Abstract
We found that the cancerous pancreas harbors a markedly more abundant microbiome compared to normal pancreas in both mice and humans, while select bacteria are differentially increased in the tumorous pancreas compared to gut. Ablation of the microbiome protects against pre-invasive and invasive PDA, whereas transfer of bacteria from PDA-bearing hosts, but not controls, reverses tumor-protection. Bacterial ablation was associated with immunogenic reprogramming of the PDA tumor microenvironment including a reduction in myeloid-derived suppressor cells and an increase in M1 macrophage differentiation, promoting Th1 differentiation of CD4+ T cells and CD8+ T cell activation. Bacterial ablation also enabled efficacy for checkpoint-targeted immunotherapy by upregulating PD-1 expression. Mechanistically, the PDA microbiome generated a tolerogenic immune program by differentially activating select toll-like receptors in monocytic cells. These data suggest that endogenous microbiota promote the crippling immune-suppression characteristic of PDA and that the microbiome has potential as a therapeutic target in the modulation of disease progression.
Keywords: Pancreatic cancer, Microbiome, Kras, Germ-free mice, T cells, Bifidobacterium
Introduction
Pancreatic ductal adenocarcinoma (PDA) is the 3rd most lethal cancer in the United States and accounts for 85% of all pancreatic malignancies (1). Peri-pancreatic inflammation is paramount for induction of oncogenesis. Innate and adaptive immune cell subsets cooperate through various mechanisms to promote tumorigenesis. We previously reported that activation of pattern recognition receptors (PRRs), which transmit inflammation in response to microbial pathogens, accelerates tumorigenesis, whereas mice deficient in select PRR signaling (including Toll-like receptor (TLR) 4, TLR7, TLR9, and Mincle) exhibit slower progression of PDA (2–6). The pro-tumorigenic effects of PRR ligation in PDA are mediated through multiple mechanisms including induction of innate and adaptive immune suppression, activation of pro-tumorigenic signaling pathways such as NF-κB, Notch, and STAT3, and activation of fibrogenic cells in the PDA tumor microenvironment (2–5). Based on these data we postulated that bacterial dysbiosis influences PDA progression.
Both microbial dysbiosis and disrupted epithelial barrier function leading to translocation of bacteriaare thought to be inducing factors in neoplastic transformation (7). The microbiome has emerged as a contributor to oncogenesis in a number of intestinal tract malignancies including laryngeal, esophageal, gastric, and colorectal cancers, as well as in primary liver cancer (8). In the aforementioned malignancies the gut microbiome is in direct contact with the at-risk organ or, in the case of liver cancer, the recipient of portal venous drainage from the intestine. However, very few reports implicate the gut microbiome in carcinomas ostensibly remote from the gastrointestinal lumen or its drainage (9,10). Moreover, the etiologic relationship between the intra-pancreatic microbiota and immune-suppressive inflammation in PDA has not been described.
We found that PDA is associated with a distinct stage-specific gut and pancreatic microbiome that drives disease progression by inducing intra-tumoral immune suppression. Conversely, targeting the microbiome markedly protected against PDA and enhanced anti-tumor immunity and susceptibility to immunotherapy. Our data suggest that elements of the microbiome may be useful in early diagnosis and risk stratification. Further, microbial-targeted therapies may reduce risk in pre-invasive disease and may be used as an adjuvant to standard therapies or in synergy with checkpoint-directed immunotherapy in invasive disease.
Results
To determine whether endoluminal gut bacteria access the pancreas, we administered fluorescently-labeled Enterococcus faecalis to WT mice via oral gavage. Bacteria migrated into the pancreas suggesting that intestinal bacteria can directly influence the pancreatic microenvironment (Figure 1a). Similar findings were observed using GFP-labeled Escherichia coli (Figure 1b). 16S rRNA FISH indicated a markedly greater presence of bacteria in both mouse and human PDA compared with normal pancreas (Figure 1c, d). qPCR analysis confirmed increased bacterial abundance in PDA compared with normal pancreas in mice and humans (Figure 1e, f). Repopulation experiments in antibiotic-treated WT mice suggested that the gut microbiome from Pdx1Cre;LsL-KrasG12D;p53R172H (KPC) mice has a higher capacity for translocation to the pancreas compared with WT gut microbiome (Figure 1g). Notably, Agrobacterium and Rhizobium, which are associated with mouse chow, were among the most abundant genera in both the WT mouse pancreas and the p48Cre;LSL-KrasG12D (KC) pancreas, further suggesting translocation (Figure S1a). To characterize the human intra-pancreatic microbiome, we performed 16S rRNA gene sequencing on PDA tumors from 12 patients. Thirteen distinct phyla were detected in human PDA. Proteobacteria (45%), Bacteroidetes (31%) and Firmicutes (22%) were most abundant and were prevalent in all samples (Figure S1b). Actinobacteria (1%) was also prevalent in all samples. Genera Pseudomonas and Elizabethkingia were highly abundant and prevalent in all human PDA specimens (Figure S1c). The bacterial composition in human PDA was distinct from that of normal human pancreas, based on assessment of clade abundances using Linear discriminant analysis Effect Size (LEfSe) (Figure S1d).
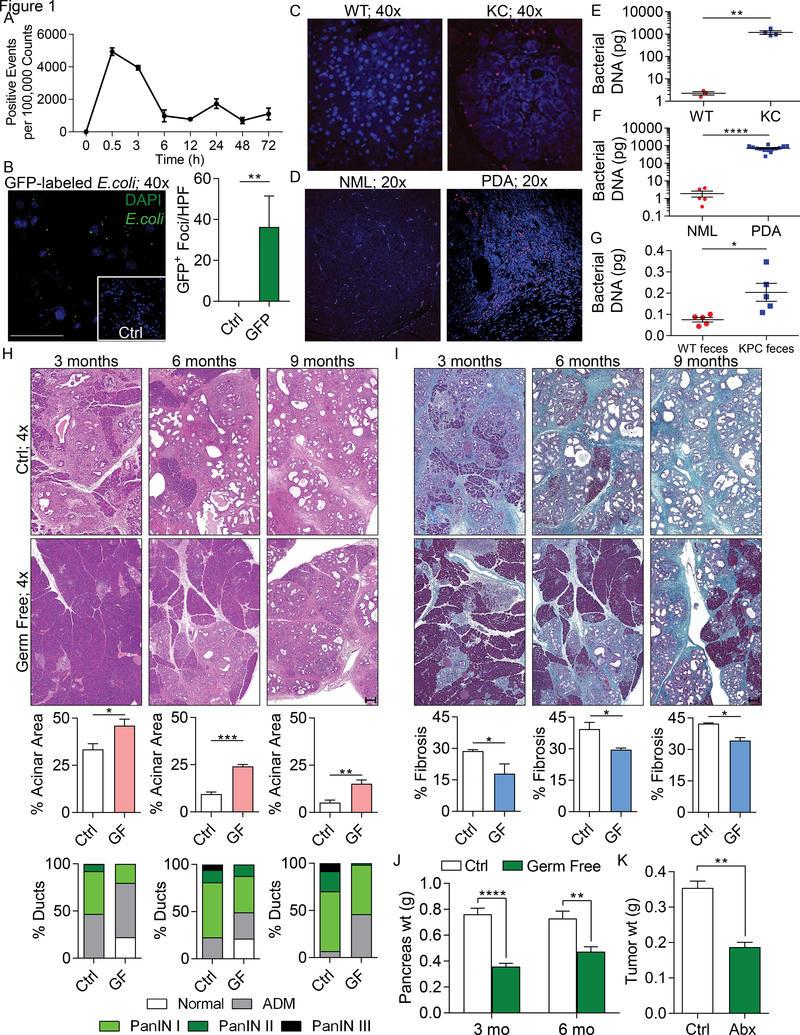
Figure 1. The tumorous pancreas has an abundant microbiome and its ablation is protective against pancreatic disease progression.
(A) WT mice were administered CFSE-labeled E. faecalis (2.5×108 CFU) via oral gavage. Pancreata were harvested and digested at the indicated timed intervals and tested for the presence of these bacteria (n=3 mice/time point). This experiment was repeated twice with similar results. (B) WT mice were administered GFP-labeled E. coli (2.5×108 CFU) via oral gavage. Pancreata were harvested at 6h and the number of GFP+ foci was determined by immune fluorescence microscopy compared to control. This experiment was repeated twice (n=3; **p<0.01; scale bar =50μm). (C) The abundance of intra-pancreatic bacteria was compared in 3-month-old WT and KC mice by FISH (n=5/group). Representative images are shown. This experiment was repeated twice. (D) The abundance of intra-pancreatic bacteria was compared in healthy individuals and age/gender/BMI matched PDA patients by FISH (n=5/group). Representative images are shown. (E) Bacterial DNA content was compared in WT and KC mice using qPCR. Each dot represents data from a single mouse pancreas. This was repeated three times (**p<0.01). (F) Bacterial DNA content was compared in healthy individuals (NML) and age/gender/BMI matched PDA patients using qPCR. Each dot represents data from a single human pancreas (****p<0.0001). (G) 8-week old WT mice were treated with an ablative oral antibiotic regimen. 3 weeks after treatment, mice were repopulated using fecal bacteria from either 3 month-old WT or KPC mice. Bacterial colonization of the pancreas was analyzed by qPCR 2 weeks after repopulation. This experiment was repeated twice (n=5/group; *p<0.05). (H-J) Control and germ-free KC mice were sacrificed at 3, 6, or 9 months of life. Representative (H) H&E- and (I) trichrome-stained sections are shown. The percentage of ducts exhibiting normal morphology, acinoductal metaplasia (ADM), or graded PanIN lesions were determined based on H&E staining. The fraction of fibrotic area per pancreas was calculated based on trichrome staining (scale bars = 200μm). (J) Pancreatic weights were recorded at 3 or 6 months of life (n=10/group; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001). (K) WT mice were treated with an ablative oral antibiotic regimen and then orthotopically inoculated with KPC-derived PDA cells. Animals were sacrificed at 3 weeks and tumor weights were recorded (n=4/group; **p<0.01). This experiment was repeated more than 5 times with similar results.
To determine whether bacteria promote the progression of pancreatic dysplasia, we employed the slowly progressive KC model of pancreatic oncogenesis. Germ-free KC mice were protected against disease progression and stromal expansion. Compared to age-matched control KC mice, germ-free cohorts exhibited delayed acinar effacement, reduced pancreatic dysplasia, diminished intra-tumoral fibrosis, and lower pancreatic weights (Figure 1h-j). Similarly, in an invasive orthotopic PDA model using KPC-derived tumor cells, WT mice treated with an ablative oral antibiotic regimen developed ~50% reduced tumor burdens (Figure 1k). Bacterial ablation was similarly protective when using Kras wild-type Pan02 cells (Figure S1e). These data imply that bacteria promote the progression of pancreatic oncogenesis in both pre-invasive and invasive models. We confirmed that our oral antibiotic regimen ablated the pancreatic microbiome (Figure S1f).
To identify longitudinal perturbations in the microbiome associated with temporal progression of pancreatic dysplasia, we serially interrogated fecal bacterial profiles in KC and WT mice over a period of nine months using 16S rRNA sequencing. Early in murine life, gut bacterial community structures in KC and WT cohorts were similar. Bacteroidetes and Firmicutes were the dominant phyla, but no significant differences were observed between cohorts. However, while Actinobacteria were present in low abundance in young WT and KC mice, they increased to ~60% abundance in the KC cohorts by week 20. Conversely, Actinobacteria did not expand in the WT cohort. Deferribacteres also increased abruptly in KC mice in weeks 28–36 (Figure S2a). At the genus level, the KC gut microbiome clustered separately from WT after week 13 (Figure S2b). Longitudinal comparisons of gut microbial communities in KC mice suggested an enrichment of Bifidobacterium at later time points (Figure S2c). Linear discriminant analysis (LDA) similarly showed that Bifidobacterium was progressively enriched in the KC cohort compared with WT from weeks 13–36 (Figure S2d). We found that Bifidobacterium pseudolongum was the most abundant Bifidobacterium species in KC mice (Figure S2e). Principal coordinate analysis (PCoA) computed using weighted UniFrac distance metrics confirmed distinct differences in gut microbial communities between WT and KC cohorts at select time points (Figure S2f, g). α-diversity analyses further suggested significant differences in taxonomy-based richness (Chao1), observed operational taxonomic units (OTUs), Shannon diversity index, and phylogeny-based diversity (PD) in the KC cohort with progressive oncogenesis (Figure S3a-d). α-diversity in gut microbial communities was also distinct between WT and KC cohorts (Figure S3e-h).
Since we showed that bacteria can migrate from the gut to the pancreas, we evaluated bacterial membership and structure in fecal samples of PDA patients (PDA; n=32) compared with matched healthy individuals (NML; n=31). At the phylum level, the gut microbiota of PDA patients and controls were each similarly dominated by Firmicutes and Bacteroidetes (Figure S4a). However, Proteobacteria, Synergistetes, and Euryarchaeota were significantly more abundant in PDA patients compared with healthy subjects. Notably, whereas Proteobacteria comprised only ~8% of gut bacteria in PDA patients (Figure S4a), this phyla constituted nearly 50% abundance in the cancerous pancreas (Figure S1b). Direct comparison of the gut and pancreas microbiomes in PDA patients for which both fecal and tumor samples were available indicated differentially increased translocation of gram negative Proteobacteria to the pancreas (Figure S4b). Pseudomonas and Elizabethkingia were the most abundant Proteobacteria genera in PDA, whereas Prevotella and Bacteroides were more abundant in the gut (Figure S4c). Comparison of the pancreas and duodenal microbiomes in KC mice also indicated enrichment of select gram negative bacteria in the pancreas (Figure S4d). LDA analysis suggested that numerous genera belonging to Firmicutes, Proteobacteria, Actinobacteria, and Bacteroidetes were expanded in the gut in human PDA (Figure S5a). Alpha diversity measures assessing the human gut microbiome suggested differences between NML and PDA cohorts based on the Abundance-based Coverage Estimator (ACE), Chao1, Observed OTUs, Shannon, Simpson, and PD indices (Figure S5b).
To test whether pathogenic bacteria promote pancreatic oncogenesis in genetically susceptible hosts, we ablated gut bacteria in KC mice using oral antibiotics and then selectively repopulated cohorts using feces derived from either WT mice or KPC mice before sacrifice at 22 weeks of life (Figure S6a). Consistent with our previous results, bacterial ablation was protective against disease progression (Figure 2a). Further, repopulation using KPC-derived feces accelerated tumor growth to baseline levels whereas repopulation with feces from age-matched WT mice failed to significantly accelerate tumorigenesis (Figure 2a). Similarly, in the germ-free KC model, repopulation using feces derived from PDA-bearing KPC mice, but not WT mice, accelerated disease progression, and mitigated the augmented T cell infiltration associated with the germ-free condition (Figure S6b and Figure 2b-d). Repopulation using B. pseudolongum similarly accelerated oncogenesis (Figure 2b-d). Using FISH we confirmed repopulation of the pancreas with B. pseudolongum, again indicating translocation of gut bacteria to the pancreas (Figure 2e).
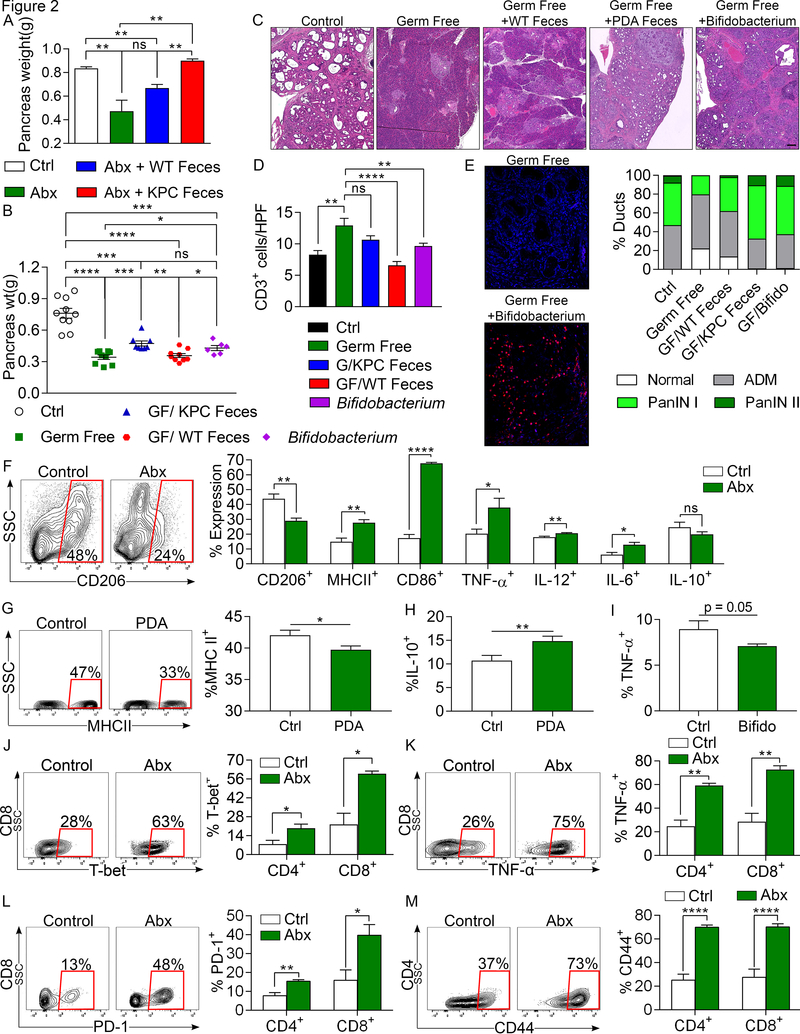
Figure 2. The microbiome in PDA-bearing hosts promotes tumor-progression and intra-tumoral immune suppression.
(A) KC mice treated with an ablative oral antibiotic regimen for 8 weeks were repopulated with i) feces from 3-month old WT mice, ii) feces from 3 month-old KPC mice, or iii) sham-repopulated (vehicle only). Mice were sacrificed 8 weeks later and pancreas weights from each cohort were compared to each other and to age-matched control KC mice that were not treated with antibiotics (n=3–4/group). This experiment was repeated three times. (B-D) The gut microbiome of germ-free 6-week-old KC mice were repopulated with feces from 3-month-old WT or KPC mice, B. pseudolongum, or sham-repopulated. Mice were sacrificed 8 weeks later. (B) Tumor weights were measured. Each point represents data from a single mouse. (C) Representative H&E-stained sections of pancreata are shown compared with age-matched non-germ free controls (scale bar =100μm). Ductal histology was quantified. (D) CD3+ T cell infiltration was determined by IHC. All repopulation experiments were repeated 3 times. (E) The gut microbiome of germ-free 6-week-old KC mice were repopulated with B. pseudolongum or sham-repopulated (n=5/group). Colonization of pancreata with B. pseudolongum was confirmed using FISH at 8 weeks. This experiment was repeated twice. (F) Control and oral antibiotic-treated WT mice were orthotopically implanted with KPC-derived tumor cells. Gr1–CD11b+F4/80+ macrophages were gated and assessed for expression of CD206, MHC II, CD86, TNF-α, IL-12, IL-6, and IL-10 (n=5/group). Macrophage profiling experiments were repeated more than 5 times (G, H) Splenic macrophages from untreated mice were harvested and cultured in vitro with cell-free extract from gut bacteria of control or KC mice. After 24h, macrophages were analyzed for expression of (G) MHC II and (H) IL-10 (n=18/group). (I) Splenic macrophages were cultured in vitro with cell-free extract from B. pseudolongum or with PBS. After 24h, macrophages were analyzed for expression of TNF-α. Macrophage polarization experiments were repeated 3 times in replicates of 5. (J-M) Control and oral antibiotic-treated WT mice were orthotopically implanted with KPC-derived tumor cells. CD4+ and CD8+ T cells were gated and tested for expression of (J) T-bet, (K) TNF-α, (L) PD-1, and (M) CD44. Representative contour plots and quantitative data are shown. Immune-phenotyping experiments were repeated more than 5 times (n=5/group; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
We postulated that the microbiome promotes PDA progression by inducing peritumoral immune suppression. Consistent with our IHC results in the KC model, we found that microbial ablation resulted in a marked increase in the fraction of intra-tumoral T cells and a reduction in the fraction of myeloid-derived suppressor cells (MDSC) in the orthotopic KPC model (Figure S6c, d). Analysis of the phenotype of tumor-associated macrophages (TAMs) suggested that microbial ablation leads to a reduction in immune-suppressive CD206+ M2-like TAMs with a concomitant increase in M1-like TAMs, expressing higher MHC II, CD86, TNF-α, IL-12, and IL-6 (Figure 2f). Analysis of chemokine expression suggested that TAMs infiltrating tumors of antibiotic-treated mice expressed increased levels of numerous M1-assoicated chemokines (Figure S6e). We found that cell-free extract from gut bacteria of PDA-bearing hosts reduced MHC II expression, but upregulated IL-10, in splenic macrophages compared with cell-free extract from gut bacteria of control mice (Figure 2g, h). Cell free extract from B. pseudolongum had similar effects at mitigating M1-differentiation of macrophages (Figure 2i). Since we reported that macrophage polarization dictates T cell immunogenicity in PDA (6,11), we postulated that bacterial ablation would activate the tumor-infiltrating T cell population. Accordingly, anti-microbial treatment resulted in an increased intra-tumoral CD8:CD4 T cell ratio (Figure S6f), which is associated with enhanced immunogenicity in PDA (12). Moreover, microbial ablation enhanced the Th1-polarization of CD4+ T cells and the acquisition of a cytotoxic CD8+ T cell phenotype as evidenced by up-regulation of T-bet (Figure 2j), TNF-α (Figure 2k), IFN-γ, and CD38 (Figure S6g, h). Both intra-tumoral CD4+ and CD8+ T cells in antibiotic-ablated mice also increased their expression of PD-1 (Figure 2l) and CD44 (Figure 2m), and CD4+ T cells expressed higher ICOS and LFA-1 (Figure S6i). T-reg differentiation was not affected by bacterial ablation (Figure S6j). Antibiotic ablation also increased intra-tumoral immunogenicity in the orthotopic Pan02 model (Figure S6k-m). Notably, repopulation of the microbiome after antibiotic ablation using feces derived from KPC mice reversed the intra-tumoral immunogenic changes associated with bacterial ablation (Figure S6n-p). Whole pancreas Nanostring array confirmed that genes associated with T cell proliferation and immune activation were upregulated in tumors of antibiotic-treated mice (Figure S7a). Collectively, these data suggest that the microbiome regulates immunogenicity in PDA.
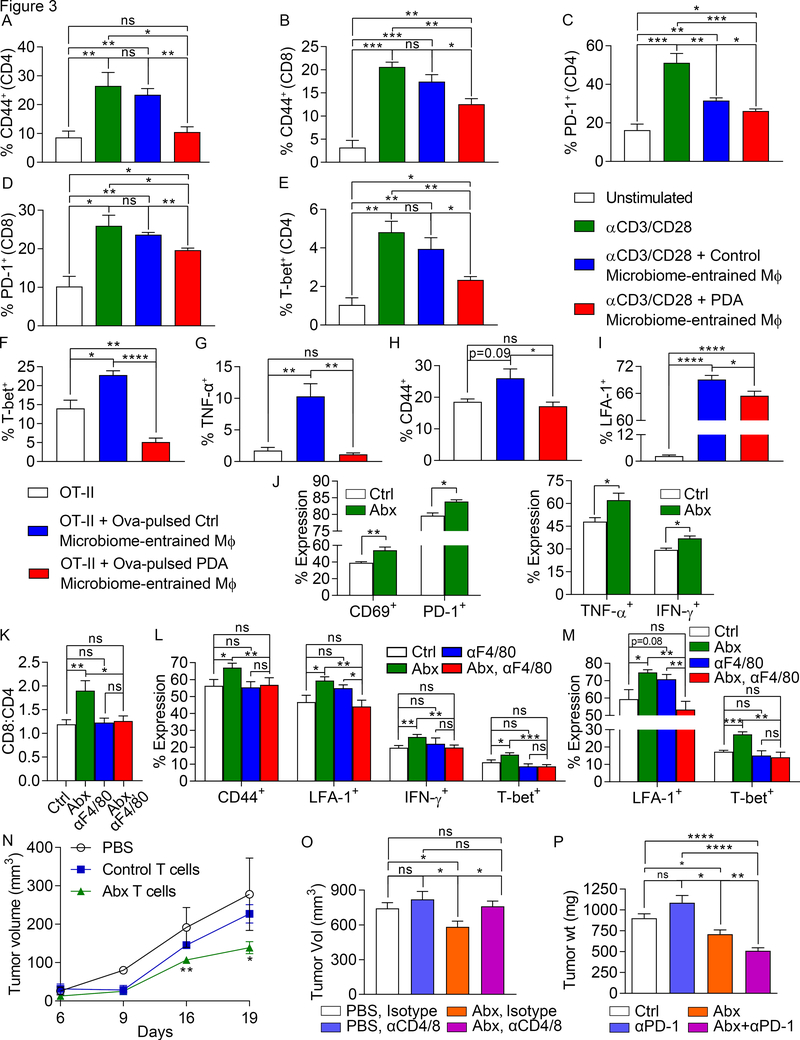
To further investigate whether microbiome-entrained macrophages mediate T cell suppression in PDA, we stimulated CD4+ and CD8+ T cells using CD3/CD28 co-ligation either alone or in the presence of splenic macrophages that had been treated with cell-free extract from gut bacteria derived from WT mice or KC mice. Macrophages entrained by gut bacterial extracts from PDA-bearing hosts mitigated CD4+ and CD8+ T cell activation, as evidenced by reduced expression of CD44 and PD-1, and prevented Th1 differentiation in CD4+ T cells, as determined by expression of T-bet (Figure 3a-e). By contrast, macrophages entrained by gut bacteria of control mice were largely non-inhibitory. We further tested whether macrophages entrained by the PDA microbiome were deficient at antigen presentation. Consistent with our previous data, Ova-pulsed macrophages treated with cell-free extract from gut bacteria derived from PDA-bearing hosts exhibited a reduced capacity to activate and induce Th1 differentiation in Ova-restricted CD4+ T cells (Figure 3f-i). Similarly, TAMs harvested from PDA tumors in antibiotic-ablated mice exhibited increased capacity to activate T cells relative to TAMs from PDA tumors in control mice (Figure 3j). Moreover, in vivo macrophage neutralization abrogated the intra-tumoral T cell activation associated with microbial ablation in PDA (Figure 3k-m).
Figure 3. The PDA microbiome promotes macrophage-mediated suppression of T cell immunity.
(A-E) Naïve splenic CD4+ and CD8+ T cells from WT mice were activated using CD3/CD28 co-ligation, either alone or in the presence of splenic macrophages that had been treated overnight with cell-free extract from gut bacteria derived from 3-month-old WT mice or KC mice. CD4+ and CD8+ T cell activation, respectively, were determined by expression of (A, B) CD44 and (C, D) PD-1. (E) CD4+ T cell differentiation was further evaluated by expression of T-bet. This experiment was repeated more than 5 times using 3–5 replicates per group. (F-I) Splenic macrophages that had been treated overnight with cell-free extract from gut bacteria derived from 3-month-old WT mice or KC mice were pulsed with Ova323–339 peptide and used to stimulate CD4+ OT-II T cells. T cell activation at 96h was determined by expression of (F) T-bet, (G) TNF-α, (H) CD44, and (I) LFA-1. This experiment was repeated 4 times using 4–5 replicates per group. (J) Control and oral antibiotic-treated WT mice bearing orthotopic KPC-derived tumor cells were sacrificed at 21 days. TAMs were FACS-sorted, loaded with Ova257–264 peptide, and used to stimulate Ova-restricted CD8+ OT-I T cells. T cell activation was determined by expression of TNF-α, IFN-γ, CD69, and PD-1. This experiment was repeated 3 times (n=5/group). (K-M) Cohorts of orthotopic PDA-bearing mice treated with oral PBS or ablative antibiotics were serially administered neutralizing αF480 or isotype control (n=10/group). Mice were sacrificed at 21 days and tumor-infiltrating T cells were analyzed for (K) the CD8:CD4 ratio, (L) CD4+ T cell expression of CD44, LFA-1, IFN-γ, and T-bet, and (M) CD8+ T cell expression of LFA-1 and T-bet. (N) PDA-infiltrating T cells from orthotopic KPC tumor-bearing mice that had been treated with an ablative oral antibiotic regimen or sham-treated were harvested on day 21 by FACS, mixed with FC1242 cells in a 1:10 ratio, and subcutaneously implanted in the flank of recipient mice. Additional controls received FC1242 cells alone. Tumor volumes were measured at serial intervals. This experiment was repeated 3 times (n=4/group). (O) Cohorts of orthotopic PDA-bearing mice treated with oral PBS or ablative antibiotics were serially administered neutralizing αCD4 and αCD8 mAbs or isotype control. Mice were sacrificed at 21 days and pancreatic tumors were weighed (n=8–9/group). T cell depletion experiments were performed more than 3 times in orthotopic PDA-bearing mice. (P) WT mice were treated with vehicle (n=9), αPD-1 (n=16), an ablative oral antibiotic regimen (n=6), or both (n=9). Mice were challenged with orthotopic KPC tumor and sacrificed at 3 weeks. Treatments were started before tumor implantation and continued until the time of sacrifice. This experiment was repeated 4 times (n=10/group; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
To definitively implicate enhanced adaptive immunity in the tumor-protection associated with microbial ablation, we harvested T cells from orthotopic KPC tumors in either control or antibiotic-treated mice and adoptively transferred the T cells to cohorts of mice challenged with subcutaneous KPC tumor. Transfer of PDA-infiltrating T cells from control mice failed to protect; however, tumor-infiltrating T cells derived from antibiotic-treated mice reduced tumor burden by ~50% (Figure 3n). Accordingly, T cell depletion abrogated the tumor-protective effects of bacterial ablation but did not affect the rate of tumor growth in PDA-bearing control mice (Figure 3o). Furthermore, since bacterial ablation upregulated PD-1 expression in intra-tumoral CD4+ and CD8+ T cells (Figure 2l), we postulated that microbial ablation would have synergistic efficacy with PD-1-directed therapies. Whereas PD-1 blockade did not protect control mice against orthotopic PDA, ablative oral antibiotics coupled with αPD-1 therapy was synergistically protective based on tumor size (Figure 3p). Combined antibiotic + αPD-1 therapy also resulted in enhanced intra-tumoral CD4+ and CD8+ T cell activation (Figure S7b, c). In addition, T cells upregulated CXCR3 and LFA-1, which were not increased in expression with either monotherapy alone (Figure S7d, e).
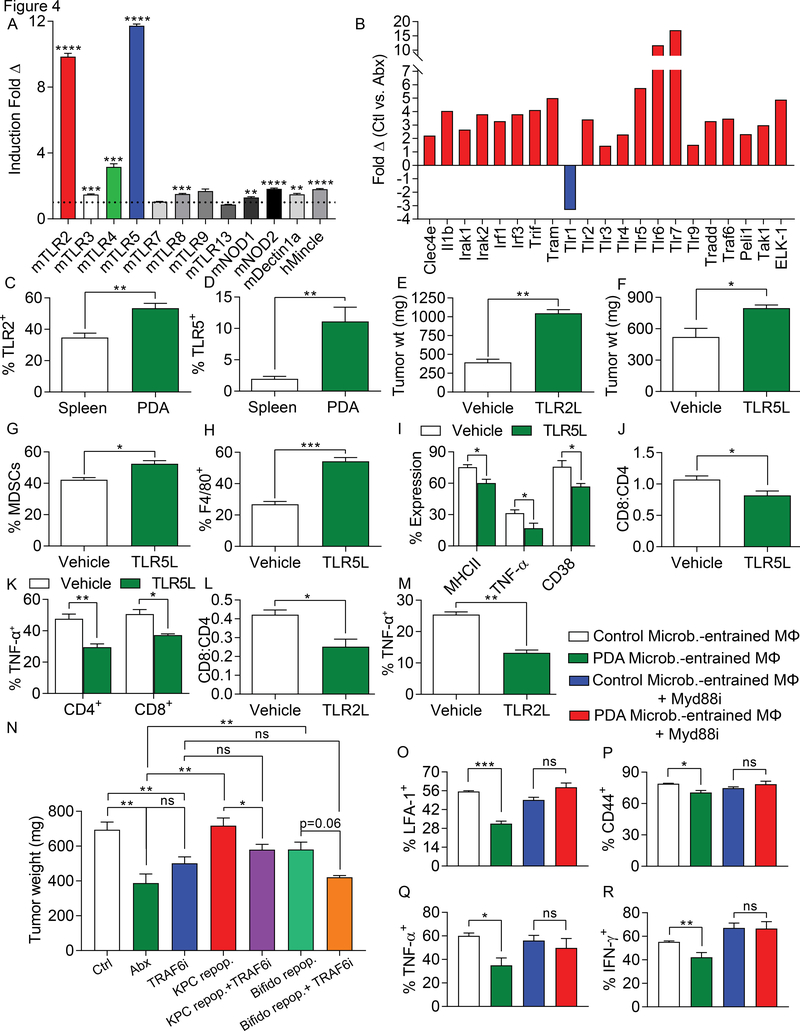
We previously reported that diverse pattern recognition receptors (PRRs), including TLR3, TLR4, TLR7, TLR9, NLRP3, Dectin-1, and Mincle, are upregulated in PDA and their activation accelerates oncogenesis via induction of innate and adaptive immune suppression (2–4,13,14). We postulated that the immune-tolerance promoted by the PDA microbiome is the result of higher PRR activation in the tumor microenvironment. Consistent with our hypothesis, we found that cell-free extract from gut bacteria derived from KC mice induced higher activation of diverse PRR reporter cell lines, most notably TLR2, TLR4, and TLR5, compared with gut bacterial extract from WT mice (Figure 4a). Further, PDA tumors in antibiotic-ablated hosts exhibited markedly lower expression of PRRs and associated signaling molecules compared to PDA tumors in control mice (Figure 4b). We confirmed that, similar to the PRRs we previously studied, expression of TLR2 and TLR5 were upregulated in macrophages in PDA by flow cytometry (Figure 4c, d), their ligation accelerated tumor growth (Figure 4e, f), and accentuated innate and adaptive immune suppression (Figure 4g-m). Moreover, in vivo inhibition of TLR signaling by blocking TRAF6 abrogated the PDA-promoting effects of repopulating antibiotic-ablated mice with KPC feces or B. pseudolongum (Figure 4n). Further, macrophages entrained by the PDA-microbiome in the context of inhibition of TLR signaling failed to suppress T cell immunogenicity, suggesting that the PDA microbiome programs TAMs via TLR signaling to induce immune tolerance (Figure 4o-r).
Figure 4. The PDA microbiome induces immune suppression via differential TLR activation.
(A) Cell-free extract from gut bacteria derived from 3 months old WT or KC mice (n=3) were tested for activation of a diverse array of PRR-specific HEK293 reporter cell lines. (B) Orthotopic KPC tumors were harvested on day 21 from control and oral antibiotic-treated WT mice. PRR-related gene expression in PDA was determined using a PCR array and performed in duplicate. Data indicates fold change in gene expression for control compared to antibiotic-treated groups. This array was repeated twice. (C) Expression of TLR2 and (D) TLR5 were tested in spleen and PDA-infiltrating macrophages from orthotopic KPC tumors (n=5). These experiments were repeated twice. (E) WT mice were orthotopically implanted with KPC-derived tumor cells and serially treated with TLR2 (Pam3CSK4) or (F) TLR5 (Flagellin) ligand or vehicle. Tumor growth was determined at 3 weeks (n=3–5/group). These experiments were repeated twice. (G-K) WT mice were orthotopically implanted with KPC-derived tumor cells and serially treated with TLR5 ligand. Tumors were harvested at 3 weeks and (G) the fraction of Gr1+CD11b+ MDSC and (H) F4/80+Gr1–CD11b+ TAM infiltration was determined by flow cytometry. (I) Expression of MHC II, TNF-α, and CD38 on TAMs were determined. (J) The CD8/CD4 T cell ratio was determined as was (K) TNF-α expression on CD4+ and CD8+ T cells (n=3–5/group). (L, M) WT mice were orthotopically implanted with KPC-derived tumor cells and serially treated with TLR2 ligand. Tumors were harvested at 3 weeks and (L) the CD8/CD4 T cell ratio and (M) TAMs expression of TNF-α were determined. These experiments were repeated twice (n=3–5/group). (N) WT mice pre-treated with an ablative oral antibiotic regimen or vehicle for 6 weeks were repopulated with i) feces from 3 month-old KPC mice (n=12), ii) B. pseudolongum (n=6), or iii) sham-repopulated (n=4). Mice were challenged with orthotopic KPC cells. Cohorts were additionally treated serially with a TRAF6 inhibitor or control. Treatments were started at the time of tumor implantation and continued until sacrifice at 21 days. Quantitative analysis of tumor weights are shown. (O-R) Splenic macrophages were entrained with extract from the gut microbiome of either WT or KC mice in the context of MyD88 inhibition or control. Macrophages were then used in αCD3/αCD28-based T cell stimulation assays. CD4+ T cell activation was determined by expression of (O) LFA-1, (P) CD44, (Q) TNF-α, and (R) IFN-γ. This experiment was repeated five times in 3–4 replicates per group with similar results (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001).
A primary uncertainty in tumor biology is the question of why oncogenesis proceeds at variable rates in hosts with similar genetic risk factors. A quintessential example of this in murine modeling of cancer is the variable tumor phenotype in KPC mice (15). We postulated that a factor driving phenotypic variance in PDA progression in genetically identical mice is the degree of bacterial dysbiosis. To address this, we prospectively collected fecal specimens from 12-week-old KPC mice, segregated them into aggressive PDA (adv-KPC) and slowly progressive (ea-KPC) disease groups based on microscopic disease progression (Figure S8a), and compared their gut microbial phenotypes to age-matched littermate WT controls. Bacteroides and Lactobacillus were among the most predominant genera in the three cohorts (Figure S8b). LDA analysis revealed that, similar to data in the KC model, numerous genera belonging to Bacteroidetes and Firmicutes and select Actinobacteria- and Deferribacteres-associated genera were more prevalent in the ea-KPC and adv-KPC cohorts compared with WT (Figure S8c, d). When we contrasted the microbial genera in ea-KPC and adv-KPC mice, we observed that Elizabethkingia, Enterobacteriaceae, and Mycoplasmataceae, were significantly overrepresented in ea-KPC, whereas Helicobacteraceae, Bacteroidales, and Mogibacteriaceae were more prevalent in adv-KPC (Figure S8e). Global relationships between microbial communities of the WT, ea-KPC, and adv-KPC cohorts were analyzed by PCoA, indicating significant variations between the cohorts and a high degree of similarity within each individual cohort (Figure S8f). Accordingly, fecal phylogenetic diversity was significantly different between the 3 murine subsets (Figure S8g). LDA analysis of the human PDA gut microbiome similarly indicated significant differences in bacterial abundances between Stage I/II and Stage IV patients (Figure S8h). Collectively, these data suggest that bacterial communities are distinct between early and advanced PDA.
Discussion
The gut microbiome is known to be important in the maintenance of homeostasis in several physiologic processes, including host energy metabolism, gut epithelial permeability, gut peptide hormone secretion, and host inflammatory state. Microbial dysbiosis is also being increasingly recognized for its role in oncogenesis (16,17). We found that germ-free mice are protected against PDA progression. Oral antibiotic administration also slowed oncogenic progression, whereas select bacterial transfer or bulk fecal transfer from PDA-bearing mice, but not control mice, accelerated tumorigenesis. These observations support a role for the microbiome in promoting disease progression. However, it is also likely that oncogenic Kras expression influences the composition and diversity of the gut and intra-pancreatic flora. Moreover, our data suggest oral antibiotics may be useful as a chemo-preventive measure for high-risk patients with advanced PanIN lesions or for individuals at increased genetic risk for PDA development. This also raises the prospect of a potential role for probiotics in mitigating PDA risk.
Inflammation is paramount for PDA development and progression. PDA is invariably preceded by and associated with a robust inflammatory cell infiltrate that has profound influences on disease progression (18). Specific intra-pancreatic leukocytic subsets can have divergent effects on tumorigenesis by either combating cancer growth via innate or antigen-restricted tumoricidal immune responses or by promoting tumor progression by inducing immune suppression (19). Th1 CD4+ T cells and CD8+ T cells mediate tumor-protection in murine models of PDA and are associated with prolonged survival in human disease (20). Conversely, we recently reported that antigen-restricted Th2-deviated CD4+ T cells promote pancreatic tumor progression in mice (4). Accordingly, intra-tumoral CD4+ Th2 cell infiltrates correlate with reduced survival in human PDA (20,21). Foxp3+ Tregs also facilitate tumor immune-escape in PDA (22). However, regulation of the balance between immunogenic and immune-suppressive myeloid and subsequent T cell differentiation in PDA is uncertain. We discovered that the microbiome is a potent modulator of the programming of the inflammatory tumor microenvironment.
We demonstrated that the distinct bacterial dysbiosis associated with PDA results in both innate and adaptive immune suppression. Depletion of the gut microbiome led to a diminution of MDSC infiltration and reprogramming of TAMs toward a tumor-protective M1-like phenotype. We recently reported that macrophage polarization dictates effector T cell phenotype in PDA (6). Accordingly, ablation of the gut microbiome accentuated Th1 polarization of CD4+ T cells and enhanced the cytotoxic phenotype of CD8+ T cells, as evidenced by high T-bet, TNF-α, and IFN-γ expression. Our findings that gut bacterial ablation induces immunogenic reprogramming of the tumor microenvironment and markedly increases PD-1 expression on effector T cells suggest that oral antibiotics in combination with checkpoint-directed immunotherapy may be an attractive strategy for experimental therapeutics in PDA patients. Targeting the microbiome has also been recently shown to synergize with cytotoxic chemotherapy in select cancers by affecting the metabolism of the chemotherapeutic agents (23).
The etiologic relationship between gut bacteria and immune-suppressive inflammation in PDA has not been previously described. Recent work by our group and others have shown that ligation of select pattern recognition receptors accelerates PDA progression whereas their inhibition or genetic deletion is protective (2–6) (17,18). Here, we demonstrate that TLR2 and TLR5 ligation promote PDA and induce innate and adaptive immune suppression. Moreover, we found that the immune-suppressive effects of PDA-associated bacterial extract on macrophages were absent in macrophages deficient in TLR signaling, suggesting that the suppressive effects of the PDA microbiome on macrophage programming is dependent on TLR ligation. Specifically, we show that the suppressive effects of PDA-microbiome entrained macrophages on T cell activation are abrogated in absence of TLR signaling. Moreover, while repopulation of antibiotic ablated mice with the KPC microbiome or with B. pseudolongum accelerated oncogenesis, this tumor-promoting effect was abated when TLR signaling was abrogated in vivo. Collectively, these data show mechanistic evidence that tumor-promoting effects of the PDA microbiome are TLR-dependent.
To improve upon the potential for biomarker discovery or development of novel therapies, it is imperative to elucidate the specific microbial taxa associated with organ-specific oncogenesis. Our bacterial translocation experiments suggest interactions between the two compartments, presumably via the pancreatic duct which is in anatomic continuity with the intestinal tract. In human adults, Bacteroidetes and Firmicutes usually dominate the intestinal microbiota, while Actinobacteria, Proteobacteria, and Verrucomicrobia represent a minor proportion (24). Conversely, Proteobacteria, Actinobacteria, Fusobacteria, and Verrucomicrobia were each higher in the gut of PDA patients as compared to healthy controls. Interestingly, Proteobacteria was also enriched in the intra-pancreatic microbiome in PDA bearing patients and was associated with advanced disease. The increased translocation of this pathogenic gram negative taxa supports the observation that ligation of select TLRs by lipopolysaccharides and flagellins can promote tolerogenic macrophage programming in the tumor microenvironment.
A group of pathogens, even at low abundance within the microbial community, can act as keystone species or signatures that support and shape community structure and membership in a manner that promotes disease pathogenesis (25). The skewed microbial structure and membership in human gut and pancreatic tissues of PDA patients and in mouse models of pancreatic cancer reiterate the possible involvement of mono- or poly-bacterial communities in the initiation and progression of PDA. We found that B. pseudolongum was differentially abundant in gut and tumor and accelerated oncogenesis in a TLR dependent manner. In addition, cell free extracts from B. pseudolongum polarized macrophages to upregulate tolerogenic cytokines including IL-10. Recent studies similarly reported a higher abundance of Bifidobacterium in the tissues of patients with colorectal adenomas (26,27). By contrast, a recent report showed that commensal B. longum conferred protection in melanoma by promoting anti-PD-L1 therapy; indicating that different species may have diverging effects in the tumor microenvironment (28).
In summary, our study elucidates the presence of a distinct gut microbiome that is associated with progressive pancreatic oncogenesis in mice. Further we show that the pancreas harbors its own microbiome that is associated with disease stage in mice and humans. We also detail the intra-pancreatic immune programming induced by the microbiome. Modulation of the PDA microbiome to augment immunotherapy is an attractive avenue for experimental therapeutics. Further, prospective studies are necessary for identification of microbial signatures with tumor specificity that may have potential for early diagnosis and risk stratification.
Methods
Animals and in vivo models
KC mice, which develop spontaneous pancreatic neoplasia by targeted expression of mutant Kras in the pancreas, were a gift from Dafna Bar-Sagi (New York University) (29). C57BL/6 (H-2Kb) mice (WT) were originally purchased from Jackson Labs (Bar Harbor, ME) and were bred in-house and crossed with the KC model after 8 generations. Littermate controls were used in experiments. Animals were housed in a specific pathogen free vivarium and fed standard mouse chow. KPC mice, which express mutant intra-pancreatic Kras and p53, were a gift from Mark Phillips (New York University) (15). OT-I and OT-II mice were purchased from Jackson Labs (Bar Harbor, ME) and bred in-house. Both male and female mice were used, but animals were sex- and age-matched in each experiment. For orthotopic tumor experiments, 8–10 week old mice were employed. No formal power analyses, randomization, exclusions, or blinding were performed. Mice were administered intra-pancreatic injections of tumor cells derived from pancreata of KPC mice (105 cells in Matrigel; generated from KPC mice as described (2); utilized within 6 months of generation; not re-authenticated during this time) and were sacrificed after 3 weeks, as previously described (2). Alternatively, mice were administered intra-pancreatic injections of Pan02 tumor cells (106 cells in Matrigel; gift of and authenticated by Daniel Meruelo, received May 2016, New York University) and sacrificed at 5 weeks. Both cell lines were commercially MAP tested by Taconic in December, 2016 via RAPIDMAP-12 panel and all results were negative. In select experiments, mice were serially treated with a neutralizing αPD-1 mAb (150μg, i.p., RMP1–14, 2x/week; Bioxcell, West Lebanon, NH). In other experiments, CD4+ T cells (GK1.5), CD8+ T cells (53–6.72), or F480+ macrophages (CI:A3–1; all Bioxcell) were depleted with neutralizing mAbs using regimens as previously described (6,30). Alternatively, mice were serially treated with TRAF6 inhibitor (66μg, i.p., 3x/week; Novus Biologicals, San Diego, CA), TLR2 (Pam3CSK4, 50μg, i.p., 3x/week) or TLR5 (Flagellin, 10μg, i.p., 2x/week; both Invivogen, San Diego, CA) ligands. For T cell transfer experiments, intra-tumoral T cells were harvested by FACS, mixed with 105 FC1242 cells in a 1:10 ratio, and subcutaneously implanted into the flank of recipient mice. Germ-free KC mice were generated by re-deriving and crossing p48Cre and LSL-KrasG12D mice in a germ-free environment at the National Gnotobiotic Rodent Resource Center (Chapel Hill, NC). Longitudinal cohort studies were conducted to monitor microbial communities throughout experiments by serially collecting specimens from littermate WT and KC or KPC mice. Fecal and tissue specimens were stored in sterile tubes at −80°C until further use.
Antibiotic treatment, fecal, and bacterial transfer experiments
To ablate the gut microbiome, 6-week-old WT or KC mice were administered an antibiotic cocktail by oral gavage daily for five consecutive days. Controls were gavaged with PBS. The oral gavage cocktail contained Vancomycin (50mg/ml; Sigma, St. Louis, MO), Neomycin (10mg/ml; Sigma), Metronidazole (100mg/ml; Santa Cruz Biotech, Dallas, TX) and Amphotericin (1mg/ml; MP Biomedicals, Santa Ana, CA), as described (31). Additionally, for the duration of the experiments, mouse drinking water was mixed with Ampicillin (1mg/ml; Santa Cruz Biotech), Vancomycin (0.5mg/ml; Sigma), Neomycin (0.5mg/ml; Sigma), Metronidazole (1mg/ml; Santa Cruz Biotech) and Amphotericin (0.5μg/ml; MP Biomedicals). In fecal transfer experiments, six fecal pellets from mice were collected and resuspended in 1 ml of PBS and 200μl of the fecal slurry was used for orogastric gavage every other day for 2 weeks. For species-specific repopulation experiments, Bifidobacterium pseudolongum (1×108/ml; Cat. #25526, American Type Culture Collection, Manassas, VA) was used to orally gavage germ-free mice at 6 weeks of age. Repopulation was confirmed by gram stain and qPCR of fecal sample and by FISH in pancreatic tissues. To assess bacterial translocation to the pancreas, WT mice were orally gavaged with 2.5 × 108 CFU of Enterococcus faecalis that were labeled with CFSE according to the manufacturer’s instructions (Invitrogen, Carlsbad, CA). Recipient mice were then serially sacrificed at 3 hour intervals, and their pancreata were harvested. Single cell suspensions of pancreata were prepared and analyzed by flow cytometry for the presence of CFSE-labeled bacteria. Alternatively, GFP-labeled Escherichia coli (2.5 × 108 CFU) were introduced via oral gavage in mice and pancreatic sections were examined at 3 hours by immune fluorescence microscopy. All experiments were approved by the New York University School of Medicine Institutional Animal Care and Use Committee (IACUC).
Murine cellular isolation, flow cytometry, and in vitro experiments
Pancreatic leukocytes were harvested from mouse PDA as described previously (6). Briefly, pancreata were resected and placed in ice-cold PBS with 1% FBS, Collagenase IV (1 mg/mL; Worthington Biochemical, Lakewood, NJ), and DNAse I (2 U/mL; Promega, Madison, WI). After mincing, tissues were incubated in the same solution at 37°C for 20 minutes with gentle shaking. Specimens were then passed through a 70 μm mesh, and centrifuged at 350g for 5 minutes. Cells were resuspended in ice-cold PBS with 1% FBS. After blocking FcγRIII/II with an anti-CD16/CD32 mAb (eBioscience, San Diego, CA), cell labeling was performed by incubating 106 cells with 1 μg of fluorescently conjugated antibody directed against murine CD44 (IM7), CD206 (C068C2), PD-1 (29F.1A12), CD3 (17A2), CD4 (RM4–5), CD8 (53–6.7), CD45 (30-F11), CD11b (M1/70), Gr1 (RB6–8C5), CD11c (N418), CD38 (90), CD86 (GL-1), MHC II (M5/114.15.2), IL-6 (MP5–20F3), IL-10 (JES5–16E3), IL-12/IL-23 p40 (C15.6), IFN-γ (XMG1.2), LFA-1 (H155–78), CXCR3 (CXCR3–173), TNFα (MP6-XT22), TLR2 (6C2), TLR5 (ACT5), ICOS (15F9; all Biolegend, San Diego, CA), T-bet (4B10), and FoxP3 (FJK-16s; both eBioscience). Cell preparation for intracellular staining was performed using the Fixation and Permeabilization Solution Kit (eBiosciences). Flow cytometry was performed on the LSR-II (BD Biosciences, Franklin Lakes, NJ). FACS-sorting was performed on the SY3200 (Sony, Tokyo, Japan). Data were analyzed using FlowJo (Treestar, Ashland, OR). Gates were based on isotype control. In select experiments, cell-free extract from 4 × 1012 CFU/ml bacteria from the gut of WT or KC (10 week-old) mice or Bifidobacterium pseudolongum (American Type Culture Collection, Manassas, VA) was used for treatment of splenic macrophages overnight before analysis of macrophage phenotype by flow cytometry. In some experiments, MyD88 inhibitory peptide (200μM, Novus Biologicals) or control were added, as we described (4). Bacterial cell-free extracts were also used to activate HEK293 PRR reporter cell lines (Invivogen), as we previously described (13). For T cell activation assays, splenic T cells were stimulated using plate-bound αCD3/αCD28 alone or in co-culture with macrophages (5:1 ratio), as we described (13). Alternatively, antigen-restricted splenic CD4+ OT-II T cells or CD8+ OT-I T cells were stimulated using antigen-presenting cells pulsed with the appropriate Ova peptide, as we reported (32). T cell activation was determined at 96h by flow cytometry. In select experiments, TAMs were cultured overnight at a concentration of 106 cells/ml before harvest of cell culture supernatant. Chemokine levels in cell culture supernatant were analyzed using the LEGENDplex bead array (Biolegend).
Histology, immunohistochemistry, and RNA analysis
For histological analysis, pancreatic specimens were fixed with 10% buffered formalin, dehydrated in ethanol, embedded with paraffin, and stained with H&E and Gomori’s Trichrome, anti-Ki67 (ab15580), anti-CD3 (ab5690, both Abcam, San Franciso, CA), and TUNEL (Promega, Madison, WI). The fraction of preserved acinar area was calculated as previously described (11). Histological data from control KC mice were previously reported (6). The fraction and number of ducts containing all grades of PanIN lesions were measured by examining 10 high-power fields (HPFs; 40X) per slide. PanINs were graded according to established criteria (33): In PanIN I ducts, the normal cuboidal pancreatic epithelial cells transition to columnar architecture (PanIN Ia) and gain polyploid morphology (PanIN Ib). PanIN II lesions are associated with loss of polarity. PanIN III lesions, or in-situ carcinoma, show cribriforming, budding off of cells, and luminal necrosis with marked cytological abnormalities, without invasion beyond the basement membrane. Pancreata from 12 week-old KPC mice were segregated based on microscopic assessment by H&E staining of the percentage of pancreatic area occupied by invasive cancer: ea-KPC designated tumors exhibited <25% of pancreatic area occupied by invasive cancer; adv-KPC tumors exhibited >75% pancreatic area replacement by invasive cancer. Pancreata of mice with 25–75% invasive PDA were excluded from analysis so as to maintain distinctness of the groups. RNA extraction from pancreatic tumors was performed using the RNeasy Mini kit (Qiagen, Germantown, MD) as per the manufacturer’s instructions. For Nanostring analysis, the nCounter mouse inflammation panel was employed using the nCounter Analysis System (both Nanostring, Seattle, Washington) (34). In selected experiments, the pre-configured “Mouse TLR Signaling Pathway RT2 Profiler PCR Array” was used as per the manufacturer’s instructions (Qiagen).
Quantitative PCR
Total bacterial DNA in the pancreatic tissue and fecal samples was determined by real-time qPCR using 16S primers. Briefly, the 10ul reaction mix contained 2X Power SYBR® Green master mix (Applied Biosystems, Foster City, CA), 100 nM of forward and reverse primers, and 15 ng sample DNA on the Bio Rad CFX384 Real-time system. The reaction was programmed as follows: Denaturation at 94° C for 10 min, 40 cycles of 94° C for 1 min, annealing at 60° C for 1 min, and elongation at 72° C for 90 sec, followed by a final elongation at 72° C for 5 mins. E. coli DNA was used to plot a standard curve to calculate bacterial DNA concentration in the sample. A previously described standard protocol was used to convert CT values of each sample to total bacterial DNA in the sample (35).
Fluorescence In Situ Hybridization (FISH)
The EUB338 16S rRNA gene probe or a Bifidobacterium specific probe labeled with the fluorophore Cy3 (extinction wavelength, 555 nm; emission wavelength, 570 nm; Molecular Probes, Eugene, OR) was used to detect the bacterial colonization within human and mouse pancreatic tissues by FISH. Fluorescence microscopic analysis was conducted with Nikon Eclipse 90i confocal microscope (Nikon, Melville, NY) using a Cy3 labeled-probe at 50 pmol/ml as described (36–38).
Human sample collection
Human fecal samples were collected from healthy volunteers and PDA patients using rectal swabs. Specimens were stored in sterile TE buffer for 16S sequencing analysis. Patients on antibiotic treatment within the past 3 months, or patients who had received neoadjuvant chemo- or radio-therapy were excluded. Human tissue samples were sterilely collected from patients at NYU Langone Medical Center. Human specimens were obtained using an Institutional Review Board (IRB) approved protocol, conducted in accordance with the Declaration of Helsinki, the Belmont Report, and U.S. Common Rule, and donors of de-identified specimens gave written informed consent. All specimens were stored at −80°C till further use. Sample sizes for human experiments were not determined based on formal power calculations.
Bacterial DNA extraction and sequencing
Pancreatic tissue samples were suspended in 500 μL sterile PBS. Samples were pretreated by vortexing for 30 seconds followed by sonication and overnight treatment with Proteinase K (2.5 μg/mL) at 55ºC, as we described (39,40). Total bacterial genomic DNA was purified from tissue and fecal samples using the MoBio Power fecal kit as per the manufacturer’s instructions (MoBio Laboratories Inc., Carlsbad, CA). DNA was quantified for concentration and purity by NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA) and stored at −20°C till further analysis. For high throughput 16S rRNA library preparation and sequencing, the V3-V4 hyper-variable region of the 16S gene was amplified from the genomic DNA of mice and human fecal and pancreatic tissues samples according to the Illumina 16S metagenomics protocol (Part # 15044223 Rev. B) (41). The purified DNA was quantified fluorometrically by Quant-iT PicoGreen assay (Molecular Probes, Inc., Eugene, Oregon, USA) in a SpectraMax M5 microplate reader (Molecular Devices, Sunnyvale, California, USA), and the concentration adjusted to 10 ng/μL for all sequencing assays. PCR was initially performed using the primer set, 341F (5’-CCTACGGGNGGCWGCAG-3’) and 805R (5’-GACTACHVGGGTATCTAATCC-3’) (41,42); each with overhang adapter sequences (IDT, Coralville, Iowa) using 2x Kapa HiFi Hotstart ReadyMix DNA polymerase (KapaBiosystems, Wilmington, MA). Samples were amplified in duplicates and purified using AMPure XP beads. Amplification was performed at 95°C (3 min), with 25 cycles of 95°C (30 sec), 55°C (30 sec), 72°C (30 sec), and final extension of 72°C (5 min). Dual indices from Illumina Nextera XT index kits (Illumina, San Diego, CA) were added to target amplicons in a second PCR using 2x Kapa HiFi Hotstart ReadyMix DNA polymerase. PCR conditions were 95°C (3 min), with 8 cycles of 95°C (30 sec), 55°C (30 sec), 72°C (30 sec), and final extension of 72°C (5 min). After each PCR cycle, AMPure XP beads purified libraries were checked for purity by nanodrop, quantified by PicoGreen assay, and size confirmed on agarose gels. Negative controls were included in all sequencing runs. Equimolar amounts of the generated libraries with dual-index were combined and quantified fluorometrically. The pooled amplicon library was denatured, diluted, and sequenced on an Illumina MiSeq platform using MiSeq Reagent Kit v3 (600 cycles) following the 2×300-bp paired-end sequencing protocol.
Phylogenetic and statistical analyses
The Illumina-generated sequence data was processed using the quantitative insights into microbial ecology software package (QIIME) v1.8.0 (43,44). Sequences with quality score of Q20 and higher were assembled for paired-ends using default parameters of PANDASEQ with a minimum overlap of 25 nucleotides and maximum of 100 nucleotides between forward and reverse reads (45,46). The sequences were de-multiplexed and quality trimmed using default parameters in QIIME script, split_libraries_fastq.py (47). The resulting total 7,040,079 sequences had an approximate read length of 500 bp. The filtered sequences were clustered into operational taxonomic units (OTUs) based on a 97% similarity threshold using UCLUST algorithm (48) and the chimeric sequences were removed by ChimeraSlayer (49). OTUs were picked by denovo OTU picking method, pick_otus.py. Representative sequences were aligned with PyNAST against Greengenes database (gg_13_8 release). Taxonomy was assigned to the identified OTUs using the basic local alignment search tool (BLAST) reference database and Greengenes taxonomy-mapping file. The script make_phylogeny.py was used to create phylogenetic trees with the FastTree program (50). Low abundance OTUs with <2 counts were removed and the remaining OTUs were used for downstream analysis. A total of 6,521,333 sequence reads (92.63%) were clustered into 11,357 OTUs (1,400,000 reads) for longitudinal mouse fecal samples; 10,710 OTUs (1,105,527 reads) for orthotopic mouse fecal samples; 248 OTUs (942,696 reads) for mouse tissue samples; 52,745 (2,706,209 reads) for human fecal samples; and 690 OTUs (366,901 reads) for human tissue samples.
The microbial relative abundance plots at all taxonomic levels were generated using biom, phyloseq, and pheatmap packages in R. OTUs with ≥0.1% abundance in at least one sample were considered for analysis and <0.1% were binned into an ‘Other’ category. The Mann-Whitney U test was used to compute significance between the health and disease cohorts, whereas significance within the samples over time was determined by Wilcoxon signed rank test in R. Alpha diversity plots, such as richness estimators (Observed OTUs, ACE, Chao1) and diversity estimators (Shannon Index, Simpson Index, Phylogenetic Diversity (PD)) were generated using R-phyloseq and vegan. The data was rarefied by random subsampling without replacement to a depth of 2,500. This was based on minimum sequences in a dataset in order to normalize the read counts between samples. Two-tailed student’s t-test was also used when two groups were compared and ANOVA for three groups. ß diversity Principal Coordinate Analysis (PCoA) plots were computed between cancer and control samples by weighted UniFrac distances and significance was assessed by Adonis test (PERMANOVA). LEfSe tool was used to identify differentially significant bacterial taxa between the cohorts with Kruskal-Wallis test (51). P values ≤0.05 were considered statistically significant.
Quality Control
For adequate quality control, we employed best practices of previously published studies (52–54). All the samples were collected using standard sterile technique. We maintained consistency in DNA extraction techniques and reagents throughout. All PCR reagents were periodically checked for environmental contaminants using 16S universal primers. All qPCR reactions had appropriate controls (without template) to exclude bacterial DNA contamination.
Statistical considerations for tumor size and immunologic analyses
Data is presented as mean +/− standard error. Statistical significance in immune phenotyping studies and in measurements of tumor size was determined by the Student’s t test using GraphPad Prism 7 (GraphPad Software, La Jolla, CA). P-values <0.05 were considered significant.
Supplementary Material
Statement of Significance.
We found that a distinct and abundant microbiome drives suppressive monocytic cellular differentiation in pancreatic cancer via selective toll-like receptor ligation leading to T cell anergy. Targeting the microbiome protects against oncogenesis, reverses intra-tumoral immune-tolerance, and enables efficacy for check-point based immunotherapy. These data have implications for understanding immune-suppression in pancreatic cancer and its reversal in the clinic.
Acknowledgments
This work was supported by NIH CA206105 (DS, GM), CA168611 (GM), CA155649 (GM), DE025992 (DS), CA180277 (XL), CA175794 (AS), P40 OD010995 (RBS), P30 DK034987 (RBS), Department of Defense Peer Reviewed Medical Research Program (GM), the Lustgarten Foundation (DS, GM), AACR-PanCan (GM), the Panpaphian Association of America (CZ), the National Pancreas Foundation (CZ), Crohn’s and Colitis Foundation of America (RBS), and the Irene and Bernard Schwartz Fellowship in GI Oncology (DD). We thank the New York University Langone Medical Center (NYU LMC) Histopathology Core Facility, the NYU LMC Flow Cytometry Core Facility, the NYU LMC Microscopy Core Facility, and the NYU LMC BioRepository Center, each supported in part by the Cancer Center Support Grant P30CA016087 and by grant UL1 TR000038 from the National Center for the Advancement of Translational Science (NCATS). KC mice were a gift of D. Bar-Sagi and KPC mice were a gift of M. Philips, both from New York University.
Grant Support: This work was supported by NIH CA206105 (DS, GM), CA168611 (GM), CA155649 (GM), DE025992 (DS), CA180277 (XL), CA175794 (AS), P40 OD010995 (RBS), P30 DK034987 (RBS), Department of Defense Peer Reviewed Medical Research Program (GM), the Lustgarten Foundation (DS, GM), AACR-PanCan (GM), the Panpaphian Association of America (CZ), the National Pancreas Foundation (CZ), Crohn’s and Colitis Foundation of America (RBS) and the Irene and Bernard Schwartz Fellowship in GI Oncology (DD).
Footnotes
Competing financial interests
None of the authors have any competing financial interests to declare.
The authors declare no potential conflicts of interest.
Data Availability
Sequence data will be available in the Sequence Read Archive (SRA) database at NCBI [SRA Submission Number: SUB3605635].
References
- 1.Siegel RL, Miller KD and Jemal A (2016) Cancer statistics, 2016. CA: a cancer journal for clinicians, 66, 7–30. [DOI] [PubMed] [Google Scholar]
- 2.Zambirinis CP, Levie E, Nguy S, Avanzi A, Barilla R, Xu Y, Seifert L, Daley D, Greco SH, Deutsch M et al. (2015) TLR9 ligation in pancreatic stellate cells promotes tumorigenesis. The Journal of experimental medicine, 212, 2077–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibrahim J, Nguyen AH, Rehman A, Ochi A, Jamal M, Graffeo CS, Henning JR, Zambirinis CP, Fallon NC, Barilla R et al. (2012) Dendritic cell populations with different concentrations of lipid regulate tolerance and immunity in mouse and human liver. Gastroenterology, 143, 1061–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ochi A, Nguyen AH, Bedrosian AS, Mushlin HM, Zarbakhsh S, Barilla R, Zambirinis CP, Fallon NC, Rehman A, Pylayeva-Gupta Y et al. (2012) MyD88 inhibition amplifies dendritic cell capacity to promote pancreatic carcinogenesis via Th2 cells. The Journal of experimental medicine, 209, 1671–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zambirinis CP, Ochi A, Barilla R, Greco S, Deutsch M and Miller G (2013) Induction of TRIF- or MYD88-dependent pathways perturbs cell cycle regulation in pancreatic cancer. Cell cycle, 12, 1153–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seifert L, Werba G, Tiwari S, Giao Ly NN, Alothman S, Alqunaibit D, Avanzi A, Barilla R, Daley D, Greco SH et al. (2016) The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature, 532, 245–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Owyang C and Wu GD (2014) The Gut Microbiome in Health and Disease. Gastroenterology, 146, 1433–1436. [DOI] [PubMed] [Google Scholar]
- 8.Plottel CS and Blaser MJ (2011) Microbiome and Malignancy. Cell host & microbe, 10, 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bultman SJ (2014) Emerging roles of the microbiome in cancer. Carcinogenesis, 35, 249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutkowski MR, Stephen TL, Svoronos N, Allegrezza MJ, Tesone AJ, Perales-Puchalt A, Brencicova E, Escovar-Fadul X, Nguyen JM, Cadungog MG et al. (2015) Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer cell, 27, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seifert L, Werba G, Tiwari S, Giao Ly NN, Nguy S, Alothman S, Alqunaibit D, Avanzi A, Daley D, Barilla R et al. (2016) Radiation Therapy Induces Macrophages to Suppress Immune Responses Against Pancreatic Tumors in Mice. Gastroenterology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ino Y, Yamazaki-Itoh R, Shimada K, Iwasaki M, Kosuge T, Kanai Y and Hiraoka N (2013) Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. British journal of cancer, 108, 914–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daley D, Mani VR, Mohan N, Akkad N, Ochi A, Heindel DW, Lee KB, Zambirinis CP, Pandian GSB, Savadkar S et al. (2017) Dectin 1 activation on macrophages by galectin 9 promotes pancreatic carcinoma and peritumoral immune tolerance. Nature medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daley D, Mani VR, Mohan N, Akkad N, Pandian G, Savadkar S, Lee KB, Torres-Hernandez A, Aykut B, Diskin B et al. (2017) NLRP3 signaling drives macrophage-induced adaptive immune suppression in pancreatic carcinoma. The Journal of experimental medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, Rustgi AK, Chang S and Tuveson DA (2005) Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer cell, 7, 469–483. [DOI] [PubMed] [Google Scholar]
- 16.Garrett WS (2015) Cancer and the microbiota. Science, 348, 80–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwabe RF and Jobin C (2013) The microbiome and cancer. Nature reviews. Cancer, 13, 800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark CE, Hingorani SR, Mick R, Combs C, Tuveson DA and Vonderheide RH (2007) Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer research, 67, 9518–9527. [DOI] [PubMed] [Google Scholar]
- 19.Zheng L, Xue J, Jaffee EM and Habtezion A (2013) Role of immune cells and immune-based therapies in pancreatitis and pancreatic ductal adenocarcinoma. Gastroenterology, 144, 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukunaga A, Miyamoto M, Cho Y, Murakami S, Kawarada Y, Oshikiri T, Kato K, Kurokawa T, Suzuoki M, Nakakubo Y et al. (2004) CD8+ tumor-infiltrating lymphocytes together with CD4+ tumor-infiltrating lymphocytes and dendritic cells improve the prognosis of patients with pancreatic adenocarcinoma. Pancreas, 28, e26–31. [DOI] [PubMed] [Google Scholar]
- 21.De Monte L, Reni M, Tassi E, Clavenna D, Papa I, Recalde H, Braga M, Di Carlo V, Doglioni C and Protti MP (2011) Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. The Journal of experimental medicine, 208, 469–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiraoka N, Onozato K, Kosuge T and Hirohashi S (2006) Prevalence of FOXP3+ regulatory T cells increases during the progression of pancreatic ductal adenocarcinoma and its premalignant lesions. Clinical cancer research : an official journal of the American Association for Cancer Research, 12, 5423–5434. [DOI] [PubMed] [Google Scholar]
- 23.Geller LT, Barzily-Rokni M, Danino T, Jonas OH, Shental N, Nejman D, Gavert N, Zwang Y, Cooper ZA, Shee K et al. (2017) Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science, 357, 1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tojo R, Suárez A, Clemente MG, de los Reyes-Gavilán CG, Margolles A, Gueimonde M and Ruas-Madiedo P (2014) Intestinal microbiota in health and disease: Role of bifidobacteria in gut homeostasis. World Journal of Gastroenterology : WJG, 20, 15163–15176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown JH, Whitham TG, Morgan Ernest SK and Gehring CA (2001) Complex Species Interactions and the Dynamics of Ecological Systems: Long-Term Experiments. Science, 293, 643–650. [DOI] [PubMed] [Google Scholar]
- 26.Nugent JL, McCoy AN, Addamo CJ, Jia W, Sandler RS and Keku TO (2014) Altered Tissue Metabolites Correlate with Microbial Dysbiosis in Colorectal Adenomas. Journal of Proteome Research, 13, 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sanapareddy N, Legge RM, Jovov B, McCoy A, Burcal L, Araujo-Perez F, Randall TA, Galanko J, Benson A, Sandler RS et al. (2012) Increased rectal microbial richness is associated with the presence of colorectal adenomas in humans. The ISME Journal, 6, 1858–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML et al. (2015) Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science, 350, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hingorani SR, Petricoin EF III, Maitra A, Rajapakse V, King C, Jacobetz MA, Ross S, Conrads TP, Veenstra TD, Hitt BA et al. (2003) Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer cell, 4, 437–450. [DOI] [PubMed] [Google Scholar]
- 30.Bedoret D, Wallemacq H, Marichal T, Desmet C, Quesada Calvo F, Henry E, Closset R, Dewals B, Thielen C, Gustin P et al. (2009) Lung interstitial macrophages alter dendritic cell functions to prevent airway allergy in mice. The Journal of clinical investigation, 119, 3723–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reikvam DH, Erofeev A, Sandvik A, Grcic V, Jahnsen FL, Gaustad P, McCoy KD, Macpherson AJ, Meza-Zepeda LA and Johansen FE (2011) Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PloS one, 6, e17996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rehman A, Hemmert KC, Ochi A, Jamal M, Henning JR, Barilla R, Quesada JP, Zambirinis CP, Tang K, Ego-Osuala M et al. (2013) Role of fatty-acid synthesis in dendritic cell generation and function. Journal of immunology, 190, 4640–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hruban RH, Adsay NV, Albores-Saavedra J, Compton C, Garrett ES, Goodman SN, Kern SE, Klimstra DS, Kloppel G, Longnecker DS et al. (2001) Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. The American journal of surgical pathology, 25, 579–586. [DOI] [PubMed] [Google Scholar]
- 34.Mi H, Muruganujan A, Casagrande JT and Thomas PD (2013) Large-scale gene function analysis with the PANTHER classification system. Nature protocols, 8, 1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ott SJ, Musfeldt M, Ullmann U, Hampe J and Schreiber S (2004) Quantification of intestinal bacterial populations by real-time PCR with a universal primer set and minor groove binder probes: a global approach to the enteric flora. Journal of clinical microbiology, 42, 2566–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sunde PT, Olsen I, Gobel UB, Theegarten D, Winter S, Debelian GJ, Tronstad L and Moter A (2003) Fluorescence in situ hybridization (FISH) for direct visualization of bacteria in periapical lesions of asymptomatic root-filled teeth. Microbiology, 149, 1095–1102. [DOI] [PubMed] [Google Scholar]
- 37.Thimm T and Tebbe CC (2003) Protocol for rapid fluorescence in situ hybridization of bacteria in cryosections of microarthropods. Applied and environmental microbiology, 69, 2875–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi YS, Kim YC, Baek KJ and Choi Y (2015) In Situ Detection of Bacteria within Paraffin-embedded Tissues Using a Digoxin-labeled DNA Probe Targeting 16S rRNA. Journal of visualized experiments : JoVE, e52836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pushalkar S, Li X, Kurago Z, Ramanathapuram LV, Matsumura S, Fleisher KE, Glickman R, Yan W, Li Y and Saxena D (2014) Oral microbiota and host innate immune response in bisphosphonate-related osteonecrosis of the jaw. In J Oral Sci, 6, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pushalkar S, Ji X, Li Y, Estilo C, Yegnanarayana R, Singh B, Li X and Saxena D (2012) Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiology, 12, 144–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M and Glöckner FO (2013) Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research, 41, e1–e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herlemann DPR, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ and Andersson AF (2011) Transitions in bacterial communities along the 2000[thinsp]km salinity gradient of the Baltic Sea. ISME J, 5, 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M et al. (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. The ISME Journal, 6, 1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navas-Molina JA, Peralta-Sánchez JM, González A, McMurdie PJ, Vázquez-Baeza Y, Xu Z, Ursell LK, Lauber C, Zhou H, Song SJ et al. (2013) Advancing our understanding of the human microbiome using QIIME. Methods in enzymology, 531, 371–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masella AP, Bartram AK, Truszkowski JM, Brown DG and Neufeld JD (2012) PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics, 13, 31–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pircalabioru G, Aviello G, Kubica M, Zhdanov A, Paclet MH, Brennan L, Hertzberger R, Papkovsky D, Bourke B and Knaus UG (2016) Defensive Mutualism Rescues NADPH Oxidase Inactivation in Gut Infection. Cell host & microbe, 19, 651–663. [DOI] [PubMed] [Google Scholar]
- 47.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nature methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- 49.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E et al. (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Research, 21, 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price MN, Dehal PS and Arkin AP (2009) FastTree: Computing Large Minimum Evolution Trees with Profiles instead of a Distance Matrix. Molecular Biology and Evolution, 26, 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS and Huttenhower C (2011) Metagenomic biomarker discovery and explanation. Genome Biology, 12, R60–R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, Knight R and Ley RE (2014) Conducting a microbiome study. Cell, 158, 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mukherjee S, Huntemann M, Ivanova N, Kyrpides NC and Pati A (2015) Large-scale contamination of microbial isolate genomes by Illumina PhiX control. Standards in genomic sciences, 10, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinha R, Abnet CC, White O, Knight R and Huttenhower C (2015) The microbiome quality control project: baseline study design and future directions. Genome biology, 16, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.