Abstract
Intramembrane proteases hydrolyze peptide bonds within the membrane as a regulatory paradigm that is conserved across all forms of cellular life. Many of these enzymes are thought to be oligomeric, and that their resulting quaternary interactions form the basis of their regulation. However, technical limitations have precluded directly determining the oligomeric state of intramembrane proteases in any membrane. Using single-molecule photobleaching, we determined the quaternary structure of 10 different rhomboid proteins (the largest superfamily of intramembrane proteases) and six unrelated control proteins in parallel detergent micelle, planar supported lipid bilayer, and whole-cell systems. Bacterial, parasitic, insect, and human rhomboid proteases and inactive rhomboid pseudoproteases all proved to be monomeric in all membrane conditions but dimeric in detergent micelles. These analyses establish that rhomboid proteins are, as a strict family rule, structurally and functionally monomeric by nature and that rhomboid dimers are unphysiological.
Introduction
Rhomboid proteases are multispanning helical membrane proteins that form a serine protease active site within a membrane bilayer (1). Rhomboid-catalyzed hydrolysis of peptides bonds releases target effector domains from the membrane as the committed step of various cell signaling pathways (2).
Classic genetic screens were first to identify rhomboid in Drosophila melanogaster as a gene necessary to relay information during embryogenesis (2). Subsequent studies revealed rhomboid to be a serine protease that activates signaling by liberating transmembrane epidermal growth factor ligand precursors (1). Under physiological conditions, rhomboid proteolysis is essential and constitutes the only limiting step for EGFR (Epidermal Growth Factor Receptor) pathway activation. In the mitochondria of yeast, a rhomboid protease regulates mitochondrial fusion, whereas rhomboid cleavage in mammals is central to a mitochondrial quality control surveillance pathway that, when dysregulated, leads to Parkinson’s disease (3). Even inactive rhomboid pseudoproteases, which have lost their catalytic residues, regulate the major cell surface sheddase ADAM17 (4, 5).
Rhomboid proteases also play central roles in unicellular organisms. Parasitic microbes deploy their rhomboid proteases to regulate adhesion to host cells: the malaria parasite uses a rhomboid protease to sever contacts with the host plasma membrane at the end of the invasion program, whereas noninvasive parasites like amoebae and trichomonads untether themselves from host epithelia through rhomboid proteolysis (6). Prokaryotic rhomboid proteases are particularly widespread, and in at least one bacterial pathogen, they facilitate cell-to-cell signaling by regulating the assembly of the twin-arginine translocon during quorum sensing (7).
Because rhomboid proteolysis is irreversible in the cell and constitutes the decisive step of key cellular processes, it has long been a mystery how rhomboid enzymes themselves are subject to regulation directly at the protein level (8). Membrane proteins commonly exist as oligomers, and their regulation has often been traced to changes in their oligomeric state. Accordingly, it has been proposed that rhomboid proteins are dimeric (9, 10, 11, 12) and that their dimeric state is important for consolidating their stability (9, 12), increasing substrate specificity (10, 12, 13), and/or regulating activity against transmembrane substrates (11, 14). However, studies of rhomboid oligomerization rely on detergents, which dissolve the membrane; assessing the oligomeric state of any intramembrane protease directly in an intact membrane bilayer has eluded experimental scrutiny.
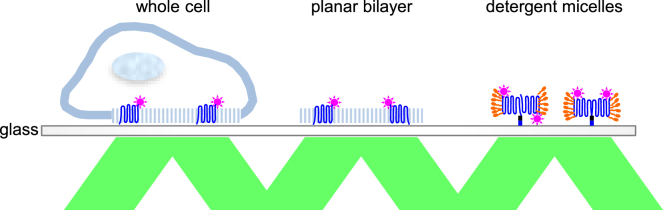
We leveraged methods for visualizing and quantifying single molecules (15, 16) to investigate the oligomeric state of rhomboid proteins directly in cells, membrane bilayers, and detergent micelles in parallel (Fig. 1). All 10 diverse rhomboid proteins that we examined were strict monomers in cells and in all membrane bilayer systems of varied composition but assumed a dimeric state in commonly used detergent micelle systems.
Figure 1.
Parallel strategy for assessing the oligomeric state of membrane proteins in whole cells, planar bilayers, and detergent micelles. Membrane proteins are diagrammed in blue, attached fluorophores are depicted as pink stars, and the detergent molecules are drawn in orange. Green parallelograms depict the incident and reflected laser light that is used to generate the evanescent excitation wave by total internal reflection fluorescence microscopy. To see this figure in color, go online.
Materials and Methods
DNA constructs
To generate Halo-tagged expression constructs, bacterial open reading frames of LacY, MsbA, GlpF, AqpZ, and GlpG were cloned into a pET21 vector containing a C-terminal Halo-His tag. DmRho4 was cloned into a pET15 vector with an N-terminal His-Halo tag. GlpG and HiGlpG were cloned into pET15, and a single N-terminal cysteine (NCys) or a C-terminal cysteine (CysC) was introduced by site-directed mutagenesis. Entire open reading frames were sequenced for all plasmids.
Protein expression and purification
To express the desired proteins, plasmids were first transformed into BL21(DE3) cells and grown at 37°C in 2 L of LB media with 100 μg/mL ampicillin while shaking at 250 rotations per minute (rpm). When the OD600 was between 0.6 and 0.7, protein expression was induced with 100 μM isopropyl-β-D-1-thiogalactopyranoside, temperature was reduced to 16°C, and cultures were allowed to grow overnight. Cells were pelleted in an Avanti JA26 XPI high-speed centrifuge (Beckman, Palo Alto, CA) at 6000 rpm, resuspended in PBS (10 mL per L of bacteria cell growth) containing a protease inhibitor cocktail (Roche, Basel, Switzerland), and lysed in a M-110A microfluidizer (Microfluidics, Westwood, MA). Cell lysates were subjected to ultracentrifugation at 60,000 rpm in a 70Ti rotor (Beckman) to pellet membranes, which were then resuspended in phosphate-buffered saline (PBS) and incubated with 2% dodecyl-β-D-maltoside (DDM) overnight. Ultracentrifugation (60,000 rpm for 30 min in a 70Ti) was used to pellet insoluble material, and the clarified supernatant was incubated with complete His-tag resin (Roche) for ∼2 h at 4°C. Resin was washed extensively, and proteins were eluted with 500 mM imidazole. Protein purity was assessed by staining proteins resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis with the BandIt protein stain and visualized with a 700 nm laser on an Odyssey infrared imager (LiCor Biosciences, Lincoln, NE). NHalo-DmRho4 was expressed in BL21(DE3) cells cultured in M9+ minimal media as described previously (17) and then purified in the same manner as described above for bacterial proteins. After purification of GlpG and HiGlpG, C- and N-terminal cysteines were labeled by incubating pure protein overnight with an equal concentration of JF549 maleimide. The next day, 10 mM dithiothreitol was added to stop the reaction, and the protein was purified on a G10 mini column.
Planar supported lipid bilayer formation
35 mm round No 1.5 glass coverslips (Warner Instruments, Hamden, CT), were cleaned by dipping in 3:1 sulfuric acid/H2O2 for 10 min and then rinsed using purified water. Proteoliposomes containing each reconstituted Halo-tagged protein were made as described previously (18) with minor modifications. 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 850757; 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, 840457; 1,2-dimyristoyl-sn-glycero-3-phosphoglycerol, 850445; 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine, 850745; and polar lipid extracts from Escherichia coli (100600) and yeast (190001) were purchased from Avanti Polar Lipids (Alabaster, AL). Desired proportions of lipids in chloroform were mixed in a pear-shaped flask, chloroform was removed with a rotary evaporator, and the resulting lipid films were placed under high vacuum overnight. Lipid films were resuspended in desired buffer and frozen and thawed five times using dry ice and a water bath, and liposomes were then extruded through a 100 nm NanoSizer extruder (T&T Scientific, Knoxville, TN). Liposomes were mixed with purified protein in detergent (0.2% DDM) to achieve a lipid/protein ratio of 1 million to 1. After 15 min, proteoliposomes were diluted to a volume of 10 mL, incubated for another 10 min, and then pelleted at 50,000 rpm for 30 min in an MLA-55 ultracentrifuge rotor (Beckman). Proteoliposomes were resuspended in 50 mM Tris, 150 mM NaCl (pH 7.4) to achieve a final concentration of 5 mM lipid. Proteoliposomes (100 μM) were then incubated at room temperature for 1 h on the polar surface of the newly cleaned glass side, spontaneously rupturing to form a planar lipid bilayer of the desired lipid and protein composition. Halo-tagged proteins were labeled by incubating with 0.5 μM halo tag ligand (HTL)-JF646 for 10 min, and excess label and proteoliposomes were removed by washing coverslips with buffer.
TIRF photobleaching and analysis
Planar supported lipid bilayers were imaged using a Nikon Eclipse Ti-E total internal reflection flurescence (TIRF) microscope (Nikon, Tokyo, Japan) with a 100×/1.49 Apo TIRF oil objective and excited with either a 561 nm or a 640 nm laser. Each planar bilayer was imaged in randomly selected areas for 1 min using 100 or 200 ms exposure times. Photobleaching experiments were analyzed using a custom program written in LabView (National Instruments, Austin, TX) kindly provided by Volker Kiessling and as described recently (19). The intensity maximum for each pixel of the first frame was used to select regions of interest. Proteins were located in this image by a single-particle detection algorithm described previously (20). The peak (central pixel) of a 6 × 6 pixel2 area around each identified center of mass were plotted as a function of time for all particles in the image series, and the number of steps in each photobleaching event were counted.
Mammalian cell culture
HEK293T cells (#CRL-11268; ATCC, Manassas, VA) were cultured in Dulbecco’s modified Eagle’s medium supplemented with L-glutamine, 10 mM Hepes (pH 7), 10 μg/mL gentamicin (Invitrogen, Carlsbad, CA), and 10% fetal bovine serum (F4135; Sigma, St. Louis, MO) at 37°C and humidified in 5% CO2. For microscopy experiments, cells were seeded onto 35 mm No. 1.5 glass-bottom dishes (MatTek, Ashland, MA) and transfected the following day with X-tremeGENE HP (Roche) and low levels (∼1 ng in 1 μg total DNA) of plasmid encoding N-terminally tagged rhomboid proteins in pHTN (Promega, Madison, WI). After 18–24 h, Halo-tagged rhomboid proteins were stained with 1 nM HTL-JF646 for 20 min and, after a brief rinse, immobilized with 4% formaldehyde in PBS before imaging.
Binding and photobleaching of protein in detergent micelles
Coverslips were custom coated with nickel by MicroSurfaces (Englewood, NJ). Pure proteins were diluted into the desired detergent buffer (either 0.1% DDM or 0.1% nonyl-glucoside) to be at a final concentration of 1 pM. Proteins with Halo tags were then labeled 1:1 with HTL-JF646 by incubating with coverslips for ∼5 min before imaging.
Results
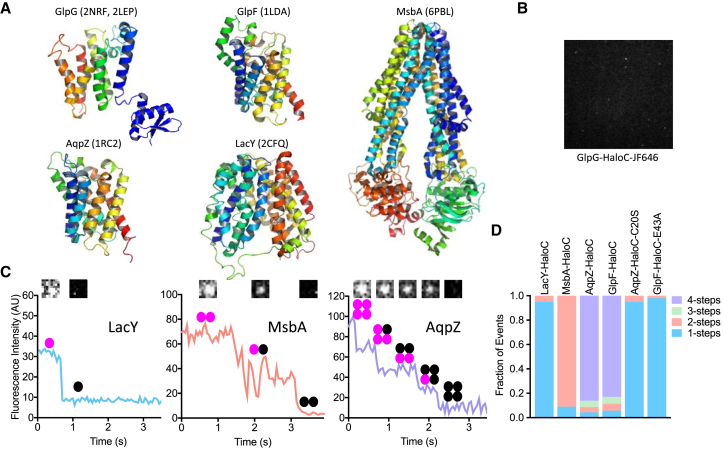
To begin developing methods to assess the oligomeric state of rhomboid proteins directly in the membrane, we first cloned a panel of well-studied bacterial membrane proteins that are known to be monomeric (LacY (21)), dimeric (MsbA (22)), or tetrameric (GlpF (23) and AqpZ (24)) (Fig. 2 A) and tagged each with the self-labeling Halo tag (25) at their C-termini. The resulting proteins were purified from E. coli, reconstituted into planar supported lipid bilayers, and labeled with bright Janelia fluor (JF) dyes (Fig. 2 B) (26). Photobleaching kinetics (Fig. 2 C) of all four proteins reflected their established oligomeric states (Fig. 2 D). Moreover, single mutations that render both AqpZ (24) and GlpF (23) monomeric yielded single-step kinetics consistent with conversion to monomers (Fig. 2 D). These data revealed that we are reliably able to assess the oligomeric state of single molecules of helical membrane proteins directly in a membrane bilayer.
Figure 2.
Quantitative photobleaching of single molecules of E. coli inner membrane proteins. (A) X-ray crystal structures depicting lateral/membrane views (periplasm up, cytosol down) of E. coli rhomboid GlpG and four other E. coli inner membrane proteins that were used as oligomer standards. Proteins are colored from blue (amino terminus) to red (carboxy terminus), and Protein Data Bank (PDB) accession numbers used to render images are included in the brackets. Note that structures of the GlpG membrane core and cytoplasmic domain that were crystallized separately are represented together in arbitrary conformation relative to each other. Minimal functional units are monomers for all except MsbA, which is dimeric, as shown. (B) A representative single-molecule TIRF microscopy image of GlpG-HaloC in a planar supported lipid bilayer. (C) Representative fluorescence intensity versus time traces of LacY-HaloC, MsbA-HaloC, and AqpZ-HaloC single molecules labeled with HTL-JF646. Insets show corresponding images and oligomer diagrams of the single-molecule spots before and after each photobleaching step. Note the distinctive steps of intensity drops as the fluorophores photobleach. (D) Quantification of photobleaching steps of bacterial proteins in supported lipid bilayers. Rare events (e.g., two-step photobleaching events in monomeric samples) are expected because molecules can be unassociated but co-occupy a space smaller than the resolution of a fluorescence microscope. To see this figure in color, go online.
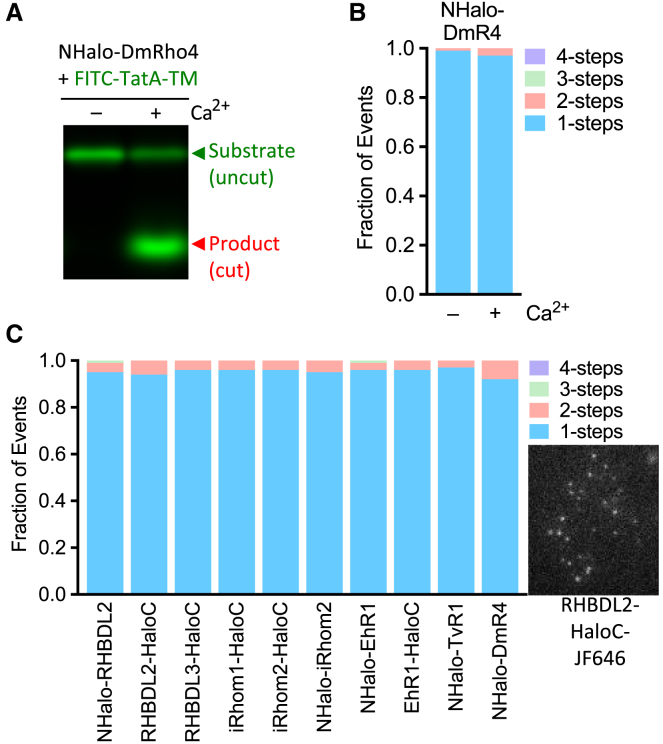
The only context in which direct regulation of a rhomboid protease has been established is D. melanogaster rhomboid-4 (DmRho4); the proteolytic activity of DmRho4 is directly and potently activated by calcium binding to its cytosolic EF hands (17). The ultimate result is lateral gate opening inside the membrane to facilitate substrate access to the internal active site. However, how this conformation change is relayed from the cytosolic domain to the membrane core and whether it is mediated through changes in oligomeric state could not be assessed without introducing detergents during coimmunoprecipitation (17). We therefore appended a Halo tag to the N-terminus of DmRho4, purified it, and reconstituted it into a planar supported lipid layer in the presence or absence of calcium. NHalo-DmRho4 activity was strongly regulated by calcium (Fig. 3 A), but single molecules of NHalo-DmRho4 photobleached in a single step, indicating only monomers were present, and calcium did not alter its monomeric state (Fig. 3 B). Therefore, DmRho4 is monomeric, and its regulation by calcium does not involve oligomerization.
Figure 3.
Eukaryotic rhomboid proteases and pseudopreoteases are monomeric in membranes. (A) Gel electrophoretic analysis of liposome-reconstituted NHalo-DmRho4 activity in the absence or presence of calcium. Shown is a fluorescence scan to reveal FITC-TatA-TM (lower band labeled in red denotes cleaved product). (B) Quantification of NHalo-DmRho4 photobleaching steps in planar bilayers in the presence or absence of calcium. (C) Quantification of photobleaching step events for six rhomboid proteins in the plasma membrane of HEK293T cells. A representative single-molecule TIRF microscopy image of RHBDL2-HaloC labeled with HTL-JF646 in HEK293T cells is shown on the right. To see this figure in color, go online.
Other eukaryotic rhomboid proteases fulfill a range of key cellular functions, and therefore their activity must be precisely regulated (2). Because in vitro assays with pure proteins have not been reported for any of these eukaryotic enzymes, we Halo-tagged human RHBDL2 and RHBDL3 rhomboid proteases, human inactive rhomboid 1 and 2 pseudoproteases, and parasitic rhomboid proteases from Trichonomas vaginalis and Entamoeba histolytica. To avoid the possibility of the Halo tag itself interfering with putative rhomboid oligomerization, we placed it on both the cytosolic (N-terminal fusions) or the extracellular side (C-terminal fusions) of a panel of rhomboid proteins. All 10 resulting constructs, including NHalo-DmRho4, again photobleached in single-step kinetics in HEK293T cells, and therefore all were only monomeric in the plasma membrane of cells (Fig. 3 C).
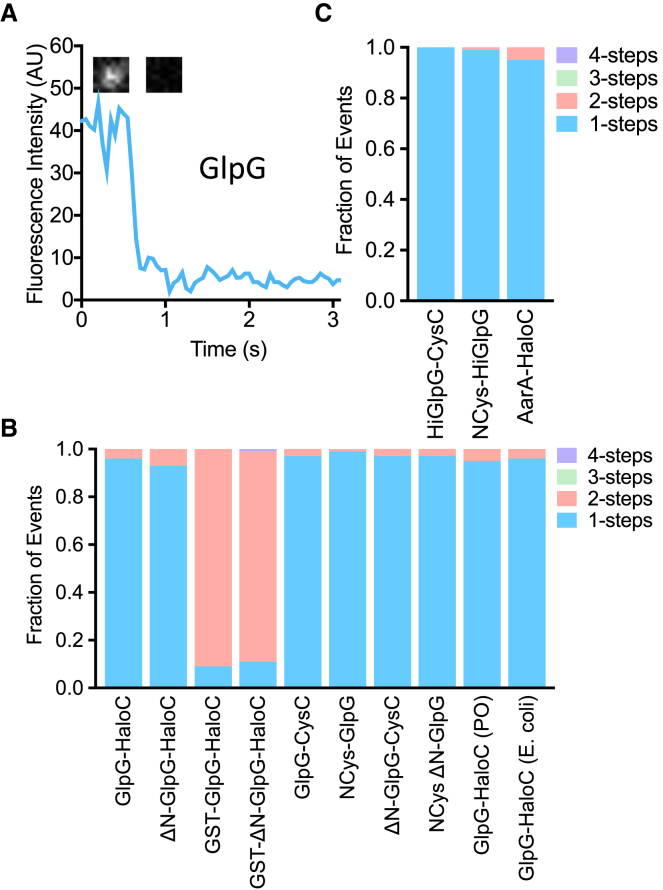
Although diverse eukaryotic rhomboid proteases all proved to be monomeric, evidence for rhomboid dimerization comes only from studies of bacterial orthologs (9, 10, 11, 13, 14). These enzymes often have a compact six-transmembrane (6TM) core (in contrast to the 7TM architecture of their eukaryotic cousins). Therefore, bacterial rhomboid proteins could have a different propensity to dimerize. In fact, the 6TM core of E. coli rhomboid GlpG has itself been crystallized in both dimeric and trimeric forms (9, 10, 11, 13, 14), and the cytosolic domain of GlpG was found to exist in both monomeric and domain-swapped dimeric forms (10, 11, 27). We therefore tagged full-length GlpG and a form missing its cytosolic domain (ΔN) with Halo at their C-termini and assessed their photobleaching kinetics. Both proteins were only monomeric in membranes (Fig. 4, A and B). GlpG has a relatively narrow hydrophobic belt that induces hydrophobic mismatch with surrounding lipids, which could contribute to dimer formation. Nevertheless, varying the thickness or composition of the membrane also did not change the monomeric nature of GlpG (Fig. 4 B).
Figure 4.
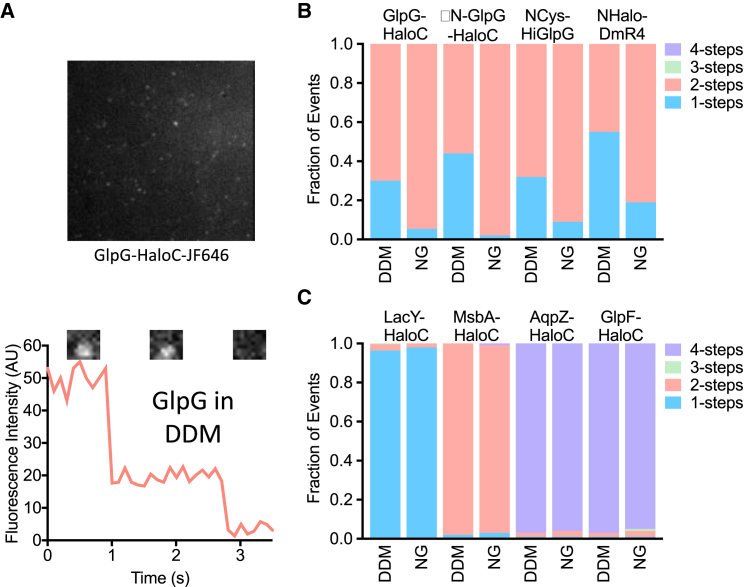
Prokaryotic rhomboid proteases are monomeric in membranes. (A) A representative fluorescence intensity versus time graph of GlpG-HaloC-JF646 in a planar lipid bilayer. Note the characteristic single-step drop. Insets show corresponding images of a single-molecule spot before and after the single photobleaching step. (B) Quantification of photobleaching step events for E. coli GlpG-HaloC labeled with HTL-JF646 or GlpG harboring a single cysteine at either its N- (NCys) or C-terminus (CysC) and labeled with maleimide-JF549. ΔN denotes removal of the first 87 cytoplasmic amino acids of GlpG, whereas GST denotes addition of the glutathione-S-transferase (GST) domain to its amino terminus. The rightmost two bars show GlpG in planar bilayers composed of 70:30 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine/1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (PO) and E. coli lipids. (C) Quantification of Haemophilus influenzae GlpG photobleaching steps is shown. HiGlpG was tagged with a single cysteine residue at either its N- (NCys) or C-terminus (CysC) and labeled with maleimide-JF549. To see this figure in color, go online.
We next evaluated whether the Halo domain itself might be interfering with dimerization by introducing a single cysteine at either the extreme N- or C-terminus of both GlpG and ΔN-GlpG. JF549 labeling through maleimide chemistry again revealed that all four proteins existed only as monomers in the membrane (Fig. 4 B). Notably, all of these proteins proved to be dimeric in detergent systems before being reconstituted into liposomes (vide infra), indicating that our labeling and detection methods can readily identify oligomers. Finally, to assess whether it was even possible for GlpG to dimerize in the membrane, we appended glutathione-S-transferase (GST), which is known to dimerize (28), to its N-terminus. GST-GlpG-Halo and GST-ΔN-GlpG-Halo proved to bleach with two-step kinetics (Fig. 4 B), revealing that both full-length GlpG and its isolated 6TM core (ΔN-GlpG) are capable of dimerizing while in the membrane, but neither does so naturally.
The putative dimerization of the rhomboid protease from Haemophilus pneumoniae has been studied most extensively (9, 14). HiGlpG contains a compact 6TM core without any extramembraneous domains. To avoid potentially disrupting formation of HiGlpG dimers with Halo, we again added a single cysteine residue at either the extreme N- or C-terminus of HiGlpG and labeled it with JF549 conjugated to a maleimide moiety. Analysis of photobleaching kinetics again revealed only monomeric species for both N- and C-terminally labeled HiGlpG (Fig. 4 C). AarA, the extensively studied 7TM rhomboid protease from Providencia stuartii whose activity was also proposed to be strongly regulated by oligomerization (14), was also only monomeric in membranes (Fig. 4 C).
Finally, evidence for dimerization of rhomboid proteins comes largely from studies in the detergent-solubilized state (analytical ultracentrifugation, size exclusion chromatography) or from immunoprecipitation from cell membranes in the presence of detergents (9, 10, 11, 13, 14). We therefore assessed whether the same rhomboid proteins that we had characterized in our planar supported lipid bilayers as monomers and our panel of control proteins are oligomeric in detergents. We deposited pure proteins harboring a hexahistidine sequence in detergent micelles onto coverslips coated with nickel nitrilotriacetic acid (NiNTA) (Fig. 5 A). Photobleaching kinetics proved to be predominantly two step, revealing that all tested rhomboid proteins are dimeric in detergent micelles (Fig. 5 B) but never in lipid bilayers (Fig. 3, B and C; Fig. 4, A–C). This observation again confirmed that our labeling and detection methods are readily able to identify rhomboid dimers when they are present. All other control proteins that we examined maintained a consistent oligomeric state both in detergent and membrane bilayer systems (Fig. 5 C).
Figure 5.
Diverse rhomboid proteases are dimeric in detergent micelles (A) A representative single-molecule TIRF microscopy image of GlpG-HaloC-His in DDM detergent micelles that was captured onto a coverslip coated with NiNTA. (B) Quantification of photobleaching step events of the indicated rhomboid proteins that were captured onto coverslips coated with NiNTA in the indicated detergent micelles (labeled above each bar; NG is nonyl-glucoside). HiGlpG-NCys was directly labeled with maleimide-JF549, whereas the other proteins were labeled via their Halo tags. (C) Quantification of the photobleaching step events of the indicated control proteins as in (B). To see this figure in color, go online.
Discussion
We implemented quantitative single-molecule methods to assess the oligomeric state of diverse rhomboid proteins in cells, planar supported lipid bilayers of various composition, and commonly used detergent micelle systems. We conclude that, as a strict family rule (with yet to be discovered exceptions), rhomboid proteins with diverse functions and across evolution are monomeric in membranes and that their structural stability, function, and regulation do not involve oligomerization.
Interestingly, although rhomboid proteins became unphysiological dimers in detergent systems, all other control proteins that we tested maintained a consistent oligomeric state in both detergent micelles (Fig. 5 C) and membranes (Fig. 2 D). A unique advantage of our methodology is its ability to interrogate the oligomeric state of virtually any membrane protein in parallel detergent micelle and membrane bilayer systems under otherwise identical conditions (Fig. 1) to reveal such intriguing and unusual behaviors. In fact, this unexpected behavior may explain why it has taken so long to establish the oligomeric state of rhomboid proteins definitively. Our approach should also prove valuable for assessing the oligomeric state of the other two intramembrane protease superfamilies whose members have recently been proposed to be oligomeric based on different criteria (29, 30). With this key concept now resolved for rhomboid, it will be important to focus effort on how intramolecular conformation changes are transduced from regulatory extracellular domains to the membrane protease core to regulate proteolytic activity (17).
Author Contributions
S.U. conceived and designed research. S.U. and A.J.B.K. made DNA expression constructs. S.U. conducted mammalian cell culture, labeling, and microscopy experiments, and A.J.B.K. performed protein purification, labeling, reconstitution, planar bilayer formation, microscopy, and data analysis. S.U. wrote the manuscript and prepared the figures, and A.J.B.K. prepared the Supporting Material. Both authors approved the final manuscript.
Acknowledgments
We very gratefully acknowledge Dr. Luke Lavis at the Janelia Research Campus of HHMI for providing the JF dyes used in this work, Rosanna P. Baker for purifying eukaryotic rhomboid proteases, Dr. Volker Kiessling for the photobleaching analysis software, and all members of the Urban Lab for their support.
This work was funded, in part, by an Innovator Award from the Discovery Fund of Johns Hopkins University (to S.U.), and award 2007-31766 from the David and Lucille Packard Foundation (to S.U.).
Editor: Joseph Falke.
Footnotes
Four tables are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)31105-6.
Supporting Material
References
- 1.Urban S., Lee J.R., Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 2.Lastun V.L., Grieve A.G., Freeman M. Substrates and physiological functions of secretase rhomboid proteases. Semin. Cell Dev. Biol. 2016;60:10–18. doi: 10.1016/j.semcdb.2016.07.033. [DOI] [PubMed] [Google Scholar]
- 3.Spinazzi M., De Strooper B. PARL: the mitochondrial rhomboid protease. Semin. Cell Dev. Biol. 2016;60:19–28. doi: 10.1016/j.semcdb.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 4.Adrain C., Zettl M., Freeman M. Tumor necrosis factor signaling requires iRhom2 to promote trafficking and activation of TACE. Science. 2012;335:225–228. doi: 10.1126/science.1214400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McIlwain D.R., Lang P.A., Mak T.W. iRhom2 regulation of TACE controls TNF-mediated protection against Listeria and responses to LPS. Science. 2012;335:229–232. doi: 10.1126/science.1214448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urban S. Making the cut: central roles of intramembrane proteolysis in pathogenic microorganisms. Nat. Rev. Microbiol. 2009;7:411–423. doi: 10.1038/nrmicro2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stevenson L.G., Strisovsky K., Rather P.N. Rhomboid protease AarA mediates quorum-sensing in Providencia stuartii by activating TatA of the twin-arginine translocase. Proc. Natl. Acad. Sci. USA. 2007;104:1003–1008. doi: 10.1073/pnas.0608140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paschkowsky S., Oestereich F., Munter L.M. Embedded in the membrane: how lipids confer activity and specificity to intramembrane proteases. J. Membr. Biol. 2017 doi: 10.1007/s00232-017-0008-5. Published online December 19, 2017. [DOI] [PubMed] [Google Scholar]
- 9.Sampathkumar P., Mak M.W., Lemieux M.J. Oligomeric state study of prokaryotic rhomboid proteases. Biochim. Biophys. Acta. 2012;1818:3090–3097. doi: 10.1016/j.bbamem.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Lazareno-Saez C., Arutyunova E., Lemieux M.J. Domain swapping in the cytoplasmic domain of the Escherichia coli rhomboid protease. J. Mol. Biol. 2013;425:1127–1142. doi: 10.1016/j.jmb.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Sherratt A.R., Blais D.R., Goto N.K. Activity-based protein profiling of the Escherichia coli GlpG rhomboid protein delineates the catalytic core. Biochemistry. 2012;51:7794–7803. doi: 10.1021/bi301087c. [DOI] [PubMed] [Google Scholar]
- 12.Cournia Z., Allen T.W., Bondar A.N. Membrane protein structure, function, and dynamics: a perspective from experiments and theory. J. Membr. Biol. 2015;248:611–640. doi: 10.1007/s00232-015-9802-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strisovsky K., Freeman M. Sharpening rhomboid specificity by dimerisation and allostery. EMBO J. 2014;33:1847–1848. doi: 10.15252/embj.201489373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arutyunova E., Panwar P., Lemieux M.J. Allosteric regulation of rhomboid intramembrane proteolysis. EMBO J. 2014;33:1869–1881. doi: 10.15252/embj.201488149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arant R.J., Ulbrich M.H. Deciphering the subunit composition of multimeric proteins by counting photobleaching steps. Chemphyschem. 2014;15:600–605. doi: 10.1002/cphc.201301092. [DOI] [PubMed] [Google Scholar]
- 16.Ulbrich M.H., Isacoff E.Y. Subunit counting in membrane-bound proteins. Nat. Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker R.P., Urban S. Cytosolic extensions directly regulate a rhomboid protease by modulating substrate gating. Nature. 2015;523:101–105. doi: 10.1038/nature14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker R.P., Urban S. An inducible reconstitution system for the real-time kinetic analysis of protease activity and inhibition inside the membrane. Methods Enzymol. 2017;584:229–253. doi: 10.1016/bs.mie.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blackburn M.R., Hubbard C., Zimmer J. Distinct reaction mechanisms for hyaluronan biosynthesis in different kingdoms of life. Glycobiology. 2018;28:108–121. doi: 10.1093/glycob/cwx096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiessling V., Crane J.M., Tamm L.K. Transbilayer effects of raft-like lipid domains in asymmetric planar bilayers measured by single molecule tracking. Biophys. J. 2006;91:3313–3326. doi: 10.1529/biophysj.106.091421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ermolova N., Guan L., Kaback H.R. Intermolecular thiol cross-linking via loops in the lactose permease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 2003;100:10187–10192. doi: 10.1073/pnas.1434239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchaklian A.H., Funk A.L., Klug C.S. Resting state conformation of the MsbA homodimer as studied by site-directed spin labeling. Biochemistry. 2004;43:8600–8606. doi: 10.1021/bi0497751. [DOI] [PubMed] [Google Scholar]
- 23.Cymer F., Schneider D. A single glutamate residue controls the oligomerization, function, and stability of the aquaglyceroporin GlpF. Biochemistry. 2010;49:279–286. doi: 10.1021/bi901660t. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt V., Sturgis J.N. Making monomeric aquaporin Z by disrupting the hydrophobic tetramer interface. ACS Omega. 2017;2:3017–3027. doi: 10.1021/acsomega.7b00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Los G.V., Encell L.P., Wood K.V. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008;3:373–382. doi: 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- 26.Grimm J.B., English B.P., Lavis L.D. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat Methods. 2015;12:244–250. doi: 10.1038/nmeth.3256. 3 p following 250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghasriani H., Kwok J.K., Goto N.K. Micelle-catalyzed domain swapping in the GlpG rhomboid protease cytoplasmic domain. Biochemistry. 2014;53:5907–5915. doi: 10.1021/bi500919v. [DOI] [PubMed] [Google Scholar]
- 28.McTigue M.A., Williams D.R., Tainer J.A. Crystal structures of a schistosomal drug and vaccine target: glutathione S-transferase from Schistosoma japonica and its complex with the leading antischistosomal drug praziquantel. J. Mol. Biol. 1995;246:21–27. doi: 10.1006/jmbi.1994.0061. [DOI] [PubMed] [Google Scholar]
- 29.Schacherl M., Gompert M., Baumann U. Crystallographic and biochemical characterization of the dimeric architecture of site-2 protease. Biochim. Biophys. Acta Biomembr. 2017;1859:1859–1871. doi: 10.1016/j.bbamem.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Zhou R., Yang G., Shi Y. Dominant negative effect of the loss-of-function γ-secretase mutants on the wild-type enzyme through heterooligomerization. Proc. Natl. Acad. Sci. USA. 2017;114:12731–12736. doi: 10.1073/pnas.1713605114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.