Abstract
Duchenne muscular dystrophy (DMD), a severe hereditary disease affecting 1 in 3,500 boys, mainly results from the deletion of exon(s), leading to a reading frameshift of the DMD gene that abrogates dystrophin protein synthesis. Pairs of sgRNAs for the Cas9 of Staphylococcus aureus were meticulously chosen to restore a normal reading frame and also produce a dystrophin protein with normally phased spectrin-like repeats (SLRs), which is not usually obtained by skipping or by deletion of complete exons. This can, however, be obtained in rare instances where the exon and intron borders of the beginning and the end of the complete deletion (patient deletion plus CRISPR-induced deletion) are at similar positions in the SLR. We used pairs of sgRNAs targeting exons 47 and 58, and a normal reading frame was restored in myoblasts derived from muscle biopsies of 4 DMD patients with different exon deletions. Restoration of the DMD reading frame and restoration of dystrophin expression were also obtained in vivo in the heart of the del52hDMD/mdx. Our results provide a proof of principle that SaCas9 could be used to edit the human DMD gene and could be considered for further development of a therapy for DMD.
Keywords: CRISPR, SaCas9, CinDel, Duchenne muscular dystrophy, genome editing
Duchêne et al. use SaCas9 to restore DMD gene reading while maintaining the correct phasing of the spectrin-like repeats that structure the central rod domain of the dystrophin protein. Eventually, such correction could lead to milder BMD with no severe symptoms.
Introduction
Duchenne muscular dystrophy (DMD) is one of the most severe hereditary diseases, affecting 1 in 3,500 newborn boys.1, 2 This X-linked disorder is caused by a mutation in the DMD gene coding for the dystrophin protein, leading to the absence of this protein in muscle fibers.3, 4 These mutations, mostly deletions of one or several exons, are responsible for a shift in the normal reading frame of the DMD gene, generating a premature stop codon leading to the loss of the dystrophin protein during the translation process. This protein is fundamental for maintaining the integrity of the sarcolemma during contraction because it allows interaction with the surrounding extracellular matrix through binding with the membrane-anchored β-dystroglycan and with the cytoskeleton through interaction with actin and microtubules.5 Notably, the dystrophin central rod domain is formed by 24 successive spectrin-like repeats (SLRs). Each of these SLRs consists of three antiparallel α helices (A, B, and C) forming a coiled-coiled structure in which each heptad contains hydrophobic amino acids in positions a and d. Interestingly, the neuronal nitric oxide synthase (nNOS) interacts with helixes A, B, and C of SLR16 and with helix A of SLR17. It has been reported that, in Becker muscular dystrophy (BMD) patients, the severity of the disease could be related to the structure of their truncated dystrophin;6 thus, we focused on creating a genomic deletion that not only restores the DMD gene reading frame but also allows the production of a dystrophin protein with SLRs correctly phased and structured.
Recent advances in the field of gene therapy offer great perspectives for the development of a curative treatment for DMD. Strategies of gene replacement that rely on the adeno-associated virus (AAV)-mediated delivery of micro-dystrophin are still undergoing development7, 8, 9 and reached the clinical trial phase, such as rAAVrh74.MHCK7.micro-dystrophin (Nationwide Children’s Hospital Columbus, Ohio, United States) or SGT-001 (University of Florida, Gainesville, Florida, United States) (https://clinicaltrials.gov/).
However, a few years ago, the discovery of the CRISPR/Cas9 system raised hope for the establishment of therapies for hereditary diseases that counter the disease at its root by editing the genome. This system was first identified as the immune system of some bacterial species, such as Streptococcus pyogenes or Staphylococcus aureus, to protect them against bacteriophage infections.10 This system has been adapted to target genomic DNA in mammals, offering possibilities to permanently edit their genome.11, 12, 13 More precisely, a single guide RNA (sgRNA) can be made to target a specific protospacer sequence as long as it is positioned next to a protospacer-adjacent motif (PAM), which is NGG for S. pyogenes and NNGRRT for S. aureus. When the Cas9/sgRNA complex binds to its protospacer, the Cas9 protein is able to generate a double-strand break (DSB) precisely three nucleotides upstream of the PAM of Cas9 of Streptococcus pyogenes (SpCas9) or the Cas9 of Staphylococcus aureus (SaCas9). Based on this precise editing capacity, we have previously shown that, by using a combination of two sgRNAs, we can generate a genomic deletion that forms a hybrid exon, 50-54, which restores dystrophin protein expression in myotubes from a patient having a deletion of exons 51 to 53.14, 15
In DMD patients, reported mutations are mostly present in a “hotspot,” including exons 45 to 55. To date, most of the research groups focus on the use of the CRISPR/Cas9 system to restore a normal reading frame through complete removal of one or several exons without taking care to restore the normal phasing of the SLR. Such an approach could lead to the formation of a dystrophin protein with an abnormally structured central rod domain, which can lead to severe BMD instead of milder BMD. Here we report an approach that covers up to 40% of all DMD mutations by creating a hybrid exon directly connecting exons 47 to exon 58 using SaCas9. Please note that we did not form a hybrid exon 44-58 or 45-58 because the dystrophin sequences of SLR R17, coded by exons 44 to 46, are necessary for the essential interaction of dystrophin with nNOS.16 Our approach will not only restore dystrophin production but will also produce a dystrophin protein with a normal SLR structure. More precisely, by using a combination of 2 sgRNAs, we created a hybrid exon, allowing us to position hydrophobic amino acids in positions a and d of the newly formed SLR. We think that, by maintaining the adequate structure of the central rod domain (i.e., the SLRs), we can not only maintain the interaction of dystrophin with elements of the dystrophin-associated proteins and with nNOS but also ensure the normal function of the dystrophin protein during contraction and relaxation of muscle fibers. To precisely produce a normally phased SLR, we used our previously characterized CRISPR-induced deletion (CinDel) method, which aims to target the exons that precede and follow the patient mutation to form a hybrid exon. We have, however, modified this approach to use the smaller SaCas9 from S. aureus, whose gene could ultimately be delivered with a pair of sgRNAs by a single AAV.17 Also, we are not targeting the exons that immediately precede and follow the patient deletion but, rather, exons 47 and 58 so that the formation of the hybrid exon 47-58 may be a therapeutic approach for any deletion, insertion, or point mutation between these two exons. Our proposed edition would thus be a treatment for roughly 40% of DMD patients.
Results
Identification and Activity Assays of Individual sgRNAs in 293T Cells
Our primary goal was to establish a strategy based on the creation of a hybrid exon, allowing correction of the DMD gene reading frame in the case of DMD patients affected by the deletion of exon 50. Thus, all possible sgRNA target sites within exons 46 to 58 were screened. We screened 51 sgRNA target sites for the SaCas9 nuclease. Because the SaCas9 nuclease induces a DSB precisely 3 nt upstream of the PAM (NNGRRT), we were able to select pairs of sgRNAs that can be combined to create a hybrid exon that restores a normal reading frame. Thus, these hybrid exons might permit the production of an internally deleted dystrophin protein. In addition, we completed our analysis by selecting pairs of sgRNAs that were also able to produce an adequate SLR. Consequently, we focused only on combinations where the hybrid junction maintained the configuration of a normal SLR, where hydrophobic amino acids are localized in positions a and d of the heptad motif (SLRs are composed of 3 α helices, each containing 7 amino acids [a to f], where hydrophobic amino acids are in locations a and d). To assess the adequate localization of those amino acids, we referred to the eDystrophin database, which provides information about the structural domains of the dystrophin protein (http://edystrophin.genouest.org/). As a result of our preliminary analysis, we focused our work on 18 sgRNAs (Table 1; Figures S1A and S1B), which can, in principle, generate 12 hybrid exons that do not only correct the reading frame of the DMD gene but also maintain the structure of the SLRs in the central rod domain (Figure S1C).
Table 1.
List of sgRNA Target Sites in Exons 46 to 58
| sgRNA No. | Exon No. | Strand | sgRNA Target Sequences, Excluding PAM (5′–3′) | sgRNA Target Sequence Position | PAM Nucleotide Position | Cut Sites in the DMD Gene | Cut Sites in the Amino Acid Sequence | sgRNA Used in the Formation of the Hybrid Exon |
|---|---|---|---|---|---|---|---|---|
| 1 | 46 | sense | TTCTCCAGGCTAGAAGAACAA | 1407207–1407227 | 1407228–1407233 | 6624 – 6225 | 2208 GAA (Glu): 2209 CAA (Gln) | 46–51; 46–53 |
| 2 | 46 | antisense | CTGCTCTTTTCCAGGTTCAAG | 1407312–1407332 | 1407306–1407311 | 6714 –6715 | 2238 CTT (Leu): 2239 GAA (Glu) | 46–53 |
| 3 | 47 | sense | GTCTGTTTCAGTTACTGGTGG | 1409686–1409706 | 1409707–1409712 | 6769 –6770 | 2257 G:TG (Val) | 47–58 |
| 4 | 47 | antisense | TCCAGTTTCATTTAATTGTTT | 1409736–1409756 | 1409730–1409735 | 6824 –6825 | 2268 AAA (Lys): 2267 CAA (Gln) | 47–58 |
| 5 | 47 | antisense | CTTATGGGAGCACTTACAAGC | 1409765–1409785 | 1409759–1409764 | 6833 – 6834 | 2278 CT:T (Leu) | 47–58 |
| 6 | 49 | antisense | TTGCTTCATTACCTTCACTGG | 1502716–1502736 | 1502710–1502715 | 7194 –7195 | 2398 CCA (Pro): 2399 GTG (Val) | 49–52; 49–53 |
| 7 | 51 | antisense | TTGTGTCACCAGAGTAACAGT | 1565282–1565302 | 1565276–1564281 | 7323 –7324 | 2441 ACT (Thr): 2442 GTT (Val) | 46–51 |
| 8 | 51 | antisense | AGTAACCACAGGTTGTGTCAC | 1565294–1565314 | 1565288-1565293 | 7335 –7336 | 2445 GTG (Val): 2446 ACA (Thr) | 46–51 |
| 9 | 52 | antisense | TTCAAATTTTGGGCAGCGGTA | 1609765–1609785 | 1609759–1609764 | 7595 –7596 | 2532 AC:C (Thr) | 47–52 |
| 10 | 52 | sense | CAAGAGGCTAGAACAATCATT | 1609802–1609822 | 1609823–1609828 | 7647 −7648 | 2549 ATC (Ile): 2550 ATT (Ile) | 49–52 |
| 11 | 53 | antisense | TTGTACTTCATCCCACTGATT | 1659891–1659911 | 1659885–1659890 | 7677 – 7678 | 2559 AAT (Asn): 2560 CAG (Gln) | 49–53 |
| 12 | 53 | sense | CTTCAGAACCGGAGGCAACAG | 1659918–1659938 | 1659939–1659944 | 7719 – 7720 | 2573 CAA (Gln): 2574 CAG (Gln) | 46–53 |
| 13 | 53 | sense | CAACAGTTGAATGAAATGTTA | 1659933–1659953 | 1659954–1659959 | 7734 –7735 | 2578 ATG (Met): 2579 TTA(Leu) | 46–53 |
| 14 | 53 | sense | GCCAAGCTTGAGTCATGGAAG | 1660017–1660037 | 1660038–1660043 | 7818 –7819 | 2606 TGG (Trp): 2607 AAG (Lys) | 46–53 |
| 15 | 53 | antisense | CTTGGTTTCTGTGATTTTCTT | 1660068–1660088 | 1660062–1660067 | 7854 – 7855 | 2618 AAG (Lys): 2619 AAA (Lys) | 46–53 |
| 16 | 58 | sense | TCATTTCACAGGCCTTCAAGA | 1860349–1860369 | 1860370–1860375 | 8554 –8555 | 2852 A:AG (Lys) | 47–58 |
| 17 | 58 | antisense | CAGAAATATTCGTACAGTCTC | 1860411–1860431 | 1860405–1860410 | 8601 –8602 | 2867 GAG (Gln): 2868 ACT (Thr) | 47–58 |
| 18 | 58 | antisense | CAATTACCTCTGGGCTCCTGG | 1860467–1860487 | 1860461–1860466 | 8657 – 8658 | 2886 GT:C (Gln) | 47–58 |
Nucleotide positions are provided with reference to the DMD gene sequence ENS00000198947 analyzed using the Benchling web tool (https://benchling.com/). Bold underlined sequences are intron sequences.
Next we designed 18 sgRNAs, which were cloned into the PX601-AAV-cytomegalovirus (CMV)::NLS-SaCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA plasmid (Addgene, 61591). These plasmids were first individually transfected in 293T cells to assess their activity. Genomic DNA was extracted 48 hr after transfection. The targeted exon, 46, 47, 49, 51, 52, 53, or 58, was amplified by PCR and submitted to a Surveyor assay for the detection of insertions or deletions (INDELs) (Figure S2A). Among the 18 sgRNAs tested, sgRNA 2-46, sgRNA 4-47, sgRNA 9-52, sgRNA 11-53, and sgRNA 15-53 showed little or no activity, whereas all remaining sgRNAs exhibited good cleavage efficiency, as observed with the Surveyor assays, which generated cleaved bands at the expected sizes. To improve the characterization of the formation of INDELs at the targeted sites, PCR products were submitted to tracking INDELs by decomposition (TIDE) analysis. The percentages of INDELs determined by this method were consistent with the Surveyor assay because samples that exhibited substantial cleavage activity also demonstrated a high rate of INDELs according to the TIDE analysis (Figure S2B).
Test of sgRNA Pairs in 293T Cells
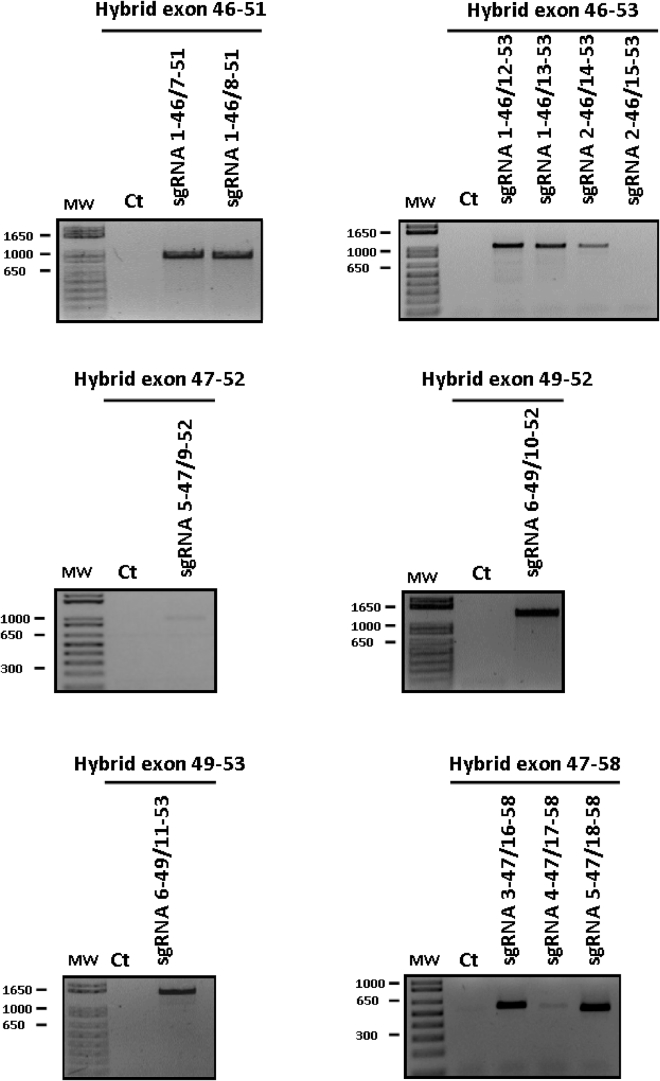
293T cells were co-transfected with pairs of sgRNAs that might not only produce a large genomic deletion but also precisely connect exons, surrounding the deletion of exon 50, to restore the reading frame and produce an adequate SLR. Thus, 12 pairs of sgRNAs (previously identified in a preliminary analysis of compatible sgRNAs) were tested. To detect the formation of the hybrid exons, PCRs were performed using the forward primer binding with the targeted exon upstream of exon 50 and the reverse primer binding with the targeted exon downstream of exon 50, previously used when sgRNAs were individually tested (Figure 1). Given that the targeted exons are separated by large introns, PCR amplification was only possible when exon deletion had occurred. The combination of sgRNAs 2-46 and 15-53 did not allow the formation of a hybrid exon 46-53; thus, no PCR product was detected. In addition, sgRNA 5-47 combined with sgRNA 9-52 and sgRNA 4-47 combined with sgRNA 17-58 were poorly effective for the formation of the hybrid exons 47-52 and 47-58, respectively. However, all remaining combinations of sgRNAs permitted the formation of hybrid exons, as observed by the detection of PCR products under these conditions.
Figure 1.
Generation of Several Hybrid Exons by Genomic Edition Using Pairs of sgRNAs
The formation of hybrid exons was tested with various pairs of previously characterized sgRNAs (Table 2) that could, in principle, restore an open reading frame for the DMD gene. The formation of the hybrid exon was confirmed by PCR amplification, (i.e., there was no PCR product when the hybrid exon was not formed). As a negative control (Ct), we used non-transfected HEK293T cells.
Analysis of Hybrid Exons
Before proceeding to further analyses, we needed to confirm that the hybrid exons that were formed were correct. Indeed, these hybrid exons should predominantly be able to restore a normal reading frame for the DMD gene. The PCR products resulting from the amplification of hybrid exons were purified and cloned into a pMiniT vector. For the analysis of the sequencing results of the hybrid exons, we counted the bacterial clones that contained the right nucleotide sequence and the bacterial clones that contained a hybrid exon where the nucleotide sequence was not exactly as expected (data not shown). To improve the characterization of these hybrid exons, we performed a modified TIDE analysis that compared a PCR product composed of 100% of the expected hybrid exon (amplified from a previously identified clone that contained a hybrid exon with the right nucleotide sequence) with a test sample amplified from the genomic DNA of treated cells (Table 2). In at least 54% of the PCR products, the nucleotide sequences of the hybrid exons were exactly as expected (Figure S3) and, thus, efficiently restored a normal reading frame in the DMD gene. Interestingly, the combination of sgRNAs 3-47 and 16-58, which resulted in a 450-kb pair (kbp) deletion, was one of the most precise pairs because 90% of the PCR products of the resulting hybrid exons 3-47/16-58 were exactly as expected. In addition, the combination of sgRNAs 5-47 and 18-58 also generated an interesting large genomic deletion, although only 68% of the PCR products contained the expected nucleotide sequence. The formation of hybrid exons 47-58 could be a treatment for about 40% of the mutations (deletion, duplications, and point mutations) identified in DMD patients. Consequently, in the next steps of our work, we chose to focus on these two combinations only.
Table 2.
Analysis of Hybrid Exon Amplicons by TIDE
| Upstream sgRNA | Downstream sgRNA | Expected Size of the Amplicon (bp) | Correct Junction (%) |
|---|---|---|---|
| 1-46 | 7-51 | 967 | 90.8 |
| 1-46 | 8-51 | 955 | 86.1 |
| 1-46 | 12-53 | 1,099 | 54.6 |
| 1-46 | 13-53 | 1,084 | 60.9 |
| 2-46 | 14-53 | 1,090 | N/A |
| 2-46 | 15-53 | 1,054 | N/A |
| 5-47 | 9-52 | 997 | N/A |
| 6-49 | 10-52 | 1,386 | 83.3 |
| 6-49 | 11-53 | 1,548 | 84.7 |
| 3-47 | 16-58 | 587 | 90.7 |
| 4-47 | 17-58 | 575 | N/A |
| 5-47 | 18-58 | 548 | 67,7 |
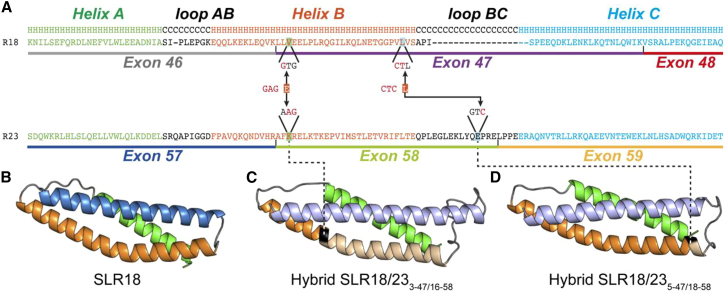
3D Spectrin-like Models Resulting from the Formation of Hybrid Exons 3-47/16-58 and 5-47/18-58
The dystrophin protein is important to maintain the integrity of the sarcolemma. The central part of this protein, called the rod domain, is made of 24 SLRs. According to homology models available from the eDystrophin website (http://edystrophin.genouest.org/), the dystrophin SLR predicted structures are composed of the typical triple coiled-coil structure; i.e., three α helices (A, B, and C) linked by two loops (AB and BC),18 as represented in Figure 2B for the integral structure of SLR18 (R18). For the structure of the resulting hybrid SLR (R18-R23) using pairs of sgRNAs (either pair 3-47/16-58 or pair 5-47/18-5), spectrin repeats R19 to R22 are removed, but the reading frame resulting in the formation of hybrid exons 47–58 are conserved, both with junction points in helix B (Figures 2C and 2D). The homology model for these two hybrid exons presents a similar length and structure as the original SLR, R18, with a shorter BC loop for hybrid exon 5-47/18-58. Thus, the homology models suggest that the dystrophin produced following the formation of hybrid exons with sgRNA pairs 3-47/16-58 or 5-47/18-58 would have a similar structure as the integral dystrophin and, therefore, should function in a similar manner. These predictive structures of hybrid SLRs confirmed that the hybrid exons 47-58 (formed with either sgRNAs 3-47 and 16-58 or with sgRNAs 5-47 and 18-58) are good candidates to produce dystrophin proteins with an adequate structure, thus increasing the potential functionality of the newly produced, internally deleted dystrophin.
Figure 2.
Structural Representations of SLR18 and Two Hybrid SLRs
(A) Primary structure alignments for SLR18 and SLR23. The amino acid sequences of the exons associated with these spectrin-like repeats are underlined in different colors. The secondary structure of the SLRs is represented above the sequences, H for helices and C for the loop segments. Structural motifs, as identified in the primary sequence alignment, are colored as follows: helix A is in green, helix B is in orange, and helix C is in blue. Loops AB and BC are in black. sgRNAs 3-47 and 5-47 are cutting in exon 47 in sequences coding for helix B. sgRNAs 16-58 and 18-58 are cutting in exon 58 in sequences coding, respectively, for helix B and the BC loop. When these sgRNAs are used in pairs (i.e., sgRNAs 3-47 with 16-58 and sgRNAs 5-47 and 18-58), the nucleotides between their cut sites are deleted, leading in both cases to the formation of the hybrid exon 47-58. Note that, because these sgRNAs are cutting within a codon, a new codon is formed at the junction site, one coding for glutamic acid (E) and the other for leucine (L), respectively, within hybrid exon 3-47/16-58 and 5-47/18-58. These hybrid exons produced, respectively, hybrid SLR3-47/16-58 and hybrid SLR5-47/18-58. (B) Homology models for integral SLR18 obtained from the eDystrophin Website (http://edystrophin.genouest.org/). (C and D) The homology models for the hybrid SLR18/233-47/16-58 (C) and hybrid SLR18/235-47/18-58 (D) were realized using the iTasser web server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/). Colors are darker for the segment derived from SLR18 and lighter for the segment derived from SLR23. The junction position is highlighted in black.
Formation of the Hybrid Exons 47-58 and Restoration of Dystrophin Protein Expression in DMD Patient Cells
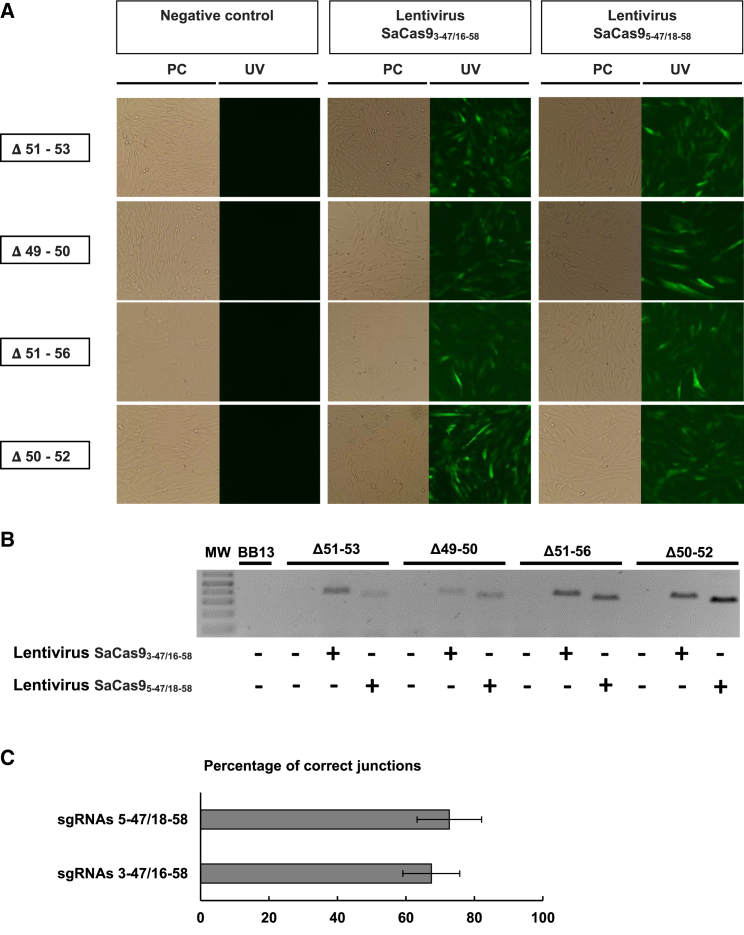
For the experiments with DMD patient cells, we only focused on the two pairs of sgRNAs that could be suitable for the highest number of DMD mutations; i.e., the combination of sgRNAs 3-47 and 16-58 and the combination of sgRNAs 5-47 and 18-58. First, transfection and nucleofection of a combination of plasmids that code for SaCas9 and for a pair of sgRNAs were tested (data not shown). Because of low transfection and nucleofection efficiencies, viral transduction of myoblasts was subsequently tested. Using integrative lentiviral vectors encoding the SaCas9 gene and a pair of sgRNAs (i.e., Lenti-SaCas93-47/16-58 and Lenti-SaCas95-47/18-58) and EGFP, myoblasts from four different DMD patients having different exon deletions (Δ49-50, Δ50-52, Δ51-53 and Δ51-56) were transduced. The transduction was evaluated by EGFP expression in myoblasts 3 days later (Figure 3A). Following proliferation of transduced cells, genomic DNA was extracted from a cell sample to assess the formation of the hybrid exons 47-58 (Figure 3B). In non-transduced myoblasts, no PCR amplification was detected because the distance between the primers is too long (i.e., 450 kbp). In all transduced cells, the hybrid exons were detected with both pairs of sgRNAs (i.e., sgRNAs 3-47 and 16-58 and sgRNAs 5-47 and 18-58). In addition, PCR products were submitted to sequencing to confirm that the hybrid exons contained the expected nucleotide sequences. We observed by TIDE analysis that the hybrid exons contained the exact nucleotide sequences in 67% and 73%, respectively, for the pairs of sgRNAs 3-47 and 16-58 and sgRNAs 5-47 and 18-58 (Figure 3C).
Figure 3.
Lentiviral Delivery of the SaCas9 Gene and Pairs of sgRNAs Restored the Expression of Dystrophin in DMD Patient Cells
Myoblasts from 4 DMD patients with different exon deletions (i.e., exons 51 to 53, exons 49 and 50, exons 51 to 56, and exons 50 to 52) were infected in 8 μg/mL of Polybrene in MB-1 medium with a third-generation lentiviral vector encoding the SaCas9 gene and a pair of sgRNAs (i.e., either the pair of sgRNAs 3-47 and 16-58 or the pair of sgRNAs 5-47 and 18-58). (A) The SaCas9 protein was immediately followed by a T2A-EGFP fragment, whose fluorescence allowed to control of the level of infection of the myoblasts. (B) PCR amplifications were performed from purified genomic DNA to detect the formation of the hybrid exons 47-58. (C) The amplicon sequences were submitted to TIDE analysis to determine the percentage of exact junction (n = 4). The error bars are SE.
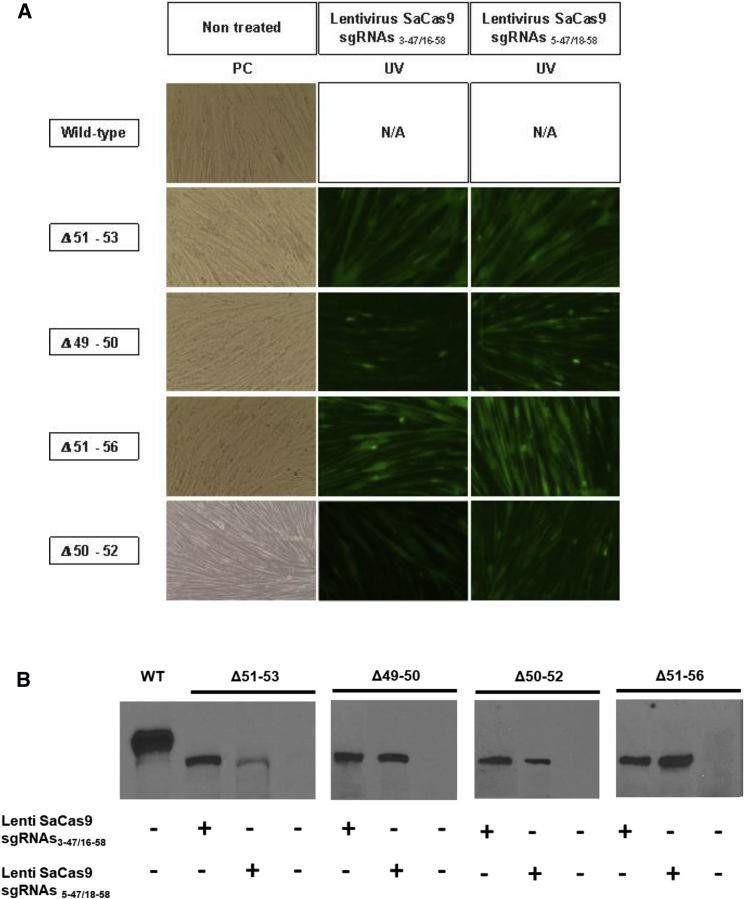
Following confirmation of the formation of the hybrid exon, myoblasts were grown until 100% confluence was reached and allowed to fuse into myotubes for at least 7 days in low-serum medium. We observed that transduced myoblasts were still able to fuse into myotubes because EGFP expression was detected in myotubes (Figure 4A). Next, the restoration of dystrophin expression was investigated with a western blot using an anti-dystrophin antibody (NCL-Dys2, Novocastra) in treated and non-treated myotubes (Figure 4B). As expected, our positive control myoblasts (derived from a healthy child) demonstrated the expression of the wild-type dystrophin (427 kDa), whereas no dystrophin was detected in all myotubes from untreated DMD myoblasts. Interestingly, all myotubes generated from DMD patient myoblasts transduced with Lenti-SaCas93-47/16-58 or with Lenti-SaCas95-47/18-58 were able to express a truncated dystrophin whose size corresponded to the targeted formation of the hybrid exon 47-58 that resulted in an ∼360-kDa dystrophin protein. These results demonstrate that it is possible to create a large genomic deletion to reframe the DMD gene and allow the de novo synthesis of dystrophin.
Figure 4.
Restoration of Dystrophin Expression in Edited Myotubes
(A) Myoblasts from a healthy subject (WT) and from 4 DMD patients with different exon deletions were infected with the lentivirus coding for SaCas9, a pair of sgRNAs, and EGPF and allowed to fuse into myotubes for at least 7 days in low-serum medium. EGFP expression allowed us to observe that myoblasts infected with the lentivirus were able to fuse into myotubes. (B) The myotube proteins were then extracted and submitted to a western blot analysis for detection of the dystrophin protein using an anti-dystrophin monoclonal antibody (NCL-Dys2, Novocastra).
Off-Target Analysis
For analysis of the possible off-target mutations, we focused our study on the best two pairs of sgRNAs (i.e., sgRNAs 3-47 and 16-58 and sgRNAs 5-47 and 18-58). The potential off-target sites were identified with the in silico tools provided on the website https://www.benchling.com/, which calculates an off-target score using an algorithm developed by Hsu et al.19 For each sgRNA, we considered the top 10 high scores of off-targets with the most permissive PAM, meaning NNGRR in addition to NNGRRT. We choose to focus only on off-targets related to annotated genes. For sgRNA 3-47, the highest off-target score identified was 2.5. However, none of the 10 off-targets with the highest scores were related to an annotated gene. sgRNA 5-47 had potential off-targets with scores below 0.8 and no annotated genes. sgRNA 16-58 showed off-target scores ranging from 7.4 to 0.8. Its highest off-target was the IL-17A gene localized in chromosome 6 (position +52190188). Interestingly, the off-target site in the IL-17A gene is localized in its intron 3 after the stop codon. Thus, even if off-target mutations would occur in this localization, they would have no effect on the IL-17A protein, implicated in immune function. In addition, sgRNA 18-58 had a potential off-target site in the TRIM67 gene localized in chromosome 1 (position −231217983), with a score of 1.8. However, the targeted sequence is located after the stop codon of the TRIM67 gene and, thus, would also have no effect on the encoded protein.
To characterize these possible off-targets, we worked with the genomic DNA of non-treated myoblasts and of myoblasts transduced with lentiviral vectors that coded for the expression of SaCas9 and for a pair of sgRNAs. The cells were cultured for 4 weeks. IL-17A and TRIM67 off-target genomic regions were amplified by PCR and submitted to a Surveyor assay (Figure S4A). No difference between untreated and treated myoblasts was observed, meaning that if off-target mutations occurred, then their frequency was below the detection limit of the Surveyor assay, which is 3% to 5% according to the manufacturer’s instructions. The PCR products were also submitted to TIDE analysis (Figure S4B). Despite a slightly higher INDELs rate with sgRNA 16-58 in the IL-17A gene, no significant difference was observed in comparison with control cells (n = 4, Student's t test, p = 0.34). In addition, sgRNA 18-58 did not significantly increased the INDELs rate in the TRIM67 gene (n = 4, Student's t test, p = 0.38).
For a deeper characterization, these possible off-target regions were submitted to deep sequencing analysis (Figure S4C). For the off-target TRIM67, no significant increase in the percentage of INDELs was detected between treated and untreated myoblasts (n = 4, paired t test, p = 0.6). However, the sequences of the off-target sites in the IL-17A gene exhibited a low but significant increase in INDELs at the site targeted by sgRNA 16-58 (n = 4, paired t test, p = 0.018). The physiological effects of these off-target mutations are probably not important because this off-target sequence is not located in an exon coding for the IL-17A protein or in a splice acceptor or a splice donor. Consequently, no physiological effect should be produced by mutation of this off-target site.
Obviously, for any further experiments in human DMD patients, one should consider to more extensively characterize off-target events using high-throughput next-generation sequencing, which should be preferentially evaluated in cells of each patient because of the genomic variations from one human to another.
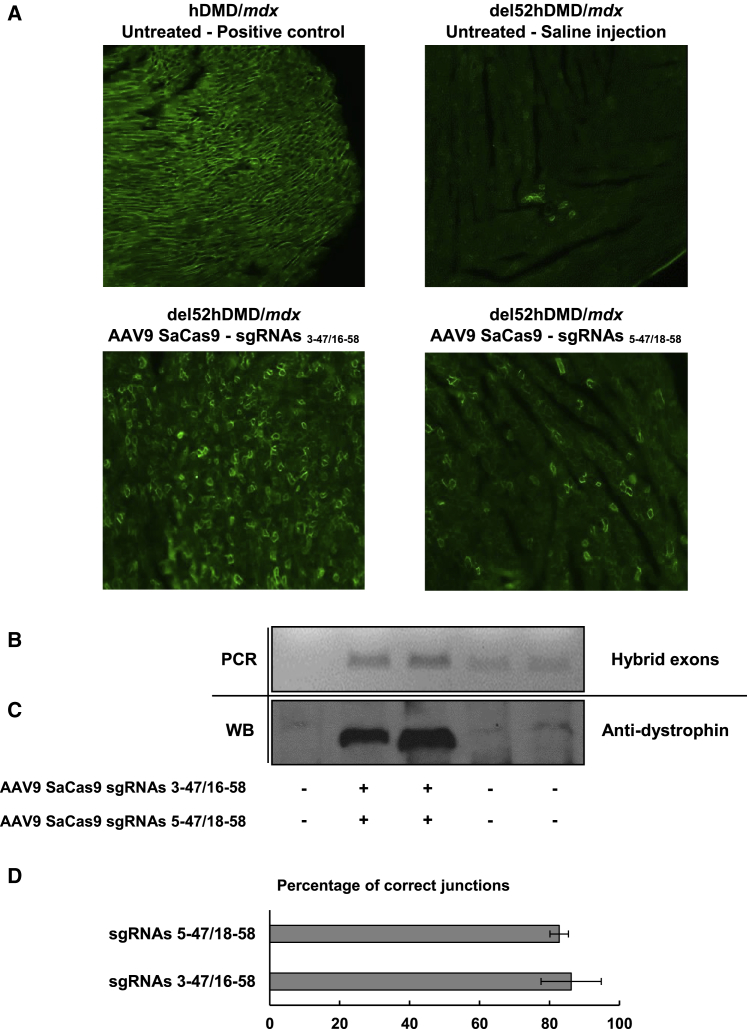
Systemic Delivery of AAV9 Partially Restores Dystrophin Expression in the Heart of del52hDMD/mdx Mice
To assess the in vivo feasibility of our approach, the systemic delivery of the CRISPR/Cas9 components was tested with AAV9 vectors in del52hDMD/mdx mice. Four- to five-week-old mice were injected with a total amount of 7.5 × 1013 vector genomes (vg)/kg; 3.75 × 1013 vg/kg of AAV9-SaCas9 coding for the SaCas9 gene combined with 3.75 × 1013 vg/kg of AAV9-sgRNA3-47/16-58 or AAV9-sgRNA5-47/18-58 coding for the production of a pair of sgRNAs that previously permitted the formation of a hybrid exon 47-58. Mice were sacrificed 6 weeks after the injection to ensure accumulation of dystrophin protein in edited cells. In saline-injected del52hDMD/mdx mice, only a few dystrophin-positive cardiomyocytes were observed by immunohistochemistry. Because our polyclonal antibody rabbit anti-dystrophin detected mouse and human dystrophin, the dystrophin-positive cardiomyocytes may be due to somatic mutations of the human or mouse DMD gene restoring the normal reading frame. This is similar to the so-called revertant skeletal muscle fibers observed in mdx mice.20 In all mice treated with a pair of sgRNAs, either sgRNAs 3-47 and 16-58 or sgRNAs 5-47 and 18-58, dystrophin expression was observed in more cardiomyocytes. Interestingly, a higher dystrophin expression level was observed by immunohistochemistry when the sgRNAs 3-47 and 16-58 were used (Figure 5A). The restoration of dystrophin expression was consistent with the detection of the hybrid exons 47-58 by PCR (Figure 5B). However, such PCR did not permit the detection of hybrid exons in the tibialis anterior or in the diaphragm of the treated del52hDMD/mdx mice. This may be due to a lower amount of viral particles in these tissues (data not shown). TIDE analysis demonstrated that the hybrid exons generated by the sgRNAs 3-47 and 16-58 harbored the expected sequence in 86% of amplicons and that those produced following treatment with sgRNA 5-47 and 18-58 contained 83% of the right nucleotide sequence (Figure 5D). Following protein extraction, western blot analysis confirmed that the de novo expressed dystrophin in the heart corresponded to the size of the internally truncated dystrophin we aimed to create (Figure 5C). Together, these results demonstrate that our approach is suitable for in vivo applications.
Figure 5.
Formation of the Hybrid Exons 3-47/16-58 and 5-47/18-58 Restored the Expression of Dystrophin Protein In Vivo in Cardiomyocytes of the del52hDMD/mdx Mouse Model
4- to 5-week-old del52hDMD/mdx mice were systemically injected in the tail vein with a total amount of 7.5 × 1013 vg/kg of AAV9s coding for SaCas9 and for a pair of sgRNAs (i.e., either sgRNAs 3-47 and 16-58 or sgRNAs 5-47 and 18-58). Mice were euthanized 6 weeks after the injection, and tissues were collected. (A) Heart cryo-sections were stained with a rabbit polyclonal anti-dystrophin antibody (ab15277, Abcam). (B) PCR confirmed formation of the hybrid exons 47-58 in the hearts of treated mice. (C) The presence of the internally truncated dystrophin protein was confirmed by western blot analysis. (D) The amplicon sequences of the hybrid exons were submitted to TIDE analysis to determine the percentage of exact junction (n = 4). The error bars are SE.
Discussion
Over the past 5 years, several CRISPR-based approaches have been investigated for the development of therapies for DMD. Single-cut genome editing relies on the alteration of splice-acceptor sequences to splice out the targeted exon to restore the normal reading frame in the mature mRNA, leading to the production of dystrophin protein.21, 22, 23, 24, 25, 26 Other groups developed strategies that rely on the complete removal of one or several exons to restore the DMD gene reading frame using a pair of sgRNAs inducing DSBs in introns.26, 27, 28 The edition of the dystrophin gene by homology-directed repair (HDR) has also been investigated by treatment of muscle stem cells followed by their engraftment or by direct delivery of nanoparticles formed by a complex between a Cas9/sgRNA and a donor DNA.26, 29, 30 This approach restored the dystrophin gene and full-length dystrophin protein, but it remains limited to in situ treatment only.
Current strategies, which rely on exon skipping or deletion of one or several complete exons, do not consider the structure of the dystrophin protein and the phasing of the SLR that results from splicing out or deletions of these exons. As reported by Nicolas et al,6 the abnormal structure of the SLRs that form the central rod domain of the dystrophin protein could be responsible for the severity and disease progression of BMD. Indeed, dystrophin from some BMD patients harboring large in-frame deletions can still be functional and lead to a milder phenotype.31 Thus, features learned from BMD patients are of major interest for research groups that develop mini- and micro-dystrophin and whose work mainly focuses on the designing of a truncated dystrophin protein, which nevertheless contains an adequate functional structural domain.32 Interestingly, it has been shown that micro-dystrophins having SLRs correctly phased improved their function in comparison with abnormal SLRs.
Several researchers have worked for many years to produce a mini or a micro-dystrophin gene that not only produces a protein but that also codes for a protein with an optimal structure to ensure good function.33, 34, 35, 36, 37, 38, 39, 40 Because an adequate structure is very important for a mini- and a micro-dystrophin gene, the structure of the dystrophin protein produced by a CRISPR/Cas9 modification should also be carefully taken into consideration. In a previous work, we used SpCas9 to induce precise deletion to form the hybrid exon 50-54 through induction of DSBs at precise genomic sites. The main advantage of SaCas9 over SpCas9 is that its smaller size could, in principle, permit insertion of its gene as well as 2 sgRNAs in only one AAV. This great advantage for an eventual clinical application would not only reduce the cost of the treatment, but, moreover, every cell infected by the unique AAV vector would have all components required to produce the hybrid exon. However, the development of such a product requires further development to obtain this all-in-one vector, which requires a promoter smaller than the CMV that allows expression of SaCas9 in skeletal muscle and cardiac cells only, along with the sgRNA promoter that exhibits enough activity.
Consequently, our research group considered that it could be highly relevant to focus on the development of a CRISPR-based therapy that not only restores the DMD gene reading frame but also aims to form a normal SLR that mimics the dystrophin protein present in BMD patients with no severe symptoms. The aim of our project was to provide proof of principle for a therapeutic approach using the CRISPR/SaCas9 technology for DMD patients harboring different mutations in their DMD gene. In a previous publication, our team demonstrated that, through the use of SpCas9, it was possible to form a hybrid exon 50-54 that not only restored the DMD gene normal reading frame but also formed a normally phased SLR, a key component of the central rod domain of the dystrophin protein. However, this strategy was only suitable for DMD patients affected by a mutation of their DMD gene between exons 50 and 54. Based on a similar approach, we aimed, in the present project, to form a hybrid exon that could provide a therapy for a higher percentage of DMD patients. In addition, we chose to design our approach using SaCas9, which is smaller than SpCas9 and could thus be delivered by a single AAV coding for the SaCas9 gene and a pair of sgRNAs.
Previous work had already demonstrated the ability of the SaCas9 nuclease and a pair of sgRNAs to edit the mdx gene in the mdx−/− mouse, a dystrophic model, by completely removing the mutated exon 23 that consequently restored the expression of dystrophin.26, 27, 41 Here we report, for the first time, the use of SaCas9 to edit the human DMD gene in myoblasts from DMD patients and in the humanized dystrophic del52hDMD/mdx mouse model. We initially aimed to identify pairs of sgRNAs that could be suitable for the treatment of DMD patients having a deletion of exon 50. Finally, we identified two pairs of sgRNAs (sgRNAs 3-47 and 16-58 and sgRNAs 5-47 and 18-58) that could be used in a therapy for all DMD patients having a deletion, an insertion, or a point mutation between exons 47 and 58. Note that we purposely did not select sgRNA pairs that would delete exon 44 or 45, which are coding for dystrophin segment binding with nNOS, because this interaction is important for normal dystrophin function and muscle fiber integrity.42, 43 Because DMD gene mutations are responsible for the muscle wasting development of fibrosis that reduces the amount of targetable muscle tissue and, thus, leads to impaired muscle contraction, this therapeutic approach should be primarily used for the treatment of young DMD patients. On the other hand, our selected pairs of sgRNAs could also be used to treat BMD patients who exhibit a severe phenotype and, thus, could be of interest for the structural improvement of the dystrophin of Becker patient with a severe phenotype. The potential of this strategy should be explored, but, to date, no mouse model harboring the severe Becker phenotype exists; thus, this precludes study of the benefits of improvement of the structure of the dystrophin protein in such patients. The sgRNA pair 3-47/16-58 seems the most promising because it allowed restoration of dystrophin protein expression more efficiently than the sgRNA 5-47/18-58 pair. sgRNA 16-58 could lead to an off-target mutation in the IL-17A gene, which could be problematic in DMD patients. Even if this off-target site is located within a non-coding segment of the IL-17A gene, strategies to limit possible adverse effects would have to be considered. A possible strategy would be to use a skeletal muscle- and cardiac tissue-specific promoter, such as the CK8e promoter developed by Amoasii et al.,21 Bengtsson et al.,26 and Martari et al.,44 which restricts the expression of the nuclease to tissues where editing is required to improve the DMD patient phenotype. In addition, the development of tools that prevent the sustained expression of CRISPR nucleases is required to restrain nuclease expression to the minimal time period required for sufficient editing of the target gene45, 46 while limiting the occurrence of off-target mutations. Here we empirically assessed the potential off-targets of our sgRNAs based on an in silico prediction tool. This tool, and other similar ones, identify off-targets based on the position of one or several mismatches in the sgRNA,19, 47 but several articles have reported that the sites of predicted off-target mutations are not always reliable.48 Ultimately, a pre-clinical study should characterize more extensively the overall off-target mutations of sgRNA 3-47 and 16-58 using unbiased high-throughput next-generation sequencing.49, 50, 51
Our follow-up study will aim to improve overall DMD gene editing in vivo in young del52hDMD/mdx mice to restore dystrophin protein expression in the diaphragm and skeletal muscle in addition to the heart. We will also assess whether the formation of hybrid exons, such as the hybrid exon 47-58, which maintains the SLR structure, results in a more substantial functional improvement in comparison with the editing of the DMD gene by the removing one or several full exons without maintaining the correct phasing of the SLR.
Materials and Methods
Identification of DNA Targets and sgRNAs
For identification of the sgRNA targets, we used the website https://www.benchling.com/. Based on the DMD gene sequence (ENSG00000198947), we identified all NNGRRT PAMs for the SaCas9 from exons 46 to 58. Oligonucleotides corresponding to the sgRNAs of interest were ordered from Integrated DNA Technologies (Coralville, IA).
Expression Vector and Cloning of sgRNAs
Selected sgRNAs were cloned into the plasmid pX601-AAV-CMV::NLS-SaCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA (PX601) (Addgene, 61591) following Zhang’s laboratory protocol. The plasmid PX601 was linearized using the BsaI restriction enzyme (New England Biolabs, Ipswich, MA), followed by purification with a gel extraction kit (Thermo Scientific, Vilnius, Lithuania). The sgRNAs of interest were then cloned into the BsaI site using the Quickligase (NEB, Ipswich, MA), followed by transformation in Top10-competent bacteria. In the plasmid pBSU6_FE_ScaffoldRSV_GFP (a gift from the Dirk Grimm Lab, Heidelberg University, Germany), oligonucleotides were sequentially cloned into the BsmbI or BbsI restriction sites. Clones were amplified in liquid Luria-Bertani (LB), and, following plasmid DNA extraction, clones were sequenced. The sequencing results were analyzed using CLC Sequence Viewer (CLC Bio).
Transfection Procedure
293T cells were grown under 5% CO2 at 37°C with DMEM supplemented with 10% fetal bovine serum (FBS) and 1% penicillin and streptomycin. The initial experiments to assess the activity of sgRNAs were made in 293T cells. A total amount of 900 ng of plasmid was transfected into cells using 3 μL of Lipofectamine 2000. 48 hr following transfection, cells were collected, and genomic DNA was extracted, purified using the phenol:chloroform method, and concentrated by salt and ethanol precipitation.
Genomic DNA Extraction and Analysis
48 hr after transfection of the pX601-AAV-CMV:NLS-SaCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA plasmid(s), the genomic DNA was extracted from 293T cells or myoblasts using a standard phenol:chloroform method. Briefly, the cell pellet was suspended in 100 μL of lysis buffer containing 10% sarcosyl and 0.5 M (pH 8) EDTA. 20 μL of proteinase K (10 mg/mL) was added. The suspension was mixed by aspirating and releasing and then incubated for 10–15 min at 55°C. The suspension was then centrifuged at 13,200 rpm for 5 min. The supernatant was collected in a new microfuge tube. One volume of phenol-chloroform was added, and, following centrifugation, the aqueous phase was recovered in a new microfuge tube. Then DNA was precipitated using a 1/10 volume of 5 M NaCl and 2 volumes of 100% ethanol, followed by 5 min centrifugation at 13,200 rpm. The pellet was washed with 70% ethanol and centrifuged, and the DNA was suspended in double-distilled water. The genomic DNA concentration was assayed with a Nanodrop spectrophotometer (Thermo Scientific, Logan, UT).
Assessment of the Formation by sgRNAs of INDELs on On-Targets and Off-Targets
Genomic regions targeted by a sgRNA (either on-target or off-target sequences) were amplified by PCR before further analysis. First, cleavage efficiency was assessed by Surveyor assay according to the manufacturer’s instructions. PCR products were then sequenced, and the resulting .ab files were used for their characterization using the TIDE tool provided at https://tide.deskgen.com.52 Amplifications of genomic DNA from untreated cells were used as control samples. For deep sequencing analysis, new sets of primers were designed to permit the amplification of PCR products no longer than 350 bp.
Assessment of the Formation of Hybrid Exons
For the characterization of the nucleotide sequence of the hybrid exons, PCR products corresponding to the amplification of these hybrid exons were cloned into the pMiniT plasmid vector of a PCR cloning kit following the manufacturer’s instructions (NEB, Ipswich, MA). After transformation into bacteria and clone growth in a liquid medium, plasmidic DNA was extracted and purified for sequencing.
The TIDE analysis tool was used to characterize of the rate of formation of perfect hybrid exons. As a control sample, hybrid exons that contained the exact nucleotide sequence, previously inserted in a pMiniT vector and sequenced, were amplified by PCR and used as a control sample for the analysis. As a test sample, pools of PCR products amplified from treated cells were used.
Lentivirus Production
A lentiviral vector that allows for the expression of SaCas9 along with 2 sgRNAs was constructed, starting from the plasmid FUGW (Addgene, 14883). First, the SaCas9 gene and a T2A-EGFP were inserted under the control of the CMV promoter. Two U6 promoters were then inserted to control the expression of two sgRNAs. A restriction site, either BbsI or BsmbI, was inserted downstream of each U6 promoter to permit the cloning of each individual sgRNA. For each lentivirus production, 4 × 106 293T cells (a packaging cell line) were plated into a 10 × 10 cm Petri dish one day prior to transfection. On the day of transfection, cells were transfected with a combination of four plasmids (Gag-Pol, Rev, vesicular stomatitis virus G protein [VSV-G], and the transfer vector) using a calcium phosphate transfection method. On the day after transfection, the medium was removed and replaced with fresh medium. 48 hr later, the medium containing the viral particles was collected. Following centrifugation to remove cell debris, the viral supernatant was concentrated and purified by ultracentrifugation at 15,000 × g for 90 min in a 20% sucrose gradient. Following supernatant removal, viral particles were suspended into cold 1× PBS.
Myoblast Transduction with the Recombinant Lentiviral Vector
Myoblasts from different DMD patients (harboring a deletion of exons 49 to 50, 50 to 52, 51 to 53, or 51 to 56) were grown under 5% CO2 at 37°C in MB-1 medium supplemented with 40 ng/mL of basic fibroblast growth factor (bFGF) and 15% FBS. For viral transduction, myoblasts were treated overnight in MB-1 medium supplemented with 8 μg/mL of Polybrene with the previously purified lentivirus. On the day after transduction, cells were washed using 1× PBS and cultured in Polybrene-free MB-1 medium. Transduction efficiency was assessed 48 hr after medium replacement by observing EGFP fluorescence.
Western Blot Analysis
After the fusion of myoblasts into myotubes, proteins were extracted using a protein lysis buffer (75 mM Tris-HCl [pH 8.0], 1 mM DTT, 1 mM PMSF, and 1% SDS). The protein concentrations were determined using an amido black assay.53 Protein samples were separated onto a 6% SDS-PAGE gel. Proteins were then transferred overnight onto a nitrocellulose blotting membrane. That membrane was blocked for 8 hr in 5% milk in 1× Tris-buffered saline (TBS) and 0.01% Tween 20 at 4°C. The primary antibody anti-dystrophin (dilution 1:50, NCL-Dys2; Novocastra, Newcastle, UK) was incubated overnight at 4°C. The secondary antibody goat anti-mouse horseradish peroxidase (HRP) (dilution 1:5,000) was applied for 1 hr. Blots were revealed using Clarity Western enhanced chemiluminescence (ECL) blotting substrates (Bio-Rad, Mississauga, ON, Canada).
Production of AAV9 Viral Vectors
All viral vectors were produced by the Molecular Tools Platform of Centre de Recherche de l'Institut Universitaire en Santé Mentale de Québec (CRIUSMQ, Québec, Canada). The AAV9 vector that encodes the SaCas9 gene under the control of the CMV promoter was produced from the plasmid PX601 (Addgene, Cambridge, MA). Two AAV9 vectors that permitted the expression of a pair of sgRNAs were produced from the plasmid pBSU6 (pBSU6_FE_Scaffold_RSV_GFP, Dirk Grimm lab, Heidelberg University, Germany).
In Vivo Experiments
Three AAV9 vectors were designed to determine the in vivo feasibility of the formation of the hybrid exons 47-58. One AAV9 permitted the expression of the SaCas9 protein under the control of the CMV promoter. The two other AAV9s allowed the production of two pairs of sgRNAs: one vector encoded sgRNA 3-47 (targeting exon 47) and sgRNA 16-58 (targeting exon 58), and the other vector encoded sgRNA 5-47 (targeting exon 47) and sgRNA 18-58 (targeting exon 58). The sgRNA expression was controlled by a human pU6 promoter. The virus that coded for the pairs of sgRNAs also coded for the expression of GFP. Six weeks after the intravenous injection, mice were sacrificed, and tissues (heart, diaphragm, tibialis anterior, and liver) were collected and incubated overnight in 30% sucrose at 4°C. Then samples were embedded into cryomatrix and flash-frozen. Samples were stored at −80°C.
Targeted Deep Sequencing and Bioinformatics Analysis
For the deep sequencing analysis, primers were redesigned to allow the amplification of PCR products of a maximum of 350 bp. Samples were sent for sequencing to the Next-Generation Sequencing Platform of McGill University (McGill University and Genome Quebec Innovation Centre, Montreal, Canada). Fastq file quality was checked using the FastQC tool (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Before characterization of the sequencing results, Fastq files were trimmed by sliding window (SLIDINGWINDOW:4:15) and minimum length (according to the expected size of the amplicons) using the algorithm Trimmomatic.54
Molecular Modeling
Homology models of the WT DMD gene corresponding to SLRs R18 and R23 were obtained from the eDystrophin website (http://edystrophin.genouest.org/).18 Primary sequence alignment was realized using the TM-Align55 (https://zhanglab.ccmb.med.umich.edu/TM-align/) web server based on the structural alignment of the R19 and R21 homology models. The homology models were realized using the iTasser web server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/)56, 57, 58, 59 using primary sequences of the hybrid exon for deletion of exons 47–58.
hDMD/mdx and del52hDMD/mdx Mouse Models
The hDMD/mdx mouse (a gift from Dr. A. Aartsma-Rus) contains the full human DMD gene (2.4 Mb) (coding for dystrophin) stably integrated into mouse chromosome 5.60 In addition, the mouse dystrophin gene (the mdx gene) contains a point mutation in exon 23 that creates a stop codon, thus abrogating the production of mouse dystrophin. del52hDMD/mdx (a gift from Dr. A. Aartsma-Rus) also contains the human DMD gene but harbors a deletion of exon 52 that abrogates synthesis of the human dystrophin protein.61 All experiments were approved by the animal care committee of the Centre Hospitalier de l’Université Laval.
Immunohistochemistry
Frozen tissues were processed with a Cryostat (Leica Biosystems, Concordia, ON, Canada) to obtain 12-μm-thick transversal sections that were placed on a glass slide pre-coated with glycine. Tissue sections were blocked for 1 hr in a blocking solution (1× PBS supplemented with 10% FBS. The primary antibody directed against dystrophin (ab15277, Abcam, Toronto, ON, Canada) diluted in the blocking solution was incubated for 1 hr. After 3 washing steps of 10 min with 1× PBS, a secondary antibody (i.e., goat anti-rabbit immunoglobulin G [IgG] Alexa Fluor 488; A-11008, RRID AB_143165, Thermo Fisher Scientific) diluted in the blocking solution was then added for another 1 hr. Next, samples were washed 3 times for 10 min with 1× PBS, and then stained samples were mounted under a covered glass slide with 1× PBS supplemented with 50% glycerol. Dystrophin expression and localization were observed by fluorescence microscopy.
Author Contributions
B.L.D. designed the experiments, performed the experiments, and wrote the manuscript. K.C. provided technical assistance for the western blots. J.-P.I.-E. assisted with the design of the experiments. J.R. provided technical assistance for molecular biology. D.L.O. provided technical assistance for lentiviral vector production. P.L. and X.B. made the model of the structure of spectrin-like repeats. J.P.T. conceived the experiments and corrected the manuscript.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
We thank Dr. Aartmus for providing the del52hDMD/mdx mouse model. This research project was supported by grants from the Canadian Institute of Health Research grant number 271774, the Foundation for Cell and Gene Therapy, and The Cell Network. B.L.D. has been supported by a fellowship from MITACs.
Footnotes
Supplemental Information includes four figures and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.08.010.
Supplemental Information
References
- 1.Emery A.E. Population frequencies of inherited neuromuscular diseases--a world survey. Neuromuscul. Disord. 1991;1:19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 2.Emery A.E. The muscular dystrophies. Lancet. 2002;359:687–695. doi: 10.1016/S0140-6736(02)07815-7. [DOI] [PubMed] [Google Scholar]
- 3.Muntoni F., Torelli S., Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/s1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 4.Bladen C.L., Salgado D., Monges S., Foncuberta M.E., Kekou K., Kosma K., Dawkins H., Lamont L., Roy A.J., Chamova T. The TREAT-NMD DMD Global Database: analysis of more than 7,000 Duchenne muscular dystrophy mutations. Hum. Mutat. 2015;36:395–402. doi: 10.1002/humu.22758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rybakova I.N., Patel J.R., Ervasti J.M. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J. Cell Biol. 2000;150:1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nicolas A., Raguénès-Nicol C., Ben Yaou R., Ameziane-Le Hir S., Chéron A., Vié V., Claustres M., Leturcq F., Delalande O., Hubert J.F., French Network of Clinical Reference Centres for Neuromuscular Diseases (CORNEMUS) Becker muscular dystrophy severity is linked to the structure of dystrophin. Hum. Mol. Genet. 2015;24:1267–1279. doi: 10.1093/hmg/ddu537. [DOI] [PubMed] [Google Scholar]
- 7.Yue Y., Pan X., Hakim C.H., Kodippili K., Zhang K., Shin J.H., Yang H.T., McDonald T., Duan D. Safe and bodywide muscle transduction in young adult Duchenne muscular dystrophy dogs with adeno-associated virus. Hum. Mol. Genet. 2015;24:5880–5890. doi: 10.1093/hmg/ddv310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nelson D.M., Lindsay A., Judge L.M., Duan D., Chamberlain J.S., Lowe D.A., Ervasti J.M. Variable rescue of microtubule and physiological phenotypes in mdx muscle expressing different miniaturized dystrophins. Hum. Mol. Genet. 2018 doi: 10.1093/hmg/ddy209. Published online June 12, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Guiner C., Servais L., Montus M., Larcher T., Fraysse B., Moullec S., Allais M., François V., Dutilleul M., Malerba A. Long-term microdystrophin gene therapy is effective in a canine model of Duchenne muscular dystrophy. Nat. Commun. 2017;8:16105. doi: 10.1038/ncomms16105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ran F.A., Cong L., Yan W.X., Scott D.A., Gootenberg J.S., Kriz A.J., Zetsche B., Shalem O., Wu X., Makarova K.S. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iyombe-Engembe J.P., Ouellet D.L., Barbeau X., Rousseau J., Chapdelaine P., Lagüe P., Tremblay J.P. Efficient Restoration of the Dystrophin Gene Reading Frame and Protein Structure in DMD Myoblasts Using the CinDel Method. Mol. Ther. Nucleic Acids. 2016;5:e283. doi: 10.1038/mtna.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duchêne B., Iyombe-Engembe J.P., Rousseau J., Tremblay J.P., Ouellet D.L. From gRNA Identification to the Restoration of Dystrophin Expression: A Dystrophin Gene Correction Strategy for Duchenne Muscular Dystrophy Mutations Using the CRISPR-Induced Deletion Method. Methods Mol. Biol. 2018;1687:267–283. doi: 10.1007/978-1-4939-7374-3_19. [DOI] [PubMed] [Google Scholar]
- 16.Molza A.E., Mangat K., Le Rumeur E., Hubert J.F., Menhart N., Delalande O. Structural Basis of Neuronal Nitric-oxide Synthase Interaction with Dystrophin Repeats 16 and 17. J. Biol. Chem. 2015;290:29531–29541. doi: 10.1074/jbc.M115.680660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedland A.E., Baral R., Singhal P., Loveluck K., Shen S., Sanchez M., Marco E., Gotta G.M., Maeder M.L., Kennedy E.M. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolas A., Lucchetti-Miganeh C., Yaou R.B., Kaplan J.C., Chelly J., Leturcq F., Barloy-Hubler F., Le Rumeur E. Assessment of the structural and functional impact of in-frame mutations of the DMD gene, using the tools included in the eDystrophin online database. Orphanet J. Rare Dis. 2012;7:45. doi: 10.1186/1750-1172-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman E.P., Morgan J.E., Watkins S.C., Partridge T.A. Somatic reversion/suppression of the mouse mdx phenotype in vivo. J. Neurol. Sci. 1990;99:9–25. doi: 10.1016/0022-510x(90)90195-s. [DOI] [PubMed] [Google Scholar]
- 21.Amoasii L., Long C., Li H., Mireault A.A., Shelton J.M., Sanchez-Ortiz E., McAnally J.R., Bhattacharyya S., Schmidt F., Grimm D. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aan8081. eaan8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Long C., Amoasii L., Mireault A.A., McAnally J.R., Li H., Sanchez-Ortiz E., Bhattacharyya S., Shelton J.M., Bassel-Duby R., Olson E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long C., Li H., Tiburcy M., Rodriguez-Caycedo C., Kyrychenko V., Zhou H., Zhang Y., Min Y.L., Shelton J.M., Mammen P.P.A. Correction of diverse muscular dystrophy mutations in human engineered heart muscle by single-site genome editing. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aap9004. eaap9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long C., McAnally J.R., Shelton J.M., Mireault A.A., Bassel-Duby R., Olson E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y., Long C., Li H., McAnally J.R., Baskin K.K., Shelton J.M., Bassel-Duby R., Olson E.N. CRISPR-Cpf1 correction of muscular dystrophy mutations in human cardiomyocytes and mice. Sci. Adv. 2017;3:e1602814. doi: 10.1126/sciadv.1602814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bengtsson N.E., Hall J.K., Odom G.L., Phelps M.P., Andrus C.R., Hawkins R.D., Hauschka S.D., Chamberlain J.R., Chamberlain J.S. Muscle-specific CRISPR/Cas9 dystrophin gene editing ameliorates pathophysiology in a mouse model for Duchenne muscular dystrophy. Nat. Commun. 2017;8:14454. doi: 10.1038/ncomms14454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson C.E., Hakim C.H., Ousterout D.G., Thakore P.I., Moreb E.A., Castellanos Rivera R.M., Madhavan S., Pan X., Ran F.A., Yan W.X. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ousterout D.G., Kabadi A.M., Thakore P.I., Majoros W.H., Reddy T.E., Gersbach C.A. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu P., Wu F., Mosenson J., Zhang H., He T.C., Wu W.S. CRISPR/Cas9-Mediated Genome Editing Corrects Dystrophin Mutation in Skeletal Muscle Stem Cells in a Mouse Model of Muscle Dystrophy. Mol. Ther. Nucleic Acids. 2017;7:31–41. doi: 10.1016/j.omtn.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee K., Conboy M., Park H.M., Jiang F., Kim H.J., Dewitt M.A., Mackley V.A., Chang K., Rao A., Skinner C. Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nat. Biomed. Eng. 2017;1:889–901. doi: 10.1038/s41551-017-0137-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yazaki M., Yoshida K., Nakamura A., Koyama J., Nanba T., Ohori N., Ikeda S. Clinical characteristics of aged Becker muscular dystrophy patients with onset after 30 years. Eur. Neurol. 1999;42:145–149. doi: 10.1159/000008089. [DOI] [PubMed] [Google Scholar]
- 32.Harper S.Q., Hauser M.A., DelloRusso C., Duan D., Crawford R.W., Phelps S.F., Harper H.A., Robinson A.S., Engelhardt J.F., Brooks S.V., Chamberlain J.S. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat. Med. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y., Duan D. Novel mini-dystrophin gene dual adeno-associated virus vectors restore neuronal nitric oxide synthase expression at the sarcolemma. Hum. Gene Ther. 2012;23:98–103. doi: 10.1089/hum.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li S., Kimura E., Ng R., Fall B.M., Meuse L., Reyes M., Faulkner J.A., Chamberlain J.S. A highly functional mini-dystrophin/GFP fusion gene for cell and gene therapy studies of Duchenne muscular dystrophy. Hum. Mol. Genet. 2006;15:1610–1622. doi: 10.1093/hmg/ddl082. [DOI] [PubMed] [Google Scholar]
- 35.Harper S.Q., Crawford R.W., DelloRusso C., Chamberlain J.S. Spectrin-like repeats from dystrophin and alpha-actinin-2 are not functionally interchangeable. Hum. Mol. Genet. 2002;11:1807–1815. doi: 10.1093/hmg/11.16.1807. [DOI] [PubMed] [Google Scholar]
- 36.Yuasa K., Sakamoto M., Miyagoe-Suzuki Y., Tanouchi A., Yamamoto H., Li J., Chamberlain J.S., Xiao X., Takeda S. Adeno-associated virus vector-mediated gene transfer into dystrophin-deficient skeletal muscles evokes enhanced immune response against the transgene product. Gene Ther. 2002;9:1576–1588. doi: 10.1038/sj.gt.3301829. [DOI] [PubMed] [Google Scholar]
- 37.Draviam R.A., Wang B., Li J., Xiao X., Watkins S.C. Mini-dystrophin efficiently incorporates into the dystrophin protein complex in living cells. J. Muscle Res. Cell Motil. 2006;27:53–67. doi: 10.1007/s10974-006-9055-8. [DOI] [PubMed] [Google Scholar]
- 38.Bostick B., Yue Y., Long C., Marschalk N., Fine D.M., Chen J., Duan D. Cardiac expression of a mini-dystrophin that normalizes skeletal muscle force only partially restores heart function in aged Mdx mice. Mol. Ther. 2009;17:253–261. doi: 10.1038/mt.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramos J., Chamberlain J.S. Gene Therapy for Duchenne muscular dystrophy. Expert Opin. Orphan Drugs. 2015;3:1255–1266. doi: 10.1517/21678707.2015.1088780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lai Y., Thomas G.D., Yue Y., Yang H.T., Li D., Long C., Judge L., Bostick B., Chamberlain J.S., Terjung R.L., Duan D. Dystrophins carrying spectrin-like repeats 16 and 17 anchor nNOS to the sarcolemma and enhance exercise performance in a mouse model of muscular dystrophy. J. Clin. Invest. 2009;119:624–635. doi: 10.1172/JCI36612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El Refaey M., Xu L., Gao Y., Canan B.D., Adesanya T.M.A., Warner S.C., Akagi K., Symer D.E., Mohler P.J., Ma J. In Vivo Genome Editing Restores Dystrophin Expression and Cardiac Function in Dystrophic Mice. Circ. Res. 2017;121:923–929. doi: 10.1161/CIRCRESAHA.117.310996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sander M., Chavoshan B., Harris S.A., Iannaccone S.T., Stull J.T., Thomas G.D., Victor R.G. Functional muscle ischemia in neuronal nitric oxide synthase-deficient skeletal muscle of children with Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA. 2000;97:13818–13823. doi: 10.1073/pnas.250379497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stamler J.S., Meissner G. Physiology of nitric oxide in skeletal muscle. Physiol. Rev. 2001;81:209–237. doi: 10.1152/physrev.2001.81.1.209. [DOI] [PubMed] [Google Scholar]
- 44.Martari M., Sagazio A., Mohamadi A., Nguyen Q., Hauschka S.D., Kim E., Salvatori R. Partial rescue of growth failure in growth hormone (GH)-deficient mice by a single injection of a double-stranded adeno-associated viral vector expressing the GH gene driven by a muscle-specific regulatory cassette. Hum. Gene Ther. 2009;20:759–766. doi: 10.1089/hum.2008.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen Y., Liu X., Zhang Y., Wang H., Ying H., Liu M., Li D., Lui K.O., Ding Q. A Self-restricted CRISPR System to Reduce Off-target Effects. Mol. Ther. 2016;24:1508–1510. doi: 10.1038/mt.2016.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai X., Chen X., Fang Q., Li J., Bai Z. Inducible CRISPR genome-editing tool: classifications and future trends. Crit. Rev. Biotechnol. 2018;38:573–586. doi: 10.1080/07388551.2017.1378999. [DOI] [PubMed] [Google Scholar]
- 47.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin F., Sánchez-Hernández S., Gutiérrez-Guerrero A., Pinedo-Gomez J., Benabdellah K. Biased and Unbiased Methods for the Detection of Off-Target Cleavage by CRISPR/Cas9: An Overview. Int. J. Mol. Sci. 2016;17:E1507. doi: 10.3390/ijms17091507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X., Wang Y., Wu X., Wang J., Wang Y., Qiu Z., Chang T., Huang H., Lin R.J., Yee J.K. Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol. 2015;33:175–178. doi: 10.1038/nbt.3127. [DOI] [PubMed] [Google Scholar]
- 50.Kim D., Bae S., Park J., Kim E., Kim S., Yu H.R., Hwang J., Kim J.I., Kim J.S. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods. 2015;12:237–243. doi: 10.1038/nmeth.3284. 1 p following 243. [DOI] [PubMed] [Google Scholar]
- 51.Tsai S.Q., Nguyen N.T., Malagon-Lopez J., Topkar V.V., Aryee M.J., Joung J.K. CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods. 2017;14:607–614. doi: 10.1038/nmeth.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brinkman E.K., Chen T., Amendola M., van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chapdelaine P., Vignola K., Fortier M.A. Protein estimation directly from SDS-PAGE loading buffer for standardization of samples from cell lysates or tissue homogenates before Western blot analysis. Biotechniques. 2001;31:478. doi: 10.2144/01313bm04. 480, 482. [DOI] [PubMed] [Google Scholar]
- 54.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang J., Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv342. W174–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J., Yan R., Roy A., Xu D., Poisson J., Zhang Y. The I-TASSER Suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.‘t Hoen P.A., de Meijer E.J., Boer J.M., Vossen R.H., Turk R., Maatman R.G., Davies K.E., van Ommen G.J., van Deutekom J.C., den Dunnen J.T. Generation and characterization of transgenic mice with the full-length human DMD gene. J. Biol. Chem. 2008;283:5899–5907. doi: 10.1074/jbc.M709410200. [DOI] [PubMed] [Google Scholar]
- 61.Veltrop M., van Vliet L., Hulsker M., Claassens J., Brouwers C., Breukel C., van der Kaa J., Linssen M.M., den Dunnen J.T., Verbeek S. A dystrophic Duchenne mouse model for testing human antisense oligonucleotides. PLoS ONE. 2018;13:e0193289. doi: 10.1371/journal.pone.0193289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.