Abstract
The human protein TDP-43 is a major component of the cellular aggregates found in amyotrophic lateral sclerosis and other neurodegenerative diseases. Insoluble cytoplasmic aggregates isolated from the brain of amyotrophic lateral sclerosis and frontotemporal lobar degeneration patients contain ubiquitinated, hyperphosphorylated, and N-terminally truncated TDP-43. Truncated fragments of TDP-43 identified from patient tissues contain part of the second RNA recognition motif (RRM2) and the disordered C-terminus, indicating that both domains can be involved in aggregation and toxicity. Here, we focus on RRM2. Using all-atom replica-averaged metadynamics simulations with NMR chemical shift restraints, we characterized the atomic structure of non-native states of RRM2, sparsely populated under native conditions. These structures reveal the exposure to the solvent of aggregation-prone peptide regions, normally buried in the native state, supporting a role in aggregation for the partially folded states of RRM2.
Introduction
The 43 kDa human transactive response DNA-binding protein (TDP-43) is a 414-amino-acid protein with two RNA recognition motifs (RRM1 and RRM2), an N-terminal dimerization domain, and a C-terminal prion-like glycine-rich domain (1). TDP-43 is a nuclear protein that is ubiquitously expressed and whose functions include RNA splicing and transcriptional repression (2, 3). In 2006, TDP-43 was identified as the main constituent of the cytoplasmic inclusions that are the hallmark of most forms of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) (4, 5). Since then, a large number of TDP-43 mutations in familial and sporadic ALS patients have been reported (ALS Online Genetics Database) (6), thus establishing a primary role for TDP-43 in ALS pathogenesis. TDP-43 forms aggregated species in vitro that display high structural similarity with TDP-43 aggregates from FTLD and ALS patients (7). Furthermore, ALS-related mutations identified in the TDP-43 gene have been reported to accelerate aggregation in vitro and to promote the formation of more numerous aggregates in vivo (7).
The cytoplasmic aggregates found in FTLD and ALS brain contain ubiquitinated, hyperphosphorylated, and N-terminally cleaved TDP-43 fragments (known as C-terminal fragments) (4, 8). TDP-43 25 kDa C-terminal fragments, identified as the pathological species in FTLD and ALS (4, 8), are generated from proteolytic cleavage within RRM2 (8, 9, 10) and consequently comprise a C-terminal part of RRM2 and the glycine-rich region. When these C-terminal fragments of TDP-43 have been expressed in cell culture, they were observed to become ubiquitinated and hyperphosphorylated and to form cytoplasmic aggregates (10, 11), recapitulating the observations made in patient-derived brain tissue (11). Moreover, cytoplasmic inclusions formed upon ectopic expression of the C-terminal truncation product of caspase cleavage were observed to be toxic in M17 neuroblastoma cells (10). To investigate the domain-specific contributions to neural toxicity in TDP-43, the aggregation propensity and toxicity of different TDP-43 constructs have been studied in various disease models. Studies of TDP-43 fragments with differing lengths showed that both the truncated RRM2 (residue 208–265) and the glycine-rich region contribute to formation of insoluble cytoplasmic aggregates in mouse neuroblastoma N2a cells, with the truncated RRM2 playing a primary role (12). In another mouse neuronal cell line (motor-neuron-derived NSC-34), the C-terminal part of RRM2 was found to be required but not sufficient for aggregation, with different TDP-43 fragments likely impairing neurite growth through different toxic mechanisms (13). In yeast, both RRM2 and the glycine-rich region are required for aggregate formation and toxicity (14). Taken together, these studies emphasize the importance of these two domains in the abnormal formation of cytoplasmic aggregates and in the pathogenic role of TDP-43 in FTLD and ALS.
TDP-43 RRM2 is an unusually stable domain, as shown by thermal and chemical denaturation studies (12, 15, 16). The solution structure of TDP-43 RRM2 (Protein Data Bank (PDB): 1WF0, Fig. S1) shows a large cluster of 12 connected hydrophobic residues—isoleucines, leucines, and valines (ILV) (17)—in the core of the domain that likely contributes to its high stability (16). Denaturant-induced unfolding studies have revealed the presence of an intermediate state on the RRM2 unfolding pathway. This intermediate state has a low, although not negligible, population under native conditions that increases with increasing denaturant concentration to a maximal value of ∼80% at 4 M guanidine hydrochloride before fully unfolding (16). This partially folded state of TDP-43, populated under native conditions, may increase the probability of cleavage—leading to the production of toxic truncated forms of TDP-43 (10, 11, 12, 13, 14, 18)—or directly contribute to aggregate formation. A structural characterization at atomic resolution of this partially folded intermediate state is not yet available, but it is required in order to establish its role in ALS and other neurodegenerative diseases by contributing to misfolding and aggregation of TDP-43. Molecular dynamics simulations have been successfully used to explore denatured states and folding intermediates of proteins (19, 20, 21, 22). The use of advanced sampling methods and the inclusion of experimental data as structural restraints allow sampling of low-populated states in shorter simulation time and provide an accurate representation of the system (23, 24, 25, 26, 27, 28, 29, 30, 31). Here, we used these approaches to characterize the structure of the partially folded state of TDP-43 RRM2.
Materials and Methods
TDP-43 RRM2 structure preparation
The structure of RRM2 of TDP-43 used in the simulation, residues 189–261, was obtained from the deposited NMR solution structure (PDB: 1WF0). Wild-type mutations S191R, G192K, and G200E were introduced in the sequence of RRM2 using the Mutator plugin (v. 1.3) of Visual Molecular Dynamics (VMD) (32). The resulting structure was solvated using GROMACS (33, 34, 35, 36) with 37,000 water molecules in a dodecahedron water box (1140 nm3), containing 5 Na+ ions to enforce charge neutrality.
NMR assignment of TDP-43 RRM2
Backbone chemical shifts of TDP-43 RRM2 at 6 M concentration of urea were collected as previously described by Mackness et al. (B.C. Mackness, B.R. Morgan, L.M. Deveau, S.V. Kathuria D.T., F.M., and J.A.Z., unpublished data) (Biological Magnetic Resonance Data Bank (BMRB): 27549).
Simulation protocol
Molecular dynamics simulations of the RRM2 of TDP-43 were performed in GROMACS using the Amber03W force field (37) with the TIP4P05 water model (38). The equations of motion were integrated using the LINear Constraint Solver constraints to allow a time step of 2 fs (39). The van der Waals interactions were implemented with a cutoff distance of 0.9 nm, and the particle mesh Ewald method was used to treat electrostatic interactions with periodic boundary conditions (40). All simulations were carried out in the canonical ensemble at constant volume, and temperature was maintained with the Nosé-Hoover thermostat (41).
A 30 ns unfolding trajectory at 450 K was preliminarily collected, starting from the solvated structure described above, to obtain four starting conformations for the replica-averaged metadynamics (RAM) simulations (42, 43). The Cα root mean-square deviation (RMSD) of the resulting structures varies between 2.5 and 3 Å. Each conformation was subsequently relaxed at 300 K for 10 ns. PLUMED2 (44) and GROMACS were used to perform the RAM simulations using chemical shifts as replica-averaged restraints (43, 45) and bias-exchange metadynamics (42). Four replicas of the system were run in parallel at 300 K with restraints from experimental data applied at each time step to the average values of the NMR chemical shifts calculated with the Camshift algorithm (46, 47). A force constant of 12 kJ/(mol ⋅ ppm2) was used, and the chemical shifts of proline, glycine, aspartate, glutamate, and histidine residues were not included in the simulations (19). The experimental chemical shifts were collected at 6 M urea, and at this concentration of urea, the intermediate state is in fast exchange with the unfolded state. Thus, the measured chemical shifts are the population-weighted average of the two states (B.C. Mackness, B.R. Morgan, L.M. Deveau, S.V. Kathuria, D.T., F.M., and J.A.Z., unpublished data). The population of the unfolded state is estimated to be less than 5% (B.C. Mackness, B.R. Morgan, L.M. Deveau, S.V. Kathuria, D.T., F.M. and J.A.Z., unpublished data). Each replica of the system employed a metadynamics approach to bias a collective variable (CV), with exchanges between the replicas attempted every 50 ps according to a replica-exchange scheme (48).
The CVs used in the simulations were calculated as described:
-
•
Total α-helical content (αC): the ALPHARMSD module of PLUMED2 was used, with a rational switching function (49) (r0 = 0.08 nm, n = 8, m = 12).
-
•
Total β-sheet content (βC): the ANTIBETARMSD module of PLUMED2 was used, with a rational switching function (49) (r0 = 0.08 nm, n = 8, m = 12).
-
•
Radius of gyration (Rg): the GYRATION module of PLUMED2 was used.
-
•
Number of hydrophobic contacts within the ILV cluster (#ILV): the CONTACTMAP module of PLUMED2 was used. The presence of each of the eighteen contacts identified between isoleucine, leucine and valine residues for the native state of the RRM2 of TDP-43 (16, 17) was evaluated using a rational switching function (49) with r0 = 0.6 nm for the distance between the Cβ atoms of each pair of residues.
Gaussians deposition was performed with an initial rate of 0.125 kJ/mol/ps, where the σ values were set to 0.46, 0.32, 0.006, and 0.34 nm, for αC, βC, Rg, and #ILV, respectively. The σ values were obtained as the average fluctuations of each CV from unbiased simulations.
RAM simulations analysis
The RAM simulations were analyzed using the METAGUI plugin of VMD (50). Configurations corresponding to the partially structured states of RRM2 were grouped in microstates by dividing the four-dimensional CV space in hypercubes (50). The free energy of the microstates was then computed with the Weighted Histogram Analysis Method technique (51, 52). These microstates were clustered (53) to identify the energy basins in the free-energy landscape of RRM2 (Fig. S4). For each conformation, the difference from the structure of the native state was calculated and shown as RMSD of the Cα in Fig. S13.
As previously described (19), the convergence of the sampling was assessed by monitoring the differences of the free energies at increasing simulation length during the simulations. After the first 570 ns, the free-energy landscapes for each CV were stable below 2 kJ/mol (Fig. S2), and thus the sampling was converged.
Hydrophobic ILV contacts for the structures resulting from the simulations were estimated as the amount of surface area buried between a pair of residues: SASAa + SASAb−SASAa + b, with a and b being ILV residues and SASA being the solvent-accessible surface area of the side chain of residue a, residue b, and residues a and b together. The presence of an ILV contact was computed using VMD as 1 if two residues have at least 45 Å2 of buried surface area and 0 otherwise (17). The probability of an ILV contact was calculated as the occurrence of the contact in the microstates of each basin.
Aggrescan3D (54) calculations were performed using the Aggrescan3D server with a distance of aggregation analysis of 5 Å. 25 structures for each basin were uploaded to the server and the average value and standard deviation of each residue were used for graphical representation.
Results and Discussion
We used RAM simulations (42, 43) with backbone chemical shift restraints and bias-exchange metadynamics to characterize the partially folded state of TDP-43 RRM2. The bias-exchange metadynamics approach employed in our simulations consisted of performing four metadynamics simulations in parallel on different replicas of the system, with each simulation biasing a different CV as was previously described (19). At each timestep, chemical shift restraints from experimental data collected for Cα, Cβ, N, and HN for the intermediate state at 6 M urea concentration (BMRB: 27549) were applied to the ensemble of the four replicas. Mackness et al. (B.C. Mackness, B.R. Morgan, L.M. Deveau, S.V. Kathuria, D.T., F.M., and J.A.Z., unpublished data) have shown that at 6 M concentration of urea, the population of this partially folded state is similar to that of the native state, ∼50%. In this study, we characterize the structure of this partially folded state of TDP-43 RRM2 that is present under native conditions but sampled with low frequency. The application of the chemical shift restraints collected for the intermediate state at 6 M urea increases the frequency of sampling of this partially folded state, but the free-energy landscape calculated from the simulations does not correspond to what would be observed under native conditions.
The use of chemical denaturant in general, and urea in particular, is a common approach to investigate protein stability and the mechanism of unfolding (55). An earlier study of the unfolding of chymotrypsin inhibitor 2 has shown that MD simulations performed in the presence of 8 M urea at 60°C or in water at 125°C (above the protein melting point) display the same unfolding mechanism (56). The two simulations collected under different unfolding conditions were reported to yield similar partially folded intermediate states, sharing the same amount of secondary structure. A greater protein nonpolar SASA, however, was reported for the conformations collected in the urea simulations than in water (56). It is reasonable, therefore, to expect that the structural features of conformations obtained from our RAM simulations will be representative of the ensemble of structures observed in aqueous solution, although the relative statistical weight of each state will not be accurate.
In another study, Mielke et al. (57) showed that reference random-coil chemical shifts collected in 8 M urea for 13Cα and 13Cβ have modest to negligible differences to those collected in aqueous solution. These results suggest that the chemical shift differences observed for TDP-43 at 6 M urea compared to native conditions arise mainly from conformational changes rather than solvent effects.
Appropriate choice of the collective variable is essential to guarantee proper sampling of the energy landscape (58, 59, 60). We chose the following CVs: the total α-helical content, the total β-sheet content, the radius of gyration (Rg), and the number of hydrophobic contacts of ILV residues. The secondary structure content and Rg were chosen because fluctuations of secondary structures and volume capture the relevant structural changes sampled by a protein (19). In addition, the amount of secondary structure and the Rg of the folding intermediate state are significantly different from that of the native state in TDP-43 RRM2 ( nm; nm) (16; Mackness, B.R. Morgan, L.M. Deveau, S.V. Kathuria, D.T., F.M., and J.A.Z., unpublished data). The fourth CV was chosen to be the number of hydrophobic contacts within the ILV cluster (#ILV contacts) because a large cluster of 12 ILV residues in RRM2 provides an essential core of folding stability in this domain (16, 17). With the approach described in the Materials and Methods, we collected a total of 2500 ns of simulation time, and we obtained a free-energy landscape within a statistical uncertainty <2 kJ/mol for free energies up to 10 kJ/mol (Fig. S2).
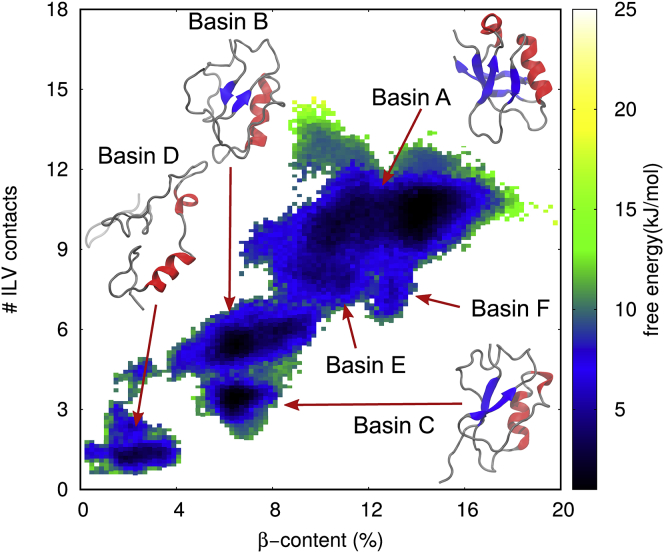
The collected simulations showed an effective sampling of the conformation space, with the resulting conformations spanning a wide range of values of the CVs: α-helical and β-strand content was sampled between 0 and 20%, Rg between 1 and 3.1 nm, and #ILV contacts between 0 and 16 (Fig. S3). From the trajectories, we calculated the free-energy landscape of TDP-43 RRM2 as a function of the four CVs (Figs. 1 and S4). The resulting free-energy landscape revealed six energy basins (Figs. 1 and S4; Table S1).
Figure 1.
Characterization of the free-energy landscape of the partially folded states of TDP-43 RRM2. The two-dimensional free-energy landscape is shown as a function of two of the four collective variables used in the RAM simulations, β-sheet content and number of hydrophobic contacts. Representative structures are shown for the basins with statistical weights greater than 10%.
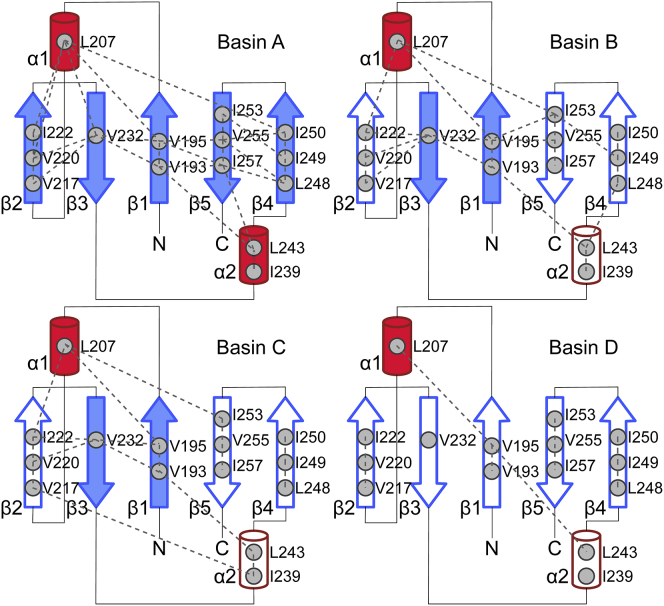
The ensemble of conformations in the major basin, identified as A, shares most of its structural features with the native state of TDP-43 RRM2, including hydrogen bonds and contacts between ILV residues of the β-sheet and between Leu 207 in helix α1 with hydrophobic residues in β1, β2, and β4 (Figs. 2 and 3; Table S1). Unlike the native state, Leu 243 in helix α2 loses interaction with β4, but it interacts with Val 193 in β1 (Figs. 2 and S5). This conformational change reflects the different hydrogen bonds pattern within α2 and thus the reduced stability of the helix compared to the native state (Fig. 3).
Figure 2.
Residual structure of the microstates in the partially folded state of TDP-43 RRM2. ILV cluster contact maps for RRM2 are displayed on the secondary structure elements of the native state of the RRM2. α-helices and β-strands are depicted as cylinders and arrows, respectively. Secondary structure elements are colored as solid if the structure is present in the microstate. Isoleucine, leucine, and valine residues are shown as gray circles, and ILV contacts are depicted as dashed lines. To view this figure in color, go online.
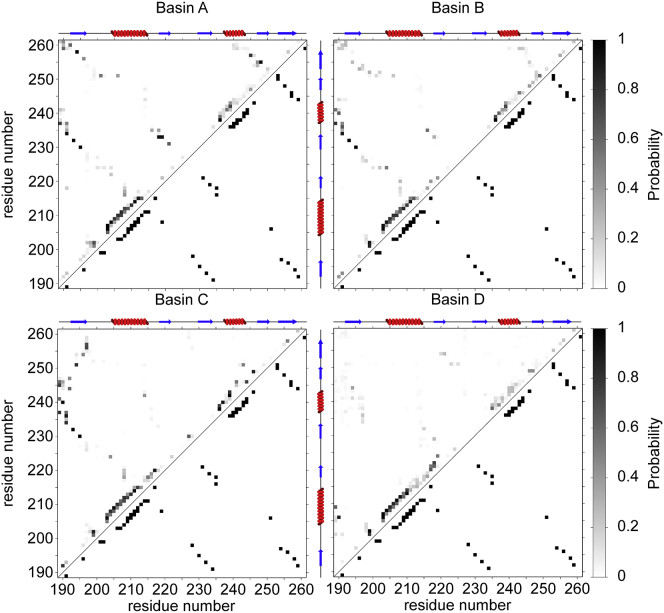
Figure 3.
Residual structure of the microstates in the partially folded state of the TDP-43 RRM2. A comparison of the average probability of hydrogen bonds formation in the microstates corresponding to the basins shown in Fig. 1 (above diagonal) with the native state (below diagonal) is shown. A schematic representation of the secondary structure elements of the native state is depicted on top of each map. To view this figure in color, go online.
Microstates from basins B and C (Fig. 1; Table S1) share several common structural features, including Rg and β-sheet content, and display an overall decreased structure and compactness compared to the native state and cluster A (Fig. 1). Both basins B and C are characterized by a structural core formed by β1, β3, and α1 (Fig. 3), with a stable β-sheet formed by β1 and β3 (Fig. 3). In cluster C, however, β4 and β5 merge into an elongated strand (Fig. 1) that only forms a marginally stable parallel pairing with β1, not supported by any hydrophobic ILV contacts (Figs. 2 and 3). A structural reorganization from β-strand to α-helix of residues 216–219, part of β2 in the native state, is observed with higher probability in basin C than B (Figs. 3 and S6).
Basin D is the most-extended and least-structured cluster of microstates (Fig. 1). The microstates in cluster D show a high degree of structural disorder, with α1 and a short non-native transient α-helix formed by residues 218–221 as the only stable secondary structural elements (Figs. 3 and S6). Long-range hydrophobic contacts within the ILV cluster are limited to the interaction of Val 195 with Leu 207 and Leu 243 (Fig. 2). Basin E and basin F represent low-populated states with no significant structural differences from cluster A (Fig. S7) but more relaxed packing, as indicated by the lower #ILV contacts and larger SASA (Fig. S8).
In summary, the free-energy landscape of TDP-43 RRM2 consists of six basins segregated in three separated regions in the CV-space (Fig. 1) with progressive loss of secondary structure and hydrophobic contacts, suggesting a likely path toward unfolding. The first region includes basins A, E and F, the microstates with the highest content of secondary structures and most similar to the native state. The second region, formed by basins B and C, represents conformations of RRM2 that maintain a compact structure (Rg ∼ 1.2 nm) and several features of the topology of the native state (α1, β1, and β3) but have lost several secondary structural elements (α2, β2, β4, and β5). The last region, basin D, contains the most extended and the least structured microstates of RRM2: with the exception of α1 and a short transient α-helix formed by residues 218–221 (Figs. 3 and S6), these microstates resemble a random coil conformation.
To validate the results of the RAM simulations, we calculated the backbone chemical shifts of each conformation using the software TALOS+ (61), a different algorithm than the one used in the RAM simulations. We observed good agreement with chemical shifts measured at 6 M urea (Fig. S9). An Rg of 2.34 ± 0.4 nm was estimated for TDP-43 RRM2 from small angle x-ray scattering data collected at 6 M urea (B.C. Mackness, B.R. Morgan, L.M. Deveau, S.V. Kathuria, D.T., F.M., and J.A.Z., unpublished data). The presence of chemical denaturant in the small angle x-ray scattering experiments is likely to favor less-compact conformations of RRM2 with a larger hydrophobic SASA than our RAM simulations collected in a traditional water box without denaturant (56). The Rg values and the calculated backbone chemical shifts for basin D, which includes the most extended conformations of RRM2, are consistent with the experimental data collected at 6 M urea (Fig. S10). We predict these extended conformations of basin D to be significantly more populated at high concentrations of denaturant.
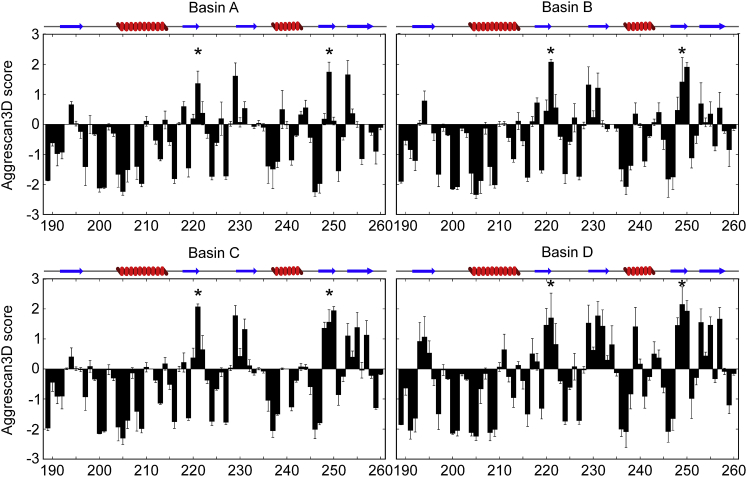
To assess a role for these partially folded states of RRM2 in TDP-43 proteinopathies, we estimated the aggregation propensity of the microstates in each basin. We employed the Aggrescan3D (54) (A3D) server to predict the aggregation propensities of each residue in the partially folded states of RRM2 corresponding to the energy basins described above. The results for the native state (Fig. S11; Table S2) show that A3D identified only two aggregation-prone residues (A3D score >1): Phe 221 and Ile 249. A3D predicted additional regions of RRM2 to have significant propensity to aggregate (Fig. 4) in the partially folded states of RRM2 (Table S2).
Figure 4.
Partially folded states of the TDP-43 RRM2 exhibit increased predicted propensity to aggregation compared to the native state. The aggregation propensity of each residue in the RRM2 is shown as the Aggrescan3D score for the microstates corresponding to the basins shown in Fig. 1. Positive values correspond to aggregation-prone residues, and negative values correspond to soluble residues. Residues predicted as aggregation-prone in the native state are identified with a ∗. A schematic representation of the secondary structure elements of the native state is depicted on top of each plot. To view this figure in color, go online.
In basin A, two additional residues are found to be aggregation prone in β3 and β5 (Fig. 4; Table S2) as a result of their increased exposure to the solvent due to the less-stable pairing between the β-strands. Conformations from basins B and C are predicted to have additional aggregation-prone residues (five in basin B and nine in basic C, respectively) in regions that are part of strands β2, β3, β4, and β5 in the native state (Fig. 4).
The increased exposure of the hydrophobic residues in cluster B and C compared to the native state reflects the reduced β-content of these microstates because strands β2, β4, and β5 are not included in the β-sheet but are sampling a random coil conformation (Figs. 2 and 3). In accordance with the lack of secondary structures in energy basin D, conformations in this cluster are predicted to have several aggregation-prone residues, otherwise structured in the native state (Fig. 4). Based on the structural analysis of the microstates identified in our RAM simulations, we observed that the hydrophobic cluster centered around Val 193, Val 195 (β1), and Val 232 (β3) (Fig. 2) is critical for the stability of basin A, B, C, E, and F. We predict that decreasing the hydrophobic moiety of these key residues through mutagenesis (i.e., V193A or V195A or V232A), thus disrupting the hydrophobic cluster, will destabilize states from basins A, B, C, E, and F more than the states of basin D and will increase the aggregation propensity of the mutant variants.
The most significant feature that emerges from the A3D analysis is the increased aggregation propensity for strands β3, β4, and β5 in the partially folded states of TDP-43 RRM2 relative to the native state. Strands β3 and β5 were found to be prone to fibril formation and able to form two-dimensional sheet-like fibrils resembling that of the amyloid fibril protofilaments in in vitro studies that used different synthesized peptide sequences from RRM2 (12). Moreover, recent work has shown that peptides corresponding to RRM2 residues 247–257 (β4 and β5) are able to form fibrils in vitro through polymorphic assemblies (62). Remarkably, the structures of the partially structured states found in this RAM simulation study share several traits with the truncated variant of RRM2 lacking strand β1 and part of helix α1 found in cytoplasmic aggregates from patient tissues (8). Indeed, the truncation of RRM2 increases the solvent accessibility of hydrophobic residues, which could serve as a platform for aggregation (21).
Conclusions
In conclusion, we employed RAM with NMR chemical shifts as experimental restraints to characterize the structures of sparsely populated and partially folded states of TDP-43 RRM2 at equilibrium with the native state. To populate significantly the partially folded states that are otherwise transient under native conditions, we used experimental data collected at high concentration of urea (6 M), and we calculated the free-energy landscape of RRM2. We determined the three-dimensional structures of six ensembles of states that correspond to six distinct energy basins in the free-energy landscape. Basins B, C, and D (Fig. 1) show the loss of several elements of secondary structure (β3, β4, β5, and α2) that can expose several cleavage sites, making the protein more susceptible to truncation (11, 63). A common feature of these partially folded conformations is the exposure to the solvent of the highly hydrophobic peptide regions located on strands β3, β4, and β5 that are aggregation-prone. Previous studies had shown that short peptides carrying the sequence of strands β3, β4, and β5 assemble under physiological conditions in long straight sheet-like fibrils (12, 62).
Our results provide, to our knowledge, new insights into the mechanisms underlying TDP-43 proteinopathies, proposing a role in fibrogenesis for the transient partially folded states of RRM2. These states may have a role in the function of TDP-43, for example, by increasing the exposure of the nuclear export signal (64) (residues 239–250; Fig. S12) buried in the native state or by facilitating interactions between the prion-like C-terminal domain and other proteins. However, upon environmental changes such as oxidative stress (65), RNA depletion (66), or TDP-43 overexpression, the usually low-populated partially folded states of RRM2 can be sampled with higher frequency, increasing the probability of generating toxic truncated forms of TDP-43 (10, 11, 12, 13, 14) and triggering self-aggregation by exposure to the solvent of the aggregation-prone strands β3, β4, and β5 (12, 62). In addition, non-native conformations of TDP-43 can be irreversibly sequestered in cytoplasmic fibrils, seeded by the pathogenic truncated C-terminal fragments of TDP-43 (67).
Author Contributions
D.T. and F.M. designed the research. D.T. performed the simulations and analysis. All authors wrote the manuscript.
Acknowledgments
We thank Drs. B. C. Mackness, O. Bilsel, and P. Robustelli for helpful discussion.
This work was funded by National Institutes of Health, United States grant GM117008 (F.M.), and the ALS Association (J.A.Z.).
Editor: David Eliezer.
Footnotes
Thirteen figures and two tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)31063-4.
Supporting Material
References
- 1.Lee E.B., Lee V.M., Trojanowski J.Q. Gains or losses: molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 2011;13:38–50. doi: 10.1038/nrn3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buratti E., Dörk T., Baralle F.E. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buratti E., Baralle F.E. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front. Biosci. 2008;13:867–878. doi: 10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 4.Neumann M., Sampathu D.M., Lee V.M. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 5.Arai T., Hasegawa M., Oda T. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 6.Abel O., Shatunov A., Al-Chalabi A. Development of a smartphone app for a genetics website: the amyotrophic lateral sclerosis online genetics database (ALSoD) JMIR Mhealth Uhealth. 2013;1:e18. doi: 10.2196/mhealth.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson B.S., Snead D., Gitler A.D. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Igaz L.M., Kwong L.K., Lee V.M. Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am. J. Pathol. 2008;173:182–194. doi: 10.2353/ajpath.2008.080003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y.J., Xu Y.F., Petrucelli L. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J. Neurosci. 2007;27:10530–10534. doi: 10.1523/JNEUROSCI.3421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y.J., Xu Y.F., Petrucelli L. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Igaz L.M., Kwong L.K., Lee V.M. Expression of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J. Biol. Chem. 2009;284:8516–8524. doi: 10.1074/jbc.M809462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y.T., Kuo P.H., Yuan H.S. The truncated C-terminal RNA recognition motif of TDP-43 protein plays a key role in forming proteinaceous aggregates. J. Biol. Chem. 2013;288:9049–9057. doi: 10.1074/jbc.M112.438564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang C., Tan W., Xu Z. The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PLoS One. 2010;5:e15878. doi: 10.1371/journal.pone.0015878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson B.S., McCaffery J.M., Gitler A.D. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo P.H., Doudeva L.G., Yuan H.S. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackness B.C., Tran M.T., Zitzewitz J.A. Folding of the RNA recognition motif (RRM) domains of the amyotrophic lateral sclerosis (ALS)-linked protein TDP-43 reveals an intermediate state. J. Biol. Chem. 2014;289:8264–8276. doi: 10.1074/jbc.M113.542779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kathuria S.V., Chan Y.H., Matthews C.R. Clusters of isoleucine, leucine, and valine side chains define cores of stability in high-energy states of globular proteins: Sequence determinants of structure and stability. Protein Sci. 2016;25:662–675. doi: 10.1002/pro.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Q., Yokoshi M., Kawahara Y. The cleavage pattern of TDP-43 determines its rate of clearance and cytotoxicity. Nat. Commun. 2015;6:6183. doi: 10.1038/ncomms7183. [DOI] [PubMed] [Google Scholar]
- 19.Camilloni C., Vendruscolo M. Statistical mechanics of the denatured state of a protein using replica-averaged metadynamics. J. Am. Chem. Soc. 2014;136:8982–8991. doi: 10.1021/ja5027584. [DOI] [PubMed] [Google Scholar]
- 20.Neudecker P., Robustelli P., Kay L.E. Structure of an intermediate state in protein folding and aggregation. Science. 2012;336:362–366. doi: 10.1126/science.1214203. [DOI] [PubMed] [Google Scholar]
- 21.Morgan B.R., Zitzewitz J.A., Massi F. Structural rearrangement upon fragmentation of the stability core of the ALS-linked protein TDP-43. Biophys. J. 2017;113:540–549. doi: 10.1016/j.bpj.2017.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le Marchand T., de Rosa M., Ricagno S. Conformational dynamics in crystals reveal the molecular bases for D76N beta-2 microglobulin aggregation propensity. Nat. Commun. 2018;9:1658. doi: 10.1038/s41467-018-04078-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Best R.B., Zhu X., Mackerell A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Best R.B., Mittal J., MacKerell A.D., Jr. Inclusion of many-body effects in the additive CHARMM protein CMAP potential results in enhanced cooperativity of α-helix and β-hairpin formation. Biophys. J. 2012;103:1045–1051. doi: 10.1016/j.bpj.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindorff-Larsen K., Maragakis P., Shaw D.E. Systematic validation of protein force fields against experimental data. PLoS One. 2012;7:e32131. doi: 10.1371/journal.pone.0032131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li D.W., Brüschweiler R. Iterative optimization of molecular mechanics force fields from NMR data of full-length proteins. J. Chem. Theory Comput. 2011;7:1773–1782. doi: 10.1021/ct200094b. [DOI] [PubMed] [Google Scholar]
- 27.Freddolino P.L., Harrison C.B., Schulten K. Challenges in protein folding simulations: timescale, representation, and analysis. Nat. Phys. 2010;6:751–758. doi: 10.1038/nphys1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Camilloni C., Cavalli A., Vendruscolo M. Replica-averaged metadynamics. J. Chem. Theory Comput. 2013;9:5610–5617. doi: 10.1021/ct4006272. [DOI] [PubMed] [Google Scholar]
- 29.Pitera J.W., Chodera J.D. On the use of experimental observations to bias simulated ensembles. J. Chem. Theory Comput. 2012;8:3445–3451. doi: 10.1021/ct300112v. [DOI] [PubMed] [Google Scholar]
- 30.Cavalli A., Camilloni C., Vendruscolo M. Molecular dynamics simulations with replica-averaged structural restraints generate structural ensembles according to the maximum entropy principle. J. Chem. Phys. 2013;138:094112. doi: 10.1063/1.4793625. [DOI] [PubMed] [Google Scholar]
- 31.Roux B., Weare J. On the statistical equivalence of restrained-ensemble simulations with the maximum entropy method. J. Chem. Phys. 2013;138:084107. doi: 10.1063/1.4792208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–28.. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 33.Pall S., Abraham M.J., Lindahl E. Springer; 2014. Tackling exascale software challenges in molecular dynamics simulations with GROMACS. In International Conference on Exascale Applications and Software; pp. 3–27. [Google Scholar]
- 34.Pronk S., Páll S., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hess B., Kutzner C., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 36.Berendsen H.J., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 37.Best R.B., Mittal J. Protein simulations with an optimized water model: cooperative helix formation and temperature-induced unfolded state collapse. J. Phys. Chem. B. 2010;114:14916–14923. doi: 10.1021/jp108618d. [DOI] [PubMed] [Google Scholar]
- 38.Abascal J.L., Vega C. A general purpose model for the condensed phases of water: TIP4P/2005. J. Chem. Phys. 2005;123:234505. doi: 10.1063/1.2121687. [DOI] [PubMed] [Google Scholar]
- 39.Hess B., Bekker H., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 40.Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 41.Evans D.J., Holian B.L. The Nosé–Hoover thermostat. J. Chem. Phys. 1985;83:4069–4074. [Google Scholar]
- 42.Piana S., Laio A. A bias-exchange approach to protein folding. J. Phys. Chem. B. 2007;111:4553–4559. doi: 10.1021/jp067873l. [DOI] [PubMed] [Google Scholar]
- 43.Camilloni C., Robustelli P., Vendruscolo M. Characterization of the conformational equilibrium between the two major substates of RNase A using NMR chemical shifts. J. Am. Chem. Soc. 2012;134:3968–3971. doi: 10.1021/ja210951z. [DOI] [PubMed] [Google Scholar]
- 44.Tribello G.A., Bonomi M., Bussi G. PLUMED 2: new feathers for an old bird. Comput. Phys. Commun. 2014;185:604–613. [Google Scholar]
- 45.Camilloni C., Cavalli A., Vendruscolo M. Assessment of the use of NMR chemical shifts as replica-averaged structural restraints in molecular dynamics simulations to characterize the dynamics of proteins. J. Phys. Chem. B. 2013;117:1838–1843. doi: 10.1021/jp3106666. [DOI] [PubMed] [Google Scholar]
- 46.Robustelli P., Kohlhoff K., Vendruscolo M. Using NMR chemical shifts as structural restraints in molecular dynamics simulations of proteins. Structure. 2010;18:923–933. doi: 10.1016/j.str.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 47.Kohlhoff K.J., Robustelli P., Vendruscolo M. Fast and accurate predictions of protein NMR chemical shifts from interatomic distances. J. Am. Chem. Soc. 2009;131:13894–13895. doi: 10.1021/ja903772t. [DOI] [PubMed] [Google Scholar]
- 48.Sugita Y., Okamoto Y. Replica-exchange molecular dynamics method for protein folding. Chem. Phys. Lett. 1999;314:141–151. [Google Scholar]
- 49.Pietrucci F., Laio A. A collective variable for the efficient exploration of protein beta-sheet structures: application to SH3 and GB1. J. Chem. Theory Comput. 2009;5:2197–2201. doi: 10.1021/ct900202f. [DOI] [PubMed] [Google Scholar]
- 50.Biarnés X., Pietrucci F., Laio A. METAGUI. A VMD interface for analyzing metadynamics and molecular dynamics simulations. Comput. Phys. Commun. 2012;183:203–211. [Google Scholar]
- 51.Marinelli F., Pietrucci F., Piana S. A kinetic model of trp-cage folding from multiple biased molecular dynamics simulations. PLoS Comput. Biol. 2009;5:e1000452. doi: 10.1371/journal.pcbi.1000452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar S., Rosenberg J.M., Kollman P.A. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The method. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 53.Rodriguez A., Laio A. Machine learning. Clustering by fast search and find of density peaks. Science. 2014;344:1492–1496. doi: 10.1126/science.1242072. [DOI] [PubMed] [Google Scholar]
- 54.Zambrano R., Jamroz M., Ventura S. AGGRESCAN3D (A3D): server for prediction of aggregation properties of protein structures. Nucleic Acids Res. 2015;43:W306–W313. doi: 10.1093/nar/gkv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pace C.N. Determination and analysis of urea and guanidine hydrochloride denaturation curves. Methods Enzymol. 1986;131:266–280. doi: 10.1016/0076-6879(86)31045-0. [DOI] [PubMed] [Google Scholar]
- 56.Bennion B.J., Daggett V. The molecular basis for the chemical denaturation of proteins by urea. Proc. Natl. Acad. Sci. USA. 2003;100:5142–5147. doi: 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mielke S.P., Krishnan V.V. Characterization of protein secondary structure from NMR chemical shifts. Prog. Nucl. Magn. Reson. Spectrosc. 2009;54:141–165. doi: 10.1016/j.pnmrs.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abrams C., Bussi G. Enhanced sampling in molecular dynamics using metadynamics, replica-exchange, and temperature-acceleration. Entropy (Basel) 2013;16:163–199. [Google Scholar]
- 59.Maragliano L., Vanden-Eijnden E. A temperature accelerated method for sampling free energy and determining reaction pathways in rare events simulations. Chem. Phys. Lett. 2006;426:168–175. [Google Scholar]
- 60.Laio A., Gervasio F.L. Metadynamics: a method to simulate rare events and reconstruct the free energy in biophysics, chemistry and material science. Rep. Prog. Phys. 2008;71:126601. [Google Scholar]
- 61.Shen Y., Delaglio F., Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guenther E.L., Ge P., Eisenberg D.S. Atomic-level evidence for packing and positional amyloid polymorphism by segment from TDP-43 RRM2. Nat. Struct. Mol. Biol. 2018;25:311–319. doi: 10.1038/s41594-018-0045-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arai T., Hasegawa M., Akiyama H. Phosphorylated and cleaved TDP-43 in ALS, FTLD and other neurodegenerative disorders and in cellular models of TDP-43 proteinopathy. Neuropathology. 2010;30:170–181. doi: 10.1111/j.1440-1789.2009.01089.x. [DOI] [PubMed] [Google Scholar]
- 64.Winton M.J., Igaz L.M., Lee V.M. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 2008;283:13302–13309. doi: 10.1074/jbc.M800342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cohen T.J., Hwang A.W., Lee V.M. Redox signalling directly regulates TDP-43 via cysteine oxidation and disulphide cross-linking. EMBO J. 2012;31:1241–1252. doi: 10.1038/emboj.2011.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pesiridis G.S., Tripathy K., Lee V.M. A “two-hit” hypothesis for inclusion formation by carboxyl-terminal fragments of TDP-43 protein linked to RNA depletion and impaired microtubule-dependent transport. J. Biol. Chem. 2011;286:18845–18855. doi: 10.1074/jbc.M111.231118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Furukawa Y., Kaneko K., Nukina N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J. Biol. Chem. 2011;286:18664–18672. doi: 10.1074/jbc.M111.231209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.