Abstract
Aims
To investigate the accuracy and reproducibility of a scoring system for cervical intraepithelial neoplasia (CIN1–3) based on immunohistochemical (IHC) biomarkers Ki-67 and p16ink4a.
Methods
115 cervical tissue specimens were reviewed by three expert gynaecopathologists and graded according to three strategies: (1) CIN grade based on H&E staining only; (2) immunoscore based on the cumulative score of Ki-67 and p16ink4a only (0–6); and (3) CIN grade based on H&E supported by non-objectified IHC 2 weeks after scoring 1 and 2. The majority consensus diagnosis of the CIN grade based on H&E supported by IHC was used as the Reference Standard. The proportion of test positives (accuracy) and the absolute agreements across pathologists (reproducibility) of the three grading strategies within each Reference Standard category were calculated.
Results
We found that immunoscoring with positivity definition 6 yielded the highest proportion of test positives for Reference Standard CIN3 (95.5%), in combination with the lowest proportion of test positives in samples with CIN1 (1.8%). The proportion of test positives for CIN3 was significantly lower for sole H&E staining (81.8%) or combined H&E and IHC grading (84.8%) with positivity definition ≥CIN3. Immunoscore 6 also yielded high absolute agreements for CIN3 and CIN1, but the absolute agreement was low for CIN2.
Conclusions
The higher accuracy and reproducibility of the immunoscore opens the possibility of a more standardised and reproducible definition of CIN grade than conventional pathology practice, allowing a more accurate comparison of CIN-based management strategies and evaluation of new biomarkers to improve the understanding of progression of precancer from human papillomavirus infection to cancer.
Keywords: cervical cancer, immunohistochemistry, HPV, Ki 67, diagnosis
Introduction
Cervical screening programmes are based on the detection and treatment of cervical precancer. Accurate colposcopy and histological assessment of cervical precursor lesions is essential to determine clinical management. Histologically cervical lesions are classified as cervical intraepithelial neoplasia (CIN) and categorised as CIN1, CIN2 or CIN3 based on the extension of immature, dysplastic cells into the squamous epithelium above the basal layer (one-third to full thickness) and the severity of cellular abnormality. CIN3 is considered the most advanced precancerous lesion. Although CIN is classified into three grades, the development of CIN to cancer is a dynamic process and represents a morphological continuum.1
The diagnosis of the pathologist is subjective and based on personal experience, making use of histomorphological features in H&E stained slides alone or in combination with immunohistochemical (IHC) findings. The heterogeneity inherent in this subjective grading of CIN results in limited reproducibility with moderate interobserver and intraobserver agreement,2 3 and consequently has effects on treatment. Generally, CIN3 is treated by excision and CIN1 is managed conservatively. However, for CIN2 management differs between clinics, that is, either excisional treatment or close surveillance, because of the high regression rate of CIN2.4 5
Due to the moderate reproducibility of CIN grading, the WHO has introduced a two-tiered grading system in 2013,6 in which the term high-grade CIN is used for CIN2 and CIN3, and low-grade CIN for CIN1. The diagnosis of high-grade squamous intraepithelial lesion (HSIL) results in excisional treatment to prevent precancers developing into cancer. Consequently, this approach results in overtreatment of potentially non-progressive or regressive lesions. The USA adopted and recently optimised this two-tiered classification system,2 whereas most European countries adopted the CIN grading system.7 In the latter, preferably CIN2 should be divided into (early productive) CIN1-like and (late transforming) CIN3-like lesions.8
An IHC or biomarker-based classification system might improve the accuracy and reproducibility of grading CIN, and hence standardisation of diagnosis. This will allow comparison of the results of clinical management between centres for clinical audit and also simpler comparison of alternative management strategies in clinical trials. Ki-67 and p16ink4a are used widely to guide CIN grading by pathologists and expression of these markers increases with increasing CIN.2 5 9–16 Ki-67 staining in suprabasal and parabasal layers is an indicator of cellular proliferation, whereas diffuse p16ink4a staining occurs when p16ink4a is overexpressed as a result of functional inactivation of retinoblastoma protein by the human papillomavirus (HPV) E7 protein. These are the consequences of a persistent cervical HPV infection and deregulation of E6 and E7 oncogene expression in proliferating cells. Deregulation of E6 and E7 oncogenes causes chromosomal instability, which is the driving force for accumulation of (epi)genetic aberrations in host cell genes and drives the progression of CIN lesions towards cervical cancer.8 17
In this study, we propose a classification system using the cumulative immunoscore value of Ki-67 and p16ink4a, compare accuracy and reproducibility of CIN grading based on this score to classical histomorphological and IHC CIN grading and show the benefits of this scoring system.
Methods
Study population
In this retrospective cross-sectional study, we selected 115 formalin-fixed paraffin-embedded cervical biopsy and large loop excision of the transformation zone (LLETZ) specimens guided by severity of the initial diagnosis (no dysplasia n=22; CIN1 n=22; CIN2 n=27; CIN3 n=22; squamous cell carcinoma (SCC) n=22) from the files of the Pathology Department of the VU University Medical Center in Amsterdam, The Netherlands. The selected tissue blocks were anonymously processed for the purposes of this study. Ethical approval was waived according to the regulations in the Netherlands.18
Immunohistochemistry
All tissue blocks were cut to provide sections of 3 µm. The first and last sections were used for H&E staining to ensure the presence of the same cervical lesion, and in-between sections were used for immunostaining with mouse monoclonal antibodies against Ki-67 antigen (DAKO, Clone MIB-1) or p16ink4a antigen (Roche, CINtec, Clone E6H4), using the automated IHC Ventana staining machine (Roche, Basel, Switzerland).
Scoring of Ki-67
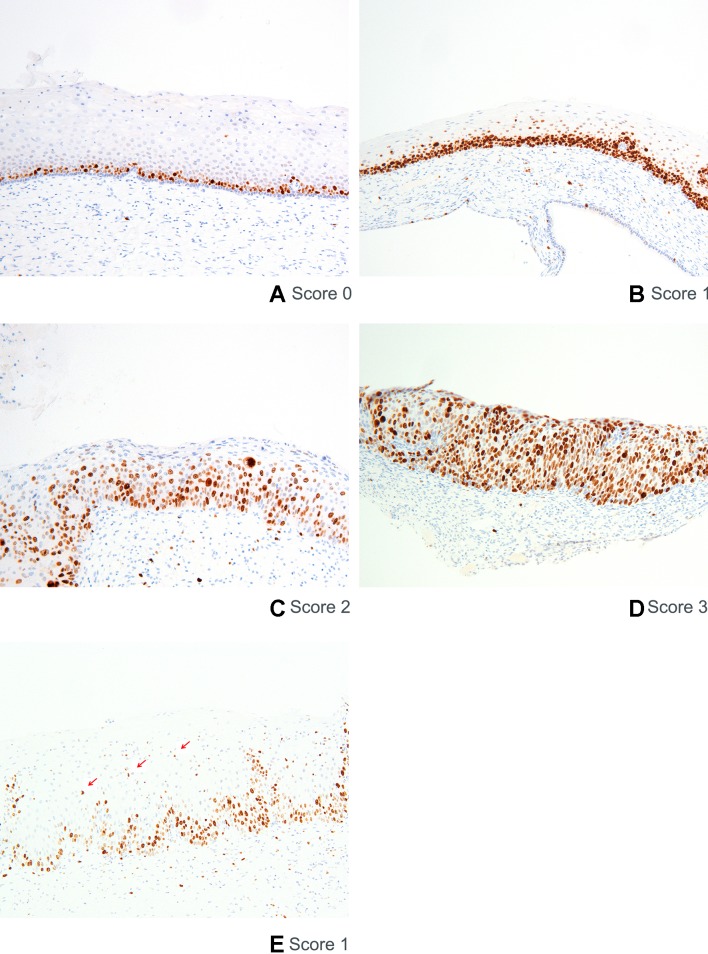
Nuclear Ki-67 staining in cells of the squamous epithelium was scored positive. Score 0 is a normal staining pattern (ie, staining of nuclei in the basal layer, figure 1A). Score 1 is defined as positive nuclei predominantly found in the lower one-third of the epithelium (figure 1B). Score 2 is defined as positive nuclei predominantly found in the lower two-thirds of the epithelium (figure 1C). Score 3 is defined as positive nuclei in more than two-thirds of the epithelium (figure 1D). The presence of a few scattered positive individual cells in the upper two-thirds layer of the epithelium in a predominant staining pattern in the lower one-third is scored as 1 (figure 1E). Also, a few scattered positive individual cells found in the upper one-third layer of the epithelium in a predominant pattern with two-thirds involvement of the epithelium is scored as 2.
Figure 1.
Scoring of Ki-67 immunostaining. Representative examples are shown for the three-tiered score system of Ki-67 immunostaining. Nuclear Ki-67 staining in cells of the squamous epithelium is scored positive. Positive nuclei in the basal layer is considered a normal staining pattern and scored as 0 (A), positive nuclei predominantly found in the lower one-third of the epithelium as 1 (B), positive nuclei predominantly found in lower two-thirds of epithelium as 2 (C), and positive nuclei in more than two-thirds of epithelium as 3 (D). An example of individual scattered positive cells found in the upper layers of the epithelium in a predominant parabasal staining pattern is shown (E).
Areas where dermal papillae narrow down the width of the epithelium cannot be scored reliably and are therefore not taken into account.
Scoring of p16ink4a
Diffuse or ‘block’ staining for p16ink4a of the cell cytoplasm or nucleus in squamous epithelium was considered positive.2 3 19 Score 0 is defined as either no p16ink4a positivity or focally scattered positive cells or small cell clusters (ie, patchy staining, figure 2A). Score 1 is defined as low intensity, diffuse positivity restricted to the lower one-third part of the epithelium (figure 2B). Score 2 is defined as continuous positivity in the lower two-thirds of the epithelium (figure 2C). Score 3 is defined as positive cells involving the full thickness of the epithelium (ie, diffuse full thickness staining, figure 2D).
Figure 2.
Scoring of p16ink4a immunostaining. Representative examples are shown for the three-tiered score system of p16ink4a immunostaining. Diffuse or ‘block’ staining for p16ink4a of the cell cytoplasm or nucleus in squamous epithelium is considered positive. No positivity or patchy staining was scored 0 (A), diffuse positivity in the lower third of the epithelium as 1 (B), diffuse positivity in the lower two-thirds of the epithelium as 2 (C), and diffuse positivity involving the full thickness of the epithelium was scored as 3 (D).
It is considered important to distinguish positive diffuse or ‘block’ p16ink4a staining from patchy or background staining.
Work procedure
H&E score
Three expert gynaecopathologists (P1, P2 and P3) from two different laboratories (MCGB, DJ and MvdS) received the H&E slides, selected the area with the most dysplastic features of the epithelium and graded the CIN lesion based on morphologic characteristics (further referred to as ‘H&E score’). All specimens were classified for H&E scoring as no dysplasia, CIN1, CIN2, CIN3 or SCC, according to international criteria.20
Ki-67 and p16ink4a immunoscore
Subsequently, the pathologists scored the Ki-67 and p16ink4a expression on a separate scoring sheet, using the scoring system described above and depicted in figures 1 and 2. This scoring sheet, including examples of the scoring, was discussed with and approved by all three pathologists prior to the start of the study. No morphologic features were taken into account in this scoring. The cumulative immunoscore of Ki-67 (scores 0–3) and p16ink4a (scores 0–3) could vary from 0 to 6, irrespective of how the score was obtained (further referred to as ‘Ki-67 and p16ink4a immunoscore’). For example, we considered a cumulative score of 5, either established by a Ki-67 score of 2 and p16ink4a score of 3, or vice versa, as identical.
Combined H&E and non-objectified IHC score
At least 2 weeks after scoring of the immunostains, each pathologist was asked to render the CIN grade now based on morphologic features in combination with the subjective interpretation of the immunostaining (further referred to as ‘combined H&E and non-objectified IHC score’).
Reference Standard
The consensus diagnosis of the combined H&E and non-objectified IHC score, based on agreement of CIN grade in at least two out of three pathologists, was used as the ‘Reference Standard’. In six lesions in which no majority CIN score was available, consensus was reached in a panel discussion with a fourth pathologist (CJLMM).
Statistical analysis
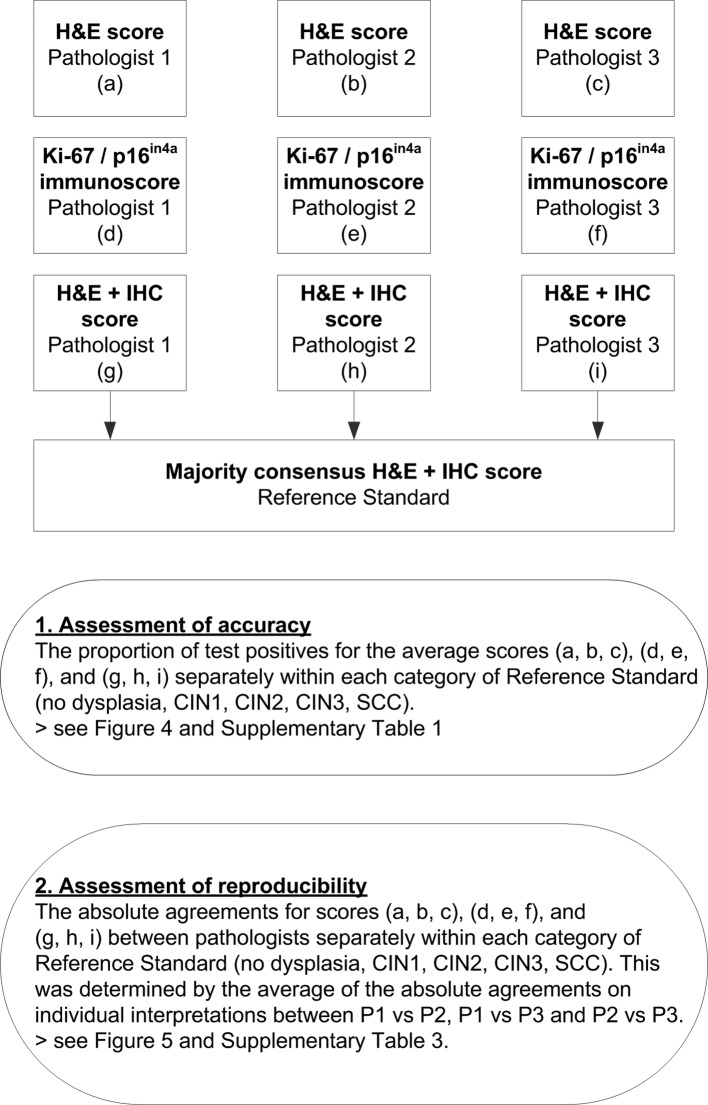
A flow chart of the statistical analysis is shown in figure 3. To assess the accuracy of a test strategy, we calculated the proportion of test positives for all CIN grading strategies separately within each of the different Reference Standard diagnoses (ie, no dysplasia, CIN1, CIN2, CIN3 and SCC). A positive status for each strategy was obtained using the following definitions: diagnosis of ≥CIN2 based on H&E scoring for strategy I; diagnosis of ≥CIN3 based on sole H&E scoring for strategy II; diagnosis of ≥CIN2 based on combined H&E and non-objectified IHC scoring for strategy III; diagnosis of ≥CIN3 based on combined H&E and non-objectified IHC scoring for strategy IV; immunoscore of ≥4 for strategy V based on Ki-67 and p16ink4 immunoscoring for strategy V; immunoscore of ≥5 based on Ki-67 and p16ink4 immunoscoring for strategy VI; and immunoscore of 6 based on Ki-67 and p16ink4 immunoscoring for strategy VII (table 1). We accounted for the scores of three pathologists by pooling the proportions of test positives of the individual pathologists. Absolute differences between the pooled proportions of test positives of each strategy per Reference Standard category were calculated with 95% CIs. If a CI did not include the value 0, the difference was considered statistically significant.
Figure 3.
Flow chart of statistical analysis. CIN, cervical intraepithelial neoplasia; IHC, immunohistochemistry; SCC, squamous cell carcinoma.
Table 1.
Overview of scoring strategies
| Strategy | Scoring | Positivity definition |
| I | H&E score | ≥CIN2 |
| II | H&E score | ≥CIN3 |
| III | Combined H&E and non-objectified IHC score | ≥CIN2 |
| IV | Combined H&E and non-objectified IHC score | ≥CIN3 |
| V | Immunoscore | ≥4 |
| VI | Immunoscore | ≥5 |
| VII | Immunoscore | 6 |
CIN, cervical intraepithelial neoplasia; IHC, immunohistochemistry.
Second, to assess the reproducibility of a test strategy, we calculated the average of absolute agreements for every CIN grading strategy (with positivity definitions of strategies I–VII, see table 1) between pathologists (P1 vs P2, P1 vs P3 and P2 vs P3) within each Reference Standard category.
Calculations were performed in SPSS (V.22) and STATA (V.14.1).
Results
Histology scoring
Different grades for H&E, H&E and IHC, Ki-67 and p16ink4a immunoscores, and the Reference Standard category by the three pathologists for the 115 cervical biopsy and LLETZ specimens are shown in table 2.
Table 2.
Outcome of different grading strategies of each individual pathologist
| P1 | P2 | P3 | Reference Standard* n |
|
| n | n | n | ||
| A. H&E scoring | ||||
| No dysplasia | 26 | 34 | 44 | |
| CIN1 | 31 | 11 | 17 | |
| CIN2 | 16 | 8 | 20 | |
| CIN3 | 20 | 40 | 12 | |
| SCC | 22 | 22 | 22 | |
| B. Ki-67 and p16ink4a immunoscore | ||||
| Immunoscore 0 | 25 | 20 | 0 | |
| Immunoscore 1 | 21 | 10 | 37 | |
| Immunoscore 2 | 4 | 9 | 6 | |
| Immunoscore 3 | 7 | 5 | 5 | |
| Immunoscore 4 | 4 | 3 | 5 | |
| Immunoscore 5 | 9 | 7 | 10 | |
| Immunoscore 6 | 45 | 61 | 52 | |
| C. Combined H&E and non-objectified IHC score | ||||
| No dysplasia | 28 | 35 | 41 | 35 |
| CIN1 | 24 | 12 | 19 | 19 |
| CIN2 | 21 | 8 | 18 | 17 |
| CIN3 | 20 | 38 | 15 | 22 |
| CIN3 | 22 | 22 | 22 | 22 |
*The Reference Standard is defined as the majority diagnosis of the combined H&E and non-objectified IHC score, based on agreement on CIN grade in two out of three pathologists (see also the Methods section).
CIN, cervical intraepithelial neoplasia; IHC, immunohistochemistry; P, pathologist; SCC, squamous cell carcinoma.
Accuracy
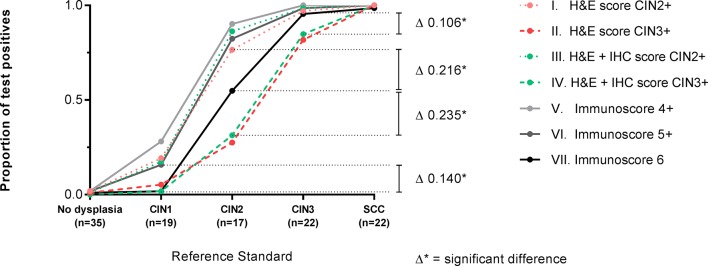
Overall, taking the proportion of test positives as a measurement of the accuracy, CIN grading based on the Ki-67 and p16ink4a immunoscore, with immunoscore 6 to define a positive status (CIN grading strategy VII), detected the highest numbers of Reference Standard CIN3, combined with the fewest CIN1 (figure 4, online supplementary table 1). Furthermore, CIN grading based solely on H&E grading compared with grading based on combined H&E and non-objectified interpretation of IHC (strategy I vs III, and strategy II vs IV) showed no significant differences in detection of all Reference Standard categories (online supplementary table 2).
Figure 4.
Accuracy of different cervical intraepithelial neoplasia (CIN) grading strategies. The proportion of test positives for the different CIN grading strategies I–VII is shown separately within each Reference Standard: no dysplasia, CIN1, CIN2, CIN3 and cervical squamous cell carcinoma (SCC). The H&E score (strategies I and II) is based solely on morphologic characteristics, the H&E+immunohistochemistry (IHC) score (strategies III and IV) is rendered after interpretation of Ki-67 and p16ink4a immunostains, and the immunoscore (strategies V, VI and VII) is the total value of the scoring for Ki-67 and p16ink4a immunostains independently scored by the pathologist. Strategy VII detects the highest number of CIN3 and the fewest CIN1. The exact values for the proportion of test positives are shown in online supplementary table 1 and the absolute differences between all point values (indicated by ∆) are shown in online supplementary table 2.
jclinpath-2018-205271supp001.pdf (313.1KB, pdf)
In more detail, within Reference Standard no dysplasia and SCC, no significant differences in accuracy (online supplementary table 2) between any of the CIN grading strategies were observed, with the proportions of test positives of all grading strategies being close to 0 and 100%, respectively.
Within Reference Standard CIN1, the accuracy of CIN grading strategy I (≥CIN2 based on sole H&E scoring), strategy III (≥CIN2 based on combined H&E and non-objectified interpretation of IHC), strategy V (immunoscore ≥4) and strategy VI (immunoscore ≥5), with proportions of test positives ranging from 17.5% to 28.1%, was significantly higher than the accuracy of strategy II (≥CIN3 based on sole H&E scoring), strategy IV (≥CIN3 based on combined H&E and non-objectified interpretation of IHC) and strategy VII (immunoscore 6), with proportions of test positives ranging from 1.7% to 5.3%.
Within Reference Standard CIN2, the accuracy of CIN grading strategies I, III, V and VI, with proportions of test positives ranging from 76.5% to 90.2%, was significantly higher than the accuracy of strategy VII, with a proportion of test positives of 54.9%. Furthermore, the accuracy of CIN grading strategies II and IV was significantly lower compared with strategy VII, with proportions of test positives of 27.5% and 31.4%.
For the detection of Reference Standard CIN3, the accuracy of CIN grading strategies I, III, V, VI and VII was near 100%, with proportions of test positives ranging from 97.0% to 100%, and significantly higher than the accuracy of strategies II and IV with proportions of test positives of 81.8% and 84.8%.
Reproducibility
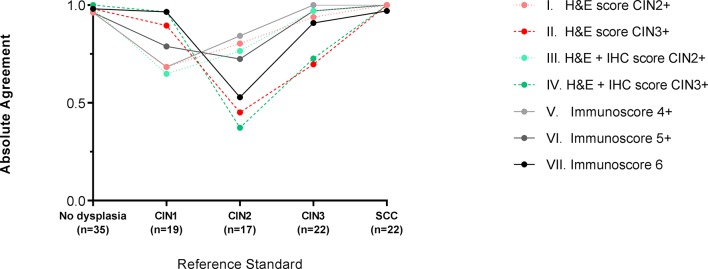
Overall, grading of CIN based on the Ki-67 and p16ink4a immunoscore with a definition for positivity of immunoscore 6 (strategy VII) showed a high absolute agreement for Reference Standard CIN3 (90.9%) in combination with a high agreement for Reference Standard CIN1 (96.5%, figure 5, online supplementary table 3). Other grading strategies with a high absolute agreement for Reference Standard CIN3 showed moderate agreement for Reference Standard CIN1. All strategies showed only moderate agreement for Reference Standard CIN2.
Figure 5.
Reproducibility of different cervical intraepithelial neoplasia (CIN) grading strategies. The absolute agreement among pathologists for the different CIN grading strategies I–VII, representing the reproducibility, is shown separately within each Reference Standard: no dysplasia, CIN1, CIN2, CIN3 and cervical squamous cell carcinoma (SCC). The H&E score (strategies I and II) is based solely on morphologic characteristics, the H&E+immunohistochemistry (IHC) score (strategies III and IV) is rendered after interpretation of Ki-67 and p16ink4a immunostains, and the immunoscore (strategies V, VI and VII) is the total value of the scoring for Ki-67 and p16ink4a immunostains independently scored by the pathologist. The exact values of absolute agreement are shown in online supplementary table 3.
In more detail, the absolute agreement between pathologists was high for Reference Standard no dysplasia and SCC (all ≥96.2%; figure 5 and online supplementary table 3). For Reference Standard CIN1, the absolute agreement was moderate to high (range 64.9%–96.5%) with highest agreement for CIN grading strategies IV and VII. For Reference Standard CIN2, the absolute agreement was moderate (range 37.3%–84.3%). For Reference Standard CIN3, the absolute agreement was high for CIN grading strategies I, III, V, VI and VII (range 90.9%–100%) and moderate for strategies II and IV (69.7% and 72.7%).
Discussion
In this cross-sectional study, we describe a simple CIN grading system based solely on a cumulative three-tiered immunoscore for biomarkers Ki-67 and p16ink4a to perform better in terms of accuracy and reproducibility, compared with the classical histological and non-objectified IHC CIN grading system. We have added Ki-67 scores to the p16ink4a immunoscore because it is essential to identify proliferative activity in p16ink4a-positive CIN. Performance of p16ink4a scoring without other markers or H&E grading is known to increase the shift from CIN1 to CIN2, resulting in overtreatment.2 Interestingly, our additional statistical analysis shows that sole p16ink4a staining has a lower accuracy for CIN3 than combined p16ink4a and Ki-67 staining (online supplementary figure 1). By using the Ki-67 and p16ink4a immunoscore, we showed that immunoscore 6 was able to detect reliably the highest number of CIN3, combined with the lowest proportion of CIN1 lesions. The proportion of test positives for CIN3 of immunoscore 6 (95.5%) was significantly higher in comparison to the classical CIN grading based on sole H&E staining (81.8% for positivity definition ≥CIN3) or combined H&E with non-objectified IHC interpretation (84.8% for positivity definition ≥CIN3). This accurate detection of CIN3 and CIN1 by immunoscore 6 indicates that a substantial proportion of classically graded CIN2 will be recategorised into CIN1 and CIN3 lesions by use of this immunoscore. Because the Ki-67 and p16ink4a immunoscore defines with more accuracy and better reproducibility the grade of the cervical lesion, it facilitates clear and accurate communication about CIN lesions between pathologists and clinicians and provides a more reliable basis to decide whether cervical treatment or a wait-and-see policy is appropriate.
Decisions on clinical management are based primarily on CIN grade. However, management guidelines of CIN2 vary substantially between and within non-US countries. Presently, gynaecologists generally will treat all CIN3 lesions, but for CIN2 lesions, depending on the size of the lesion and patient’s preference and age, both treatment and a wait-and-see policy are widely advocated.21–23 The CIN grading system presented in this paper can be used as a proposal for further research to validate our results and to achieve more standardisation in CIN management.
This proposal is based on the most accurate CIN grading strategy found in this study to detect CIN3 (treatment) and CIN1 (no treatment). For clinical use we propose to first define a CIN grade (CIN1–3) based on an H&E staining. Then the immunoscore of a CIN lesion should be reported. Based on the immunoscore treatment can be defined: suggesting no treatment for lesions with an immunoscore 0–3, a wait-and-see policy for lesions with an immunoscore 4 and 5, and excisional treatment for lesions with an immunoscore 6. Thus, treatment is based on the most accurate and reproducible CIN grading. Especially for CIN2 where only 55% had an immunoscore 6, use of the immunoscore would direct clinical management objectively and separate this group into lesions requiring close follow-up and lesions requiring treatment. Additional studies with a large number of samples should validate these data, and give insights on management suggestions before implementation in clinical practice can be recommended.
In the recently published US Lower Anogenital Squamous Terminology Standardization Project for HPV-Associated Lesions (LAST) guidelines, Darragh et al extensively investigated the most optimal classification strategy for the grading of genital intraepithelial neoplasia.2 They further optimised the Bethesda classification for dividing genital lesions in HSILs and low-grade squamous intraepithelial lesions. Thereby they dissuade classically three-tiered CIN grading and recommend p16ink4a staining only for CIN2+ lesions where the pathologist is in doubt. Positivity for p16ink4a of at least one-third of the epithelium supports the diagnosis of HSIL, and treatment of all HSILs is recommended.2 As more than half of CIN2 lesions and a substantial number of CIN3 lesions will regress,4 23–26 treatment of all HSIL results in considerable overtreatment which has major consequences, especially for women in their fertile age, in terms of cervical morbidity and preterm delivery.27 The Ki-67 and p16ink4a immunoscore could benefit patients in reducing the current practice of overtreatment by excising or ablating all CIN2 that is known to include productive, regressive and progressive lesions.
The Ki-67 and p16ink4a scoring system has some limitations. It still involves microscopic evaluation of a biopsy with its attendant sampling error, and interpretation of immunostaining may be difficult in some cases. However, by strict definition of the different scores and addressing difficulties in scoring as described in this article, potential scoring problems have been reduced to very low levels.
To define the Reference Standard score, we used the consensus diagnosis of CIN based on the combined H&E score with non-objectified Ki-67 and p16ink4a interpretation. This was done because the use of H&E in conjunction with these IHC markers has the highest accuracy.19 28–30 To prevent that the Ki-67 and p16ink4a immunoscore could have influenced the CIN grade based on H&E and non-objectified Ki-67 and p16ink4a score, at least 2 weeks were present between the scoring of Ki-67 and p16ink4a, and the grading of CIN based on combined H&E and non-objectified Ki-67 and p16ink4a interpretation.
A further limitation is that expression of Ki-67 and p16ink4a represents only part of the complex process of progression of CIN to cancer. Reduction or loss of completion of oncogenic HPV life cycle as, for instance, indicated by loss or reduced expression of HPV E4 protein in CIN3 is also potentially relevant for progression of CIN and for treatment decisions especially in CIN2.31 32 In addition, the presence of hypermethylation of promotor regions in host cell genes (ie, CADM1, MAL, miR124-2 and FAM19A4) is associated with CIN2/3 lesions with a high short-term progression risk for cervical cancer.8 33 The evaluation of HPV E4 protein staining in CIN, classified by the immunoscore system and the presence of hypermethylation of these genes, is presently under investigation. Preliminary results seem to confirm that, in HPV-positive lesions, E4 staining decreases from immunoscore 0 to 4, whereas hypermethylation starts to be present in immunoscore 4 lesions with highest values in immunoscore 6 lesions.34 Collectively, the immunoscore system seems to provide a classification system that could be useful and important in studying the role of these additional markers in improving management of CIN.
In conclusion, the grading of CIN by this simple Ki-67 and p16ink4a immunoscore system shows a higher accuracy and better reproducibility than the classical CIN grading system, especially for CIN3 (treatment) and CIN1 (no treatment). Due to the optimisation of CIN3 and CIN1 diagnoses, a division of classical CIN2 into these categories can be made. Validation in large study numbers, preferably in a prospective trial in which also other biomarkers such as E4 and hypermethylation of host cell genes are taken into account, is needed. Furthermore, the Ki-67 and p16ink4a immunoscore system better reflects where in the biological trajectory of development of cervical cancer through infection and precancer the cervical lesion is situated. This new grading system might provide a basis for development of standardisation in the diagnosis of CIN and clinical management in women with cervical precancer.
Take home messages.
Grading of cervical intraepithelial neoplasia (CIN) by a simple Ki-67 and p16ink4a immunoscore system has a higher accuracy and reproducibility compared with current CIN grading.
By use of the Ki-67 and p16ink4a immunoscore, most of classical CIN2 can be divided into more accurately graded CIN1 and CIN3.
Use of the Ki-67 and p16ink4a immunoscore for CIN grading allows better evaluation of the role of new biomarkers in the development of cervical cancer.
Acknowledgments
We gratefully acknowledge all the research staff and technicians of the Department of Pathology VU University Medical Center.
Footnotes
AL and WWK contributed equally.
Handling editor: Cheok Soon Lee.
Contributors: MZ and CJLMM have set up the trial. MZ, AL, WWK, MCGB, DJ, MvdS, DAMH, RDMS, PJFS, JB, WGVQ and CJLMM were involved in data collection. MZ and HB performed the statistical analysis. MZ managed the database. MZ, MCGB, DJ and CJLMM drafted the manuscript. All authors critically reviewed the manuscript and approved the final version. All authors had full access to all of the data in the study and can take responsibility for the integrity of the data and the accuracy of the data analysis and believe that the manuscript represents honest work. CJLMM affirms that the manuscript is an honest, accurate and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as planned have been explained.
Funding: The authors have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Competing interests: DAMH, PJFS, RDMS and CJLMM are minority shareholders of Self-screen, a spin-off company of VUmc, of which CJLMM is part-time director since September 2017. Self-screen holds patents related to the work (ie, hrHPV test and methylation markers for cervical cancer screening). DAMH serves occasionally on the scientific advisory board of Pfizer. PJFS has been on the speakers bureau of Roche diagnostics, Gen-Probe, Abbott, Qiagen and Seegene and has been a consultant for Crucell. JB received travel support from DDL Diagnostic Laboratory, speakers’ fees from Qiagen and consultancy fees from Roche, GlaxoSmithKline and Merck/SPMSD; all JB’s fees were collected by his employer. WGVQ is shareholder of DDL Diagnostic Laboratory. CJLMM has received speakers’ fee from Qiagen and SPMSD/Merck, served occasionally on the scientific advisory board (expert meeting) of Qiagen and SPMSD/Merck and has been by occasion consultant for Qiagen. CJLMM has a very small number of shares in Qiagen, and was minority shareholder of Diassay until April 2016. All other authors have no conflict of interest to declare.
Patient consent: Not required.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Heatley MK. How should we grade CIN? Histopathology 2002;40:377–80. 10.1046/j.1365-2559.2002.01365.x [DOI] [PubMed] [Google Scholar]
- 2. Darragh TM, Colgan TJ, Cox JT, et al. The lower anogenital squamous terminology standardization project for HPV-Associated Lesions: background and consensus recommendations from the college of american pathologists and the american society for colposcopy and cervical pathology. J Low Genit Tract Dis 2012;16:205–42. 10.1097/LGT.0b013e31825c31dd [DOI] [PubMed] [Google Scholar]
- 3. Herrington CS. The terminology of pre-invasive cervical lesions in the UK cervical screening programme. Cytopathology 2015;26:346–50. 10.1111/cyt.12307 [DOI] [PubMed] [Google Scholar]
- 4. Ostör AG. Natural history of cervical intraepithelial neoplasia: a critical review. Int J Gynecol Pathol 1993;12:186–92. [PubMed] [Google Scholar]
- 5. Baak JP, Kruse AJ, Robboy SJ, et al. Dynamic behavioural interpretation of cervical intraepithelial neoplasia with molecular biomarkers. J Clin Pathol 2006;59:1017–28. 10.1136/jcp.2005.027839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. WHO Guidelines for Screening and Treatment of Precancerous Lesions for Cervical Cancer Prevention. Geneva 2013. [PubMed] [Google Scholar]
- 7. von Karsa L, Arbyn M, De Vuyst H, et al. European guidelines for quality assurance in cervical cancer screening. Summary of the supplements on HPV screening and vaccination. Papillomavirus Res 2015;1:22–31. 10.1016/j.pvr.2015.06.006 [DOI] [Google Scholar]
- 8. Steenbergen RD, Snijders PJ, Heideman DA, et al. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer 2014;14:395–405. 10.1038/nrc3728 [DOI] [PubMed] [Google Scholar]
- 9. Bulten J, van der Laak JA, Gemmink JH, et al. MIB1, a promising marker for the classification of cervical intraepithelial neoplasia. J Pathol 1996;178:268–73. [DOI] [PubMed] [Google Scholar]
- 10. Kruse AJ, Baak JP, de Bruin PC, et al. Ki-67 immunoquantitation in cervical intraepithelial neoplasia (CIN): a sensitive marker for grading. J Pathol 2001;193:48–54. [DOI] [PubMed] [Google Scholar]
- 11. Dray M, Russell P, Dalrymple C, et al. p16(INK4a) as a complementary marker of high-grade intraepithelial lesions of the uterine cervix. I: Experience with squamous lesions in 189 consecutive cervical biopsies. Pathology 2005;37:112–24. 10.1080/00313020500058607 [DOI] [PubMed] [Google Scholar]
- 12. Doorbar J. Papillomavirus life cycle organization and biomarker selection. Dis Markers 2007;23:297–313. 10.1155/2007/613150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Conesa-Zamora P, Doménech-Peris A, Ortiz-Reina S, et al. Immunohistochemical evaluation of ProEx C in human papillomavirus-induced lesions of the cervix. J Clin Pathol 2009;62:159–62. 10.1136/jcp.2008.061408 [DOI] [PubMed] [Google Scholar]
- 14. Koeneman MM, Kruitwagen RF, Nijman HW, et al. Natural history of high-grade cervical intraepithelial neoplasia: a review of prognostic biomarkers. Expert Rev Mol Diagn 2015;15:527–46. 10.1586/14737159.2015.1012068 [DOI] [PubMed] [Google Scholar]
- 15. Mills AM, Paquette C, Castle PE, et al. Risk stratification by p16 immunostaining of CIN1 biopsies: a retrospective study of patients from the quadrivalent HPV vaccine trials. Am J Surg Pathol 2015;39:611–7. 10.1097/PAS.0000000000000374 [DOI] [PubMed] [Google Scholar]
- 16. Natl. Guidel. ’Cervical Intraepithelial Neoplasia': NVOG. Inhoudsopgave, 2004. [Google Scholar]
- 17. Korzeniewski N, Spardy N, Duensing A, et al. Genomic instability and cancer: lessons learned from human papillomaviruses. Cancer Lett 2011;305:113–22. 10.1016/j.canlet.2010.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Federation of Biomedical Scientific Societies. Human Tissue and Medical Research: Code of Conduct for responsible use. 2011. https://www.federa.org/sites/default/files/images/print_version_code_of_conduct_english.pdf
- 19. Dijkstra MG, Heideman DA, de Roy SC, et al. p16(INK4a) immunostaining as an alternative to histology review for reliable grading of cervical intraepithelial lesions. J Clin Pathol 2010;63:972–7. 10.1136/jcp.2010.078634 [DOI] [PubMed] [Google Scholar]
- 20. Wright TW, Ronnett BM, Kurman RJ, et al. Precancerous lesions of the cervix In: Kurman RJ, Ellenson LH, Ronnett BM, eds Blaustein’s Pathology of the Female Genital Tract, 2011. [Google Scholar]
- 21. Martin CM, O’Leary JJ. Histology of cervical intraepithelial neoplasia and the role of biomarkers. Best Pract Res Clin Obstet Gynaecol 2011;25:605–15. 10.1016/j.bpobgyn.2011.04.005 [DOI] [PubMed] [Google Scholar]
- 22. Castle PE, Stoler MH, Solomon D, et al. The relationship of community biopsy-diagnosed cervical intraepithelial neoplasia grade 2 to the quality control pathology-reviewed diagnoses: an ALTS report. Am J Clin Pathol 2007;127:805–15. 10.1309/PT3PNC1QL2F4D2VL [DOI] [PubMed] [Google Scholar]
- 23. Castle PE, Schiffman M, Wheeler CM, et al. Evidence for frequent regression of cervical intraepithelial neoplasia-grade 2. Obstet Gynecol 2009;113:18–25. 10.1097/AOG.0b013e31818f5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trimble CL, Piantadosi S, Gravitt P, et al. Spontaneous regression of high-grade cervical dysplasia: effects of human papillomavirus type and HLA phenotype. Clin Cancer Res 2005;11:4717–23. 10.1158/1078-0432.CCR-04-2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ovestad IT, Gudlaugsson E, Skaland I, et al. The impact of epithelial biomarkers, local immune response and human papillomavirus genotype in the regression of cervical intraepithelial neoplasia grades 2-3. J Clin Pathol 2011;64:303–7. 10.1136/jcp.2010.083626 [DOI] [PubMed] [Google Scholar]
- 26. Munk AC, Gudlaugsson E, Ovestad IT, et al. Interaction of epithelial biomarkers, local immune response and condom use in cervical intraepithelial neoplasia 2-3 regression. Gynecol Oncol 2012;127:489–94. 10.1016/j.ygyno.2012.09.010 [DOI] [PubMed] [Google Scholar]
- 27. Kyrgiou M, Mitra A, Arbyn M, et al. Fertility and early pregnancy outcomes after treatment for cervical intraepithelial neoplasia: systematic review and meta-analysis. BMJ 2014;349:g6192 10.1136/bmj.g6192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Horn LC, Reichert A, Oster A, et al. Immunostaining for p16INK4a used as a conjunctive tool improves interobserver agreement of the histologic diagnosis of cervical intraepithelial neoplasia. Am J Surg Pathol 2008;32:502–12. 10.1097/PAS.0b013e31815ac420 [DOI] [PubMed] [Google Scholar]
- 29. Bergeron C, Ordi J, Schmidt D, et al. Conjunctive p16INK4a testing significantly increases accuracy in diagnosing high-grade cervical intraepithelial neoplasia. Am J Clin Pathol 2010;133:395–406. 10.1309/AJCPXSVCDZ3D5MZM [DOI] [PubMed] [Google Scholar]
- 30. Bergeron C, Ronco G, Reuschenbach M, et al. The clinical impact of using p16(INK4a) immunochemistry in cervical histopathology and cytology: an update of recent developments. Int J Cancer 2015;136:2741–51. 10.1002/ijc.28900 [DOI] [PubMed] [Google Scholar]
- 31. Griffin H, Soneji Y, Van Baars R, et al. Stratification of HPV-induced cervical pathology using the virally encoded molecular marker E4 in combination with p16 or MCM. Mod Pathol 2015;28:977–93. 10.1038/modpathol.2015.52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Baars R, Griffin H, Wu Z, et al. Investigating diagnostic problems of CIN1 and CIN2 Associated With High-risk HPV by Combining the Novel Molecular Biomarker PanHPVE4 With P16INK4a. Am J Surg Pathol 2015;39:1518–28. 10.1097/PAS.0000000000000498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bierkens M, Hesselink AT, Meijer CJ, et al. CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer 2013;133:1293–9. 10.1002/ijc.28138 [DOI] [PubMed] [Google Scholar]
- 34. van Zummeren M, Kremer WW, Leeman A, et al. HPV E4 expression and DNA hypermethylation of CADM1, MAL, and miR124-2 genes in cervical cancer and precursor lesions. Modern Pathology. In Press 2018. 10.1038/s41379-018-0101-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jclinpath-2018-205271supp001.pdf (313.1KB, pdf)