Abstract
Background
We have previously conducted the Sirolimus and Autophagy Inhibition in LAM (SAIL) trial, a phase 1 dose-escalation study of the combination of sirolimus and hydroxychloroquine in patients with lymphangioleiomyomatosis (LAM). The goal of the present study was to analyze sera from the SAIL trial to identify novel biomarkers that could shed light into disease pathogenesis and response to therapy.
Methods
We used the DiscoveryMAP platform from Rules Based Medicine to simultaneously measure 279 analytes in sera collected at each visit from subjects enrolled in the SAIL trial. We used longitudinal regression and pathway analysis to examine analyte rate of change and corresponding effect on lung function and to identify networks and potential nodes of interest.
Results
A total of 222 analytes were included in the analysis. We identified 32 analytes that changed over the treatment period of the study. Pathway analysis revealed enrichment in cytokine-receptor interaction and mechanistic/mammalian target of rapamycin-related pathways, in addition to seemingly unrelated processes such as rheumatoid arthritis. Search Tool for the Retrieval of Interacting Genes/Proteins analysis identified two hubs centered around acetyl-CoA carboxylase alpha and beta and coagulation factor II. In addition, we identified vascular endothelial growth factor receptor-3 and CCL21 as molecules significantly associated with changes in FEV1 during the study period.
Conclusions
We performed a large-scale analyte study in sera of women with LAM and identified potential markers that could be linked to disease pathogenesis, lung injury, and therapeutic response. These data will enable future investigation into the specific roles of these molecules in LAM.
Trial Registry
ClinicalTrials.gov; No. NCT01687179; URL: www.clinicaltrials.gov).
Key Words: autophagy, biomarkers, LAM, lung function, TOR
Abbreviations: BDNF, brain-derived neurotrophic factor; GO, gene ontology; HCQ, hydroxychloroquine; HD, high-dose; KEGG, Kyoto Encyclopedia of Genes and Genomes; LAM, lymphangioleiomyomatosis; LD, low-dose; MMP, matrix metalloproteinase; mTOR, mechanistic/mammalian target of rapamycin; TSC, tuberous sclerosis complex; VEGF-D, vascular endothelial growth factor-D; VEGFR-3, vascular endothelial growth factor receptor-3
Lymphangioleiomyomatosis (LAM) is a rare cystic lung disease that affects women, usually with onset during the childbearing years. LAM is characterized by biallelic loss-of-function mutations in the TSC1 or TSC2 gene1 within smooth muscle-like “LAM cells.” The mechanistic/mammalian target of rapamycin (mTOR) complex is activated in LAM cells.2 Rapamycin is US Food and Drug Administration–approved for the treatment of LAM on the basis of the landmark Multicenter International Lymphangioleiomyomatosis Efficacy and Safety of Sirolimus trial, which showed that rapamycin stabilized the rate of lung function decline, but continuous use is required for sustained effect.3
mTORC1 is a key inhibitor of autophagy; therefore, tuberous sclerosis complex (TSC)-deficient LAM cells are predicted to have low levels of autophagy. By inhibiting mTORC1, rapamycin strongly induces autophagy, which has been shown to induce pro-survival effects in TSC-deficient cells.4 In vitro and in vivo data support the hypothesis that the combination of autophagy inhibition (hydroxychloroquine [HCQ]) and mTORC1 inhibition (rapamycin) can result in the death of TSC2-deficient cells.4 We have previously performed the Sirolimus and Autophagy Inhibition in LAM (SAIL) phase 1 dose escalation study of a combination of sirolimus and HCQ for the treatment of LAM.5 The study enrolled 13 subjects, with 3 receiving low-dose HCQ (200 mg) and 10 receiving 400 mg HCQ.5 The SAIL trial met the primary end point of safety, and a secondary end point suggested potential efficacy in stabilizing lung function 6 months after stopping combination therapy. Serum samples were collected from subjects enrolled in SAIL during all study visits with the exploratory goal of identifying biomarkers of disease and response to therapy.
The study of peripheral blood in patients with LAM has previously identified multiple molecules6 that are differentially present in the sera of women with LAM. The most important of these is vascular endothelial growth factor (VEGF)-D, a diagnostic biomarker of LAM in clinical use7, 8, 9 and a candidate biomarker of therapeutic response to rapamycin.10 To date, however, no large-scale studies of sera from women with LAM have been reported.
In this study, we measured the concentrations of 279 serum proteins in samples collected from subjects enrolled in the SAIL trial at every study visit (seven visits total). We hypothesized that this large-scale study of analytes would identify biomarkers that shed new light on disease pathogenesis and others that indicate response to therapy. We further hypothesized that using network medicine approaches would unravel previously unsuspected important molecular pathways, enhancing our understanding of LAM pathogenesis.
Background and Study Population
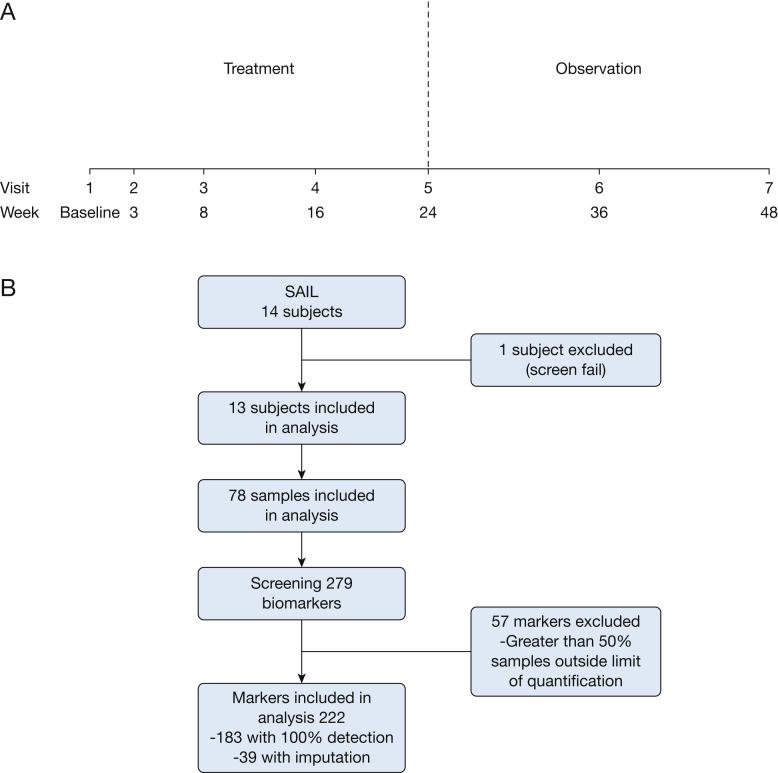
The study population was composed of participants in the SAIL trial, the design of which has been described previously (e-Table 1).5 Study visits are summarized in Figure 1. The study was designed by the investigators and the protocol was approved by the Department of Defense and the Institutional Review Boards at Brigham and Women’s Hospital and the NHLBI (IRB#2012P000669).
Figure 1.
Study procedure. A, Trial design and timing of sample collection. B, Analytes selected for analysis. SAIL = Sirolimus and Autophagy Inhibition for Patients With Lymphangioleiomyomatosis.
Analytes Measurements
Serum was collected from every enrolled subject at each study visit. Analytes were measured using the DiscoveryMAP platform (Myriad RBM).
Statistical Analysis
One hundred eighty-three analytes were measurable in all samples. An additional 39 proteins had ≤50% missing values, which were imputed; therefore, a total of 222 proteins were included in the analysis.
The R programming language (version 3.3.2) was used for all statistical analyses.11 A heat map displaying analyte levels at baseline, end of treatment, and end of study was generated using the R package gplots (version 3.0.1),12 with clustering performed using the built-in R function hclust and scaling of analyte levels per subject achieved with the R package mousetrap (version 3.1.0).11, 13 All repeated-measure analysis of variance and longitudinal regression modeling was performed using the R package lmerTest (version 2.0.33). Ontology and network analyses were run using DAVID (version 6.8)14, 15 and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; version 10.5).16
Results
Patient Population and Analyte Measurements
Study design and characteristics of study subjects have been previously published5 and are detailed in Figure 1 and e-Table 1. A total of 79 sera were submitted for analysis from 14 subjects; one subject screen failed and that sample was not included in the analysis. One hundred eighty-three analytes were measurable in every sample (Fig 1 and e-Table 2).
Relationship Between Baseline Study Subject Characteristics and Analyte Levels
We evaluated baseline analyte levels per the different population characteristics. We found that baseline use of supplemental O2 was associated with 15 significantly regulated proteins (P < .05). These differences however, were not present when adjusted for multiple testing (q > 0.05). Menopausal status is known to be associated with decreased rates of lung function decline in LAM.17, 18 We found that, in addition to higher levels of follicle-stimulating hormone and luteinizing hormone, women who were postmenopausal had significantly lower levels of macrophage colony stimulating factor 1 compared with premenopausal women with LAM (Table 1). Gene ontology (GO) terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with supplemental oxygen use and menopausal status are listed in e-Tables 3 and 4, respectively.
Table 1.
Baseline Analyte Level Comparisons per Population Characteristic
| Analyte | Supplemental Oxygen |
P | q | |
|---|---|---|---|---|
| Yes (n = 6) | No (n = 7) | |||
| Galectin 3 | 0.93 ± 0.04 | 0.71 ± 0.05 | .006 | 0.55 |
| MCSF | −0.27 ± 0.05 | −0.51 ± 0.05 | .007 | 0.55 |
| KLK7 | 3.09 ± 0.07 | 3.37 ± 0.05 | .01 | 0.55 |
| ADM | 0.49 ± 0.06 | 0.29 ± 0.03 | .02 | 0.55 |
| Gelsolin | 1.35 ± 0.02 | 1.46 ± 0.03 | .02 | 0.55 |
| GDF15 | −0.30 ± 0.04 | −0.44 ± 0.03 | .02 | 0.55 |
| AB40 | −0.30 ± 0.04 | −0.50 ± 0.06 | .02 | 0.55 |
| LH | 1.27 ± 0.13 | 0.73 ± 0.15 | .02 | 0.55 |
| Cystatin B | 1.25 ± 0.08 | 0.98 ± 0.06 | .02 | 0.55 |
| FSH | 1.64 ± 0.17 | 0.95 ± 0.21 | .03 | 0.55 |
| HSP70 | 1.07 ± 0.06 | 0.82 ± 0.07 | .03 | 0.55 |
| TRAILR3 | 1.21 ± 0.06 | 0.99 ± 0.06 | .03 | 0.55 |
| ADAMTS8 | 2.12 ± 0.02 | 2.23 ± 0.04 | .03 | 0.57 |
| FAS | 1.41 ± 0.05 | 1.27 ± 0.02 | .04 | 0.62 |
| TNC | 2.92 ± 0.10 | 2.56 ± 0.12 | .04 | 0.62 |
| Postmenopause |
P | q | ||
|---|---|---|---|---|
| Yes (n = 7) | No (n = 6) | |||
| FSH | 1.84 ± 0.07 | 0.78 ± 0.10 | < .001 | < 0.001 |
| LH | 1.41 ± 0.06 | 0.62 ± 0.08 | < .001 | 0.001 |
| MCSF | −0.24 ± 0.03 | −0.53 ± 0.04 | < .001 | 0.02 |
| HSP70 | 1.11 ± 0.04 | 0.79 ± 0.06 | .001 | 0.08 |
| BAFF | 2.62 ± 0.03 | 2.43 ± 0.04 | .003 | 0.14 |
| Galectin 3 | 0.93 ± 0.04 | 0.71 ± 0.05 | .005 | 0.17 |
| MMP9 total | 3.24 ± 0.03 | 3.08 ± 0.04 | .009 | 0.28 |
| PARC | 2.17 ± 0.07 | 1.86 ± 0.08 | .01 | 0.39 |
| SCF | 2.72 ± 0.05 | 2.54 ± 0.04 | .02 | 0.39 |
| Immunoglobulin M | 0.21 ± 0.11 | 0.56 ± 0.04 | .02 | 0.42 |
| HE4 | 3.02 ± 0.06 | 2.76 ± 0.08 | .02 | 0.42 |
| EPO | 1.19 ± 0.09 | 0.90 ± 0.07 | .03 | 0.42 |
| GDF15 | −0.30 ± 0.04 | −0.44 ± 0.04 | .03 | 0.42 |
| Osteopontin | 0.91 ± 0.17 | 0.42 ± 0.07 | .03 | 0.42 |
| GIP | −0.72 ± 0.23 | −1.40 ± 0.10 | .03 | 0.42 |
| tPA | 0.19 ± 0.06 | 0.02 ± 0.04 | .04 | 0.42 |
| IL-23 | 0.80 ± 0.02 | 0.70 ± 0.04 | .04 | 0.42 |
| Cystatin B | 1.24 ± 0.08 | 0.99 ± 0.06 | .04 | 0.42 |
| TSH | 0.02 ± 0.12 | 0.35 ± 0.08 | .04 | 0.42 |
| KLK7 | 3.11 ± 0.09 | 3.35 ± 0.05 | .04 | 0.42 |
| MCP2 | 1.74 ± 0.06 | 1.53 ± 0.07 | .04 | 0.42 |
| YKL40 | 1.62 ± 0.08 | 1.39 ± 0.07 | .04 | 0.42 |
| OPG | 0.86 ± 0.03 | 0.74 ± 0.04 | .046 | 0.42 |
| TM | 0.58 ± 0.03 | 0.49 ± 0.02 | .046 | 0.42 |
| IL-18 | 2.53 ± 0.06 | 2.36 ± 0.03 | .0497 | 0.43 |
AB40 = beta amyloid 1 40; ADAMTS8 = a disintegrin and metalloproteinase with thrombospondin motifs 8; ADM = adrenomedullin; BAFF = B-cell activating factor; EPO = erythropoietin; FAS = FASLG receptor; FDR = false discovery rate; FSH = follicle-stimulating hormone; GDF15 = growth differentiation factor 15; GIP = gastric inhibitory polypeptide; HSP70 = heat shock protein 70; KLK7 = kallikrein 7; LH = luteinizing hormone; MCP = monocyte chemotactic protein; MCSF = macrophage colony-stimulating factor 1; MMP9 = matrix metalloproteinase 9; OPG = osteoprotegerin; PARC = pulmonary and activation regulated chemokine; SCF = stem cell factor; TM = thrombomodulin; TNC = tenascin C; tPA = tissue type plasminogen activator; TRAILR3 = tumor necrosis factor–related apoptosis-inducing ligand receptor 3; TSH = thyroid-stimulating hormone.
Analytes Expression in Study Population and Changes Over Treatment Period
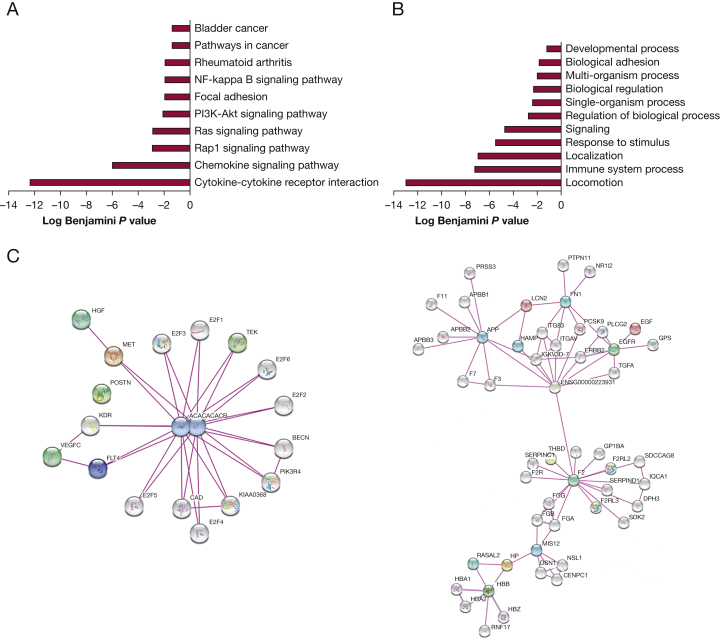
After controlling for multiple hypothesis testing, 32 analytes were found to significantly change over the treatment period (24 weeks: visits 1-5) (Table 2, Fig 2). The most significantly changed were pulmonary and activation regulated chemokine or CCL18 and, not surprisingly, VEGF-D. KEGG and GO enrichment analyses were performed on this set of analytes (q < 0.05). The KEGG results demonstrate an enrichment in cytokine pathways and pathways involved in mTORC1 signaling (Rap/Ras/PI3K) in addition to other seemingly unrelated diseases such as rheumatoid arthritis (Fig 3A). The GO term results are centered around processes related to chemotaxis and immune system (Fig 3B). Finally, all the treatment-regulated analytes were subjected to network analysis using STRING.19 Two notable hub proteins were identified: (1) acetyl-CoA carboxylase alpha and beta, which together form the acetyl-CoA carboxylase complex, a critical step in lipid biosynthesis and known mTORC1 downstream target20 (e-Fig 1, Fig 3C) and (2) coagulation factor II or thrombin, which is involved in wound healing, blood homeostasis, and inflammation and has previously been shown to induce endothelial cell proliferation through p70S6K-dependent pathways.21
Table 2.
Analytes Significantly Changed Over the Treatment Period (Week 24)
| Analyte | P | q |
|---|---|---|
| PARC | < .001 | < 0.001 |
| VEGF-D | < .001 | < 0.001 |
| SPD | < .001 | < 0.001 |
| TIE 2 | < .001 | < 0.001 |
| TRAILR3 | < .001 | < 0.001 |
| Eotaxin 2 | < .001 | < 0.001 |
| MIP1 beta | < .001 | < 0.001 |
| P selectin | < .001 | 0.001 |
| Resistin | < .001 | 0.004 |
| EN-RAGE | < .001 | 0.005 |
| Folate receptor gamma | < .001 | 0.007 |
| GDF 15 | < .001 | 0.007 |
| Periostin | < .001 | 0.008 |
| MMP9 total | < .001 | 0.009 |
| SDF1 | < .001 | 0.009 |
| TATI | < .001 | 0.01 |
| TFF3 | .001 | 0.01 |
| Thrombomodulin | .001 | 0.02 |
| Vaspin | .001 | 0.02 |
| NGAL | .002 | 0.02 |
| CCL21 | .002 | 0.03 |
| CD5L | .003 | 0.03 |
| VEGFR-3 | .003 | 0.03 |
| CCL15 | .003 | 0.03 |
| TARC | .004 | 0.03 |
| EGF | .004 | 0.03 |
| HGFR | .004 | 0.03 |
| Haptoglobin | .006 | 0.048 |
| CEA | .006 | 0.048 |
| PECAM 1 | .007 | 0.048 |
| ENA 78 | .007 | 0.048 |
| SHBG | .007 | 0.048 |
CCL = CC motif chemokine; CD5L = CD5 antigen like; CEA = carcinoembryonic antigen; EGF = epidermal growth factor; ENA 78 = epithelial-derived neutrophil activating protein 78; HGFR = hepatocyte growth factor receptor; MIP1 beta = macrophage inflammatory protein 1 beta; PECAM1 = platelet endothelial cell adhesion molecule; SDF1 = stromal cell–derived factor 1; SHBG = sex hormone–binding globulin; SPD = pulmonary surfactant associated protein D; TARC = thymus and activation regulated chemokine; TATI = pancreatic secretory trypsin inhibitor; TFF3 = Trefoil factor 3; TIE 2 = tyrosine kinase with Ig and EGF homology domains 2; Vaspin = visceral adipose tissue derived serpin A12; VEGF-D = vascular endothelial growth factor D; VEGFR-3 = vascular endothelial growth factor receptor-3.
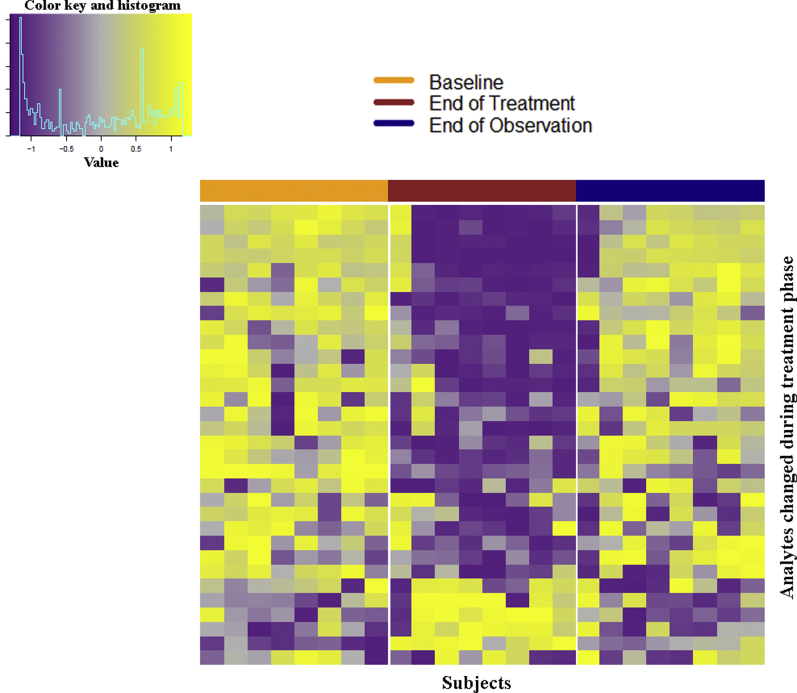
Figure 2.
Peripheral blood proteins changed with combination therapy with sirolimus and hydroxychloroquine. Heat map of analytes that significantly changed during the treatment period for subjects with analyte measurements at baseline, the end of treatment (week 24), and the end of observation (week 48). Columns: individual patient measurements at weeks 0, 24, and 48; rows: analytes that changed significantly between weeks 0 and 24 by repeated measures analysis of variance. Log10-transformed analyte levels were scaled separately for each patient across the three visits of interest. Increased shades of yellow, increased relative to subject mean; increased shades of purple, decreased relative to subject mean; gray, unchanged relative to subject mean. Analytes were clustered hierarchically and subjects were clustered hierarchically and separately within each visit.
Figure 3.
Network analysis of treatment-dependent analytes. Analysis of 32 analytes differentially changed over time and multiple visits between baseline and end of treatment. STRING did not recognize the analyte PECAM1, so it was not included in the network. A, KEGG pathways. B, GO terms enriched in this group of analytes. C, STRING analysis shows a hub centered around acetyl-CoA carboxylase alpha and beta, and F2. F2 = coagulation factor II; GO = gene ontology; KEGG = Kyoto Encyclopedia of Genes and Genomes; PECAM1 = platelet endothelial cell adhesion molecule 1.
Analyte Changes as Predictors of Lung Function
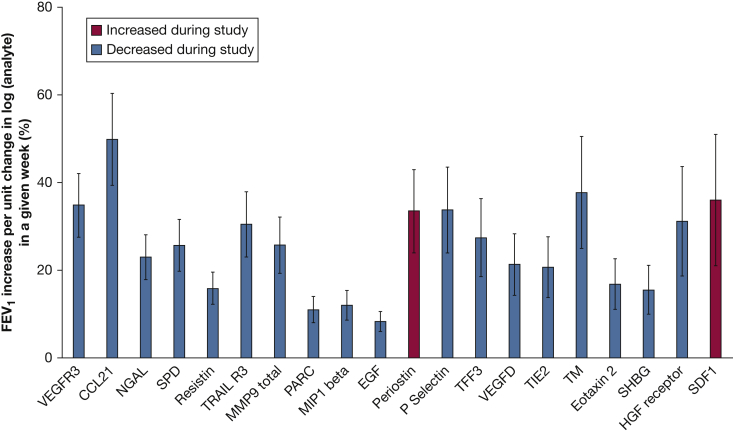
We examined the relationship between analyte and FEV1 changes over the treatment period. Of the 32 analytes that changed over visits 1 through 5, changes in VEGF-receptor (VEGFR)-3 and CCL21 were associated with changes in % FEV1 over the treatment period (baseline to week 24 [q < 0.05]). When comparing two subjects with the same baseline % FEV1 and holding time constant, a subject with a 1-unit lower log VEGFR-3 was estimated to have a % FEV1 that was 38.7 ± 8.2 higher, and a 1-unit lower log CCL21 was estimated to result in a % FEV1 that was 56.8 ± 13.8 % higher.
We further examined the ability of analytes to predict lung function over the entire study period: baseline to 48 weeks. Of the 32 analytes that significantly changed with treatment, 20 correlated with FEV1 over the duration of the study (Table 3, Fig 4). Again, CCL21 and VEGFR-3 were the most significantly associated with FEV1 (% predicted). CCL21 decreased in response to combination therapy, and, when holding time constant, a subject with a 1-log unit lower CCL21 level was estimated to have a 49.8 ± 10.5 higher FEV1 % predicted. On the other hand, Periostin increased after treatment, and a subject with a 1-log unit higher Periostin level was estimated to have a FEV1 % predicted that was 33.5 ± 9.5 higher. Changes in VEGF-D levels were associated with changes in lung function over the course of the entire study, with every log unit decrease in VEGF-D associated with a 21.3 ± 7.0 increase in FEV1 % predicted when holding time constant.
Table 3.
Increase in FEV1 % for Every Log Unit Change in Indicated Analyte in a Given Week When Analyzing FEV1 Over Entire Study Duration
| Analyte | Increase in %FEV1 (Mean ± SEM) | P | q |
|---|---|---|---|
| VEGFR3 | 34.8 ± 7.3 | < .001 | < 0.001 |
| CCL21 | 49.8 ± 10.5 | < .001 | < 0.001 |
| NGAL | 23.0 ± 5.1 | < .001 | < 0.001 |
| SPD | 25.7 ± 5.9 | < .001 | < 0.001 |
| Resistin | 15.9 ± 3.6 | < .001 | < 0.001 |
| TRAIL R3 | 30.5 ± 7.4 | < .001 | < 0.001 |
| MMP9 total | 25.8 ± 6.5 | < .001 | 0.001 |
| PARC | 11.0 ± 3.0 | < .001 | 0.003 |
| MIP1 beta | 12.0 ± 3.4 | .001 | 0.003 |
| EGF | 8.4 ± 2.3 | .001 | 0.003 |
| Periostin | 33.5 ± 9.5 | .001 | 0.003 |
| P Selectin | 33.8 ± 9.8 | .001 | 0.003 |
| TFF3 | 27.5 ± 8.9 | .004 | 0.009 |
| VEGFD | 21.3 ± 7.0 | .004 | 0.009 |
| TIE2 | 20.7 ± 6.9 | .005 | 0.01 |
| TM | 37.7 ± 12.8 | .005 | 0.01 |
| Eotaxin 2 | 16.9 ± 5.7 | .006 | 0.01 |
| SHBG | 15.6 ± 5.6 | .008 | 0.01 |
| HGFR | 31.2 ± 12.5 | .02 | 0.03 |
| SDF1 | 36.0 ± 15.0 | .02 | 0.03 |
See Table 2 legend for expansion of abbreviations.
Figure 4.
Analytes significantly associated with changes in % FEV1 over entire study. Estimated increase in % FEV1 given a 1-log unit change in the analyte when comparing two individuals with the same baseline FEV1 in a given week (mean ± standard error). Analytes are ordered from left to right on the basis of decreasing association significance. Analytes that increased during the study (red) and decreased during the study (blue) are included. MMP = matrix metalloproteinase 9; NGAL = neutrophil gelatinase-associated lipocalin; PARK = pulmonary and activation regulated chemokine; SHBG = sex hormone–binding globulin; SPD = pulmonary surfactant–associated protein D; VEGF-D = vascular endothelial growth factor-D.
Early Changes in Analyte Levels and Prediction of Lung Function
For the design of future clinical trials,22 it is important to identify the earliest analyte changes associated with changes in lung function; therefore, we analyzed the earliest treatment time point (week 3) for evidence of changes in the analytes. Of the 222 analytes included in the analysis, 38 were changed as early as week 3 of therapy by paired t test (q < 0.05) (Table 4). CCL18 and matrix metalloproteinase 9 (MMP9) featured the most significant changes, decreasing by 48.6% and 31.2%, respectively. Mean CCL18 level continued to decrease throughout the treatment period before increasing after treatment cessation. Mean MMP9 level stabilized during treatment and increased during the observation period. VEGF-D significantly decreased by 12.7% at week 3. After correction for multiple testing, no analyte changes by week 3 correlated with FEV1 over the treatment period (baseline to 24 weeks); however, we found that early changes (between visits 1 and 2) in TFF3 and CD5L were significantly associated with FEV1 over the entire study period (baseline to week 48). When comparing two subjects with the same baseline FEV1, a 1-unit decrease in the log difference (or log of the fold change) in TFF3 and CD5L between week 3 and baseline corresponded to an additional increase of 2.9 ± 0.7 and 2.3 ± 0.7% predicted post-bronchodilator weekly rate of FEV1 change, respectively. In other words, FEV1 after 24 weeks of treatment was estimated to be approximately 69.8% and 56.3% higher if the log difference between week 3 and baseline was 1 unit lower for TFF3 and CD5L, respectively.
Table 4.
Early Changes (Week 3) in Analyte Levels
| Analyte | Mean Log Difference Visit 2 and 1 Mean ± SEM |
P | q |
|---|---|---|---|
| PARC | −0.31 ± 0.03 | < .001 | < 0.001 |
| MMP9 total | −0.17 ± 0.02 | < .001 | < 0.001 |
| Resistin | −0.29 ± 0.04 | < .001 | < 0.001 |
| SPD | −0.18 ± 0.02 | < .001 | < 0.001 |
| Eotaxin 2 | −0.15 ± 0.02 | < .001 | 0.001 |
| TIE 2 | −0.19 ± 0.03 | < .001 | 0.002 |
| SDF1 | 0.08 ± 0.01 | < .001 | 0.002 |
| Periostin | 0.10 ± 0.02 | < .001 | 0.002 |
| MCP2 | −0.14 ± 0.02 | < .001 | 0.002 |
| TATI | −0.11 ± 0.02 | < .001 | 0.002 |
| IL-18 | −0.16 ± 0.03 | < .001 | 0.004 |
| MIP1 beta | −0.28 ± 0.05 | < .001 | 0.004 |
| NGAL | −0.17 ± 0.03 | < .001 | 0.004 |
| Tumor necrosis factor ligand superfamily member 12 (Tweak) | 0.09 ± 0.02 | < .001 | 0.004 |
| TRAIL R3 | −0.18 ± 0.03 | < .001 | 0.004 |
| ENA 78 | −0.17 ± 0.03 | < .001 | 0.004 |
| Angiopoietin 1 | −0.14 ± 0.03 | < .001 | 0.004 |
| CD5L | −0.13 ± 0.03 | < .001 | 0.004 |
| Protein S100 A6 | −0.21 ± 0.05 | < .001 | 0.009 |
| CCL15 | −0.08 ± 0.02 | < .001 | 0.01 |
| GDF15 | −0.12 ± 0.03 | .001 | 0.01 |
| CEA | 0.10 ± 0.02 | .001 | 0.01 |
| TARC | 0.14 ± 0.03 | .002 | 0.01 |
| VEGF-D | −0.10 ± 0.02 | .002 | 0.02 |
| Haptoglobin | 0.14 ± 0.03 | .002 | 0.02 |
| EN-RAGE | −0.31 ± 0.08 | .002 | 0.02 |
| Fibulin 1C | 0.16 ± 0.04 | .002 | 0.02 |
| Lactoferrin | −0.23 ± 0.06 | .003 | 0.02 |
| P selectin | −0.12 ± 0.03 | .003 | 0.02 |
| TFF3 | −0.13 ± 0.03 | .003 | 0.02 |
| Urokinase type plasminogen activator receptor | −0.14 ± 0.04 | .005 | 0.03 |
| MCP4 | −0.08 ± 0.02 | .005 | 0.03 |
| B2M | −0.07 ± 0.02 | .005 | 0.04 |
| Thrombomodulin | −0.07 ± 0.02 | .006 | 0.04 |
| Cystatin B | −0.12 ± 0.04 | .006 | 0.04 |
| Lectin-like oxidized LDL receptor 1 | −0.20 ± 0.06 | .006 | 0.04 |
| CEACAM1 | −0.08 ± 0.02 | .007 | 0.04 |
| SHBG | −0.16 ± 0.05 | .008 | 0.05 |
Durable Effects of Combination Therapy on Analyte Levels
The SAIL trial was designed with 6 months of therapy and 6 months of observation. When the entire study period was examined, we found that 68 analytes significantly changed over time (e-Table 5). Of these 68 analytes, 46 (68%) normalized to baseline levels during the observation period.
When we examined the 32 analytes that changed during the study period, we found that 14 remained stable during the observation period, which could be an indication of a durable response to combination therapy (e-Table 6). Notably, CCL21 and VEGFR-3 levels remained unchanged during the observation period, whereas VEGF-D levels increased after treatment cessation.
Analysis of Low- vs High-Dose HCQ
To gain insights into changes resulting from increasing doses of HCQ, we evaluated subjects who received low-dose (LD, 200 mg, n = 3) vs those who received higher dose (HD, 400 mg, n = 10) HCQ during the treatment phase of the study. We found that 17 analytes showed different regulation between low and higher dose HCQ (e-Table 7), suggesting perhaps that these changes were due to the combinatorial effects of rapamycin and HCQ treatments and not driven by mTORC1 inhibition alone. KEGG pathway analysis revealed enrichments for cytokine receptor interaction and signaling as well as hematopoietic cell lineage and Toll-like receptor signaling. A STRING analysis demonstrated an organized network centered around actin-related protein 2, which plays an important role in the actin cytoskeleton and autophagosome formation.23
For these 17 analytes, we additionally compared LD and HD subjects more directly by computing protein level differences from baseline at each visit separately for LD and HD subjects and performing t tests directly between LD and HD subjects at each visit. Vitronectin levels were consistently different from baseline levels in the HD subjects for visits 4 through 7 (P < .05), with average levels decreasing through the study period, whereas there were no differences from baseline in the LD group. In addition, except for week 8 (visit 3), vitronectin levels were significantly lower in HD subjects compared with LD subjects (P < .05).
Visfatin levels in the HD group decreased over the study period and were consistently different from baseline at each study visit except for visit 7 (P < .05). Moreover, these levels were significantly lower than the LD group at the end of observation (visit 7; HD 0.57 ± 0.04 vs LD: 0.96 ± 0.06, P < .05).
Changes in Autophagy-Associated Analytes
To examine the potential effect of HCQ on autophagy, we cross-referenced the 222 analytes included in the analysis with a database of known autophagy-related human genes, identifying eight analytes: AXL receptor tyrosine kinase, brain-derived neurotrophic factor (BDNF), cathepsin D, epidermal growth factor receptor (EGFR), human epidermal growth factor receptor 2, insulin, receptor tyrosine protein kinase erbB3, and soluble superoxide dismutase 1. Of these analytes, only BDNF levels changed significantly by repeated measures analysis of variance for all subjects (q < 0.01 for entire study duration, P < .05 for treatment period only). Furthermore, we investigated differences in these analyte levels comparing subjects that received the low dose of HCQ and those that received the high dose. Regardless of HCQ dosage, BDNF levels consistently decreased in comparison to baseline throughout the treatment period (LD: visits 4 and 5; HD: visits 2-5, P < .05), with HD subjects only exhibiting significantly lower BDNF levels than LD subjects at week 36 (visit 6, HD: 1.27 ± 0.02 vs LD: 1.40 ± 0.01, P < .05). EGFR levels were significantly lower in HD than LD subjects at week 24 (visit 5, HD: 0.38 ± 0.01 vs LD: 0.50 ± 0.02, P < .05) and week 36 (visit 6, HD: 0.41 ± 0.01 vs LD: 0.51 ± 0.01, P < .05), although they were also significantly lower at baseline (HD: 0.39 ± 0.02 vs LD: 0.53 ± 0.01, P < .05).
Discussion
Biomarker identification is critical for the management of patients with lung disease, especially biomarkers that occur prior to changes in FEV1 or FVC, or so-called predictive biomarkers. Identifying markers of response to therapy can change our approaches for patient care, allowing a more personalized approach to therapy. In addition, and of importance, the identification of biomarkers of response to therapy could allow a change in the design of clinical trials, with earlier quantitative end points. This is especially important in a rare lung disease such as LAM, because fewer subjects would need to be enrolled and the trial could be conducted more efficiently vs trials undertaken on the basis of lung function end points.22 Biomarkers of treatment effect that occur before changes in lung function could allow doses to be “personalized.” Finally, the identification of biomarkers could potentially provide valuable insights into disease pathogenesis.
Our approach of using a large-scale analytes study led to the identification of novel, previously unsuspected candidate biomarkers of response to therapy in LAM, which could potentially be linked to cellular responses and disease pathogenesis. For example, we identified 32 analytes that changed during the treatment period. A network analysis identified acetyl-CoA carboxylase complex as one of the central nodes regulating these analytes. Acetyl-CoA carboxylase complex plays important roles in fatty acid synthesis and is a known downstream target of mTORC1.20 Surprisingly, another central node that we identified is coagulation factor 2, known to be regulated by mTORC1 but previously not shown to potentially play important roles in disease pathogenesis in LAM; however, the coagulation cascade has been shown to contribute to pathogenesis of lung disease,24 specifically idiopathic pulmonary fibrosis.25
We identified VEGFR-3 and CCL21 as predictors of lung function decline, molecules that are likely involved in disease pathogenesis because both are intimately involved in lymphatic endothelial biology. VEGFR-3, a tyrosine kinase receptor and the canonical receptor for VEGF-D,26 is expressed on the cell surface of LAM cells.27, 28 Of note, a pan-tyrosine kinase inhibitor has previously been shown to be a viable therapeutic target in animal models of LAM.29 Soluble VEGFR-3, a potent inhibitor of lymphangiogenesis,30, 31 has previously been described as a biomarker in melanoma.32 In our studies, levels of soluble VEGFR-3 were predictive of lung function decline, with decreases in soluble VEGFR-3 levels leading to improved FEV1. The relationship of these findings to the pathogenesis of LAM remains to be investigated.
CCL21, the second predictor of lung function decline, a chemokine secreted by lymphatic endothelial cells, plays an important role in immune tolerance33 and has previously been shown to upregulate VEGF-D in cancer cells through the ERK pathway.34 The role of CCL21 in LAM pathogenesis is not known; however, it has been shown to be a predictor of lung involvement in systemic lupus erythematous.35
In addition, we found that early changes in TFF3 and CD5L/AIM could predict changes in lung function. TFF3 has previously been shown to be increased in a variety of lung diseases including idiopathic pulmonary fibrosis, sarcoidosis, and COPD.36 TFF3 has not been previously studied in TSC or LAM; however, its activity in the brain can be blocked by rapamycin, indicating an important role for the mTORC1 pathway in TFF3 signaling.37 TFF3 is likely a marker of epithelial injury, a phenomenon poorly studied in LAM. TFF3, if validated as a marker of early therapeutic response, could be used in the design of future clinical trials, potentially limiting time spent on study drug for those patients unresponsive to treatment.
CD5L/AIM38 is expressed on inflamed tissue macrophages. This soluble protein plays important roles in lipid homeostasis and the control of inflammatory responses. In Th17 cells, CD5L is necessary for the regulation of pathogenicity with loss of CD5L resulting in pathogenic Th17 and autoimmunity.39 On the other hand, AIM-/- mice demonstrate an accelerated resolution of inflammation after LPS administration, likely due to enhanced macrophage efferocytosis of apoptotic neutrophils.40 CD5L-induced macrophage anti-inflammatory response is mediated by autophagy.41 CD36, a known receptor for CD5L, is an established target of mTORC1,42 likely through HIF-1α–dependent mechanisms.43 These changes in CD5L could be related to either sirolimus, HCQ, or the combination. Furthermore, CD5L is a critical regulator of lipid mediators in health and in acute lung injury,40 in addition to being a key regulator of intracellular lipidomic profile and cholesterol biosynthesis.39 The data provide further evidence related to the importance of the lipid biosynthesis pathways in response to therapy and perhaps disease pathogenesis.
The SAIL trial examined the combination of mTORC1 inhibition and autophagy inhibition. In an effort to understand changes linked to inhibition of autophagy rather than mTORC1, we identified through network analysis actin-related protein 2, a molecule involved in autophagy, as a potential upstream node explaining the differential response between lower and higher dose HCQ. Furthermore, we found that levels of BDNF, a known autophagy mediator via the PI3K/Akt/mTOR pathway,44 significantly decreased over the course of the treatment period in all subjects, with subjects receiving the higher HCQ dose having a lower BDNF level at week 36. In addition, visfatin levels were found to be significantly lower by the end of observation in subjects receiving the high-dose HCQ, whereas vitronectin levels were significantly lower in subjects taking high-dose vs low-dose HCQ in all but one study visit. Visfatin/NAMPT has been shown to enhance autophagy in cerebral ischemia,45 whereas to our knowledge, vitronectin has not yet been implicated in autophagy. On the basis of these findings, BDNF, visfatin, and vitronectin could be indicative of successful autophagy inhibition; therefore, the levels of these analytes could be monitored in future trials to appropriately estimate a safe observation duration post drug withdrawal.
Our study has several limitations, including: (1) the lack of a control group that was treated with sirolimus alone, so that more granular distinction could be made as to analytes that were modulated by the combination of HCQ and sirolimus vs sirolimus alone; (2) the lack of a placebo group, which prevented a direct correlation between analyte changes and outcomes; (3) the lack of a validation cohort; and (4) the duration of the study period (treatment and observation) may not have been long enough to adequately assess response to sirolimus and HCQ treatment. Internal controls such as high follicle-stimulating hormone and luteinizing hormone levels in menopausal subjects and the identification of VEGF-D in our analysis provide a measure of reassurance regarding our samples and analyses.
In summary, we present the first large-scale study of analytes in sera of women with LAM. We identified markers that could be linked to disease pathogenesis, lung injury, and therapeutic response. These data will allow future investigations into the role of these specific molecules in disease, and, after validation in other cohorts, could perhaps serve as end points of future clinical trials and/or enable personalized approaches to the therapy of LAM.
Acknowledgment
Author contributions: Study design and first draft of the manuscript: A. M. L., J. M., E. P. H., and S. E.-C. Acquisition, analysis, or interpretation of the data: all authors. Critical revision of the manuscript for important intellectual content and approval of last draft: all authors. E. P. H. and S. E.-C. had full access to the data and have final responsibility for decision to submit for publication.
Financial/nonfinancial disclosures: None declared.
Role of sponsors: The funding source did not play any role in study design, data collection, and analysis or decision to publish.
Other contributions: We are indebted to patients and their families for their participation in this study. We also thank The LAM Foundation and the Tuberous Sclerosis Alliance for patient referral. We thank the Data Safety Monitoring Board Members and Medical Monitor, as well as the clinical trials support staffs at the BWH and the Office of the Clinical Director, National Heart, Lung, and Blood Institute, National Institutes of Health.
Additional information: The e-Appendix, e-Figure, and e-Tables can be found in the Supplemental Materials section of the online article.
Footnotes
FUNDING/SUPPORT: This study was supported by the Department of Defense [Grant W81XWH-12-1-0578] to E. P. H.] and in part by the Intramural Research Program, National Institutes of Health, National Heart, Lung, and Blood Institute.
Supplementary Data
References
- 1.Carsillo T., Astrinidis A., Henske E.P. Mutations in the tuberous sclerosis complex gene TSC2 are a cause of sporadic pulmonary lymphangioleiomyomatosis. Proc Natl Acad Sci U S A. 2000;97(11):6085–6090. doi: 10.1073/pnas.97.11.6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henske E.P., McCormack F.X. Lymphangioleiomyomatosis - a wolf in sheep's clothing. J Clin Invest. 2012;122(11):3807–3816. doi: 10.1172/JCI58709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCormack F.X., Inoue Y., Moss J. Efficacy and safety of sirolimus in lymphangioleiomyomatosis. N Engl J Med. 2011;364(17):1595–1606. doi: 10.1056/NEJMoa1100391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parkhitko A., Myachina F., Morrison T.A. Tumorigenesis in tuberous sclerosis complex is autophagy and p62/sequestosome 1 (SQSTM1)-dependent. Proc Natl Acad Sci U S A. 2011;108(30):12455–12460. doi: 10.1073/pnas.1104361108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Chemaly S., Taveira-Dasilva A., Goldberg H.J. Sirolimus and autophagy inhibition in lymphangioleiomyomatosis: results of a phase I clinical trial. Chest. 2017;151(6):1302–1310. doi: 10.1016/j.chest.2017.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nijmeh J., El-Chemaly S., Henske E.P. Emerging biomarkers of lymphangioleiomyomatosis. Expert Rev Respir Med. 2018;12(2):95–102. doi: 10.1080/17476348.2018.1409622. [DOI] [PubMed] [Google Scholar]
- 7.Young L.R., Inoue Y., McCormack F.X. Diagnostic potential of serum VEGF-D for lymphangioleiomyomatosis. N Engl J Med. 2008;358(2):199–200. doi: 10.1056/NEJMc0707517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young L.R., Vandyke R., Gulleman P.M. Serum vascular endothelial growth factor-D prospectively distinguishes lymphangioleiomyomatosis from other diseases. Chest. 2010;138(3):674–681. doi: 10.1378/chest.10-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui Y., Steagall W.K., Lamattina A.M. Aberrant SYK kinase signaling is essential for tumorigenesis induced by TSC2 inactivation. Cancer Res. 2017;77(6):1492–1502. doi: 10.1158/0008-5472.CAN-16-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young L., Lee H.S., Inoue Y. Serum VEGF-D a concentration as a biomarker of lymphangioleiomyomatosis severity and treatment response: a prospective analysis of the Multicenter International Lymphangioleiomyomatosis Efficacy of Sirolimus (MILES) trial. Lancet Respir Med. 2013;1(6):445–452. doi: 10.1016/S2213-2600(13)70090-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Team RC. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. Accessed March 14, 2018.
- 12.Warnes GR, Bolker B, Bonebakker L, et al. gplots: various R Programming tools for plotting data. R package version 3.0.1. https://CRAN.R-project.org/package=gplots. Accessed March 14, 2018.
- 13.Kieslich PJ, Wulff DU, Henninger F, Haslbeck JMB. mousetrap: process and analyze mouse-tracking data. R package version 3.1.0. 2017; https://CRAN.R-project.org/package=mousetrap. Accessed March 14, 2018.
- 14.Huang da W, Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D., Morris J.H., Cook H. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu C., Lee H.S., Pappas G.P. A phase II clinical trial of an aromatase inhibitor for postmenopausal women with lymphangioleiomyomatosis. Ann Am Thorac Soc. 2017;14(6):919–928. doi: 10.1513/AnnalsATS.201610-824OC. [DOI] [PubMed] [Google Scholar]
- 18.El-Chemaly S., Henske E.P. Towards personalised therapy for lymphangioleiomyomatosis: lessons from cancer. Eur Respir Rev. 2014;23(131):30–35. doi: 10.1183/09059180.00008813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snel B., Lehmann G., Bork P., Huynen M.A. STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 2000;28(18):3442–3444. doi: 10.1093/nar/28.18.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown N.F., Stefanovic-Racic M., Sipula I.J., Perdomo G. The mammalian target of rapamycin regulates lipid metabolism in primary cultures of rat hepatocytes. Metabolism. 2007;56(11):1500–1507. doi: 10.1016/j.metabol.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 21.Parrales A., Lopez E., Lee-Rivera I., Lopez-Colome A.M. ERK1/2-dependent activation of mTOR/mTORC1/p70S6K regulates thrombin-induced RPE cell proliferation. Cell Signal. 2013;25(4):829–838. doi: 10.1016/j.cellsig.2012.12.023. [DOI] [PubMed] [Google Scholar]
- 22.El-Chemaly S., Henske E.P. The next breakthrough in LAM clinical trials may be their design: challenges in design and execution of future LAM clinical trials. Expert Rev Respir Med. 2015;9(2):195–204. doi: 10.1586/17476348.2015.1024663. [DOI] [PubMed] [Google Scholar]
- 23.Coutts A.S., La Thangue N.B. Actin nucleation by WH2 domains at the autophagosome. Nat Commun. 2015;6:7888. doi: 10.1038/ncomms8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chambers R.C., Scotton C.J. Coagulation cascade proteinases in lung injury and fibrosis. Proc Am Thorac Soc. 2012;9(3):96–101. doi: 10.1513/pats.201201-006AW. [DOI] [PubMed] [Google Scholar]
- 25.Scotton C.J., Krupiczojc M.A., Konigshoff M. Increased local expression of coagulation factor X contributes to the fibrotic response in human and murine lung injury. J Clin Invest. 2009;119(9):2550–2563. doi: 10.1172/JCI33288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bailey E., Cui Y., Casey A. Pulmonary vasculopathy associated with FIGF gene mutation. Am J Pathol. 2017;187(1):25–32. doi: 10.1016/j.ajpath.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davis J.M., Hyjek E., Husain A.N., Shen L., Jones J., Schuger L.A. Lymphatic endothelial differentiation in pulmonary lymphangioleiomyomatosis cells. J Histochem Cytochem. 2013;61(8):580–590. doi: 10.1369/0022155413489311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Issaka R.B., Oommen S., Gupta S.K. Vascular endothelial growth factors C and D induces proliferation of lymphangioleiomyomatosis cells through autocrine crosstalk with endothelium. Am J Pathol. 2009;175(4):1410–1420. doi: 10.2353/ajpath.2009.080830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atochina-Vasserman E.N., Abramova E., James M.L. Pharmacological targeting of VEGFR signaling with axitinib inhibits Tsc2-null lesion growth in the mouse model of lymphangioleiomyomatosis. Am J Physiol Lung Cell Mol Physiol. 2015;309(12):L1447–L1454. doi: 10.1152/ajplung.00262.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makinen T., Jussila L., Veikkola T. Inhibition of lymphangiogenesis with resulting lymphedema in transgenic mice expressing soluble VEGF receptor-3. Nat Med. 2001;7(2):199–205. doi: 10.1038/84651. [DOI] [PubMed] [Google Scholar]
- 31.Singh N., Tiem M., Watkins R. Soluble vascular endothelial growth factor receptor 3 is essential for corneal alymphaticity. Blood. 2013;121(20):4242–4249. doi: 10.1182/blood-2012-08-453043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mouawad R., Spano J.P., Comperat E., Capron F., Khayat D. Tumoural expression and circulating level of VEGFR-3 (Flt-4) in metastatic melanoma patients: correlation with clinical parameters and outcome. Eur J Cancer. 2009;45(8):1407–1414. doi: 10.1016/j.ejca.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 33.Kozai M., Kubo Y., Katakai T. Essential role of CCL21 in establishment of central self-tolerance in T cells. J Exp Med. 2017;214(7):1925–1935. doi: 10.1084/jem.20161864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun L., Zhang Q., Li Y., Tang N., Qiu X. CCL21/CCR7 up-regulate vascular endothelial growth factor-D expression via ERK pathway in human non-small cell lung cancer cells. Int J Clin Exp Pathol. 2015;8(12):15729–15738. [PMC free article] [PubMed] [Google Scholar]
- 35.Odler B., Bikov A., Streizig J. CCL21 and IP-10 as blood biomarkers for pulmonary involvement in systemic lupus erythematosus patients. Lupus. 2017;26(6):572–579. doi: 10.1177/0961203316668418. [DOI] [PubMed] [Google Scholar]
- 36.Doubkova M., Karpisek M., Mazoch J., Skrickova J., Doubek M. Prognostic significance of surfactant protein A, surfactant protein D, Clara cell protein 16, S100 protein, trefoil factor 3, and prostatic secretory protein 94 in idiopathic pulmonary fibrosis, sarcoidosis, and chronic pulmonary obstructive disease. Sarcoidosis Vasc Diffuse Lung Dis. 2016;33(3):224–234. [PubMed] [Google Scholar]
- 37.Luo Y.X., Han H., Shao J. mTOR signalling in the nucleus accumbens shell is critical for augmented effect of TFF3 on behavioural response to cocaine. Sci Rep. 2016;6:27895. doi: 10.1038/srep27895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanjurjo L., Aran G., Roher N., Valledor A.F., Sarrias M.R. AIM/CD5L: a key protein in the control of immune homeostasis and inflammatory disease. J Leukoc Biol. 2015;98(2):173–184. doi: 10.1189/jlb.3RU0215-074R. [DOI] [PubMed] [Google Scholar]
- 39.Wang C., Yosef N., Gaublomme J. CD5L/AIM regulates lipid biosynthesis and restrains Th17 cell pathogenicity. Cell. 2015;163(6):1413–1427. doi: 10.1016/j.cell.2015.10.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura H., Suzuki M., Konno S. Orchestrating Role of Apoptosis inhibitor of macrophage in the resolution of acute lung injury. J Immunol. 2017;199(11):3870–3882. doi: 10.4049/jimmunol.1601798. [DOI] [PubMed] [Google Scholar]
- 41.Sanjurjo L., Amezaga N., Aran G. The human CD5L/AIM-CD36 axis: a novel autophagy inducer in macrophages that modulates inflammatory responses. Autophagy. 2015;11(3):487–502. doi: 10.1080/15548627.2015.1017183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C., Hu L., Zhao L. Inflammatory stress increases hepatic CD36 translational efficiency via activation of the mTOR signalling pathway. PLoS One. 2014;9(7):e103071. doi: 10.1371/journal.pone.0103071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mwaikambo B.R., Yang C., Chemtob S., Hardy P. Hypoxia up-regulates CD36 expression and function via hypoxia-inducible factor-1- and phosphatidylinositol 3-kinase-dependent mechanisms. J Biol Chem. 2009;284(39):26695–26707. doi: 10.1074/jbc.M109.033480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen A., Xiong L.J., Tong Y., Mao M. Neuroprotective effect of brain-derived neurotrophic factor mediated by autophagy through the PI3K/Akt/mTOR pathway. Mol Med Rep. 2013;8(4):1011–1016. doi: 10.3892/mmr.2013.1628. [DOI] [PubMed] [Google Scholar]
- 45.Wang P., Guan Y.F., Du H., Zhai Q.W., Su D.F., Miao C.Y. Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy. 2012;8(1):77–87. doi: 10.4161/auto.8.1.18274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.