Abstract
New concepts are reviewed in Cannabis systematics, including phylogenetics and nomenclature. The family Cannabaceae now includes Cannabis, Humulus, and eight genera formerly in the Celtidaceae. Grouping Cannabis, Humulus, and Celtis actually goes back 250 years. Print fossil of the extinct genus Dorofeevia (=Humularia) reveals that Cannabis lost a sibling perhaps 20 million years ago (mya). Cannabis print fossils are rare (n=3 worldwide), making it difficult to determine when and where she evolved. A molecular clock analysis with chloroplast DNA (cpDNA) suggests Cannabis and Humulus diverged 27.8 mya. Microfossil (fossil pollen) data point to a center of origin in the northeastern Tibetan Plateau. Fossil pollen indicates that Cannabis dispersed to Europe by 1.8–1.2 mya. Mapping pollen distribution over time suggests that European Cannabis went through repeated genetic bottlenecks, when the population shrank during range contractions. Genetic drift in this population likely initiated allopatric differences between European Cannabis sativa (cannabidiol [CBD]>Δ9-tetrahydrocannabinol [THC]) and Asian Cannabis indica (THC>CBD). DNA barcode analysis supports the separation of these taxa at a subspecies level, and recognizing the formal nomenclature of C. sativa subsp. sativa and C. sativa subsp. indica. Herbarium specimens reveal that field botanists during the 18th–20th centuries applied these names to their collections rather capriciously. This may have skewed taxonomic determinations by Vavilov and Schultes, ultimately giving rise to today's vernacular taxonomy of “Sativa” and “Indica,” which totally misaligns with formal C. sativa and C. indica. Ubiquitous interbreeding and hybridization of “Sativa” and “Indica” has rendered their distinctions almost meaningless.
Keywords: : Cannabaceae, Cannabis sativa, center of origin, barcode, molecular clock, palynology
Introduction
Taxonomy includes classification (the identification and categorization of organisms) and nomenclature (the naming and describing of organisms). Taxonomy, in the light of evolution, becomes systematics: the evolutional relationships among living things. Classification, in the light of evolution, becomes phylogenetics: the genealogical study of relationships among individuals and groups in a nested hierarchy.
This review of Cannabis systematics will consist of four sections: (1) the family Cannabaceae with the recent addition of former Celtidaceae; (2) the genus Cannabis, and when and where she evolved; (3) the species Cannabis sativa, including two subspecies: C. sativa subsp. sativa and C. sativa subsp. indica; (4). the vernacular taxonomy of “Sativa” and “Indica.”
The Family Cannabaceae
The family Cannabaceae currently consists of Cannabis and Humulus, plus eight genera formerly in the Celtidaceae: Celtis, Pteroceltis, Aphananthe, Chaetachme, Gironniera, Lozanella, Trema, and Parasponia.1 Some botanists combine Parasponia and Trema, but Parasponia species uniquely form nitrogen-fixing nodules in symbiosis with rhizobial bacteria. This trait is shared only by legumes. In contrast, other Cannabaceae form a symbiosis with arbuscular mycorrhizal fungi, including Cannabis.2 Family Cannabaceae now includes about 170 species.
Cesalpino3 first elucidated taxonomic affinities between Cannabis and her sister genus Humulus in 1583. Before him, botanists classified Cannabis with phylogenetically unrelated plants based on leaf shape, human usage, and other totally artificial characters. Cesalpino was an Aristotelian essentialist; he reasoned that plants should be classified by the morphology of their most essential functions—reproduction (flowers and fruits), and nutrition (xylem and phloem).
Schultes4 summarizes, “the earliest trend in taxonomic works was to include Cannabis in the Urticaceae; that in the last half of the last century [19th] and the early part of this century [20th], most authorities favoured the Moraceae; that the modern tendency appears to maintain the family Cannabaceae as separate from these.” Schultes is widely quoted or paraphrased, but he presented a simplified history of taxonomy.
Early taxonomists lumped together members of the Urticaceae, Moraceae, and Cannabaceae, and referred to these amalgamated entities by a variety of names. For example, Adanson5 lumped 11 genera in 1763: Cannabis, Humulus, Celtis, two Urticaceae genera, four Moraceae genera, and two unrelated genera. His accuracy (percentage of genera now placed in Cannabaceae, Urticaceae, or Moraceae) was 9 out of 11, or 82%. Some other early concepts are presented in Table 1.
Table 1.
Some Early Plant Families into Which Cannabis and Humulus Have Been Classified, Listed Chronologically
| Author (date) | Family name (and subfamily limited to Cannabis and Humulus if designated) | Number of genera in family (and subfamily where designated) now classified in Cannabaceae-Urticaceae-Moraceae-Celtidaceae-other, with percentage accuracy |
|---|---|---|
| Adanson (1763)5 | Castaneaceae Section III | 2-2-4-1-2, 82% |
| Linnaeus (1764)6 | Scabridae | 2-3-3-1-4, 69% |
| Lamarck (1788)7 | Figuiers (Moraceae) Section II | 2-4-0-0-1, 86% |
| de Jussieu (1789)8 | Urticae Section II | 2-7-2-0-2, 85% |
| Batsch (1802)9 | Scabridae | 2-7-5-0-5, 74% |
| (section Exalbuminosa) | (2-0-0-0-0, 100%) | |
| Martynov (1820)10 | Cannabaceae | 2-0-0-0-0, 100% |
| Blume (1825)11 | Urticeae | 1-3-7-1-5, 71% |
| (section Cannabineae) | (1-0-0-0-0, 100%) | |
| Gaudichaud-Beaupré (1826)12 | Urticeae | 2-26-10-1-9, 81% |
| (section Cannabineae) | (2-0-0-0-0, 100%) | |
| Nees von Esenbeck et al. (1835)13 | Urticaceae | 2-2-2-1-1, 88% |
| (tribe Cannabinae) | (2-0-0-0-0, 100%) | |
| Lindley (1836)14 | Urticaceae | 2-27-18-0-15, 76% |
| (subfam. Cannabineae) | (2-0-0-0-0, 100%) | |
| Endlicher (1837)15 | Cannabineae | 2-0-0-0-0, 100% |
| Lindley (1846)16 | Cannabinaceae | 2-0-0-0-0, 100% |
| Bentham and Hooker (1880)17 | Urticaceae | 2-44-48-8-8, 97% |
| (tribe Cannabineae) | (2-0-0-0-0, 100%) | |
| Engler and Prantl (1889)18 | Moraceae | 2-6-46-0-0, 100% |
| (subfam. Cannaboideae) | (2-0-0-0-0, 100%) | |
| Cronquist (1968)19 | Cannabaceae | 2-0-0-0-0, 100% |
| Angiosperm Phylogeny Group (2003)1 | Cannabaceae | 2-0-0-0-8, 100% |
Adanson5 was more accurate than Linnaeus,6 but Lamarck7 outdid them both—although only Adanson accurately combined Cannabis, Humulus, and Celtis. Lamarck7 first placed Cannabis and Humulus in Moraceae (although using French instead of Latin, as Figuiers), and de Jussieu8 first placed Cannabis and Humulus in Urticaceae (spelling it Urticae).
Several 18th century taxonomists followed Adanson and grouped Cannabis, Humulus, and Celtis,11–13 but Adanson's concept lost recognition thereafter. Batsch9 first segregated Cannabis and Humulus into their own subfamily. Martynov10 elevated the pair to a family rank, and coined the name Cannabaceae. Subsequent botanists, unaware of Martynov, coined Cannabineae15 and Cannabinaceae.16 These incorrect spellings still appear in the literature. Others continued to place Cannabis into the Urticaceae or Moraceae.17,18 Not until the mid-20th century has Cannabaceae come into common usage.19
In 2003, Angiosperm Phylogeny Group1 merged Cannabaceae with Celtidaceae based on genetic (DNA) evidence. The merger is morphologically counterintuitive—Cannabis and Humulus species are herbal plants, whereas the Celtidaceae are woody trees. Yang et al.20 provide the latest analysis, utilizing four chloroplast DNA (cpDNA) genes (trnL-trnF, rbcL, atpB-rbcL, and rps16). Their results strongly support this expanded family as a monophyletic group. Cannabis and Humulus form a clade that nests within former Celtidaceae genera. Naming the family Celtidaceae might better reflect its members, but the name Cannabaceae is older and therefore holds nomenclatural priority.
Genetic (DNA) data should be congruent with phenotypic characters (morphology, phytochemistry, host–parasite relationships, etc.). Congruency can be tested with a procedure called character mapping, also known as ancestral reconstruction—assessing phenotypic evolution by laying phenotypic characters upon a phylogenetic tree. Yang et al.20 reconstructed eight ancestral characters in the Cannabaceae.
Cannabis and Humulus shared only three of the eight ancestral characters: triporate pollen grains, imbricate flower aestivation, and a persistent perianth (domesticated Cannabis has a deciduous perianth). Ancestral stipule arrangement—extrapetiolar stipules—was expressed by Cannabis, with a shift to interpetiolar stipules in Humulus. An ancestral seed coat character—a seed coat lacking microscopic holes—was expressed by Cannabis, with a shift to a seed coat with holes in Humulus. Three other Cannabaceae ancestral characters were not shared by Cannabis and Humulus: monoecy (Cannabis and Humulus are dioecious), alternate leaf arrangement (Cannabis and Humulus have both alternate and opposite leaves), and fleshy drupes (Cannabis and Humulus have achenes).
In their younger days, Cannabis and Humulus lost a sibling genus. Dorofeev21 described and illustrated fruits of two species in an extinct genus: Humularia reticulata and Humulus tymensis. Dorofeev found both species in central Siberia, and dated them to the Oligocene Epoch, 33.9–23.03 million years ago (mya). Collinson22 re-examined Dorofeev's fossils, and confirmed that they represent an extinct taxon within Cannabaceae. Grudzinskaya23 assigned a new (legitimate) name, Dorofeevia, to the extinct genus.
The Genus Cannabis
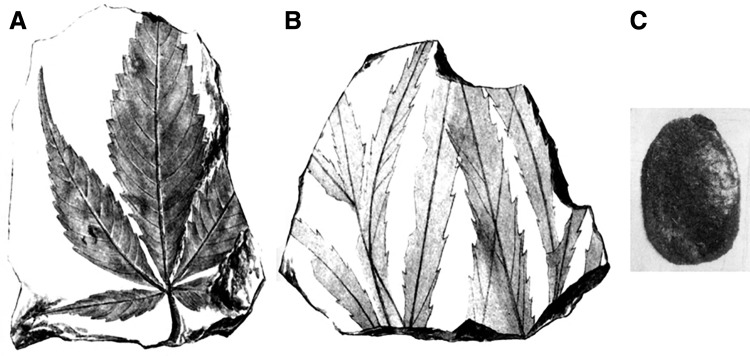
Accurately determining when Cannabis evolved is difficult, because the genus lacks a robust print fossil record (impressions of leaves or fruits in rocks). Friedrich24 found fossil leaves in Germany that he named Cannabis oligocaenica (Fig. 1A, B). Friedrich did not date his fossil, but his species epithet refers to the Oligocene Epoch. Palamarev25 identified a fossil seed (achene) as “Cannabis sp.” in Bulgaria (Fig. 1C). He dated the fossil to the “Pontian age,” 7.3–5.3 mya, which is the end of the Miocene Epoch (23.03–5.33 mya).
FIG. 1.
Macrofossils identified as Cannabis (not to scale). (A, B) Are from Friedrich,28 and (C) Is from Palamarev.25 (A, B) Reproduced from a publication whose copyright has expired; (C) Reproduced courtesy of Vladimir Bozukov, Bulgarian Academy of Sciences.
Dorofeev26 described and illustrated a fossil seed, “Cannabis sp.,” from the Miocene Epoch, found in Siberia. He had previously assigned the fossil to Humulus lupulus.27 Dorofeev21 changed his mind again, and reidentified the fossil as an extinct species, Humulus irtyshensis. He described three other extinct Humulus species, all from Siberia: H. strumulosus, dating to the Oligocene,21 H. minimus, also from the Oligocene,21 and H. rotundatus, dating to the Miocene.27
MacGinitie29 found leaf prints in Florissant, Colorado, that he named Vitus florissantella. He subsequently renamed one of the fossils Humulus florissantella.30 The fossil lacks diagnostic fruits; assigning the leaf to either Vitus or Humulus is debatable. The Florissant fossil bed has been dated to 34.07 mya using 40Ar/39Ar radiometric dating.31
For organisms like Cannabis that lack a good fossil record, a “molecular clock” can estimate when they diverged from other organisms. The molecular clock uses DNA to measure time, because DNA accumulates random mutations at a fairly constant rate. Some species might evolve at different rates, however, so computer algorithms now allow for variable rates between lineages in a phylogenetic tree, and calibrate the clock with fossil dates of related plants.
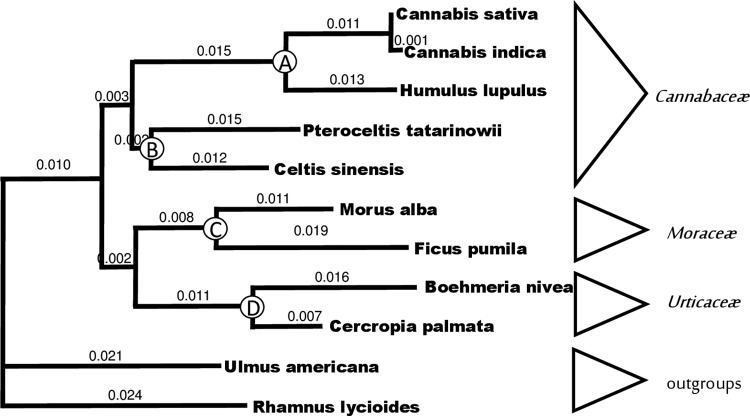
McPartland and Guy32 used a variable rate-smoothing algorithm, calibrated with four fossil records. They constructed a phylogenetic tree using cpDNA sequences (rbcL+trnL-trnF intergenic spacer data obtained from Gilmore,33 and other sequences from Genbank). ClustalX (version 2.0) was used to build a multiple sequence alignment. A maximum likelihood phylogenetic tree was constructed with PAUP* (version 4.0b10), using Modeltest 3.06 to select an optimal ML model. Divergence dates were estimated with r8s (version 1.70, nonparametric algorithm).
The clock was calibrated with fossils of Humulus (node A, 28–16 mya34), Celtis (node B, 65–56 mya35), Morus and Ficus (node C, 56–34 mya22), and Boehmeria (node D, 60–34 mya22,34). The phylogenetic tree with branch lengths and calibration nodes appears in Figure 2. They estimated that Humulus and Cannabis diverged from a common ancestor 27.8 mya. C. sativa and Cannabis indica diverged 1.05 million years ago, but this was not published because the taxa differ at only one nucleotide site.
FIG. 2.
Phylogenetic tree used for calculating divergence dates.
Every species occupies an indigenous geographical area, its native range. An Iraqi agronomist named Ibn Wahshīyah first speculated upon the native range of C. sativa in 904 AD. He proposed that šāhdānaj (C. sativa) came from India or perhaps China.36 Starting with Linnaeus, the native range of a cultivated plant has been deduced by locating its congeners, or wild relatives. Finding wild relatives is complicated by the fact that C. sativa easily escapes cultivation, and reacquires wild-type characteristics (a.k.a., it becomes naturalized, and survives as a “feral escape”). C. sativa reacquires wild-type characteristics in as little as 50 generations (years).37
Linnaeus38 knew C. sativa as a cultivated plant in Europe, so he deduced its native range was elsewhere: India Orientali (encompassing the Indian subcontinent, southeastern Asia, and the Malay Archipelago), Japonia (Japan), and Malabaria (the Malabar coast of southwest India). Some botanists considered C. sativa native to Europe, rather than Asia.39,40 Winterschmidt41 recognized two species, C. sativa and Cannabis chinensis, with their native ranges in Russland and Ostindien, respectively. Lamarck42 recognized two species, C. sativa and C. indica. He suggested that C. sativa grew croît naturellement in Persia and presque naturalisée in Europe, whereas C. indica originated in India.
Within a species's native range lies its center of origin, from whence it dispersed. De Candolle43 offered Central Asia as the C. sativa center of origin. “The species has been found wild, beyond a doubt, to the south of the Caspian Sea (Azerbaijan, Iran, Turkmenistan), near the Irtysch (Irtysh River, arising in the Altai, flowing through Kazakhstan to western Siberia), in the desert of Kirghiz (the Kazakh steppe), beyond Lake Baikal in Dahuria (Transbaikal). The antiquity of the cultivation of hemp in China leads me to believe that its area extended further to the east…” He added that C. sativa grew “almost wild in Persia,” but he doubted it was indigenous there, “since in that case the Greeks and the Hebrews would have known of it at an earlier period.” C. sativa was not native to the Levant, because the ancient Egyptians and Hebrews did not know hemp.
Due to the paucity of Cannabis print fossils, paleobotanists have turned to microfossils, in the form of fossil pollen or subfossil (noncarbonized) pollen. Hundreds of fossil pollen studies (FPSs) have reported Cannabis or Humulus pollen. Many paleobotanists, confronted with the morphological similarities between Cannabis and Humulus pollen grains, resort to collective names, for example, Cannabis/Humulus or Cannabaceae. These aggregate data can be dissected by using ecological proxies, instead of grain morphology. Cannabis/Humulus pollen in a steppe assemblage (occurring with Poaceae, Artemisia, and Chenopodiaceae pollen) is consistent with wild-type Cannabis. Cannabis/Humulus pollen in a mesophytic forest assemblage (occurring with Alnus, Salix, and Populus pollen) is consistent with Humulus.44
A meta-analysis of 88 FPSs in Asia used these methods.45 The oldest pollen in a steppe assemblage was located in Níngxià Province, China, and dated to 19.6 mya. A map of the FPSs constructed with geographical information system (GIS) software identified the northeastern Tibetan Plateau as the Cannabis center of origin. This geographical region, during the Oligocene, agrees with two hypotheses regarding the evolution of cannabinoid biosynthesis: (1) Cannabinoids protect plants from ultraviolet light (UVB) at higher altitudes, generated by the Tibetan uplift. (2) Cannabinoids deter vertebrate herbivores—the expansion of steppe during the Oligocene led to the evolution of Central Asian animals that feed on Cannabis today, such as Ungulates (horses), Rodentia (some families of rats, mice, and hamsters), Lagomorpha (rabbits and pikas), and Columbiformes (pigeons and doves).
A meta-analysis of 479 FPSs in Europe44 addressed the generally accepted concept that the dispersal of C. sativa from Asia to Europe depended upon human transport. The Scythians are often implicated, ca. 700 BCE.43 The meta-analysis identified Cannabis pollen in Europe that predated the Scythians, and even predated Homo sapiens. The oldest pollen dated to the Eopleistocene (1.8–1.2 mya), and came from the Upper Volga River basin in Russia. A prehistoric dispersal to Europe should not come as a surprise, because three other Cannabaceae genera spread through Eurasia, even to North America: Celtis,46 Humulus,46 and Pteroceltis.47
During the Last Glacial Maximum, northern Europe was covered by ice, with a southern margin between 52° N (midland England) and 56° (north of Moscow). Tree species retreated to glacial refugia in southern Europe, south of around 45° N. Between these latitudes, GIS mapping revealed a belt of Cannabis pollen spanning Europe.44
Following the warm and wet Holocene Climactic Optimum (7–6 kya), forests replaced steppe, and Cannabis retreated to steppe refugia in the Pontic steppe and the Mediterranean coast.44 This pattern was repeated through several cycles of stadials (ice ages) and interstadials (warmer, wetter periods). Cannabis went through repeated “genetic bottlenecks,” when the population shrank to small numbers during range contractions. Small populations experience genetic drift, where new genotypes arise randomly. Conventional wisdom states that differences between C. sativa and C. indica are due to human selection, and therefore not “natural.” Instead, FPSs suggests that genetic drift initiated allopatric differences between European C. sativa and Asian C. indica.
The Species C. sativa
The publication of Linnaeus's Species Plantarum in 175348 is treated as the starting point of botanical nomenclature, hence C. sativa has nomenclatural priority. Thirty-two years later, Lamarck42 coined C. indica for plants with provenance from India, Southeast Asia, and South Africa. Lamarck's description of C. indica differed from his description of C. sativa by eight “very distinct” morphological characters in stalks, branching habitus, leaflets, and flowers. Lamarck also described chemotaxonomic differences: C. indica produced a strong odor, and was psychoactive, “The principal effect of this plant consists of going to the head, disrupting the brain, where it produces a sort of drunkenness that makes one forget one's sorrows, and produces a strong gaiety.”
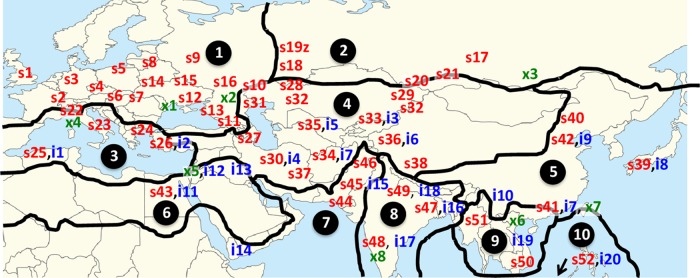
Linnaeus's disciples soon rendered subjective decisions regarding Lamarck's concept of a polyspecific genus. Persoon49 reduced Lamarck's species to C. sativa β indica. Persoon used Greek letters to indicate a varietal rank.50 There is evidence of cultural bias influencing these taxonomic decisions, arising from personality cults surrounding Linnaeus and Lamarck.51 It is expected that the distributions of C. sativa and C. indica (at species or subspecies rank) should occupy separate floristic regions. The distribution of plants that field botanists identified as C. sativa or C. indica in the 18th–19th centuries is presented in Figure 3.
FIG. 3.
Taxon names applied by field botanists. Locations of “C. sativa” labeled alphanumerically, s1, s2, and so on. Locations of “C. indica” labeled alphanumerically, i1, i2, and so on. Locations of other Cannabis taxa labeled alphanumerically, x1, x2, and so on. Base map shows the boundaries of 10 floristic regions (Takhtajan 1986). Map adapted from McPartland and Guy,51 which links alphanumerical sites with literature citations.
The map shows no hint of endemic distribution in specific floristic regions, for either C. sativa or C. indica. Field botanists assigned names according to their cultural biases. In general, Linnaeus's disciples from Scandinavia and Great Britain used “C. sativa.” Lamarck's disciples from France and Francophone Russia used “C. indica” or coined new taxa, such as C. chinensis and Cannabis gigantea. Reports of “C. sativa” in the Indian floristic region (region 8 in Fig. 3) are particularly striking. Examining field botanists' collections in 15 worldwide herbaria verified erroneous determinations.51
C. sativa and C. indica can be separated by morphology (C. sativa is taller with a fibrous stalk, whereas C. indica is shorter with a woody stalk), by phytochemistry (C. sativa has a cannabinoid ratio of Δ9-tetrahydrocannabinol (THC)>cannabidiol [CBD], whereas C. indica has a ratio of THC<CBD), and by differences in their original geographic range (C. sativa in Europe and C. indica in Asia). The taxonomic rank at which we separate C. sativa and C. indica is the primary issue in Cannabis taxonomy: are they different subspecies,52 or different species?53,54
Taxonomic ranks are relative levels within a hierarchy, and notoriously subjective. Darwin55 could not reconcile the continuous process of evolution with the discrete concept of taxonomic ranks. “I was much struck how entirely vague and arbitrary is the distinction between species and varieties.” Generally, different species are reproductively isolated and cannot hybridize, but this is not always true in plants. For example, Brassica napus (rapeseed) can hybridize with Brassica rapa (a weed); genetically modified B. napus has spread transgenic glyphosate resistance to its weedy relative.56
“DNA barcodes” make the question of rank less “vague and arbitrary.” The Consortium for the Barcode of Life uses the mitochondrial COI gene as a DNA barcode to identify animal species. Herbert57 proposed a 2.7% difference between two COI sequences as the threshold for flagging genetically divergent specimens as distinct species. The low rate of nucleotide substitutions in plant mitochondrial genes precludes the use of COI as a plant barcode. Numerous barcodes have been proposed for plants. Kress and Erickson58 reported sequence divergences (“barcode gaps”) between plant species: a species threshold of 5.7% for ITS1, 2.7% for trnH-psbA, 2.1% for rpoB2, 1.4% for rpoC1, and 1.3% for rbcL.
McPartland and Guy59 used barcode gaps in five sequences (rbcL, matK, trnH-psbA, trnL-trnF, and ITS1) to place the Cannabis question of rank in context with other plants. Pairwise alignments of sequences were made with BLAST. Differences between aligned sequences were quantified by tallying the number of nucleotide nonidentities as a percentage of the total BLAST alignment. These calculations were repeated with four groups of plants. All sequences were obtained from GenBank (Table 2).
Table 2.
Pairwise Barcode Gaps in Five Groups of Plants
| Taxon comparison | rbcL (%) | matK (%) | trnH-psbA (%) | trnL-trnF (%) | ITS (%) | Mean % ±SEM | |
|---|---|---|---|---|---|---|---|
| 1 | Apples and oranges | 8.6 | 18.2 | 19.0 | 26.6 | 18.0 | 18.07±2.869 |
| 2 | Solanum lycopericum and Solanum tuberosum | 1.00 | 1.07 | 5.67 | 0.40 | 11.6 | 3.95±2.134 |
| Humulus lupulus and Humulus japonicus | 1.03 | 1.13 | 3.18 | 1.0 | 10.7 | 3.41±1.869 | |
| Trema orientalis and Trema micrantha | 0.72 | 0.73 | Missing | 0.61 | 8.55 | 2.65±1.966 | |
| Fagopyrum esculentum and Fagopyrum tataricum | 0.50 | 2.76 | Missing | 0.12 | 7.27 | 2.66±1.642 | |
| Panax ginseng and Panax pseudoginseng | 1.25 | 1.22 | 2.69 | 1.11 | 5.57 | 2.37±0.852 | |
| 3 | Raphanus sativus and Raphanus raphanistrum | 0.0 | 0.0 | 0.0 | 3.54 | 2.84 | 1.28±0.789 |
| Populus alba and Populus trichocarpa | 0.73 | 0.12 | 2.95 | 0.0 | 2.45 | 1.25±0.610 | |
| Oryza sativa and Oryza rufipogon | 0.0 | 0.1 | 0.89 | 0.0 | 3.4 | 0.879±0.652 | |
| Gossypium hirsutum and Gossypium barbadense | 0.21 | 0.26 | 2.09 | 0.61 | 0.55 | 0.743±0.346 | |
| 4 | Camellia sinensis var. sinensis and C. sinensis var. assamica | 0.0 | Missing | 0.0 | 1.22 | 1.90 | 0.780±0.471 |
| O. sativa subsp. indica and O. sativa subsp. japonica | 0.01 | 0.0 | 1.16 | 0.0 | 1.30 | 0.494±0.301 | |
| Brassica oleracea var. capitata and B. oleracea var. italica | 0.40 | Missing | 0.0 | 0.79 | 0.50 | 0.406±0.257 | |
| Acorus calamus var. calamus and A. calamus var. americanus | 0.0 | 0.0 | 0.0 | 0.0 | 1.52 | 0.304±0.304 | |
| Curcumis melo subsp. melo and C. melo subsp. agrestis | 0.0 | 0.24 | Missing | 0.63 | 0.16 | 0.258±0.134 | |
| 5 | Cannabis sativa and Cannabis indica | 0.08 | 0 | 0.40 | 0.15 | 1.4 | 0.406±0.257 |
Apples and oranges—different genera and different species—express a mean barcode gap of 18.07% over the five sequences. The mean barcode gap in five pairs of related species that cannot hybridize (group 2 in Table 2) is 3.0%±0.3%, which is analogous to the 2.7% threshold for COI in animal species. The mean barcode gap in five pairs of closely related species that can hybridize (group 3) is 1.0%±0.1%. The mean barcode gap in five pairs of plants classified at the rank of subspecies or variety (group 4) is 0.43%±0.1%.
For C. sativa and C. indica, the mean barcode gap is 0.406±0.257. This difference nearly equals the mean of plants at the rank of subspecies or variety. C. sativa and C. indica should not be considered different species. The proper nomenclature is C. sativa subsp. sativa and C. sativa subsp. indica.52
Small and Cronquist52 erected a formal botanical nomenclature for C. sativa that has not been replaced. Their taxonomic concept is relatively simple: a two-step hierarchic classification system. The first step recognizes two subspecies based on THC content in dried female flowering tops, with 0.3% THC as the dividing point. The second step recognizes two varieties within each subspecies, based on their domestication phase:
C. sativa subsp. sativa var. sativa (low THC, with domestication traits)
C. sativa subsp. sativa var. spontanea (low THC, wild-type traits)
C. sativa subsp. indica var. indica (high THC, domestication traits)
C. sativa subsp. indica var. kafiristanica (high THC, wild-type traits)
The protologs of these four varieties are reviewed elsewhere, including their original descriptions, synonymies, and photographs of the four herbarium type specimens.51 That review compared and critiqued other taxonomic models by Vavilov, Schultes, de Meijer, and Hillig. Small's model adheres closest to protolog data (with C. indica treated as a subspecies). Importantly, Small and Cronquist52 noted that C. sativa subsp. sativa var. spontanea Vavilov 192260 has nomenclatural priority over C. sativa var. ruderalis Janischevsky 1924.61 Regarding the latter taxon, Janischevsky,61 also offered an alternative rank at the species level, C. ruderalis, but added, “I am inclined to consider it a well marked variety.”
Vernacular Taxonomy: “Sativa,” “Indica,” and “Ruderalis”
A folk taxonomy of drug-type plants, “Sativa” and “Indica,” has entangled and subsumed the nomenclature of C. sativa and C. indica. Thousands of websites generalize about the morphological, phytochemical, organoleptic, and clinical properties of these plants. “Sativa” is recommended for treating depression, headaches, nausea, and loss of appetite; it causes a stimulating and energizing type of psychoactivity. “Indica” is recommended for treating insomnia, pain, inflammation, muscle spasms, epilepsy, and glaucoma; it causes a relaxing and sedating psychoactivity.
“Sativa” plants produce more THC than CBD, and a terpenoid profile that smells “herbal” or “sweet.” “Indica” plants produce more CBD than “Sativa,” with a THC-to-CBD ratio closer to 1:1. “Indica” terpenoids impart an acrid or “skunky” aroma. Clarke62 first described the unique organoleptic properties of “Indica” plants, as a “slow flat dreary high.”
Small63 noted that “Sativa” and “Indica” were “quite inconsistent” with formal nomenclature, because C. sativa subsp. sativa should strictly apply to nonintoxicant plants. Conflating formal and vernacular taxonomy has begun to muddle otherwise excellent studies that worked with “Sativa” but latinized the taxon as C. sativa. This confusion even appeared in the distinguished journal Nature.64 “Sativa” and “Indica” written in quotation marks mean different things than C. sativa and C. indica written in italics.
McPartland et al.65 derided the inaccuracy of vernacular taxonomy, followed by others.54,63,66 Some experts propose jettisoning all vernacular names in favor of a metabolomics classification, “from cultivar to chemovar.”67,68 The parade of mistakes leading to “Sativa” and “Indica” is detailed elsewhere.51
Briefly, Vavilov69 assigned the taxon C. sativa to plants cultivated in Afghanistan for hashish. This concept departed from Linnaeus's protolog of C. sativa as a fiber-type, nonintoxicant plant from Europe. Vavilov69 coined a new taxon, C. indica var. afghanica, for plants with obovate leaflets, medium height, and profuse branching. Some botanists argue that afghanica designates a wild-type plant. But Vavilov's descriptions, illustrations, and other evidence indicate that afghanica was a feral escape of cultivated plants.51,52
Schultes et al.70 assigned the taxon C. indica to Afghani plants, and described the taxon having broad, oblanceolate leaflets, densely branched, more or less conical in shape, and very short (< 1.3 m). Designating these plants as C. indica was faulty; Lamarck was entirely unfamiliar with Afghani Cannabis. Lamarck's protolog of C. indica describes plants that are relatively tall, laxly branched, and with narrow leaflets.
Anderson71 repeated the errors by Vavilov and Schultes. He typified C. indica with plants that Schultes described in Afghanistan. He assigned C. sativa to plants consistent with Lamarck's C. indica. Anderson illustrated these concepts in a line drawing (Fig. 4). This illustration has become pervasive on the web as the poster child of vernacular nomenclature.
FIG. 4.
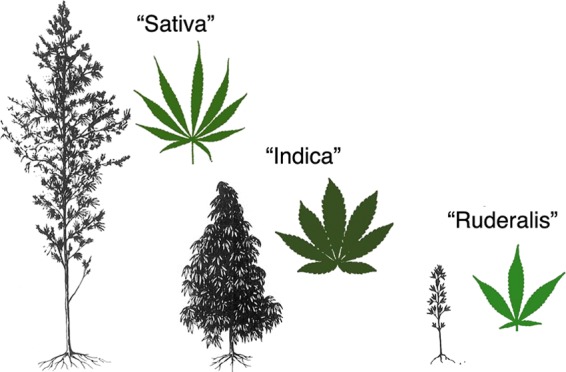
Cannabis vernacular taxonomy, image adapted from Anderson,71 courtesy of the Harvard University Herbaria and Botany Libraries.
De Meijer and van Soest72 introduced the vernacular taxonomy to peer-reviewed literature: “Indica” refers to plants with broad leaflets, compact habit, and early maturation, typified by plants from Afghanistan. “Sativa” refers to plants with narrow leaflets, slender and tall habit, and late maturation, typified by plants from India and their descendants in Thailand, South and East Africa, Colombia, and Mexico.
Categorizing cannabis as either “Sativa” and “Indica” has become an exercise in futility. Ubiquitous interbreeding and hybridization renders their distinction meaningless. The arbitrariness of these designations is illustrated by “AK-47,” a hybrid that won “Best Sativa” in the 1999 Cannabis Cup, and won “Best Indica” four years later.73 More than 30 years ago, unhybridized plants of Indian heritage and Afghani landraces were already difficult to obtain.62 Hybridization has largely obliterated population differences, “especially between the two kinds of fiber forms and between the two kinds of marijuana forms.”74
Schultes et al.70 made another taxonomic error. He eschewed Vavilov's taxon, C. sativa var. spontanea, in favor of Janischevsky's later synonym, Cannabis ruderalis. He then departed from Janischevsky's concept of C. ruderalis by applying the taxon to extremely short (≤0.61 m), unbranched plants with broad leaflets from Central Asia, instead of Janischevsky's relatively tall, laxly branched plants with narrow leaflets from southeastern Europe (Janischevsky described plants up to 2.1 m tall). Anderson71 illustrated a plant consistent with Schultes, not Janischevsky. One of the first seed bank catalogs illustrated “Ruderalis” plants growing near the Hungary-Ukraine border.75 The photos of “Ruderalis” show plants with strong apical dominance and little branching. These traits are consistent with a spontaneous escape of cultivated hemp, and depart from concepts by both Vavilov and Janischevsky.
In today's vernacular taxonomy, “Ruderalis” is applied to plants that exhibit one to three characteristics: CBD≅THC, wild-type morphology, or early flowering (sometimes called “autoflowering,” that is, day-neutral, flowering not induced by light cycle). Some authors have tried to reconcile “Sativa” and “Indica” with formal C. sativa and C. indica. McPartland et al.65 noted that Afghani plants were mislabeled “Indica.” They reassigned “Indica” to Vavilov's taxon, at species rank (Cannabis afghanica) or varietal rank (C. sativa var. afghanica).
In summary, reconciling the vernacular and formal nomenclatures: “Sativa” is really indica, “Indica” is actually afghanica, and “Ruderalis” is usually sativa. All three are varieties of one species, C. sativa L.
Abbreviations Used
- CBD
cannabidiol
- cpDNA
chloroplast DNA
- FPSs
fossil pollen studies
- GIS
geographical information system
- THC
Δ9-tetrahydrocannabinol
Acknowledgments
Vladimir Bozukov and the Harvard University Herbaria and Botany Libraries gave permissions to reproduce images. The curators of 15 herbaria authorized access to specimens: B (Berlin), BM (British Museum, London), BPI (Beltsville, MD), CUP (Cornel University), F (Field Museum, Chicago), ECON (Economic botany, Harvard), GH (Gray, Harvard), IND (Bloomington, IN), K (Kew, London), LE (St. Petersburg, Russia), LINN (Linnaeus, London), NY (Bronx, New York), P (Paris), PH (Philadelphia), US (Smithsonian, Washington DC), WIR (Vavilov Institute, St. Petersburg).
Author Disclosure Statement
No competing financial interests exist.
Cite this article as: McPartland JM (2018) Cannabis systematics at the levels of family, genus, and species, Cannabis and Cannabinoid Research 3:1, 203–212, DOI: 10.1089/can.2018.0039.
References
- 1.Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436 [Google Scholar]
- 2.McPartland JM, Cubeta MA. New species, combinations, host associations and location records of fungi associated with hemp (Cannabis sativa). Mycol Res. 1997;101:853–857 [Google Scholar]
- 3.Cesalpino A. De plantis libri XVI Andreae Caesalpini Aretini. Georgium apud Marescott: Florence, 1583 [Google Scholar]
- 4.Schultes RE. Random thoughts and queries on the botany of Cannabis. In: Joyce CRB, Curry Sh, (eds.) The botany and chemistry of Cannabis. J. & A. Churchill: London, 1970,. pp. 11–38. [Google Scholar]
- 5.Adanson M. Familles des Plantes. Vincent: Paris, 1763 [Google Scholar]
- 6.Linnaeus C. General plantarum, 6th ed Laurentii Salvii: Holmiae, 1764 [Google Scholar]
- 7.Lamarck JB. Encyclopédie Méthodique, Botanique, Tome second, Part 2. Panckoucke: Paris, 1788, pp. 401–774 [Google Scholar]
- 8.de Jussieu AL. Genera plantarum secundum ordines naturales disposita. Herissant & Barrois: Paris, 1789 [Google Scholar]
- 9.Batsch AJGK. Tabula affinitatum regni vegetabilis. Landes-Industrie-Comptoir:. Vinariae, 1802 [Google Scholar]
- 10.Martynov II. Техно-ботанический словарь на латинском и русском языках Imperial Russian Academy: St. Petersburg, 1820 [Google Scholar]
- 11.Blume CL. Bijdragen tot de flora van Nederlandsch Indië, 10de Stuk. Ter Lands Drukkerij: Batavia, 1825 [Google Scholar]
- 12.Gaudichaud-Beaupré C. Voyage autour du monde, Livre Premier. Pillet Aìne: Paris, 1826 [Google Scholar]
- 13.Nees von Esenbeck TFL, Spenner FKL, Endlicher SL, et al. . Genera plantarum florae germanicae, vol. IV Henry & Cohen: Bonn, 1835 [Google Scholar]
- 14.Lindley J. A natural system of botany; or, a systematic view of the organisation, natural affinities, and geographical distribution of the whole vegetable kingdom. Longman, Rees, Orme, Brown, Green and Longman: London, 1836 [Google Scholar]
- 15.Endlicher S. Genera plantarum. Beck: Vienna, 1837 [Google Scholar]
- 16.Lindley J. The vegetable kingdom. Bradbury & Evans: London, 1846 [Google Scholar]
- 17.Bentham G, Hooker JD. Urticaceae. In: Genera plantarum, vol. 3 1880, pp. 341–395 [Google Scholar]
- 18.Engler A, Prantl K. Die natürlichen Pflanzenfamilien, Teil III, Apteilung I. Vilhelm EngelmanL: Leipzig, 1889 [Google Scholar]
- 19.Cronquist A. The evolution and classification of flowering plants. Houghton Mifflin Co.: Boston, 1968 [Google Scholar]
- 20.Yang MQ, van Velzen R, Bakker FT, et al. . Molecular phylogenetics and character evolution of Cannabaceae. Taxon. 2013;62:473–485 [Google Scholar]
- 21.Dorofeev PI. Cannabaceae. : Takhtajan AL. (ed.), Ископаемые цветковые растения России и сопредельных государств, Т T. 2 [Fossil flowering plants of Russia and neighboring states, Vol. 2]. Izd-vo Nauka: Leningrad, 1982, pp. 43–48. [Google Scholar]
- 22.Collinson ME. The fossil history of the Moraceae, Urticaceae (including Cecropiaceae), and Cannabaceae. In: Crane PR, Blackmore S. (eds.), Evolution, systematics, and fossil history of the Hamamelidae,Volume 2 Clarendon Press: Oxford, 1989,. pp. 319–339. [Google Scholar]
- 23.Grudzinskaya IA. К систематике семейства Cannabaceae [To systematics of family Cannabaceae]. Bot Z. 1988;73:589–593 [Google Scholar]
- 24.Friedrich PA. Cannabis oligocaenica nov. spec. In: Beiträge zur Kenntniss der Tertiärflora der Provinz Sachsen. Schropp: Berlin, 1883, pp. 165–166 [Google Scholar]
- 25.Palamarev E. Неогенската карпофлора на Мелнишкия басейн [Neogene carpoflora from the Melnik Basin]. 1982;16:3–43 [Google Scholar]
- 26.Dorofeev PI. Миоценовая флора Мамонтовой горы на Алдане [Miocene flora of the Mammoth mountain on the Aldan]. Izd-vo Akademia nauk SSSR: Leningrad; 1969 [Google Scholar]
- 27.Dorofeev PI. Третичные флоры Западной Сибири [Tertiary flora of Western Siberia]. Izd-vo Akademia nauk SSSR: Leningrad, 1963 [Google Scholar]
- 28.Friedrich PA. Atlas zu den Abhandlungen zur geologischen Specialkarte von Preussen den Thüringischen Staaten, Band IV, Heft 3. Schropp: Berlin, 1883 [Google Scholar]
- 29.MacGinitie HD. Fossil plants of the Florissant beds, Colorado. Carnegie Inst Wash Publ. 1953;599:1–198 [Google Scholar]
- 30.MacGinitie HD. The Eocene Green River flora of northwestern Colorado and northwestern Utah. University of California Press: Berkeley, 1969 [Google Scholar]
- 31.Meyer HW. The fossils of Florissant. Smithsonian Books: Washington DC, 2003 [Google Scholar]
- 32.McPartland JM, Guy GW. THC synthase in Cannabis has undergone accelerated evolution and positive selection pressure. In: Proceedings of the 20th Annual Symposium on the Cannabinoids. International Cannabinoid Research Society: Research Triangle Park, NC, 2010,. p. 43 [Google Scholar]
- 33.Gilmore S, Peakall R, Robertson J. Organelle DNA haplotypes reflect crop-use characteristics and geographic origins of Cannabis sativa. Forensic Sci Int. 2007;172:179–190 [DOI] [PubMed] [Google Scholar]
- 34.Tiffney BH. Fruit and seed dispersal and the evolution of the Hamamelidae. Ann Mo Bot Gard. 1986;73:394–416 [Google Scholar]
- 35.Manchester SR, Akhmetiev MA, Kodrul TM. Leaves and fruits of Celtis aspera (Newberry) comb. nov. (Celtidaceae) from the Paleocene of North America and Eastern Asia. Intern J Plant Sci. 2002;163:725–736 [Google Scholar]
- 36.Hämeen-Anttila J. The last pagans of Iraq: Ibn Wahshīyah and his Nabatean Agriculture. Brill Academic Publishers: Leiden; 2006 [Google Scholar]
- 37.Small E. Morphological variation of achenes of Cannabis. Can J Bot. 1975;53:978–987 [Google Scholar]
- 38.Linnaeus C. Hortus Cliffortianus. Amsterdam, 1737 [Google Scholar]
- 39.Thiébaut de Berneaud A. Chanvre. In: Guérin-Méneville FE, ed. Dictionnaire pittorosque d'histoire naturelle et des phénomènes de la nature, Tome deuxième. De Cosson: Paris, 1835, pp. 87–89 [Google Scholar]
- 40.Keppen T. Догадка о происхожденіи большинства индоевропейскихъ названій конопли. Журнал Министерства народнаго просвěщенія 1886;245:73–86 [Google Scholar]
- 41.Winterschmidt JS. Naturgetreue Darstellung aller inn- und ausländischen Material-Samen und getrockneten früchte. Herausgeber: Nürnberg, 1818 [Google Scholar]
- 42.Lamarck JB. Encyclopédie Méthodique, Botanique, Tome premier, part 2. Panckoucke: Paris, 1785, pp. 345–752 [Google Scholar]
- 43.de Candolle ALP. Origine des Plantes Cultivées. Baillière: Paris, 1883 [Google Scholar]
- 44.McPartland JM, Guy GW, Hegman W. Cannabis is indigenous to Europe and cultivation began during the Copper or Bronze age: a probabilistic synthesis of fossil pollen studies. Veg Hist Archaeobot. 2018;27:635–648 [Google Scholar]
- 45.McPartland JM, Guy GW. Cannabis may have evolved in the northeastern Tibetan Plateau, based on an interdisplinary study of genetics, fossil pollen, and ecology. In: Proceedings of the 26th Annual Symposium on the Cannabinoids. International Cannabinoid Research Society: Research Triangle Park, NC, 2016, p. 61 [Google Scholar]
- 46.Gray A. Diagnostic characters of new species of phanerogamous plants collected in Japan by Charles Wright, Botanist of the U.S. North Pacific Exploring Expedition. With observation upon the relations of the Japanese flora of that of North America. Mem Amer Acad Arts Sci. 1859;6:377–453 [Google Scholar]
- 47.Manchester SR, Chen ZD, Lu AM, et al. . Eastern Asian endemic seed plant genera and their paleogeographic history throughout the northern hemisphere. J Syst Evol. 2009;47:1–42 [Google Scholar]
- 48.Linnaeus C. Species Plantarum, Tomus II. Laurentii Salvii: Holmiae, 1753 [Google Scholar]
- 49.Persoon CH. Synopsis plantarum seu enchiridium botanicum, Pars secunda. Treuttel & Würtz: Parisiis, 1807 [Google Scholar]
- 50.McPartland JM. Persoon: a phanerogamic footnote. Mycotaxon. 1992;45:257–258 [Google Scholar]
- 51.McPartland JM, Guy GW. Models of Cannabis taxonomy: a systematic review. Bot Rev. 2017;83:327–381 [Google Scholar]
- 52.Small E, Cronquist A. A practical and natural taxonomy for Cannabis. Taxon. 1976;25:405–435 [Google Scholar]
- 53.Hillig KW. A multivariate analysis of allozyme variation in 93 Cannabis accessions from the VIR germplasm collection. J Industr Hemp. 2004;9:5–22 [Google Scholar]
- 54.Clarke RC, Merlin MD. Cannabis evolution and ethnobotany. University of California Press: Berkeley, 2013 [Google Scholar]
- 55.Darwin CR. On the origin of species. John Murray: London, 1859 [Google Scholar]
- 56.Warwick SI, Légère A, Simard MJ, et al. . Do escaped transgenes persist in nature? The case of an herbicide resistance transgene in a weedy Brassica rapa population. Mol Ecol. 2008;17:1387–1395 [DOI] [PubMed] [Google Scholar]
- 57.Hebert PD, Stoeckle MY, Zemlak TS, et al. . Identification of birds through DNA barcodes. PLoS Biol. 2004;2:e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One. 2007;2:e508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McPartland JM, Guy GW. A question of rank: using DNA bar codes to classify Cannabis sativa and Cannabis indica. In: Proceedings of the 24th Annual Symposium on the Cannabinoids. International Cannabinoid Research Society: Research Triangle Park, NC, 2014,. p. 54 [Google Scholar]
- 60.Vavilov NI. Полевые культуры Юго-Востока [Field crops of the southeast]. Труды по прикладной ботанике генетике и селекции . 1922;13(Suppl 23):147–148 [Google Scholar]
- 61.Janischevsky DE. Форма конопли на сорных местах в Юго-Восточной России [A form of Cannabis in wild areas of south-eastern Russia]. Ученые записки Саратовского государственного университета имени Н. Γ. Чернышевского . 1924;2:3–17 [Google Scholar]
- 62.Clarke RC. Cannabis evolution. Masters thesis Indiana University, Bloomington, 1987. [Google Scholar]
- 63.Small E. Cannabis as a source of medicinals, nutraceuticals, and functional foods. In: Acharya SN, Thomas JE, (eds.) Advances in medicinal plant research. Research Signpost: Trivandrum, Kerala, India, 2007, pp. 1–39 [Google Scholar]
- 64.Gould J. The Cannabis crop. Nature. 2015;525:S2–SS3 [DOI] [PubMed] [Google Scholar]
- 65.McPartland JM, Clarke RC, Watson DP. Hemp diseases and pests—management and biological control. CABI Publishing: Wallingford, United Kingdom, 2000 [Google Scholar]
- 66.Piomelli D, Russo EB. The Cannabis sativa versus Cannabis indica debate: an interview with Ethan Russo, MD. Cannabis Cannabinoid Res. 2016;1:44–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hazekamp A, Fischedick JT. Cannabis—from cultivar to chemovar. Drug Test Anal. 2012;4:660–667 [DOI] [PubMed] [Google Scholar]
- 68.Hazekamp A, Tejkalová K, Papadimitriou S. Cannabis: from cultivar to chemovar II—a metabolomics approach to Cannabis classification. Cannabis Cannabinoid Res. 2016;1:202–215 [Google Scholar]
- 69.Vavilov NI, Bukinich DD. Konopli. Труды по прикладной ботанике генетике и селекции 1929;33(Suppl):380–382 [Google Scholar]
- 70.Schultes RE, Klein WM, Plowman T, et al. . Cannabis: an example of taxonomic neglect. Harv Univ Botanical Musem Leaflets 1974;23:337–367 [Google Scholar]
- 71.Anderson LC. Leaf variation among Cannabis species from a controlled garden. Harv Univ Botanical Musem Leaflets 1980;28:61–69 [Google Scholar]
- 72.de Meijer EPM, van Soest LJM. The CPRS Cannabis germplasm collection. Euphytica. 1992;62:201–211 [Google Scholar]
- 73.McPartland JM. Cannabis sativa and Cannabis indica versus “Sativa” and “Indica.” In: Chandra S, Lata H, ElSohly MA. (eds.). Cannabis sativa: Botany and Biotechnology. Springer International: Cham, Switzerland, 2017, pp. 101–121 [Google Scholar]
- 74.Small E. Cannabis: a complete guide. CRC Press: Boca Raton, FL, 2017 [Google Scholar]
- 75.Schoenmakers N. The seed bank 1986/1987 catalogue. Drukkerij Dukenburg: Nijmegen, The Netherlands, 1986 [Google Scholar]