Abstract
Adipocytes play a pivotal role in the regulation of energy metabolism. While white adipocyte stores energy, brown adipocyte dissipates energy by producing heat. In addition, another type of heat-producing adipocyte, beige adipocyte, emerges in white adipose tissue in response to chronic coldness. This phenotypic adaptation to the cold environment is considered to be attributed to the epigenetic modifications. Histone methylation is a chemically stable epigenetic modification and thus a proper mechanism for long-lasting cellular memory. Several histone methyl-modifying enzymes such as EHMT1, JMJD1A, JMJD3, and LSD1 are reported to be involved in the beige adipose cell fate determination. Among these, a histone demethylase JMJD1A senses cold environment by being phosphorylated at S265 in response to β-adrenergic receptor stimulation. Phosphorylated JMJD1A regulates both acute and cold thermogenesis. Under acute coldness, phosphorylated JMJD1A forms a complex with chromatin remodeler SWI/SNF and DNA-bound PPARγ, which recruits JMJD1A to the target genomic regions in brown adipocyte. This complex formation, in turn, induces the expression of target genes by bringing the enhancer and the promoter into close proximity. During chronic coldness, phosphorylated JMJD1A regulates beige adipogenesis through a two-step mechanism. In the first step, phosphorylated JMJD1A is recruited to the regulatory regions of target genes by forming a complex with PRDM16, PGC1α, and DNA-bound PPARγ. In the second step, JMJD1A demethylates histone H3K9me2 and induces stable expression of beige-selective genes. The phenotypic analyses of Jmjd1a-null mice and non-phosphorylated mutant S265A Jmjd1a knock-in mice indicate that JMJD1A is a potential therapeutic target for the treatment of obesity-related diseases including metabolic syndrome and type 2 diabetes.
Keywords: BAT, Beige adiopocyte, JMJD1A, PPARγ, Histone methylation, Epigenetics
Introduction
Epigenetic mechanisms modulate gene expression without alterations in the genomic sequence. Therefore, epigenetic regulation of gene transcription would be an appropriate mechanism for cells to flexibly adapt to the environmental changes. Such epigenetic mechanisms include methylation of DNA, posttranslational modifications of histone, and non-coding RNAs. Histones are subject to various posttranslational modifications including acetylation, methylation, phosphorylation, ubiquitination, and acylation. Most (if not all) of these covalent modifications of histones are reversible. Among the diverse histone modifications, methylation of histone-lysine residue is chemically stable and had been considered to be static and irreversible until the discovery of the histone demethylase lysine demethylase 1 (LSD1) [1] and JmjC domain-containing histone demethylases [2, 3]. The chemical stability of histone methylation marks would contribute to prolonged cellular memory.
Adipose tissue plays important roles in maintaining energy homeostasis. White adipose tissue (WAT) stores excess energy as the form of triglycerides, while brown adipose tissue (BAT) dissipates energy in the form of heat (reviewed in [4]). Recently, it is widely recognized that there are multiple types of thermogenic adipose tissues, i.e., brown adipose tissue and beige (or brite) adipose tissue. BAT is abundant in the intrascapular region of infants and the size of BAT gradually reduces during the growth toward adult humans. On the other hand, the beige adipose tissue is induced in WAT in response to environmental cues such as prolonged coldness, adrenergic stimulation, and the treatment with a ligand for peroxisome proliferator-activated receptor-γ. Brown adipocytes are differentiated from Myf5-expressing dermomyotome precursors from which myotomes are also differentiated [5, 6]. Beige adipocyte emerges either from the preadipocyte expressing both platelet-derived growth factor receptor-α (PDGFRα) and stem cells antigen 1 (SCA1) or from the smooth muscle-like cells expressing myosin heavy chain 11 (MYH11+) (reviewed in [4]). Thus, the distinct precursor cells differentiate into the thermogenic adipocytes. Beige adipogenesis is an adaptation mechanism by which the capacity of cellular heat production is temporally increased under the chronic cold environment. Once the cold stress is removed, beige adipocytes return to WAT. These long-lasting but reversible phenotypic changes of adipose tissues are considered to be attributed to epigenetic regulation of gene transcription in response to cold environment [4]. However, the mechanism of sensing coldness by the epigenetic factors and subsequent regulation of transcription of beige-selective genes are still not well understood.
Regulations of beige adipogenesis by histone methyl-modifying enzymes
There are several lines of evidence that beige adipogenesis is regulated by histone methylases and demethylases: euchromatic histone-lysine N-methyltransferase 1 (EHMT1) (also known as GLP), Jumonji domain-containing 1A (JMJD1A) (also known as JHDM2A or KDM3A), JMJD3 (also known as KDM6B), and LSD1 (also known as KDM1A or AOF2) (Table 1).
Table 1.
Histone methyl-modifying enzymes involved in the regulation of beige adipogenesis
| Histone methyl-modifying enzymes | Activity | Roles in beige adipocyte | Refs |
|---|---|---|---|
| EHMT1 (GLP1) | H3K9 methyltransferase | Promotion of beige adipocyte (adipose tissue-specific Ehmt1 knockout mice and Ehmt1-null white adipocytes show reduced expressions of beige-selective genes) | [7] |
| JMJD1A (JHDM2A, KDM3A) | H3K9me1/2 demethylase | Promotion of beige adipocyte (inducing beige adipogenesis by being recruited to the regions of beige-selective genes in response to β-ADR and, in turn demethylating H3K9 in the regions) | [19] |
| JMJD3 (KDM6B) | H3K27me2/3 demethylase | Promotion of beige adipocyte (inducing beige adipogenesis by resolving bivalency of H3K4me3 and H3K27me3) | [10] |
| LSD1 (KDM1A, AOF2) | Demethylase of H3K4me1/2 and H3K9me1/2 | Maintenance of beige adipocytes (reducing age-related transition of beige adipocyte to white adipocyte), Promotion of beige adipocyte (being required for beige induction by Zfp516) | [15, 16] |
EHMT1 regulates browning of subcutaneous WAT and differentiation and function of BAT [7]. Although the detailed mechanism by which EHMT1 regulates beige adipogenesis is not completely elucidated, a dual mechanism of EHMT1 regulation of BAT cell fate and function may be partially applicable. EHMT1 on one hand catalyzes the methylation of histone H3K9 [8] which is a signature of heterochromatin and thereby induces chromatin condensation and gene silencing. Thus, EHTM1 represses the expressions of muscle-selective genes through its methyltransferase activity. EHMT1 alternatively activates BAT-selective genes through stabilizing PRDM16 protein which is a potential regulator of beige adipogenesis. In addition, adipose tissue-specific Ehmt1 null mice presented less than half levels of mRNA expression of beige-selective genes in response to β3-adrenergic receptor (β3-ADR) stimulation [7]. Requirement of EHMT1 for beige adipogenesis is also revealed by the cell-autonomous study using stromal vascular fractions isolated from the inguinal WAT of Ehmt1flox/flox mice subsequently infected with Cre-recombinase expressing adenovirus.
JMJD3 is a histone H3K27me2/me3 demethylase [9] and promotes beige adipogenesis by resolving bivalency of active H3K4me3 and repressive H3K27me3 in primary inguinal WAT preadipocytes [10]. The bivalent domain of H3K4me3 and H3K27me3 silences the beige-selective genes in primary white adipocytes and at the same time keeps them poised for future activation [11, 12]. The mice over-expressing Jmjd3 transgene under the control of Fabp4 promoter showed higher expression of thermogenic genes such as Ucp1 and Cidea in primary inguinal white adipocytes (iWAT) as well as in BAT compared to their control littermates [10]. The enzyme-dependent regulation of beige adipogenesis by JMJD3 was confirmed by producing another line of transgenic mice which express the demethylation-defective point mutant H1388A of JMJD3. The expressions of Ucp1 were slightly reduced in BAT and reduced to a half level in iWAT in these mice compared to their control.
LSD1 is a demethylase of histone H3K4me1/2 and H3K9me1/2. There are several lines of evidence that LSD1 regulates BAT function and also mediates beige adipogenesis. The adipose tissue-specific LSD1-deficient mice produced by crossing an adiponectin–Cre line with an LSD1 floxed line shows reduced BAT thermogenesis [13, 14]. The age-related transition of beige adipocytes to white adipocytes is rescued by the adipocyte-specific expression of LSD1 through maintaining the expression of PPARα [15]. In addition, the beige adipogenesis induced by a transcription factor Zfp516 requires its interaction with LSD1 [16]. Taken together, there is a link between LSD1 and beige adipogenesis, although the direct regulating mechanism of beige adipogenesis by LSD1 is not clearly elucidated.
JMJD1A is a demethylase of histone H3K9me1/2 [17]. Notably, the sophisticated mechanisms by which JMJD1A regulates acute and chronic thermogenesis have been precisely elucidated [18, 19].
Mice lacking JMJD1A are a model of metabolic syndrome
The development of obesity and its associated disease referred to as metabolic syndrome is influenced by the environmental factors. Because histone methylation is an appropriate molecular mechanism for recording environmental cues in the cells, it had been speculated that histone-modifying enzymes would play a role in the development of metabolic syndrome. JMJD1A is the first histone-modifying enzyme of which deficient mice are reported to show metabolic syndrome [20, 21]. JMJD1A is a JmjC domain-containing Fe(II)- and α-ketoglutarate (αKG)-dependent dioxygenase which specifically demethylates histone H3K9me1 and H3K9me2. Human JMJD1A is comprised of 1321 amino acids (a.a.) and contains JmjC domain and zinc finger domain, both of which are indispensable for enzyme activity of JMJD1A [17]. Synthetic protein of partial JMJD1A (489–1321 a.a. of hJMJD1A), which contains both JmjC domain and zinc finger domain, presents the enzymatic activity comparable to the full-length protein. The single amino acid substitution at the cofactor binding site, H1120Y, in JmjC domain of the partial JMJD1A protein disrupts its enzymatic activity. Jmjd1a is expressed ubiquitously in mouse and highly in testis [20]. The functional roles of JMJD1A include regulating spermatogenesis [22], sex determination [23], development and maintenance of cancer [9, 24], stem cell self-renewal [25], and regulating energy metabolism. Multiple lines of JMJD1A-null mice produced separately by two groups present similar phenotypes of metabolic defects [20, 21]. JMJD1A-null mice show adult-onset obesity. The body weight of JMJD1A-null mice is comparable to the control mice at the age of 4 weeks, whereas it becomes significantly more at the weeks of 8. Plasma profiling showed higher plasma levels of triglycerides, cholesterol, glucose, and insulin in JMJD1A-null mice compared to their control mice. In addition, insulin insensitivity of JMJD1A-null mice was demonstrated by glucose tolerance test and insulin tolerance test. Interestingly, JMJD1A-null mice showed lower body temperature compared to their control mice in response to overnight fasting or acute cold exposure [18, 20, 21]. Functional analysis using metabolic cage revealed that JMJD1A-null mice show time-dependent higher respiratory quotient which indicates lower fat usage in these mice [20]. Together, JMJD1A-null mice show defects in energy metabolism. Considering that the cold-induced thermogenesis utilizes energy source at a high rate [26, 27], lower body temperature of JMJD1A-null mice in response to cold exposure as well as fasting is possibly due to the defect in utilization of energy source. In addition to the energy utilization defect and lower body temperature under the cold and fasting conditions, JMJD1A-null mice showed functional defects in the BAT. Physiologically, catecholamine is secreted from the sympathetic nerve ending in response to cold exposure, binds to β-ADR of brown adipocytes, and induces lipolysis and the expression of UCP1 which plays a pivotal role in producing heat by uncoupling cellular respiration and mitochondrial ATP synthesis [4]. This signaling cascade is artificially activated by the treatment of β-ADR agonist such as isoproterenol (ISO), a non-selective β-ADR agonist. Further investigation using a brown adipose cell line HIB1B in which Jmjd1a expression is knocked down (KD) using short-hairpin RNA revealed that the ISO-induced expression of Ucp1 is reduced in Jmjd1a-KD HIB1B cells compared to the control cells [21]. This reduction of Ucp1 expression in Jmjd1a knockdown cells was partially rescued by the overexpression of JMJD1A. It is claimed that the mechanism by which JMJD1A regulates Ucp1 expression in BAT is due to maintaining H3K9me2 at low levels in the enhancer region of Ucp1 through enhanced recruitment of transcription factors [i.e., PPARγ, retinoid X receptor α (RXRα), and, Arf2] and co-activators [i.e., PPARγ-coactivator 1 − α (PGC1α), CPB/P300, and Src1] [21]. This claim is considered based on the fact that JMJD1A is a demethylase of histone H3K9. However, it is somehow a bit confusing, because brown adipocytes are already differentiated to the thermogenic adipocytes in which chromatin conformation in the region of thermogenic genes would be in an “open” state, and thus, it would not be critical to maintain low levels of H3K9me2 at the enhancer regions of thermogenic genes.
Demethylation of H3K9me2 during beige adipogenesis
Homeotherms need to generate heat to maintain their body temperature. There are two types of thermogenesis: shivering and non-shivering thermogenesis. While shivering of skeletal muscle contributes to produce heat, non-shivering thermogenesis occurs by uncoupling the processes of proton-gradient generation and ATP synthesis. BAT and beige adipose tissues play important roles in non-shivering thermogenesis. They increase heat production in response to both acute and chronic cold exposure. BAT produces heat in response to acute cold exposure (~ hours in mice), while the induction of heat production by beige adipogenesis in WAT occurs after prolonged cold exposure (~ weeks in mice) [4]. The expression of thermogenic genes such as Ucp1 is constitutively high in BAT, but very low in subcutaneous WAT (scWAT). The constitutively high expression of Ucp1 in BAT is associated with the lower levels of H3K9me2 in the enhancer and promoter regions of Ucp1 in comparison with subcutaneous WAT [19]. A similar trend is observed in total histone in BAT compared to scWAT. Consistent with these observations, the induction of Ucp1 in BAT in response to acute cold exposure is not associated with the further reduction from the already “low” levels of H3K9me2 in the enhancer and promoter regions of Ucp1 [19]. These facts suggest that the induction of Ucp1 expression in BAT under acute coldness would not be critically regulated by demethylation of H3K9me2, but regulated by a mechanism independent of histone demethylation.
Regulation of Ucp1 expression in BAT by phosphorylation of JMJD1A
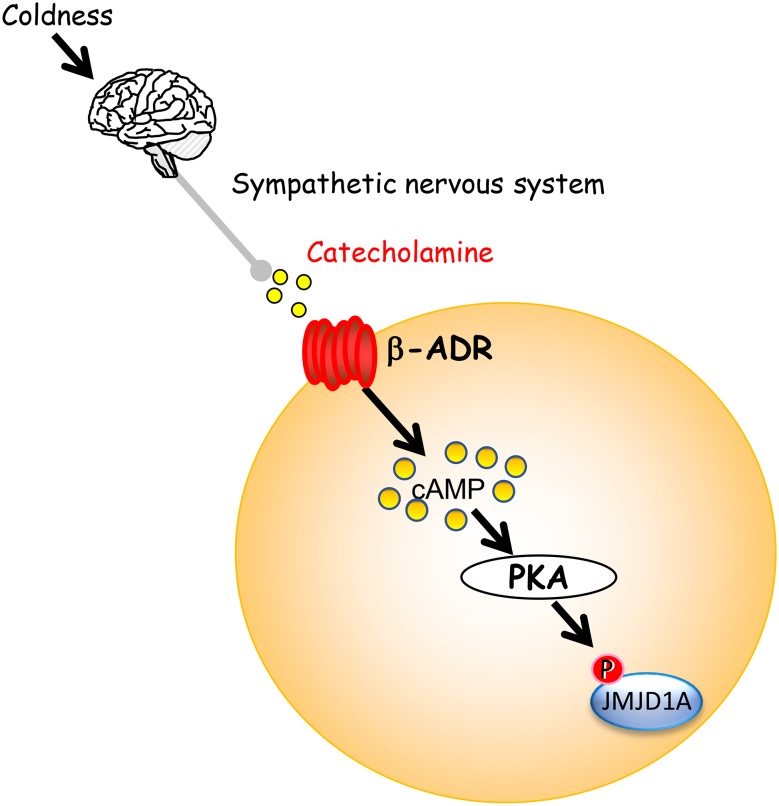
In response to acute coldness, the β-ADR is stimulated in brown adipocytes via the sympathetic nervous system and activates adenylyl cyclase (AC), which, in turn, leads to increase in the cAMP levels, activates PKA, and induces Ucp1 expression and lipolysis (Fig. 1). Impaired regulation of body temperature of the JMJD1A-null mice under acute cold exposure and fasting suggested that JMJD1A has a link with the βADR–cAMP–PKA signaling pathway. Mass spectrometry and immunoblot analysis using JMJD1A antibody revealed that PKA phosphorylates JMJD1A at serine 265, which also physiologically occurs in BAT of the mice under cold stress or β-ADR stimulation [18] (Fig. 1). In addition, the combination of the ISO-dependent genome-wide binding analysis of JMJD1A using ChIP-seq and the transcriptome analysis by microarray revealed Ucp1 and Adrb1, encoding β1-ADR, as the target genes of JMJD1A during acute coldness. To confirm the impact of phosphorylation of JMJD1A at S265 on gene transcription, either wild-type (WT) or non-phosphorylated mutant (S265A) of human JMJD1A was retrovirally transduced to the immortalized mouse primary brown adipocytes (iBAT) in which the expression of endogenous JMJD1A is knocked down by short-hairpin RNA (iBATsh) [18]. The gene expressions of both Ucp1 and Adrb1 were induced in response to ISO stimulation of βADR in WT-JMJD1A-iBATsh, while the induction was severely repressed in S265A-JMJD1A-iBATsh. Collectively, the cold-induced expression of Ucp1 and Adrb1 in BAT is mediated by the phosphorylation of JMJD1A at S265. Remembering that the partial amino acid sequence of JMJD1A (489–1321 a.a.) is sufficient for its enzyme activity, it is conceivable that phosphorylated JMJD1A regulates the expressions of target genes through the mechanism independent of its enzyme activity. Indeed, in vitro enzyme activity assays using both the HTRF–FRET and the MALDI-TOF/MS reveled that the activity of JMJD1A was not affected by the phosphorylation at S265. It is also notable that the mutations of the key residue for the demethylase activity of JMJD1A (H1120Y or H1120F) did not alter the ISO-induced expression of Ucp1 and Adrb1 in iBATsh [18]. These results collectively support the concept that demethylation of H3K9 is not indispensable for the induction of Ucp1 expression during acute thermogenesis in the BAT. This is the clear difference from the requirement of demethylation of H3K9me2 during beige adipogenesis as described below [19]. In general, phosphorylation of an enzyme regulates its enzyme activity, subcellular localization, or complex formation with other proteins. Although phosphorylation of JMJD1A does not affect its enzyme activity or subcellular localization in iBAT, it is critical for complex formation. Proteomic analysis using a highly specific monoclonal JMJD1A antibody revealed the ISO-induced JMJD1A binding with SWI/SNF components such as ARID1A (BAF250a), BRG1 (SMRACA4), and BAF60b (SMRD2), and RNA polymerase II. In addition, JMJD1A binds to PPARγ through SWI/SNF [18]. The ISO-induced formation of JMJD1A–SWI/SNF–PPARγ complex functionally regulates gene transcription, so that the series of iBAT lines in which the expression of each component of SWI/SNF is knocked down show reduced expression of Ucp1 and Adrb1 (Fig. 2).
Fig. 1.
Cold-sensing system by phosphorylation of JMJD1A. Environmental coldness activates the sympathetic nervous system and catecholamine is secreted from the nerve ending. Catecholamine activates β-ADR, which, in turn, increases cAMP, activates PKA and phosphorylates histone H3K9 demethylase JMJD1A at S265
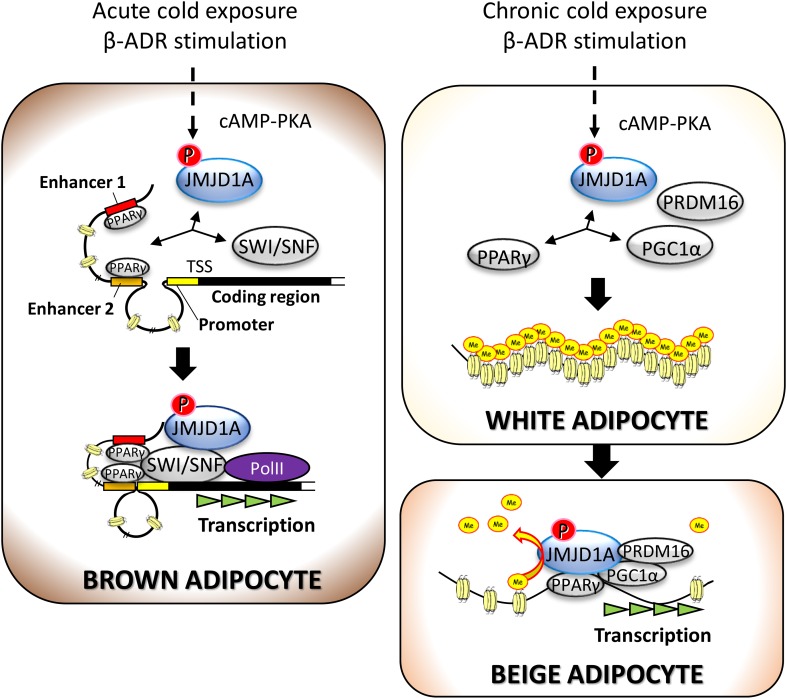
Fig. 2.
Phosphorylated JMJD1A mediates both acute heat production in BAT and chronic beige adipogenesis in WAT. Acute thermogenesis in brown adipocytes occurs by phosphorylated JMJD1A-dependent complex formation with SWI/SNF chromatin remodeler and PPARγ bound to the specific DNA sequence called the response element. The P-JMJD1A–SWI/SNF–PPARγ complex induces thermogenic gene expression by facilitating long-range chromatin remodeling to induce the proximity of the enhancer and promoter (left). The long-lasting chronic coldness causes beige adipogenesis by inducing the expressions of beige-selective genes by a two-step mechanism. First, the phosphorylated JMJD1A at S265 is recruited to the regulatory regions of beige-selective genes by forming a complex with PRDM16, PGC1α, and PPARγ (right top). Second, the recruited JMJD1A demethylates H3K9me2 and induces the expression of beige-selective genes (right bottom)
SWI/SNF is a chromatin remodeler complex which, by getting enhancer and promoter closer, facilitates long-range chromatin interactions to induce gene expression. Therefore, the complex formation of JMJD1A with SWI/SNF is considered to change chromatin conformation. The combination of multiple ChIP-seq analysis using the antibodies against H3K27Ac, H3K4me3, ARID1A, BRG1, or JMJD1A revealed the ISO-induced recruitment of JMJD1A and SWI/SNF to both the H3K27Ac-enriched active enhancers and the H3K4me3-enriched promoter of Ucp1 and Adrb1 [18]. Notably, the enhancers and the promoter are in close proximity in BAT and brown adipocytes and get even closer in response to the cold exposure or the treatment with ISO or an activator of adenylate cyclase forskolin as determined by performing chromosome conformation capture (3C). The induction of proximity was not observed in S265A–JMJD1A–iBATsh and iBATs in which the Brg1 expression is suppressed by shRNA. These results indicate that the cold-induced β-ADR signal facilitates long-range DNA looping by forming the JMJD1A–SWI/SNF–PPARγ complex and induces the proximity of the enhancers and the promoter, which in turn, activates gene transcription of Adrb1 and Ucp1 by facilitating the recruitment of RNA polymerase (Fig. 2). In the different aspects, JMJD1A is recruited to the specific target genomic locus through forming a complex with SWI/SNF and PPARγ which binds to the specific DNA sequence called the PPAR response element. The similar mechanism that JMJD1A regulates the proximity of enhancer and promoter is reported in human umbilical vein endothelial cells (HUVEC) in response to hypoxia [28]. Another well-known example of transcriptional regulation by forming a long-range chromatin loop is the direct interaction of the promoters of IRX3 and its remote enhancer in the non-coding region in FTO [29, 30]. The regulation of IRX3 also correlates with beige adipogenesis and obesity, while it is still controversial if IRX3 positively or negatively regulates thermogenesis [29–32].
Physiological relevance of phosphorylation at S265 of JMJD1A to acute thermogenesis was determined by generating the knock-in mouse with a serine 265-to-alanine mutation (Jmjd1a-S265AKI/KI) [19]. Similar to the observation in Jmjd1a-KO mice, Jmjd1a-S265AKI/KI mice showed intolerance to acute cold exposure relative to Jmjd1a-+/+ mice in maintaining their body temperature. Cell-autonomous study of the primary adipocytes obtained from Jmjd1a–S265AKI/KI mice showed the reduced mitochondrial respiration. In conclusion, JMJD1A senses cold exposure by being phosphorylated in response to β-ADR stimulation and regulates thermogenic gene expression through chromatin remodeling to induce acute non-shivering thermogenesis in BAT.
JMJD1A mediates beige adipogenesis
While thermogenesis in BAT occurs acutely in response to cold exposure, beige adipogenesis in subcutaneous WAT and following heat production is a chronic adaptation mechanism which occurs under the long-lasting cold exposure. An underlying mechanism for the acute induction of thermogenic genes in BAT is the remodeling of chromatin structure which is associated with reduced levels of H3K9me2 in the enhancer and promoter regions [19] (Fig. 2). On the contrary, the transcription of thermogenic genes is firmly silenced in scWAT, which is associated with higher levels of H3K9me2 in the regulatory regions of thermogenic genes, and induced only when beige adipogenesis occurs during chronic coldness [19]. Indeed, the higher levels of H3K9me2 in scWAT decreased after chronic cold exposure (4 °C for 1 week). Furthermore, the induced expressions of beige-selective genes are maintained during chronic coldness and also after removing chronic coldness. The low levels of H3K9me2 in the regulatory regions of beige-selective genes are considered to contribute to maintain the beige adipocyte phenotype. Thus, the low and high levels of H3K9me2 play roles in stabilizing the characteristics of white adipocytes and beige adipocytes, respectively.
Cell-autonomous study using immortalized beige adipocytes which are derived from scWAT (im-scWAT) revealed that the beige adipogenesis is associated with the reduction of H3K9me2 in the regulatory regions of beige-selective genes such as Ucp1, Cidea, and Ppara [19]. In addition, beige adipogenesis under chronic cold condition is severely repressed in Jmjd1a–KO mice. The reduction of beige adipogenesis in Jmjd1a–KO mice is associated with higher levels of H3K9me2 in the regulatory regions of beige-selective genes. These data indicated that JMJD1A mediates beige adipogenesis through its enzyme activity.
Notably, the similar reduction of beige adipogenesis is observed in Jmjd1a–S265AKI/KI mice. The recruitment of JMJD1A to the regulatory genomic regions of beige-selective genes increases toward the end of beige adipocyte differentiation of the immortalized subcutaneous WAT cells (im-scWATs) and is also positively and negatively regulated by the treatment with ISO and a β-blocker propranolol, respectively [19]. im-scWATs, in which endogenous Jmjd1a is knocked down and non-phosphorylated mutant (S265A) of human JMJD1A is retrovirally transduced (S265A-JMJD1A im-scWATs), showed reduced expressions of beige-selective genes and reduced recruitment of JMJD1A to their genomic loci, which is associated with reduced thermogenesis by flux analysis in comparison with wild-type (WT)-JMJD1A im-scWATs. Together, the phosphorylation-dependent recruitment of JMJD1A to its target gene loci is predominantly required for demethylation of H3K9me2 in the regulatory regions of the beige-selective genes and inducing their transcription (Fig. 2). Hence, the induction of phosphorylation of JMJD1A would be a potential approach for the artificial induction of beige adipogenesis. Indeed, immortalized scWATs transduced with S265D-phosphomimetic JMJD1A (S265D-hJMJD1A im-scWATs) showed increased expressions of beige-selective genes and mitochondrial respiration, which were reduced to the level comparable to WT-JMJD1A im-scWATs in the im-scWATs transduced with the double mutant JMJD1A with both S265D and a mutation of catalytic histidine to tyrosine (S265D-H1120Y-JMJD1A im-scWATs) [19]. Thus, the regulation of beige adipogenesis by phosphorylated JMJD1A requires both the recruitment to the target gene loci and enzyme activity. Presumably, the recruitment of phosphorylated JMJD1A to the beige-selective gene loci is prerequisite for the demethylation of H3K9me2 to induce the transcription.
Of note, PRDM16 and PGC1α as well as PPARγ bind to phosphorylated JMJD1A in scWAT. PRDM16 and PGC1α are both transcriptional co-regulators which bind each other and potently promote beige adipogenesis by mediating beige-selective gene expression. In addition, chronic treatment with a ligand for PPARγ as well as cold-induced β-ADR signaling are the external cues that potently promote beige adipogenesis (Fig. 3). The phosphorylated JMJD1A would play a central role as a signal sensing mediator to form a complex of transcriptional co-regulators and activates transcription of beige-selective genes (Figs. 2, 3).
Fig. 3.
JMJD1A as an integrator of the multiple induction signals of beige adipogenesis. Beige adipogenesis is induced by chronic environmental cold exposure, stimulation of β-ADR signal, and the chronic exposure to ligands for PPARγ. JMJD1A is phosphorylated in response to cold-induced β-ADR signal and forms a complex with PPARγ during beige adipogenesis. Thus, it is considered that JMJD1A plays a pivotal role in integrating extra-cellular signals to induce the expressions of beige-selective genes
A two-step regulation mechanism of adipocyte thermogenesis by JMJD1A
Histone demethylase JMJD1A senses coldness by being phosphorylated at S265 in response to β-ADR stimulation and regulates adipocyte thermogenesis by acute and chronic mechanisms. In response to acute cold exposure, JMJD1A is phosphorylated in BAT and induces the expression of its target genes by forming a complex with SWI/SNF and PPARγ and remodels chromatin structure to induce the proximity of the enhancer(s) and the promoter of thermogenic genes. During chronic cold exposure, phosphorylated JMJD1A in scWAT regulates the expressions of beige-selective genes through a two-step mechanism. In the first step, JMJD1A is phosphorylated and recruited to the regulatory regions of beige-selective genes by forming a complex with the co-regulatory factors such as PRDM16, PGC1α, and PPARγ, all of which play a pivotal role in the development of beige adipocytes. The recruitment of JMJD1A to the target gene loci is prerequisite for the second step. The second step is the enzymatic demethylation of H3K9me2 which has a role to maintain cellular memory. The consequent reduction of the levels of H3K9me2 in the regions of beige-selective genes contribute to the stable expression of the genes to maintain the characteristics of beige adipocytes. While the acute coldness induces the phosphorylation of JMJD1A and following expression of thermogenic genes within a few hours in BAT, the processes of beige adipogenesis in scWAT requires long-lasting (~ a week) cold exposure. The molecular mechanism of delayed response of the demethylase activity after the phosphorylation of JMJD1A is still an open question. There are several putative mechanisms involved in this regulation. For example, an additional signal cascade is to be activated for the formation of a transcriptional regulatory complex. Such a signal may induce a posttranslational modification of a component(s) of the protein complex and/or JMJD1A at alternative amino acid residue(s). It is also possible that the protein levels of transcriptional co-regulators such as PRDM16 and PGC1α need to be up-regulated by reducing their protein degradation and/or by inducing their transcription. In addition, the levels of cofactors may affect the enzyme activity. JmjC domain-containing proteins including JMJD1A require Fe2+ and α-Ketoglutarate (α-KG) as cofactors for their enzyme activity [4]. αKG is an intermediate metabolite of Krebs cycle and also linked to glutamine metabolism. The increased metabolic rate in the heat production under the cold environment may result in the accumulation of intracellular αKG, which as a cofactor increases the enzyme activity of JMJD1A and leads to demethylation of H3K9me2.
In summary, JMJD1A is an important player in the regulation of energy metabolism especially in the sensing and adaptation to the environmental changes. JMJD1A-KO mice present phenotypes similar to metabolic syndrome [20]. In addition, high fat diet-fed JMJD1A-S265A-KI mice acclimated to chronic coldness showed impaired insulin sensitivity and decreased phosphorylation levels of Akt in subcutaneous WAT, BAT, and skeletal muscles [19]. Thus, the recent advances in the epigenetic roles of histone modifications in thermogenesis suggest artificial activation of JMJD1A as a potential therapeutic approach for obesity-related diseases such as metabolic syndrome and diabetes.
Conclusions and future perspectives
Histone methylation is an appropriate mechanism for long-lasting cellular memory. Several histone methyl-modifying enzymes such as EHMT1, JMJD1A, JMJD3, and LSD1 have been reported to be involved in the regulation of beige adipogenesis. However, the precise epigenetic roles of these enzymes in beige adipogenesis are not yet fully understood. It is speculated that other methyl-modifying enzymes would emerge as novel epigenetic regulators in beige adipogenesis. In addition, the mechanism of interaction of these histone-modifying enzymes with other transcriptional regulators is still an open question. It is also to be elucidated if the functionally correlated multiple histone marks such as bivalent modifications of H3K4/K27me3 and recently reported H3K4/K9me3 [12] would play a role in beige adipogenesis. The epigenetic mechanisms of beige adipogenesis would be an interesting and promising research area that is expected to unveil novel molecular mechanisms of regulation of the adipose cell fate as well as to present potential therapeutic approaches for obesity-related diseases.
Acknowledgements
A summary of this review was presented in the Lilly Award Lecture at the 61st Japan Diabetes Society 2018, Tokyo, Japan. The author would like to express sincere gratitude to Dr. Juro Sakai for his mentoring, his colleagues and collaborators for their helpful support during performing the projects, and Dr. Hiroshi Shibata for critical reading of the manuscript. The author is supported by JSPS KAKENHI (Grant Numbers 18H04796, 17H03631, 25291002), Astellas Foundation for Research on Metabolic Disorders, the Novartis Foundation (Japan) for the Promotion of Science, the Tokyo Biochemical Research Foundation, the Naito Foundation, the Ichiro Kanehara Foundation, Japan Diabetes Foundation, Suzuken Memorial Foundation, and Kao Research Council for the Study of Healthcare Science.
Conflict of interest
The author declares no competing interests.
Ethics policy
This article does not contain any experimental studies with human or animal subjects.
References
- 1.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;7:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 2.Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;7:801–806. doi: 10.1016/S0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, Kouzarides T. Reversing histone methylation. Nature. 2005;7054:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- 4.Inagaki T, Sakai J, Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat Rev Mol Cell Biol. 2016;8:480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seale P, Bjork B, Yang W, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;7207:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kajimura S, Seale P, Kubota K, et al. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;7259:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ohno H, Shinoda K, Ohyama K, et al. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature. 2013;7478:163–167. doi: 10.1038/nature12652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ogawa H, Ishiguro K, Gaubatz S, et al. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;5570:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- 9.Kaukonen R, Mai A, Georgiadou M, et al. Normal stroma suppresses cancer cell proliferation via mechanosensitive regulation of JMJD1a-mediated transcription. Nat Commun. 2016;7:12237. doi: 10.1038/ncomms12237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan D, Huang L, Zhu LJ, et al. Jmjd3-mediated H3K27me3 dynamics orchestrate brown fat development and regulate white fat plasticity. Dev Cell. 2015;5:568–583. doi: 10.1016/j.devcel.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;2:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 12.Matsumura Y, Nakaki R, Inagaki T, et al. H3K4/H3K9me3 bivalent chromatin domains targeted by lineage-specific DNA methylation pauses adipocyte differentiation. Mol Cell. 2015;4:584–596. doi: 10.1016/j.molcel.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 13.Zeng X, Jedrychowski MP, Chen Y, et al. Lysine-specific demethylase 1 promotes brown adipose tissue thermogenesis via repressing glucocorticoid activation. Genes Dev. 2016;16:1822–1836. doi: 10.1101/gad.285312.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin JZ, Farmer SR. LSD1-a pivotal epigenetic regulator of brown and beige fat differentiation and homeostasis. Genes Dev. 2016;16:1793–1795. doi: 10.1101/gad.288720.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duteil D, Tosic M, Willmann D, et al. Lsd1 prevents age-programed loss of beige adipocytes. Proc Natl Acad Sci USA. 2017;20:5265–5270. doi: 10.1073/pnas.1702641114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambeat A, Gulyaeva O, Dempersmier J, et al. LSD1 interacts with Zfp516 to promote UCP1 transcription and brown fat program. Cell Rep. 2016;11:2536–2549. doi: 10.1016/j.celrep.2016.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamane K, Toumazou C, Tsukada Y, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;3:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 18.Abe Y, Rozqie R, Matsumura Y, et al. JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat Commun. 2015;6:7052. doi: 10.1038/ncomms8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abe Y, Fujiwara Y, Takahashi H, et al. Histone demethylase JMJD1A coordinates acute and chronic adaptation to cold stress via thermogenic phospho-switch. Nat Commun. 2018;9:1566. doi: 10.1038/s41467-018-03868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inagaki T, Tachibana M, Magoori K, et al. Obesity and metabolic syndrome in histone demethylase JHDM2a-deficient mice. Genes Cells. 2009;8:991–1001. doi: 10.1111/j.1365-2443.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 21.Tateishi K, Okada Y, Kallin EM, et al. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;7239:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada Y, Scott G, Ray MK, et al. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;7166:119–123. doi: 10.1038/nature06236. [DOI] [PubMed] [Google Scholar]
- 23.Kuroki S, Matoba S, Akiyoshi M, et al. Epigenetic regulation of mouse sex determination by the histone demethylase Jmjd1a. Science. 2013;6150:1106–1109. doi: 10.1126/science.1239864. [DOI] [PubMed] [Google Scholar]
- 24.Fan L, Peng G, Sahgal N, et al. Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival. Oncogene. 2016;19:2441–2452. doi: 10.1038/onc.2015.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh YH, Zhang W, Chen X, et al. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;20:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;1:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 27.Cannon B, Nedergaard J. Metabolic consequences of the presence or absence of the thermogenic capacity of brown adipose tissue in mice (and probably in humans) Int J Obes. 2010;34:S7–S16. doi: 10.1038/ijo.2010.177. [DOI] [PubMed] [Google Scholar]
- 28.Mimura I, Nangaku M, Kanki Y, et al. Dynamic change of chromatin conformation in response to hypoxia enhances the expression of GLUT3 (SLC2A3) by cooperative interaction of hypoxia-inducible factor 1 and KDM3A. Mol Cell Biol. 2012;15:3018–3032. doi: 10.1128/MCB.06643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smemo S, Tena JJ, Kim KH, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;7492:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Claussnitzer M, Dankel SN, Kim KH, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;10:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou Y, Lu P, Shi J, et al. IRX3 promotes the browning of white adipocytes and its rare variants are associated with human obesity risk. EBioMedicine. 2017;24:64–75. doi: 10.1016/j.ebiom.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inagaki T. Regulations of adipocyte phenotype and obesity by IRX3. positive or negative? EBioMedicine. 2017;24:7–8. doi: 10.1016/j.ebiom.2017.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]