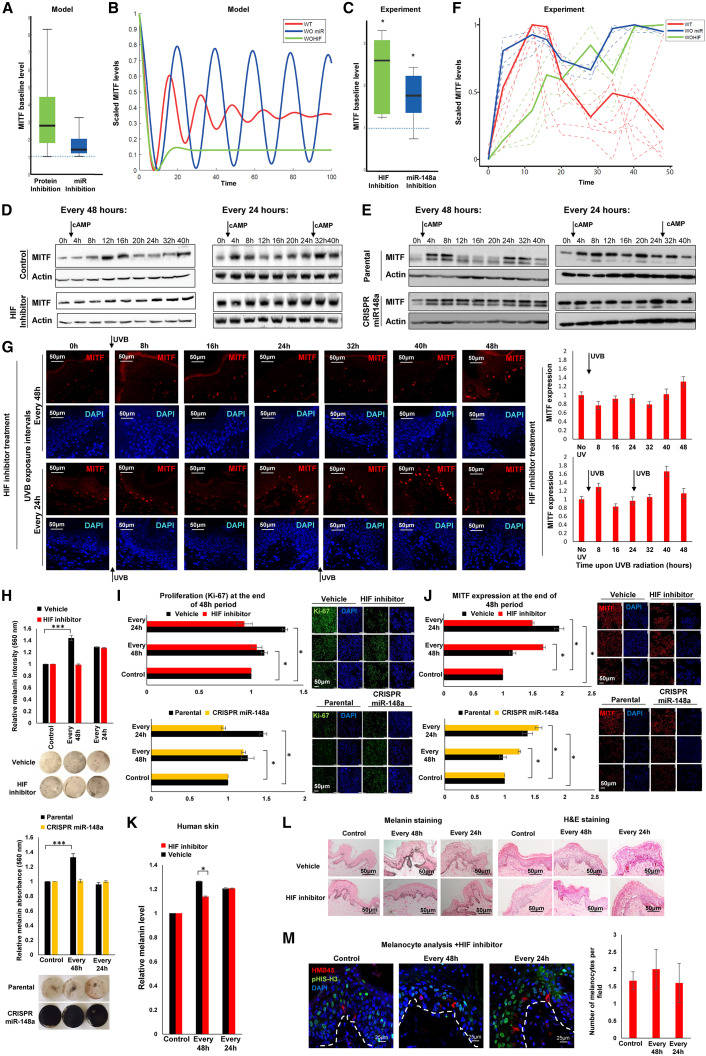
Summary
Skin sun exposure induces two protection programs: stress responses and pigmentation, the former within minutes and the latter only hours afterward. Although serving the same physiological purpose, it is not known whether and how these programs are coordinated. Here, we report that UVB exposure every other day induces significantly more skin pigmentation than the higher frequency of daily exposure, without an associated increase in stress responses. Using mathematical modeling and empirical studies, we show that the melanocyte master regulator, MITF, serves to synchronize stress responses and pigmentation and, furthermore, functions as a UV-protection timer via damped oscillatory dynamics, thereby conferring a trade-off between the two programs. MITF oscillations are controlled by multiple negative regulatory loops, one at the transcriptional level involving HIF1α and another post-transcriptional loop involving microRNA-148a. These findings support trait linkage between the two skin protection programs, which, we speculate, arose during furless skin evolution to minimize skin damage.
Keywords: UVB radiation, skin pigmentation, skin proliferation, MITF dynamics, trait linkage
Graphical Abstract
Highlights
-
•
UV exposure frequency reveals a trade-off between skin protection programs
-
•
MITF dynamics synchronize skin stress responses and pigmentation
-
•
MITF serves as a UV-protection timer
-
•
Two negative regulatory loops involving miR-148a and HIF1α underlie MITF dynamics
Malcov-Brog et al. report that UVB exposure every other day induces more skin pigmentation than daily exposure. This reveals a trade-off between the two skin protection programs, pigmentation and stress response. MITF synchronizes skin protection programs and functions as a UV-protection timer via damped oscillatory dynamics, controlled by multiple negative regulatory loops.
Introduction
Two protection programs are activated upon sun exposure with different timescales. In the short term, within minutes, there is the UV radiation stress response that involves proliferation, inflammation, DNA repair, and immune system recruitment (Clydesdale et al., 2001). In the longer term, within hours and up to days of exposure, there is production of pigment in epidermal melanocytes (Coelho et al., 2009), which physically protects DNA against UV-induced mutations and serves to prepare the skin for the next UV exposure (D’Orazio et al., 2006). Pigment production initiates hours after radiation (Coelho et al., 2009); however, the mechanism regulating this delay is unknown. Further, although these two programs serve the same physiological purpose, but on different timescales, it is not clear how their action is synchronized.
It has already been shown that there is crosstalk between keratinocytes and melanocytes within the epidermis upon UV radiation (Oren and Bartek, 2007). Keratinocytes produce and secrete melanocyte-stimulating hormone (αMSH), which binds to the G-protein-coupled receptor melanocortin 1 receptor (MC1R) expressed at the melanocyte surface. Activation of MC1R leads to activation of adenylate cyclase, which, in turn, increases cAMP levels and activates PKA in melanocytes. Once activated, PKA phosphorylates the transcription factor CREB, triggering recruitment of CBP/p300 and transcription of CREB’s target genes, including the melanocyte master regulator, microphthalmia-associated transcription factor (MITF) (Oren and Bartek, 2007). In summary, a cascade of events lead to MITF activation, a master regulator that controls production of melanin pigment and its transfer to surrounding keratinocytes to provide physical protection against UV-induced DNA damage (D’Orazio et al., 2006). Notably, MITF has also been shown to upregulate the expression of genes involved in survival and proliferation (Oren and Bartek, 2007), raising the possibility that it is involved in the activation of both UV protection programs.
Adaptive morphological changes are considered to evolve by alterations in specific developmental programs during organism development (Carroll, 2008). A common selective pressure can result in different programs independently altering, or, in some cases, it can result in trait linkage, whereby a shared regulation evolves, presumably because coordinated action confers an adaptive advantage. For example, independently regulated programs were shown to control beak width and length in Darwin’s finches, the joint outcome of which is presumed to confer niche specialization (Mallarino et al., 2012); whereas in teleost fish, tooth and cusp numbers exhibit linkage, controlled by a single genetic locus, BMP4 (Plikus et al., 2005). Trait linkage typically involves a trade-off in that at least one of the linked programs functions sub-optimally, though this cost is usually hidden, as its revelation requires a particular configuration (genotype and/or environment) not necessarily observed in the population.
Here, using both human and mouse skin models, we demonstrate that daily UV exposure yields reduced pigmentation compared to exposure every other day, even when the total amount of UV is normalized. In light of this surprising observation, we discovered that there is a tightly controlled temporal relationship between the stress response and pigmentation skin protection programs that is governed by MITF expression dynamics. Further, using mathematical modeling and complementary validation experiments, we delineated that MITF dynamics in response to UVB exposure are the result of a double-negative regulatory loop at the transcription and translation levels. Daily exposure disrupts the dynamics of MITF, disturbing the sequential ordering of its downstream regulatory pathway, resulting in a protection system that is sensitive to the frequency rather than the total amount of UV exposure. Taken together, for the first time, these data reveal a linkage between the stress response and pigmentation skin protection programs, with MITF serving as a synchronizing UV-protection timer that dictates their trade-off.
Results
The Frequency of UVB Exposure Dictates a Trade-off between Skin Protection Programs
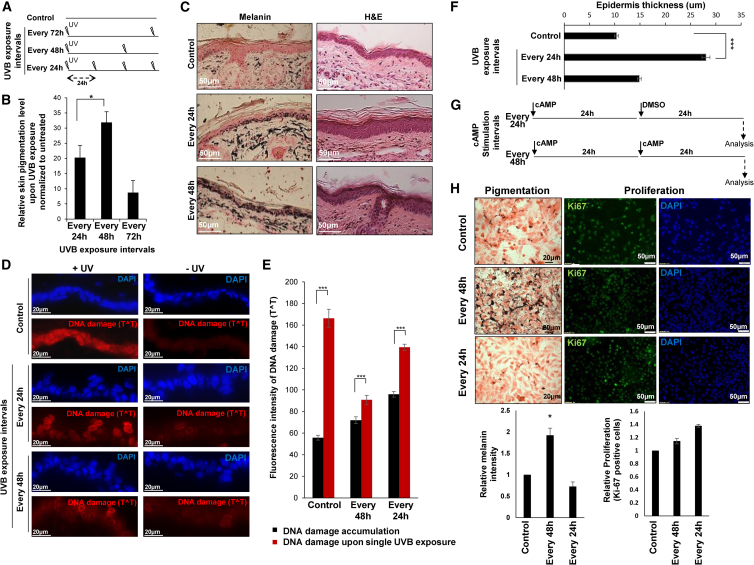
To investigate the regulation of skin responses to UV radiation, we exposed mice to UVB radiation (50 mJ/cm2) at periodicities of 24, 48, or 72 hr for 60 days using the FDA-recommended photo-therapy protocol (Nolan et al., 2010) (Figure 1A). We expected to observe a positive correlation between exposure frequency and accumulation of pigment. Remarkably, we observed that UVB exposure every 48 hr induced significantly more skin pigmentation than any of the other periodicities (Figures 1B and 1C, left panel), even when we controlled for total UV dosage (Figure S1A). Since skin pigmentation provides protection against UV-induced DNA damage (D’Orazio et al., 2006), we examined whether mice exposed to UV at a 48-hr periodicity exhibited higher skin protection. To this end, we quantified thymine dimers (TˆT), which are a common UVB-induced DNA lesion (Regan et al., 1968). Indeed, mice exposed to UVB at a 48-hr periodicity exhibited less DNA damage than mice from the other treatment groups, even when values were normalized to the accumulated DNA damage (Figures 1D, 1E, S1B, and S1C). Although pigmentation was increased in mice exposed to UVB at a 48-hr periodicity, as compared to a 24-hr periodicity, we noted that epidermis thickness exhibited the opposite pattern (Figures 1C [right panel], 1F, and S1D). No apoptotic epidermal keratinocytes (sunburned cells; Bayerl et al., 1995, D’Orazio et al., 2006) were found, demonstrating that radiation was below the erythema level (Figures S1E and S1F).
Figure 1.
The Frequency of UVB Exposure Dictates a Trade-off between Skin Protection Programs
(A) Schematic presentation of the experimental procedure. Mouse skin was UVB irradiated every 24, 48, or 72 hr or untreated (Control).
(B) Melanin level of C57BL/6 mice skin upon 60 days of UVB radiation (50 mJ/cm2) at indicated periodicity normalized to untreated controls; Error bars indicate SEM. ∗p < 0.05 (n = 4).
(C) Fontana-Masson (melanin) and H&E staining of ear sections from the representative mice after 60 days of treatment. Scale bars, 50 μm.
(D) DNA damage analysis (see STAR Methods) by Thymine dimer (TˆT) staining (red) of ear sections. DAPI staining (cell nuclei) appears in blue. Left: DNA-damage level immediately upon a single UVB dose (50 mJ/cm2) (+UV). Right: the accumulated DNA damage of ears that did not received the final UV dose (−UV). Scale bars, 20 μm.
(E) DNA damage (TˆT) quantification. Error bars indicate SEM. ∗∗∗p < 0.001 (n = 4 mice; n = 10 images from each section).
(F) Epidermal thickness quantification of the mice skin treated as in (A). Error bars represent SEM. ∗∗∗p < 0.001 (n = 6).
(G) Schematic of the 24-hr- or 48-hr-interval cAMP treatments in cell culture.
(H) Fontana-Masson, Ki67, and DAPI (cell nuclei) staining (n = 6) of melanoma cells upon single or double cAMP stimulation. Vehicle-treated cells were used as a control. Error bars represent SEM. ∗p < 0.05. Scale bars, 20 μm for pigmentation and 50 μm for proliferation.
See also Figure S1.
To corroborate these unexpected observations, we used the MNT-1 melanoma cell line, a well-established model for studying pigmentation and normal melanogenesis mechanisms (Kushimoto et al., 2001), which has a transcriptome highly comparable to that of normal melanocytes (Hoek et al., 2004). Upon UV radiation, DNA damage in keratinocytes induces P53 activation, which upregulates αMSH expression (Oren and Bartek, 2007). αMSH is secreted and interacts with melanocytes, leading to cyclic AMP (cAMP) elevation, which results in transcriptional activation of MITF (Oren and Bartek, 2007). When using an in vivo system, either human or mouse skin, UVB radiation induces keratinocytes to produce αMSH and to release it toward melanocytes. αMSH will activate cAMP cascade in melanocyte, leading to pigment production (Oren and Bartek, 2007). When we want to directly activate melanocyte in culture, the keratinocyte presence is missed; therefore, melanocyte will be stimulated directly with cAMP, which will mimic the UVB effect, as we have done before (Levy et al., 2010b). Consistent with our previous results, stimulation with cAMP every 48 hr induced more pigmentation and less proliferation, compared to stimulation every 24 hr (Figures 1G, 1H, and S1G). These data further support that the skin is sensitive to the periodicity of UVB exposure rather than to the total amount of radiation. Taken together, these observations uncover that, at the 48-hr periodicity, the stress response does not correlate with the pigmentation response to UVB exposure, which led us to hypothesize that there may be linkage between these two UV protection programs, with the particular condition of 48-hr periodicity revealing the trade-off.
Skin Protection Programs Have a Sequential Dynamic of Expression
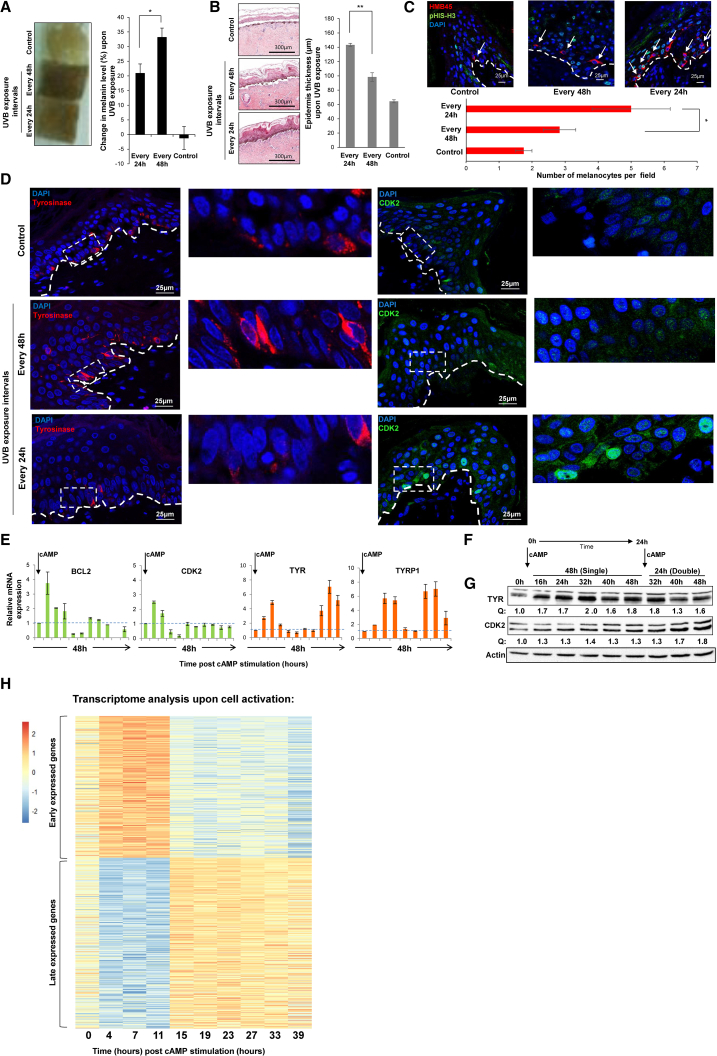
In line with previous results, human skin exposed to 250 mJ/cm2 UVB every 48 hr exhibited significantly greater pigmentation (Figures 2A and S2A) and lower epidermal thickness (Figure 2B), compared to samples treated every 24 hr. Additionally, significantly more melanocytes were observed when skin was exposed to UVB in 24-hr intervals than in 48-hr intervals (Figure 2C). Next, we evaluated the expression of representative genes for skin proliferation and pigmentation; namely, CDK2 and TYR, respectively (Levy and Fisher, 2011), at the protein level. Focusing on the basal epidermis where melanocytes are located, we observed induced expression of TYR, 32 hr after a first and single dose of radiation, which was suppressed by additional UVB exposure separated from the first one by 24 hr (Figure 2D, left panel). In contrast, CDK2 expression demonstrated induced expression after the first UVB exposure, which further increased after the second exposure (Figure 2D, right panel, and Figure S2B). These results show that expression dynamics of representative stress and pigmentation proteins mirror the phenotypes of proliferation and pigmentation.
Figure 2.
Skin Protection Programs Have a Sequential Dynamic of Expression
For a Figure360 author presentation of Figure 2, see https://doi.org/10.1016/j.molcel.2018.09.022.
(A) Bright image of human skin after 10 days of exposure to UVB (250 mJ/cm2) at indicated intervals. Graph represents quantification of the changes in melanin levels normalized to day 0. Error bars represent SEM. ∗p < 0.05 (n = 5 donors).
(B) H&E staining of human skin as described in (A). Graph represents the epidermal thickness quantification. Error bars represent SEM. ∗∗p < 0.01 (n = 1 representative donor per 55 segments).
(C) Immunofluorescence staining of melanocytes (HMB45) and proliferation marker (Phospho Histone H3) following UVB (250 mJ/cm2) exposure of human skin at the indicated intervals. Graph represents melanocyte number per field quantification. Untreated skin was used as a control. Error bars represent SEM. ∗p < 0.05. Scale bars, 25 μm.
(D) Immunofluorescence analysis of TYR (red) and CDK2 (green) upon single or double UVB exposure (200 mJ/cm2) of human skin. Control samples were not irradiated. Samples were irradiated once at time 0 or once at time 0 and again at 24 hr. Analysis was performed at 32 hr after the first dose. Scale bars, 25 μm.
(E) Proliferation (BCL2 and CDK2) and pigmentation (TYR and TYRP1) gene expression level upon single cAMP stimulation of MNT-1 cells, approximately every 5 hr. Data were normalized to actin. Error bars represent SEM of the technical replicates (n = 3).
(F) Experimental design: MNT-1 cells were treated with cAMP at time 0 (single) or treated twice at time 0 and 24 hr after first stimulation (double).
(G) Protein levels of TYR and CDK2 were determined at indicated times. Quantification of protein amount normalized to actin (Q) is indicated.
(H) Scaled expression (Z scores) of the early-induced (top) and late-induced (bottom) genes in melanocytes at baseline and at the indicated time points post-cAMP stimulation.
See also Figure S2 and Tables S1 and S2.
We used the MNT-1 melanoma cell line to corroborate the different gene-expression dynamics of the two skin protection programs. Initially, we examined mRNA expression of BCL2 and CDK2 as representative genes regulating survival and that of TYR and TYRP1 as representative pigmentation genes (Levy and Fisher, 2011) approximately every 5 hr during the 48-hr period of cAMP treatment. Both survival and pigmentation genes peaked approximately 10 hr following a single stimulation with cAMP (Figure 2E). However, in the case of the pigmentation genes, we observed a second higher peak that was not observed for the survival genes (Figure 2E). Notably, a second stimulation resulted in elevated expression of CDK2 and decreased TYR expression, as compared to the levels of these proteins in the absence of a second stimulation (Figures 2F and 2G). To examine gene-expression dynamics systematically, we analyzed the whole transcriptome 48 hr following cAMP stimulation at 10 time points. Two clear waves of early (n = 833) and late (n = 1,015) genes (false discovery rate [FDR] ≤ 0.05) were induced upon stimulation (Figures 2H, S2C, and S2D; Tables S1 and S2), indicating a differential temporal regulation of distinct gene-expression programs.
This analysis further supports the hypothesis that both survival and pigmentation programs are initiated following UV exposure yet exhibit distinct expression dynamics with different time windows, suggesting the existence of a master regulator or regulators that synchronize the two programs and dictates skin response to UVB exposure periodicity.
UVB Exposure Periodicity Directly Affects MITF Dynamics Behavior
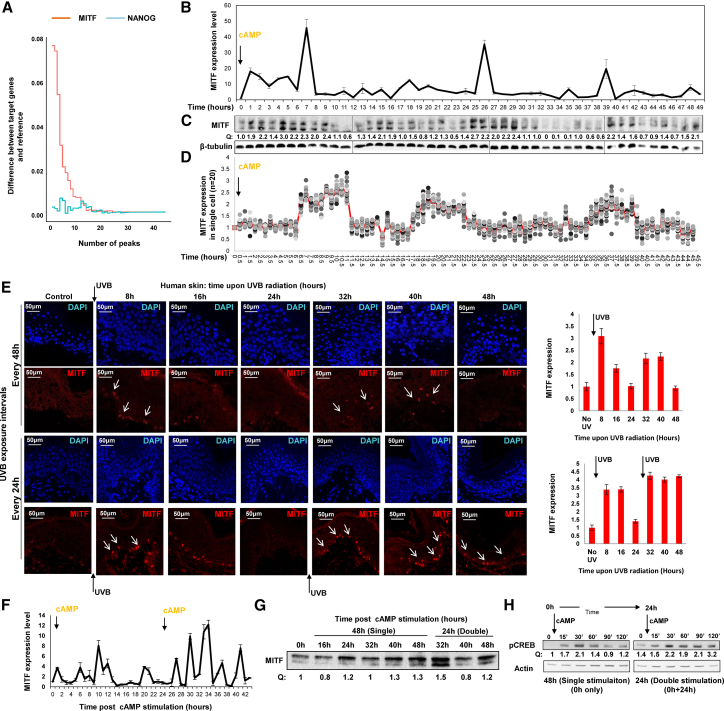
To identify the master regulator, we searched for a common upstream regulator of the early and late genes by Ingenuity Pathway Analysis and identified a significant enrichment for MITF, the key regulator of the melanocyte lineage (Levy and Fisher, 2011; p = 0.0012). Indeed, an assessment of the genomic occupancy of MITF on the promoters of the early and late genes based on chromatin immunoprecipitation sequencing (ChIP-seq) analysis (Webster et al., 2014) revealed a significant enrichment of MITF binding at these sites (ps = 0.0001 and 0.00021, respectively, Figure 3A), an enrichment that was not observed for NANOG, an unrelated control regulator (ps = 0.32 and 0.56, respectively; Table S3). To provide further support that MITF is acting as a master regulator of the UV skin protection programs, we monitored MITF dynamics during the first 48 hr following stimulation of MNT-1 cells with a single dose of cAMP. Interestingly, MITF exhibited a damped oscillatory expression pattern during the first 48 hr following the single stimulation, at both the mRNA and protein levels (Figures 3B and 3C), yet did not exhibit a dependency on the circadian clock in the absence of stimulation (Figure S3A). To exclude the possibility that the observed dampened behavior of MITF is due to non-synchronization between cells, we analyzed MITF levels in single cells every 30 min after initiation of cAMP stimulation over 48 hr. Indeed, MITF oscillatory dynamics were dampened at the single-cell level (Figures 3D and S3B). We then measured the expression of MITF in human skin upon UVB exposure every 48 hr or 24 hr. Similarly we observed a damped oscillatory pattern of expression (Figures 3E, upper panel, and S3C). Next, we examined MITF expression in MNT-1 cells after two stimulations, by means every 24 hr, by re-analyzing the samples from Figures 2F and 2G. Following the sequential 24-hr stimulations, MITF exhibited more numerous and pronounced peaks, as compared to after the first stimulation, both at the mRNA and protein levels (Figures 3F and 3G; actin was used as a loading control, as presented in Figure 2G). Similar disruption to MITF dynamics was observed in human skin UVB irradiated every 24 hr (Figure 3E, lower panels).
Figure 3.
UVB Exposure Periodicity Directly Affects MITF Dynamics Behavior
(A) The difference in the number of MITF (pink) and Nanog (blue) peaks detected in the promoters of early and late genes relative to all genes.
(B and C) MNT-1 cells were treated with a single dose of cAMP, and MITF mRNA (B) and protein (C) were quantified hourly for 48 hr. mRNA was normalized to actin, and β-tubulin was used as a loading control. Error bars represent SEM of the technical replicates (n = 3).
(D) MNT-1 cells were treated as in (C), and samples were fixed every 30 min for 48 hr (images are shown in Figure S3B). Graph represents quantification of MITF fluorescence intensity in single cells (n = 20).
(E) Immunofluorescence analysis of MITF expression (arrows) in human skin at the indicated time points after UVB treatment (250 mJ/cm2) at the 24-hr or 48-hr intervals. DAPI-stained nuclei appear in blue. Error bars represent SEM. ∗p < 0.05 (n = 8 cells in each time point). Scale bars, 50 μm.
(F) MITF mRNA expression in MNT-1 cells upon every 24-hr cAMP stimulation normalized to GAPDH. Error bars represent SEM of the technical replicates (n = 2).
(G) MITF protein levels at the indicated time points in cells treated as in (F). Actin was used as a loading control, as in Figure 2G. Quantification of protein amount normalized to actin (Q) is indicated.
(H) Cells were treated with cAMP at 24-hr or 48-hr intervals. Phospho-CREB protein levels were measured at the indicated times post-cAMP treatment. Quantification of protein amount normalized to actin (Q) is indicated.
Lastly, to confirm the role of MITF in the waves of gene expression in response to UV, we examined whether an upstream regulator of MITF—namely, CREB—exhibits waves of activity. Upon UVB radiation, αMSH interacts with the G-protein-coupled receptor MC1R on melanocytes, leading to activation of the cAMP/CREB signaling pathway, and CREB phosphorylation transcriptionally indicates the expression of MITF (Khaled et al., 2010). Therefore, we measured the phospho-CREB level upon cAMP treatment as an indicator of MITF transcriptional activation (Khaled et al., 2010), immediately after cAMP activation and at 24 hr post-activation. CREB phosphorylation levels were maintained longer after the second stimulation, as compared to after the first stimulation (Figures 3H, S3D, and S3E), suggesting that cells “remember” that there was an activation 24 hr prior. The different patterns of CREB phosphorylation upon a single stimulation versus sequential stimulations reflect different MITF oscillatory behaviors. Taken together, these data support a model whereby MITF serves as a timer of the UV-protection programs, controlling the skin response to UV radiation as a function of the periodicity or schedule of the exposure.
Two Layers of Negative Regulatory Loops Determine MITF Damped Oscillatory Behavior
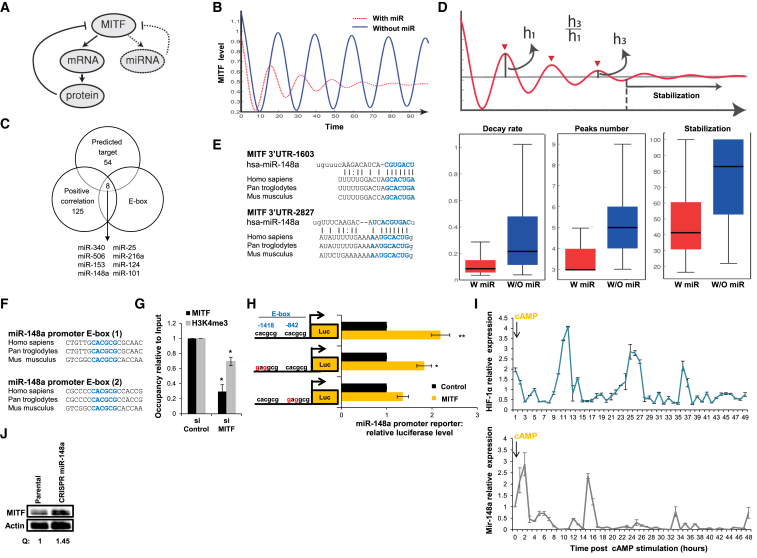
To further understand MITF regulation, we analyzed all known predicted and validated MITF regulators using Ingenuity Pathway Analysis (Table S4) and generated a mathematical model that simulates MITF dynamics under their regulation (Figure 4A, solid lines). For this, we used two proteins: HIF1α and phosphodiesterase 4D (PDE4D); both are induced by MITF at the transcription level (Busca et al., 2005) while negatively regulating its activity either by a direct repression (for HIF1α; Feige et al., 2011) or by cAMP degradation (for PDE4D; Khaled et al., 2010). We simulated the dynamics of this regulatory circuitry using over 2 million uniformly sampled rate constants (STAR Methods) and observed a damped oscillatory pattern under a broad range of parameter sets (Figure S4A), indicating the robustness of MITF oscillation under our model (Figure 4B, blue line). However, for many parameter sets, oscillation dampening was minor as compared to the experimental result (Figure 3). Thus, we looked for additional regulation layers that fit the experimental observation.
Figure 4.
Two Layers of Negative Regulatory Loops Determine MITF Damped Oscillatory Behavior
(A) Mathematical model depicting MITF negative regulation by PDE4D and HIF1α (bold, solid lines indicate that transcription and translation are required) and miRNAs (dashed lines indicate that only transcription is required).
(B) A simulation of MITF expression dynamics as derived from the mathematical model under negative regulation of protein only (blue) or a combination of protein and miRNA (red).
(C) Overlap between 62 miRNAs predicted to target MITF (http://www.targetscan.org), 133 miRNAs in positive correlation with MITF (Bell et al., 2014), and miRNAs that include E-box in their promoter.
(D) Upper panel: an illustration depicting the dynamical features used for comparison between the reference (protein-derived regulation) and the reference under an additional miRNA regulation. Lower panels indicate oscillation features: decay rate (corresponding to the ratio between the third and first peaks, left panel), number of peaks (middle panel), and time to reach stabilization (right panel) under negative regulation of either protein (blue) or a combination of protein and miRNA (red).
(E) Predicted binding site for miR-148a in the MITF 3′UTR sequence.
(F) Two conserved MITF DNA binding sites (E-boxes) in miR-148a promoter sequence.
(G) ChIP was performed on extracts from WM3682 cells transfected with siMITF or siControl. Protein:chromatin-crosslinked complexes were precipitated with the indicated antibodies. PCR primers spanning the region encoding the miR-148a promoter were used. The data show promoter occupancy relative to input. Error bars represent SEM. ∗p < 0.05 (n = 3).
(H) Expression of miR-148a promoter reporter with WT or mutated E-box regions upon MITF cDNA (MITF) or empty vector (control) expression in HEK293T cells. Firefly luciferase activity was normalized to Renilla luciferase activity. Fold changes relative to control are indicated. Error bars represent SEM. ∗p < 0.05; ∗∗p < 0.01 (n = 3).
(I) HIF1α and miR-148a levels were quantified in MNT-1 samples collected hourly for 48 hr upon cAMP stimulation. Levels were normalized to actin and U6, respectively. Error bars represent SEM of technical replicates (n = 3).
(J) MITF protein levels in parental WT MNT-1 cells (control) and the miR-148a-deleted MNT1 cell line. Actin was used as a loading control. Quantification of protein amount normalized to actin (Q) is indicated.
MITF is also known to regulate the transcription of microRNAs (miRNAs) (Golan et al., 2015, Levy et al., 2010a). miRNAs are involved in fine-tuning numerous biological systems through post-transcriptional downregulation of their targets (Hornstein and Shomron, 2006). We identified eight miRNAs that have the potential to generate a negative regulatory loop with MITF (Figure 4C; Table S4); in agreement with MITF being their predicted target (TargetScan), they are positively correlated with MITF (Bell et al., 2014) and have E-box in their promoter, the conserved MITF consensus binding sequences (5′-CATGTG-3′; 5′-CACGCG-3′, E-box elements). Therefore, we introduced miRNA negative regulation into the model (Figure 4A, dashed lines) and investigated its impact on MITF expression dynamics. Of note, unlike the negative regulation exerted by HIF1α and PDE4D, which requires transcription and translation of the effectors, the proposed negative regulation exerted by miRNA does not require translation and, thus, is expected to be more rapid. Similar to the original model, the revised model also produced robust damped oscillations (Figures 4B, dashed red line, and S4B). However, we found that the expected oscillations in the revised model exhibited significantly higher decay rates, fewer peaks, and shorter stabilization periods, as compared to those in the original model (Figures 4D and S4C), observations that matched the experimental results. We further hypothesized that such features would be beneficial, as long-lasting expression of the survival program is detrimental. We thus sought to validate experimentally whether this additional negative-feedback loop is involved in regulation of the UV-protection system.
To identify the miRNAs that are directly involved in the regulation of MITF dumped oscillatory dynamics, we screened for the biological effect of each candidate miRNA (Figure 4C; Table S4). We examine how these miRNAs affect MITF protein level (Figure S4D) and MITF 3′UTR expression (Figure S4E) and how MITF overexpression or downregulation affects their expression (Figures S4F–S4H). Among the tested miRNAs, miR-148a appears as the best candidate for generating a negative regulatory loop with MITF. Notably, the human MITF genomic region includes two conserved miR-148a binding sites in the 3′ UTR (Figure 4E). miR-148a was shown to downregulate MITF expression in mice (Haflidadóttir et al., 2010, Porstner et al., 2015), and in a separate study, MITF was noted as occupying the miR-148a promoter (Ozsolak et al., 2008). To further establish that MITF induces miR-148a transcription, we inspected the human miR-148a promoter (∼1.8 kb upstream of the transcription start site) and identified two conserved MITF consensus binding sequences (5′-CACGCG-3′, E-box elements; Figure 4F). ChIP analysis validated that MITF occupies the miR-148a promoter (Figure 4G) and not downstream of the miRNA (Figure S4I). Furthermore, MITF depletion was accompanied by a decrease in histone 3 trimethylation at lysine 4 (H3K4me3) over the miR-148a promoter (Figure 4G). Since trimethylation is an epigenetic marker of transcriptionally active chromatin (Li et al., 2007), this observation lends further support to the premise that MITF activates miR-148a transcription. Lastly, to demonstrate direct regulation of miR-148a by MITF, we analyzed expression of a luciferase reporter gene under the control of a wild-type (WT) or mutated miR-148a promoter. MITF-dependent luciferase expression was substantially decreased by mutation of the second E-box element (Figure 4H). Furthermore MITF and miR-148a were found to positively correlate in a snapshot analysis (Figures S4J and S4K), strengthening our premise that MITF regulates miR-148a expression. We conclude that MITF directly regulates the expression of miR-148a.
Next, we measured HIF1α and miR-148a during the first 48 hr following stimulation of MNT-1 cells with a single dose of cAMP. We observed a damped oscillatory expression pattern for HIF1α and miR-148a (Figure 4I) that was MITF dependent (Figure S4L), supporting the hypothesis that miR-148a expression, indeed, plays a role in the MITF regulatory circuity. Lastly, we generated an MNT-1 cell line deleted for miR-148a using CRISPR-Cas9 (Figures S4M and S4N) and observed an upregulation of MITF levels, as compared to the parental WT line (Figure 4J). Taken together, these data indicate that MITF is negatively regulated by miR-148a, revealing, for the first time, the existence of a feedback loop at the post-transcriptional level that governs MITF expression.
Regulation of Skin Protection Systems by External Disruption of MITF Dynamics
To further understand how MITF dynamics control differential activation of gene-expression programs, we used our mathematical model, which predicted that MITF de-repression by either miRNA or HIF1α inhibition should result in an increase in MITF baseline levels (Figure 5A). Additionally, the model predicted differential effects on MITF dynamics stemming from the lack of regulation by the two different inhibitory arms (Figures 5B and S5A). Specifically, HIF1α inhibition was predicted to result in the loss of MITF oscillatory behavior, whereas miRNA inhibition was predicted to yield an augmented oscillatory dynamics (Figures 5B and S5A, left). To validate these predictions experimentally, we altered either miR-148a by a CRISPR line or HIF1α expression pharmacologically (500 nM FM19G11) (Figure S5B) and examined MITF expression longitudinally following cAMP stimulation. As the model predicted, MITF baseline levels increased in the presence of either inhibition (compare “0h” time points in Figures 5C, S5C, and S5D). Moreover, under HIF1α inhibition, MITF levels gradually increased in a non-oscillatory manner (Figures 5D, 5F, and 5G, top), whereas under miRNA inhibition, MITF-level oscillations were undampened (Figures 5E, 5F, and S5D). We further analyzed the effect of the inhibitors on MITF dynamics in the context of multiple rounds of cAMP stimulations. Notably, in these experiments, MITF dynamics are interrupted twice, by the inhibition of its regulators and by the additional stimulation. First, to clarify the effect of the inhibition of its regulators and dual stimulation, we quantified MITF expression levels under either inhibition of the regulatory arms following single or double stimulation (Figures 5D, 5E, and S5A, right panel); MITF expression levels were scaled and averaged across biological replicates (Figure S5E; conditions are color coded to the left). Hierarchical clustering analysis of the resulting MITF dynamics showed a high similarity between MITF expression under HIF1α inhibition under a single or dual stimulation, demonstrating the prominent effect of this inhibition to overcome the dual-stimulation-induced effects. MITF dynamics demonstrated similar disruption in human skin, in the presence of HIF1α inhibition and upon double UVB exposure (Figure 5G, bottom). In contrast, miR-148a inhibition and the control were clustered together under the repetitive stimulation, establishing the similarity of dual-stimulation effect on MITF dynamics that was less sensitive to the inhibition (Figure S5E). To further demonstrate the effect of repetitive stimulation on MITF dynamics, we added a simulation and statistical analyses of the dynamic features of MITF under this condition, as compared to a single stimulation (Figure S5F). These analyses demonstrated the expected enhanced oscillatory dynamics of MITF following double stimulation, as reflected by both the number of the oscillations and their amplitude (Figure S5G).
Figure 5.
Regulation of Skin Protection Systems by External Disruption of MITF Dynamics
(A) Fold change of MITF baseline under inhibition of either protein (green) or miRNA (blue) relative to the baseline value without any inhibition, as predicted by the mathematical model.
(B) A representative simulation of MITF dynamics under normal conditions (red) or under either HIF (green) or miRNA (blue) inhibitions. WO, without.
(C) Fold change of MITF baseline under inhibition of either HIF1α (green) or miR-148a (blue) relative to the baseline value without any inhibition, as observed experimentally. ∗p = 0.05 and ∗p = 0.03 for HIF1α and miR-148a inhibition, respectively; paired t test.
(D) MITF protein levels in WM3682 cells treated with vehicle or HIF1α inhibitor FM19G11 at the indicated times following 24-hr- or 48-hr-interval cAMP stimulation. Actin was used as a loading control.
(E) MITF protein levels in parental WT MNT-1 cells or the CRISPR-miR-148a MNT-1 cells at the indicated times following 24-hr- or 48-hr-interval cAMP stimulation. Actin was used as a loading control.
(F) Scaled MITF protein expression level dynamics under normal conditions (red) or under either HIF (green) or miRNA (blue) inhibitions. Dashed lines correspond to individual experiments, whereas bold lines correspond to the 75% quantile of MITF levels across all experiments.
(G) Human skin was treated with vehicle (control) or with FM19G11 (HIF1α inhibitor) and was exposed to UVB at 24-hr or 48-hr intervals. MITF immunofluorescence was analyzed (red) every 8 hr and quantified in the graphs on the right. DAPI-stained nuclei appear in blue. Error bars represent SEM (n = 8 cells in each time point). Scale bars, 50 μm.
(H–J) WM3682 cells were treated with vehicle or HIF1α inhibitor FM19G11 and parental WT MNT-1, or the CRISPR-miR-148a MNT-1 cells were stimulated with cAMP every 24 hr or 48 hr, fixed, and analyzed for (H) melanin levels (upper and lower panels; Fontana-Masson staining), (I) proliferation (Ki67), and (J) MITF levels. Graphs represent quantifications of the three experiments. Error bars represent SEM. ∗∗∗p < 0.001, ∗p < 0.05 (n = 3). Scale bars, 50 μm.
(K) Human skin was treated with vehicle or HIF1α inhibitor FM19G11 (30 mM) and subjected to no UV or to 24-hr or 48-hr UVB intervals (50 and 250 mJ/cm2, respectively). Graph represents quantification of skin pigmentation. Error bars represent SEM. ∗p < 0.05 (n = 2).
(L) Fontana-Masson (melanin) and H&E staining of the human skin, treated as in (K).
(M) Immunofluorescence staining of melanocytes (HMB45) and a proliferation marker (Phospho Histone H3) following UVB (250 mJ/cm2) exposure of human skin at the indicated intervals. Graph represents melanocyte number per field quantification. An untreated skin sample was used as a control. Error bars represent SEM. Scale bars, 25 μm.
See also Figure S5.
We have established that inhibition of MITF regulators, at either the protein or the miR level, significantly disrupts their dynamic behavior, in multiple rounds of UVB exposure. We further examined whether these changes in dynamics can be observed phenotypically. Melanoma cells in the presence or absence of HIF1α or miR-148a were stimulated with cAMP every 48 hr or every 24 hr, followed by phenotypic analysis at 48 hr after first stimulation. Under no inhibition, pigment level was significantly higher when cells were stimulated every 48 hr with cAMP rather than every 24 hr (Figures 1H and 5H). However, under inhibition, this elevated pigmentation was not observed. Notably, under miR-148a inhibition, cells were highly pigmented, due to very high MITF and TYR levels (Figures S5H and S5I), and we cannot conclude that cAMP exposure does not alter pigmentation, because pigmentation is already at the maximum levels. Furthermore, proliferation rate, as measured by Ki67-positive cells, was higher upon double cAMP stimulation (every 24 hr) compared to stimulation every 48 hr only in the presence of HIF1α and miR-148a (Figures 5I and S5J). Next, we analyzed MITF levels in the cell, at 48 hr after first stimulation. Upon single stimulation (every 48 hr), MITF levels return to the original level, while additional stimulation at 24 hr after the first stimulation, resulted in higher MITF levels in the cell (Figures 5J and S5K). In the absence of HIF1α or miR-148a, the system does not return to its baseline level, i.e., MITF levels remain high, under either single (every 48 hr) or double (every 24 hr) cAMP stimulations (Figure 5J). Consistent with the results obtained in culture, UV exposure of human and mouse skin, under the frequency of every 48 hr, resulted in higher skin pigmentation, compared to that from exposure every 24 hr (Figures 1B, 1C, and 2A). When HIF1α inhibitor was applied on the skin, prior to UV exposure, the higher skin pigmentation upon 48-hr frequencies was significantly decreased (Figures 5K, 5L, S5L, S5M, and S5N). On the other hand, upon HIF1α inhibitor application, no difference in melanocyte proliferation was observed under different UV exposure scheduling (Figure 5M). Taken together, our data demonstrate that, in normal conditions (a 48-hr interval), MITF exhibits dumped oscillatory dynamics upon UV exposure, resulting in an increased skin pigmentation. When MITF dynamics is perturbed by either additional UV exposure, 24 hr after the first exposure, or inhibition of its regulators, cell fate is disrupted as well. These data suggest that MITF dynamics regulates pigmentation and proliferation and that MITF serves as a UV-protection timer controlling the trade-off between the two skin protection systems.
Discussion
Using mathematical models and empirical studies, we show here that the transcription factor MITF, known as the central regulator of the melanocyte lineage, acts as a UV-protection timer, driving a temporal regulatory cascade following UV exposure that activates, first, cell survival and then the pigmentation programs. This programmatic synchronization is achieved by the damped oscillatory expression pattern of MITF and results in the linkage of two beneficial phenotypic traits and in the benefit of their ordered expression. Notably, while the inhibitory effect of each identified component (HIF1α and miR-148a) on MITF is known, their combinatorial effect through different inhibition rates on MITF dynamics is novel, resulting in a tremendous effect on the phenotype and activation of different programs. The expression dynamics of other master regulators have been shown to determine phenotype; for example, the transcription factors p53 (Purvis et al., 2012) and nuclear factor κB (NF-κB) (Hoffmann et al., 2002). It might be that dynamic activity of transcription factors is an advantage when the system needs to adjust rapidly to consistent exposure to stressors such as UVB.
The molecular mechanisms of how transcription factor dynamics control differential activation of gene programs—and, thus, cell fate—remain largely unknown (Yosef and Regev, 2011). One possibility is that MITF has differential affinities for different promoters due to DNA sequences or structures. This notion would explain why, immediately after stimulation, the high levels of MITF induced expression of the genes involved in cell-cycle regulation, whereas toward the end of the 48-hr period post-stimulation, low levels of MITF failed to activate these early genes. Pigmentation genes have high affinity for MITF, and only small amounts of MITF are required for their expression. Pigmentation genes are not efficiently expressed immediately after stimulation, however. It was previously demonstrated that H3K27 acetylation is abundant over pigmentation gene bodies (Laurette et al., 2015), and we postulated that pigmentation genes require epigenetic modification in order to be activated.
We examined H3K27 acetylation at loci of validated MITF target genes (Levy and Fisher, 2011) and found that H3K27 acetylation patterns were distinct for pigmentation-related and cell-cycle-related genes (Figures S5O and S5P). In the primary human melanocytes evaluated, the pigmentation gene bodies had significantly higher levels of H3K27 acetylation than cell-cycle gene bodies known to correlate with transcription (Rajagopal et al., 2014). Pigmentation gene expression in the same cells were expressed at significantly higher levels than cell-cycle genes (Figure S5Q). This suggests that pigmentation genes require epigenetic modification of their gene bodies in order to be expressed.
The H3K27 acetylation is regulated by BRG1, a member of the SWF/SNI chromatin remodeling complex (Laurette et al., 2015). BRG1 specifically facilitates MITF transcriptional activation of the pigmentation program but not the pro-survival program (Keenen et al., 2010). We found that BRG1 levels were upregulated at late time points post-cAMP stimulation, while additional cAMP stimulation at 24 hr disrupted BRG1 upregulation (Figure S5R). The BRG1 pattern of expression mirrors the late expression of pigmentation genes induced by MITF. This suggests that BRG1 facilitates MITF-induced expression of pigmentation genes by inducing their H3K27 acetylation after stimulation, resulting in their late expression. A further investigation of BRG1 activity post-melanocyte activation is needed.
Another possible explanation is that MITF levels might influence the equilibrium between homo- and heterodimers (Steingrímsson et al., 2004), and different complexes may dictate differential gene expression. Further, it might be that only at a certain threshold of MITF concentration will a cell respond by activating a specific gene set, as was shown previously (Yosef and Regev, 2011).
Taken together, evidence suggests that transcription factor dynamics influences cell fate (Hoffmann et al., 2002, Nelson et al., 2004, Purvis et al., 2012, Yosef and Regev, 2011), although further understanding of the molecular mechanism that regulates gene-expression dynamics is needed. Understanding transcription factor dynamics will enable optimal scheduling of drug treatments. For example, it has been demonstrated that the effective timing of chemotherapy treatment depends on the dynamics of P53 (Chen et al., 2016). Future studies are required to delineate exactly how the distinct sets of programmatic genes are differentially controlled by MITF.
From an evolutionary perspective, the phenotypic trait linkage conferred by MITF showcases a theoretical concept in developmental evolution; namely, that optimizing the phenotypic coupling of multiple programs can come at the cost of being less optimal (i.e., reduced fitness) in one or more of the individual coupled programs (Carroll, 2008). In this case, tightly controlled synchronization of the stress response and pigmentation programs is at the cost of optimal induction of pigmentation when there is daily UV exposure; the system may have developed as optimal for a 48-hr periodicity of UV exposure. Vitamin D is necessary for the maintenance of calcium homeostasis and bone metabolism and is also important for a healthy immune system and prevention of cancer (Wacker and Holick, 2013). High levels of vitamin D are found in the serum of volunteers 48 hr after a single UV exposure (Clemens et al., 1982). This suggests that, as for pigmentation, vitamin D production is reduced when UV exposure is daily, and these adaptations to the frequency of sun exposure may have arisen during furless skin evolution.
Skin cancer incidence increases every year (Rogers et al., 2015). UV exposure is a major risk factor for all common cutaneous malignancies (Rogers et al., 2015). MITF has been shown to play a key role in melanoma development, the most deadly type of human skin cancer (Bell and Levy, 2011). It will be interesting to learn whether the dynamic oscillations of MITF are retained under malignant transformation and whether they affect disease progression.
Finally, our work offers an immediate translational opportunity for sunless induction of skin pigmentation (Figure 5), which directly provides protection from UV-induced DNA damage.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse anti Thymine Dimer clone KTM53 | Kamiya | Cat#MC-062; RRID: AB_1233355 |
| Rabbit anti CDK2 clone M2 | Santa Cruz | Cat#SC-163; RRID: AB_631215 |
| Mouse anti Tyrosinase clone T311 | Santa Cruz | Cat#SC-20035; RRID: AB_628420 |
| Mouse anti MITF clone C5 | Laboratory of David Fisher | N/A |
| Rabbit anti pCREB (Ser133) | Cell signaling | Cat#9191; RRID: AB_331606 |
| Mouse anti actin clone C4 | Santa Cruz | Cat#sc-47778; RRID: AB_2714189 |
| Rabbit anti Ki-67 clone D3B5 | Cell signaling | Cat#11882S; RRID: AB_2687824 |
| Mouse anti CDK2 clone 2D9 | LSBio | Cat#C337371; RRID: AB_2728767 |
| Rabbit anti CDK2 clone E304 | Abcam | Cat#ab32147; RRID: AB_726775 |
| Mouse anti HMB45 + M2-7C10 + M2-9E3 | Abcam | Cat#ab732; RRID: AB_305844 |
| Mouse anti β-tubulin | Sigma-Aldrich | Cat#T4026; RRID: AB_477577 |
| Rabbit anti H3K4me3 | Laboratory of David Fisher | N/A |
| Rabbit Anti-Histone H3 (phospho S10) | Abcam | Cat#ab5176; RRID: AB_304763 |
| Donkey anti mouse Alexa 488-conjugated secondary antibody | Life Technologies | Cat# A-11008; RRID: AB_143165 |
| Goat anti rabbit Alexa-594-conjugated secondary antibody | Life Technologies | Cat# A-21203; RRID: AB_141633 |
| Biological Samples | ||
| Human skin | Resected remnant skin from Wolfson Medical Center | approved Helsinki (0015-16-WOMC) |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Forskolin | Sigma-Aldrich | F6886-10MG |
| FM19G11 | Sigma-Aldrich | F8807-5MG |
| VECTASHIELD Antifade Mounting Medium with DAPI | Vector | H-1200 |
| Fluoroshield with DAPI | Sigma-Aldrich | F6057 |
| Deposited Data | ||
| Original imaging data in Mendeley Data | This paper | https://dx.doi.org/10.17632/jsmb8ymk94.1 |
| Gene-expression profiling of MNT-1 melanoma line upon treatment with cAMP | This paper | GEO: GSE114764 |
| Experimental Models: Cell Lines | ||
| Human: MNT-1 | Laboratory of Vincent J. Hearing; Virador et al., 2001. | RRID: CVCL_5624 |
| Human: WM3682 | Laboratory of Levi A. Garraway; Golan et al., 2015 | RRID: CVCL_AP78 |
| Human: WM3526 | Laboratory of Levi A. Garraway; Golan et al., 2015 | N/A |
| Human: WM3314 | Laboratory of Levi A. Garraway; Golan et al., 2015 | N/A |
| Human: 451LU | Laboratory of Levi A. Garraway; Golan et al., 2015 | RRID: CVCL_6357 |
| Human: A375 | Laboratory of Levi A. Garraway; Golan et al., 2015 | RRID: CVCL_0132 |
| Human: WM1716 | Laboratory of Levi A. Garraway; Golan et al., 2015 | RRID: CVCL_AP82 |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 | Harlan Laboratories | Cat#1BL/626 |
| Oligonucleotides | ||
| siMITF | Ambion | Cat#AM16204 |
| Mimic miR-148a | Ambion | ID: MC10263 Cat#4464066 |
| Mimic miR-101 | Ambion | ID: MC11414 |
| Mimic miR-216a | Ambion | ID: MC24316 |
| Mimic miR-506 | Ambion | ID: MC10709 |
| Mimic miR-25 | Ambion | ID: MC12401 |
| Mimic miR-124 | Ambion | ID: MC11154 |
| Mimic miR-340 | Ambion | ID: MC12670 |
| SiControl | Ambion | ID: AM4635 |
| Sequences of all oligonucleotides are detailed in Table S5. | This paper | N/A |
| Recombinant DNA | ||
| PGL4.11-148a promoter-Luciferase-clone 1 | This paper | N/A |
| 3′ UTR MITF Luciferase reporter | GeneCopoeia | N/A |
| pcDNA MITF expression plasmid | Laboratory of David Fisher | N/A |
| Software and Algorithms | ||
| MATLAB R2017B | MathWorks | https://www.mathworks.com/ |
| R3.4.3 | CRAN | https://cran.r-project.org/ |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Carmit Levy (carmitlevy@post.tau.ac.il).
Experimental Model and Subject Details
All experiments involving human samples have been approved by the E. Wolfson Medical Center, Israel (Helsinki Ethical approval no. 0015-16-WOMC).
All animal experiments were conducted with approval from the University of Tel Aviv Institutional Animal Care and Use Committee (01-16-38).
Mice
All experiments were carried out in accordance with guidelines of the Tel Aviv University Institutional Animal Care and Use Committee. A total of 16 Female mice were used, 5 control and 11 UVB-treated. C57BL/6 mice at the age of 8 weeks were purchased from Harlan Labs. Mice were exposed to different intervals of UV exposure for a period of 21-60 days.
Human Skin
Human skin was resected remnant skin was grown on a keratinocyte-based serum-free semi-solid medium (0.33% agarose) absorbed in 10% FBS DMEM for 10 days. Every 48h the skin was transferred to fresh semi-solid medium. Human skin was incubated in 37°C, humidified incubators supplemented with 5% CO2 not including the time of the UV exposure treatment.
Cell Culture
The melanoma cell line MNT-1 (RRID: CVCL_5624) was provided by V. Hearing (National Institutes of Health, Washington, DC, USA). WM3314, WM3682 (RRID: CVCL_AP78), A375 (RRID: CVCL_0132), WM1716 (RRID: CVCL_AP82), and 451LU (RRID: CVCL_6357) melanoma cells were kindly given by Dr. Levi A. Garraway (Dana-Farber Cancer Institute, Boston, MA, USA). Cells were cultured in DMEM medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin/L-glutamine. Minimal media was made with DMEM medium supplemented with 0%–1% FBS and 1% penicillin/streptomycin/L-glutamine. The sex of the cells was not determined for this study.
Method Details
Genome Editing
CRISPR guides flanking miR-148a were designed using crispr.mit.edu software from the Zhang lab (Cong et al., 2013). Selected guides, targeting regions upstream (gttctaatctgaggacgggt) and downstream (ccaattcccttgaagcgggt) of miR-148a were cloned separately into pL-CRISPR.SFFV.GFP (a kind gift from Benjamin Ebert; Addgene plasmid # 57827). Both vectors were co-transfected for 48h into MNT-1 melanoma cells using Fugen HD (Promega) according to manufacturer’s specifications, before single cell sorting into 96 well dishes. Once the cultures were established, the clones were screened by sequencing of the miR-148a locus. Generation of CRISPR line was more successful in MNT-1 line compare to other melanoma lines.
UV Exposure
Human and mouse skin was exposed to UVB in an XX-15 stand equipped with 15-W, 302-nm UVB bulbs (Ultra-Violet Products, UK) at doses of 200 or 50 mJ cm−2 UVB, respectively. Animals were exposed to ultraviolet irradiation in a manually made chamber that was designed to allow freedom of movement during irradiation. UV emission was measured with the use of a “UVX radiometer” (Ultra-Violet Products) equipped with a UVB-measuring head. The delivered doses were calibrated for UVB emittance.
Reagents
In order to mimic a key step in melanocyte pigmentation process we stimulated the cells with Forskolin, which directly activates adenyl cyclase, thus induces an increase of cellular cAMP level (Levy et al., 2010b). Forskolin (Sigma-Aldrich) dissolved in DMSO and added to the cells at a final concentration of 20 μM. After incubation of 30 min, cells were washed with PBS and serum-depleted media was added. HIF1α inhibitor, FM19G11 (Sigma-Aldrich) was dissolved in DMSO and was added to the cells at a final concentration of 500 nM 2 h prior to Forskolin stimulation and for the whole period of the respective experiment.
Plasmids and Cloning
An 1800-nt fragment of the human miR-148a promoter upstream of the transcription start site was amplified from human genomic DNA. The fragment was digested with NheI-BglII and inserted into pGL4.11 basic vector (Promega) upstream of the luciferase reporter gene. Site-directed mutagenesis was performed on two MITF binding sites at positions 1417 and 841 nucleotides upstream of the transcription start site (primers sequences are given in Table S5). pcDNA3-MITF-HA expression vector was a gift from Dr. David. E. Fisher (Harvard Medical School, Boston, MA, USA). pEZX-MT01 3′ UTR MITF luciferase plasmid (GeneCopoeia) was a generous gift from Dr. Dror Avni (Sheba Medical Center, Ramat Gan, Israel).
Transfection and Luciferase Reporter Assay
For luciferase assays, constructs of miR-148a promoter (wild-type and two mutated E-box), MITF promoter and MITF 3′ UTR were transfected using jetPEI Reagent according to the manufacturer’s protocol. Cell lysates were prepared 48-72h after the transfection, followed by activity measurement of firefly luciferase, using the Dual Luciferase kit (Promega) according to the manufacturer’s recommendations. The promoter activity and MITF 3′ UTR inhibition were normalized to the constitutively expressed Renilla.
Oligonucleotide Transfection
MiR-148a mimic, control miRNA mimic, MITF siRNA, and control siRNA were transfected into the cells using HiPerFect (QIAGEN) according to the manufacturer’s protocols. Cells were transfected twice with 100 pmol of oligonucleotide per well at 24h intervals. Transfected cells were assayed 24h after the second transfection. In the co-transfection of miR-148a mimic and 3′UTR MITF constructs, cells were first transfected with the miR-148a mimic or control miRNA mimic and then 24h later with 1 μg pEZX-MT01 3′ UTR MITF. Transfected cells were assayed 24h after the second transfection.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described previously (Levy et al., 2010b). The immunoprecipitations were performed with MITF rabbit polyclonal antibody (kindly provided by Dr. David. E. Fisher, Harvard Medical School), H3K4 rabbit polyclonal antibody (Cell Signaling, 9725), and normal rabbit IgG (Santa Cruz Biotechnology, sc-2027) as control antibody. qRT-PCR was carried out using primers listed in Table S5.
RNA Purification and qRT-PCR
Total RNA was purified using Trizol (Ambion) according to manufacturer’s instructions. RNA was quantified by measuring OD260/280. For qRT-PCR analysis of mature miR-148a, 10 ng of total RNA was used in a TaqMan miRNA assay according to the manufacturer’s protocols (Applied Biosystems). Expression levels were normalized to RNU6B (U6) expression. For mRNA analyses, RNA was subjected to qScript cDNA Synthesis Kit (Quanta) and FastStart Universal SYBR® Green FastMix® (Quanta). Relative expression was normalized to actin. Means plus and minus standard errors (SEM) are presented. All primer sequences are shown in Table S5.
Microarray Analysis
Single-stranded cDNA was generated from the amplified cRNA with the WT cDNA Synthesis Kit (Affymetrix) and then fragmented and labeled with the WT Terminal Labeling Kit (Affymetrix). Samples were hybridized with Clarioum S Human Arrays (Affymetrix) and scanned at the HUJI Microarray Core Facility. Array scanning was performed according to the manufacturer’s instructions (Affymetrix). Raw data were processed in R for background correction and RMA normalization. Normalized expression data were log-transformed. To understand gene-expression dynamics following MITF stimulation, we applied diffusion-map algorithm on z-scaled expression data (destiny package), which revealed two major groups of samples of early and late time points that were separated by the first diffusion component (Figure S2B). To discover the peaks of expression for each gene over 48h, we first calculated the expression range for each gene as the difference between the maximal and the minimal expression levels across all time points post-stimulation. Then, we identified the peaks in expression as the local maxima of expression with magnitude (calculated as the difference from the previous local minima) more than 10% of the expression range. We report per peak the expression levels, the peak magnitude scaled to the expression range, and the earliest and latest time points of increase in expression levels (Table S2).
H3K27ac Acetylation Profiles of Human Primary Melanocyte
The bed file of H3K27ac ChIP-seq analysis of foreskin melanocyte from skin01 (A18462) (GEO: GSM1127072) (Laurette et al., 2015) was converted to a bam file, using bedToBam in BEDTools (Quinlan and Hall, 2010), and data were sorted and indexed using Samtools (http://www.htslib.org). A plot of average profiles of H3K27ac in genomic regions of the whole genome, pigmentation genes, and cell-cycle genes was generated using the program ngs.plot (Shen et al., 2014). Validated MITF target genes (Levy and Fisher, 2011) that were tested were cell cycle genes (BCL2, MET, TBX2, CDKN1A, and CYB5R3) and pigmentation genes (TYR, TYRP, MLANA, TYRO, DCT, TRPM1, and PMEL).
RNA-Seq of Human Primary Melanocytes
Left reads numbering 59,288,029 from the bed file of RNA-seq analysis of foreskin melanocyte from skin01 (A04596) (GEO accession GSM751276) were converted to a bam file, followed by sorting and indexing. Reads of each gene were counted using HTSeq (Anders et al., 2015), considering the reads as unstranded. Reads per kilobase per million mapped reads (RPKM) were calculated for each gene by the formula: RPKM = numReads / (geneLength/1000 ∗ totalNumReads/1,000,000). Gene lengths were calculated by an in-house script that sums the exons of the transcripts of each gene using the Ilumina iGenomes (https://support.illumina.com/sequencing/sequencing_software/igenome.html) hg19 genes.gtf annotation file. Gene size was taken as the length of the longest transcript of each gene. Genes with RPKM < 0.2 were excluded, leaving 14,102 expressed genes.
In order to check if the mean RPKM of the seven pigmentation genes is statistically significant, 1,000,000 random sets of seven genes were sampled from the 14,102 genes and the mean of each set was calculated. The p value for the mean of the pigmentation gene set was calculated as a fraction of the random sets means with at least the same value as that of the pigmentation set (p value = 0.0005 p value = 0.000001, when excluding or including PMEL in the mean of the pigmentation set, receptively).
MITF mRNA Expression Regulators
Genes that regulate MITF mRNA expression were retrieved from using Ingenuity Pathways Analysis (http://www.ingenuity.com/index.html) by growing a network around MITF using experimental and high confidence networking data, as well as only edges with expression relationship type.
Western Blot
The cells were lysed as previously described (Levy et al., 2010b). Membrane were exposed overnight to antibodies targeting MITF (C5; kindly provided by Dr. David. E. Fisher, Harvard Medical School) pCREB (Cell Signaling Cat#9198, RRID: AB_331606), tyrosinase (Santa Cruz Biotechnology Cat#sc-20035, RRID: AB_628420), CDK2 (Santa Cruz Biotechnology Cat#sc-163, RRID: AB_631215). β-tubulin mouse monoclonal, (Sigma-Aldrich Cat#T4026, RRID:AB_477577), and actin (Santa Cruz Biotechnology Cat#sc-47778, RRID: AB_2714189). Proteins were visualized with SuperSignal Chemiluminescent Substrates (Pierce) using horseradish peroxidase-conjugated anti-mouse (Vector, PI2000) or anti-rabbit (Vector, PI1000) secondary antibody.
miRNA/mRNA Correlation Analysis
Paired miRNA/mRNA expression data were generated as described previously (Bell et al., 2014). The correlation between MITF expression and each miRNA was calculated using Pearson’s coefficient.
Immunofluorescence Histology
Skin samples were biopsied at indicated time points after UV exposure and were immediately placed in 4% paraformaldehyde for overnight at 4°C, washed with PBS and were then paraffin embedded. Sections were cut at 8-10 μm, dried at 37°C, deparaffinized in xylene, hydrated in a graded series of ethanol, and subjected to microwave EDTA antigen retrieval. Samples were blocked with 5% BSA, 0.5% Tween-20 in PBS. Slides were incubated with antibodies to detect thymine-dimer (Kamiya Biomedical Cat#MC-062, RRID: AB_1233355), MITF (C5, kindly provided by Dr. David. E. Fisher), tyrosinase (Santa Cruz Biotechnology Cat#sc-20035, RRID: AB_628420), Cdk2 (LSBio Cat#C337371, temporary RRID: AB_2728767; Abcam Cat#ab32147; RRID:AB_726775), phospho-histone H3 (Abcam Cat#ab5176, RRID:AB_304763), HMB45 (Abcam Cat#ab732, RRID:305844) and Ki-67 (Cell Signaling Cat#11882S, RRID: AB_2687824) antibodies. Staining was performed by incubating with Alexa 488- (Life Technologies Cat# A-11008, RRID:AB_143165) or Alexa-594- (Life Technologies Cat# A-21203, RRID:AB_141633) conjugated secondary antibodies. Nuclear staining was performed with DAPI. Images were obtained at × 20 or × 40 magnification using fluorescence microscopy (Nikon) or at × 63 magnification using Leica SP8 confocal microscopy. Quantification analysis was done by ImageJ. At least 10 cells for each condition or time point were quantified. For quantification of protein expression level, florescence intensity of the indicated protein and DAPI in the same cell were quantified; at least 25 cells were analyzed per condition. Data represent the ratio of protein intensity normalized to DAPI.
Mathematical Model
General
We generated a mathematical model depicting the dynamical behavior of the MITF. For simplicity, we considered all the proteins-induced negative regulation of MITF as one arm consisting of transcription and translation stages. miRNA doesn’t undergo a translation stage, therefore represents a more immediate negative regulation. Since the timing of miRNA negative regulation differs from the protein negative regulation, miRNA negative regulation was considered as a distinct regulatory loop that doesn’t include a translation process.
Equations
The model’s equations are:
The dynamic variables of the system are: x, , y and z, denoting the levels of MITF, proteins’ mRNA, proteins and miRNA, respectively. The dynamic parameters of the system may be divided into three classes: production rates, degradation rate, and inhibition constants. , , and denote the production rates of x, , y, and z, respectively. Here we assume that the production rates of mRNA and miRNA are similar, therefore in our model: Natural degradation rates of the dynamic variables are , , , and for x, , y, and z, respectively. Note that we assume that mRNA does not undergo degradation but only translation, therefore mRNA degradation rate is equal to protein production rate. We further assumed that degradation rate of miRNA is similar to the degradation rate of mRNA, therefore: . We modeled the negative regulation of y and z on x by a saturating Michaelis-Menten function. Accordingly, the inhibition constants of the system represent the affinity of the inhibitor for its substrate and the degradation rate of the inhibitor-substrate complex. and denote y-, and z- dependent x degradation, respectively. Accordingly, and denote y-x and z-x dissociation rates, respectively. For simplicity, we assumed that the dissociation rate of z from x is similar to the dissociation rate of y from x, therefore: .
Simulations
To derive the dynamic behavior of the system, we simulated it under a broad range of dynamic parameters. We hypothesized that the relationships between the parameters rather than their absolute values, determine the dynamic behavior of the system. As the system uses 8 dynamic parameters, there are 8! possible ways to order them. For each possible ordering of the parameters set, we randomly sampled 50 parameters set from (0,1) interval and ordered them according to the desired order, resulting with 2,016,000 parameter sets. For each parameter set, we simulated the model during a [0,100] time interval following a single stimulation. All simulations were performed in MATLAB using the ode45 function.
Initial Conditions
Steady-state levels of all dynamic variables in the system are exclusively dictated by the parameter sets. Therefore, we reasoned that one cannot use the same initial condition for all parameters sets. To achieve similar initial conditions for all simulations, we used initial conditions that depended on the steady-state levels of the system. We calculated steady-state levels of all dynamic variable by solving the equation: , yielding the following equations:
MITF initial level was set to twice its steady-state value, while the initial levels of miRNA, mRNA and protein were set to their steady-state levels. Formally, the initial conditions for each parameters was set to: .
Simulation Analysis
To detect MITF peaks, we used MATLAB function: “findpeaks”. To avoid spurious peaks, we considered all the peaks whose prominence was more than 1% of MITF steady-state level. MITF fluctuative behavior was defined as at least 3 peaks during the [0,100] time interval. We defined “stabilization time” as the earliest time after which MITF was “trapped” inside the 1% interval around its steady-state level. We calculated it as the last time that MITF levels exceeded the 1% interval from the steady-state. We defined “decay rate” as the fold-change between the third and the first peaks. The peak values were normalized to MITF steady-state level.
Analysis of the System without miRNA or Protein Regulation
To determine the dynamic behavior of the system without miRNA or protein regulation, we used the same dynamic and steady-state equations with either or , respectively.
Inhibition Analysis
To analyze the effects of inhibition of either regulatory arm, we focused on two features of the system: baseline MITF and MITF dynamic behavior, namely: oscillations. We analyzed only the parameter sets that exhibited more than two peaks under natural conditions (2 inhibitory regulatory loops), and satisfy that the ratio between the degradation rates: , corresponding to the cases where the inhibition effect of miRNA on MITF is not negligible relative to protein inhibition effect. We derived the baseline levels of MITF analytically from the model’s equations under inhibition of either regulatory arm alone. To analyze the effects of inhibitions of either regulatory arm on MITF fluctuations, we calculated MITF levels at peaks timing (calculated under normal conditions) relative to their levels at the same time under regulatory inhibition. For this analysis, MITF levels were normalized to its steady-state levels.
Dual Stimulation
We showed experimentally that in the second stimulation CREB phosphorylation is substantially enhanced compared with the first stimulation. Hence, we checked how increased MITF production rate affects MITF dynamic behavior while all the other dynamic parameters were kept stable. The initial condition of the system was similar to the case without increased production. To determine the robustness of the increased production effects, we simulated the system using a broad range of increased MITF production rates. We examined three features related to MITF dynamic behavior, number of peaks, height of the first peak, and peaks frequency, and compared them to the same features under baseline MITF production rate.
Quantification and Statistical Analysis
Statistical Analysis
Standard parametric t tests were applied and standard errors were calculated for each dataset with n > 2. p values < 0.05 were considered significant.
Quantification of Melanin Intensity
Cells were seeded according to the experiment, fixed in 4% PFA and stained for melanin with fontana masson as per the manufacturer’s protocol (Abcam, ab150669). The images were captured in the bright field microscope (Nikon, Japan). The color balance of the images were adjusted in Photoshop using color balance, black, and white functions to avoid interference of cells in quantification (only melanin was highlighted). The images were then quantified for the intensity of the melanin stained areas using ImageJ software. The data represent the average of three randomly selected fields. The mean values SEM are displayed with p values calculated by paired t test.
DAPI Staining Quantification
The cells fixed and processed for the immunofluorescence were used stained with DAPI and counted under X20 magnification. Using ImageJ, the DAPI stained images were converted to greyscale (8 bit) and correct threshold was set. The particles that merged were separated by cutting them apart and adding a 1-pixel-thick line using the ‘watershed’ function. The ‘analyze particles’ function was used to determine the cell count ranging from pixels 100-infinity. The data represent the average of nine randomly selected fields.
Epidermis Thickness Quantification in Human and Mouse Section
Human and mouse sections were stained for Hematoxylin/Eosin staining. Images were taken using Nikon fluorescence microscopy (mouse) or Aperio slide scanner (human) (X20) and quantitatively analyzed using ImageJ. An average number of 10 segments were measured from each field, with five fields analyzed for each group.
DNA Damage Quantification
At the end of the UV treatment period, one ear was covered with a UV shield sticker, and the other ear was exposed. Mice were exposed to a single UVB dose (50 mJ cm−2), followed by immediate skin harvesting. DNA damage was measured in both ears, the covered one (which will give the accumulated DNA damage level upon all the UVB treated period) and the uncovered one (which will provide knowledge of how much the skin is protected against UV exposure). Mouse ears were immunostained for DNA damage quantified by measurement of thymine dimers (Kamiya Biomedical, MC-062) antibody. Thymine dimers are the common UVB-induced DNA lesion (Regan et al., 1968). Ten representative cells from each section were marked (Figure S1B), and the red florescence intensity was measured using ImageJ software. Graphs show average values.
Skin Pigmentation and Erythema Quantification
Skin reflective colorimetric measurements were performed with a DSM II colorMeter (Cortex Technology). The instrument was calibrated using the white standard background provided by the manufacturer prior the measurements. Each skin sample was measured 5 times and values were averaged. Pigment level of mouse ears was measured at the end of the exposure period (21-60 days) and normalized to control UV-free mouse (n = 4 mice in each group). Pigment level of human skin was measured at the end of the exposure period (10 days). In each measurement by the instrument the pigmentation level is received as well as the erythema level.
Data and Software Availability
The accession number for the gene-expression profiling data of MNT-1 melanoma cell line upon treatment with cAMP reported in this paper is GEO: GSE114764
The accession number for the original imaging data reported in this paper is deposite in Mendeley Data: https://dx.doi.org/10.17632/jsmb8ymk94.1.
Acknowledgments
The authors gratefully thank Drs. Meenhard Herlyn and Levi Garraway for supplying melanoma cultures. C.L. gratefully thanks Dr. Ana Moshkovsky and Prof. Noga Kronfeld-Schor for useful discussions and Yuval and Omer Levy for exponential joy. C.L. acknowledges grant support from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 726225); the I-CORE Gene Regulation in Complex Human Disease Center no. 41/11; Israel Science Foundation (ISF) grant 129/13; Fritz Thyssen Stiftung; the Israel Cancer Research Fund (ICRF) grant 2011-706-RCDA; the Salomea (Mika) and Herman Berger Z”L Foundation; and the United States-Israel Binational Science Foundation (BSF) grant 2011172. H.M.-B. gratefully acknowledges the Shtacher Family award; The Naomi Foundation, through the Tel Aviv University GRTF Program; Dr. Eli Fisher for his generous scholarship; and Yoav Brog for the joint journey. Research by S.S.S-O. and A.A. was supported by Israel Science Foundation (ISF) grant 1365/12. C.L. and S.S.S.-O. thank the Kavli Frontiers of Science Program for providing the opportunity for this collaboration. Research by M.K. and A.N. was supported by the Gustave Roussy foundation and Natixis. A.N. was supported by a fellowship from the French ministry of higher education and research.
Author Contributions
H.M.-B. designed the experimental approach, performed the majority of the experimental work, analyzed data, and participated in the writing of the manuscript. A.A. performed the mathematical modeling and transcriptome analyses, and participated in the writing of the manuscript. T.G. cloned the miR-148 promoter and participated in the writing of the manuscript. I.D. performed the ChIP-seq analysis. T.G., S.P., A.N., and D.S. performed in vitro phenotypic analysis. F.N. performed in vivo phenotypic analysis. L.T. and C.C. generated the miR-148a CRISPR-CAS9 cell line. T.P., J.F., Y.S., P.G., and R.B. provided human skin samples. Y.T. and D.R. performed phylogenetic profiling. Y.N. and S.E. performed the H3K27 acetylation analysis and MITF regulators. M.K. developed the hypothesis, designed the genomic and pigmentation experimental approaches, and wrote the manuscript. S.S.S.-O. developed the hypothesis, coordinated the mathematical modeling and genomic analyses, and wrote the manuscript. C.L. developed the hypothesis, designed the experimental approach, coordinated the project, and wrote the manuscript.
Declaration of Interests
The authors declare no competing financial interests. S.S. Shen-Orr is a co-founder of CytoReason, holds equity in it, and is a member of its board.
Published: October 25, 2018
Footnotes
Supplemental Information includes five figures and five tables and can be found with this article online at https://doi.org/10.1016/j.molcel.2018.09.022.
Contributor Information
Mehdi Khaled, Email: mehdi.khaled@gustaveroussy.fr.
Shai S. Shen-Orr, Email: shenorr@technion.ac.il.
Carmit Levy, Email: carmitlevy@post.tau.ac.il.
Supplemental Information
References
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayerl C., Taake S., Moll I., Jung E.G. Characterization of sunburn cells after exposure to ultraviolet light. Photodermatol. Photoimmunol. Photomed. 1995;11:149–154. doi: 10.1111/j.1600-0781.1995.tb00157.x. [DOI] [PubMed] [Google Scholar]
- Bell R.E., Levy C. The three M’s: melanoma, microphthalmia-associated transcription factor and microRNA. Pigment Cell Melanoma Res. 2011;24:1088–1106. doi: 10.1111/j.1755-148X.2011.00931.x. [DOI] [PubMed] [Google Scholar]
- Bell R.E., Khaled M., Netanely D., Schubert S., Golan T., Buxbaum A., Janas M.M., Postolsky B., Goldberg M.S., Shamir R. Transcription factor/microRNA axis blocks melanoma invasion program by miR-211 targeting NUAK1. J. Invest. Dermatol. 2014;134:441–451. doi: 10.1038/jid.2013.340. [DOI] [PubMed] [Google Scholar]
- Busca R., Berra E., Gaggioli C., Khaled M., Bille K., Marchetti B., Thyss R., Fitsialos G., Larribere L., Bertolotto C. Hypoxia-inducible factor 1α is a new target of microphthalmia-associated transcription factor (MITF) in melanoma cells. J. Cell. Biol. 2005;170:49–59. doi: 10.1083/jcb.200501067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll S.B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Chen S.H., Forrester W., Lahav G. Schedule-dependent interaction between anticancer treatments. Science. 2016;351:1204–1208. doi: 10.1126/science.aac5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens T.L., Adams J.S., Henderson S.L., Holick M.F. Increased skin pigment reduces the capacity of skin to synthesise vitamin D3. Lancet. 1982;1:74–76. doi: 10.1016/s0140-6736(82)90214-8. [DOI] [PubMed] [Google Scholar]
- Clydesdale G.J., Dandie G.W., Muller H.K. Ultraviolet light induced injury: immunological and inflammatory effects. Immunol. Cell Biol. 2001;79:547–568. doi: 10.1046/j.1440-1711.2001.01047.x. [DOI] [PubMed] [Google Scholar]
- Coelho S.G., Choi W., Brenner M., Miyamura Y., Yamaguchi Y., Wolber R., Smuda C., Batzer J., Kolbe L., Ito S. Short- and long-term effects of UV radiation on the pigmentation of human skin. J. Investig. Dermatol. Symp. Proc. 2009;14:32–35. doi: 10.1038/jidsymp.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Orazio J.A., Nobuhisa T., Cui R., Arya M., Spry M., Wakamatsu K., Igras V., Kunisada T., Granter S.R., Nishimura E.K. Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature. 2006;443:340–344. doi: 10.1038/nature05098. [DOI] [PubMed] [Google Scholar]
- Feige E., Yokoyama S., Levy C., Khaled M., Igras V., Lin R.J., Lee S., Widlund H.R., Granter S.R., Kung A.L. Hypoxia-induced transcriptional repression of the melanoma-associated oncogene MITF. Proc. Natl. Acad. Sci USA. 2011;108:E924–E933. doi: 10.1073/pnas.1106351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan T., Messer A.R., Amitai-Lange A., Melamed Z., Ohana R., Bell R.E., Kapitansky O., Lerman G., Greenberger S., Khaled M. Interactions of melanoma cells with distal keratinocytes trigger metastasis via Notch signaling inhibition of MITF. Mol. Cell. 2015;59:664–676. doi: 10.1016/j.molcel.2015.06.028. [DOI] [PubMed] [Google Scholar]
- Haflidadóttir B.S., Bergsteinsdóttir K., Praetorius C., Steingrímsson E. miR-148 regulates Mitf in melanoma cells. PLoS ONE. 2010;5:e11574. doi: 10.1371/journal.pone.0011574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek K., Rimm D.L., Williams K.R., Zhao H., Ariyan S., Lin A., Kluger H.M., Berger A.J., Cheng E., Trombetta E.S. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Levchenko A., Scott M.L., Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- Hornstein E., Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38(Suppl):S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Keenen B., Qi H., Saladi S.V., Yeung M., de la Serna I.L. Heterogeneous SWI/SNF chromatin remodeling complexes promote expression of microphthalmia-associated transcription factor target genes in melanoma. Oncogene. 2010;29:81–92. doi: 10.1038/onc.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled M., Levy C., Fisher D.E. Control of melanocyte differentiation by a MITF-PDE4D3 homeostatic circuit. Genes Dev. 2010;24:2276–2281. doi: 10.1101/gad.1937710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushimoto T., Basrur V., Valencia J., Matsunaga J., Vieira W.D., Ferrans V.J., Muller J., Appella E., Hearing V.J. A model for melanosome biogenesis based on the purification and analysis of early melanosomes. Proc. Natl. Acad. Sci. USA. 2001;98:10698–10703. doi: 10.1073/pnas.191184798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurette P., Strub T., Koludrovic D., Keime C., Le Gras S., Seberg H., Van Otterloo E., Imrichova H., Siddaway R., Aerts S. Transcription factor MITF and remodeller BRG1 define chromatin organisation at regulatory elements in melanoma cells. eLife. 2015;4:e06857. doi: 10.7554/eLife.06857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C., Fisher D.E. Dual roles of lineage restricted transcription factors: the case of MITF in melanocytes. Transcription. 2011;2:19–22. doi: 10.4161/trns.2.1.13650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C., Khaled M., Iliopoulos D., Janas M.M., Schubert S., Pinner S., Chen P.H., Li S., Fletcher A.L., Yokoyama S. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol. Cell. 2010;40:841–849. doi: 10.1016/j.molcel.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy C., Khaled M., Robinson K.C., Veguilla R.A., Chen P.H., Yokoyama S., Makino E., Lu J., Larue L., Beermann F. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Mallarino R., Campàs O., Fritz J.A., Burns K.J., Weeks O.G., Brenner M.P., Abzhanov A. Closely related bird species demonstrate flexibility between beak morphology and underlying developmental programs. Proc. Natl. Acad. Sci. USA. 2012;109:16222–16227. doi: 10.1073/pnas.1206205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson D.E., Ihekwaba A.E., Elliott M., Johnson J.R., Gibney C.A., Foreman B.E., Nelson G., See V., Horton C.A., Spiller D.G. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- Nolan B.V., Yentzer B.A., Feldman S.R. A review of home phototherapy for psoriasis. Dermatol. Online J. 2010;16:1. [PubMed] [Google Scholar]
- Oren M., Bartek J. The sunny side of p53. Cell. 2007;128:826–828. doi: 10.1016/j.cell.2007.02.027. [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Poling L.L., Wang Z., Liu H., Liu X.S., Roeder R.G., Zhang X., Song J.S., Fisher D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus M.V., Zeichner-David M., Mayer J.A., Reyna J., Bringas P., Thewissen J.G., Snead M.L., Chai Y., Chuong C.M. Morphoregulation of teeth: modulating the number, size, shape and differentiation by tuning Bmp activity. Evol. Dev. 2005;7:440–457. doi: 10.1111/j.1525-142X.2005.05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porstner M., Winkelmann R., Daum P., Schmid J., Pracht K., Côrte-Real J., Schreiber S., Haftmann C., Brandl A., Mashreghi M.F. miR-148a promotes plasma cell differentiation and targets the germinal center transcription factors Mitf and Bach2. Eur. J. Immunol. 2015;45:1206–1215. doi: 10.1002/eji.201444637. [DOI] [PubMed] [Google Scholar]
- Purvis J.E., Karhohs K.W., Mock C., Batchelor E., Loewer A., Lahav G. p53 dynamics control cell fate. Science. 2012;336:1440–1444. doi: 10.1126/science.1218351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopal N., Ernst J., Ray P., Wu J., Zhang M., Kellis M., Ren B. Distinct and predictive histone lysine acetylation patterns at promoters, enhancers, and gene bodies. G3 (Bethesda) 2014;4:2051–2063. doi: 10.1534/g3.114.013565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regan J.D., Trosko J.E., Carrier W.L. Evidence for excision of ultraviolet-induced pyrimidine dimers from the DNA of human cells in vitro. Biophys. J. 1968;8:319–325. doi: 10.1016/S0006-3495(68)86490-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers H.W., Weinstock M.A., Feldman S.R., Coldiron B.M. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the U.S. population, 2012. JAMA Dermatol. 2015;151:1081–1086. doi: 10.1001/jamadermatol.2015.1187. [DOI] [PubMed] [Google Scholar]
- Shen L., Shao N., Liu X., Nestler E. ngs.plot: quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrímsson E., Copeland N.G., Jenkins N.A. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Virador V., Matsunaga N., Matsunaga J., Valencia J., Oldham R.J., Kameyama K., Peck G.L., Ferrans V.J., Vieira W.D., Abdel-Malek Z.A. Production of melanocyte-specific antibodies to human melanosomal proteins: expression patterns in normal human skin and in cutaneous pigmented lesions. Pigment Cell Res. 2001;14:289–297. doi: 10.1034/j.1600-0749.2001.140410.x. [DOI] [PubMed] [Google Scholar]
- Wacker M., Holick M.F. Sunlight and vitamin D: a global perspective for health. Dermatoendocrinol. 2013;5:51–108. doi: 10.4161/derm.24494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster D.E., Barajas B., Bussat R.T., Yan K.J., Neela P.H., Flockhart R.J., Kovalski J., Zehnder A., Khavari P.A. Enhancer-targeted genome editing selectively blocks innate resistance to oncokinase inhibition. Genome Res. 2014;24:751–760. doi: 10.1101/gr.166231.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef N., Regev A. Impulse control: temporal dynamics in gene transcription. Cell. 2011;144:886–896. doi: 10.1016/j.cell.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.