Summary.
We propose a curve-based Riemannian geometric approach for general shape-based statistical analyses of tumours obtained from radiologic images. A key component of the framework is a suitable metric that enables comparisons of tumour shapes, provides tools for computing descriptive statistics and implementing principal component analysis on the space of tumour shapes and allows for a rich class of continuous deformations of a tumour shape. The utility of the framework is illustrated through specific statistical tasks on a data set of radiologic images of patients diagnosed with glioblastoma multiforme, a malignant brain tumour with poor prognosis. In particular, our analysis discovers two patient clusters with very different survival, subtype and genomic characteristics. Furthermore, it is demonstrated that adding tumour shape information to survival models containing clinical and genomic variables results in a significant increase in predictive power.
Keywords: Clustering, Glioblastoma multiforme, Magnetic resonance imaging, Shape manifold, Survival analysis
1. Introduction
There is intensive worldwide interest in preventing, detecting and treating cancer. Radiologic tools for detecting and treating cancer play central roles in disease management and surveillance. Technological advances in imaging equipment and techniques, and development of stage-specific methods for cancer, make medical imaging an indispensable tool for clinicians to monitor various cancers (Gutman et al., 2013). Clinical decision making, particularly for the brain, is routinely made on the basis of radiological image-based features in a magnetic resonance image (MRI). The three main analytical tasks in such settings, each with its own set of challenges, are
segmentation of the tumour region from the MRI,
characterization of the tumour via its shape, volume or other features and
development of prognostic models that link MRI features with genomic and clinical variables.
In this paper, we focus primarily on tasks (b) and (c). Brain tumour characterization is not straightforward because the tissue surrounding the tumour is often heterogeneous in spatial and imaging profiles (Krabbe et al., 1997) and sometimes overlaps with normal tissues (Provenzale et al., 2006). For example, it is extremely difficult to distinguish between primary central nervous system lymphoma and high grade glioma by using MRIs (Liu et al., 2011). Integrating volumetric and morphological features of tumours that are obtained from MRIs with clinical and genomic variables is usually based on non-objective numerical summaries of the features generated by experts. Thus, it is difficult to ascertain the reliability and reproducibility of such studies, and to generalize to different clinical settings.
The biological process governing tumour growth generates artefacts that can assist in the above-described tasks. A tumour normally originates from a single cell, and, as it proliferates in size, it exhibits heterogeneity in physiological and shape-related features (Marusyk et al., 2012). Both intertumour and intratumour heterogeneity are critical for characterizing tumours (De Sousa et al., 2013). Interpatient tumour heterogeneity can be quantified by morphological characteristics such as the shape and size of the tumour (McLendon et al., 2008), in addition to the genomic and clinical characteristics of a patient.
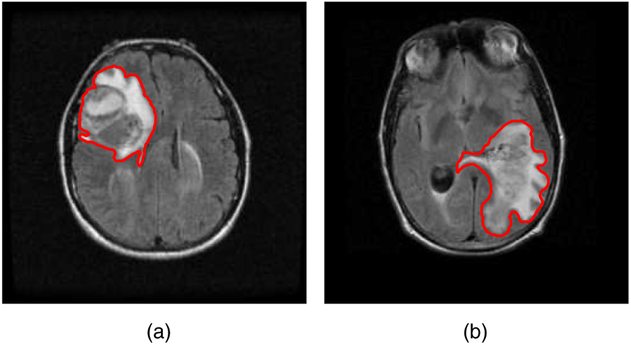
The relevance of tumour shape in characterizing tumour heterogeneity is linked to its growth process. Intrinsic brain tumours tend to evolve along tracts of white matter, altering the tracts in complex ways that include infiltration, displacement and disruption (Goldberg-Zimring et al., 2005). It is conceivable that new insight into patterns of tumour growth and invasion in the brain can be obtained through a better understanding of the shape and evolution of the tumour. Tumour shape is significantly influenced by the location in the brain and other anatomical constraints—in some places it might infiltrate and in others displace the fibre tracts. Irregular or spiculated shapes suggest an anisotropic structure of the underlying white matter; spherical or regular shapes imply a lack of structural or anatomical restrictions. The size of the tumour evidently affects its shape, especially in the presence of anatomical restrictions. It is reasonable to theorize that a better understanding of the relationship between the tumour’s shape and size, and histopathological factors related to the brain tumour would enhance the understanding of the tumour’s biological growth process; this would not only enable better prognosis but also potentially predict the likelihood of therapeutic success. For example, Fig. 1 shows two semiautomated segmentations of T2-weighted fluid-attenuated inversion recovery brain axial MRIs of patients diagnosed with glioblastoma multiforme (GBM), which is also known as grade IV glioma, with survival times of longer than 50 months (Fig. 1(a)) and shorter than 1 month (Fig. 1(b)). The tumour shape for the patient with longer survival appears to be more regular or spherical than the irregular shape corresponding to the patient with a short survival; the tumour sizes appear to be quite different as well. Evidently, the tumour locations for the two patients are different, which influences both size and shape.
Fig. 1.
T2-weighted fluid-attenuated inversion recovery MRIs of two patients diagnosed with GBM, with survival times (a) longer than 50 months and (b) shorter than 1 month: the segmented tumour is marked in red
Although the potential importance of tumour shape as a prognostic biomarker has been recognized (Goldberg-Zimring et al., 2005; McLendon et al., 2008), there is a striking paucity of progress in this direction. This is primarily due to the difficulty of representing and integrating tumour shape into existing statistical models. Current approaches that incorporate the information of a segmented brain tumour’s shape and size in models for tumour characterization and classification are based on subjective features provided by experts such as tumour circularity or sphericity and irregularity, and numerical summaries such as surface-to-volume ratio, total tumour area and entropy of the radial distribution of boundary pixels (Krabbe et al., 1997). Such radiological features are only indicative of tumour shape and do not fully characterize the shape. Furthermore, the subjective nature of the features ensures that statistical inference that is founded on them will suffer from a lack of reproducibility and reliability. In a recent paper exploring the predictive power of MRI features in the context of GBM, Gutman et al. (2013) stated that (page 568):
‘…it is often challenging to extract objective information for scientific analysis from prose statements of imaging features by neuroradiologists who typically use idiosyncratic vocabulary’.
Gutman et al. (2013) used various measures of agreement of ordinal and numerical values of neuroimaging features such as size and percentage of necrosis suggested by three expert radiologists, and they noted that volumetric and morphological information of the GBM tumour is informative for characterizing its biological growth process.
1.1. Statistical challenges and contributions
We can circumvent issues that are associated with qualitative and quantitative summaries of tumour shape by quantifying and utilizing information about the entire tumour shape. This extension, however, is not straightforward. Viewed statistically, tumour shape is a non-Euclidean object residing on (a quotient space of) some non-linear manifold. Thus, appropriate representation of a tumour shape should naturally employ statistical methods for non-Euclidean data objects. Motivated by this need, we focus on examining the utility of the two-dimensional shape of GBM tumours obtained from a single brain axial imaging slice with the largest tumour area in two contexts:
for detection of intertumour heterogeneity and
for evaluation of its association with molecular (genomic) profiles and survival times of patients who are diagnosed with GBM.
The methods that we employ are broadly applicable to various tumour types. Recent studies of scalar-on-image regression models in neuroimaging data applications incorporated the entire image (see, for example, Reiss and Ogden (2010), Li et al. (2015) and Goldsmith et al. (2013)); such methods are not applicable in the current setting since MRIs of GBM tumours cannot even be coregistered.
We model the two-dimensional tumour shapes as properties of parametric curves in , which provides the flexibility to accommodate uncertainty regarding landmarks and other curve features. In particular, we adapt the geometric framework for statistical shape analysis of closed curves that was proposed by Srivastava et al. (2011). In summary, our main contributions are as follows.
We define a suitable shape space that captures relevant information pertaining to tumour shapes represented as closed curves given by their outlines in two-dimensional MRIs.
We define notions of a geodesic path and distance between tumour shapes, and an average tumour shape; we also perform shape-based principal component analysis (SPCA) to identify and visualize principal directions of variation in a sample of tumour shapes.
We illustrate the utility of the developed tools in clustering GBM tumour shapes, and other inferential tasks such as two-sample testing and survival time modelling.
We develop a coherent statistical representation of the tumour shape and use a geometric framework to implement tasks such as clustering and integrating tumour shape as a potential prognostic factor in statistical models that are commonly used in oncology studies. We find the motivation in the GBM data set (Section 2), for which issues about the use of MRI features have been recognized but not satisfactorily addressed. We examine statistical methods to integrate the tumour shape with genomic and clinical features of GBM and investigate associations between them; this can subsequently accelerate effective personalized therapeutic strategies for cancer development and progression. Note that the method presented is more general and can be applied to other cancers and imaging modalities as well.
The rest of this paper is organized as follows. First, in Section 2, we introduce the GBM data set. In Section 3, we provide statistical tools for analysing tumour shapes under an elastic framework. In particular, we focus on comparing and averaging tumour shapes, and summarizing shape variability in a sample of tumours. Section 4 considers specific statistical tasks on the GBM data set including clustering, hypothesis testing and survival modelling. Section 5 provides a short discussion and directions for future work.
2. Description of glioblastoma multiforme data set
GBM, which is the most common malignant brain tumour that is found in adults, is a morphologically heterogeneous disease. Despite recent medical advancements, the prognosis for most patients with GBM is extremely poor. In the USA alone, 12 000 new cases are being diagnosed every year (http://www.abta.org/about-us/news/brain-tumour-statistics/), among which less than 10% survive 5 years after diagnosis (Tutt, 2011). The median survival time for GBM patients is about 12 months (McLendon et al., 2008). Biological features that differentiate GBM from any other grade of tumour include hypoxia and pseudopalisading necrosis, and proliferation of blood vessels near the tumour.
For our study, we collated MRIs with linked genomic and clinical data from 63 patients who consented under the Cancer Genome Atlas protocols (http://cancergenome.nih.gov/). The data from presurgical T1-weighted post-contrast (T1) and T2-weighted fluid-attenuated inversion recovery (T2) MRIs for these patients were obtained from the Cancer Imaging Archive (http://www.cancerimagingarchive.net/). The data set comprising survival times and clinical and genomic variables was obtained from cBioPortal (http://www.cbioportal.org/).
The imaging data set is a subset of a larger patient cohort that contains information on the linked clinical and genomic variables. For clinical variables, we used the survival times of the patients and Karnofsky performance scores KPS (Karnofsky and Burchenal, 1949). KPS indicates the ability of cancer patients to perform simple tasks (Crooks et al., 1991) and is widely used to assess quality of life during disease diagnosis and treatment. Recent investigations have identified four different subtypes of GBM: classical, mesenchymal, neural and proneural, each of which is characterized by different molecular alterations (Verhaak et al., 2010). We also curated the information about these subtypes of GBM and some well-characterized GBM driver genes (Frattini et al., 2013): DDIT3, EGFR, KIT, MDM4, PDGFRA, PIK3CA and PTEN. Biologically, a gene is known as a driver gene when there is a mutation along with DNA level changes (amplifications or deletions). The full tumour volumes from T1 and T2 MRIs were also recorded for each patient. Preprocessing of images including details of segmentation, a more detailed description of the clinical and genomic covariates, and the demographics corresponding to the clinical covariates are presented in Section 1 of the on-line supplementary material.
3. Quantifying variability in tumour shapes: a geometric approach
Some issues that are associated with characterizing tumours in MRIs can be alleviated through a suitable representation, which should be sufficiently versatile to accommodate various subjective evaluations by neuroradiologists and, at the same time, be mathematically and statistically well defined to facilitate various inferential tasks. Shape analysis based on landmarks (finite collections of ordered points) (Dryden and Mardia, 1998) is not sufficiently flexible in this context since the tumours rarely have landmark features as such. Even if present, identifying tumour landmarks is difficult and may require subjective assessment. A natural way to represent a tumour is to use a two-dimensional curve that corresponds to its boundary, which allows for uncertainty in all landmark locations.
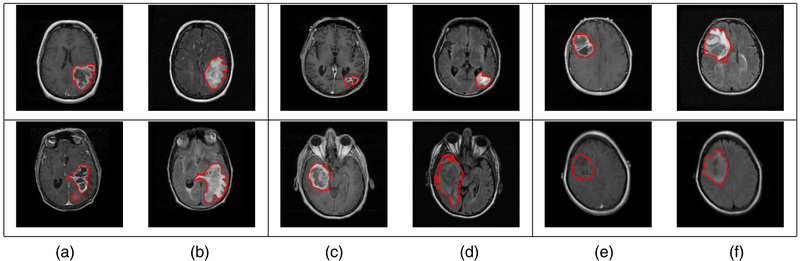
We adopt the shape definition of Srivastava et al. (2011) that is particularly attractive in the current context (see Joshi et al. (2007), Srivastava et al. (2011) and Kurtek et al. (2012) for details). While describing the tools, we concurrently illustrate their usage on the GBM data set. To obtain an idea of this problem’s complexity, we display a few examples of tumour contours overlaid on the corresponding T1 and T2 MRI slices in Fig. 2 (each row represents a different patient). The tumour shapes are heterogeneous, and at first glance it is difficult to ascertain any relationship between tumour shapes and survival times. To obtain insight into possible relationships between tumour shapes and outcomes, more sophisticated approaches are required. Throughout this section, we use the word metric to refer to a Riemannian metric (i.e. an inner product in tangent spaces) and distance to refer to the measure of differences between objects.
Fig. 2.
Examples of manually segmented tumour contours overlaid on the (a), (c), (e) T1- and (b), (d), (f) T2-images for patients with (a), (b) short (less than 1 month), (c), (d) medium (about 15 months) and (e), (f) long (more than 50 months) survival: each row represents a different patient (tumour)
3.1. Representation of tumour shape and elastic metric
The tumour shapes should be invariant to translation and rotation. Scaling might be considered important and can easily be incorporated in our framework. Denote a parameterized, planar, closed curve representing the outline of a tumour by a function . Since the tumour outline is a closed curve, it is natural to parameterize it by using the unit circle domain , instead of an interval. There are several possibilities for representing β for shape analysis. One can simply use the x- and y-co-ordinate functions of β; another possibility is to parameterize β by using the arc length and to compute the angle that makes with the x-axis (here, t is the curve parameter) (Klassen and Srivastava, 2006). For an overview of the various possible representations, and associated properties of shape spaces, see Bauer et al. (2014).
The choice of a metric on the tumour shape space is vital for comparing two shapes. Unlike typical problems in shape analysis, no template shape is available when considering tumours. In this context, it is imperative that the metric capture all possible deformations that match one tumour shape to another. One candidate metric is the elastic metric, which is defined as follows. Suppose that is the speed function and is the angle function. Consider two tangent vectors (small perturbations) (δpi, δθi), i = 1, 2, in the tangent space of (p, θ). The elastic metric (Mio et al., 2007) is defined as
| (1) |
for constants a, b > 0. The first term in equation (1) measures variations in the speed function (i.e. how fast the tumour outline is traversed), whereas the second term measures the variation in the direction of the unit tangent vectors via the standard Euclidean inner product between δθ1 and δθ2 (denoted by ⟨·, ·⟩); a and b provide the relative weights for the two terms. In other words, the first term captures the amount of stretching and the second term captures the amount of bending that are required to deform one tumour shape into another. Both terms are needed to generate natural deformations between tumour shapes. However, choosing a and b is difficult and problem dependent.
An important source of variation is the choice of parameterization of the tumour contours. This is a nuisance parameter when comparing tumour shapes, since the choice of parameterization is arbitrary and shape preserving, i.e. the tumour contour can be reparameterized in many different ways, but its shape remains unchanged. A common approach in the shape analysis literature is to normalize curve parameterizations to arc length to ensure that traversal along the curve is at unit speed. Under this scenario, only bending deformations are allowed, which often results in suboptimal point correspondences across shapes (Mio et al., 2007). We describe how it is possible not only to employ the elastic metric efficiently, but also to ensure that the resulting geodesic distance is invariant to the choice of parameterization. Unless otherwise stated, all curves are parameterized via arc length.
3.1.1. Square-root velocity function
Let is an orientation preserving diffeomorphism} be the group of reparameterization functions and orientation imply clockwise or counterclockwise traversal of the contour (i.e. γ is an invertible function that maps the unit circle to itself and preserves direction). The reparameterization of a tumour curve β, termed the action of Γ on the space of curves, is given by composition: (β, γ) = β ο γ. The chief issue with using the popular -metric is that the distance between two tumour contours β1 and β2 is not preserved under the action of Γ: ∥β1 – β2 ≠ ∥β1 ο γ – β2 ο γ∥ for a general γ ∈ Γ. In other words, the action of Γ on the space of tumour curves is not isometric, which means that a comparison of two tumour shapes depends on their parameterizations.
A proposed solution (Joshi et al., 2007; Srivastava et al., 2011; Kurtek et al., 2012) is to use a different representation of curves called the square-root velocity function (SRVF), which is given by , where ∣·∣ is the standard Euclidean norm in . This representation is convenient because it is automatically translation invariant. Conversely, β can be reconstructed from q up to a translation. If a tumour curve β is reparameterized to β ο γ, then its SRVF changes from q to .
The main reasons for using the SRVF for tumour shape analysis are that
the complicated but desirable elastic metric reduces to the standard -metric with and b = 1, allowing for both bending and stretching of tumour shapes and
∥q1 – q2∥=∥(q1, γ)–(q2, γ)∥, for all γ ∈ Γ, allowing for parameterization invariant analysis of tumour shapes.
If invariance to scale is required, each tumour shape can be rescaled to unit length. After rescaling, , i.e. the representation space of all SRVFs is a Hilbert sphere. For tumour shapes, the size of the tumour is often important, and the variability in tumour shape due to scale differences is considered to be important as well. In the GBM data example, we decouple tumour shape and size and consider them individually as covariates in the survival models. For a closed curve, which characterizes the tumour contours that we are studying, the corresponding SRVF satisfies the additional closure condition . Thus, the space of all unit length, planar, closed tumour curves, represented by their SRVFs, is given by , , and is called the preshape space.
3.1.2. Geodesic paths and distances in the elastic shape space
In the absence of a template tumour shape, it is critical to visualize deformations or changes in tumour shape. The choice of the elastic metric and the SRVF of two tumour shapes make it possible to compute natural geodesic paths and distances between them; as a consequence, we can visually examine the meaningful deformations of one tumour shape that transforms it into the other by traversing the geodesic path. This is potentially useful to radiologists for assessing possible changes in tumour morphology, thereby facilitating targeted interventions.
3.1.2.1. Preshape space with parameterization and rotation variability.
The preshape space is a non-linear submanifold of the Hilbert sphere due to the closure condition. It becomes a Riemannian manifold with the standard -metric, , where (i.e. v1 and v2 are elements of the tangent space to at q; they are often referred to as shooting vectors) and the inner product in the integrand is the standard Euclidean inner product in . The task of computing geodesic paths between any two elements is accomplished numerically, using an algorithm called path straightening that was introduced by Klassen and Srivastava (2006) and adapted to the SRVF representation by Srivastava et al. (2011). This algorithm initializes a path in connecting q1 and q2, and iteratively ‘straightens’ it until it becomes a geodesic. The geodesic distance is then simply the length of the geodesic path. Section 4 of the on-line supplementary material provides a list of algorithms that were used in this work along with relevant references. The issue with is that it contains contributions from two nuisance sources of variation. The distance between two tumour outlines is non-zero when they are within
a rotation and/or
a reparameterization of each other.
3.1.2.2. Shape space accounting for parameterization and rotation variability.
To remedy the issues with the preshape geodesic distance between two tumour shapes, it needs to be computed while accounting for all possible
rotations and
reparameterizations of one tumour shape to register it optimally to the other.
This is achieved as follows.
Let the special orthogonal group SO(2) be the set of 2 × 2 rotation matrices. For a tumour contour β and a rotation O ∈ SO(2), the SRVF of the rotated curve Oβ is given by Oq. Thus, to unify all elements in that denote the same tumour shape, we define equivalence classes of the type . Each such equivalence class [q] is associated with a unique tumour shape and vice versa. The set of all equivalence classes is called the shape space and is the quotient space . The distance can be used to define a distance between tumour shapes on according to
| (2) |
The geodesic distance is now the elastic distance on the space of tumour shapes and is invariant to rotation and reparameterization; as a consequence, all possible deformations pertaining to stretching and bending of tumour shapes are captured. Moreover, is bounded above by π/2, thereby offering a natural scale for comparing tumour shapes. In practice, the minimization in the definition of is performed by sampling each curve with a large number of points, and then recursively applying singular value decomposition to find the optimal rotation and the dynamic programming algorithm with an additional seed search to find the optimal reparameterization.
3.1.2.3. Illustrative examples.
We present multiple simulated and real data examples comparing non-elastic geodesic paths and distances (we optimize only over rotations and the seed placement but not the full reparameterization group) to the proposed elastic versions computed in the shape space. The points along the geodesic path between two tumour shapes should be viewed as the possible deformations transforming one tumour shape into the other. Since, in contrast with elastic shape analysis, the non-elastic framework does not allow stretching and compression deformations, we observe some unnatural shapes appearing along the geodesic paths in that case.
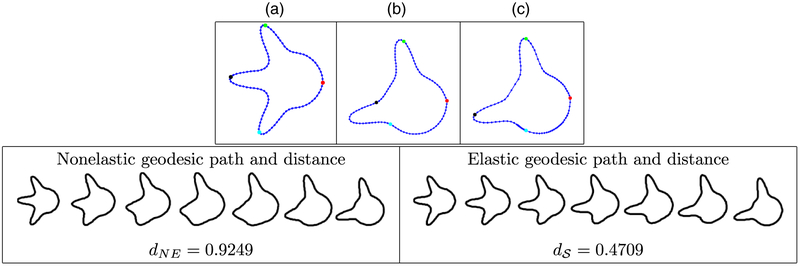
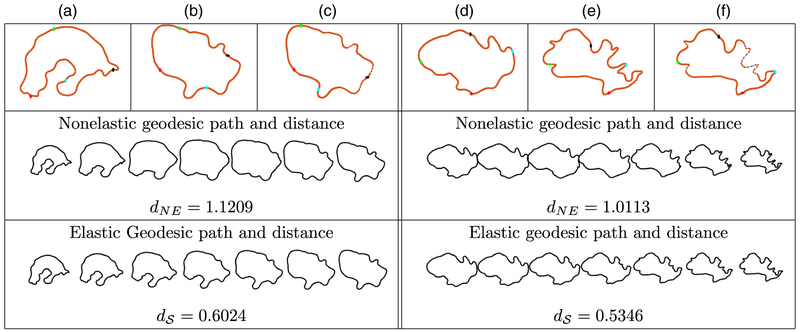
We first illustrate our approach on two simulated curves that are ‘toy’ tumour shapes. The curves were generated to reflect the protrusion-type behaviour of real GBM tumours, and were both initially parameterized with respect to their arc lengths. This example is shown in Fig. 3. First, with the given arc length parameterizations, the geometric features on the two curves do not match. This can be seen from the four coloured points. Fig. 3(a) shows the first simulated tumour outline where the green, black and cyan points correspond to three peaks. Fig. 3(b) shows the second tumour shape, where the green point corresponds to a peak whereas the other two do not. This results in an unnatural non-elastic geodesic deformation between these two shapes, where two of the peaks on the first shape are distorted to form the second peak on the second shape; the resulting distance is dNE = 0.9249. Under the elastic framework on the shape space , the optimal reparameterization can match the first two peaks across the two curves very well (the green and black points). Of course, there is no counterpart to the third peak on the second curve (the cyan point). This results in a natural deformation where the two matched peaks are preserved along the geodesic path whereas the third simply grows; the resulting distance is = 0.4709 (nearly a 50% decrease). We hypothesize that improvements such as that in this simulated example are extremely important in capturing natural variability in GBM tumour shapes. On visual inspection, the observed tumour contours have many geometric structures such as the peaks in this example. This motivates the use of the elastic shape analysis framework for studying GBM tumours.
Fig. 3.
Comparison of two simulated tumour shapes (we show four coloured points of correspondence for improved visualization; the resulting geodesic paths are sampled uniformly by using seven points (NE, non-elastic)): (a) curve with three protruding peaks; (b) curve with two protruding peaks before reparameterization (uniform spacing of points); (c) the same as (b) after reparameterization (optimal non-uniform spacing of points)
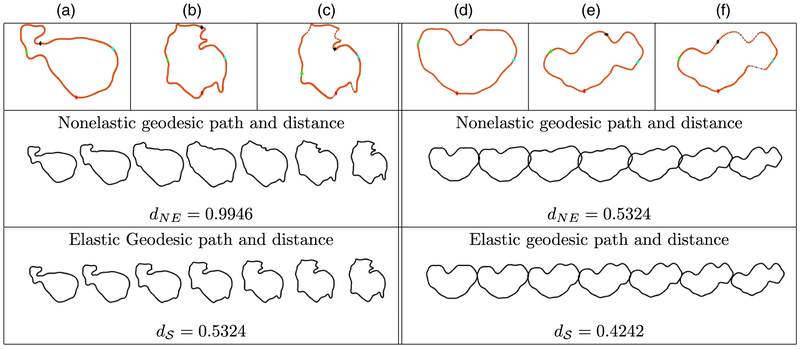
Next, we illustrate the elastic representation, alignment and computation of geodesic paths and distances between GBM tumour shapes corresponding to patients with different survival times; Fig. 4 presents two examples for the T1-modality, whereas Fig. 5 considers the T2-modality. In all examples, we have marked four corresponding points in red, green, black and cyan, and show the stretching and compression of points along the tumour curve due to optimization over Γ. The benefit of using the elastic framework becomes apparent when computing and visualizing geodesic paths between the tumour shapes: the points along the path represent tumour shapes that are elastically deformed in a natural way and preserve important shape features of the tumours. Indeed, when we allow non-uniform spacing of points along the curves, the geodesic deformation is improved because of an improved matching of geometric features across the tumour shapes. For example, for the T1-example in Figs 4(a)–4(c), the deformations along the geodesic path defined through the distance are natural in the following sense: the highly concave geometric feature of both tumours is nicely preserved along the geodesic path; this is not true in the non-elastic case. At the same time, other local geometric features in the form of concave and convex curve segments are preserved along the elastic shape geodesic. This is also clearly evident in the two examples that are shown for the T2-modality in Fig. 5. These geodesic path improvements are accompanied by significant distance reductions between the non-elastic (dNE) and elastic () frameworks; such improvements are even more drastic when we consider statistical modelling of tumour shapes. The examples presented thus support our proposal for the use of elastic shape analysis of GBM tumours for association with patient survival and genomic variables.
Fig. 4.
(a), (b), (c) Comparison of T1 tumour shapes for patients with survival times of 14.3 and 29.2 monthsand (d), (e), (f) comparison of T1 tumour shapes for patients with survival times of 8.8 and 48.6 months (we show four coloured points of correspondence for improved visualization; the resulting geodesic paths are sampled uniformly by using seven points (NE, non-elastic)): (a), (d) curve representing the first tumour; (b), (e) curve representing the second tumour before reparameterization (uniform spacing of points); (c), (f) same as (b) and (e) after reparameterization (optimal non-uniform spacing of points)
Fig. 5.
(a), (b), (c) Comparison of T2 tumour shapes for patients with survival times of 2.69 and 13.3 months and (d), (e), (f) comparison of T2 tumour shapes for patients with survival times of 6.14 and 0.72 months: the descriptions of the panels are the same as in Fig. 4
3.2. Statistical summaries of tumour shapes
Hereafter, our analyses focus on the shape space and the distance . However, we illustrate the resulting differences in the statistical summaries under non-elastic and elastic shape analysis. We define and illustrate computations of a mean tumour shape and covariance of a sample of tumour shapes, both defined with respect to . Consequently, we demonstrate how SPCA can be applied to explore and visualize the directions of variation in tumour shape based on patient level information. Identifying such directions can be useful in understanding the most likely deformations of the tumour shapes and can be potentially used to monitor the disease and for targeted therapeutic interventions.
3.2.1. Mean and covariance
Under the SRVF framework, the shape space is a (quotient space of a) non-linear submanifold of the Hilbert sphere, which is equipped with a Riemannian structure under the -metric. We first introduce some notation. Let be the SRVFs of two tumour preshapes and v ∈ Tq1 (). Then, the maps and are the exponential and inverse exponential maps respectively (Srivastava et al., 2011).
Let {β1,…, βη} denote a sample of given tumour contours and {q1,…, qn} be their corresponding SRVFs. Then, the Karcher (Frechet) mean tumour shape is defined as
| (3) |
A general gradient-based approach for finding this mean is provided in Le (2001) and Dryden and Mardia (1998). The Karcher mean is actually an entire equivalence class of curves. For the remainder of our analysis, we select one element of this class . One could also use the more robust geometric median as an alternative representative shape (Fletcher et al., 2009); for simplicity, we do not consider this case in the current work.
The general computation of the covariance around the estimated shape mean is as follows. Let , i = 1,…, n, denote the shooting vectors from the mean shape to each of the shapes in the given data. This first involves an optimal rotation O* and optimal reparameterization γ* of each qi, resulting in , to register it to the mean shape . Then, the covariance kernel can be defined as a function given by
In practice, since the curves must be sampled with a finite number of points, say m, the resulting covariance matrices are finite dimensional. Often, the observation size n is much less than m and, consequently, n controls the degree of variability in the stochastic model.
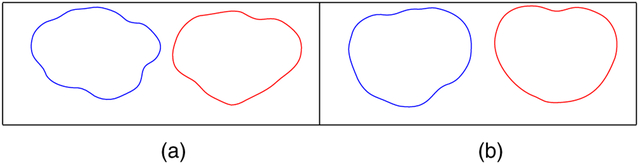
Fig. 6 displays a comparison of elastic and non-elastic shape averages for the T1 and T2 tumour shapes in our data set. In both cases, the elastic approach provides averages that have sharper geometric features than those provided by the non-elastic method. Thus, elastic shape analysis better summarizes the data, as most of the tumour shapes have multiple convex and concave characteristics. In other words, when we ignore the registration of points across curves (as in non-elastic analysis), shape features tend to average out, and the resulting shape means appear to be ‘oversmoothed’.
Fig. 6.
Comparison of elastic (blue) and non-elastic (red) shape averages of (a) T1 and (b) T2 tumour shapes
3.2.2. Shape-based principal component analysis
We explore dominant directions of variation in a sample of tumour shapes with an efficient basis for by using traditional PCA (which is also referred to as tangent PCA). Although we could also use the principal geodesic analysis that was developed in Fletcher et al. (2003) for the same purpose, we choose the simpler tangent PCA method for data analysis in this work. Let be the observed tangent data matrix with n observations and m sample points in on each tangent, i.e. each column of V is , i = 1,…, n, stacked into a long vector. Let be the resulting covariance matrix and let K = UΣUT be its singular value decomposition. The submatrix that is formed by the first r columns of U, which is called , spans the r-dimensional principal subspace of the observed shapes and provides the observations of the principal coefficients as . Thus, each original tumour shape can be represented by using a finite set of principal coefficients acting as Euclidean co-ordinates. These coefficients can then be used in a survival model for prediction as shown later.
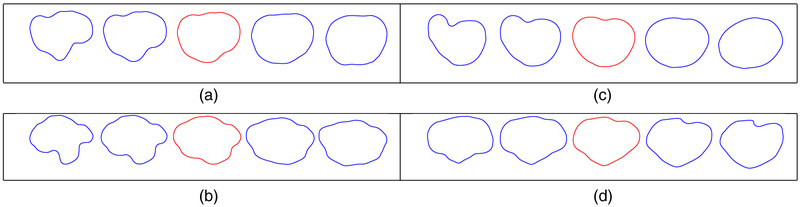
Fig. 7 displays the first principal direction of variation for T1 and T2 GBM tumour shapes; visualization of principal directions of shape variability in anatomical structures is an effective and common qualitative assessment (Shen et al., 2009; Epifanio and Ventura-Campos, 2014). For each case, we compare the elastic and non-elastic methods. The elastic principal paths capture more geometric features and are better at representing the overall variability in the tumour shapes. We compute the overall variance for each SPCA model as 7.86 (elastic) and 12.74 (non-elastic) for T1 tumours, and 13.43 (elastic) and 27.68 (non-elastic) for T2 tumours. The elastic models are more compact and provide a more efficient Euclidean representation of the tumour shapes in terms of the principal coefficients. Note that, because of a high level of heterogeneity of the tumour shapes, over 30 elastic SPCA components are needed to explain more than 95% of the variance. In section 2 of the on-line supplementary material, we additionally show that the elastic approach provides more natural results in the context of SPCA-based shape modelling and reconstruction.
Fig. 7.
Comparison of (a), (b) elastic and (c), (d) non-elastic principal directions of variation for (a), (c) T1 and (b), (d) T2 tumour shapes: in each example, we display the path within 2 standard deviations of the mean (red)
4. Shape-based clustering, testing and survival analysis in glioblastoma multiforme
The elastic framework for analysing tumour shapes enables us to perform a variety of estimation and inferential statistical tasks. In particular, SPCA of tumours provides the possibility of devising methods based on principal coefficients, which can be profitably viewed as Euclidean features or summaries of the tumour shape for inclusion in regression models. Using a data set of MRIs of GBM brain tumours, we applied clustering, two-sample testing and survival modelling to illustrate the advantages that are associated with the elastic representation of tumour shapes and the related geometric framework in the context of assessing patient survival and association with genomic or clinical variables. From here on, we perform statistical analysis via the elastic framework only.
4.1. Clustering of glioblastoma multiforme tumour shapes
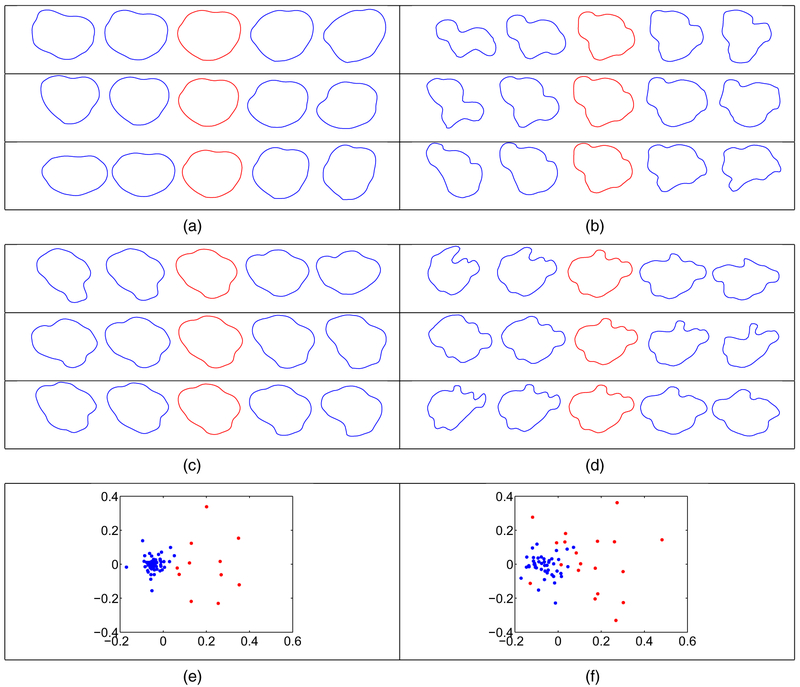
As a first unsupervised task, we performed hierarchical clustering of T1 and T2 tumour shapes by using the elastic shape distance. We first calculated the pairwise distance matrix and then used complete linkage to separate the shapes into two clusters for each modality (motivated by short versus long survival and supported by cluster visualization; see Figs 8(e) and 8(f)). To visualize the variability in each cluster better, we performed clusterwise SPCA and plotted the three principal directions of variation in each cluster for the T1- and T2-modalities in Fig. 8. We also report the cumulative variance in each cluster in Table 1. For both modalities, the variance in cluster 1 is much smaller than the variance in cluster 2. This can also be seen in the principal directions of variation; the shapes that are shown along cluster 1 directions (including the mean shape) are smoother and more circular.
Fig. 8.
Clusterwise principal directions of variation for T1 and T2 tumour shapes (in each example, we display the path within 2 standard deviations of the mean (red)): (a) T1 cluster 1; (b) T1 cluster 2; (c) T2 cluster 1; (d) T2 cluster 2; (e) multi-dimensional scaling plot of the T1 tumour shape data ( , cluster 1;
, cluster 1;  , cluster 2); (f) multi-dimensional scaling plot of the T2 tumour shape data (
, cluster 2); (f) multi-dimensional scaling plot of the T2 tumour shape data ( , cluster 1;
, cluster 1;  , cluster 2)
, cluster 2)
Table 1.
Cumulative variance of the clusterwise SPCA models for the T1 and T2 tumour data
| Cluster | Cumulative variance for T1 |
Cumulative variance for T2 |
|---|---|---|
| 1 | 5.51 | 10.23 |
| 2 | 14.74 | 17.95 |
We present a multi-dimensional scaling plot of the data in Figs 8(e) and 8(f), which confirm that cluster 2 is much more variable than cluster 1. Furthermore, the separability of the clusters is very good for both modalities, suggesting that the choice of two clusters is appropriate in this setting. In Table 2, we provide the mean and median survival times that are associated with the clusters, computed by using tumour shape data in each modality. First, the T1-modality provides better discrimination between survival times than does the T2-modality. Furthermore, for both modalities, we see that the mean and median survival times are higher in cluster 1, which contains much lower cumulative variance. This suggests that cluster 1 is more homogeneous, which is associated with longer survival times; cluster 2 is more heterogeneous and is associated with shorter survival times. This can also be attributed to the general morphological structure of tumours in the two clusters. The tumours in cluster 1 are often smoother and more circular than those in cluster 2, which are more irregular. It is this irregularity that is indicative of a more severe and infiltrative tumour with blurred margins and, as a result, shorter survival times. Note that the mean difference in survival times between cluster 1 and cluster 2 computed by using T1 tumour shapes is 6.8 months, which is large compared with the 12-month median survival time in GBM.
Table 2.
Summaries of clusterwise survival for the T1 and T2 tumour data
| Cluster | Survival (months) for T1 (mean) |
Survival (months) for T1 (median) |
Survival (months) for T2 (mean) |
Survival (months) for T2 (median) |
|---|---|---|---|---|
| 1 | 18.8 | 14.4 | 18.2 | 14.2 |
| 2 | 12.0 | 10.8 | 16.3 | 13.3 |
| Difference | 6.8 | 3.6 | 1.9 | 0.9 |
4.1.1. Cluster validation via enrichment
We use the concept of Bayesian cluster enrichment to study the association between the computed clusters, the tumour subtypes and other genomic covariates. In this approach, we want to compare the relative occurrence of a specific dichotomous covariate (with label 0 for no occurrence and 1 for occurrence) across the two clusters. To develop a Bayesian model for this, let θ1 ∈ [0,1] and θ2 ∈ [0,1] denote respectively the true proportion of 1s and 0s in cluster 1; let y1 and y2 denote respectively the observed number of 1s and 0s in cluster 1. Then, y1 ~ binomial (n1, θ1) and y2 ~ binomial(n2, θ2), where n1 is the total number of 1s and n2 is the total number of 0s. Consider a beta(1,1) prior on the true proportions θ1 and θ2. Since the beta distribution is conjugate for the binomial distribution, the posterior distribution is of the same family as the prior; the resulting posterior distributions for θ1 and θ2 are given by πθ1 (θ1 ∣y1, n1) ~ beta(y1 +1, n1 – y1 + 1) and πθ2(θ2∣y2, n2) ~ beta(y2 + 1, n2 – y2 + 1). We generate a large number of samples from the two posteriors πθ1 and πθ2, and approximate the true probability P(θ1 > θ2) by using Monte Carlo methods. We refer to this approximate quantity as the enrichment probability. The intuition behind this approach is as follows. If the computed clusters are not associated with the dichotomous covariate of interest, the resulting posteriors for θ1 and θ2 should be very similar. This in turn results in a Monte Carlo estimate of P(θ1 > θ2) that is close to 0.5, or no enrichment. In contrast, when the two posteriors are drastically different, the Monte Carlo estimate of P(θ1 > θ2) would be either very close to 1 (if y1 is much larger than y2) or 0 (if y1 is much smaller than y2). These two scenarios constitute high enrichment of the covariate in one of the two computed clusters (a given covariate can be enriched in only one cluster at a time).
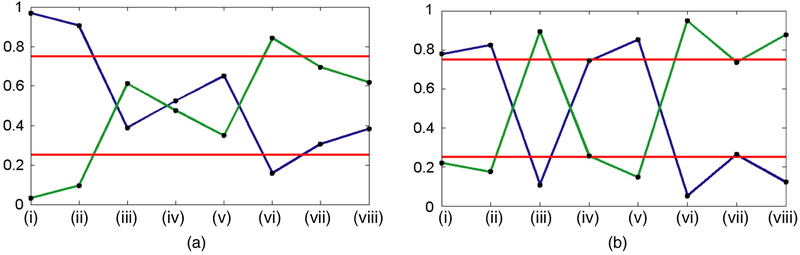
We present enrichment plots in Fig. 9. Each plot shows the enrichment probabilities as a line plot with high and low cut-offs in the form of horizontal lines at 0.75 and 0.25. We note the following trends from the enrichment plots. The classical and mesenchymal tumour subtypes are enriched in cluster 1 for both modalities. The proneural tumour subtype is enriched in cluster 2 for the T2-modality. Interestingly, the mesenchymal subtype, which is a very aggressive form of GBM, was enriched in the cluster with higher survival. However, on closer examination, there was an equal number of mesenchymal and non-mesenchymal subtypes in cluster 1 for both modalities (the enrichment probability was mostly driven by the arrangement in cluster 2). Furthermore, the patients in cluster 1 with the mesenchymal subtype had lower survival than their non-mesenchymal counterparts (by about 1.5 months).
Fig. 9.
Enrichment plots for the (a) T1- and (b) T2-modalities ( , 0.75 and 0.25 cut-offs for ‘high’ enrichment in cluster 1 (blue) and cluster 2 (green)): (i) classical; (ii) mesenchymal; (iii) proneural; (iv) EGFR; v) MDM4; (vi) PDGFRA; (vii) PIK3CA; (viii) PTEN
, 0.75 and 0.25 cut-offs for ‘high’ enrichment in cluster 1 (blue) and cluster 2 (green)): (i) classical; (ii) mesenchymal; (iii) proneural; (iv) EGFR; v) MDM4; (vi) PDGFRA; (vii) PIK3CA; (viii) PTEN
The enrichment plots for both imaging modalities display results that are consistent with some of the well-characterized genomic signatures in GBM. We note the following strong associations between tumour subtypes and driver gene mutations that have also been found in other studies (McNamara et al., 2013; Verhaak et al., 2010):
proneural subtype and PDGFRA gene mutation (in T2) and
classical and mesenchymal subtypes and EGFR gene mutation (in T2).
EGFR mutation is a common molecular signature of GBM. It promotes proliferation of the tumour, which is associated with classical and mesenchymal subtypes (Fischer and Aldape, 2010). PDGFRA also plays an important role in cell proliferation and migration, and angiogenesis. Unlike EGFR, this gene was found to be mutated in high amounts in the proneural subtype of GBM tumours only (Verhaak et al., 2010).
4.2. Permutation test for difference in tumour shape means
The distance between two tumour shapes opens up the possibility of a distance-based non-parametric two-sample test for differences in mean tumour shapes. To ascertain the association between tumour shapes and survival times of GBM patients, we dichotomize the data on the basis of four different survival cut-offs that have been examined in the literature (Nebert, 2000; Affronti et al., 2009; Mazurowski et al., 2013): 12, 13, 14 and 15 months. Under the null hypothesis that the two groups have equal mean shapes, a permutation test analogous to the case of landmark-based shape analysis (Dryden and Mardia, 1998) can be constructed under no assumptions on the distributions of the two groups. For each cut-off, we calculate the test statistic, which is the shape distance between the Karcher mean estimates for the two groups based on the given data. The distribution of this test statistic under the null hypothesis is not easily determined. Thus, we employ a permutation test by combining shapes from both samples (survival labels are exchangeable under the null hypothesis). We use 1000 random permutations of the labels to generate the distribution of the test statistic.
The resulting p-values for the T1- and T2-modalities, and all the cut-offs, are presented in Table 3. Based on our test statistic, there is a significant difference between T1 mean tumour shapes at the 0.05-level only at the 13-month cut-off. For the T2 tumour shapes, there is a highly significant shape mean difference for the 14- and 15-month cut-offs. The results clearly depend on the choice of the cut-off; nevertheless, this result provides support for our hypothesis that tumour shape features can be useful in survival analysis in GBM studies. We use only the mean shape information in this hypothesis test, although we expect that the covariance information is also useful. We demonstrate how that can be achieved by using a principal coefficient representation of tumour shapes in subsequent survival modelling.
Table 3.
Permutation test results for TI and T2 tumour shapes†
| Survival cut-off (months) |
T1 p-value | T2 p-value |
|---|---|---|
| 12 | 0.511 | 0.134 |
| 13 | 0.039 | 0.426 |
| 14 | 0.712 | <0.001 |
| 15 | 0.841 | <0.001 |
p-values smaller than 0.05 are indicated by italics.
4.3. Survival model adjusted for tumour shape
Next, we ascertain the utility of augmenting clinical and genetic information with imaging information when modelling survival probabilities of GBM patients. In particular, we investigate the association between the shape of a tumour and survival times (with censoring), in the presence of genetic and clinical covariates, using the geometry-based elastic shape method. On performing SPCA in the shape space , each tumour shape is represented in the principal directions of variation basis via its principal coefficients, which can be used as predictors in a survival model. Geodesic paths that are constructed by using principal shooting vectors allow for the possibility of traversing the principal directions of shape variation and monitoring changes in the shape of a tumour. It is customary to choose a handful of principal directions that explain most of the shape variability; however, since is infinite dimensional, and it is unclear how one can interpret the directions in the context of tumour shapes, we propose to use all available directions to capture maximal information from the data. Indeed, it may be that a direction corresponding to a small (in magnitude) eigenvalue represents a physiologically important tumour shape deformation. To incorporate all information from the images, we perform separate SPCA on tumours that were obtained from both T1- and T2-MRIs, and collate the principal coefficients from each imaging modality. Employing all available shape principal coefficients translates to a large number of imaging-based shape predictors in a potential survival model, necessitating dimension reduction through variable selection.
To assess whether incorporating imaging covariates, through principal tumour shape coefficients, improves discriminatory power of the survival model, we compare three nested models:
M1, a model with a set of clinical covariates C;
M2, a model with clinical and a set of genetic covariates G;
M3, a model with clinical, genetic and a set of imaging covariates I in the form of shape principal coefficients;
note that M1 ⊂ M2 ⊂ M3 where A ⊂ B denotes that model A is nested within model B.
The clinical covariate KPS contains a few missing values; we impute a value of 80 for those cases as advised by Gutman et al. (2013). A proportional hazards model (Cox, 1972), hereafter referred to as the Cox model, is used as the de facto model underlying models M1, M2 and M3, modelling the survival times of the patients in the presence of clinical, genetic and imaging predictors. Note that M1 is defined as the Cox model with C, M2 is defined as the Cox model with C ∪ G, and M3 is defined as the Cox model with C ∪ G ∪ I. Importantly, model M3, with a large number of tumour shape principal coefficients as predictors (62 each for T1 and T2), is fitted to the data by penalizing the negative log-likelihood by using a lasso penalty. Furthermore, we use leave-one-out cross-validation to determine the value of the penalty parameter. Specifically, if η is the vector of coefficients, then model M3 is fitted by solving the optimization problem minη[−log-partial likelihood of M3] + λ∣η∣1, where ∣η∣1 is the -norm of η. We use the R package glmnet by Friedman et al. (2011) for our implementation of model M3 with leave-one-out cross-validation. The set I is then redefined to contain only the principal coefficients with non-zero regression coefficients obtained from this lasso regression.
4.3.1. Significant directions of shape variation and other results
Next, we focus on the results of fitting the threemodels. Using the lasso penalty formodel M3, we first identify the principal tumour shape coefficients with non-zero regression coefficients, owing to the lack of a generally accepted way of testing for significance within the lasso framework (see recent work by Lockhart et al. (2014)). We uncover six principal directions of variation from T1 tumour shapes and five from T2 tumour shapes, when adjusted for the presence of predictors in C and G. The 11 coefficients comprise the operative new set I. One can visualize deformations of the Karcher mean tumour shape by following the vector field along the geodesic in the directions that are represented by the significant principal coefficients in the survival model. Such plots can potentially be used by neuroradiologists to visualize and make qualitative statements about deviations from the mean shape relating to increased or decreased chances of survival, when adjusting for clinical and genetic factors. We provide these displays in Section 3 of the on-line supplementary material for both modalities. The shapes become more irregular as we traverse the significant principal directions in the direction of a decreased chance of survival. The higher principal directions show global deformations that introduce a high level of non-smoothness, which are indicative of a protrusion of the tumours into neighbouring structures.
Results from fitting the three Cox models are given in Table 4. Gutman et al. (2013) found significant association between the clinical covariate KPS and survival time, adjusting for other numerical radiological summaries; this agrees with our results for all three models. Although the KPS-score is measured on a scale of 0–100, the only distinct values in our data set were 60, 80 and 100 along with missing values for 12 patients. As a measure of the ability to perform activities of daily living, the KPS-scores influence the survival time only indirectly and, in this data set, they complement the influence of the tumour shape principal coefficients. Since tumour volume was recorded for each patient from T1- and T2-images, we considered the shapes of tumour outlines rather than shapes and sizes. The size of the tumour was included in the model as a separate covariate through the tumour volume. It is known that tumours with EGFR mutations are larger than tumours with other mutations (Hatanpaa et al., 2010). In our analyses, EGFR and tumour volumes from both T1- and T2-images were not found to correlate significantly with survival time in the presence of tumour shape information. This finding is at odds with that of Gutman et al. (2013) where lesion size was used. It is known that older patients with GBM show high EGFR amplification. However, the variable EGFR informs us only if a mutation has occurred, not amplification. The age of a patient who is diagnosed with GBM is known to influence the survival time (Weller and Wick, 2011). Older age is typically used as a surrogate marker for change in the biology of GBM. The mean age in our data set was 56.33 years; the variable Age appeared to have significant correlation with survival time in all three models, and the inclusion of tumour shape information did not alter that.
Table 4.
Results from fitting Cox models M1, M2 and M3†
| Model | Predictors significant at 0.05 |
C-index 1 (Harrell et al., 1982) |
C-index 2 (Gömen and Heller, 2005) |
|---|---|---|---|
| M1 | Age, KPS | 0.641 | 0.652 |
| Clinical | |||
| M2 | Age, KPS | 0.722 | 0.728 |
| Clinical + genetic | DDIT3, PIKC3A | ||
| M3 | Age, KPS, DDIT3 | 0.859 | 0.841 |
| Clinical + genetic + imaging | 11 principal component shape coefficients |
Predictors significant at the 5% level are tabulated, and the two concordance indices are reported.
The discriminatory power of models M1, M2 and M3 was compared by using their concordance indices (C-indices), which are defined as the proportion of all pairs of patients whose predicted survival times are correctly ordered among all patients who can actually be ordered. For comparison, we use the C-index that was proposed by Harrell et al. (1982, 1984), and another version of it based on a U-statistic (Gömen and Heller, 2005). The C-indices (which were obtained through both methods) for model M3 are significantly higher than the C-indices for models M2 and M1. This indicates a clear benefit in incorporating tumour shape predictors in the form of principal coefficients in a survival model to obtain good discriminatory power. The Kaplan–Meier estimates of the survival functions for the three models, along with a description, are provided in section 3 of the on-line supplementary material.
In summary, among the driver genes that are known to be significant in GBM studies, only DDIT3 appears to have a significant correlation with the survival time of a patient when adjusted for the effect of tumour shape. Mutation of the driver gene DDIT3 appears to be associated with low survival probability (see Fig. 6 in the on-line supplementary material); it is known to regulate the glioma pathway through unregulated genes indirectly. Our analyses indicate that the shape of the tumour captures sufficient information about the individual relationships between each of the driver genes and survival time. A deeper study of the relationships between the shape of the tumour and driver genes is well worth exploring.
5. Discussion and future work
The use of shape analysis in medical imaging has been proposed before in other disease domains; we refer the reader to chapter 17 of Tofts (2003) and references within, for a good review. The shape of specific anatomical structures in the brain has been successfully used in multiple-sclerosis studies by Goldberg-Zimring et al. (1998), who based the analysis of shape on a few shape indices of the lesion. Landmark-based techniques using Procrustes averaging were used to study schizophrenia by DeQuardo et al. (1996). However, landmark and descriptor-based methods are not directly applicable to oncology because of multiple issues mentioned in this paper. In this work, we provide a comprehensive, Riemannian geometric solution to this problem that provides tools for various statistical analyses of tumour shapes. The benefits of this framework are clear:
it provides an elastic metric to measure interpretable shape deformations,
it defines a formal mathematical and statistical framework and
it provides tools for shape alignment, comparison, summarization, clustering, classification, hypothesis testing and other tasks.
We demonstrate these benefits through a detailed study of tumour shapes in the context of GBM. The method proposed can be readily extended to any cancer and/or other imaging modalities with similar data characteristics and scientific questions.
The focus of this paper is on two-dimensional tumour shapes obtained from the segmented tumour of a single axial slice of the brain with largest tumour area. The influence of the location and anisotropic nature of the white matter tracts on the shape of the tumour can be better assessed with three-dimensional shape analysis, which is currently in progress. The geometric framework that is presented in this paper allows for the extension to three-dimensional shapes (via square-root normal fields (Jermyn et al., 2012)), which would enable us to capture the full elastic shape of the tumour. However, studying parameterized surfaces in this context is difficult because of the large shape heterogeneity of the tumours. Except for the work of Goldberg-Zimring et al. (2005), who used spherical harmonic functions to model the three-dimensional shape of a tumour (akin to non-elastic analysis of tumour shapes), there is a lack of progress in this direction.
One way to view the proposed survival model is within the context that is offered by regression with functional predictors. The parametric closed curve representing a tumour shape predictor can be viewed as an element of the preshape space , which is a submanifold of and not a vector space. Current approaches with functional predictors using basis representations of the tumour shape or the coefficient function, or both, are hence inapplicable (see Morris (2015) for a detailed review).
The geometric framework that is used in this paper enables us to perform PCA on the space of tumour shapes under a Riemannian metric. The physiological interpretation of the principal directions, however, is unclear and much work remains to be done in this direction. Construction of a set of basis functions for the tangent space of a tumour shape that captures the biologically relevant deformations of the shape would be particularly useful; this requires significant input from clinicians in the form of prior shape information. The deformations that are observed in the tumour shape as we move away from the mean along the direction of decreased survival are striking; the shape appears to become more spiculated, which is consistent with the heuristic understanding of the seriousness of an irregularly shaped tumour. The visualization that is afforded within our framework, in our opinion, can profitably be used by neuroradiologists for initial non-invasive diagnoses. An alternative approach would be to use sparse PCA methods to model the variability in tumour shapes, which has recently proven useful in generating results that are clinically interpretable (Sjöstrand et al., 2007).
Applying the survival model M3 to the GBM data set, we uncovered several potentially interesting relationships between the shape of the tumour (expressed through the principal coefficients) and driver genes. This merits further consideration, and the implementation of our methods on other GBM data sets would offer more insight. Biological validation of the correlations between the two can significantly impact targeted personalized treatment strategies for GBM patients. Importantly, prognostic biomarkers of the transition time from a low grade to a malignant glioma can be determined.
Supplementary Material
Acknowledgements
We thank Joonsang Lee, Juan Martinez, Shivali Narang and Ganesh Rao for their help with processing the MRIs. All four authors were partially supported by National Institutes of Health grant R01-CA214955. SK and KB were partially supported by National Science Foundation grant DMS 1613054. SK was also partially supported by National Science Foundation grant CCF 1740761. AR was supported by Cancer Center Support Grant Bioinformatics shared resource P30-CA016672, an institutional research grant from the MD Anderson Cancer Center, a research scholar grant from the American Cancer Society (RSG-16-005-01) and a career development award from the MD Anderson Brain Tumor ‘Specialized program of research excellence’. VBs work was supported by National Institutes of Health grants R01-CA194391 and R01160736, National Science Foundation grant 1463233, and a Cancer Center Support Grant from the National Institutes of Health–National Cancer Institute (grant P30-CA016672).
Footnotes
Supporting information
Additional ‘supporting information’ may be found in the on-line version of this article:
‘Supplementary material for radiologic image-based statistical shape analysis of brain tumors’.
Contributor Information
Karthik Bharath, University of Nottingham, UK.
Sebastian Kurtek, Ohio State University, Columbus, USA.
Arvind Rao, University of Texas MD Anderson Cancer Center, Houston, USA.
Veerabhadran Baladandayuthapani, University of Texas MD Anderson Cancer Center, Houston, USA.
References
- Affronti ML, Heery CR, Herndon JE, Rich JN, Reardon DA, Desjardins A, Vredenburgh JJ, Friedman AH, Bigner DD and Friedman HS (2009) Overall survival of newly diagnosed glioblastoma patients receiving carmustine wafers followed by radiation and concurrent temozolomide plus rotational multiagent chemotherapy. Cancer, 115, 3501–3511. [DOI] [PubMed] [Google Scholar]
- Bauer M, Bruveris M and Michor PW (2014) Overview of the geometries of shape spaces and diffeomorphism groups. J. Math. Imgng Visn, 50, 60–97. [Google Scholar]
- Cox DR (1972) Regression models and life-tables (with discussion). J. R. Statist. Soc. B, 34, 187–220. [Google Scholar]
- Crooks V, Waller S, Smith T and Hahn TJ (1991) The use of Karnofsky Performance Scale in determining outcomes and risk in geriatric outpatients. J. Gerontol, 46, 139–144. [DOI] [PubMed] [Google Scholar]
- DeQuardo JR, Bookstein FL, Green WDK, Grunberg JA and Tandon R (1996) Spatial relationships of neuroanatomic landmarks in schizophrenia. Psychiatr. Res. Neurimgng, 67, 311–318. [PubMed] [Google Scholar]
- De Sousa FEM, Vermeulen L, Fessler E and Medema JP (2013) Cancer heterogeneity—a multifaceted view. Eur. Molec. Biol. Organizn Rep, 14, 686–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden IL and Mardia KV (1998) Statistical Shape Analysis. Chichester: Wiley. [Google Scholar]
- Epifanio I and Ventura-Campos N (2014) Hippocampal shape analysis in Alzheimer’s disease using functional data analysis. Statist. Med, 33, 867–880. [DOI] [PubMed] [Google Scholar]
- Fischer I and Aldape K (2010) Molecular tools: biology, prognosis, and therapeutic triage. Neurimgng Clin. Nth Am, 20, 273–282. [DOI] [PubMed] [Google Scholar]
- Fletcher PT, Lu C and Joshi SC (2003) Statistics of shape via principal geodesic analysis on Lie groups In Proc. Conf. Computer Vision and Pattern Recognition, pp. 95–101. New York: Institute of Electrical and Electronics Engineers. [Google Scholar]
- Fletcher PT, Venkatasubramanian S and Joshi SC (2009) The geometric median on Riemannian manifolds with application to robust atlas estimation. Neuroimage, 45, S143–S152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, Danussi C, Dolgalev I, Porrati P, Pellegatta S, Heguy A, Gupta G, Pisapia DJ, Canoll P, Bruce JN, McLendon RE, Yan H, Aldape K, Finocchiaro G, Mikkelsen T, Privé GG, Bigner DD, Lasorella A, Rabadan R and Iavarone A (2013) The integrated landscape of driver genomic alterations in glioblastoma. Nat. Genet, 45, 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T and Tibshirani R (2011) Regularization paths for Cox’s proportional hazards model via coordinate descent. J. Statist. Softwr, 39, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg-Zimring D, Achiron A, Miron S, Faibel M and Azhari H (1998) Automated detection and characterization of multiple sclerosis lesions in brain MR images. Magn. Reson. Imgng, 16, 311–318. [DOI] [PubMed] [Google Scholar]
- Goldberg-Zimring D, Talos I-F, Bhagwat JG, Haker SJ, Black PM and Zou KH (2005) Statistical validation of brain tumor shape approximation via spherical harmonics for image-guided neurosurgery. Acad. Radiol, 12, 459–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith J, Huang L and Crainiceanu CM (2013) Smooth scalar-on-image regression via spatial Bayesian variable selection. J. Computnl Graph. Statist, 23, 46–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gömen M and Heller G (2005) Concordance probability and discriminatory power in proportional hazards regression. Biometrika, 92, 965–970. [Google Scholar]
- Gutman DA, Cooper LAD, Hwang SN, Holder CA, Gao J, Aurora TD, Dunn WD Jr, Scarpace L, Mikkelsen T, Jain R, Wintermark M, Jilwan M, Raghavan P, Huang E, Clifford RJ, Mongkolwat P, Kleper V, Freymann J, Kirby J, Zinn PO, Moreno CS, Jaffe C, Colen R, Rubin DL, Saltz J, Flanders A and Brat DJ (2013) MR imaging predictors of molecular profile and survival. Radiology, 267, 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrell FE, Califf R, Pryor D, Lee K and Rosati R (1982) Evaluating the yield of medical tests. J. Am. Statist. Ass, 77, 2543–2546. [PubMed] [Google Scholar]
- Harrell FE, Califf R, Pryor D and Rosati R (1984) Regression modelling strategies for improved prognostic prediction. Statist. Med, 3, 143–152. [DOI] [PubMed] [Google Scholar]
- Hatanpaa KJ, Burma S, Zhao D and Habib AA (2010) Epidermal growthfactor receptorin glioma: signal transduction, neuropathology, imaging, and radioresistance. Neoplasia, 12, 675–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermyn IH, Kurtek S, Klassen E and Srivastava A (2012) Elastic shape matching of parameterized surfaces using square root normal fields In Proc. Eur. Conf. Computer Vision (eds Fitzgibbon A, Lazebnik S, Perona P, Sato Y and Schmid C), pp. 804–817. Berlin: Springer. [Google Scholar]
- Joshi SH, Klassen E, Srivastava A and Jermyn IH (2007) A novel representation for Riemannian analysis of elastic curves in In Proc. Conf. Computer Vision and Pattern Recognition, pp. 1–7. New York: Institute of Electrical and Electronics Engineers. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnofsky DA and Burchenal JH (1949) The Clinical Evaluation of Chemotherapeutic Agents in Cancer. New York: Columbia University Press. [Google Scholar]
- Klassen E and Srivastava A (2006) Geodesics between 3D closed curves using path-straightening. In Proc. Eur. Conf. Computer Vision, pp. 95–106. [Google Scholar]
- Krabbe K, Gideon P, Wagn P, Hansen U, Thomsen C and Madsen F (1997) MR diffusion imaging of human intracranial tumors. Neuroradiology, 39, 483–489. [DOI] [PubMed] [Google Scholar]
- Kurtek S, Srivastava G, Klassen E and Ding Z (2012) Statistical modeling of curves using shapes and related features. J. Am. Statist. Ass, 1θ7, 1152–1165. [Google Scholar]
- Le H (2001) Locating Frechet means with application to shape spaces. Adv. Appl. Probab, 33, 324–338. [Google Scholar]
- Li F, Zhang T, Wang Q, Gonzalez M, Maresh EL and Coan JA (2015) Spatial Bayesian variable selection and grouping for high-dimensional scalar-on-image regression. Ann. Appl. Statist, 9, 687–713. [Google Scholar]
- Liu Y-H, Muftah M, Das T, Bai L, Robson K and Auer D (2011) Classification of MR images based on Gabor wavelet analysis. J. Med. Biol. Engng, 32, 22–28. [Google Scholar]
- Lockhart R, Taylor J, Tibshirani RJ and Tibshirani R (2014) A significance test for the lasso. Ann. Statist, 42, 413–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, Almendro V and Polyak K (2012) Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer, 12, 323–334. [DOI] [PubMed] [Google Scholar]
- Mazurowski MA, Desjardins A and Malof JM (2013) Imaging descriptors improve the predictive power of survival models for glioblastoma patients. Neuro-oncology, 15, 1389–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis GM, Olson JJ, Mikkelsen T, Lehman N, Aldape K, Yung WK, Bogler O, Weinstein JN, VandenBerg S, Berger M, Prados M, Muzny D, Morgan M, Scherer S, Sabo A, Nazareth L, Lewis L, Hall O, Zhu Y, Ren Y, Alvi O, Yao J, Hawes A, Jhangiani S, Fowler G, San Lucas A, Kovar C, Cree A, Dinh H, Santibanez J, Joshi V, Gonzalez-Garay ML, Miller CA, Milosavljevic A, Donehower L, Wheeler DA, Gibbs RA, Cibulskis R, Sougnez C, Fennell T, Mahan S, Wilkinson J, Ziaugra L, Onofrio R, Bloom T, Nicol R, Ardlie K, Baldwin J, Gabriel S, Lander ES, Ding L, Fulton RS, McLellan MD, Wallis J, Larson DE, Shi X, Abbott R, Fulton L, Chen K, Koboldt DC, Wendl MC, Meyer R, Tang Y, Lin L, Osborne JR, Dunford-Shore BH, Miner TL, Delehaunty K, Markovic C, Swift G, Courtney W, Pohl C, Abbott S, Hawkins A, Leong S, Haipek C, Schmidt H, Wiechert M, Vickery T, Scott S, Dooling DJ, Chinwalla A, Weinstock GM, Mardis ER, Wilson RK, Getz G, Winckler W, Verhaak RG, Lawrence MS, O’Kelly M, Robinson J, Alexe G, Beroukhim R, Carter S, Chiang D, Gould J, Gupta S, Korn J, Mermel C, Mesirov J, Monti S, Nguyen H, Parkin M, Reich M, Stransky N, Weir BA, Garraway L, Golub T, Meyerson M, Chin L, Protopopov A, Zhang J, Perna I, Aronson S, Sathiamoorthy N, Ren G, Yao J, Wiedemeyer WR, Kim H, Kong SW, Xiao Y, Kohane IS, Seidman J, Park PJ, Kucherlapati R, Laird PW, Cope L, Herman JG, Weisenberger DJ, Pan F, Van den Berg D, Van Neste, L., Yi JM, Schuebel KE, Baylin SB, Absher DM, Li JZ, Southwick A, Brady S, Aggarwal A, Chung T, Sherlock G, Brooks JD, Myers RM, Spellman PT, Purdom E, Jakkula LR, Lapuk AV, Marr H, Dorton S, Choi YG, Han J, Ray A, Wang V, Durinck S, Robinson M, Wang NJ, Vranizan K, Peng V, Van Name E, Fontenay GV, Ngai J, Conboy JG, Parvin B, Feiler HS, Speed TP, Gray JW, Brennan C, Socci ND, Olshen A, Taylor BS, Lash A, Schultz N, Reva B, Antipin Y, Stukalov A, Gross B, Cerami E, Wang WQ, Qin LX, Seshan VE, Villafania L, Cavatore M, Borsu L, Viale A, Gerald W, Sander C, Ladanyi M, Perou CM, Hayes DN, Topal MD, Hoadley KA, Qi Y, Balu S, Shi Y, Wu J, Penny R, Bittner M, Shelton T, Lenkiewicz E, Morris S, Beasley D, Sanders S, Kahn A, Sfeir R, Chen J, Nassau D, Feng L, Hickey E, Barker A, Gerhard DS, Vockley J, Compton C, Vaught J, Fielding P, Ferguson ML, Schaefer C, Zhang J, Madhavan S, Buetow KH, Collins F, Good P, Guyer M, Ozenberger B, Peterson J and Thomson E (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature, 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara MG, Sahebjam S and Mason WP (2013) Emerging biomarkers in glioblastoma. Cancers, 5, 1103–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio W, Srivastava A and Joshi SH (2007) On shape of plane elastic curves. Int. J. Comput. Visn, 73, 307–324. [Google Scholar]
- Morris JS (2015) Functional regression. A. Rev. Statist. Appl, 2, 321–359. [Google Scholar]
- Nebert DW (2000) Extreme discordant phenotype methodology: an intuitive approach to clinical pharmacogenetics. Eur. J. Pharmcol, 410, 107–120. [DOI] [PubMed] [Google Scholar]
- Provenzale JM, Mukundan S and Baroriak DP (2006) Diffusion-weighted and perfusion MR imaging for brain tumor characterization and assessment of treatment. Radiology, 239, 632–649. [DOI] [PubMed] [Google Scholar]
- Reiss PT and Ogden RT (2010) Functional generalized linear models with images as predictors. Biometrics, 66, 61–69. [DOI] [PubMed] [Google Scholar]
- Shen L, Farid H and McPeek MA (2009) Modeling three-dimensional morphological structures using spherical harmonics. Evolution, 63, 1003–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöstrand K, Rostrup E, Ryberg C, Larsen R, Studholme C, Baezner H, Ferro J, Fazelias F, Pantoni L, Inziteri D and Waldemar G (2007) Sparse decomposition and modeling of anatomical shape variation. IEEE Trans. Med. Imgng, 26, 1625–1635. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Klassen E, Joshi SH and Jermyn IH (2011) Shape analysis of elastic curves in Euclidean spaces. IEEE Trans. Pattn Anal. Mach. Intell, 33, 1415–1428. [DOI] [PubMed] [Google Scholar]
- Tofts P (2003) Quantitative MRI of the Brain. Chichester: Wiley. [Google Scholar]
- Tutt B (2011) Glioblastoma cure remains elusive despite treatment advances. Oncology, 56, no. 3. [Google Scholar]
- Verhaak G W R. Hoadley, K. A, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelley M, Tamayo P, Weir BA, Gabrie S, Windder W, Gupta S, Jakkula L, Feiler H, Hodgson JG, James CD, Sarkaria JN, Brennan C, Kahn A, Spellman PT, Wilson RK, Speed TP, Gray JW, Meyerson M, Getz G, Perou CM, Hayes DN and Cancer Genome Atlas Research Network (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell, 17, 98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M and Wick W (2011) Are we ready to demystify age in glioblastoma?, or does older age matter in glioblastoma? Neuro-oncology, 13, 365–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.