, USA). The mRNA levels of target genes were normalized to GAPDH mRNA levels which showed no expression changes from viral vector injections.
TDZD-8 and Rat Ethanol Self Administration Studies
Operant chambers (Coulbourn Instruments, Allentown, PA) were fitted with 100 μl dipper cups and two levers, each with a cue light above. Presses on the active lever resulted in delivery of reinforcer paired with a tone and illumination of the corresponding cue light. Presses on inactive lever produced no programmed consequence. Ethanol naive rats were trained to respond for ethanol reinforcers via sucrose-fading procedure [27] as described in Supplementary Materials. Once rats were reliably responding for ethanol on an FR3 schedule (one month of 20% EtOH responding, at least 0.5 g/kg EtOH consumption in the last 5 days and no more than 20% variability in responding in the last 5 days), pretreatment with TDZD-8 (LKT Laboratories, Inc, St Paul MN) commenced. Doses (vehicle, 1, 3, and 10 mg/kg) were tested in a within subject, Latin square design so each rat received each dose of the compound. Testing occurred once a week on Thursdays and all rats received saline habituation injections the three days prior to test days. All doses were dissolved in 10% DMSO (Sigma-Aldrich, St Louis, MO) and were administered via intra-peritoneal injection. Rats were returned to their home cages after injections, for 30 min, prior to the start of the operant test session. Upon completion of tests examining the effects of TDZD-8 on fixed ratio responding, the effects of TDZD-8 on a progressive ratio schedule of reinforcement were determined. Progressive ratio experiments followed the same procedure as the fixed ratio pre-treatment studies except that each successive reinforcer was delivered after an exponential number of lever presses were completed according to the formula: (response requirement = 5reinforcer(s) previously earned × 0.1 – 5). Operant sessions would time out after 15 min of inactivity. After each session, the number of reinforcers received and the number of active and inactive lever presses were recorded. In the progressive ratio studies breakpoint was also recorded. A second cohort of rats were trained as a control group. These rats underwent the same operant training and test experimental sessions, except these rats received only sucrose (1.5% w/v) as a reinforcer and had no exposure to ethanol throughout the entire study.
Abstinence model of relapse-like behavior
After completion of the FR and PR tasks, rats entered a 3-week abstinence period upon completion of the fixed and progressive ratio tests with TDZD-8. Animals remained in their home cages for 3 weeks and did not enter the operant chambers or receive ethanol or sucrose during this period. Upon completion of the abstinence period, rats were once again pretreated with TDZD-8 (3 mg/kg) or vehicle and placed in operant chambers. The progressive ratio session was identical to that described earlier with the exception that ethanol or sucrose reinforcers were not available. Cue lights and tones were active and active lever presses were recorded.
GSK3B-network set-based association analysis for alcohol dependence
Human homologs of genes correlating with Gsk3b in ethanol-induced microarray expression change in mouse mPFC were selected for association analysis for alcohol dependence using data provided by the Collaborative Study on the Genetics of Alcoholism (COGA) [28] as summarized in Supplementary Materials. Groups of genes having a graded Pearson correlation (≥0.80, ≥0.85, ≥0.90, and ≥0.95) with S-scores of Gsk3b responses to ethanol were assessed for SNPs conferring risk for alcohol dependence, with a p-value < 0.05 defined as significant in these set-based analyses. SNPs located within genes, as well as SNPs 2Kb up and downstream from the genes were included. These same criteria were used for analyses on individual genes (Table S1), but with a slightly stricter p-value of <0.01 defined as risk-conferring. p-values of SNP sets were determined empirically by random resampling (10,000 permutations).
Statistical analyses
Data were expressed as mean ± SEM and were analyzed parametrically. Two-way repeated measures ANOVAs were used for all analyses of drinking behavior, with Tukey’s post hoc to compare all means, and Sidak’s multiple comparisons correction for effect of genotype at a given ethanol concentration. One-way ANOVAs with Tukey’s post hoc tests were used to assess multiple doses of ethanol on GSK3B phosphorylation and multiple doses of TDZD-8 on operant conditioning responses. Unpaired two-tailed t-tests were used to analyze light-dark box assays, q-RT-PCR, ELISA, and single-dose operant conditioning data. In the last case, Welch’s correction was used to account for unequal variances. Values of p < 0.05 are considered statistically significant.
Results
Acute ethanol inhibits GSK3B activity and decreases mRNA expression in PFC
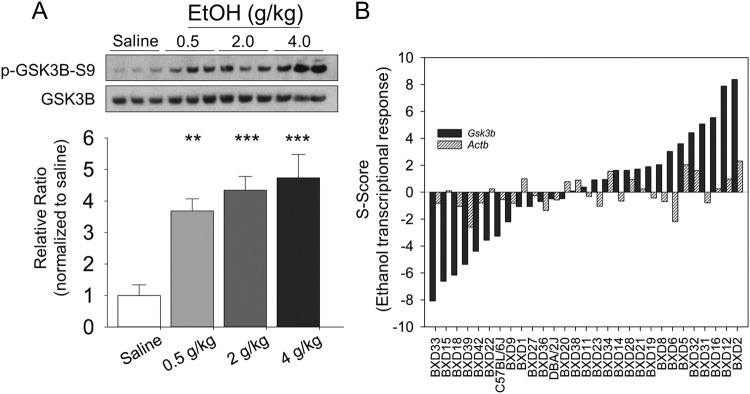
Our previous microarray studies showed that acute ethanol regulated Gsk3b mRNA in PFC as part of a highly ethanol-responsive gene network [4]. Prior to detailed studies on Gsk3b modulation of ethanol behaviors, we further confirmed ethanol regulation of Gsk3b in PFC at the mRNA and protein level. Acute ethanol treatment markedly increased phosphorylation of the Ser-9 inhibitory site on GSK3B in PFC of C57BL/6J mice (F3,17 = 12.94, p < 0.001) 30 min after ethanol injection (Fig. 1a). Tukey’s post hoc analysis showed all doses of ethanol differed from saline (p < 0.01) but not from one another. These data are consistent with the post-translational inhibition of GSK3B by ethanol in mPFC.
Fig. 1.
GSK3B in medial prefrontal cortex is highly responsive to acute ethanol. a GSK3B protein in C57BL/6J mouse mPFC is phosphorylated at its inhibitory Serine-9 residue following i.p. injections of 0.5, 2.0, and 4.0 g/kg ethanol. Tissue for protein analyses was collected 30 min after injection. Representative Western blots of p-GSK3B-S9 and total GSK3B protein shown at top. Quantified intensity values (n = 5–6/dose) shown at bottom, with treatment effect significant at p < 0.001 by one-way ANOVA. Tukey’s post hoc testing revealed each dose differed significantly from saline control: **p < 0.01, ***p < 0.001. b Transcriptional response of Gsk3b RNA shows dynamic and robust response 4 h after injection of ethanol (1.8 g/kg). Direction of expression change varies across BXD panel, indicating genetic regulation of genomic response. Gsk3b 3’UTR transcript (probeset 1437001_at) shown in black, with exonic Actb (probeset 1436722_a_at) shown for reference (grey)
A selective examination of Gsk3b transcript levels from a prior microarray study of BXD mice [4], confirmed that ethanol treatment (1.8 g/kg, i.p., 4 h) altered Gsk3b mRNA expression (Fig. 1b). Direction of expression change varied across the BXD panel, indicating genetic background modulation of the Gsk3b ethanol response. In C57BL/6J mice, acute ethanol strongly decreased Gsk3b mRNA expression.
GSK3B overexpression increases ethanol consumption
We then directly tested a role for mPFC GSK3B in ethanol consumption by stereotactic over-expression with AAV viral vectors in C57BL/6J mice. Using initial in vivo imaging, we verified hrGFP emission up to 8 weeks following AAV-GSK3B transduction (Fig. S1A). GSK3B overexpression was confirmed at the mRNA (t4 = 6.78, p = 0.0025) and protein level (t4 = 3.05, p = 0.038) by quantitative RT-PCR, ELISA and immunofluorescence (Fig. S1B–E). GSK3B over-expression was largely limited to NeuN-positive neurons of mPFC (Fig. S1B, E).
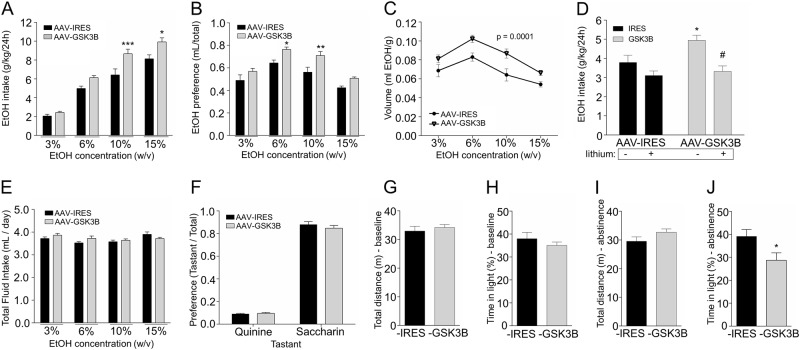
GSK3B over-expression in mPFC produced a significant increase in mean daily ethanol consumption (g/kg/day) using a progressive ethanol dosing regimen (Fig. 2a, F1,28 = 20.68, p < 0.0001 for viral genotype). AAV-GSK3B mice consumed larger (~30–40%) amounts of ethanol (g/kg/day) at 10% (p < 0.001) and 15% (p < 0.01) w/v ethanol concentrations compared to control AAV-IRES mice. Viral genotype also significantly altered mean ethanol preference ratio (Fig. 2b, F1,28 = 19.34, p = 0.0001), with AAV-GSK3B mice displaying an enhanced ethanol preference during access to 6% (p < 0.05) and 10% (p < 0.01) ethanol solutions. We further assessed ethanol drinking behavior as a function of ethanol concentration, wherein mean volume consumed of ethanol-containing solution was used as a proxy for reinforcers delivered. This showed an inverted U-shape dose-effect curve peaking at the 6% w/v ethanol (Fig. 2c), and with GSK3B mice consistently displaced to higher values than controls. Two-way ANOVA revealed a significant effect of viral genotype on mean volume of ethanol-containing solution per body weight (F1,28 = 20.44, p = 0.0001).
Fig. 2.
GSK3B-overexpressing mice show a lithium-sensitive increased ethanol consumption, increased ethanol preference, and increased anxiety-like behavior following abstinence. a Compared to IRES mice, AAV-GSK3B mice (n = 14–16) show significantly increased ethanol consumption (g/kg/day) during free access, two-bottle choice at 10% (***p = 0.0005) and 15% (*p = 0.0147) ethanol concentrations. b AAV-GSK3B mice show increased ethanol preference (EtOH/total fluid) during two-bottle choice at 6% (*p = 0.028) and 10% (**p = 0.0039) ethanol concentrations. c AAV-GSK3B shifts the dose-response relationship between ethanol concentration and mL consumed significantly upward (p = 0.0001). d Addition of Lithium diet significantly decreased ethanol consumption in AAV-GSK3B mice (n = 7–8 per group, #treatment effect of lithium within AAV-GSK3B mice, p = 0.0021; *genotype effect in untreated mice, p = 0.0451 vs. IRES). Treatment effect of lithium was not significant within AAV-IRES mice (p = 0.398) and there was no interaction effect of genotype × lithium. e, f AAV-GSK3B and AAV-IRES injected mice did not differ in total fluid intake or taste preferences: e Average total fluid intake across each of 5 days of access to multiple concentrations of ethanol (n = 14–16/group, p > 0.05), f Quinine and saccharin preferences, as determined by volume consumed over total fluid consumption (n = 14–16/group, p > 0.05). g Basal locomotor activity (n = 10–12/group, p > 0.05) as measured by distance traveled (cm) in both chambers, during a 10 min test session in LD chamber. h Baseline anxiety-like behavior (n = 10–12/group, p > 0.05) as measured by %time spent in the light chamber during 10-min light-dark test. i 24 h after ethanol abstinence, AAV-IRES and AAV-GSK3B (n = 7–8/group) did not differ in total distance traveled. j AAV-GSK3B mice showed a significantly decreased time spent in the light (*p = 0.0365), indicating increased anxiety-like behavior during abstinence from ethanol
In contrast to differences in ethanol intake, viral genotype did not alter total fluid consumption (Fig. 2e, F1,28 = 0.433, p = 0.5158). Furthermore, there was no significant difference between AAV-GSK3B and AAV-IRES mice in two-bottle choice voluntary consumption of saccharin (0.015% w/v) or quinine (0.05 mM) (Fig. 2f, tsaccharin, df=28 = 0.92, p = 0.36; tquinine, df=28 = 0.45, p = 0.66), suggesting taste preferences were not altered [29]. Blood ethanol levels were also not different between AAV-GSK3B and AAV-IRES mice at 60 or 120 min after injection (i.p.) with 2.0 g/kg ethanol (Figure S2B, F1,4 = 1.48, p = 0.29), demonstrating no pharmacokinetic differences between viral genotypes.
To further investigate the role of GSK3B activity in this procedure, in a separate experiment we introduced lithium, a well validated GSK3B inhibitor [30, 31], into diets for half of each experimental group for 4 weeks while receiving access to ethanol (10% w/v) and water (Fig. 2d). Lithium treatment (F1,26 = 15.51, p = 0.0005) and viral genotype (F1,26 = 5.483, p = 0.027) both significantly altered mean ethanol consumption (g/kg/day), while the interaction term was not significant (F1,26 = 2.58, p = 0.12). By post hoc testing, lithium significantly decreased ethanol consumption in AAV-GSK3B mice relative to standard chow (p < 0.01). AAV-IRES mice fed lithium showed a non-significant decrease in ethanol consumption (95% CI: 0.4908 to −1.859 g/kg difference in mean consumption). The lack of significant interaction indicates that lithium did not specifically decrease drinking in the context of GSK3B overexpression, but rather that both overexpression and lithium provided significant individual contributions to ethanol consumption. Serum lithium levels did not differ significantly (F1,2 = 0.11, p = 0.771) between AAV-GSK3B (mean = 0.65 mmol/L) and AAV-IRES mice (mean = 0.70 mmol/L) fed lithium diets. Lithium treated-mice were within the human therapeutic serum levels of 0.6 and 1 mmol/L and well below the lowest toxic dose of 1.2 mmol/L [32]. Mice fed a standard chow had average serum lithium levels <0.3 mmol/L.
As acute sensitivity to ethanol is known to correlate with risk for AUD in humans [33] and ethanol consumption in some animal models [34], we assessed whether GSK3B over-expression might alter behavioral responses to acute ethanol. AAV-GSK3B mice did not differ significantly from AAV-IRES mice in duration of ethanol-induced (3.6 g/kg, i.p.) loss-of-righting reflex (LORR; Figure S2A, t24 = 0.278, p = 0.78), suggesting no effect on the high-dose sedative properties of ethanol.
GSK3B overexpression increases anxiety-like behavior during ethanol abstinence
Human clinical studies suggest that stress and anxiety, particularly following during ethanol abstinence, are important contributing factors in AUD and to the high relapse rate in alcoholic patients [35]. We therefore assessed whether GSK3B over-expression altered anxiety-like behavior in the light-dark conflict model [36] 24 h after withdrawal from the twenty consecutive days of daily 2-bottle choice, escalating ethanol (3–15% w/v) access. As shown in Fig. 2i, j, AAV-GSK3B mice spent significantly less time in the light (t13 = 2.33, p = 0.037) compared to AAV-IRES mice, despite no significant difference in total distance traveled (t13 = 0.30, p = 0.77) during a 10-min test session. A separate cohort of mice tested for baseline anxiety-like behavior under ethanol-naive conditions showed no effect of viral genotype on either percent time in the light or total distance traveled (Fig. 2g, h, t%time, df=18 = 0.97, p = 0.35; tdistance, df=18 = 0.63, p = 0.54). These findings suggest that GSK3B over-expression may increase anxiety-like behavior specifically under the condition of ethanol abstinence.
GSK3B overexpression alters BDNF gene expression
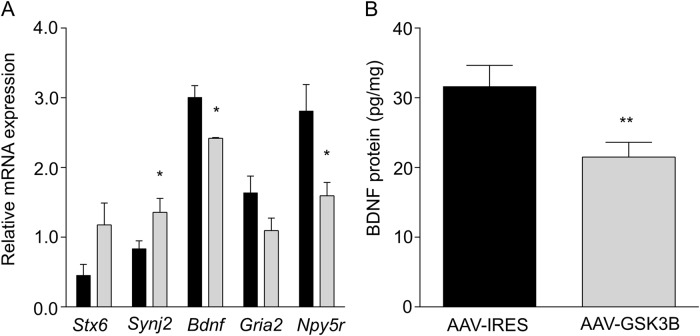
Pilot microarray studies were performed on individual mouse mPFC samples (n = 3/group) 3 weeks following virus injection to assist in selecting downstream synaptic targets possibly responding to GSK3B over-expression (see Supplementary Materials, Table S2). From genes showing nominal expression differences between AAV-GSK3B and AAV-IRES control mice (Table S2) and published literature on mechanisms of GSK3B in synaptic plasticity or genes regulating ethanol consumption [37–41], we selected the following potential downstream candidates for further study: syntaxin 6 (Stx6), synaptojanin 2 (Synj2), brain-derived neurotrophic factor (Bdnf), ionotropic glutamate receptor subunit AMPA2 (Gria2), and neuropeptide Y receptor Y5 (Npy5r). By q-RT-PCR, Synj2 expression was found to be significantly up-regulated (t8 = 2.47, p = 0.039) and Bdnf (t4 = 3.49, p = 0.025) and Npy5r (t8 = 2.45, p = 0.040) significantly down-regulated in mPFC of AAV-GSK3B mice (Fig. 3a). Stx6 and Gria2 were not altered by GSK3B overexpression in mPFC (tStx6, df=8 = 2.29, p = 0.051, tGria2, df=8 = 1.65). Of these genes, Bdnf has been strongly implicated in the literature as interacting with GSK3B [12, 42] and with mPFC-specific regulation of ethanol drinking behavior [21, 43]. We therefore selected BDNF protein for subsequent ELISA analysis. In agreement with the q-RT-PCR results, BDNF protein was significantly decreased in mPFC of AAV-GSK3B mice (Fig. 3b, t4 = 4.78, p = 0.0088). The direction of BDNF modulation by GSK3B over-expression is consistent with changes in Bdnf expression contributing to increased ethanol consumption in mouse models [21, 43].
Fig. 3.
GSK3B over-expression leads to expression changes in synaptic signaling genes. a Relative levels of Stx6, Synj2, Bdnf, Gria2, and Npy5r in AAV-IRES (black) and AAV-GSK3B (grey) mouse mPFC, determined by q-RT-PCR (*p < 0.05, n = 4–6 per group). b Decreased BDNF protein was confirmed in mPFC of AAV-GSK3B mice by ELISA (**p < 0.01, n = 3/group)
Pharmacologic GSK3B inhibition decreases operant ethanol self-administration and relapse-like behavior
To complement gain-of-function and lithium studies above, and possibly implicate new pharmacologic agents for treatment of AUD, we studied the actions of systemic GSK3B pharmacological inhibition on positive reinforcing properties of ethanol. Ethanol-naive Wistar rats were trained to lever press for ethanol (20% v/v) until operant responding was stable. The highly selective GSK3B inhibitor TDZD-8 [44] was tested for its effect on ethanol-maintained responding under both fixed-ratio (FR) and progressive-ratio (PR) reinforcement schedules.
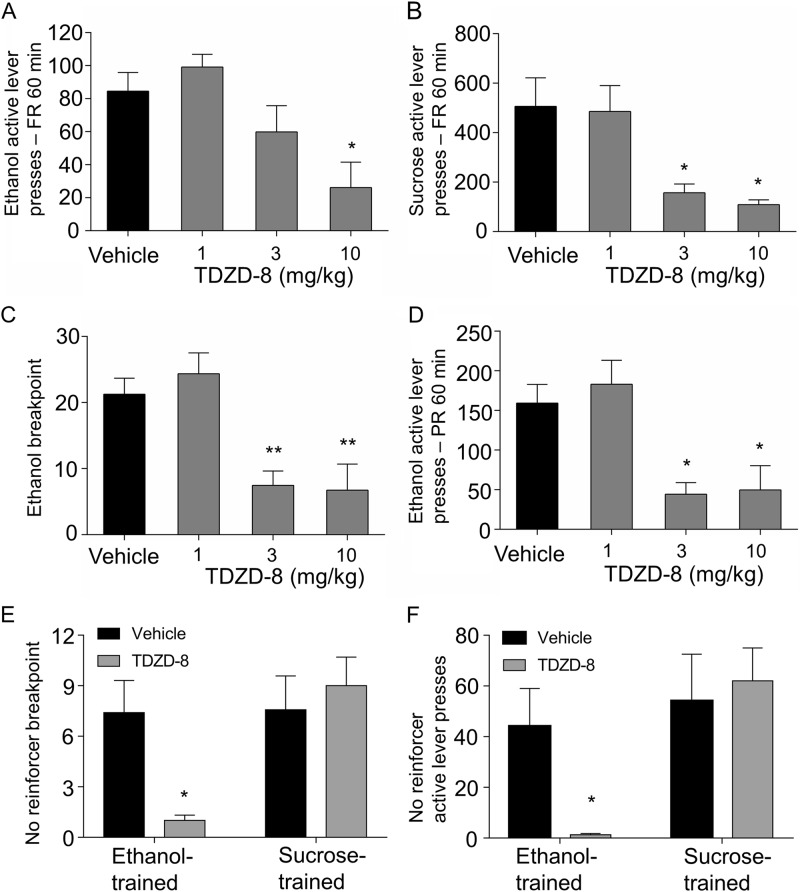
TDZD-8 significantly attenuated responding on the ethanol-paired lever in an FR schedule of reinforcement (Fig. 4a, F3,30 = 8.89, p = 0.0002). The 10 mg/kg dose decreased active lever pressing compared to vehicle (p < 0.05). To complement lever press data, we calculated ethanol consumption during FR sessions for reinforcers consumed with confirmed licks as described in Supplementary Materials. TDZD-8 treatment decreased ethanol consumption (g/kg/session) is shown in Figure S3 (F3,15 = 8.52, p = 0.0015). Under a PR schedule of reinforcement, TDZD-8 had a marked impact on breakpoint, a measure of motivation to self-administer (Fig. 4c, F3,30 = 11.85, p < 0.0001), with both the 3 mg/kg (p < 0.01) and 10 mg/kg (p < 0.01) doses associated with significant reduction vs. vehicle. TDZD-8 also significantly attenuated overall active lever presses during the PR session (Fig. 4d, F3,30 = 10.52, p < 0.0001) at both the 3 mg/kg (p < 0.05) and 10 mg/kg (p < 0.05) doses. To determine if the effects of TDZD-8 were specific to ethanol, a second cohort was trained to lever press for sucrose (1.5%). TDZD-8 (3 mg/kg and 10 mg/kg) significantly attenuated responding on the sucrose-paired lever on an FR schedule of reinforcement (Fig. 4b, F3,21 = 8.34, p = 0.0008). Together, these data suggest a possible effect of TDZD-8 on general reward-seeking behavior in non-abstinent rats.
Fig. 4.
Operant behavioral studies of GSK3B inhibitor TDZD-8, before and after 3 week abstinence period. a TDZD-8 dose-dependently decreased active lever pressing for a 20% ethanol reinforcer (n = 11, *p < 0.05). b TDZD-8 dose-dependently decreased active lever pressing for a 1.5% (w/v) sucrose reinforce (n = 8, *p < 0.05). c On a PR schedule 3 mg/kg and 10 mg/kg TDZD-8 decreased breakpoint (BP) for 20% ethanol reinforcer (n = 11, **p < 0.01). d TDZD-8 also dose-dependently decreased total active lever presses (n = 11, *p < 0.05) for ethanol reinforcer over the PR test session. e, f Following 3 weeks of ethanol abstinence, rats trained for ethanol or sucrose operant self-administration were re-introduced to operant chambers with previously ethanol/sucrose-paired light and tone stimuli in the absence of reinforcer. e 3 mg/kg dose of TDZD-8 decreased ethanol breakpoint but not sucrose breakpoint. f Number of active lever presses before timeout (15 min of inactivity) during the re-introduction to the chambers also showed significant effect of 3 mg/kg TDZD-8 in the ethanol but not sucrose trained animals (n = 6–7 per group, *p < 0.05 by unpaired t-test)
Next, we sought to determine if TDZD-8 would impact ethanol administration following abstinence, as relapse to ethanol seeking is a major occurrence in alcoholics. Thus, we tested for cue-driven motivation to self-administer reinforcer after protracted abstinence, in ethanol or sucrose trained rats (Fig. 4e, f). A PR schedule was used to assess motivated reinforcer-seeking in the absence of reinforcer [45]. TDZD-8 showed a striking attenuation of breakpoint in ethanol trained animals (Fig. 4e, t = 3.065, p = 0.0375) but no effect on breakpoint in sucrose trained animals (Fig. 4f, t = 0.46, p = 0.68). The TDZD-8 specific effect on relapse-like behavior in abstinent ethanol-seeking animals was also reflected in total active lever presses (Fig. 4f, t = 2.83, p = 0.047) while sucrose-seeking animals showed no difference in active lever presses (Fig. 4f, t = 0.42, p = 0.70) over the duration of the test session. Thus, TDZD-8 inhibition of GSK3B appeared to show specific effects on the motivation to seek ethanol during abstinence. However, future operant studies will need to supplement measures of consumption (see Fig. S3) with BEC, as Blegen et al. [46] have recently shown that operant responding does not necessarily correlate well with BEC.
Gsk3b-centric ethanol-responsive gene network is associated with risk for alcohol dependence
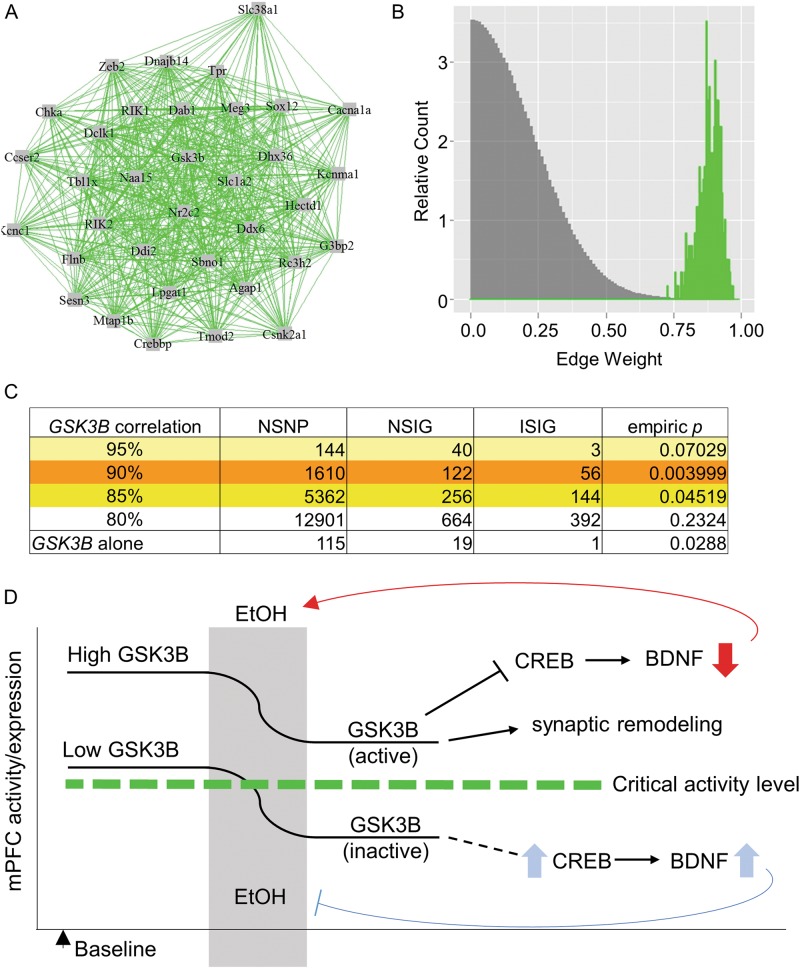
We previously identified Gsk3b as a highly-interconnected gene in a gene network enriched for ethanol-responsive genes in mPFC [4]. From these data, prior knowledge on the role of GSK3B in synaptic plasticity and affective disorders, and our results above on GSK3B modulation of ethanol consumption in mice, we hypothesized that a Gsk3b-centric gene expression correlation network would show genetic association with alcohol dependence in humans. We therefore investigated the correlation structure of the Gsk3b ethanol-responsive network from our prior microarray data [4] on acute ethanol-induced expression changes across BXD mouse strains, using a measure of ethanol-responsiveness (S-scores; see 'Methods') to identify genes similar to Gsk3b in terms of ethanol regulation. Analysis of individual probeset (gene) correlations with Gsk3b expression following acute ethanol treatment in mPFC (n = 29 mouse strains) showed that 34 genes had correlations ≥90% with Gsk3b. There was a highly unique degree of interconnectivity in this Gsk3b network (Fig. 5a) compared to randomly chosen similar sized networks from the rest of the genome (Fig. 5b). This confirmed the concept that Gsk3b is a central member of a unique gene network in mouse mPFC that is strongly co-regulated by ethanol.
Fig. 5.
A Gsk3b-centered expression network in mouse mPFC is enriched for SNPs conferring increased risk for alcohol dependence in human GWAS data. a Network of genes with S-score, a measure of ethanol-responsiveness, correlation of r ≥ 0.9 with Gsk3b, found in the mPFC of BXD mice. b Comparative histogram of WGCNA-calculated edge weights (gene–gene expression correlations) of the 35 genes in the Gsk3b network (green) and edge weight values of 10,000 permutations of randomly selected groups of 35 genes (grey) from the BXD mPFC microarray. c SNP-set level analyses of human GWAS (COGA dataset, n = 1205 cases, 700 controls) were performed on groups of homologous genes. Results are shown for genes at multiple levels of correlation (80–95%) with Gsk3b in mouse mPFC. The result of a SNP-set analysis within the GSK3B gene alone is also given. NSNP = total number of genotyped SNPs in given gene or geneset; NSIG = number of significantly risk-conferring SNPs; ISIG = number of independent (non-LD) risk-conferring SNPs. Empiric p values were computed by 10,000 permutations of phenotypes. d Putative model by which mPFC GSK3B expression/activity may predispose to increased alcohol consumption and eventual dependence. Left side of diagram indicates a basal state prior to any ethanol exposure. We propose that high GSK3B activity is associated with high risk of increased ethanol consumption. GSK3B inhibition following ethanol exposure might normally serve to self-regulate ethanol consumption, such as through increased cortical BDNF which has been previously been shown to moderate drinking. With increased total GSK3B or increased basal GSK3B activity, there may be increased residual activity following a given ethanol exposure, allowing for continued GSK3B-mediated inhibition of BDNF, promoting the transition to increased consumption
We next queried gene-based human association data from the Collaborative Study on the Genetics of Alcoholism (COGA) project [47, 48] to determine if the uniquely correlated Gsk3b ethanol-responsive network contained more genes having polymorphisms associated with alcohol dependence in human GWAS studies than would be expected by chance for gene sets of similar sizes. The above mouse expression correlation analysis was used to create gene sets based on ≥95, 90, 85, and 80% correlation with Gsk3b S-score measurement of ethanol responsivity. Set-based analyses revealed significant enrichment of alleles conferring risk for alcohol dependence in the groups of human homolog genes correlating ≥85% and ≥90% with Gsk3b in the mouse BXD study. The latter of these human homolog groups showed the strongest significance, p = 0.004 (Fig. 5c). Additionally, set-based analyses of SNPs within individual genes showed significant enrichment of GSK3B (p = 0.029), DNAJB14 (p = 0.025), G3BP2 (p = 0.017), and KCNMA1 (p = 0.0096) (Table S1). Together, these mouse functional genomic and human genetic results implicate the Gsk3b network both as having a potentially central role in behavioral responses to ethanol and as a possible risk factor for alcohol dependence.
Discussion
The present study was motivated by our observation that acute ethanol alters expression of a highly-interconnected network of genes centered around Gsk3b in mPFC of recombinant inbred mice and prior literature suggesting a role of Gsk3b in brain responses to ethanol or other drugs of abuse. We over-expressed GSK3B in mPFC of mice and inhibited its activity in rats to study its potential role in ethanol consumption and motivation. Our studies revealed substantial effects of Gsk3b gene expression and GSK3B activity on ethanol consumption behaviors. Downstream of Gsk3b overexpression, changes in other genes in mPFC reveal potential mechanisms for these effects, most notably decreased Bdnf expression. Further, we found the Gsk3b co-regulated gene network, as well as GSK3B itself, to be significantly enriched for risk-conferring SNPs in human alcoholic patients. Taken together, these results provide evidence for an ethanol-responsive gene network in risk for alcohol dependence and a potential therapeutic target in GSK3B.
Relationship between ethanol and prefrontal cortical GSK3B activity
Acute ethanol robustly phosphorylated GSK3B at its inhibitory Ser-9 residue at doses from 0.5 to 4.0 g/kg in mPFC and also downregulated Gsk3b mRNA expression in C57BL/6J mice. These results are consistent with prior published studies showing that acute ethanol increased Ser-9 phosphorylation in rat PFC [19] and mouse NAC [17]. However, viral-mediated Gsk3b overexpression in mouse mPFC significantly increased ethanol consumption and preference. As seen in Fig. 2c, there was an upward shift in the behavioral response of ethanol drinking across concentrations. This finding suggests that increased GSK3B activity augments the usual behavioral response of consumption. If acute ethanol-induced inhibition of PFC GSK3B normally provides a satiety-like (or self-regulatory) response, it may be that overexpression of GSK3B increases resistance to this response, thereby increasing ethanol consumption (see Fig. 5d). We are assuming these actions of GSK3B occur in neurons of cingulate cortex, since our AAV viral vector over-expression was highly localized to NeuN positive cells in that area (Fig. S1E). However, a major goal of ongoing studies is to identify the neuronal subtype(s) in which GSK3B activity may be driving ethanol-related behaviors. The increase in ethanol drinking with GSK3B over-expression was likely not related to changes in the sedative/hypnotic effects of ethanol, as we observed no difference in ethanol LORR. A limitation of our LORR and ethanol-drinking studies is that BEC was not measured to confirm that differences in behavior were paralleled by differences in BEC. However, differences in ethanol intake are usually highly correlated with consequent differences in BEC.
The observation that lithium, a GSK3B inhibitor [30], decreased ethanol consumption in the GSK3B-overexpressing mice provides support that the increased ethanol consumption was indeed due to increased GSK3B activity, and not off-target effects of the overexpression vector. Although lithium is thought to perhaps have multiple biological actions, its role as an inhibitor of GSK3b is widely published. However, we reinforced these GSK3B over-expression and lithium data with studies discussed below using the more selective drug, TDZD-8. Lithium also produced a significant main effect on ethanol consumption, but not an interaction effect with genotype or significant effect on IRES injected animals, possibly due to statistical power issues. This suggests, as would be expected, that lithium could decrease ethanol consumption in both controls as Gsk3b over-expressing animals if these studies were repeated with greater numbers. However, our goal was to show that a Gsk3b inhibitor would decrease the ethanol consumption caused by AAV-GSK3B injection.
Despite no differences in anxiety-like behavior or total locomotor behavior at baseline, after a 20-day period of ethanol access, animals overexpressing Gsk3b exhibited an increase in anxiety-like behavior during ethanol abstinence. This suggests that higher Gsk3b expression in mPFC may act to sensitize the brain to the anxiety-like effects of ethanol abstinence, possibly serving as a motivational factor for increasing ethanol self-administration. Similarly, inhibition of GSK3B in mPFC may contribute to anxiolytic-like effects of ethanol. In support of this hypothesis, selective deletion of Gsk3b in mouse forebrain has been shown to have anxiolytic-like effects [16]. Furthermore, our laboratory has recently identified the gene encoding a GSK3B-interacting protein, Ninein, as a strong candidate gene for an ethanol-anxiolysis quantitative trait locus identified on mouse Chr 12 [49]. However, these findings on possible increases in anxiety-like behavior following ethanol abstinence with GSK3B over-expression require future studies to clarify the interaction between ethanol consumption, GSK3B levels, and anxiety.
Our finding that elevating PFC Gsk3b expression decreased BDNF protein and mRNA (Fig. 3) could have particular mechanistic significance, given substantial evidence of a GSK3B-BDNF relationship in the literature: BDNF has been found to inhibit GSK3B activity via PI3K signaling [14], while active GSK3B has been found to decrease Bdnf mRNA levels [50]. Reduced expression of BDNF has been strongly implicated as a modulator of progressive ethanol consumption in both mouse and rat models, specifically in the mPFC [21, 43]. Thus, decreased BDNF represents a likely mechanism by which increased GSK3B increases ethanol consumption, although the specific pathway remains to be elucidated. Active GSK3B has been found to inhibit DNA-binding ability of the BDNF-transcription factor CREB, thus representing a hypothesized mechanism (Fig. 5) for the GSK3B-BDNF effect seen here. However further studies are necessary to directly test the roles of BDNF and CREB.
Using a pilot microarray study and subsequent q-RT-PCR validation, we found altered expression of additional plasticity related genes in mouse mPFC: Synj2 and Npy5r (Fig. 3). There was also a trend toward decreased Stx6 expression (p < 0.1). Generally these findings provide evidence that altered GSK3B expression may play a significant role in ethanol-related synaptic remodeling, consistent with its emergent role as a mediator of plasticity [11]. Npy5r may also particularly contribute to ethanol-related behaviors, as neuropeptide Y (NPY) levels are inversely related to ethanol consumption in mice and rats [51, 52], NPY expression is decreased in mPFC of postmortem tissue from alcoholics [53], and SNPs in the Y2R or Y5R receptor genes have been associated with alcohol dependence or withdrawal [54]. However, further studies would be needed to clarify the potential role of NPY receptor signaling in the modulation of ethanol consumption by GSK3B.
Pre-clinical and potential translational implications
Results with a selective GSK3B inhibitor in ethanol operant self-administration studies in rats provided cross-species validation of the mouse viral vector over-expression findings. While we used lithium as an initial pharmacologic tool in mouse viral vector studies due to the extensive animal model and human literature on lithium as an inhibitor of GSK3B, we used the more specific GSK3B inhibitor TDZD-8 in pre-clinical studies of rats utilizing systemic delivery, to better model a possible pharmacotherapeutic effect. Pilot studies on TDZD-8 in mice showed a species-selective toxicity with some lethality (10%) for unknown reasons. We had no lethality or any other evidence of toxicity in pilot studies in rats and thus chose to use that rodent species for the TDZD-8 studies, in addition to operant ethanol self-administration being more widely studied in rats. TDZD-8 decreased the motivation to self-administer ethanol and sucrose, indicating inhibition of reward-seeking. Following abstinence from reinforcer, TDZD-8 nearly eliminated cue-driven relapse-like behavior in rats trained to respond to ethanol, while showing no effect in rats trained to respond for sucrose. This suggests a degree of specificity to GSK3B activity in the context of ethanol-seeking post abstinence, which may have particular relevance for the role of GSK3B in treatment of relapse.
While GSK3B is ubiquitously expressed, systemic administration of a TDZD drug is a necessary preclinical test given drug administration limitations in humans. If a contributing factor to alcohol use is a primary imbalance of GSK3B (i.e., over activity) in the mPFC, then systemic administration would be expected to exert its effects through action here, albeit with a risk of off-target effects. Another caveat about this experiment is the negative effect on sucrose as well as ethanol seeking, indicating general anti-reward properties and possible anhedonia. This may limit clinical utility of GSK3B inhibitors and alternate targets in the GSK3B network should be considered. However it should be noted that a current first-line treatment for AUD, naltrexone, also exerts negative effects on responding for natural reinforcers [55]. Highly specific GSK3B inhibitors such as TDZD-8 may allow for GSK3B inhibition with reduced side effects, and thus could have significant promise as novel AUD therapeutics, particularly in regard to relapse. It is worth noting that tideglusib, a highly selective and CNS penetrant derivative of TDZD-8, is currently being investigated in human trials as a treatment for neurodegenerative diseases such as Alzheimer’s disease and has shown no reported toxicity other than transient liver function abnormalities [56]. Thus, such TDZD-8 derivatives appear safe for use in humans.
Our results on the role of GSK3B in modulating ethanol consumption in rodents are further strengthened by our findings that a network of genes, whose ethanol responsiveness correlates strongly with Gsk3b, is significantly associated with risk for alcohol dependence (Fig. 5). This network included other genes previously identified as key mediators of ethanol-induced signaling and behavior, namely Kcnma1 [57]. We posit that the tightly coordinated ethanol regulation of gene expression in this Gsk3b-related network suggests an important role in neuroadaptation to ethanol and that some portion of the risk for alcohol dependence conferred by variants in individual genes and the network as a whole may be due to differences in individuals’ initial ethanol response, given that low level of response is a known predictor of future alcoholism [33]. In general support of this concept, two recent reports suggest GSK3B is a potential modulator of chronic ethanol adaptations in signaling systems involving Kcnma1 [58] and dopamine D2 receptors [59]. Just because Gsk3b was a highly-interconnected gene in our BXD study, this does not necessarily imply that ethanol-responsive genes correlated with Gsk3b are regulated by Gsk3b itself. For example, although our data suggests that Bdnf and Npy5r may be altered by GSK3B viral vector over-expression, these genes were not found in the network co-regulated by ethanol with Gsk3b. This suggests a possible divergence between the mechanisms regulating the ethanol responsiveness of the Gsk3b-centric network, and those downstream of GSK3B that impact ethanol consumption.
Taken together, our findings strongly implicate GSK3B as a key signaling molecule modulating ethanol consumption, a potential risk factor for alcohol dependence, and as a possible pharmacotherapeutic target for treatment of AUD. Future studies on GSK3B as a therapeutic target, and the downstream mechanisms mediating GSK3B actions on ethanol consumption, are clearly warranted.
Electronic supplementary material
Acknowledgements
This work is dedicated to the memory of Dr. Matthew Riley of the National Institute for Alcohol Abuse and Alcoholism (NIAAA), who consistently encouraged and challenged the investigators involved in this work. We thank Dr. Kenneth S. Kendler for reviewing the manuscript, Dr. Steve Negus for discussions on behavioral economics analysis and members of the Miles laboratory for many helpful discussions during the course of this work. Additional support was provided by the COGA Consortium for the use of genotype data. A full acknowledgement of COGA investigators is included in Supplementary Methods and Materials. The Collaborative Study on the Genetics of Alcoholism (COGA), Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes ten different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, A. Goate, J. Rice, K. Bucholz); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy), Howard University (R. Taylor) and Virginia Commonwealth University (D. Dick). Other COGA collaborators include: L. Bauer (University of Connecticut); D. Koller, S. O’Connor, L. Wetherill, X. Xuei (Indiana University); Grace Chan (University of Iowa); N. Manz, M. Rangaswamy (SUNY Downstate); A. Hinrichs, J. Rohrbaugh, J-C Wang (Washington University in St. Louis); A. Brooks (Rutgers University); and F. Aliev (Virginia Commonwealth University). A. Parsian and M. Reilly are the NIAAA Staff Collaborators. COGA investigators continue to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owe a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). Funding support for GWAS genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the National Institute on Alcohol Abuse and Alcoholism, the NIH GEI (U01HG004438), and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). Kim Doheny and Elizabeth Pugh from CIDR and Justin Paschall from the NCBI dbGaP staff provided valuable assistance with genotyping and quality control in developing the dataset available at dbGaP. This work was supported by NIAAA grants U01AA016667 (MFM), P50 AA022537 (DD, SB, MFM), K02 AA018755 (DD) and F30 AA024382 (ADV).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Andrew van der Vaart, Xianfang Meng
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41386-018-0202-x).
References
- 1.Sacks JJ, Gonzales KR, Bouchery EE, Tomedi LE, Brewer RD. 2010 National and state costs of excessive alcohol consumption. Am J Prev Med. 2015;49:e73–9. doi: 10.1016/j.amepre.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 2.Salvatore JE, et al. Polygenic scores predict alcohol problems in an independent sample and show moderation by the environment. Genes. 2014;5:330–46. doi: 10.3390/genes5020330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kerns RT, et al. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–66. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolen AR, et al. Genetic dissection of acute ethanol responsive gene networks in prefrontal cortex: functional and mechanistic implications. PLoS ONE. 2012;7:e33575. doi: 10.1371/journal.pone.0033575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Vaart AD, et al. The allostatic impact of chronic ethanol on gene expression: A genetic analysis of chronic intermittent ethanol treatment in the BXD cohort. Alcohol. 2017;58:93–106. doi: 10.1016/j.alcohol.2016.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harris GJ, et al. Frontal white matter and cingulum diffusion tensor imaging deficits in alcoholism. Alcohol Clin Exp Res. 2008;32:1001–13. doi: 10.1111/j.1530-0277.2008.00661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalivas PW, Volkow N, Seamans J. Neuron. 2005;45:647–50. doi: 10.1016/j.neuron.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 8.George O, et al. Recruitment of medial prefrontal cortex neurons during alcohol withdrawal predicts cognitive impairment and excessive alcohol drinking. Proc Natl Acad Sci USA. 2012;109:18156–61. doi: 10.1073/pnas.1116523109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woodgett JR, Davison MT, Cohen P. The calmodulin-dependent glycogen synthase kinase from rabbit skeletal muscle. Purification, subunit structure and substrate specificity. Eur J Biochem. 1983;136:481–7. doi: 10.1111/j.1432-1033.1983.tb07766.x. [DOI] [PubMed] [Google Scholar]
- 10.Woodgett JR. Molecular cloning and expression of glycogen synthase kinase-3/factor A. EMBO J. 1990;9:2431–8. doi: 10.1002/j.1460-2075.1990.tb07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peineau S, et al. The role of GSK-3 in synaptic plasticity. Br J Pharmacol. 2008;153(Suppl 1):S428–37. doi: 10.1038/bjp.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rui Y, et al. Activity-dependent regulation of dendritic growth and maintenance by glycogen synthase kinase 3beta. Nat Commun. 2013;4:2628. doi: 10.1038/ncomms3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sereno L, et al. A novel GSK-3beta inhibitor reduces Alzheimer’s pathology and rescues neuronal loss in vivo. Neurobiol Dis. 2009;35:359–67. doi: 10.1016/j.nbd.2009.05.025. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Jope RS. Is glycogen synthase kinase-3 a central modulator in mood regulation? Neuropsychopharmacology. 2010;35:2143–54. doi: 10.1038/npp.2010.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beaulieu JM, et al. Lithium antagonizes dopamine-dependent behaviors mediated by an AKT/glycogen synthase kinase 3 signaling cascade. Proc Natl Acad Sci USA. 2004;101:5099–104. doi: 10.1073/pnas.0307921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latapy C, Rioux V, Guitton MJ, Beaulieu JM. Selective deletion of forebrain glycogen synthase kinase 3beta reveals a central role in serotonin-sensitive anxiety and social behaviour. Philos Trans R Soc Lond B Biol Sci. 2012;367:2460–74. doi: 10.1098/rstb.2012.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neasta J, Ben Hamida S, Yowell QV, Carnicella S, Ron D. AKT signaling pathway in the nucleus accumbens mediates excessive alcohol drinking behaviors. Biol Psychiatry. 2011;70:575–82. doi: 10.1016/j.biopsych.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, F., et al., mTORC1-dependent translation of collapsin response mediator protein-2 drives neuroadaptations underlying excessive alcohol-drinking behaviors. Mol Psychiatry, 2017;22:89-101. 10.1038/mp.2016.12 [DOI] [PMC free article] [PubMed]
- 19.Neznanova O, et al. Acute ethanol challenge inhibits glycogen synthase kinase-3beta in the rat prefrontal cortex. Int J Neuropsychopharmacol. 2009;12:275–80. doi: 10.1017/S1461145708009620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mai L, Jope RS, Li X. BDNF-mediated signal transduction is modulated by GSK3beta and mood stabilizing agents. J Neurochem. 2002;82:75–83. doi: 10.1046/j.1471-4159.2002.00939.x. [DOI] [PubMed] [Google Scholar]
- 21.Darcq E, et al. MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Mol Psychiatry. 2015;20:1219–31. doi: 10.1038/mp.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grusser SM, et al. Cue-induced activation of the striatum and medial prefrontal cortex is associated with subsequent relapse in abstinent alcoholics. Psychopharmacology. 2004;175:296–302. doi: 10.1007/s00213-004-1828-4. [DOI] [PubMed] [Google Scholar]
- 23.Seo D, Sinha R. The neurobiology of alcohol craving and relapse. Handb Clin Neurol. 2014;125:355–68. doi: 10.1016/B978-0-444-62619-6.00021-5. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy RE, Archer KJ, Miles MF. Empirical validation of the S-Score algorithm in the analysis of gene expression data. BMC Bioinforma. 2006;7:154. doi: 10.1186/1471-2105-7-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang L, Wang L, Ravindranathan A, Miles MF. A new algorithm for analysis of oligonucleotide arrays: application to expression profiling in mouse brain regions. J Mol Biol. 2002;317:225–35. doi: 10.1006/jmbi.2001.5350. [DOI] [PubMed] [Google Scholar]
- 26.Meliska CJ, Bartke A, McGlacken G, Jensen RA. Ethanol, nicotine, amphetamine, and aspartame consumption and preferences in C57BL/6 and DBA/2 mice. Pharmacol Biochem Behav. 1995;50:619–26. doi: 10.1016/0091-3057(94)00354-8. [DOI] [PubMed] [Google Scholar]
- 27.Tolliver GA, Sadeghi KG, Samson HH. Ethanol preference following the sucrose-fading initiation procedure. Alcohol. 1988;5:9–13. doi: 10.1016/0741-8329(88)90036-5. [DOI] [PubMed] [Google Scholar]
- 28.Edenberg HJ, et al. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–52. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bachmanov AA, Tordoff MG, Beauchamp GK. Ethanol consumption and taste preferences in C57BL/6ByJ and 129/J mice. Alcohol Clin Exp Res. 1996;20:201–6. doi: 10.1111/j.1530-0277.1996.tb01630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O’Brien WT, Klein PS. Validating GSK3 as an in vivo target of lithium action. Biochem Soc Trans. 2009;37:1133–8. doi: 10.1042/BST0371133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chou CH, et al. GSK3beta regulates Bcl2L12 and Bcl2L12A anti-apoptosis signaling in glioblastoma and is inhibited by LiCl. Cell Cycle. 2012;11:532–42. doi: 10.4161/cc.11.3.19051. [DOI] [PubMed] [Google Scholar]
- 32.Hopkins HS, Gelenberg AJ. Serum lithium levels and the outcome of maintenance therapy of bipolar disorder. Bipolar Disord. 2000;2:174–9. doi: 10.1034/j.1399-5618.2000.020304.x. [DOI] [PubMed] [Google Scholar]
- 33.Schuckit MA. Low level of response to alcohol as a predictor of future alcoholism. Am J Psychiatry. 1994;151:184–9. doi: 10.1176/ajp.151.2.184. [DOI] [PubMed] [Google Scholar]
- 34.Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- 35.Driessen M, et al. The course of anxiety, depression and drinking behaviours after completed detoxification in alcoholics with and without comorbid anxiety and depressive disorders. Alcohol Alcohol. 2001;36:249–55. doi: 10.1093/alcalc/36.3.249. [DOI] [PubMed] [Google Scholar]
- 36.Kliethermes CL, Cronise K, Crabbe JC. Anxiety-like behavior in mice in two apparatuses during withdrawal from chronic ethanol vapor inhalation. Alcohol Clin Exp Res. 2004;28:1012–9. doi: 10.1097/01.ALC.0000131976.40428.8F. [DOI] [PubMed] [Google Scholar]
- 37.Zhu LQ, et al. GSK-3 beta inhibits presynaptic vesicle exocytosis by phosphorylating P/Q-type calcium channel and interrupting SNARE complex formation. J Neurosci. 2010;30:3624–33. doi: 10.1523/JNEUROSCI.5223-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smillie KJ, Cousin MA. The Role of GSK3 in Presynaptic Function. Int J Alzheimers Dis. 2011;2011:263673. doi: 10.4061/2011/263673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei J, Liu W, Yan Z. Regulation of AMPA receptor trafficking and function by glycogen synthase kinase 3. J Biol Chem. 2010;285:26369–76. doi: 10.1074/jbc.M110.121376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeon SJ, et al. Activation of adenosine A2A receptor up-regulates BDNF expression in rat primary cortical neurons. Neurochem Res. 2011;36:2259–69. doi: 10.1007/s11064-011-0550-y. [DOI] [PubMed] [Google Scholar]
- 41.Silva AP, et al. Up-regulation of neuropeptide Y levels and modulation of glutamate release through neuropeptide Y receptors in the hippocampus of kainate-induced epileptic rats. J Neurochem. 2005;93:163–70. doi: 10.1111/j.1471-4159.2004.03005.x. [DOI] [PubMed] [Google Scholar]
- 42.Yang C, et al. The combined effects of the BDNF and GSK3B genes modulate the relationship between negative life events and major depressive disorder. Brain Res. 2010;1355:1–6. doi: 10.1016/j.brainres.2010.07.079. [DOI] [PubMed] [Google Scholar]
- 43.Tapocik JD, et al. microRNA-206 in rat medial prefrontal cortex regulates BDNF expression and alcohol drinking. J Neurosci. 2014;34:4581–8. doi: 10.1523/JNEUROSCI.0445-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez A, Alonso M, Castro A, Perez C, Moreno FJ. First non-ATP competitive glycogen synthase kinase 3 beta (GSK-3beta) inhibitors: thiadiazolidinones (TDZD) as potential drugs for the treatment of Alzheimer’s disease. J Med Chem. 2002;45:1292–9. doi: 10.1021/jm011020u. [DOI] [PubMed] [Google Scholar]
- 45.Bowers MS, et al. Nucleus accumbens AGS3 expression drives ethanol seeking through G betagamma. Proc Natl Acad Sci USA. 2008;105:12533–8. doi: 10.1073/pnas.0706999105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blegen MB, et al. Alcohol operant self-administration: Investigating how alcohol-seeking behaviors predict drinking in mice using two operant approaches. Alcohol. 2018;67:23–36. doi: 10.1016/j.alcohol.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bierut LJ, et al. Defining alcohol-related phenotypes in humans. The Collaborative Study on the Genetics of Alcoholism. Alcohol Res Health. 2002;26:208–13. [PMC free article] [PubMed] [Google Scholar]
- 48.Bierut LJ, et al. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci USA. 2010;107:5082–7. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Putman AH, et al. Identification of quantitative trait loci and candidate genes for an anxiolytic-like response to ethanol in BXD recombinant inbred strains. Genes Brain Behav. 2016;15:367–81. doi: 10.1111/gbb.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yasuda S, Liang MH, Marinova Z, Yahyavi A, Chuang DM. The mood stabilizers lithium and valproate selectively activate the promoter IV of brain-derived neurotrophic factor in neurons. Mol Psychiatry. 2009;14:51–9. doi: 10.1038/sj.mp.4002099. [DOI] [PubMed] [Google Scholar]
- 51.Ehlers CL, et al. Neuropeptide Y levels in ethanol-naive alcohol-preferring and nonpreferring rats and in Wistar rats after ethanol exposure. Alcohol Clin Exp Res. 1998;22:1778–82. doi: 10.1111/j.1530-0277.1998.tb03979.x. [DOI] [PubMed] [Google Scholar]
- 52.Thiele TE, Marsh DJ, Ste Marie L, Bernstein IL, Palmiter RD. Ethanol consumption and resistance are inversely related to neuropeptide Y levels. Nature. 1998;396:366–9. doi: 10.1038/24614. [DOI] [PubMed] [Google Scholar]
- 53.Mayfield RD, et al. Patterns of gene expression are altered in the frontal and motor cortices of human alcoholics. J Neurochem. 2002;81:802–13. doi: 10.1046/j.1471-4159.2002.00860.x. [DOI] [PubMed] [Google Scholar]
- 54.Wetherill L, et al. Neuropeptide Y receptor genes are associated with alcohol dependence, alcohol withdrawal phenotypes, and cocaine dependence. Alcohol Clin Exp Res. 2008;32:2031–40. doi: 10.1111/j.1530-0277.2008.00790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bienkowski P, Kostowski W, Koros E. Ethanol-reinforced behaviour in the rat: effects of naltrexone. Eur J Pharmacol. 1999;374:321–7. doi: 10.1016/S0014-2999(99)00245-9. [DOI] [PubMed] [Google Scholar]
- 56.Lovestone S, et al. A phase II trial of tideglusib in Alzheimer’s disease. J Alzheimer’s Dis. 2015;45:75–88. doi: 10.3233/JAD-141959. [DOI] [PubMed] [Google Scholar]
- 57.Davies AG, et al. A central role of the BK potassium channel in behavioral responses to ethanol in C. elegans. Cell. 2003;115:655–66. doi: 10.1016/S0092-8674(03)00979-6. [DOI] [PubMed] [Google Scholar]
- 58.Velazquez-Marrero C, et al. Alcohol Regulates BK Surface Expression via Wnt/beta-Catenin Signaling. J Neurosci. 2016;36:10625–39. doi: 10.1523/JNEUROSCI.0491-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cheng Y, et al. Distinct synaptic strengthening of the striatal direct and indirect pathways drives alcohol consumption. Biol Psychiatry.10.1016/j.biopsych.2016.05.016 (2016). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.