SUMMARY
Tracheal and esophageal disorders are prevalent in humans and are difficult to accurately model in mice. We therefore established a three-dimensional organoid model of esophageal development through directed differentiation of human pluripotent stem cells. Sequential manipulation of BMP, WNT, and RA signaling pathways was required to pattern definitive endoderm into foregut, anterior foregut (AFG), and dorsal AFG spheroids. Dorsal AFG spheroids grown in a 3D matrix formed human esophageal organoids (HEOs), and HEO cells could be transitioned into two-dimensional cultures and grown as esophageal organotypic rafts. In both configurations, esophageal tissues had proliferative basal progenitors and a differentiated stratified squamous epithelium. Using HEO cultures to model human esophageal birth defects, we identified that Sox2 promotes esophageal specification in part through repressing Wnt signaling in dorsal AFG and promoting survival. Consistently, Sox2 ablation in mice causes esophageal agenesis. Thus, HEOs present a powerful platform for modeling human pathologies and tissue engineering.
eTOC
Trisno et al have generated human esophageal organoids (HEOs) through the directed differentiation of pluripotent stem cells. HEOs contain esophageal progenitors and a differentiated stratified squamous epithelium. Using HEOs to model foregut development revealed that SOX2 regulates NKX2–1 through modulation of Wnt signaling.
Graphical Abstract

INTRODUCTION
The esophagus actively facilitates the passing of food from the oral cavity and pharynx to the stomach. It consists of a stratified squamous epithelium, muscle layers, and an enteric nervous system to sense stretch and control peristalsis. Congenital diseases such as esophageal atresia are caused by gene mutations that result in luminal narrowing or discontinuity. Other diseases affect the esophagus later in life, such as esophageal carcinoma, eosinophilic esophagitis, achalasia and other motility disorders. There are substantial differences in tissue architecture between mouse and human esophagus, emphasizing the importance of having human esophageal tissue models for research.
Human tissue organoids, differentiated from pluripotent stem cells (PSCs) or obtained directly from organs, have proven to be excellent models of tissue physiology and pathology (McCauley and Wells, 2017). In general, the process of converting PSCs into organ cell types relies on step-wise differentiation that recapitulates organogenesis, including formation of definitive endoderm (DE), anteriorposterior patterning into foregut, midgut, and hindgut, organ specification, and differentiation into organ specific lineages. This approach has been used to generate human anterior and posterior endoderm organoids including respiratory, gastric, small intestine and colon (Chen et al., 2017; Dye et al., 2015, 2016, McCracken et al., 2014, 2017; Múnera et al., 2017; Spence et al., 2011). However, human PSCderived esophageal tissues have not been reported. Dual BMP and TGFβ inhibition after DE induction generates anterior foregut (AFG); however, this yielded a mix of tissues including pharyngeal, esophageal and respiratory endoderm (Green et al., 2011; Kearns et al., 2013; Longmire et al., 2012). This suggests that a more refined patterning approach based on pathways that control esophageal development is required to direct differentiation of PSCs specifically into esophagus.
Several signaling pathways guide differentiation and morphogenesis of the developing esophagus. The esophageal epithelium derives from definitive endoderm (DE), a 2-dimensional sheet of cells that forms during gastrulation (Zorn and Wells, 2007). The DE then is patterned along the anteriorposterior axis by Wnt, BMP and FGF signaling and forms a primitive gut tube, divided broadly into the foregut, midgut, and hindgut (Dessimoz et al., 2006; McLin et al., 2007; Stevens et al., 2017; Zorn and Wells, 2009). The foregut is further patterned into posterior foregut by retinoic acid (RA) (Bayha et al., 2009; Niederreither et al., 1999; Wang et al., 2006). The anterior foregut (AFG) gives rise to the esophagus and respiratory tract. Respiratory specification in response to Wnt and BMP activation results in expression of the transcription factor Nkx2–1 whereas inhibition of BMP in the dorsal foregut promotes development of Sox2-expressing esophageal epithelium (Domyan et al., 2011; Goss et al., 2009; HarrisJohnson et al., 2009; Que et al., 2006; Rankin et al., 2016). The esophagus starts as a simple cuboidal epithelium but develops into a stratified squamous epithelium that expresses multiple keratin proteins and a basal layer that expresses Sox2 and p63 (Rosekrans et al., 2015; Zhang et al., 2016).
Here, we temporally manipulated the above signaling pathways to differentiate human PSCs into esophageal organoids. Following DE formation, we identified that precise temporal manipulation of BMP, WNT, and RA pathways direct formation of AFG spheroids. Consistent with in vivo data, AFG spheroids acquired a respiratory fate through activation of WNT and BMP pathways whereas BMP inhibition promoted formation of dorsal foregut spheroids that upon continued growth for 1–2 months formed human esophageal organoids (HEOs). HEOs contained stratified squamous epithelium, with distinct basal and luminal cell layers, and harbored proliferative esophageal progenitors that could be expanded and differentiated into esophageal epithelium in organotypic raft cultures. We used HEOs, in parallel with mouse embryos, to identify molecular pathways that are affected by SOX2 loss-of-function, one cause of esophageal atresia in humans and mice (Domyan et al., 2011; Que et al., 2007). While reduced Sox2 function leads to esophageal atresia in mice, complete loss of Sox2 in mouse foregut endoderm results in esophageal agenesis. Loss of SOX2 function and transcriptional profiling of human and mouse foregut identified that SOX2 regulates the dorsal expression of Wnt antagonists such as SFRP2, suggesting that SOX2 represses the ability of Wnt to induce a respiratory fate in the dorsal foregut. In conclusion, HEOs provide a complementary platform to study human esophageal organogenesis, birth defects, and disease.
RESULTS
Wnt and retinoic acid signaling control anterior versus posterior foregut fate
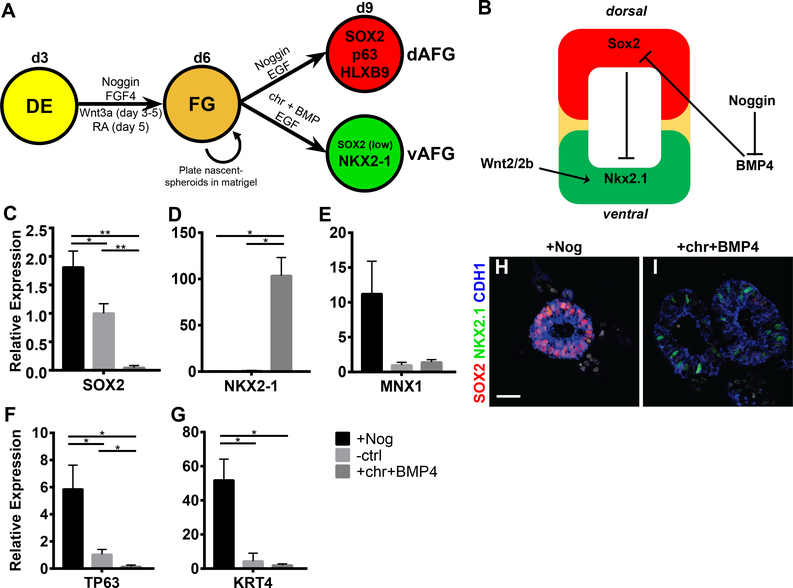
To generate foregut derivatives, hPSCs were first induced into DE and then three-dimensional (3D) SOX2-expressing foregut spheroids as previously described (Figure 1) (D’Amour et al., 2005; Dye et al., 2016; McCracken et al., 2014, 2017). In attempting to generate esophageal organoids, the primary challenge was to generate foregut tissue of the correct regional identity. Endoderm patterning is regulated by differential BMP, WNT and RA signaling, where the highest levels of activation of these pathways promoting a mid- and hindgut fate and lower levels promoting a foregut fate (Bayha et al., 2009; Davenport et al., 2016; Matt et al., 2003; McLin et al., 2007; Tiso et al., 2002; Wang et al., 2006). Based on our previous studies indicating that the duration of signaling is critical for differentiation, we tested the effects of duration of Wnt activation during foregut spheroid formation on anterior-posterior identity (Spence et al., 2011). We found that shorter duration of chiron, a canonical Wnt pathway activator (through GSK3β inhibition), or Wnt3a treatment following DE formation resulted in formation of anterior foregut (AFG) spheroids expressing HNF1β and SOX2, with low levels of posterior foregut markers PROX1 and HNF6 (Figure 1A-E,S1A-G). HNF1β is not expressed in pharyngeal endoderm indicating that AFG spheroids were not pharyngeal (Figure 1C-E,S1B). CDX2, a mid/hindgut marker, is not expressed (Figure 1B,D,E). From this, we concluded that the regional identity of these foregut spheroids (HNF1B+/SOX2+, PROX1-/HNF6-) are distal to the pharynx and proximal to the posterior foregut.
Figure 1:
Specifying anterior foregut fate by modulating Wnt and retinoic acid signaling during foregut spheroid development.
(A) The experimental protocol to pattern foregut spheroids along anterior-posterior axis by manipulating the duration of Wnt activation (chiron - chr). (B-C) qPCR analysis of varying chiron treatment duration on patterning of foregut spheroids as measured by (B) the foregut marker SOX2 and mid/hindgut marker CDX2, and (C) the anterior foregut (AFG) marker HNF1B, and the posterior foregut markers PROX1 and HNF6. (D-E) Whole-mount immunofluorescence (IF) analysis with HNF1B, SOX2 and CTNNB1 of nascent spheroids (day 6) treated with 1 day (D) and 3 days (E) of chiron. (F) The experimental protocol to pattern foregut spheroids along anterior-posterior axis using retinoic acid (RA). (G) Effects of varying the duration of RA treatment on 3-day-old foregut spheroids as measured by SOX2, TP63 (ΔN isoform), GATA4, and PDX1. (J-K) IF analysis on early esophageal markers SOX2 and p63 in untreated spheroids (I), and spheroids treated with RA for 1 day (J) or 4 days (K). (H) Quantification of the percent of SOX2+ and p63+ epithelial cells per spheroid. Scale bar = 25μm. See quantification and statistical analysis section for details. See also Figure S1 and S2.
Four days of RA treatment is also known to posteriorize foregut spheroids (McCracken et al., 2014), and loss of RA signaling results in abnormal development of posterior foregut organs (Bayha et al., 2009; Wang et al., 2006). We therefore investigated whether shortening the duration of RA signaling in our foregut cultures would promote a more anterior fate. Foregut cultures treated with RA for 4 days express posterior foregut markers, GATA4 and PDX1, whereas 1 day of RA treatment resulted in spheroids that express TP63, a marker expressed in the developing esophagus (Fig. 1F-K,S1O-R). Cultures lacking RA, or containing DEAB, an aldehyde dehydrogenase inhibitor (blocking RA synthesis) yielded spheroids with minimal TP63 expression and increased levels of the pharyngeal markers PAX9 and OTX2 (Figure 1G-K,S1F-G; Figure S1S-V). Together, our data suggest that brief activation of RA promotes foregut regional identity consistent with the presumptive esophageal domain.
Anterior foregut spheroids are competent to form esophagus or respiratory lineages.
The presumptive esophageal/respiratory region of the foregut is patterned along the dorsalventral (D-V) axis, resulting in specification of the esophageal and respiratory fates respectively. We predicted that AFG spheroids would respond to D-V patterning cues to acquire either an esophageal or respiratory fate. Studies in vertebrate embryos demonstrate that BMP and Wnt signaling promote a respiratory fate (NKX2–1+ SOX2-), while Noggin-mediated inhibition of BMP signaling in the dorsal foregut tube is required for esophageal development (Domyan et al., 2011; Fausett et al., 2014; Goss et al., 2009; Harris-Johnson et al., 2009; Que et al., 2006). Thus, we treated day 6 AFG spheroids with either 3 days of chiron and BMP4 or alternatively with Noggin to inhibit BMP signaling (Figure 2A-B). Treatment with chiron+BMP4 resulted in induction of NKX2–1 and repression of SOX2, whereas treatment with Noggin dorsalized spheroids, as marked by elevated levels of SOX2, MNX1, KRT4, and TP63 (Figure 2C-I) (Daniely et al., 2004; Sherwood et al., 2009). Taken together, inhibition of BMP signaling in cultures of AFG spheroids promoted a dorsal anterior foregut identity.
Figure 2:
Anterior foregut spheroids have esophageal-respiratory competence.
(A) Schematic depicting experimental protocol to pattern AFG spheroids along the dorsal-ventral axis. (B) Current simplified model of the cues guiding dorsal-ventral patterning of the AFG of mouse and frog embryos. (C-G) qPCR analysis of 3-day-old spheroids (day 9) treated for 3 days with Noggin, untreated (ctrl), or chiron and BMP4 (10ng/mL) using dorsal markers SOX2 and MNX1 (C+E), the respiratory marker NKX2–1 (D), ΔN splice variant of TP63 (F), and the stratified squamous epithelium marker KRT4 (G). (H-I) IF staining for SOX2, NKX2–1, CDH1, and nuclei (DAPI) in Noggin (H) versus chiron+BMP4 (I) treated spheroids. Scale bar = 25μm. See quantification and statistical analysis section for details. See also Figure S3.
Formation of esophageal organoids with a stratified squamous epithelium
To determine if dorsally-patterned AFG spheroids were competent to grow into esophageal organoids, we cultured them suspended in matrigel with EGF alone or in the context of manipulation of other pathways predicted to promote esophageal development including Wnt activation (chiron), extended BMP inhibition (Noggin), activation of hedgehog (SAG, a smoothened agonist), and FGF10. While most of these manipulations had no effect on growth (data not shown), we found that addition of FGF10 to cultures from day 6 to day 13 resulted in improved efficiency of spheroid to organoid outgrowth (Figure S4A-E) and did not affect the patterning and differentiation into esophageal organoids (data not shown).
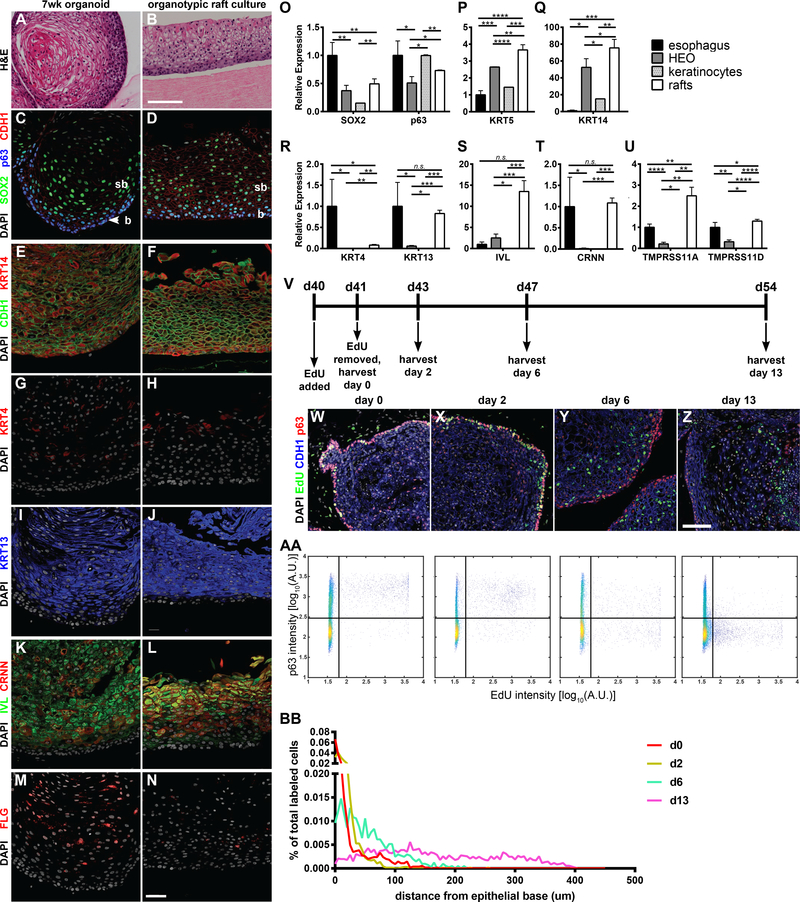
Next, we compared the morphologic and molecular development of putative human esophageal organoids (HEOs) to normal development of the embryonic mouse esophagus. During embryonic growth and development, the esophagus transitions from simple cuboidal epithelium at E12.5 to a multilayered/stratified epithelium between E14.5 and E17.5 (Figure 3G,3J,3M,3P,S4F-O,S4R) (Chen et al., 2012). Similarly, over the course of one month, HEOs expanded in size from approximately 50μm to 200–400μm in diameter (Figure 3B-F). Moreover, the organoid epithelium transitioned from a simple epithelium that was largely SOX2, p63 double-positive to a multilayered epithelium expressing markers of esophageal stratified squamous epithelium (Figure S4H-Q,S4S). At one month, HEOs expressed basal markers p63 and KRT14 as well as the suprabasal marker KRT13 (Figure 3H,3K,3N,3Q). This expression pattern is similar to an E17.5 esophagus composed of multilayered epithelium (Figure 3G,3J,3M,3P).
Figure 3:
Dorsal anterior foregut spheroids form organoids comprised of a stratified squamous epithelium that expresses esophageal markers. </p.>(A) Schematic depicting differentiation of DE into human esophageal organoids (HEOs). (B-F) Brightfield images depicting growth of nascent spheroids into HEOs. (G-R) Comparison of E17.5 esophagi. (G,J,M,K) to 1- and 2-month-old HEOs (H-I,K-L,N-O,Q-R), by IF analysis of the transcription factors Sox2 and p63 (G-I), epithelial markers Krt8 versus Krt14 (J-O), and the suprabasal marker Krt13 (P-R). (S-V) qPCR analysis of the identity and maturation of esophageal organoids at 1- and 2- months of age compared to human gastric and intestinal organoids (HGO and HIO) and pediatric esophageal biopsies by the stratified squamous epithelial markers p63, KRT5, KRT13, IVL, CRNN. (W) Unsupervised hierarchical clustering of 2 month HEOs compared to various biopsies of the GI tract. (X) Principal component analysis of 1 month old HIOs, HGOs, and HEOs. (Y) Heat map of log2-transformed normalized TPM values of selected genes (esophageal, gastric, intestinal) averaged across replicates. SSE = stratified squamous epithelium; b = basal; sb = suprabasal. Scale bar = 500μm (B-F), 50μm (G-L), 100μm (O-R), and 25μm (O’-R’). See quantification and statistical analysis section for details. See also Figure S4.
One month HEOs were still relatively immature as evidenced by broad SOX2 epithelial expression and expression of KRT8, a marker of immature esophagus (Figure 3J,S4L-M). We therefore extended the culture period out to 2 months, which resulted in additional growth and formation of a stratified epithelium that lacked KRT8. We observed robust expression of KRT14 throughout the basal layer and KRT13 and IVL throughout the differentiated suprabasal layers, demonstrating the presence of a stratified squamous epithelium (Figure 3I,3L,3O,3R). Histologically, 2 month HEOs had a more defined basal layer as well as squamous cells suprabasally, with no evidence of cornification (Figure 4A). The epithelial morphology of HEOs was easily distinguished from other organoids including gastric (HGO), intestinal (HIO) or colonic organoids, with each organoid having an epithelial morphology that is unique to that organ type (McCracken et al., 2014, 2017; Múnera et al., 2017; Spence et al., 2011).
Figure 4:
HEOs contain progenitors that give rise to differentiated stratified squamous epithelium.
(A-B) H&E staining comparing 7 week HEOs to organotypic rafts generated using HEO-derived from keratinocytes. (C-N) Comparison of 7 week HEOs to organotypic rafts by IF analysis of transcription factors SOX2 and p63 (C-D), basal marker KRT14 (E-F), suprabasal keratins KRT4 (G-H) and KRT13 (IJ), and differentiated markers IVL, CRNN, and FLG (K-N). (O-U) qPCR analysis of esophageal biopsies, 7 week HEOs, HEO-derived keratinocytes, and organotypic rafts for SOX2 and TP63 (O), KRT5 (P), KRT14 (Q), KRT4 and KRT13 (R), IVL (S), CRNN (T), and esophageal specific markers TMPRSS11A/D (U). (V) Protocol for EdU pulse-chase labeling experiment in HEOs. (W-Z) IF images of HEOs at various time-points post-labeling. (AA-BB) Analysis of IF images using a 2D histogram of P63 intensity versus EdU intensity. (AA) and a 1D histogram of percent of total EdU labeled cells versus distance from the epithelial base (BB). b = basal; sb = suprabasal. Scale bar = 50μm (C-N), 100μm (A-B,S-V). See quantification and statistical analysis section for details. See also Figure S5.
To show that HEOs were human esophageal epithelium, we compared 1 and 2 month HEOs to human esophageal biopsies as well as 1 month HGOs and HIOs by qPCR. Key stratified squamous epithelial markers p63, KRT5, KRT13, IVL, and CRNN were expressed highest in human esophageal biopsies and 2 month HEOs, whereas HGOs and HIOs negligibly expressed these transcripts (Figure 3SV). Additionally, we compared the entire transcriptome of HEOs with that of other human epithelial tissues isolated from esophagus, lung, skin, stomach, small intestine, colon, HGOs, and HIOs. Clustering analyses of RNA sequencing data revealed that HEOs are most closely related to human esophagus and EPC2, an esophageal keratinocyte cell line, as opposed to the stomach, small intestine, and colon (Figure 3W). We used principal component analyses to compare HEOs with HIOs and HGOs and found that even one month HEOs were entirely distinct from the other gastrointestinal organoids (Figure 3X). A comparison of markers of esophagus, skin, stomach, and colon revealed that HEOs were highly similar to human esophagus, reaffirming our qPCR analysis. While there was significant overlap between skin and esophagus, there were also distinct differences including KRT1 in the skin and KRT4 and 13 in the esophagus. Of note, HEOs did not express gastric or intestinal markers TFF2, CLDN18, GATA4, PDX1, CDX2, and CDH17 (Figure 3Y).
Given that growth of PSC-derived organoids in vivo has been shown to promote further maturation and function, we utilized three different transplantation-based approaches to study HEOs in vivo. We first engrafted 1 month old HEOs into the kidney capsule of immunodeficient (NSG) mice and allowed them to grow for 8 weeks, which resulted in maturation in a subset of the organoids transplanted (2/5) (Figure S5A-F). We used two other transplantation approaches that were unsuccessful: seeding HEOs onto biodegradable PEG scaffolds and transplanting them into the fat pad of mice; or engrafting HEOs into the forestomach of NSG mice (data not shown) (Dye et al., 2016). Overall, the generation of spheroids and organoid outgrowth is robust across various ES and iPS lines, with each generation resulting in a large majority of organoids expressing stratified squamous markers (Figure S4T-CC). Together, our data demonstrate that PSC-derived dorsal foregut spheroids form esophageal organoids with a well-differentiated, non-keratinized stratified squamous epithelium.
HEOs contain progenitors capable of reconstituting a stratified squamous epithelium
The esophagus contains basal progenitors that can give rise to all of the differentiated stratified layers (DeWard et al., 2014; Doupe et al., 2012; Kalabis et al., 2008). This property allows esophageal cells to be isolated, expanded in culture, and then re-differentiated into a stratified squamous epithelium. We first tested if HEOs contain esophageal progenitors by enzymatically dissociating 5 week HEOs into single cells, expanding them in monolayer cultures, and then testing their ability to re-differentiate into a stratified squamous epithelium using an organotypic raft culture method (Hoskins et al., 2009). After 14 days of organotypic culture, the HEO-derived keratinocytes gave rise to a non-keratinized stratified squamous epithelium, expressing the appropriate keratins and differentiated markers IVL, CRNN, FLG. (Figure 4A-N). HEOs, HEO-derived keratinocytes and organotypic raft cultures all expressed high levels of the basal markers p63, KRT5, and KRT14 (Figure 4O-Q), whereas organotypic rafts were the most differentiated, expressing CRNN, IVL, KRT13, TMPRSS11A and D at levels comparable to human esophageal biopsies (Figure 4R-U). However, efforts to expand these progenitors long-term in 3D organoid cultures were unsuccessful. Dissociated organoids re-plated in 3D-matrigel grew over several weeks, maintained patterning (SOX2+ p63+), and were passaged (re-dissociated) several times (Figure S5G-H,S5O,S5Q-R). However, passaging efficiency diminished over time, and we were unable to induce differentiation or stratification in these passaged organoids (Figure S5I-N,S5P). This result is similar to esophageal progenitors derived from human esophagus, demonstrating a general inability to culture these cells long-term (Kasagi et al., 2018).
Another method to study basal progenitor differentiation is through pulse-chase labeling of proliferating basal progenitors and following labeled cells as they differentiate into stratified layers over time. We labeled proliferating cells in day 40 HEOs by one day treatment of EdU and analyzed them immediately (day 0) or following a chase period for 2, 6, and 13 days post-label (Figure 4V-BB). EdU labeled cells initially appeared in p63-expressing basal cells, but over time, these cells moved into the suprabasal compartment, lost p63 expression, and were eventually shed into the lumen by 13 days postlabel (Figure 4W-BB). We conclude that HEOs contain basal progenitors that differentiate and migrate into the stratified layers, similar to esophageal epithelium.
Using HEOs and mouse genetics to identify mechanisms of esophageal development
While establishing a PSC-derived HEO model system is an important advance, we wanted to demonstrate that HEOs could be used to study human development and disease. We chose to use HEOs, in parallel with two well-established vertebrate model systems, mouse and Xenopus, to model esophageal birth defects by studying loss-of-function of SOX2. First, we first determined the consequences of complete loss of Sox2, since partial loss of SOX2 function in humans and mice leads to partial loss of the esophagus (atresia) (OMIM 206900, Fantes et al., 2003; Que et al., 2007; Williamson et al., 2006). We generated two mouse models to inducibly delete Sox2 in the foregut endoderm prior to initiation of esophageal development (FoxA2CreER ;Sox2fl/fl and Sox2CreER/fl) (Arnold et al., 2011; Park et al., 2008; Shaham et al., 2009). In both models, early deletion of Sox2 in the foregut resulted in complete esophageal agenesis, with the foregut region between the pharynx and stomach remaining as one tube lacking p63 and broadly expressing the respiratory marker Nkx2–1 (Figure 5C-F,S6A-D). The lung buds were largely similar to control embryos, and cell polarity was unaffected (Figure S6E-F). Deletion of Sox2 after initiation of esophageal development resulted in partial loss of esophageal tissue, with regions of the esophagus being severely hypoplastic at E11.5. In some regions, the esophagus is only 2–3 cells wide, remains a simple cuboidal epithelium, lacks p63 expression, and expresses Nkx2–1 in some cells (Figure 5I-L). These data suggest that Sox2 function is required for initiating esophageal development whereas loss of Sox2 one day later results in reduced esophageal tissue and identity.
Figure 5:
Early endodermal deletion of Sox2 results in esophageal agenesis in mouse.
(A-D) IF analysis for Sox2 and Nkx2–1 in control embryos (Sox2fl/fl) and Sox2 conditional endodermal knockout embryos (Sox2-DE-LOF, FoxA2CreER;Sox2fl/fl) from pregnant dams gavaged with tamoxifen at 6.5dpc. Embryo sections at E9.5 (A-B) and whole-mount IF at E11.5 (C-D) in which the image is masked highlight the endoderm. (E-F) IF images of sections with the relative section indicated in the whole-mount images (C-D) for Nkx2–1 (E) and p63 (F). Insets show only the Sox2 channel (left) and the green/right (Nkx2–1 or p63) channel. (G-H) Analysis of cell death by cleaved Caspase 3 staining in E10.5 Sox2 cKO (Sox2CreER/fl) embryos from pregnant dams gavaged at 8.5dpc. The boxed region is magnified and shown in (G’-H’), with the endoderm is outlined in white and displays only the cleaved Caspase 3. (I-L) IF analysis of E11.5 mouse control and Sox2 cKO embryos (Sox2CreER/fl) from pregnant dams gavaged at 9.5dpc. (I and J) Whole-mount IF for Nkx2–1 and Foxa2 of the foregut from a side and frontal projection. (K and L) Sections of the E11.5 foregut corresponding to their relative position in the whole-mount IF projections (I-J), stained for Nkx2–1 (K) and p63 (J), with the yellow arrowhead pointing at the mutant esophagus. Scale bar = 50μm in all IF sections, and 100μm in all IF whole-mount projections. See quantification and statistical analysis section for details. fg = foregut, dfg = dorsal foregut, vfg = ventral foregut, eso = esophagus, tr = trachea, br = bronchi, st = stomach. See also Figure S6.
To investigate if esophageal agenesis was caused by changes in cell death and proliferation, or merely absence of septation from the common foregut, we analyzed E10.5 embryos at the start of septation. Sox2CreER/fl embryos had increased cleaved Caspase 3 staining in the dorsal foregut at the level where separation would normally be occurring, suggesting that cells of the presumptive esophagus were undergoing cell death (Figure 5G-H,S6U). Proliferation, as marked by Ki67+ cells, was unchanged (Figure S6V). In both control and Sox2 knockout foreguts, there appears to be a point where the epithelium narrows midway along the dorsal-ventral axis, suggesting that the epithelium is attempting to separate into the two tubes regardless of the presence of Sox2 protein (Figure S6M-T). The results in mouse demonstrate Sox2 is required in the foregut for esophageal development, survival and for restricting Nkx2–1 to the ventral/respiratory domain.
BMP-independent roles for Sox2 in repressing Nkx2–1 expression
We next wanted to exploit the in vitro advantages of human and Xenopus foregut cultures to mechanistically explore how Sox2 initiates esophageal development. From previous studies, Sox2 is believed to repress respiratory (ventral) development and promote esophageal (dorsal) development. In order to promote ventral identity, BMP signaling is believed to repress Sox2 in the ventral foregut, thus allowing for Wnt-mediated induction of Nkx2–1 expression (Domyan et al., 2011). We tested if the sole function of BMP is to inhibit Sox2 by inhibiting Sox2 and then activating Wnt, which is predicted to be sufficient to activate Nkx2–1 in the absence of BMP. Knocking down sox2 using morpholino injections in Xenopus endoderm explants and activating canonical Wnt signaling with Bio did not activate nkx2–1 expression in the absence of BMP4 (Figure 6A-B, 6E-F). However, treatment with Bio and BMP4 expanded the nkx2–1 domain upon sox2 knock down as it was in the mouse Sox2 knockout (Figure 6CD). These data suggest two things: one, BMP signaling is required for Nkx2–1 expression independent of Sox2 inhibition; and two, Sox2 is required for repressing ectopic Nkx2–1 expression outside of the respiratory domain.
Figure 6:
Sox2 represses the respiratory fate and promotes the dorsal (esophageal) lineage.
(A-F) In situ hybridization for nkx2–1 of control (A,C,E) or Sox2 MO-injected (B,D,F) Xenopus endoderm explants analyzed at stage NF35 treated with Bio (GSK3β inhibitor) and Bio+BMP4. (G) Schematic depicting experimental protocol to generate human dorsal (Noggin) and ventral (BMP) AFG cultures. +SOX2 indicates tet-inducible SOX2, while -SOX2 indicates SOX2 CRISPRi. (H-N) Analysis of day 9 AFG cultures patterned along the dorsal-ventral axis, with or without SOX2 knockdown in the dorsal cultures using Dox-inducible CRISPRi on day 3–9; (H-K) IF staining of cultures for SOX2 and NKX2–1 and quantification in (L). (M-N) qPCR analysis for SOX2 and NKX2–1 in response to these patterning conditions. (O-U) Doxycycline-induced expression of exogenous SOX2 in ventral cultures on day 8 and analysis on day 9. (O-R) IF staining of cultures for NKX2–1 and HA-SOX2; and (S-T) qPCR analysis for SOX2 and NKX2–1 in response to patterning conditions. Scale bar = 50 μm for IF images, and 200 μm for Xenopus explant images. See quantification and statistical analysis section for details.
To determine if human SOX2 is required to prevent ectopic expression of NKX2–1, we used an iPSC line to inducibly express a repressor form of CRISPR protein that represses transcription at the SOX2 locus (CRISPRi-SOX2) (Mandegar et al., 2016). SOX2 knockdown in human dorsal anterior foregut cultures (dAFG) resulted in ectopic expression of NKX2–1 mRNA and protein (Figure 6G-J, 6L-M). Optimal NKX2–1 induction in ventral anterior foregut (vAFG) was still dependent on the presence of BMP (Figure 6K-N). To determine if SOX2 expression was sufficient to repress NKX2–1, we generated a stable tet-inducible hPSC line to express HA-tagged SOX2 in the ventral foregut during respiratory induction. Expression of SOX2 in the ventral foregut resulted in significant downregulation of NKX2–1 mRNA and protein (Figure 6Q-T). Together, these data suggest that BMP has additional functions for respiratory induction and confirm that in all contexts Wnt signaling is required for NKX2–1 expression. In addition, SOX2 expression is sufficient to repress NKX2–1 expression through unknown mechanisms.
Sox2 regulates expression of Wnt antagonists during dorsal-ventral patterning
The ease of manipulation and scalability of foregut cultures are ideally suited for “omic” approaches. We therefore took an RNA sequencing-based approach to identify genes that are regulated by SOX2 and/or BMP signaling during dorsal-ventral (esophageal-respiratory) patterning. Principal component analysis (PCA) identified that the largest groups of regulated genes were for dorsal-ventral patterning (± BMP4 or Noggin) and for SOX2-regulated genes (± dox-SOX2 CRISPRi) (Figure S7A). Moreover, SOX2 regulates a distinct set of genes in dorsal (Noggin) cultures as compared to ventral (BMP4) cultures, as indicated in the cluster heatmap and in the PCA along principal component axis 1 (Figure 7A,S7A). The use of either BMP4 or Noggin resulted in the expected changes in dorsal-ventral patterning markers, such as upregulation of NKX2–1 and repression of SOX2, MNX1, KRT4, PAX9 in the ventral (BMP-high) cultures. The loss of SOX2, however, resulted in many transcriptional changes in the dorsal foregut and relatively few in the ventral foregut, including increased expression of NKX2–1 and reduced expression of FOXE1, NTN1, and GDNF (Figure 7A-B, 7D,S7B, Table S3).
Figure 7:
Sox2 regulates expression of secreted Wnt antagonists and Wnt signaling activity in the dorsal foregut endoderm.
(A) Clustered heatmap of differentially expressed genes from RNA sequencing of day 9 dorsal (+Noggin) or ventral (+BMP4) AFG cultures with (+dox) and without SOX2 CRISPR interference (CRISPRi). (B) Venn diagram analysis of genes upregulated in dorsal and ventral cultures compared to genes that are elevated or decreased following SOX2 knockdown by CRISPRi. (C) Gene ontology (GO) term analysis on biological processes for genes positively regulated by SOX2. (D) Number of genes enriched in dorsal and ventral cultures and whether their expression was SOX2-dependent. (E) Gene set enrichment analysis of the gene ontology term “Regulation of Wnt signaling pathway”, red indicating higher expression while blue indicates low expression. (F-G) In situ hybridization for the Wnt-responsive gene Axin2 on E9.5 mouse anterior foreguts in (F) control (Sox2fl/fl) and (G) Sox2-DE-LOF (FoxA2CreER;Sox2fl/fl) embryos. (H-I) In situ hybridization for Axin2 in E10.5 mouse embryonic foregut of (H) control (Sox2fl/+) and (I) Sox2 cKO (Sox2CreER/fl) embryos taken from dams gavaged at 8.5dpc. Numbers of embryos analyzed is shown in the upper left. Boxed regions (F-I) highlight the dorsal foregut region. (J) qPCR analysis for AXIN2 in day 9 dorsal and ventral foregut cultures with or without SOX2 exogenously expressed. (K) Plotted TPM values for Wnt antagonists SFRP1, SFRP2, and DKK1 from RNA-seq of AFG cultures. (L) Proposed model on role of Sox2 in dorsal-ventral patterning of the anterior foregut. Scale bar = 100μm. See materials and methods & quantification and statistical analysis section for details. See also Figure S7 and Table S3.
Of the dorsal and ventral genes, there were transcripts that changed in response to the CRISPRi-SOX2 (either elevated or reduced) and genes that were not (SOX2-independent). For example, of the 542 genes enriched in the dorsal foregut, 75.6% (410 genes) are SOX2-independent. 17.2% (93 genes) in the dorsal foregut were downregulated upon SOX2 knockdown, and we refer to these as “positively regulated by SOX2”. There were 39 transcripts that were elevated in response to SOX2 knockdown, suggesting that these are “negatively regulated by SOX2”. In the ventral cultures, 374 genes are upregulated by BMP treatment, and of these, 38 were decreased and 5 were increased in response to loss of SOX2 (Figure 7D). Not surprisingly, more genes were regulated by SOX2 in the dorsal foregut as ventral SOX2 expression is already significantly downregulated in response to BMP. We used intersectional analysis to identify genes that BMP likely regulates through repression of SOX2 and found 46 genes (12.3%) that are both upregulated by both BMP treatment and SOX2 knockdown (“Genes negatively regulated by SOX2”). We also found 81 genes (14.9%) that are both downregulated by both BMP treatment and SOX2 knockdown (“Genes positively regulated by SOX2”) (Figure 7B). In addition, >80% of BMP-regulated transcripts were unchanged in response to SOX2 knockdown, consistent with the conclusion that BMP has a role in ventral foregut specification independent of SOX2 repression.
Performing gene ontology analysis of all 404 unique genes whose expression was reduced in dAFG in response to SOX2 knockdown yielded in many significant gene ontology terms, including two terms involving the Wnt signaling pathway (Figure 7C). Gene set enrichment analysis also found several Wnt signaling components to be significantly altered in response to SOX2 knockdown (Figure 7E). The secreted canonical Wnt signaling inhibitors SFRP1, SFRP2, DKK1, were all downregulated upon SOX2 loss (Figure 7E,K). Moreover, over-expression of SOX2 in ventral cultures upregulated SFRP2, which is consistent with published ChIP-seq data in hPSC-derived mesendoderm and endoderm showing a SOX2 binding peak at the SFRP2 locus (Figure S7C-D) (Tsankov et al., 2015).
Since SOX2 positively regulates expression of Wnt antagonists, we hypothesized that SOX2 may inhibit canonical Wnt signaling in the dorsal foregut. To investigate this, we deleted Sox2 from the mouse foregut using two genetic models, Foxa2CreER;Sox2fl/fl and Sox2CreER/fl and measured canonical Wnt/β-catenin activity by analyzing expression of the Wnt target gene Axin2 (Jho et al., 2002; Lustig et al., 2002). In situ hybridization revealed high levels of Axin2 mRNA in the ventral foregut endoderm and low levels in the dorsal foregut endoderm of control embryos. In contrast, the dorsal foregut had increased Axin2 staining when Sox2 was deleted (Figure 7F-I). Similarly, AXIN2 transcript levels were reduced in human ventral foregut cultures with SOX2 exogenously expressed (Figure 6G,7J). In addition to increased expression of canonical Wnt genes, the Nkx2–1 expression domain was expanded into the dorsal foregut (Figure 5E,S6C-D). Together with the data from human foregut cultures, we propose a model in which Sox2 positively regulates expression of secreted Wnt antagonists in the dorsal foregut, which represses canonical Wnt signaling in the dorsal foregut and restricts expression of Nkx2–1 to the ventral foregut.
DISCUSSION
Generation of HEOs and organotypic raft cultures has been described from primary esophageal cells and cell lines (Andl et al., 2003; Kalabis et al., 2012; Kasagi et al., 2018). In addition, human PSCderived anterior foregut (AFG) endoderm cultures give rise to a heterogeneous mix of multiple AFG derivatives (Green et al., 2011; Kearns et al., 2013; Longmire et al., 2012). To enrich for esophageal endoderm with this sort of approach, one must rely on cell sorting and subsequent culture, as achieved by Zhang et al. (manuscript co-submitted with this one). Alternatively, directed differentiation into specific foregut derivatives, like the esophagus, has benefited from a more granular recapitulation of early organ development. Here, we have differentiated human PSCs specifically into HEOs using a step-wise manner approximating DE formation, foregut patterning and morphogenesis, AFG patterning into the presumptive esophageal-respiratory domain, and finally dorsal foregut patterning. This approach gradually restricts endodermal differentiation potential such that one is left with dorsal AFG endoderm that grows out into esophageal organoids.
One challenge was to find conditions that generate the respiratory-esophageal anterior region of the foregut but not the anterior-most pharyngeal region. BMP inhibition is essential for foregut specification, and we found that transient Wnt and RA activation patterns foregut into esophagealrespiratory rather than pharyngeal endoderm. Moreover, we find that 1 day of RA promotes expression of TP63 and KRT4 and not posterior foregut markers GATA4 and PDX1. This effect of RA could be direct as RA promotes expression of KRT4 and TP63 in keratinocytes (Bamberger et al., 2002). Due to the lack of specific esophageal markers, we heavily relied on the presence or absence of regionally expressed markers to determine early anterior foregut endoderm identity and exclude pharyngeal, respiratory, hepatic, pancreatic, and gastric endoderm.
We also used a functional assay to show that we had generated the esophageal-respiratory region of the foregut. This anterior-posterior level of the foregut should be competent to give rise to the esophagus and respiratory lineage. We showed that AFG spheroids could respond to respiratory-inducing signals (BMP4 and Wnt activation by chiron) by upregulating NKX2–1. Conversely, repression of BMP signaling dorsalizes spheroids based on expression of SOX2, TP63, and MNX1. Interestingly, in our culture conditions, addition of a TGFβ inhibitor during foregut induction causes an increase in posterior foregut markers and reduces upregulation of NKX2–1, in contrast to other protocols (Figure S3), exemplifying how timing and combinatorial signaling pathways manipulation can result in different outcomes.
The final proof of esophageal lineage commitment from dorsal foregut spheroids was their growth into three-dimensional HEOs with a stratified squamous epithelium expressing regional keratins. Upon extended culture or in vivo transplantation, HEOs significantly increase in maturity, both morphologically and by analysis of later-stage esophageal markers (IVL, CRNN, FLG). Additionally, HEOs could be dissociated, expanded as keratinocytes, and differentiated into stratified squamous epithelium in organotypic raft cultures, demonstrating that HEOs have basal progenitor cells similar to human esophagus (Doupe et al., 2012; Kalabis et al., 2008). In fact, the expression level of differentiation markers in organotypic raft cultures approached that of human esophagus. Generation of fully differentiated and mature cell types from human PSCs has been a challenge across organ systems, and our data suggest that PSC-derived esophageal epithelium is among the most highly differentiated tissues derived to date.
HEOs will undoubtedly facilitate studies of human esophageal disease. As one example, we showed how HEOs model human esophageal birth defects. Since SOX2 mutations can cause esophageal atresia in mice and humans, we used HEOs to identify how SOX2 may control human esophageal development since the mechanism underlying its action was unclear (Fantes et al., 2003; Que et al., 2007; Williamson et al., 2006). We first identified transcriptional changes that occur upon loss of SOX2. The current model suggests that the primary role of BMP signaling is to repress Sox2, which represses Nkx2–1; however, we have identified that BMP signaling modulates many transcriptional changes independently of SOX2, suggesting that the current model is oversimplified (Domyan et al., 2011; Rankin et al., 2012) (Figure 2B). Moreover, our data suggest that SOX2 represses canonical Wnt signaling and promotes dorsal endoderm survival. In other contexts, Sox2 can both repress and promote Wnt signaling by a variety of mechanisms including direct binding to TCF/LEF as well as regulating secreted Wnt antagonists (Chen et al., 2008; Kormish et al., 2010; Li et al., 2016; Sinner et al., 2007; Zhou et al., 2016). A role for secreted Wnt antagonists, Sfrp1 and Sfrp2, in tracheoesophageal septation has been shown using Barx1 knockout mice (Woo et al., 2011). In human foregut cultures, SOX2 also regulates transcript levels of SFRP2, and loss of SOX2 causes increased Wnt activity in the dorsal foregut. From this, we conclude that SOX2 restricts the respiratory lineage from the dorsal foregut endoderm, possibly by repressing canonical Wnt signaling.
In summary, we have developed a method to generate human PSC-derived HEOs based on temporal manipulation of signals that pattern the early endoderm and foregut. HEO development is strikingly similar to mouse esophageal development and results in a patterned stratified squamous epithelium. We used human foregut cultures and genetic approaches in mice and frogs to identify molecular pathways that are regulated by Sox2 during dorsal-ventral patterning and esophageal specification. We identified that in both humans and mice, SOX2 represses Wnt activity and that failure to do so results in inappropriate dorsal activation of the respiratory program. Thus, HEOs provide a powerful model to study esophageal development and disease.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to the Lead Contact James M. Wells (james.wells@cchmc.org).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All mice and frogs were housed in the animal facility at Cincinnati Children’s Hospital Medical Center (CCHMC) in accordance with NIH Guidelines for the Care and Use of Laboratory animals. Animals were maintained on a 12 hour light-dark cycle with access to water and standard chow ad libitum. Wild-type and mutant mice and Xenopus laevis were used for studies on foregut and esophageal embryonic development. The sexes of the embryos were not determined. Male immune deficient NSG (NOD.CgPrkdcscidIl2rgtm1Wjl/SzJ) mice, aged 8–16 weeks old, were used for transplantation experiments. Healthy animals were used for all experiments. All experiments were performed under the approval of the Institutional Animal Care and Use Committee of CCHMC (protocols IACUC2016–0004 and IACUC20160059).
Human ESC/IPSC
Human embryonic stem cell (ESC) line H1 (WA01) were purchased from WiCell. Unmodified iPSC lines 65.8, 72.3, and 263.10 were generated and obtained from either the CCHMC Pluripotent Stem Cell Facilities and approved by the institutional review board at CCHMC. CRISPR interference iPSC lines (WTC11 genetic background) were generated and obtained from the Conklin lab at University of California, San Francisco (Mandegar et al., 2016). The H1 line is male, iPSC65.8 line is female, iPSC72.3 line is male, the iPSC263.10 line is male, and the SOX2 CRISPR interference line (CRISPRi-SOX2) is male. All the iPSC lines were checked for and determined to have a normal karyotype, and iPSC65.8 and iPSC72.3 have been tested with an in vivo teratoma assay.
Human Biopsy Tissue
Human esophageal tissue was collected at time of endoscopy in pediatric patients (all male, ages 3 to 13 years old) that consented to provide esophageal biopsy specimens for research purposes, which is approved by the Institutional Review Board of Cincinnati Children’s Hospital Medical Center (protocol 2008–0090). Samples were used as positive controls for esophageal tissue identity by RNA quantification.
METHOD DETAILS:
Experimental Design
To confirm the reproducibility of generating and patterning the dorsal anterior foregut spheroids from pluripotent stem cells, this method has been used by at least 2 other investigators in the Wells lab. No specific strategy for randomization was used, nor were the investigators blinded to the identity of the samples. No statistical methods were used to determine the sample size. With the exception of an instance where Sox2 knockout in the embryonic foregut by tamoxifen gavage was unsuccessful, no data or samples were excluded from analysis.
Pluripotent stem cell lines and maintenance
Both human embryonic and induced pluripotent stem cells (hESCs and hiPSCs) were maintained on feeder-free cultures. Cells are plated on hESC-qualified Matrigel (BD Biosciences, San Jose, CA) and maintained at 37°C with 5% CO2 with daily replacement of mTeSR1 media (STEMCELL Technologies, Vancouver, Canada); cells were passaged routinely every 4 days using Dispase (STEMCELL Technologies). The H1 HA-tagged SOX2 dox-inducible line was generated by cloning the human SOX2 ORF into pINDUCER20 (Addgene #44012, Meerbrey et al., 2011), generating lentivirus with help of the Viral Vector Core Facility at CCHMC, and transducing hESCs with 2 μL of virus; this line was maintained on selection with mTeSR1 and G418 (500 μg mL-1, ThermoFisher Scientific).
Differentiation of anterior foregut cultures and spheroids
Confluent hPSC cultures were treated with Acutase (STEMCELL Technologies) to resuspend as single cells in mTeSR1 and Y-27632 (10 μM, Tocris) and plated on Matrigel. On the following day, differentiation into definitive endoderm was carried out as previously described (McCracken et al., 2014). Briefly, cells were treated with Activin A (100 ng mL-1, R&D systems, Minneapolis, MN) and BMP4 (50 ng mL-1, R&D systems) on the first day in RPMI 1640 media (Life Technologies). Cells in the following two days were treated with only Activin A (100 ng mL-1) in RPMI 1640 with increasing concentrations 0.2% and 2% of HyClone defined fetal bovine serum (dFBS, GE Healthcare Life Sciences).
For anterior foregut monolayer cultures, cells were treated for 3 days in Noggin (200 ng mL-1) in RPMI 1640 with 2% dFBS, with all-trans retinoic acid (2 μM, Sigma, St. Louis, MO) the 3rd day.
Alternately, for the generation of anterior foregut spheroids, from definitive endoderm, cells were treated with FGF4 (500 ng mL-1, R&D systems), Noggin (200 ng mL-1) for 3 days in RPMI 1640 with 2% dFBS. Additional factors were tested during this time (described in results), such as CHIR99021 (“chiron” or “chr”, 2 μM, Tocris), Wnt3a (500 ng mL-1, R&D systems), SB431542 (10 μM, Tocris), DEAB (10 μM, Sigma), and retinoic acid (2 μM).
Three-dimensional culture and differentiation of anterior foregut spheroids into human esophageal organoids
Anterior foregut spheroids were transferred into 50 μL droplets of Matrigel, and are cultured for 3–58 days in the base (“Gut”) media of Advanced DMEM/F12 (ThermoFisher Scientific) supplemented with B27 supplement (1X, ThermoFisher Scientific), N2 supplement (1X, ThermoFisher Scientific), HEPES (13mM, ThermoFisher Scientific), L-Glutamine (2 mM ThermoFisher Scientific), penicillin / streptomycin (1X, ThermoFisher Scientific), and EGF (100 ng mL-1, R&D systems). In addition to this base media, the first three days were supplemented with Noggin (200 ng mL-1), FGF10 (50 ng mL-1), and CultureOne supplement (1X, ThermoFisher Scientific). FGF10 and CultureOne supplementation is continued until the end of the first week in three-dimensional culture. Media was replaced every 3–4 days. For EdU labeling, media was supplemented with EdU (10μM Invitrogen) for a defined period of time, and was removed by washing with sterile PBS twice before replacing with media without EdU.
Keratinocyte and Organotypic Raft Culture
Day 41 HEOs were dissociated using TrypLE Select (Gibco) at 37C for 30–40 minutes, during which they triturated with a 22½ & 27½ gauge needle. After dissociation, cells were reconstituted in complete keratinocyte serum free media (K-SFM, Gibco) supplemented with Y-27632 (10 μM), EGF (10 ng mL-1), and penicillin / streptomycin (1X) and subsequently plated onto collagen IV (Sigma) coated plates (1.5 μg cm-2) at approximately 1.5 x 104 cells cm-2. After reaching 90% confluency, HEO-derived keratinocytes were dissociated to single-cells with TrypLE Select and transferred into organotypic rafts cultures. Organotypic rafts were generated as previously described with minor modifications (Hoskins et al., 2009). Briefly, 1.2×106 HEO-derived keratinocytes were plated on a 24mm collagen matrix (rat tail, EMD Millipore) harboring embedded mouse fibroblasts (J2–3T3 cells). Rafts were initially cultured for 4 days with the addition of Y-27632 (10 μM) prior to exposure to the liquid-air interface to generate a stratified epithelium. After 14 days, rafts were fixed in 4% PFA and embedded into paraffin. Sections were stained with H&E and examined for histopathology by routine microscopy.
Mouse models
All animal experiments performed were approved by the Institutional Animal Care and Use Committee (IACUC) of Cincinnati Children’s Hospital Medical Center (CCHMC). FoxA2CreER mice were obtained from Anne Moon’s lab (Park et al., 2008), Sox2fl/fl mice were obtained from Richard Lang’s lab (Shaham et al., 2009), and Sox2CreER (stock #017593, Arnold et al., 2011) were obtained from The Jackson Laboratory. Mice were housed at the CCHMC animal facility, and timed matings were used to obtain embryos at the relevant stages. Pregnant dams were gavaged at various stages with tamoxifen at 0.12mg/g mouse to activate the CreER at appropriate stages. Specifically, pregnant dams in FoxA2CreER experiments were gavaged at 6.5dpc to achieve efficient recombination. In Sox2CreER experiments, pregnant dams were gavaged at 8.5dpc to knockout Sox2 prior to tracheoesophageal separation, and 9.5dpc to knockout Sox2 during/after tracheoesophageal separation.
Xenopus experiments
Xenopus laevis adults were purchased from the Nasco (Fort Atkinson, WI), and housed according to CCHMC IACUC protocols. Ovulation, in vitro fertilization, and de-jellying of embryos were performed as described (Sive et al., 2000). A mixture of previously validated Sox2 morpholinos (MOs; Van Raay et al., 2005) targeting the 5’UTR (Sox2-UTR MO) and the ATG start codon (Sox2-ATG MO) were injected at the 8-cell stage into each vegetal blastomere (2ng total MO per blastomere, 8ng total per embryo) to target endoderm. MOs were synthesized and purchased from GeneTools. Equal amount of control MO was used in control injections.
For Xenopus explant studies, stage NF20 foregut endoderm tissue was micro-dissected in 1X MBS (Modified Barth’s Saline; Sive et al., 2000) + 50 ug/mL gentamycin sulfate (MP Biochemicals), +4% Ficoll400 (Sigma) and explants were then cultured in 0.5X MBS + 0.1% Fatty Acid Free BSA (Fisher) + 50 ug/mL gentamycin sulfate with or without the following concentrations of small molecules or recombinant proteins from stages NF25-NF38 (approximately 48 hours): 3.5 μM Bio (Tocris), 50ng/mL recombinant human BMP4 (R&D systems).
In situ hybridiziation
In situ hybridization on mouse sections was performed by generating DIG-labeled probes from linearized mouse cDNA plasmids. Probes were allowed to hybridize overnight at 65°C, and on the next day, probes were thoroughly washed before blocking and incubating with anti-DIG alkaline phosphatase antibody (Sigma) in a 1:5,000 dilution in MAB buffer (maleic acid buffer, 100mM Maleic acid, 150mM NaCl, pH7.5) + 10% heat-inactivated lamb serum (Gibco) + 2% blocking reagent (Sigma) overnight at 4°C. Several washes were done before developing the slides using BM purple. In situ hybridization of Xenopus explants was performed mostly as described in (Sive et al., 2000). Briefly, embryos and explants were fixed overnight at 40C in MEMFA (0.1M MOPS, 2 mM EGTA, 1mM MgSO4, 3.7% formaldehyde), dehydrated directly into 100% ethanol, and stored at -20°C. The following minor modifications to the in situ protocol were used: proteinase K (ThermoFisher) on day 1 was used at 2ug/mL for 10 minutes on explants; the RNAse A step was omitted on day 2; and finally the anti-DIG-alkaline phosphatase antibody was used at a 1:5,000 dilution in MAB buffer + 10% heat-inactivated lamb serum + 2% blocking reagent on day2/3.
In situ hybridization on whole-mount Xenopus embryos was performed by generating anti-sense DIG labeled nkx2–1 in-situ probe was generated using linearized plasmid full-length nkx2–1 cDNA template (Small et al 2000; XbaI for linearization, T7 to synthesize antisense RNA) with the 10X DIG RNA labeling mix (Sigma) according to manufacturer’s instructions.
Immunofluoresence analysis
Tissue cultures were fixed with 4% paraformaldehyde at either room temperature for 15 minutes, 4°C for 2 hours for cryosectioning, or 4°C overnight for paraffin embedding/sectioning and mouse embryos. For paraffin embedding and sectioning, following fixation, tissues were dehydrated and embedded into paraffin blocks. Afterwards, paraffin-sectioned slides were deparaffinized and subjected to antigen retrieval in 10mM sodium citrate for 30 minutes prior to staining. For cryosectioning, tissues were thoroughly washed in PBS, left in 30% sucrose overnight, embedded in OCT compound (VWR), and sectioned at a thickness of 8μm. Commonly, slides were then permeabilized with 0.5% TritonX-100 in PBS for 10 minutes, blocked in 5% normal donkey serum (Jackson ImmunoResearch) for 1 hour, and incubated in primary antibody overnight at 4°C. On the next day, slides were thoroughly washed in PBS, incubated in secondary antibody (at 1:500) for 1 hour, and then thoroughly washed again. For EdU visualization, we used the Click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen) prior to blocking. For wholemount immunofluorescence staining, embryos were placed in 100% methanol immediately after fixation. Embryos were then permeabilized with Dent’s Bleach (4:1:1 MeOH:DMSO:30% H2O2) for 2 hours at room temperature, rehydrated with methanol washes, and blocked for several hours at room temperature to overnight at 4°C. Primary antibodies were applied and embryos incubated overnight at 4°C. Then, embryos were thoroughly washed in 0.1% TritonX-100 in PBS before incubating in secondary antibodies overnight at 4°C. Finally, embryos were washed again, dehydrated with methanol washes, and cleared with Murray’s Clear (2:1 benzyl benzoate : benzyl alcohol, Sigma) at least 15 minutes prior to imaging. For a list of antibodies and dilutions used, please see Table S2
RNA isolation and qPCR
Spheroid and organoids were harvested in total, including the embedding matrigel. Collagen plugs from the organotypic raft cultures were first removed from the transwell on day 14, and subsequently harvested in total. Total RNA was isolated using the NucleoSpin RNA kit (Macherey-Nagel) and reverse transcribed to cDNA using the SuperScript VILO cDNA synthesis kit (ThermoFisher Scientific). For qRT-PCR, we used Quantitect SYBR-Green master mix (Qiagen) and ran the reaction on a QuantStudio 6 machine (ThermoFisher Scientific). For a list of primers used, please see Table S1.
RNA sequencing and analysis
Whole-transcriptome RNA sequencing of anterior foregut cultures and HEOs (n=3 per condition or time point) was performed by the DNA sequencing and Genotyping Core Facility on an Illumina Hi-Seq 2500 platform from a Poly(A) and TruSeq library generated from isolated total RNA. RNA sequencing parameters were 75bp single-end sequencing at a depth of 10M reads per samples. Fastq read files for each sample were obtained and then aligned using the Computational Suite for Bioinformaticians and Biologists version 2.1 (CSBB-v2.1, https://sourceforge.net/projects/csbb-v2–1/). Raw transcript counts and normalized transcripts per million (TPM) values were obtained and analyzed for differential expression with CSBB-v2.1 and for Gene Set Enrichment Analysis (GSEA, Subramanian et al., 2005). For differential expression, statistical and biological significance was set at P<0.05, FDR<0.05, log fold-change>1, with a minimum of 3 transcript counts in 3 of the 6 samples. For heatmap visualization and hierarchical clustering analysis, Morpheus (https://software.broadinstitute.org/morpheus/) was used.
Anterior foregut transcriptome analyses were cross-referenced with SOX2 and SMAD1 ChIP-seq peaks from GEO sets (GSE61475, Tsankov et al., 2015; GSE47058, Watanabe et al., 2014) using HOMER to obtain lists of genes whose expression is potentially regulated by these transcription factors. Peak cutoff distance was set at 50kb from the transcription start site of any particular gene.
HEO analyses were compared to previously published RNA-seq samples on in vitro generated organoids (intestine and gastric), EPC2 cultures, and biopsies from the ENCODE Roadmap project, which including the following tissues: skin, esophagus, small intestine, stomach, colon, and lung. To compare in-house data with public data, we used Upper Quantile [between-Lane Normalization] from EDASEQ [http://bioconductor.org/packages/release/bioc/vignettes/EDASeq/inst/doc/EDASeq.pdf]. We used to CSBB’s [Computational Suite for Bioinformaticians and Biologists] version 3.0 [https://github.com/csbbcompbio/CSBB-v3.0] UpperQuantile module. We generated a matrix of expression of genes across in-house and public samples and quantile-normalized using CSBB-v3.0’s UpperQuantile module. Then, we log2 transformed the quantile normalized matrix in R. Log2 Transformed matrix was used for all downstream analysis.
We also used SVA [https://bioconductor.org/packages/release/bioc/vignettes/sva/inst/doc/sva.pdf] on the log2 transformed – quantile normalized matrix to check if there are any latent variables / surrogate variables to correct. We found no surrogate variables to correct for. This approach gave us confidence that Upper-Quantile normalization followed by Log2 transform is robust enough to remove batch and sequencing effects from the data.
QUANTIFICATION AND STATISTICAL ANALYSIS
For experiments involving spheroid patterning, organoid outgrowth, and raft experiments, “n” represents the number of replicates performed in each experiment (1 well of 3–7 organoids or 30–50 spheroids were collected for each replicate in matrigel culture, all samples from 1 well of a organotypic raft culture are considered a single replicate). For animal experiments, “n” represents the number of embryos analyzed. All data quantification is represented as the mean ± SD. To compare the various conditions tested in spheroid patterning and organoid outgrowth, t-tests with 2-tailed distribution not assuming equal (i.e. unequal) variance was used in Microsoft Excel, where *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, and ****p ≤ 0.0001.
Details for quantification and statistical analysis for figures 1–7
Figure 1: For 1H, a minimum of 20 spheroids from two experiments were assessed. For all qPCR results, the data is representative of a minimum of 2 separate experiments with n=3 wells (50–100 spheroids in each well) for each experiment. RA experiments were replicated in both H1 and iPS263.10 cell lines.
Figure 2: The data is representative of 2 separate experiments with n=3 wells (averaging 30–50 spheroids per well) in each experiment utilizing the H1 hESC line.
Figure 3: Generation of organoids is representative of 40+ experiments across 4 hES and iPS cell lines: H1, iPS 65.8, iPS 72.3, iPS 263.10. The qPCR data is representative of 2 separate experiments with n=3 wells (3–12 organoids per well) and were compared to n=5 patient biopsy samples.
Figure 4: n=2–4 wells for the HEOs to organotypic raft culture experiment. n=6–10 organoids at each time-point for the EdU experiment. n=5 patient esophageal biopsy samples. Experiments were done in the H1 hESC line.
Figure 5: For Sox2-DE-LOF embryos, n=3 embryos of each genotype at E9.5, and n=2 embryos for each analysis type for each genotype at E11.5. For Sox2 driven Sox2 cKO embryos, n=3 embryos analyzed for IF of each stage of tamoxifen administration and corresponding stage harvest.
Figure 6: All data from the human PSC-derived cultures are representative of 3 separate experiments with n=3 wells for each condition per experiment.
DATA AND SOFTWARE AVAILABILITY
The accession number for the data generated in this paper is Gene Expression Omnibus (GEO): GSE112886. This includes the organoid outgrowth comparison (1 versus 2 month human esophageal organoids, Figure 3) as well as the SOX2-knockdown experiment in dorsal and ventral anterior foregut cultures (Figure 7 and Figure S7).
ChIP-seq data of SOX2 and SMAD1 ChIP-seq peaks were downloaded from the public database GEO with accession numbers GSE61475 (Tsankov et al., 2015) and GSE47058 (Watanabe et al., 2014), respectively.
RNA-seq data of biopsies and EPC2 cultures were downloaded from the public database GEO. The accession numbers for the samples are: GSM1120313 and GSM1120314 (small intestine), GSM1010946 and GSM1120308 (lung), GSM1120307 and GSM11010960 (stomach), GSM1120315 and GSM1010974 (large intestine), GSM1010956 and GSM1120303 (esophagus), GSM2343841 and GSM234564 (lower leg skin), and GSM1592609-GSM1592611 (EPC2 day 0 cultures). The complete RNA-seq processing pipeline was done using Computational Suite for Bioinformaticians and Biologists version 2.1 (CSBBv2.1) and is available at https://sourceforge.net/projects/csbb-v2–1/.
Supplementary Material
Table S3: List of differentially expressed genes enriched in dorsal (Noggin-treated) versus ventral (BMPtreated) cultures with or without SOX2 knockdown in human anterior foregut cultures. Related to Figure 7.
Table S1: List of primers used for qPCR analysis. Related to Figures 1–7.
Table S2: List of antibodies used for immunofluorescence staining. Related to Figures 1–7.
Highlights.
Sequential Wnt, RA, and BMP signaling are required to pattern dorsal anterior foregut
Cultured dorsal anterior foregut spheroids develop into esophageal organoids (HEOs)
HEOs contain basal esophageal progenitors and stratified squamous epithelium
Sox2 is sufficient to repress the respiratory fate by suppressing Wnt signaling
ACKNOWLEDGEMENTS
We would like to thank the members of the Wells and Zorn laboratories for reagents and feedback. We also thank Chris Mayhew and Amy Pitstick from the Pluripotent Stem Cell Facility, Matt Kofron and the Confocal Imaging Core. We thank the Jiang lab for providing the Axin2 and Sfrp2 in situ probes and Lonnie Shea and Briana Dye for providing the PEG scaffolds. Finally, we would like to thank Yana Zavros for help with the orthotopic transplantation experiments. This work was supported by P01 HD093363–01, R37 A1045898, and T32 GM-063483.
Footnotes
DECLARATION OF INTERESTS
JMW and SLT have a pending patent application, number 62/570,182, related to this work that describes a method for generating esophageal tissue from human pluripotent stem cells. All other authors have no potential conflicts of interests to declare.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Andl CD, Mizushima T, Nakagawa H, Oyama K, Harada H, Chruma K, Herlyn M, and Rustgi AK (2003). Epidermal growth factor receptor mediates increased cell proliferation, migration, and aggregation in esophageal keratinocytes in vitro and in vivo. Journal of Biological Chemistry, 278(3), 1824–1830. 10.1074/jbc.M209148200 [DOI] [PubMed] [Google Scholar]
- Arnold K, Sarkar A, Yram MA, Polo JM, Bronson R, Sengupta S, Seandel M, Geijsen N, and Hochedlinger K (2011). Sox2+ Adult Stem and Progenitor Cells Are Important for Tissue Regeneration and Survival of Mice. Cell Stem Cell, 9(4), 317–329. 10.1016/j.stem.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamberger C, Pollet D, and Schmale H (2002). Retinoic acid inhibits downregulation of DeltaNp63alpha expression during terminal differentiation of human primary keratinocytes. The Journal of Investigative Dermatology, 118(1), 133–8. 10.1046/j.0022-202x.2001.01649.x [DOI] [PubMed] [Google Scholar]
- Bayha E, Jørgensen MC, Serup P, and Grapin-Botton A (2009). Retinoic acid signaling organizes endodermal organ specification along the entire antero-posterior axis. PloS One, 4(6), e5845 10.1371/journal.pone.0005845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Li J, Li H, Hu Y, Tevebaugh W, Yamamoto M, Que J, and Chen X (2012). Transcript profiling identifies dynamic gene expression patterns and an important role for Nrf2/Keap1 pathway in the developing mouse esophagus. PloS One, 7(5), e36504 10.1371/journal.pone.0036504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-W, Huang SX, de Carvalho ALRT, Ho S-H, Islam MN, Volpi S, Notarangelo LD, Ciancanelli M, Casanova J-L, Bhattacharya J, Liang AF, et al. (2017). A three-dimensional model of human lung development and disease from pluripotent stem cells. Nature Cell Biology, 19(5), 542–549. 10.1038/ncb3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Shi L, Zhang L, Li R, Liang J, Yu W, Sun L, Yang X, Wang Y, Zhang Y, and Shang Y (2008). The Molecular Mechanism Governing the Oncogenic Potential of SOX2 in Breast Cancer. Journal of Biological Chemistry, 283(26), 17969–17978. 10.1074/jbc.M802917200 [DOI] [PubMed] [Google Scholar]
- D’Amour K.a, Agulnick AD, Eliazer S, Kelly OG, Kroon E, and Baetge EE (2005). Efficient differentiation of human embryonic stem cells to definitive endoderm. Nature Biotechnology, 23(12), 1534–1541. 10.1038/nbt1163 [DOI] [PubMed] [Google Scholar]
- Daniely Y, Liao G, Dixon D, Linnoila RI, Lori A, Randell SH, Oren M, and Jetten AM (2004). Critical role of p63 in the development of a normal esophageal and tracheobronchial epithelium. American Journal of Physiology. Cell Physiology, 287(1), C171–C181. 10.1152/ajpcell.00226.2003 [DOI] [PubMed] [Google Scholar]
- Davenport C, Diekmann U, Budde I, Detering N, and Naujok O (2016). The Anterior-Posterior Patterning of Definitive Endoderm Generated from Human Embryonic Stem Cells Depends on the Differential Signaling of Retinoic Acid, Wnt- and BMP-Signaling. Stem Cells, 34(11), 2635–2647. 10.1002/stem.2428 [DOI] [PubMed] [Google Scholar]
- Dessimoz J, Opoka R, Kordich JJ, Grapin-Botton A, and Wells JM (2006). FGF signaling is necessary for establishing gut tube domains along the anterior-posterior axis in vivo. Mechanisms of Development, 123(1), 42–55. 10.1016/j.mod.2005.10.001 [DOI] [PubMed] [Google Scholar]
- DeWard AD, Cramer J, and Lagasse E (2014). Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Reports, 9(2), 701–11. 10.1016/j.celrep.2014.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domyan ET, Ferretti E, Throckmorton K, Mishina Y, Nicolis SK, and Sun X (2011). Signaling through BMP receptors promotes respiratory identity in the foregut via repression of Sox2. Development (Cambridge, England), 138(5), 971–981. 10.1242/dev.053694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupe DP, Alcolea MP, Roshan A, Zhang G, Klein a. M., Simons BD, and Jones PH (2012). A Single Progenitor Population Switches Behavior to Maintain and Repair Esophageal Epithelium. Science, 337(6098), 1091–1093. 10.1126/science.1218835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BR, Dedhia PH, Miller AJ, Nagy MS, White ES, Shea LD, and Spence JR (2016). A bioengineered niche promotes in vivo engraftment and maturation of pluripotent stem cell derived human lung organoids. ELife, 5(September2016). 10.7554/eLife.19732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BR, Hill DR, Ferguson MA, Tsai Y-H, Nagy MS, Dyal R, Wells JM, Mayhew CN, Nattiv R, Klein OD, White ES, et al. (2015). In vitro generation of human pluripotent stem cell derived lung organoids. ELife, 4, e05098 10.7554/eLife.05098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantes J, Ragge NK, Lynch S-A, McGill NI, Collin JRO, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, and FitzPatrick DR (2003). Mutations in SOX2 cause anophthalmia. Nature Genetics, 33(4), 461–463. 10.1038/ng1120 [DOI] [PubMed] [Google Scholar]
- Fausett SR, Brunet LJ, and Klingensmith J (2014). BMP antagonism by Noggin is required in presumptive notochord cells for mammalian foregut morphogenesis. Developmental Biology, 391(1), 111–24. 10.1016/j.ydbio.2014.02.008 [DOI] [PubMed] [Google Scholar]
- Goss AM, Tian Y, Tsukiyama T, Cohen ED, Zhou D, Lu MM, Yamaguchi TP, and Morrisey EE (2009). Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Developmental Cell, 17(2), 290–8. 10.1016/j.devcel.2009.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green MD, Chen A, Nostro M-C, d’Souza SL, Schaniel C, Lemischka IR, Gouon-Evans V, Keller G, and Snoeck H-W (2011). Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nature Biotechnology, 29(3), 267–72. 10.1038/nbt.1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Johnson KS, Domyan ET, Vezina CM, and Sun X (2009). beta-Catenin promotes respiratory progenitor identity in mouse foregut. Proceedings of the National Academy of Sciences of the United States of America, 106(38), 16287–92. 10.1073/pnas.0902274106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoskins EE, Morris TA, Higginbotham JM, Spardy N, Cha E, Kelly P, Williams DA, Wikenheiser-Brokamp KA, Duensing S, and Wells SI (2009). Fanconi anemia deficiency stimulates HPV-associated hyperplastic growth in organotypic epithelial raft culture. Oncogene, 28(5), 674–685. 10.1038/onc.2008.416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jho E, Zhang T, Domon C, Joo C-K, Freund J-N, and Costantini F (2002). Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Molecular and Cellular Biology, 22(4), 1172–83. 10.1128/MCB.22.4.1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalabis J, Oyama K, Okawa T, Nakagawa H, Michaylira CZ, Stairs DB, Figueiredo J, Mahmood U, Diehl JA, Herlyn M, and Rustgi AK (2008). A subpopulation of mouse esophageal basal cells has properties of stem cells with the capacity for self-renewal and lineage specification. Journal of Clinical Investigation, (118), 3860–3869. 10.1172/JCI35012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalabis J, Wong GS, Vega ME, Natsuizaka M, Robertson ES, Herlyn M, Nakagawa H, and Rustgi AK (2012). Isolation and characterization of mouse and human esophageal epithelial cells in 3D organotypic culture. Nature Protocols, 7(2), 235–246. 10.1038/nprot.2011.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasagi Y, Chandramouleeswaran PM, Whelan KA, Tanaka K, Giroux V, Sharma M, Wang J, Benitez AJ, DeMarshall M, Tobias JW, Hamilton KE, et al. (2018). The Esophageal Organoid System Reveals Functional Interplay Between Notch and Cytokines in Reactive Epithelial Changes. Cmgh, 5(3), 333–352. 10.1016/j.jcmgh.2017.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns N.a, Genga RMJ, Ziller M, Kapinas K, Peters H, Brehm M.a, Meissner A, and Maehr R (2013). Generation of organized anterior foregut epithelia from pluripotent stem cells using small molecules. Stem Cell Research, 11(3), 1003–12. 10.1016/j.scr.2013.06.007 [DOI] [PubMed] [Google Scholar]
- Kormish JD, Sinner D, and Zorn AM (2010). Interactions between SOX factors and Wnt/betacatenin signaling in development and disease. Developmental Dynamics : An Official Publication of the American Association of Anatomists, 239(August 2009), 56–68. 10.1002/dvdy.22046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhou H, Fu X, and Xiao R (2016). Directed differentiation of human embryonic stem cells into keratinocyte progenitors in vitro: an attempt with promise of clinical use. In Vitro Cellular & Developmental Biology - Animal, 52(8), 885–893. 10.1007/s11626-016-0024-2 [DOI] [PubMed] [Google Scholar]
- Longmire T.a , Ikonomou L, Hawkins F, Christodoulou C, Cao Y, Jean JC, Kwok LW, Mou H, Rajagopal J, Shen SS, Dowton A.a, et al. (2012). Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell, 10(4), 398–411. 10.1016/j.stem.2012.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig B, Jerchow B, Sachs M, Weiler S, Pietsch T, Karsten U, van de Wetering M, Clevers H, Schlag PM, Birchmeier W, and Behrens J (2002). Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Molecular and Cellular Biology, 22(4), 1184–93. 10.1128/MCB.22.4.1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, Chan AH, Miyaoka Y, Holmes K, Spencer CI, Judge LM, et al. (2016). CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell, 18(4), 541–553. 10.1016/j.stem.2016.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matt N, Ghyselinck NB, Wendling O, Chambon P, and Mark M (2003). Retinoic acid-induced developmental defects are mediated by RARβ/RXR heterodimers in the pharyngeal endoderm. Development, 130(10), 2083–2093. 10.1242/dev.00428 [DOI] [PubMed] [Google Scholar]
- McCauley HA, and Wells JM (2017). Pluripotent stem cell-derived organoids: using principles of developmental biology to grow human tissues in a dish. Development, 144(6), 958–962. 10.1242/dev.140731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Aihara E, Martin B, Crawford CM, Broda T, Treguier J, Zhang X, Shannon JM, Montrose MH, and Wells JM (2017). Wnt/β-catenin promotes gastric fundus specification in mice and humans. Nature, 541(7636), 182–187. 10.1038/nature21021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken KW, Catá EM, Crawford CM, Sinagoga KL, Schumacher M, Rockich BE, Tsai Y-H, Mayhew CN, Spence JR, Zavros Y, and Wells JM (2014). Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature, 516(7531), 400–404. 10.1038/nature13863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLin V.a, Rankin S. a, and Zorn AM (2007). Repression of Wnt/beta-catenin signaling in the anterior endoderm is essential for liver and pancreas development. Development (Cambridge, England), 134(12), 2207–17. 10.1242/dev.001230 [DOI] [PubMed] [Google Scholar]
- Meerbrey KL, Hu G, Kessler JD, Roarty K, Li MZ, Fang JE, Herschkowitz JI, Burrows AE, Ciccia A, Sun T, Schmitt EM, et al. (2011). The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proceedings of the National Academy of Sciences of the United States of America, 108(9), 3665–3670. 10.1073/pnas.1019736108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Múnera JO, Sundaram N, Rankin SA, Hill D, Watson C, Mahe M, Vallance JE, Shroyer NF, Sinagoga KL, Zarzoso-Lacoste A, Hudson JR, et al. (2017). Differentiation of Human Pluripotent Stem Cells into Colonic Organoids via Transient Activation of BMP Signaling. Cell Stem Cell, 21(1), 51–64.e6. 10.1016/j.stem.2017.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dolle P, and Chambon P (1999). Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nature Genetics, 21(4), 444–448. 10.1038/7788 [DOI] [PubMed] [Google Scholar]
- Park EJ, Sun X, Nichol P, Saijoh Y, Martin JF, and Moon AM (2008). System for tamoxifeninducible expression of Cre-recombinase from the Foxa2 locus in mice. Developmental Dynamics, 237(2), 447–453. 10.1002/dvdy.21415 [DOI] [PubMed] [Google Scholar]
- Que J, Choi M, Ziel JW, Klingensmith J, and Hogan BLM (2006). Morphogenesis of the trachea and esophagus: current players and new roles for noggin and Bmps. Differentiation, 74(7), 422–437. 10.1111/j.1432-0436.2006.00096.x [DOI] [PubMed] [Google Scholar]
- Que J, Okubo T, Goldenring JR, Nam K-T, Kurotani R, Morrisey EE, Taranova O, Pevny LH, and Hogan BLM (2007). Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development (Cambridge, England), 134(13), 2521–31. 10.1242/dev.003855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, Han L, McCracken KW, Kenny AP, Anglin CT, Grigg EA, Crawford CM, Wells JM, Shannon JM, and Zorn AM (2016). A Retinoic Acid-Hedgehog Cascade Coordinates Mesoderm-Inducing Signals and Endoderm Competence during Lung Specification. Cell Reports, 16(1), 66–78. 10.1016/J.CELREP.2016.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin S.a, Gallas AL, Neto A, Gómez-Skarmeta JL, and Zorn AM (2012). Suppression of Bmp4 signaling by the zinc-finger repressors Osr1 and Osr2 is required for Wnt/β-catenin-mediated lung specification in Xenopus. Development (Cambridge, England), 139(16), 3010–20. 10.1242/dev.078220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosekrans SL, Baan B, Muncan V, and van den Brink GR (2015). Esophageal development and epithelial homeostasis. American Journal of Physiology - Gastrointestinal and Liver Physiology, 309(4), G216–228. 10.1152/ajpgi.00088.2015 [DOI] [PubMed] [Google Scholar]
- Shaham O, Smith AN, Robinson ML, Taketo MM, Lang R.a, and Ashery-Padan R (2009). Pax6 is essential for lens fiber cell differentiation. Development (Cambridge, England), 136(15), 2567–2578. 10.1242/dev.032888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Chen T-YA, and Melton D (2009). Transcriptional dynamics of endodermal organ formation. Developmental Dynamics : An Official Publication of the American Association of Anatomists, 238(1), 29–42. 10.1002/dvdy.21810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SJ, Jonatan D, Zorn AM, and Wells JM (2007). Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Molecular and Cellular Biology, 27(22), 7802–7815. 10.1128/MCB.02179-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive H, Grainger R, and Harland R (2000). Early Development of Xenopus laevis: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Spence JR, Mayhew CN, Rankin S.a, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF, and Wells JM (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature, 470(7332), 105–109. 10.1038/nature09691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens ML, Chaturvedi P, Rankin SA, Macdonald M, Jagannathan S, Yukawa M, Barski A, and Zorn AM (2017). Genomic integration of Wnt/β-catenin and BMP/Smad1 signaling coordinates foregut and hindgut transcriptional programs. Development, 144(7), 1283–1295. 10.1242/dev.145789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences, 102(43), 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiso N, Filippi A, Pauls S, Bortolussi M, and Argenton F (2002). BMP signalling regulates anteroposterior endoderm patterning in zebrafish. Mechanisms of Development, 118(1–2), 29–37. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12351167 [DOI] [PubMed] [Google Scholar]
- Tsankov AM, Gu H, Akopian V, Ziller MJ, Donaghey J, Amit I, Gnirke A, and Meissner A (2015). Transcription factor binding dynamics during human ES cell differentiation. Nature, 518(7539), 344–9. 10.1038/nature14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raay TJ, Moore KB, Iordanova I, Steele M, Jamrich M, Harris WA, and Vetter ML (2005). Frizzled 5 signaling governs the neural potential of progenitors in the developing Xenopus retina. Neuron, 46(1), 23–36. 10.1016/j.neuron.2005.02.023 [DOI] [PubMed] [Google Scholar]
- Wang Z, Dollé P, Cardoso WV, and Niederreither K (2006). Retinoic acid regulates morphogenesis and patterning of posterior foregut derivatives. Developmental Biology, 297(2), 433–445. 10.1016/j.ydbio2006.05.019 [DOI] [PubMed] [Google Scholar]
- Watanabe H, Ma Q, Peng S, Adelmant G, Swain D, Song W, Fox C, Francis JM, Pedamallu CS, DeLuca DS, Brooks AN, et al. (2014). SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. Journal of Clinical Investigation, 124(4), 1636–1645. 10.1172/JCI71545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson K. a., Hever AM, Rainger J, Rogers RC, Magee A, Fiedler Z, Keng WT, Sharkey FH, McGill N, Hill CJ, Schneider A, et al. (2006). Mutations in SOX2 cause anophthalmiaesophageal-genital (AEG) syndrome. Human Molecular Genetics, 15(9), 1413–1422. 10.1093/hmg/ddl064 [DOI] [PubMed] [Google Scholar]
- Woo J, Miletich I, Kim B-M, Sharpe PT, and Shivdasani R. a. (2011). Barx1-mediated inhibition of Wnt signaling in the mouse thoracic foregut controls tracheo-esophageal septation and epithelial differentiation. PloS One, 6(7), e22493 10.1371/journal.pone.0022493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jiang M, Kim E, Lin S, Liu K, Lan X, and Que J (2016). Development and Stem Cells of the Esophagus. Seminars in Cell & Developmental Biology, 66, 25–35. 10.1016/j.semcdb.2016.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C, Yang X, Sun Y, Yu H, Zhang Y, and Jin Y (2016). Comprehensive profiling reveals mechanisms of SOX2-mediated cell fate specification in human ESCs and NPCs. Cell Research, 26(2), 171–189. 10.1038/cr.2016.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, and Wells JM (2007). Molecular Basis of Vertebrate Endoderm Development. In International Review of Cytology (Vol. 259, pp. 49–111). 10.1016/S0074-7696(06)59002-3 [DOI] [PubMed] [Google Scholar]
- Zorn AM, and Wells JM (2009). Vertebrate Endoderm Development and Organ Formation. Annual Review of Cell and Developmental Biology, 25(1), 221–251. 10.1146/annurev.cellbio.042308.113344 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3: List of differentially expressed genes enriched in dorsal (Noggin-treated) versus ventral (BMPtreated) cultures with or without SOX2 knockdown in human anterior foregut cultures. Related to Figure 7.
Table S1: List of primers used for qPCR analysis. Related to Figures 1–7.
Table S2: List of antibodies used for immunofluorescence staining. Related to Figures 1–7.