Abstract
Background and aims: HOXD antisense growth-associated long noncoding RNA (HOXD-AS1) was reported to be upregulated in various cancers, such as gastric cancer, hepatocellular carcinoma, colorectal cancer, and glioma. Here, we conducted a meta-analysis and The Cancer Genome Atlas data review to investigate the clinicopathologic and prognostic value of HOXD-AS1 in patients with malignant tumors.
Materials and methods: Systematic literatures were searched from PubMed, Medline, Cochrane Library, Web of Science, EMBASE database, Ovid, Chinese CNKI, and the Chinese WanFang database. The role of HOXD-AS1 in cancers was evaluated by pooled ORs and HRs with 95% CIs. The Cancer Genome Atlas dataset was used to explore the prognostic value of HOXD-AS1 in various cancers.
Results
Fifteen studies with 1,678 patients were included in this meta-analysis. The results indicated that HOXD-AS1 was associated with tumor size, differentiation, lymph node metastasis, and TNM stage. Moreover, the high HOXD-AS1 expression indicated a poor overall survival (OS) rate and can be an independent predictive factor for OS. The TCGA dataset, which included 9,502 cancer patients, showed that the expression of HOXD-AS1 was related to poor OS and disease-free survival. We also analyzed the prognostic role in different kinds of cancers such as digestive cancers, female reproductive system cancers, respiratory system cancers, and urinary system cancers.
Conclusion
This study demonstrated that HOXD-AS1 was closely correlated with tumor size, lymph node metastasis, distant metastasis, and TNM stage, and an increased HOXD-AS1 expression could be a reliable prognostic biomarker in human cancers. However, more studies are needed to confirm this conclusion.
Keywords: lncRNA HOXD-AS1, neoplasm, prognosis, meta-analysis, TCGA cohort
Introduction
Cancer has become one of the deadliest and most prevalent diseases throughout the world – there were 4,292,000 new cancer patients and 2,814,000 cancer-related deaths in People’s Republic of China in 2015.1,2 According to the American Cancer Society, there were about 1,700,000 new cancer patients and 3,000,000 cancer-related deaths in America in 2017.3 Although there is currently a rapid development of science and technology, in addition to there being an increasing number of treatments for tumors, the prognosis of cancer is still poor, mainly due to the lack of specific biomarkers for the diagnosis of tumors.4,5 Once the tumor is diagnosed, it has already progressed to the middle and late stages, causing the best treatment opportunity to be lost. Accordingly, it is of utmost importance to find the specific cancer biomarker.
Long noncoding RNA (lncRNA), which does not have the ability to encode proteins, is RNA with a transcription length of >200 bp. It was once thought to only be a by-product of RNA polymerase II transcription – a noise and garbage of gene transcription – and does not have specific biological functions.6,7 In recent years, an increasing number of studies have shown that lncRNA plays an important role in the proliferation, apoptosis, invasion, metastasis, and drug resistance of various types of tumors.8 Moreover, lncRNA has been reported to be a tumor-specific prognostic biomarker for cancers. Previous meta-analyses indicated that PCAT-1,9 CRNDE,10 ZEB1-AS1,11 and PVT1 are related to the clinicopathologic features and prognosis of tumors,12 and lncRNA may be a reliable predictive biomarker for tumors. HOXD cluster antisense RNA 1 (HOXD-AS1), also known as HAGLR, is a noncoding RNA that is transcribed in an antisense manner from the HOXD cluster on human chromosome 2q31.2 and is evolutionarily conserved.13 More and more studies have reported that HOXD-AS1 was overexpressed in various tumors, such as hepatocel-lular carcinoma,14 gastric cancer,15 colorectal cancer,16 and glioma,17 and is associated with tumor proliferation, invasion, and metastasis. Further, there is increasing evidence that high expression of HOXD-AS1 is related to clinicopathologic features such as tumor size, lymph node metastasis, differentiation and TNM stage, and prognosis.18,19 However, seeing as how the current studies included a limited sample size and the results of clinicopathologic features were inconsistent, a meta-analysis of existing literatures was conducted to investigate the relationship between HOXD-AS1 and clinicopathologic characteristics and prognosis. The TCGA dataset was also used to analyze the prognostic value of HOXD-AS1 in different tumors.
Materials and methods
Search strategy and study selection
PubMed, Medline, Cochrane Library, Web of Science, EMBASE database, Ovid, Chinese CNKI, and Chinese WanFang databases were searched in this study, the research ended on August 10, 2018, with data from any time prior to that being eligible for inclusion. The key words for searches were “HOXD-AS1” OR “HAGLR” OR “HOXD cluster antisense RNA 1” OR “Long noncoding RNA HOXD cluster antisense RNA 1” OR “lncRNA HOXD-AS1” AND “cancers” OR “neoplasm.”
In this meta-analysis, the inclusion criteria were as follows: 1) the expression of HOXD-AS1 in cancer patients was examined in tumor tissues by qRT-PCR or RNA-seq dataset; 2) the patients in the literature had a definitive diagnosis of cancer and the relationship between HOXD-AS1 and clinicopathologic or survival information was described; 3) HRs for overall survival (OS) were provided or could be calculated by survival curves; and 4) if it was a repeated study, the most recent paper was included.
The exclusion criteria were as follows: 1) non-human subject studies; 2) the HRs cannot be calculated based on the data in articles; 3) reviews, case reports, letters, editorials, conference reports, and laboratory articles.
Data extraction and quality assessment
Two investigators (FZ and XC) independently searched and assessed the literatures according to the aforementioned criteria. The first author, year of publication, country, cutoff value, number of cases, clinicopathologic features, and OS data were included in the extracted data. We used Newcastle– Ottawa Scale criteria to assess the quality of studies; when the Newcastle–Ottawa Scale (NOS) score was six or higher, these articles were considered as high quality, otherwise it was seen as a low-quality study.
Public data and tools
This study meets the publication guidelines provided by TCGA (http://cancergenome.nih.gov/publications/publicationguidelines). TCGA Data portal (https://portal.gdc.cancer.gov) and UCSC Xena project (https://xena.ucsc.edu) were used to extract the RNAseqV2 and clinical data. GEPIA was used to analyze the data as described by Liang et al.20 Differential expression analysis was performed through one-way ANOVA, and survival analysis was calculated by Kaplan–Meier method and logrank test; the HR and 95% CI are shown in the figure of Kaplan–Meier curves.
Statistical analysis
STATA 14.2 software (StataCorp LLC, College Station, TX, USA) was used to calculate the pooled ORs and HRs with 95% CIs. Engauge Digitizer 10.0 software was used to extract the survival data from Kaplan–Meier curves. The relationship between HOXD-AS1 and clinicopathologic characteristics (gender [male vs female], age [older vs younger], tumor size [larger vs smaller], differentiation [low vs high + moderate], lymph node metastasis [yes vs no], TNM stage [I+II vs III+IV]), and OS was calculated, where the fixed or random effects models were used when I2 was >50% or <50%. If the 95% CI did not overlap 1, the pooled HR or OR was considered to be statistically significant. Funnel plots and Begg’s test were used to evaluate the potential publication bias, and the sensitivity analysis and subgroup analysis were performed to examine the source of heterogeneity and stability of results.
Results
Study identification and characteristics
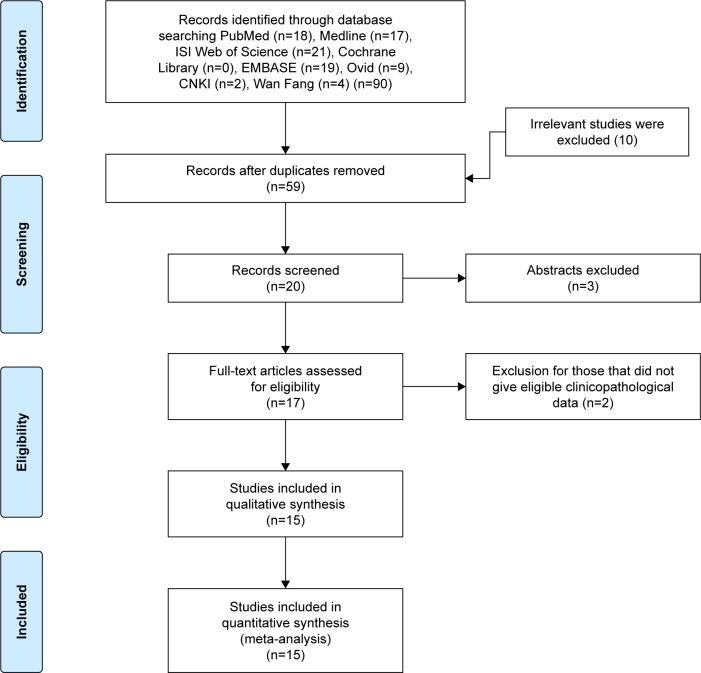
As shown in Figure 1, a total of 89 potential studies were collected from the database, and 69 studies were removed as these studies were non-HOXD-AS1-related, duplicated, or did not involve tests in tumor tissues after their titles and abstracts were reviewed; a total of 20 studies were evaluated by reading the full text and five studies were excluded because of having insufficient information. Fifteen eligible studies14–16,18,19,21–30 with 1,678 patients were ultimately included in this meta-analysis. As shown in Table 1, these studies were published from 2015 to 2018 and had sample sizes ranging from 25 to 369; all the studies were from People’s Republic of China, and 14 studies were published in English, while one was written in Chinese. All the studies had high qualities according to the NOS score.
Figure 1.
Flow diagram of study selection.
Table 1.
Characteristics of studies included in the meta-analysis
| Study | Year | Country | Sample size | Tumor type | Cutoff value | Laboratory method | Gender male (+/−)/female (+/−) | Age old (+/−)/young (+/−) | Tumor size big (+/−)/small (+/−) | Differentiation low (+/−)/high and moderate (+/−) | Lymph node metastasis yes (+/−)/no (+/−) | UICC stage I, II (+/−)/III, IV (+/−) | Survival information | HR | NOS score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| Lu S | 2017 | People’s Republic of China | 47 | HCC | Median | qRT-PCR | 15/19 | 16/5 | 11/22 | 26/3 | NA | 10/9 | NA | NA | 6 |
| 5/8 | 18/8 | 7/7 | 8/10 | 24/4 | |||||||||||
| Li JF | 2016 | People’s Republic of China | 50 | BC | NA | qRT-PCR | 19/17 | 12/7 | 24/8 | 9/13 | 5/1 | 9/20 | NA | NA | 6 |
| 9/5 | 19/17 | 6/12 | 20/18 | 29/15 | 15/6 | ||||||||||
| Gu P | 2017 | People’s Republic of China | 374 | PC | NA | RNA-Seq | NA | 124/81 | NA | NA | 36/10 | 89/72 | OS | 1.47 (1.01–2.14) (R) | 8 |
| 105/58 | 168/102 | 141/66 | |||||||||||||
| Lu CW | 2017 | People’s Republic of China | 60 | NSCLC | NA | qRT-PCR | 12/19 | 9/18 | 11/23 | NA | 17/12 | 10/32 | OS | 1.37 (1.11–3.07) (C) | 6 |
| 10/19 | 13/20 | 11/15 | 5/26 | 12/6 | |||||||||||
| Wang H | 2017 | People’s Republic of China | 120 | HCC | Median | qRT-PCR | 8/13 | 25/24 | 27/23 | NA | NA | 54/60 | OS | 1.64 (1.09–2.47) (R) | 8 |
| 52/45 | 35/36 | 33/37 | 6/0 | ||||||||||||
| Wang QH | 2017 | People’s Republic of China | 87 | NSCLC | Median | qRT-PCR | 29/21 | 28/24 | 30/12 | NA | NA | 18/22 | OS | 1.92 (1.20–3.71) (C) | 8 |
| 21/17 | 21/14 | 19/26 | 31/16 | ||||||||||||
| Hu YC | 2017 | People’s Republic of China | 122 | CC | NA | qRT-PCR | NA | 36/25 | NA | NA | 18/5 | 27/41 | NA | NA | 6 |
| 31/30 | 49/50 | 40/14 | |||||||||||||
| Zhang HL | 2017 | People’s Republic of China | 25 | Melanoma | Median | qRT-PCR | NA | NA | NA | NA | NA | NA | OS | 2.28 (1.30–5.78) (C) | 6 |
| Zhang Y | 2017 | People’s Republic of China | 43 | EOC | Median | qRT-PCR | NA | 13/14 | 12/13 | NA | 8/2 | 3/10 | OS | 2.01 (1.06–5.53) (C) | 6 |
| 9/7 | 8/10 | 14/19 | 19/11 | ||||||||||||
| Zheng L | 2017 | People’s Republic of China | 104 | GC | Median | qRT-PCR | 33/24 | 28/25 | 39/16 | NA | 34/16 | 19/31 | NA | NA | 7 |
| 26/21 | 31/20 | 20/29 | 25/29 | 40/14 | |||||||||||
| Qu Y | 2018 | People’s Republic of China | 46 | OS | Median | qRT-PCR | 13/9 | NA | 11/7 | 31/21 | 10/1 | 17/19 | OS | 1.37 (1.03–5.94) (C) | 7 |
| 14/10 | 16/12 | 28/18 | 17/18 | 10/0 | |||||||||||
| Wang YY | 2018 | People’s Republic of China | 369 | OS | Median | RNA-Seq | NA | NA | NA | NA | NA | NA | OS | 1.68 (1.01–2.79) (C) | 7 |
| Li X | 2018 | People’s Republic of China | 136 | CRC | Median | qRT-PCR | 38/40 | 38/34 | 37/28 | 35/10 | 46/45 | 14/28 | OS | 1.67 (1.55–2.61) (C) | 8 |
| 30/28 | 30/34 | 31/40 | 33/58 | 22/23 | 54/40 | ||||||||||
| Xia H | 2018 | People’s Republic of China | 52 | NSCLC | Median | qRT-PCR | 14/14 | 17/15 | 11/7 | NA | 11/1 | 16/22 | OS | 1.54 (1.01–2.75) (R) | 8 |
| 14/10 | 11/9 | 17/17 | 17/23 | 12/2 | |||||||||||
| Gu WF | 2018 | People’s Republic of China | 43 | OS | Median | qRT-PCR | 15/11 | 10/13 | 19/9 | NA | NA | 4/10 | OS | 1.81 (1.03–5.18) (C) | 6 |
| 9/8 | 10/13 | 19/9 | NA | NA | 4/10 | OS | 1.81 (1.03–5.18) (C) | 6 | |||||||
Abbreviations: BC, bladder cancer; C, HR was estimated by curve; CC, cervical cancer; CRC, colorectal cancer; EOC, epithelial ovarian cancer; GC, gastric cancer; HCC, hepatocellular carcinoma; NSCLC, non-small-cell lung cancer; OS, osteosarcoma; PC, prostate cancer; R, HR was reported.
Association between HOXD-AS1 and clinicopathologic characteristics
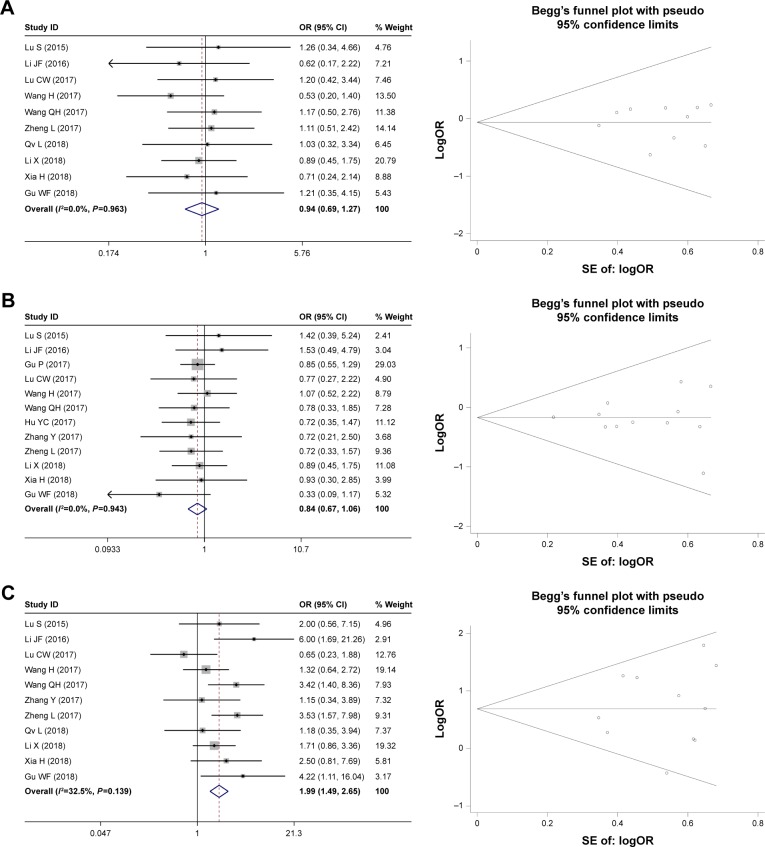
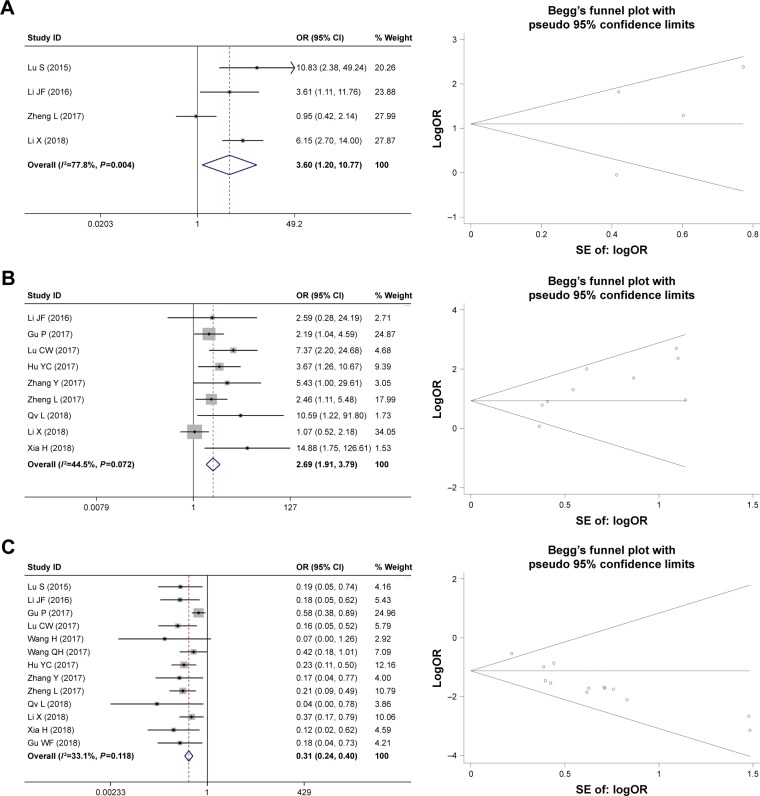
In this meta-analysis, the pooled ORs with 95% CI are shown in Table 2. Eleven studies reporting 788 patients assessed the relationship between HOXD-AS1 expression and tumor size; the results showed that the high expression of HOXD-AS1 was associated with tumor size (large:small, OR = 1.99, 95% CI = [1.49, 2.65], P<0.001, fixed effect). Similarly, four studies containing 331 patients with differentiation were included. The analysis showed that a higher expression of HOXD-AS1 was related to low differentiation (low:high + moderate, OR = 3.02, 95% CI = [1.90, 4.81], P<0.001, random effect). Further, the pooled ORs also demonstrated that HOXD-AS1 was correlated with lymph node metastasis (yes:no, OR = 2.69, 95% CI = [1.91, 3.79], P<0.001, fixed effect) and advanced TNM stage (I+II:III+IV, OR = 0.31, 95% CI = [0.24, 0.40], P<0.001, fixed effect). However, the pooled results demonstrated that the expression of HOXD-AS1 was not related to gender (male:female, OR = 0.94, 95% CI = [0.69, 1.27], P=0.677, fixed effect) or age (older:younger, OR = 0.84, 95% CI = [0.67, 2.65], P<0.001, fixed effect). Therefore, our results indicated that the high HOXD-AS1 expression significantly increased the risk of worse clinicopathologic features. Begg’s funnel plot was performed to evaluate publication bias (Figures 2 and 3). There was no publication bias for gender (P=0.858), age (P=0.732), tumor size (P=0.648), differentiation (P=0.734), lymph node metastasis (P=0.076), or TNM stage (P=0.057).
Table 2.
LncRNA HOXD-AS1 clinicopathologic features for cancers
| Heterogeneity
| |||||||
|---|---|---|---|---|---|---|---|
| Clinicopathologic features | No of studies | No of patients | Pooled OR (95% CI) | PHet | I2 (%) | P-value | Model used |
|
| |||||||
| Gender | 10 | 743 | 0.94 (0.69, 1.27) | 0.963 | 0.0 | 0.677 | Fixed |
| Age | 12 | 1,237 | 0.84 (0.67, 1.06) | 0.943 | 0.0 | 0.141 | Fixed |
| Tumor size | 11 | 788 | 1.99 (1.49, 2.65) | 0.139 | 32.5 | ,0.001 | Fixed |
| Differentiation | 4 | 331 | 3.02 (1.90, 4.81) | 0.005 | 77.8 | ,0.001 | Random |
| Lymph node metastasis | 9 | 919 | 2.69 (1.91, 3.79) | 0.072 | 44.5 | ,0.001 | Fixed |
| TNM stage | 13 | 1,268 | 0.31 (0.24, 0.40) | 0.118 | 33.1 | ,0.001 | Fixed |
Abbreviations: Fixed, fixed-effects model; NOS, Newcastle–Ottawa Scale; PHet, probability of heterogeneity; Random, random-effects model.
Figure 2.
Forest plot and Begg’s publication bias plots of studies evaluating the relationship between HOXD-AS1 expression and clinicopathologic features.
Note: (A) Gender, (B) age, and (C) tumor size.
Figure 3.
Begg’s publication bias plots and the forest plot of published articles evaluating the relationship between HOXD-AS1 expression and (A) differentiation, (B) lymph node metastasis, and (C) TNM stage.
Note: Weights are from random effects analysis.
Association between HOXD-AS1 and OS rate
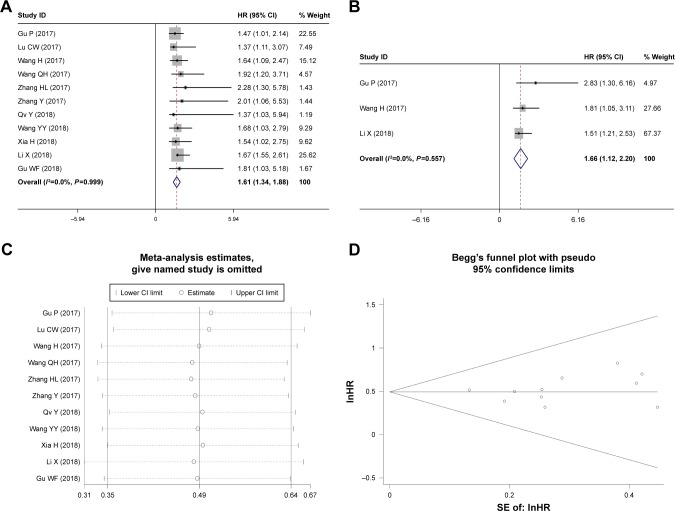
Eleven studies reporting 1,349 patients with OS were included according to different expressions of HOXD1-AS1. The fixed-effects model was used since no significant heterogeneity was observed; the pooled HRs indicated that the high HOXD-AS1 expression was related with a worse survival (HR = 1.61, 95% CI = [1.34, 1.88], P<0.001, fixed effect, Figure 4A).
Figure 4.
Forest plot of studies evaluating (A) the relationship between HOXD-AS1 expression and overall survival (OS) rate, (B) independent prognostic value, (C) Begg’s publication bias plots of OS, and (D) sensitivity analysis for OS.
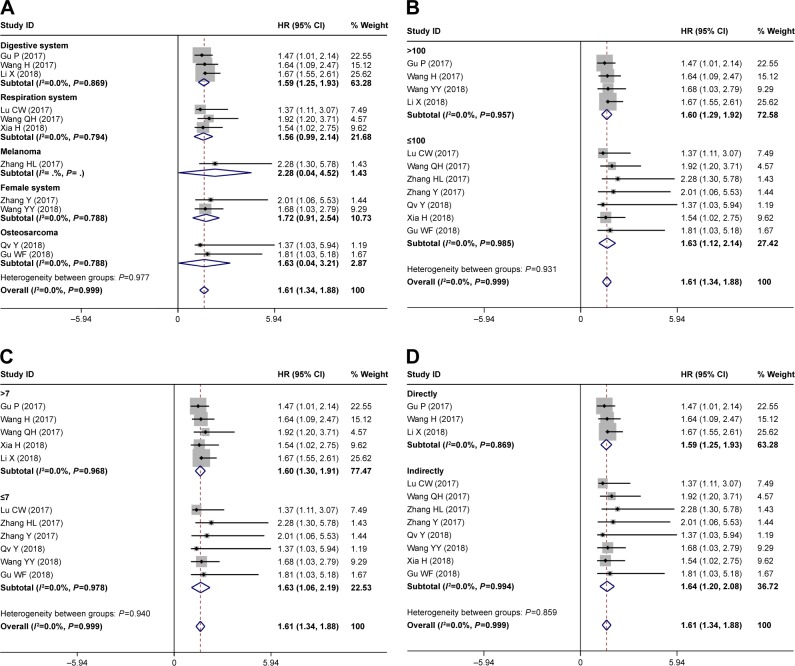
To further explore the relationship between HOXD-AS1 and OS, subgroup analyses were performed based on the tumor type, sample size, NOS score, and HR estimation method. As shown in Table 3, all of the subgroup analyses demonstrated that the expression of HOXD-AS1 was related to worse OS according to sample size (Figure 5B), NOS score (Figure 5C), and HR estimation method (Figure 5D). However, the subgroup analysis for tumor type indicated that the expression of HOXD-AS1 was only related to a worse OS in the digestive system (HR = 1.59, 95% CI = [1.25, 1.93], P<0.001, fixed effect, Figure 5A). It should be noted that HOXD-AS1 was not correlated with OS in the respiration system, melanoma, female system, or osteosarcoma, indicating that more studies should be included to verify this conclusion. The robustness of the pooled results was evaluated by sensitivity analysis – the results (Figure 4C) indicated that the results were reliable. Begg’s funnel plot was performed to evaluate publication bias; there was no publication bias for OS (P=0.436, Figure 4D).
Table 3.
Subgroup analysis of overall survival by tumor type, NOS score, sample size
| Subgroups | No of studies | No of patients | Pooled HR (95% CI) | PHet | I2 (%) | P-value |
|---|---|---|---|---|---|---|
|
| ||||||
| Tumor type | ||||||
| Respiratory system | 2 | 137 | 1.56 (0.99, 2.14) | 0.794 | 0.0 | >0.05 |
| Digestive system | 3 | 624 | 1.59 (1.25, 1.93) | 0.869 | 0.0 | <0.05 |
| Female reproductive system | 2 | 412 | 1.73 (0.90, 2.55) | 0.788 | 0.0 | >0.05 |
| Osteosarcoma | 2 | 89 | 1.63 (0.04, 3.21) | 0.788 | 0.0 | >0.05 |
| Melanoma | 1 | 25 | 1.72 (0.04, 3.21) | – | – | >0.05 |
| Sample size | ||||||
| ≤100 | 7 | 356 | 1.63 (1.34, 1.88) | 0.985 | 0.0 | <0.05 |
| >100 | 4 | 993 | 1.60 (1.29, 1.92) | 0.957 | 0.0 | <0.05 |
| NOS score | ||||||
| ≤7 | 6 | 586 | 1.63 (1.06, 2.19) | 0.978 | 0.0 | <0.05 |
| >7 | 5 | 763 | 1.60 (1.30, 1.91) | 0.968 | 0.0 | <0.05 |
| HR estimation method | ||||||
| Directly | 3 | 624 | 1.59 (1.25, 1.93) | 0.869 | 0.0 | <0.05 |
| Indirectly | 8 | 725 | 1.64 (1.20, 1.88) | 0.994 | 0.0 | <0.05 |
Abbreviation: NOS, Newcastle–Ottawa Scale.
Figure 5.
(A) Forest plots of subgroup analyses for OS by tumor type, (B) subgroup analysis by sample size, (C) subgroup analysis by Newcastle–Ottawa Scale score and (D) HR estimation method.
As shown in Figure 4B, three studies with 624 patients were included in this meta-analysis to explore the independent prognostic role of HOXD-AS1 for cancer patients. The pooled results reported that the expression of HOXD-AS1 was an independent prognostic factor for the OS of patients (HR = 1.66, 95% CI = [1.12, 2.20], P<0.001, fixed effect, Figure 5B). There was no publication bias according to Begg’s funnel plot (P=0.296).
Validation of the results in TCGA dataset
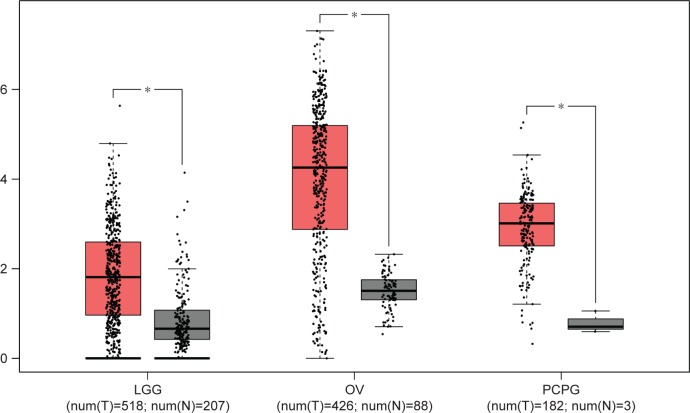
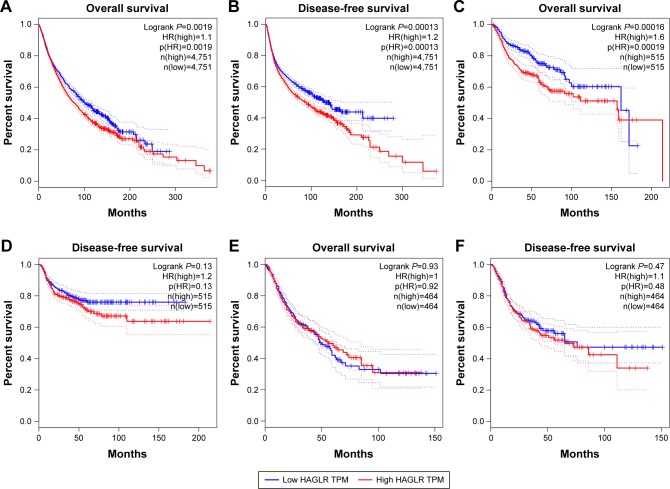
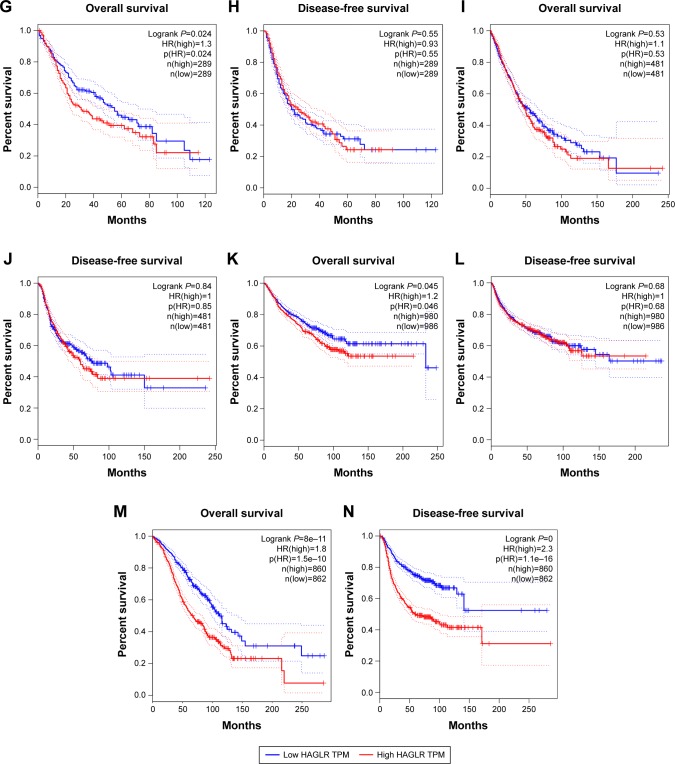
We first explored the expression of HOXD-AS1 in all kinds of related cancers using data from TCGA. As shown in Figure 6, HOXD-AS1 was also found to be overexpressed in brain lower grade glioma, ovarian serous cystadenocarcinoma, and pheochromocytoma and paraganglioma (|Log2FC|cutoff >1, q-value <0.01). We then merged the expression data and OS data (disease-free survival [DFS] data) of cancers from all of the TCGA dataset from GEPIA, which includes the respiratory system, urinary system, female reproductive system, blood system, and digestive system. Those included contain adrenocortical carcinoma, bladder urothelial carcinoma, breast invasive carcinoma, cervical squamous cell carcinoma and endocervical adenocarcinoma, cholangiocarcinoma, colon adenocarcinoma, lymphoid neoplasm diffuse large B-cell lymphoma, esophageal carcinoma, glioblastoma multiforme, head and neck squamous cell carcinoma, kidney chromophobe, kidney renal clear cell carcinoma, kidney renal papillary cell carcinoma, acute myeloid leukemia, brain lower grade glioma, liver hepatocellular carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, mesothelioma, ovarian serous cystadenocarcinoma, pancreatic adenocarcinoma, pheochromocytoma and paraganglioma, prostate adenocarcinoma, rectum adenocarcinoma, sarcoma, skin cutaneous melanoma, stomach adenocarcinoma, testicular germ cell tumors, thyroid carcinoma, thymoma, uterine corpus endometrial carcinoma, uterine carcinosarcoma, and uveal melanoma. These included 9,502 cancer patients, which were then divided into high or low expression groups based on the median HOXD-AS1 expression. The results indicated that the high expression of HOXD-AS1 denoted a worse OS (Figure 7A) and DFS (Figure 7B), confirming that an upregulated expression of HOXD-AS1 was associated with OS and DFS in cancer patients. The role of HOXD-AS1 in different kinds of cancers was then analyzed. As shown in Figure 7, the expression of HOXD-AS1 was related to OS in the head and neck cancer system (Figure 7C), hepatobiliary and pancreatic system (Figure 7I), urinary system (Figure 7K), and female reproductive system (Figure 7M). However, the HOXD-AS1 expression was not associated with OS in gastrointestinal tumors (Figure 7E) or the respiratory system (Figure 7I). The high expression of HOXD-AS1 was not significantly associated with poor DFS in head and neck cancer system (Figure 7D), hepatobiliary and pancreatic system (Figure 7F), gastrointestinal tumors (Figure 7H), respiratory system (Figure 7J), or urinary system (Figure 7L). On the contrary, the expression of HOXD-AS1 was associated with DFS in the female reproductive system (Figure 7N).
Figure 6.
The expression levels of HOXD-AS1 in LGG (brain lower grade glioma), OV (ovarian serous cystadenocarcinoma), and PCPG (pheochromocytoma and paraganglimoma).
Notes: “*” indicates a log2FC value >1 and P-value <0.01 in The Cancer Genome Atlas cohort.
Figure 7.
Validation of HOXD-AS1 in TCGA cohort.
Notes: (A) Overall survival plots of HOXD-AS1 in TCGA cohort (n=9,502, logrank P<0.01). (B) Disease-free survival plots of HOXD-AS1 in TCGA cohort (n=9,502, logrank P<0.01). (C) Overall survival plots of HOXD-AS1 in TCGA cohort in head and neck tumor (n=1,030, logrank <0.01). (D) Disease-free survival plots of TCGA cohort in head and neck tumor (n=1,030, logrank P=0.13). (E) Overall survival plots of HOXD-AS1 in TCGA cohort in the gastrointestinal tumor system (n=928, logrank =0.93). (F) Disease-free survival plots of TCGA cohort in gastrointestinal tumors (n=928, logrank P=0.47). (G) Overall survival plots of HOXD-AS1 in the TCGA cohort in hepatobiliary and pancreatic system (n=578, logrank =0.024). (H) Disease-free survival plots of TCGA cohort in the hepatobiliary and pancreatic system (n=578, logrank P=0.55). (I) Overall survival plots of HOXD-AS1 in TCGA cohort in the respiratory system (n=962, logrank =0.53). (J) Disease-free survival plots of TCGA cohort in the respiratory system (n=962, logrank P=0.84). (K) Overall survival plots of HOXD-AS1 in TCGA cohort in the urinary system (n=1,966, logrank =0.045). (L) Disease-free survival plots of TCGA cohort in the urinary system (n=1,966, logrank =0.68). (M) Overall survival plots of HOXD-AS1 in TCGA cohort in the female reproductive system (n=1,762, logrank <0.01). (N) Disease-free survival plots of TCGA cohort in the female reproductive system (n=1,762, logrank <0.01).
Abbreviation: TCGA, The Cancer Genome Atlas.
Discussion
Accumulating evidence has shown that HOXD-AS1 was upregulated in various cancers. A meta-analysis was performed to clarify the association between HOXD-AS1 and clinicopathologic and prognostic values in cancers. The results indicated that the risk of lymph node metastasis in high expression was 2.69 times higher than those in low HOXD-AS1 expression. Similarly, the risk of developing into lower differentiation, larger tumor size, and advanced TNM stage were higher than those in the low expression. The pooled results showed that the high expression of HOXD-AS1 was related to a worse OS, and HOXD-AS1 could be an independent role for the prognosis of cancers. This meta-analysis is the first review to assess the relationship between HOXD-AS1 and the prognosis of patients in different kinds of cancers. We used the TCGA dataset to explore the prognostic value of HOXD-AS1 in cancers, with the results suggesting that the expression of HOXD-AS1 may serve as a reliable biomarker for the prognosis of cancers.
Previous studies also explore the mechanism of HOXD-AS1 in different kinds of cancers,31 such as cell proliferation, invasion, metastasis, and apoptosis. In the digestive system, Zheng et al15 reported that the downregulation of HOXD-AS1 inhibited the cell growth by inactivating the janus kinase 2/signal transducer and activator of transcription 3 pathway in vitro and in vivo in gastric cancer. In hepatocellular carcinoma, Lu et al31 found that HOXD-AS1 promoted HCC metastasis through a HOXD-AS1/miR-19a/ARHGAP11A signal axis. Further, Wang et al14 indicated that the knockdown of HOXD-AS1 inhibited migration and invasion, and HOXD-AS1 could competitively bind to miR-130a-3p to prevent SOX4. In colorectal cancer, Li et al16 demonstrated that HOXD-AS1 could function as a competing endogenous RNA for miR-217 and promote CRC progression and metastasis. In the female reproductive system, Hu et al25 reported that the downregulation of HOXD-AS1 could suppress the cell growth of cervical cancer via the Ras/ERK signaling pathway. Chi et al32 found that HOXD-AS1 could act as a ceRNA of miR-130a-3p to upregulate the expression of ZEB1, enhancing the chemoresistance of cisplatin-resistant cervical cancer cells. To continue, Wang et al28 indicated that HOXD-AS1 could competitively bind to miR-608 and regulate the expression of frizzled family receptor 4 (FZD4) to promote cell proliferation, invasion, and migration of ovarian cancer. In the respiratory system, Wang et al24 found that HOXD-AS1 could negatively regulate the expression of miR-147a and positively regulate the expression of pRB, which can promote the proliferation of non-small-cell lung cancer. Xia et al29 reported that miR-133b was a downstream target of HOXD-AS1, and a knockdown of HOXD-AS1 could inhibit the proliferation, migration, and invasion of non-small-cell lung cancer cells. The researchers also explored the mechanism of HOXD-AS1 in prostate cancer,22 bladder cancer,18 glioma,17 and osteosarcoma,27 and similar results were reported. All the studies indicated that HOXD-AS1 could play an oncogenic role in the progression of cancer. The mechanism of HOXD-AS1 in different kinds of cancers was summarized in Table 4.
Table 4.
Summary of HOXD-AS1 with their potential targets, pathways, and related microRNAs
| Cancer type | Expression | Functional role | Related microRNAs | Downstream molecules | Protein binding | Signaling pathway |
|---|---|---|---|---|---|---|
|
| ||||||
| Bladder cancer | Upregulation | Cell proliferation, migration, apoptosis | / | / | / | / |
| Gastric cancer | Upregulation | Cell growth | / | p-JAK2 p-STAT3 | / | JAK/STAT pathway |
| Prostate cancer | Upregulation | Cell proliferation | / | PLK1/AURKA/FOXM1/UBE2C/CCNA2 | WDR5 | / |
| Hepatocellular carcinoma | Upregulation | Cell metastasis, apoptosis | miR-19a/miR-130a-3p | ARHGAP11A/EZH2/MMP2 | SOX4 | / |
| NSCLC | Upregulation | Cell proliferation, apoptosis, migration, invasion | miR-147a/miR-133b | / | pRB/MMP-9 | / |
| Cervical cancer | Upregulation | Cell growth | / | p-ERK1/2 | / | Ras/ERK signaling |
| Epithelial ovarian cancer | Upregulation | Cell proliferation, invasion, EMT | miR-133a-3p | E-cadherin/vimentin/β-catenin/cyclin-D1/c-myc | / | Wnt/β-catenin signaling |
| Melanoma | Upregulation | Cell proliferation, invasion | / | / | RUNX3 | / |
| Glioma | Upregulation | Migration, invasion | miR-130a | / | E2F8 | / |
| Ovarian cancer | Upregulation | Cell proliferation, migration, invasion | miR-608 | / | FZD4 | / |
| Osteosarcoma | Upregulation | Cell proliferation, migration, invasion, G0/G1 phase arrest | / | MMP-2/Bcl-2/cyclinD1/STAT3/EZH2 | P57 | / |
| Colorectal cancer | Upregulation | Cell proliferation, cell invasion, EMT and stem cell formation | miR-217 | AEG-1/EZH2 | / | / |
Notes: “/” indicates data not available.
Abbreviation: EMT, epithelial-mesenchymal transition.
There are some limitations in this meta-analysis. First, all the studies included were from People’s Republic of China, so the results may only be suitable for Chinese or Asian populations. The TCGA dataset makes up this disadvantage, though it should be noted that some of the results of the published articles and TCGA dataset were inconsistent, such as with the role of HOXD-AS1 in digestive tumors, which needs additional published articles to explore this conclusion. Second, the number of patients and tumor types included in this meta-analysis are limited, so more studies with larger sample sizes and various cancers should be included to support the results. Third, the HRs in many articles are not specifically provided, so we extracted the survival information from K-M curves, potentially creating some errors. Fourth, the TCGA dataset and subgroup analysis of OS revealed that the expression of HOXD-AS1 was not significantly associated with OS and DFS in every kind of tumor, so more studies should be included to verify these results.
Conclusion
Despite the above limitations, this meta-analysis concluded that the high expression of HOXD-AS1 was associated with large tumor size, lower differentiation, increased lymph node metastasis, and advanced TNM stage. Further, HOXD-AS1 was associated with a poor OS and DFS, indicating that HOXD-AS1 may be a potential prognostic biomarker for cancer patients, though more studies with larger sample sizes and various tumor types are needed.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 4.Avila MA, Berasain C, Sangro B, Prieto J. New therapies for hepatocellular carcinoma. Oncogene. 2006;25(27):3866–3884. doi: 10.1038/sj.onc.1209550. [DOI] [PubMed] [Google Scholar]
- 5.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Sotillo E, Thomas-Tikhonenko A. The long reach of noncoding RNAs. Nat Genet. 2011;43(7):616–617. doi: 10.1038/ng.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23(13):1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang C, Qi Z, Ge H, et al. Long non-coding RNA PCAT-1 in human cancers: a meta-analysis. Clin Chim Acta. 2018;480:47–55. doi: 10.1016/j.cca.2018.01.043. [DOI] [PubMed] [Google Scholar]
- 10.Liang C, Zhang B, Ge H, Xu Y, Li G, Wu J. Long non-coding RNA CRNDE as a potential prognostic biomarker in solid tumors: a meta-analysis. Clin Chim Acta. 2018;481:99–107. doi: 10.1016/j.cca.2018.02.039. [DOI] [PubMed] [Google Scholar]
- 11.Liang C, Liu J, Ge H, Xu Y, Li G, Wu J. The clinicopathological and prognostic value of long non-coding RNA ZEB1-AS1 in solid tumors: a meta-analysis. Clin Chim Acta. 2018;484:91–98. doi: 10.1016/j.cca.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 12.Liu C, Jin J, Liang D, et al. Long noncoding RNA PVT1 as a novel predictor of metastasis, clinicopathological characteristics and prognosis in human cancers: a meta-analysis. Pathol Oncol Res. 2018 Aug 6; doi: 10.1007/s12253-018-0451-3. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yarmishyn AA, Batagov AO, Tan JZ, et al. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genomics. 2014;15(Suppl 9):S7. doi: 10.1186/1471-2164-15-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Huo X, Yang XR, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. doi: 10.1186/s12943-017-0680-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng L, Chen J, Zhou Z, He Z. Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer cell growth via inactivating the JAK2/STAT3 pathway. Tumour Biol. 2017;39(5):1010428317705335. doi: 10.1177/1010428317705335. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Zhao X, Yang B, et al. Long non-coding RNA HOXD-AS1 promotes tumor progression and predicts poor prognosis in colorectal cancer. Int J Oncol. 2018;53(1):21–32. doi: 10.3892/ijo.2018.4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Zhao F, Cui D, et al. HOXD-AS1/miR-130a sponge regulates glioma development by targeting E2F8. Int J Cancer. 2018;142(11):2313–2322. doi: 10.1002/ijc.31262. [DOI] [PubMed] [Google Scholar]
- 18.Li J, Zhuang C, Liu Y, et al. Synthetic tetracycline-controllable shRNA targeting long non-coding RNA HOXD-AS1 inhibits the progression of bladder cancer. J Exp Clin Cancer Res. 2016;35(1):99. doi: 10.1186/s13046-016-0372-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang H, Bai M, Zeng A, Si L, Yu N, Wang X. LncRNA HOXD-AS1 promotes melanoma cell proliferation and invasion by suppressing RUNX3 expression. Am J Cancer Res. 2017;7(12):2526–2535. [PMC free article] [PubMed] [Google Scholar]
- 20.Liang J, Wei X, Liu Z, et al. Long noncoding RNA CYTOR in cancer: a TCGA data review. Clin Chim Acta. 2018;483:227–233. doi: 10.1016/j.cca.2018.05.010. [DOI] [PubMed] [Google Scholar]
- 21.Shan L. Research on the Mechanism of Long Noncoding RNA HOXD-AS1 in Hepatocellular Carcinoma [Thesis] China: The Second Military Medical University; 2015. Chinese. [Google Scholar]
- 22.Gu P, Chen X, Xie R, et al. LncRNA HOXD-AS1 regulates proliferation and chemo-resistance of castration-resistant prostate cancer via recruiting WDR5. Mol Ther. 2017;25(8):1959–1973. doi: 10.1016/j.ymthe.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu C, Ma J, Cai D. Increased HAGLR expression promotes non-small cell lung cancer proliferation and invasion via enhanced de novo lipo-genesis. Tumour Biol. 2017;39(4):1010428317697574. doi: 10.1177/1010428317697574. [DOI] [PubMed] [Google Scholar]
- 24.Wang Q, Jiang S, Song A, et al. HOXD-AS1 functions as an oncogenic ceRNA to promote NSCLC cell progression by sequestering miR-147a. Onco Targets Ther. 2017;10:4753–4763. doi: 10.2147/OTT.S143787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu YC, Wang AM, Lu JK, Cen R, Liu LL. Long noncoding RNA HOXD-AS1 regulates proliferation of cervical cancer cells by activating Ras/ERK signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(22):5049–5055. doi: 10.26355/eurrev_201711_13817. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Y, Dun Y, Zhou S, Huang XH. LncRNA HOXD-AS1 promotes epithelial ovarian cancer cells proliferation and invasion by targeting miR-133a-3p and activating Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2017;96:1216–1221. doi: 10.1016/j.biopha.2017.11.096. [DOI] [PubMed] [Google Scholar]
- 27.Qu Y, Zheng S, Kang M, et al. Knockdown of long non-coding RNA HOXD-AS1 inhibits the progression of osteosarcoma. Biomed Pharmacother. 2018;98:899–906. doi: 10.1016/j.biopha.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Zhang W, Wang Y, Wang S. HOXD-AS1 promotes cell proliferation, migration and invasion through miR-608/FZD4 axis in ovarian cancer. Am J Cancer Res. 2018;8(1):170–182. [PMC free article] [PubMed] [Google Scholar]
- 29.Xia H, Jing H, Li Y, Lv X. Long noncoding RNA HOXD-AS1 promotes non-small cell lung cancer migration and invasion through regulating miR-133b/MMP9 axis. Biomed Pharmacother. 2018;106:156–162. doi: 10.1016/j.biopha.2018.06.073. [DOI] [PubMed] [Google Scholar]
- 30.Gu W, Zhang E, Song L, et al. Long noncoding RNA HOXD-AS1 aggravates osteosarcoma carcinogenesis through epigenetically inhibiting p57 via EZH2. Biomed Pharmacother. 2018;106:890–895. doi: 10.1016/j.biopha.2018.06.173. [DOI] [PubMed] [Google Scholar]
- 31.Lu S, Zhou J, Sun Y, et al. The noncoding RNA HOXD-AS1 is a critical regulator of the metastasis and apoptosis phenotype in human hepatocellular carcinoma. Mol Cancer. 2017;16(1):125. doi: 10.1186/s12943-017-0676-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chi C, Mao M, Shen Z, Chen Y, Chen J, Hou W. HOXD-AS1 exerts oncogenic functions and promotes chemoresistance in cisplatin-resistant cervical cancer cells. Hum Gene Ther. 2018 Sep 05; doi: 10.1089/hum.2017.256. Epub. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]