ABSTRACT
Aggregatibacter actinomycetemcomitans and Aggregatibacter aphrophilus belong to the HACEK group of fastidious Gram-negative organisms, a recognized cause of infective endocarditis. A. actinomycetemcomitans is also implicated in aggressive forms of periodontitis. We demonstrated that A. aphrophilus strains, as A. actinomycetemcomitans are ubiquitously serum resistant. Both species encode two Outer membrane protein A paralogues, here denoted OmpA1 and OmpA2. As their respective pangenomes contain several OmpA1 and OmpA2 alleles, they represent potential genotypic markers. A naturally competent strain of A. actinomycetemcomitans and A. aphrophilus, respectively were used to elucidate if OmpA1 and OmpA2 contribute to serum resistance. Whereas OmpA1 was critical for survival of A. actinomycetemcomitans D7SS in 50% normal human serum (NHS), serum resistant ompA1 mutants were fortuitously obtained, expressing enhanced levels of OmpA2. Similarly, OmpA1 rather than OmpA2 was a major contributor to serum resistance of A. aphrophilus HK83. Far-Western blot revealed that OmpA1AA, OmpA2AA, and OmpA1AP can bind to C4-binding protein, an inhibitor of classical and mannose-binding lectin (MBL) complement activation. Indeed, ompA1 mutants were susceptible to these pathways, but also to alternative complement activation. This may at least partly reflect a compromised outer membrane integrity but is also consistent with alternative mechanisms involved in OmpA-mediated serum resistance.
KEYWORDS: Aggregatibacter actinomycetemcomitans, Aggregatibacter aphrophilus, serum resistance, outer membrane protein A
Introduction
The HACEK group of fastidious Gram-negative organisms is a recognized cause of infective endocarditis, responsible for 1.4 to 3% of cases [1], with the genus Aggregatibacter now being the dominant etiology of HACEK endocarditis [2]. Colonization by the human oral bacterium, Aggregatibacter actinomycetemcomitans is strongly associated with aggressive forms of periodontitis in adolescents and young adults [3,4]. Aggregatibacter aphrophilus is a closely related organism (14.7–23.7% difference in gene content relative to A. actinomycetemcomitans [5]), which belongs to the indigenous human oral microflora. Infective endocarditis and cerebral abscesses are the most frequent invasive A. aphrophilus infections, whereas this organism is widely considered to have low virulence in periodontitis [2].
A. actinomycetemcomitans exhibits large genetic diversity, and serotypes form genetically divergent lineages [4]. Highly leukotoxic A. actinomycetemcomitans genotypes, JP2 and cagE, respectively (serotype b) are strongly linked to periodontal attachment loss progression in North and West African adolescents [3,6]. A. actinomycetemcomitans produces outer membrane vesicles (OMVs), which have been demonstrated to internalize into host cells and act as a trigger of innate immunity [7]. The systemic role of A. actinomycetemcomitans, in addition to its involvement in endocarditis, includes its association with cases of soft tissue abscesses, and osteomyelitis [8], and the species can be detected in atheromatous plaque [9]. It is not known if there are specific genotypes of A. actinomycetemcomitans (or A. aphrophilus) that are more prone to translocate to the peripheral circulation.
Serum resistance is an important pathogenicity determinant of Gram-negative bacteria that enter the bloodstream and cause infection, allowing their evasion of the innate defenses present in serum, including complement and antimicrobial peptides. Resistance to complement-mediated killing by human serum appears to be important for A. actinomycetemcomitans virulence, and it is suggested to be a common trait among strains of this species [10,11]. It is not known whether serum resistance is also frequent among A. aphrophilus strains. Mechanisms of bacterial resistance against complement-mediated killing include the production of protective extracellular polysaccharide capsules, and expression of factors that inhibit or interfere with the complement cascade [12]. Outer membrane integrity is important for serum resistance of Gram-negative bacteria, and in a number of species, outer membrane proteins (OMPs) have been shown to be associated with serum resistance, e.g., Ail [13], OmpW [14], PagC [15], and OmpA [16,17]. OmpA protein family members represent key components in the structural integrity of the outer membrane of bacteria and have several described pathogenic roles [18–20].
Hence, OmpA inhibition offers a strategy to combat virulence of Gram-negative organisms [21]. C4b-binding protein (C4bp) is a major inhibitor of the classical and mannose-binding lectin (MBL) pathways of the complement system [22]. Evidence has been presented that upon interacting with C4bp, Escherichia coli OmpA inhibits the classical complement activation cascade [23,24]. On the other hand, In Acinetobacter baumanii, OmpA was shown to enhance serum resistance via trapping of the alternative complement inhibitor Factor H [16].
Among the OMPs identified in A. actinomycetemcomitans, Omp100 (also known as ApiA) was earlier demonstrated to be important for serum resistance in some serotype b, and d strains, and to bind to the alternative pathway regulator Factor H in vitro [10,25]. Evidently, A. actinomycetemcomitans OMPs are immunoreactive in the human host [26]. As presence of antibodies towards OMPs is a known trigger of classical complement activation [27], serum resistance of Aggregatibacter spp. would be expected to include mechanisms blocking this activation. A ≈ 35-kDa, 346-amino acid heat-modifiable OmpA-like protein (also known as Omp29, and Omp34) is the most abundant A. actinomycetemcomitans surface protein, and a major component of outer membrane vesicles [26,28]. The A. actinomycetemcomitans OmpA-like protein is associated with the bacterial entry into gingival epithelial cells [29], however, its role in serum resistance has not been elucidated. Whether A. aphrophilus OMPs, hitherto only subjected to a preliminary characterization [30], may possibly contribute to serum resistance is not known. The aim of the present work was to investigate if OmpA proteins play a role in serum resistance in A. actinomycetemcomitans and A. aphrophilus.
Materials and methods
Ethics considerations
All procedures were conducted in accordance with the guidelines of the local ethics committee at the Medical Faculty of Umeå University, which are in compliance with the Declaration of Helsinki (64th WMA General Assembly, Fortaleza, October 2013). For the assays using normal human serum, blood was sampled from healthy volunteers after informed consent.
Bacterial strains and growth conditions
A. actinomycetemcomitans strain D7SS is a naturally genetic competent, smooth-colony derivative of D7S (serotype a), which was originally isolated from a patient with aggressive periodontal disease [31]. Mutant A. actinomycetemcomitans derivatives, i.e., D7SS ompA1::spe [Sper], D7S ompA1::spe [Sper], D7SS ompA2::kan [Kmr], D7SS ompA1::spe, ompA2::kan [Sper, Kmr], D7SS omp100::kan [Kmr], and D7SS omp100::kan, ompA1::spe [Kmr, Sper] were generated in the present work. CCUG 3715 and NJ8700 are type strains of A. aphrophilus [32,33]. The naturally genetic competent A. aphrophilus strains HK83 (CCUG 49494), and CCUG 11575 were originally sampled from saliva, and a brain abscess, respectively [33]. DNA from NJ8700 was used to transform HK83 and CCUG11575 into a V factor-independent growth phenotype, following a described procedure [33]. Mutant derivatives of HK83, i.e., HK83 ompA1::spe [Sper], HK83 ompA2::kan [Kmr], and HK83 ompA1::spe, ompA2::kan [Sper, Kmr] were generated in the present work. Strains AHI-3151, IH-90256, and IH-90274 are part of the collection of clinical isolates of A. aphrophilus in our laboratory, established by Dr. Sirkka Asikainen. The A. aphrophilus strains 4 Aap-K, 12 Aap-K, 13 Aap-K, 21 Aap-K, 29 Aap-K, 30 Aap-K, 32 Aap-K, and 53 Aap-K belong to our bacterial strain collection at the clinical laboratory, Oral Microbiology. The A. actinomycetemcomitans and A. aphrophilus strains were routinely cultivated in air supplemented with 5% CO2, at 37°C, on blood agar plates (5% defibrinated horse blood, 5 mg hemin/l, 10 mg Vitamin K/l, Columbia agar base). Alternatively, for transformation assays, the strains were grown on Trypticase soy broth supplemented with 0.1% yeast extract, 5% heat-inactivated horse serum, and 1.5% agar (sTSB agar), and when needed, supplemented with 100 μg/ml (final concentration) spectinomycin, or kanamycin. E. coli K-12 laboratory strain DH5α was used for maintenance of plasmids and was cultured aerobically at 37°C in Luria-Bertani (LB) broth, or on LB broth solidified with 1.5% (w/v) agar.
Construction of outer membrane protein gene replacement mutants in A. actinomycetemcomitans and A. aphrophilus
A PCR-based approach following standard cloning procedures was used to construct gene replacement mutants in naturally competent strains of A. actinomycetemcomitans (D7SS), and A. aphrophilus (HK83). For A. actinomycetemcomitans, the strain D7S-1 complete genome (GenBank accession CP003496) was used as reference in oligonucleotide synthesis. For A. aphrophilus, strain NJ8700 was used as reference genome (GenBank accession CP001607). Whole genome sequence data of the A. aphrophilus model strain HK83 was kindly communicated by Niels Nørskov-Lauritsen and has been deposited at DDBJ/ENA/GenBank under the accession QMGS00000000. Sequences of the oligonucleotide primers used for mutant construction and outer membrane protein gene database entries are listed in Table 1. In brief, PCR fragments flanking the target genes were amplified. The PCR primers contained BamHI or SalI restriction sites, allowing ligation of the PCR fragments to flank either the spectinomycin resistance gene from plasmid pK-Spe [34], or the kanamycin resistance gene from pUC4K [35]. Ligation products were then used to transform D7SS and HK83 on agar plates using procedures described earlier [31]. Confirmation of allelic replacements and the orientation of the inserted resistance cassette were done by DNA sequencing and PCR. For this we used a target gene-specific F1, and R2 oligonucleotide primer (Table 1), respectively in combination with a primer specific for the appropriate antibiotic determinant, i.e., spectinomycin (Spe1: 5ʹ-CCACTCTCAACTCCTGATCC-3ʹ) or kanamycin (H7R: 5ʹ- GGACGGCGGCTTTGTTGAATAAATCG-3ʹ).
Table 1.
Target ompA1 and ompA2 genes in the A. actinomycetemcomitans and A. aphrophilus reference strains D7S-1 and NJ8700, respectively, and oligonucleotide primers used for the generation of allelic replacement mutants.
| Target gene GenBank accession number, and definition in database | Oligonucleotidea (F: forward; R: reverse) |
PCR product length (bp)b |
|---|---|---|
| Primers for A. actinomycetemcomitans | ||
|
ompA1AA c AFI86243 ‘membrane protein’ |
F1: 5ʹ- CTGCTTCACAAATAAAGGCGAGGGAG-3ʹ R1: 5ʹ- GATTGCAGTTGTCGACATTTTGATGATCC-3ʹ F2: 5ʹ-GAAATCGCTGTCGACGGTACTAAATA-3ʹ R2: 5ʹ-GCTCACGGCGGCAATCAATAAC-3’ |
1,295 1,287 |
|
ompA2AA d AFI86283 ‘membrane protein’ |
F1: 5ʹ-GAATCCCACGTCACCGTGCC-3ʹ R1: 5ʹ-GTAGCTAATGGGATCCCAGTTTTTTTC-3ʹ F2: 5ʹ-GTGACACGGATCCAGGTCGCAAAG-3ʹ R2: 5ʹ-CCGACAGTGGAATGTACGAAAACTAC-3ʹ |
1,269 1,061 |
|
omp100e AFI 86,288 ‘membrane protein’ |
F1: 5ʹ-GCATGGCGTCCAATAAACCTTG-3ʹ R1: 5ʹ-GTTTAAATAATGGATCCGTCATAATTCA-3ʹ F2: 5ʹ-CTATAACGTCGGATCCAACTTTGAGTG-3ʹ R2: 5ʹ-GCATGGTTGGAACGCTTCTTACAC-3ʹ |
1,386 1,208 |
| Primers for A. aphrophilus | ||
|
ompA1AP f ACS96969 ‘outer membrane protein A’ |
F1: 5ʹ-GTCGGATTTGACCGCACTTGTGTC-3ʹ R1: 5ʹ-GATAGCTAATGCGAGTCGACTTTTTTTCA-3ʹ F2: 5ʹ-GTAGAAATCGCTGTCGACGGTAGCAA −3ʹ R2: 5ʹ- GAG AAT CGG GAA AGG TCA CGG CT-3’ |
1,081 1,280 |
|
ompA2AP g ACS97183 ‘outer membrane protein A’ |
F1: 5ʹ-GGGAGCAGAGTGAGCAGGTG-3ʹ R1: 5ʹ-CAGCTAATGGGATCCTAGTTTTTTTC F2: 5ʹ-GAAATTGCAGTAAATGGGGATCCATAATT-3ʹ R2: 5ʹ-CAACTGACTCAACTCATCGAACAG-3’ |
1,328 1,322 |
aPrimers introducing BamHI (GGATCC) and SalI (GTCGAC) restriction sites (sequences in underlined bold) as indicated.
bPredicted based on genome sequences of D7S-1 and NJ8700, respectively.
cAlso referred to as Omp29, Omp34, and OmpA-like protein.
dEncoded protein exhibits ≈ 76% amino acid identity with OmpA1AA
eAlso referred to as ApiA.
fEncoded protein exhibits ≈ 83% amino acid identity with OmpA1AA
gEncoded protein exhibits ≈ 71% amino acid identity with OmpA1AA
SDS-PAGE and Western blot analysis
The procedures used for SDS-PAGE and Western blot analysis have been described previously [36]. Gels were stained using Coomassie Brilliant Blue R-250 (Bio-Rad) or Pierce® Silver Stain Kit (Thermo Fisher Scientific). Selected protein bands after Coomassie- or Silver-staining were excised from gels and subject to liquid chromatography tandem mass spectrometry (LC-MS/MS) at the Proteomics Core Facility at Chemical Biological Centre, Umeå University and Swedish University of Agricultural Sciences. For Western blot, we used a rabbit polyclonal antiserum specific for E. coli OmpA [37] (final dilution 1:10,000), and a patient serum from a periodontitis subject, which exhibits strong reactivity to A. actinomycetemcomitans antigens [38] (1:2,000). As secondary antibody, anti-rabbit or anti-human horseradish peroxidase (HRP)-conjugate was used (Jackson ImmunoResearch, Newmarket, UK) (1:10,000). Immunoreactive bands were visualized using Clarity™ Western ECL Substrate (Bio-Rad) or SuperSignal® West Pico Chemiluminescent Substrate (Thermo Fisher Scientific), and the ChemiDoc™ XRS + System (Bio-Rad).
Far-Western blot analysis
To detect binding of the complement regulator, C4b-binding protein to OmpA, the immunoblot membrane was blocked in tris-buffered saline, 0.1% tween 20 (TBS-T) with milk (5% w/v) overnight at 4°C, and then incubated with 1 μg/ml (final dilution) human recombinant C4bp (Abcam) in TBS-T with milk (0.5% w/v) for 2 h at room temperature. After washing four times with TBS-T, the membrane was incubated with a rabbit polyclonal antiserum specific for human C4bp (Abcam) (final dilution 1:1,500). After four washes with TBS-T, anti-rabbit HRP-conjugate (Jackson ImmunoResearch, Newmarket, UK) was used at 1:10,000, and immune detection was then performed as described above.
Extraction of outer membrane proteins
Outer membrane protein bacterial fractions were isolated essentially as described earlier [36]. A. actinomycetemcomitans or A. aphrophilus cells were harvested in 10 mM HEPES (pH 7.4) from an average of four blood agar plates. After centrifugation (40,000 × g; 20 min, 4°C), bacterial pellets were resuspended in HEPES and subject to sonication (30-second pulses; 30% amplitude). After removal of intact cells and cell debris (1,700 × g; 20 min, 4°C), supernatants were subject to ultracentrifugation (100,000 × g; 1 h, 4°C) using an SW60Ti rotor (Beckman Instruments Inc.). Pellets were resuspended in 2% (w/v) sodium-lauryl-sarcosinate (Sigma-Aldrich) in HEPES, and after sequential steps of ultracentrifugation (100,000 × g; 1 h, 4°C; SW60Ti) pellets were resuspended in buffer A (1% [w/v] n-octyl-β-D-glucopyranoside [Sigma-Aldrich] in 50 mM Tris, 5 mM EDTA, pH 8.0), in buffer A containing 0.5M NaCl, and finally in 20 mM sodium phosphate buffer (pH 7.5) containing 0.1% (w/v) SDS.
Isolation of outer membrane vesicles
As an alternative approach to obtain bacterial fractions enriched in OmpA proteins, OMVs were isolated from A. actinomycetemcomitans and A. aphrophilus cells harvested from an average of 10 blood agar plates, using ultracentrifugation as described earlier [7]. OMV pellets were washed twice with PBS (85,000 × g; 2 h, 4°C) using a 70 Ti rotor (Beckman Instruments Inc.), and then used as the OMV preparations. The yield of OMVs was estimated by quantifying vesicle preparations for protein content using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific). To assess the uniformity of OMV preparations, samples were validated by atomic force microscopy (AFM), and SDS-PAGE. OMVs were also checked for absence of bacterial contamination by cultivating small aliquots on blood agar plates in air supplemented with 5% CO2, at 37°C for 3 days.
Atomic force microscopy
For AFM, bacterial cells or OMV preparations were diluted with ultrapure water (Millipore) and placed onto a freshly cleaved mica surface. Samples were incubated for 5 min at room temperature, washed with ultrapure water, and then placed in a desiccator for ~2 h in order to dry. The samples were finally magnified through a Nanoscope V Atomic Force Microscope (Bruker AXS GmbH, Karlsruhe, Germany), using tapping mode. Final images were plane fitted in both the x and y axes and are presented in amplitude mode.
Electron microscopy
Analyses were carried out at the Umeå Core Facility for Electron Microscopy (UCEM). For transmission electron microscopy of OMV preparations, 3.5 µl of samples was applied to glow discharged formvar and carbon coated Cu-grids. The grids were washed and negatively stained in 1.5% Uranyl acetate for 2 × 15 sec. Samples were examined with a Talos 120C transmission electron microscope (FEI, Eindhoven, The Netherlands) operating at 120 kV. Micrographs were acquired with a Ceta 16M CCD camera (FEI, Eindhoven, The Netherlands) using TEM Image & Analysis software version 4.14 (FEI, Eindhoven, The Netherlands). For scanning electron microscopy, small pieces of agar containing bacterial colonies were fixed in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) at 4°C overnight, then dehydrated in graded series of ethanol, critical point dried, and metal-coated (4 nm). The morphology of the samples was analyzed with a field-emission scanning electron microscope (Zeiss Merlin FESEM).
Bacterial serum killing assay
To monitor the sensitivity of bacterial strains to normal human serum (NHS), NHS was taken from healthy volunteers. We largely followed procedures described previously [10,27], using A. actinomycetemcomitans, and A. aphrophilus strains grown on agar. In brief, prior to being used in the assays, bacteria were harvested and suspensions were adjusted to 1.0 × 109 cells/ml in PBS. Reaction mixtures contained 105 μl NHS, 95 μl PBS, and 10 μl bacterial suspension and were incubated at 37°C for 2 h. Cell survival was assessed by plating serial dilutions on agar. Heat-inactivated (56°C, 30 min) NHS (HI-NHS) was used as control. Survival rates were calculated as the ratio (%) of colony forming units (CFU) NHS/HI-NHS. When needed to distinguish between the classical and MBL, and the alternative pathway of complement activation, serum killing assays were performed in the presence of 5 mM MgCl2, and 10 mM EGTA (Mg2+/EGTA), which selectively permits the alternative pathway only [39]. To obtain NHS depleted from the MBL pathway, serum was filtered through D-mannose-agarose beads (Sigma-Aldrich, St. Louis, USA), as described previously [27].
Statistical analysis and image processing
The statistical significance of the data was calculated using two-tailed Student’s t-test. The level of statistical significance was set to P < 0.05, based on at least three independent experiments unless otherwise stated. Data are expressed as means ± standard errors of the means (SEM). Images for figures were assembled using Adobe Photoshop CS6 or Microsoft PowerPoint.
Results
Both A. actinomycetemcomitans and A. aphrophilus encode two OmpA proteins
To assess the prevalence of ompA genes in these species, we screened all whole genome sequences of A. actinomycetemcomitans strains (n = 38) and A. aphrophilus (n = 7) available in the National Center for Biotechnology (NCBI) database in September 2017. The result of this in silico analysis is summarized in Supplementary Table 1. Of the A. actinomycetemcomitans strains, 37 were found to encode the major OmpA protein (in the present work referred to as OmpA1AA). Strain SA3733 (serotype d), encoded a partial OmpA1 sequence. In total, six OmpA1AA alleles were identified exhibiting 99–100% amino acid identity to the strain D7S protein. A 356-amino acid protein sharing approximately 76% amino acid identity with OmpA1AA is in this work referred to as OmpA2AA. The database searches revealed OmpA2AA to be encoded by 37 out of the 38 A. actinomycetemcomitans genomes (not identified only in SA3733). In total, six OmpA2AA alleles were identified exhibiting 97–100% amino acid identity to the strain D7S protein. As there were allele combinations specific for some serotypes, e.g., OmpA1AA-4 and OmpA2AA-4 in serotype b, these gene and protein sequences may represent potential genotyping markers. According to the database searches, A. aphrophilus, similar to A. actinomycetemcomitans encodes two OmpA paralogues. These proteins (346 and 363–366 amino acids, respectively) exhibit ≈ 74% amino acid identity, and we have in this study referred to them as OmpA1AP and OmpA2AP. OmpA1AP was encoded in all A. aphrophilus genomes, and in total three alleles were identified exhibiting 96–100% amino acid identity to the strain NJ8700 protein. A gene encoding OmpA2AP was identified in all A. aphrophilus genomes, except ATCC7901 and in total five alleles were identified exhibiting 96–100% amino acid identity to the strain NJ8700 protein. Thus, a large majority of A. actinomycetemcomitans and A. aphrophilus strains encode two OmpA proteins.
OmpA1 is important for serum resistance in A. actinomycetemcomitans strains D7SS and D7S
To investigate if OmpA1AA contributes to serum resistance, the ompA1AA gene was subject to allelic replacement in strains D7S and D7SS. The abolished expression of OmpA1 in the mutants was confirmed by SDS-PAGE and Western blot, analyzing OMP (Figure 1(a,c)), and OMV preparations (Figure 1(b, d, and e)), respectively. These analyses revealed distinct protein bands at approximately 35 and 25 kDa, respectively, which were also confirmed to represent OmpA1AA using LC-MS/MS, and which were absent in the ompA1 mutant derivatives. According to serum killing assays using 50% NHS (Figure 2), strain D7SS was completely serum resistant (survival rate determined to 161.3%) whereas its ompA1 mutant derivative exhibited high susceptibility (survival rate 8.0%). Essentially similar survival rates were obtained when assessing the rough-colony D7SS parental strain, D7S (161.4% ± 21.9% [SEM]), and its ompA1 mutant derivative (1.1% ± 0.9% [SEM]). In contrast to the observations with ompA1, inactivation of omp100 in strain D7SS did not result in a low survival rate (106.6% ± 19.8% [SEM]). Lack of Omp100 production in the mutant was confirmed by Western blot (Figure 1(e)). Consistent with a major role of OmpA1 in serum resistance, inactivation of the ompA1 gene in the D7SS omp100 mutant led to a severely reduced survival rate, 3.1% (Figure 2). Based on these results together, we concluded that OmpA1 was a major contributor to the serum resistance of A. actinomycetemcomitans D7SS, and its parental strain D7S.
Figure 1.

SDS-PAGE analysis of outer membrane protein, and outer membrane vesicle preparations obtained from A. actinomycetemcomitans strains. OMP preparations were analyzed using Coomassie-staining (a), and Western blot using a polyclonal antiserum specific for E. coli OmpA (c). OMV preparations were analyzed using Silver-staining (b), and Western blot using a polyclonal antiserum specific for E. coli OmpA (d), or an A. actinomycetemcomitans-responsive serum from a periodontitis subject (e). Samples from the following strains are shown in the panels: D7SS (lane 1), D7SS omp100 ompA1 (lane 2 in panel a–d), D7SS ompA1 (lane 3), D7SS ompA1 R1 (lane 4), D7SS ompA1 ompA2 (lane 5), and D7SS omp100 (lane 2 in panel e). Selected protein bands are indicated with arrows. Sizes (kDa) of the proteins in the pre-stained molecular weight marker (M) are indicated along the left side.
Figure 2.
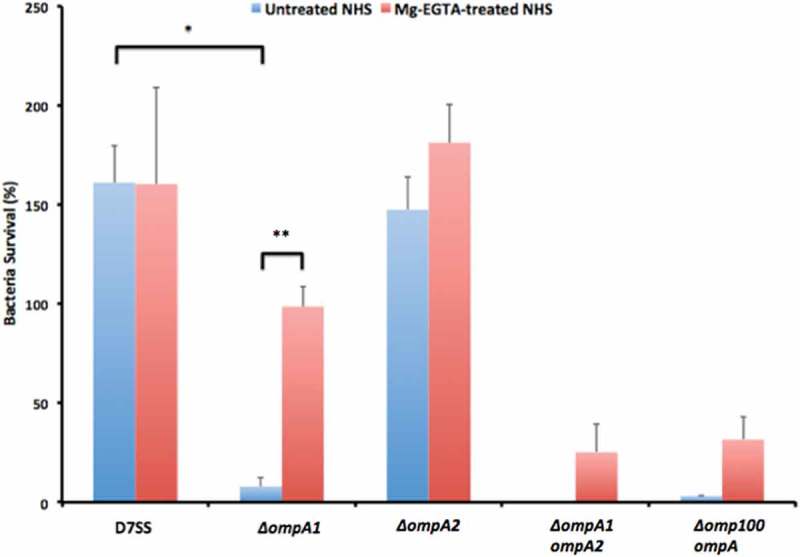
Analysis of the serum survival of A. actinomycetemcomitans strain D7SS and its mutant derivatives. 1.0 × 109 bacterial cells were incubated in 50% normal human serum (NHS), or 50% heat-inactivated (HI)-NHS at 37°C for 2 h. The assay was performed in the absence (blue bars) or presence (red bars) of Mg2+/EGTA. Bacterial serum survival was determined by viable count and expressed as ratio (%) of CFU in NHS/HI-NHS. Shown are means ± SEM from at least three independent experiments, with the exception of the D7SS ompA1 ompA2 double mutant, which was assessed twice. *P < 0.001, D7SS vs. D7SS ompA1 in the absence of Mg2+/EGTA; **P < 0.001, D7SS ompA1 in the absence vs. presence of Mg2+/EGTA.
A. actinomycetemcomitans strain D7SS ompA1 mutant derivatives regained serum resistance upon enhanced production of OmpA2
We made attempts to clone ompA1AA behind the leukotoxin promoter using the pVT1503 A. actinomycetemcomitans shuttle vector, and the same approach as for the complementation of bilRI mutants [40]. Unfortunately, this resulted solely in clones with a truncated C-terminus (data not shown), suggesting that overexpression of OmpA1 was toxic to the E. coli laboratory strain used. Nonetheless, colonies of the D7SS ompA1AA mutant were fortuitously obtained after the incubation in 50% NHS. After isolation of such clones from independent experiments, they were found to exhibit a serum survival rate, comparable to D7SS. Interestingly, according to SDS-PAGE analysis of OMV preparations from one selected serum resistant ompA1 mutant clone, denoted D7SS ompA1-R1 (survival rate 186.5% ± 62.1% [SEM]), there were two enhanced protein bands at approximate sizes very similar to OmpA1, i.e. ≈ 35 and ≈ 25 kDa (Figure 1(b,d)). These proteins, which appeared to be produced at a low level in the serum-sensitive D7SS ompA1 mutant derivative, were subject to LC-MS/MS analysis. This revealed that both protein bands corresponded to OmpA2AA. Thus, we concluded that the protein identified in the lower molecular weight band likely represented a degradation product of full length OmpA2. To investigate if serum resistant D7SS ompA1 derivatives may represent a heterogeneous population of mutants, OMV preparations from five additional clones were analyzed by SDS-PAGE (Supplementary Figure 1). This revealed upregulated OmpA2 production in four of the tested clones, suggesting that this is a common characteristic among D7SS ompA1 mutants surviving in 50% NHS. The role of OmpA2 was evaluated in D7SS ompA1, and in D7SS ompA1-R1, by allelic replacement of ompA2AA. As judged by the very low to non-existing serum survival rates of the double mutants, D7SS ompA1 ompA2 (0%), and D7SS ompA1-R1 ompA2 (1.8% ± 0.6% [SEM]), OmpA2 indeed contributed to serum resistance. In contrast, allelic replacement of ompA2AA in an ompA1AA+ gene background did not result in an apparent decrease in serum resistance (Figure 2). The survival rates of the wild-type strain and its mutant derivatives were similar in heat-inactivated 50% NHS (data not shown), whereas there was a somewhat reduced growth rate of the D7SS ompA1 single and ompA1 ompA2 double mutants in liquid cultures (Supplementary Figure 2). Electron microscopy revealed that OMV preparations from both D7SS and D7SS ompA1 ompA2 contained vesicles of various sizes (diameter approximately 30–200 nm) (Figure 3(a-b)). However, larger membrane vesicle-like structures were occasionally observed projecting from the cell surface of the double mutant, which was not observed assessing the wild-type (Figure 3(c-d)). This suggests that the serum sensitivity was accompanied by at least partially impaired membrane stability. Based on these results together, we concluded that in the absence of OmpA1, serum resistance of A. actinomycetemcomitans strain D7SS could be regained via increased production of OmpA2. As judged by DNA sequencing, background mechanisms why OmpA2 appeared to be produced at enhanced levels in the serum resistant D7SS ompA1 mutant clones did not include alterations in the ompA2 promoter and upstream region, as amplified using the primers ompA2AA F1 and ompA2AA R1 (data not shown).
Figure 3.

Electron micrographs of A. actinomycetemcomitans strains and their released OMVs. Transmission electron micrographs of OMVs released from D7SS (a), and D7SS ompA1 ompA2 (b). Scanning electron micrographs of D7SS (c), and D7SS ompA1 ompA2 (d) cultivated on agar. The arrows in panel d indicate examples of the larger membrane vesicle-like structures occasionally found projecting from the cell surface of the double mutant. Bars = 200 μm.
Serum resistance in A. aphrophilus strains
Prompted by our findings with A. actinomycetemcomitans, we wanted to investigate if OmpA proteins may contribute to serum resistance in an additional species of this genus, i.e., the close relative A. aphrophilus. In serum killing assays using 50% NHS, we screened our available (n = 15) A. aphrophilus strains in our collections, observing that they displayed moderate to high levels of serum resistance (Supplementary Table 2). Albeit this initial screening suggested that the degrees of serum resistance may vary among strains, it supported the notion that serum resistance may be ubiquitous also among A. aphrophilus strains. Strain HK83 was selected for genetic analysis as it is a naturally competent strain, facilitating the genetic analysis.
OmpA protein expression in A. aphrophilus strain HK83
Analysis of whole genome sequencing data of A. aphrophilus strain HK83 confirmed that it encodes OmpA1AP and OmpA2AP, both sharing approximately 96% amino acid identity with the corresponding proteins of the A. aphrophilus reference strain NJ8700. We noted that the OmpA1AP protein sequence of HK83 is identical to that of A. aphrophilus strains ATCC 7901 and W10433 (accession numbers OBY53818 and AKU63521, respectively), and that the HK83 OmpA2AP protein is identical to that of strain W10433 (accession number AKU63719). Strain HK83 was used to generate ompA1AP and ompA2AP single, and ompA1AP ompA2AP double mutants, respectively. To estimate the relative levels of produced OmpA1AP and OmpA2AP in HK83, SDS-PAGE and Western blot were used to analyze OMP preparations (Figure 4(a,c)). In addition, OMV preparations were assessed (Figure 4(b,d)) as A. aphrophilus strains were found to produce outer membrane vesicles, at a level similar to A. actinomycetemcomitans (Figure 5). These analyses revealed distinct protein bands at approximately 35 and 25 kDa, respectively, which were confirmed to represent OmpA1AP using LC-MS/MS, and which were absent in the ompA1AP mutant derivatives (Figure 4). This observation is consistent with the notion that OmpA1AP was the major OmpA protein produced under the growth conditions used. Moreover, SDS-PAGE revealed two protein bands with somewhat increased intensity, at approximately 37 kDa in OMP and OMV preparations from the HK83 ompA1APompA2AP double mutant (Figure 4(a,b)). According to LC-MS/MS analysis of these protein bands, the slightly larger-sized protein was identified as OmpC (AKU63902), a homologue to A. actinomycetemcomitans Omp39 (≈ 73% amino acid identity). This may represent compensatory expression of OmpC due to lack of OmpA1AP and OmpA2AP. The second protein band was identified as a methyl-galactoside ABC transporter substrate-binding protein (OBY51871).
Figure 4.

SDS-PAGE analysis of outer membrane protein, and outer membrane vesicle preparations obtained from A. aphrophilus strains. OMP preparations were analyzed with Coomassie-staining (a), and Western blot using a polyclonal antiserum specific for E. coli OmpA (c). OMV preparations were analyzed with Silver-staining (b), and Western blot using a polyclonal antiserum specific for E. coli OmpA (d). Samples from the following strains are shown in each panel: HK83 (lane 1), HK83 ompA1 (lane 2), HK83 ompA2 (lane 3), and HK83 ompA1 ompA2 (lane 4). Selected protein bands are indicated with arrows. Sizes (kDa) of the proteins in the pre-stained molecular weight marker (M) are indicated along the left side.
Figure 5.

Release of OMVs from A. aphrophilus strains. Atomic force micrographs of bacterial strains cultivated on agar. A. aphrophilus strains HK83 (a and c), CCUG 3715 (b), and HK83 ompA1 ompA2 (d), which produces large numbers of OMVs relative to HK83. Arrows indicate examples of the released A. aphrophilus vesicles in a and b. Bars = 300 nm (a, b), and 500 nm (c, d).
OmpA1 is important for the serum resistance of A. aphrophilus strain HK83
To elucidate if OmpA1AP and/or OmpA2AP might be involved in A. aphrophilus serum resistance, strain HK83 and its ompA1 and ompA2 single, and ompA1 ompA2 double mutants were assessed in serum killing experiments using 50% NHS. The ompA1 mutant exhibited a survival rate of 12.1%, i.e. almost four times lower than the parental strain, HK83 (Figure 6), clearly supporting that OmpA1AP was contributing to serum resistance. Similar to the findings with A. actinomycetemcomitans strain D7SS, inactivation of ompA2AP in HK83 did not significantly affect the serum survival. However, serum resistance was almost abolished in the ompA1 ompA2 double mutant (Figure 6), indicating that OmpA2AP contributed to the serum resistance in this background. The survival rates of the wild-type strain and its mutant derivatives were similar in heat-inactivated NHS (data not shown), whereas like the A. actinomycetemcomitans strains, there was a somewhat reduced growth rate of the HK83 ompA1 single and ompA1 ompA2 double mutants in liquid cultures (Supplementary Figure 2). As judged by the hypervesiculation phenotype of HK83 ompA1 ompA2 revealed by AFM (Figure 5(d)), its serum sensitivity was most likely accompanied by a disruption of the outer membrane integrity. Analogously to our findings with the A. actinomycetemcomitans strain, clones of the HK83 ompA1 mutant were fortuitously obtained after the incubation in 50% NHS, which after isolation were found to exhibit serum resistance at a level comparable to the parental strain. As judged by SDS-PAGE analysis of OMV preparations from selected ompA1 mutant clones with enhanced serum resistance, there was no noticeable difference in the levels of outer membrane proteins relative to the parental strain (data not shown). One clone with enhanced serum resistance, HK83 ompA1-R1 (survival rate 56.1% ± 12.6% [SEM]), was subject to inactivation of ompA2AP. As this resulted in essentially abolished serum survival (< 1%), we concluded that OmpA2 contributed to the serum resistance in this background.
Figure 6.

Analysis of the serum survival of A. aphrophilus strain HK83 and its mutant derivatives. 1.0 × 109 bacterial cells were incubated in 50% normal human serum (NHS), or 50% heat-inactivated (HI)-NHS at 37°C for 2 h. The assay was performed in the absence (blue bars) or presence (red bars) of Mg2+/EGTA. Bacterial serum survival was determined by viable count and expressed as ratio (%) of CFU in NHS/HI-NHS. Shown are means ± SEM from at least three independent experiments. *P < 0.04, HK83 vs HK83 ompA1 for both conditions.
Complement activation pathways effective in elimination of serum-sensitive OmpA mutant derivatives
To initially investigate mechanisms for OmpA-mediated serum resistance, we assessed which pathways of complement activation were effective in elimination of serum-sensitive ompA1 single and ompA1 ompA2 double mutant strains of the two species. To this end, serum killing assays were performed in the presence of Mg2+/EGTA, which allow alternative complement activation but inhibits the classical and MBL pathways (Figure 2). Whereas blocking the classical and MBL pathways had no effect on survival of the serum resistant A. actinomycetemcomitans strain D7SS, its ompA1 mutant displayed a largely increased survival rate (66.0%). Comparably, in the presence of Mg2+/EGTA there was much enhanced survival rates of the D7SS ompA1 ompA2 (25.3%), and ompA1 omp100 (31.7%), double mutants, respectively, and in serum-killing assays using MBL-depleted NHS, the survival rate of the D7SS ompA1 mutant was relatively high, i.e. 77.9% ± 27.6% [SEM]). This together supports that the serum susceptibility of the ompA1AA mutant derivatives to a relatively large extent was mediated via classical/MBL complement activation. However, as in the presence of Mg2+/EGTA, none of the serum-sensitive D7SS mutant derivatives exhibited a survival rate corresponding to that of their respective parental strains (Figure 2), we concluded that they were also susceptible to alternative complement activation.
In the presence of Mg2+/EGTA the serum survival rate of the A. aphrophilus wild-type strain, HK83, and its ompA2 derivative was 71.5% and 45.3%, respectively, consistent with a partial susceptibility to both classical/MBL and alternative complement activation (Figure 6). Likewise, although there was enhanced survival of the HK83 ompA1 mutant in the presence of Mg2+/EGTA, its survival rate was still as low as 19.3%, whereas the HK83 ompA1 ompA2 double mutant was essentially serum-sensitive. Moreover, the survival rate of the HK83 ompA1 mutant was enhanced when MBL-depleted serum was used (39.3% ± 13.9% [SEM]). Hence, taken together, these results support the notion that the classical, MBL, and alternative complement activation pathways were all involved in the elimination of the serum-sensitive ompA mutant derivatives tested. This suggests that alternate mechanisms may be involved in OmpA-mediated serum resistance.
OmpA-dependent trapping of C4b binding protein (C4bp)
To investigate possible mechanisms how the Aggregatibacter OmpA proteins may protect the bacterial cells from complement-mediated killing, we investigated whether OmpA1AA and OmpA1AP, similar to E. coli OmpA [24], might be able to bind to C4bp. For this, OMVs obtained from A. actinomycetemcomitans D7SS and A. aphrophilus HK83, and their respective ompA1 mutants, were assessed by Far-Western analysis using recombinant human C4bp, and polyclonal anti-C4bp, and anti-OmpA antibodies, respectively. The detection using the anti-C4bp antibody indicated that there was binding of C4bp as represented by a protein band on the membrane at a position corresponding to the size of OmpA, which was observed in the case of both wild-type strains (Figure 7(a)). This observation was confirmed using the anti-OmpA antibody (Figure 7(a)). In contrast, there was no such C4bp binding detected assessing the OMV samples from the ompA1AA and ompA1AP mutant, respectively (Figure 7(a)). Moreover, also OmpA2AA was found to bind to C4bp, assessing OMVs from the serum resistant strain D7SS ompA1-R1 (Figure 7(b)). These results together are consistent with the notion that both A. actinomycetemcomitans and A. aphrophilus can exhibit OmpA-dependent binding of C4bp.
Figure 7.
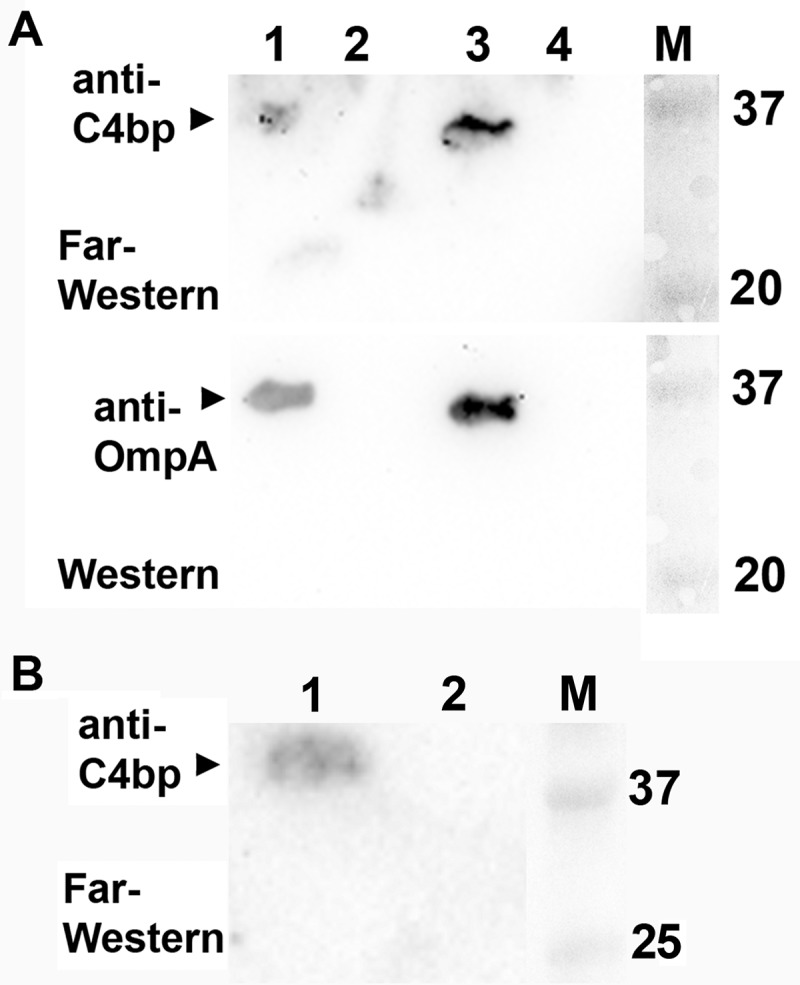
Far-Western analysis to detect binding of OmpA1AA and OmpA1AP to C4b-binding protein (C4bp). (a) OMVs obtained from A. actinomycetemcomitans D7SS (lane 1), and D7SS ompA1 (lane 2), and from A. aphrophilus HK83 (lane 3), and HK83 ompA1 (lane 4) were subjected to SDS-PAGE. For Far-Western analysis, the membrane was first incubated with C4bp (1 μg/ml final dilution) prior to immunodetection using an anti-C4bp polyclonal antibody. OmpA was detected using a polyclonal antiserum specific for E. coli OmpA. (b) Far-Western analysis of OMVs obtained from A. actinomycetemcomitans D7SS ompA1-R1 (lane 1), and D7SS ompA1 (lane 2), respectively. Sizes (kDa) of the proteins in the pre-stained molecular weight marker (M) are indicated along the right side.
Discussion
In the present work we have used naturally competent model strains of A. actinomycetemcomitans (D7SS; serotype a), and its close relative A. aphrophilus (HK83) to demonstrate the role of OmpA proteins in serum resistance of these organisms.
The observation that most whole genome-sequenced A. actinomycetemcomitans and A. aphrophilus strains encode two OmpA paralogues is in accordance with findings with a number of other Gram-negative species, e.g., Aeromonas salmonicida, Bacteroides fragilis, Haemophilus ducreyi, and Porphyromonas gingivalis [19,41–43]. We have referred to the major (i.e., the most abundant) OmpA protein of A. actinomycetemcomitans and A. aphrophilus as OmpA1AA and OmpA1AP, respectively, for consistence with studies on the other organisms encoding two OmpA paralogues. The observation that the pangenomes of these two species encode several alleles of OmpA1 and OmpA2, respectively, suggests that they represent potential genotypic markers, similar to the ompA sequences in Chlamydia trachomatis and Pasteurella multocida [44,45].
Based on previous studies with A. actinomycetemcomitans [10,46], we typically assessed the serum survival in 50% NHS. The observation that A. aphrophilus strains, similar to A. actinomycetemcomitans are ubiquitously resistant to killing by normal human serum is consistent with their association with extra-oral infections such as infective endocarditis and cerebral abscesses [2]. A role of OmpA proteins in serum resistance of A. actinomycetemcomitans and A. aphrophilus is in agreement with findings with a number of other bacterial species, including A. baumanii and E. coli [16,17]. In both Aggregatibacter species tested in the present work, the major OmpA protein, OmpA1 was important for the serum survival, albeit not essential, as a low frequency of the ompA1AA and ompA1AP mutant derivatives survived in the presence of 50% NHS. The interesting finding that ompA1AA mutants fortuitously regained their serum resistance upon increased OmpA2AA production, supported that OmpA2AA here acted as a functional homologue to OmpA1AA, which is consistent with the high degree of amino acid identity of these proteins, and with previous observations of compensatory OMP expression upon loss of major outer membrane porins [47]. Similarly, our present work suggests that there was compensatory production of OmpC and/or ABC transporter substrate protein in the ompA1APompA2AP double mutant, which would be in accordance with the role of homologues to these proteins in bacterial serum resistance [48,49].
The finding that no clones could be obtained expressing the full length OmpA1AA protein for mutant complementation, but rather a C-terminally truncated form, implies that overexpression of OmpA1 may be deleterious to the E. coli laboratory strain used. The phenomenon that OmpA of some bacterial species can be toxic in E. coli strains has earlier been described e.g., with OmpA of Haemophilus influenzae, Mycobacterium tuberculosis, and Rhodopseudomonas blastica [50–52]. In line with the essentially abolished serum survival of the ompA1 ompA2 double mutants of both species, EM and AFM, respectively revealed that they exhibited larger membrane-vesicle like structures projecting from the cell surface, and/or a hypervesiculation phenotype, consistent with earlier observations with ompA mutants exhibiting impaired membrane stability [19,53–56], and reduced growth [57].
In this work, we have also attempted to investigate putative mechanism(s) for OmpA-mediated protection of the bacterial cells from complement-mediated killing. Similar to earlier findings with Klebsiella pneumoniae [58], we observed that several complement activation pathways appeared to be effective in the elimination of serum-sensitive ompA1 single, and ompA1 ompA2 double mutant derivatives of A. actinomycetemcomitans and A. aphrophilus. This may, at least partly reflect a compromised outer membrane integrity in these mutants, which would be consistent with their somewhat reduced growth rates, observed in liquid cultures. However, evidently, OmpA proteins have been demonstrated to interact with serum components that regulate both the classical/MBL, and alternative pathways of complement activation, respectively [16,23,24]. Our finding that A. actinomycetemcomitans OmpA1 and OmpA2, and A. aphrophilus OmpA1 could bind to the classical/MBL pathway negative regulator, C4bp is consistent with observations with E. coli OmpA. It remains, however to be elucidated to which extent this interaction per se plays an active role in conferring serum resistance, similar to what was demonstrated with E. coli OmpA [23,24]. In contrast to earlier findings with a couple of A. actinomycetemcomitans serotype b and d strains [10,25], inactivation of the omp100 gene locus in the serotype a strain D7SS did not render the cells serum-susceptible. The reason for this discrepancy is not known but may reflect the relative levels of produced OMPs in the different strains under the conditions used. Such a scenario is plausible as judged by observations with Salmonella Enteritidis that the patterns of OMP expression vary among strains exhibiting different degrees of serum sensitivity [59]. The notion that A. actinomycetemcomitans and A. aphrophilus may exploit alternate mechanisms to evade complement-mediated killing is supported by recent findings that strains display different patterns of complement activation pathway component consumption in vitro [60]. Moreover, it has been demonstrated that human serum induces global stress responses to varying degrees in A. actinomycetemcomitans strains [46]. Thus, it is likely that serum resistance in both Aggregatibacter species, similar to A. baumanii [48], is highly complex, relying on a large number of gene products.
In summary, the present work has revealed the importance of primarily OmpA1, but also OmpA2 in serum resistance of A. actinomycetemcomitans and A. aphrophilus. The recognition of bacterial products mediating serum resistance represents an approach to vaccine and drug development. The existence of different alleles of OmpA1 and OmpA2 among strains in these species can possibly be utilized in the development of strain-specific vaccines and agents that reduce the serum resistance.
Funding Statement
This work was supported by TUA grants from the County Council of Västerbotten, Sweden (to J.O.), by funds from Insamlingsstiftelsen, Medical Faculty, Umeå University (to S.N.W. and J.O.), Swedish Research Council (to S.N.W.), and Svenska Tandläkare-sällskapet and Thuréus Foundation (to M.L.).
Acknowledgments
We are grateful to Elisabeth Granström, Arvid Dahlstrand Rudin, and John Burstedt for valuable technical assistance. We wish to thank Dr. Sirkka Asikainen for kindly providing the A. aphrophilus strains AHI-3151, CCUG3715, IH-90256, IH-90274, and NJ8700. Dr. Niels Nørskov-Lauritsen is thanked for kindly sending us the A. aphrophilus strains CCUG11575 and HK83, and for communicating HK83 whole genome sequence data. This work was supported by TUA grants from the County Council of Västerbotten, Sweden (to J.O.), by funds from Insamlingsstiftelsen, Medical Faculty, Umeå University (to S.N.W. and J.O.), Swedish Research Council (to S.N.W.), and Svenska Tandläkare-sällskapet and Thuréus Foundation (to M.L.).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplementary material can be accessed here.
References
- [1].Chambers ST, Murdoch D, Morris A, et al. HACEK infective endocarditis: characteristics and outcomes from a large, multi-national cohort. PLoS One. 2013;8(5):e63181 PubMed PMID: 23690995; PubMed Central PMCID: PMC3656887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Norskov-Lauritsen N.Classification, identification, and clinical significance of haemophilus and aggregatibacter species with host specificity for humans. Clin Microbiol Rev. 2014;27(2):214–14. PubMed PMID: 24696434; PubMed Central PMCID: PMC3993099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Haubek D, Ennibi OK, Poulsen K, et al. Risk of aggressive periodontitis in adolescent carriers of the JP2 clone of Aggregatibacter (Actinobacillus) actinomycetemcomitans in Morocco: a prospective longitudinal cohort study. Lancet. 2008;371(9608):237–242. PubMed PMID: 18207019. [DOI] [PubMed] [Google Scholar]
- [4].Henderson B, Ward JM, Ready D. Aggregatibacter (Actinobacillus) actinomycetemcomitans: a triple A* periodontopathogen? Periodontol 2000. 2010;54(1):78–105. PubMed PMID: 20712635. [DOI] [PubMed] [Google Scholar]
- [5].Kittichotirat W, Bumgarner RE, Chen C. Evolutionary divergence of Aggregatibacter actinomycetemcomitans. J Dent Res. 2016;95(1):94–101. PubMed PMID: 26420795; PubMed Central PMCID: PMC4700661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Johansson A, Claesson R, Höglund Åberg C, et al. The cagE gene sequence as a diagnostic marker to identify JP2 and non-JP2 highly leukotoxic Aggregatibacter actinomycetemcomitans serotype b strains. J Periodontal Res. 2017. PubMed PMID: 28397250 DOI: 10.1111/jre.12462 [DOI] [PubMed] [Google Scholar]
- [7].Thay B, Damm A, Kufer TA, et al. Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1- and NOD2-dependent NF-κB activation. Infect Immun. 2014;82(10):4034–4046. PubMed PMID: 25024364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].van Winkelhoff AJ, Slots J. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis in nonoral infections. Periodontol 2000. 1999;20:122–135. PubMed PMID: 10522225. [DOI] [PubMed] [Google Scholar]
- [9].Bale BF, Doneen AL, Vigerust DJ. High-risk periodontal pathogens contribute to the pathogenesis of atherosclerosis. Postgrad Medical J. 2017;93(1098):215–220. PubMed PMID: 27899684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Asakawa R, Komatsuzawa H, Kawai T, et al. Outer membrane protein 100, a versatile virulence factor of Actinobacillus actinomycetemcomitans. Mol Microbiol. 2003;50(4):1125–1139. PubMed PMID: 14622404. [DOI] [PubMed] [Google Scholar]
- [11].Sundqvist G, Johansson E. Bactericidal effect of pooled human serum on Bacteroides melaninogenicus, Bacteroides asaccharolyticus and Actinobacillus actinomycetemcomitans. Scand J Dent Res. 1982;90(1):29–36. PubMed PMID: 6123149. [DOI] [PubMed] [Google Scholar]
- [12].Abreu AG, Barbosa AS. How Escherichia coli circumvent complement-mediated killing. Front Immunol. 2017;8:452 PubMed PMID: 28473832; PubMed Central PMCID: PMC5397495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Bliska JB, Falkow S. Bacterial resistance to complement killing mediated by the Ail protein of Yersinia enterocolitica. Proc Natl Acad Sci USA. 1992;89(8):3561–3565. PubMed PMID: 1565652; PubMed Central PMCID: PMC48908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Li W, Wen L, Li C, et al. Contribution of the outer membrane protein OmpW in Escherichia coli to complement resistance from binding to factor H. Microb Pathog. 2016;98:57–62. PubMed PMID: 27364548. [DOI] [PubMed] [Google Scholar]
- [15].Nishio M, Okada N, Miki T, et al. Identification of the outer-membrane protein PagC required for the serum resistance phenotype in Salmonella enterica serovar Choleraesuis. Microbiology. 2005;151(Pt 3):863–873. PubMed PMID: 15758232. [DOI] [PubMed] [Google Scholar]
- [16].Kim SW, Choi CH, Moon DC, et al. Serum resistance of Acinetobacter baumannii through the binding of factor H to outer membrane proteins. FEMS Microbiol Lett. 2009;301(2):224–231. PubMed PMID: 19878322. [DOI] [PubMed] [Google Scholar]
- [17].Weiser JN, Gotschlich EC. Outer membrane protein A (OmpA) contributes to serum resistance and pathogenicity of Escherichia coli K-1. Infect Immun. 1991;59(7):2252–2258. PubMed PMID: 1646768; PubMed Central PMCID: PMC258003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Confer AW, Ayalew S. The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet Microbiol. 2013;163(3–4):207–222. PubMed PMID: 22986056. [DOI] [PubMed] [Google Scholar]
- [19].Iwami J, Murakami Y, Nagano K, et al. Further evidence that major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis stabilize bacterial cells. Oral Microbiol Immunol. 2007;22(5):356–360. PubMed PMID: 17803635. [DOI] [PubMed] [Google Scholar]
- [20].Sonntag I, Schwarz H, Hirota Y, et al. Cell envelope and shape of Escherichia coli: multiple mutants missing the outer membrane lipoprotein and other major outer membrane proteins. J Bacteriol. 1978;136(1):280–285. PubMed PMID: 361695; PubMed Central PMCID: PMC218658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Vila-Farres X, Parra-Millan R, Sanchez-Encinales V, et al. Combating virulence of Gram-negative bacilli by OmpA inhibition. Sci Rep. 2017;7(1):14683 PubMed PMID: 29089624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Suankratay C, Mold C, Zhang Y, et al. Mechanism of complement-dependent haemolysis via the lectin pathway: role of the complement regulatory proteins. Clin Exp Immunol. 1999;117(3):442–448. Epub 1999/ 09/01.PubMed PMID: 10469045; PubMed Central PMCID: PMCPMC1905373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Prasadarao NV, Blom AM, Villoutreix BO, et al. A novel interaction of outer membrane protein A with C4b binding protein mediates serum resistance of Escherichia coli K1. J Immunol. 2002;169(11):6352–6360. PubMed PMID: 12444142. [DOI] [PubMed] [Google Scholar]
- [24].Wooster DG, Maruvada R, Blom AM, et al. Logarithmic phase Escherichia coli K1 efficiently avoids serum killing by promoting C4bp-mediated C3b and C4b degradation. Immunology. 2006;117(4):482–493. PubMed PMID: 16556262; PubMed Central PMCID: PMC1564124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc Natl Acad Sci USA. 2009;106(5):1578–1583. PubMed PMID: 19164580; PubMed Central PMCID: PMC2629492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Komatsuzawa H, Asakawa R, Kawai T, et al. Identification of six major outer membrane proteins from Actinobacillus actinomycetemcomitans. Gene. 2002;288(1–2):195–201. PubMed PMID: 12034509. [DOI] [PubMed] [Google Scholar]
- [27].Aung KM, Sjöström AE, von Pawel-Rammingen U, et al. Naturally occurring IgG antibodies provide innate protection against Vibrio cholerae bacteremia by recognition of the outer membrane protein U. J Innate Immun. 2016;8(3):269–283. PubMed PMID: 26934383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kato S, Kowashi Y, Demuth DR. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb Pathog. 2002;32(1):1–13. PubMed PMID: 11782116. [DOI] [PubMed] [Google Scholar]
- [29].Kajiya M, Komatsuzawa H, Papantonakis A, et al. Aggregatibacter actinomycetemcomitans Omp29 is associated with bacterial entry to gingival epithelial cells by F-actin rearrangement. PLoS One. 2011;6(4):e18287 PubMed PMID: 21559515; PubMed Central PMCID: PMC3084700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bolstad AI, Kristoffersen T, Olsen I, et al. Outer membrane proteins of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus studied by SDS-PAGE and immunoblotting. Oral Microbiol Immunol. 1990;5(3):155–161. PubMed PMID: 2080070. [DOI] [PubMed] [Google Scholar]
- [31].Wang Y, Goodman SD, Redfield RJ, et al. Natural transformation and DNA uptake signal sequences in Actinobacillus actinomycetemcomitans. J Bacteriol. 2002;184(13):3442–3449. PubMed PMID: 12057937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Di Bonaventura MP, DeSalle R, Pop M, et al. Complete genome sequence of Aggregatibacter (Haemophilus) aphrophilus NJ8700. J Bacteriol. 2009;191(14):4693–4694. PubMed PMID: 19447908; PubMed Central PMCID: PMC2704734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Norskov-Lauritsen N, Kilian M. Reclassification of Actinobacillus actinomycetemcomitans, Haemophilus aphrophilus, Haemophilus paraphrophilus and Haemophilus segnis as Aggregatibacter actinomycetemcomitans gen. nov., comb. nov., Aggregatibacter aphrophilus comb. nov. and Aggregatibacter segnis comb. nov., and emended description of Aggregatibacter aphrophilus to include V factor-dependent and V factor-independent isolates. Int J Syst Evol Microbiol. 2006;56(Pt 9):2135–2146. PubMed PMID: 16957111. [DOI] [PubMed] [Google Scholar]
- [34].LeBlanc DJ, Lee LN, Inamine JM. Cloning and nucleotide base sequence analysis of a spectinomycin adenyltransferase AAD(9) determinant from Enterococcus faecalis. Antimicrob Agents Chemother. 1991;35(9):1804–1810. PubMed PMID: 1659306; PubMed Central PMCID: PMC245272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19(3):259–268. PubMed PMID: 6295879. [DOI] [PubMed] [Google Scholar]
- [36].Paul-Satyaseela M, Karched M, Bian Z, et al. Immunoproteomics of Actinobacillus actinomycetemcomitans outer-membrane proteins reveal a highly immunoreactive peptidoglycan-associated lipoprotein. J Med Microbiol. 2006;55(Pt 7):931–942. PubMed PMID: 16772422. [DOI] [PubMed] [Google Scholar]
- [37].Henning U, Schwarz H, Chen R. Radioimmunological screening method for specific membrane proteins. Anal Biochem. 1979;97(1):153–157. PubMed PMID: 384844. [DOI] [PubMed] [Google Scholar]
- [38].Brage M, Holmlund A, Johansson A. Humoral immune response to Aggregatibacter actinomycetemcomitans leukotoxin. J Periodontal Res. 2011;46(2):170–175. PubMed PMID: 21118415. [DOI] [PubMed] [Google Scholar]
- [39].Des Prez RM, Bryan CS, Hawiger J, et al. Function of the classical and alternate pathways of human complement in serum treated with ethylene glycol tetraacetic acid and MgCl2-ethylene glycol tetraacetic acid. Infect Immun. 1975;11(6):1235–1243. PubMed PMID: 806523; PubMed Central PMCID: PMC415205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ahlstrand T, Tuominen H, Beklen A, et al. A novel intrinsically disordered outer membrane lipoprotein of Aggregatibacter actinomycetemcomitans binds various cytokines and plays a role in biofilm response to interleukin-1β and interleukin-8. Virulence. 2017;8(2):115–134. Epub 2016/ 07/28. PubMed PMID: 27459270; PubMed Central PMCID: PMCPMC5383217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Costello GM, Vipond R, MacIntyre S. Aeromonas salmonicida possesses two genes encoding homologs of the major outer membrane protein, OmpA. J Bacteriol. 1996;178(6):1623–1630. PubMed PMID: 8626290; PubMed Central PMCID: PMC177847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Klesney-Tait J, Hiltke TJ, Maciver I, et al. The major outer membrane protein of Haemophilus ducreyi consists of two OmpA homologs. J Bacteriol. 1997;179(5):1764–1773. PubMed PMID: 9045839; PubMed Central PMCID: PMC178892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wexler HM, Tenorio E, Pumbwe L. Characteristics of Bacteroides fragilis lacking the major outer membrane protein, OmpA. Microbiology. 2009;155(Pt 8):2694–2706. PubMed PMID: 19497947. [DOI] [PubMed] [Google Scholar]
- [44].Stephens RS, Sanchez-Pescador R, Wagar EA, et al. Diversity of Chlamydia trachomatis major outer membrane protein genes. J Bacteriol. 1987;169(9):3879–3885. PubMed PMID: 3040664; PubMed Central PMCID: PMC213681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].E-Kobon T, Leeanan R, Pannoi S, et al. OmpA protein sequence-based typing and virulence-associated gene profiles of Pasteurella multocida isolates associated with bovine haemorrhagic septicaemia and porcine pneumonic pasteurellosis in Thailand. BMC Vet Res. 2017;13(1):243 PubMed PMID: 28814302; PubMed Central PMCID: PMC5559837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tang-Siegel G, Bumgarner R, Ruiz T, et al. Human serum-specific activation of alternative sigma factors, the stress responders in Aggregatibacter actinomycetemcomitans. PLoS One. 2016;11(8):e0160018 Epub 2016/08/05. PubMed PMID: 27490177; PubMed Central PMCID: PMCPMC4973924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Provenzano D, Lauriano CM, Klose KE. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J Bacteriol. 2001;183(12):3652–3662. PubMed PMID: 11371530; PubMed Central PMCID: PMC95243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Sanchez-Larrayoz AF, Elhosseiny NM, Chevrette MG, et al. Complexity of complement resistance factors expressed by Acinetobacter baumannii needed for survival in human serum. J Immunol. 2017. PubMed PMID: 28855313 DOI: 10.4049/jimmunol.1700877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Skorek K, Raczkowska A, Dudek B, et al. Regulatory protein OmpR influences the serum resistance of Yersinia enterocolitica O:9 by modifying the structure of the outer membrane. PLoS One. 2013;8(11):e79525 Epub 2013/ 11/22. PubMed PMID: 24260242; PubMed Central PMCID: PMCPMC3834241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Pullen JK, Liang SM, Blake MS, et al. Production of Haemophilus influenzae type-b porin in Escherichia coli and its folding into the trimeric form. Gene. 1995;152(1):85–88. Epub 1995/01/11. PubMed PMID: 7828934. [DOI] [PubMed] [Google Scholar]
- [51].Schmid B, Kromer M, Schulz GE. Expression of porin from Rhodopseudomonas blastica in Escherichia coli inclusion bodies and folding into exact native structure. FEBS Lett. 1996;381(1–2):111–114. Epub 1996/02/26. PubMed PMID: 8641415. [DOI] [PubMed] [Google Scholar]
- [52].Senaratne RH, Mobasheri H, Papavinasasundaram KG, et al. Expression of a gene for a porin-like protein of the OmpA family from Mycobacterium tuberculosis H37Rv. J Bacteriol. 1998;180(14):3541–3547. Epub 1998/07/11.PubMed PMID: 9657995; PubMed Central PMCID: PMCPMC107320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kulp AJ, Sun B, Ai T, et al. Genome-wide assessment of outer membrane vesicle production in Escherichia coli. PLoS One. 2015;10(9):e0139200 PubMed PMID: 26406465; PubMed Central PMCID: PMC4583269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Moon DC, Choi CH, Lee JH, et al. Acinetobacter baumannii outer membrane protein A modulates the biogenesis of outer membrane vesicles. J Microbiol. 2012;50(1):155–160. PubMed PMID: 22367951. [DOI] [PubMed] [Google Scholar]
- [55].Song T, Mika F, Lindmark B, et al. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol. 2008;70(1):100–111. PubMed PMID: 18681937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Valeru SP, Shanan S, Alossimi H, et al. Lack of outer membrane protein A enhances the release of outer membrane vesicles and survival of Vibrio cholerae and suppresses viability of Acanthamoeba castellanii. Int J Microbiol. 2014;2014:610190 PubMed PMID: 24799908; PubMed Central PMCID: PMC3995163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Manning PA, Pugsley AP, Reeves P. Defective growth functions in mutants of Escherichia coli K12 lacking a major outer membrane protein. J Mol Biol. 1977;116(2):285–300. Epub 1977/10/25. PubMed PMID: 340698. [DOI] [PubMed] [Google Scholar]
- [58].Alberti S, Marques G, Camprubi S, et al. C1q binding and activation of the complement classical pathway by Klebsiella pneumoniae outer membrane proteins. Infect Immun. 1993;61(3):852–860. Epub 1993/03/01.PubMed PMID: 8432605; PubMed Central PMCID: PMCPMC302811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Dudek B, Krzyzewska E, Kapczynska K, et al. Proteomic analysis of outer membrane proteins from Salmonella Enteritidis strains with different sensitivity to human serum. PLoS One. 2016;11(10):e0164069 PubMed PMID: 27695090; PubMed Central PMCID: PMC5047454 provided clinical bacterial strains used in experiments in the present work. There are no patents, products in development or marketed products to declare. This does not alter our adherence to PLOS ONE policies on sharing data and materials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Damgaard C, Reinholdt J, Palarasah Y, et al. In vitro complement activation, adherence to red blood cells and induction of mononuclear cell cytokine production by four strains of Aggregatibacter actinomycetemcomitans with different fimbriation and expression of leukotoxin. J Periodontal Res. 2017;52(3):485–496. PubMed PMID: 27663487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


