Abstract
Genome-scale sequencing creates vast amounts of genomic data, increasing the challenge of clinical sequence variant interpretation. The demand for high-quality interpretation requires multiple specialties to join forces to accelerate the interpretation of sequence variant pathogenicity. With over 600 international members including clinicians, researchers, and laboratory diagnosticians, the Clinical Genome Resource (ClinGen), funded by the National Institutes of Health (NIH), is forming expert groups to systematically evaluate variants in clinically relevant genes. Here, we describe the first ClinGen Variant Curation Expert Panels (VCEPs), development of consistent and streamlined processes for establishing new VCEPs, and creation of standard operating procedures (SOPs) for VCEPs to define application of the ACMG/AMP guidelines for sequence variant interpretation in specific genes or diseases. Additionally, ClinGen has created user interfaces to enhance reliability of curation and a Sequence Variant Interpretation Working Group (SVI WG) to harmonize guideline specifications and ensure consistency between groups. The expansion of VCEPs represents the primary mechanism by which curation of a substantial fraction of genomic variants can be accelerated and ultimately undertaken systematically and comprehensively. We welcome groups to utilize our resources and become involved in our effort to create a publicly accessible, centralized resource for clinically relevant genes and variants.
Keywords: ClinGen, ClinVar, Expert Panels, ACMG/AMP guidelines, sequence variant interpretation, biocuration
Background
Genome-scale sequencing is increasingly being integrated into clinical care, though many challenges remain to full implementation of genomics in a clinical setting (Han et al., 2017; Manolio et al., 2013). Rapidly evolving sequencing technologies and increasing gene-disease associations in the literature are concomitantly increasing the demand for high-quality interpretation of sequence variant pathogenicity (Bowdin et al., 2016; Chong et al., 2015). Recent papers have shown that discordance between laboratories on variant classifications is often due to siloes of inaccessible data and a lack of uniform classification methods (Amendola et al., 2016; Bean & Hegde, 2016; Harrison et al., 2017), highlighting the need for a global repository of sequence variant data that has been consistently and transparently curated and expertly reviewed.
The Standards and Guidelines for the Interpretation of Sequence Variants developed by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) (Richards et al., 2015) provide a five-tier nomenclature for assertions about genetic variants with respect to Mendelian disorders: pathogenic (P), likely pathogenic (LP), uncertain significance (VUS), likely benign (LB), and benign (B). The expanding endorsement of the ACMG/AMP guidelines by international clinical laboratories has been important for fostering consistency and harmonization of sequence variant assessment within the clinical genetics community (Azzariti et al., 2018; Ellard et al., 2017; Ghouse et al., 2018; Harrison et al., 2017; Lebo et al., 2017). Many sources have highlighted the ability of this standardized interpretation approach to enable laboratories to resolve most discrepancies under a common framework (Amendola et al., 2016; Garber et al., 2016; Harrison et al., 2017; Lebo et al., 2017). However, since the ACMG/AMP guidelines were initially developed to be broadly applicable across different domains (and thus necessarily generic), there is a need to specify assertion criteria based on the unique features of particular genes or domains of interest (Gelb et al., 2018; Ghouse et al., 2018; Kelly et al., 2018).
A core goal of the Clinical Genome Resource (ClinGen) (Rehm et al., 2015) is expert interpretation of variants, which is accomplished by convening Variant Curation Expert Panels (VCEPs) that focus on a gene or group of genes. The VCEPS are tasked with providing specifications to the ACMG/AMP guidelines for their individual genes or diseases, interpreting variants according to these rules, and publishing the interpretations through NCBI’s publicly available ClinVar database (Landrum et al., 2016). In addition, the gene/disease level criteria specifications can then also be used by the community to enhance consistency in the interpretation of variants in these genes. To enable users of ClinVar to understand the level of evidence for each variant submission, ClinGen and ClinVar developed a rating system to represent the level of review supporting each assertion (ClinVar, 2017). VCEPs represent one of the highest levels of review in ClinVar, with a correspondingly high level of confidence in their assertions; therefore, applicants are expected to fulfill a rigorous set of requirements, including establishing a multi-institutional membership with diverse expertise, following a focused scope of work, developing a work schedule, identifying and resolving conflicts of interest, and providing detailed criteria for variant assessment (ClinGen, 2018).
Expert Panels that are launched as part of ClinGen are required to follow a timeline and accomplish additional requirements as part of the ClinGen approval process. During the initial phase of ClinGen (Nov. 2013 – Sept. 2017), several Expert Panels were formed, mostly within the context of overarching Clinical Domain Working Groups (CDWGs) that make up the ClinGen curation ecosystem (Milko et al., 2018). The first VCEPs organized by ClinGen included Inherited Cardiomyopathy/MYH7 (Cardiovascular CDWG) (Kelly et al., 2018), PAH (Inborn Errors of Metabolism CDWG), PTEN (Hereditary Cancer CDWG), and RASopathy (Gelb et al., 2018), the latter was established as a “stand alone” VCEP not associated with a CDWG. The experiences of these initial VCEPs contributed significantly to the development of formal requirements for the establishment of subsequent VCEPs and the stepwise approval process described below.
ClinGen intentionally did not specify a uniform approach to these early VCEPs to provide maximum flexibility and room for experimentation in this early phase of the effort. Consequently, VCEPs developed and piloted different organizational strategies to review, discuss, and decide on specifications of the ACMG/AMP guidelines for individual genes or diseases. Simultaneously, ClinGen established a Sequence Variant Interpretation Working Group (SVI WG) tasked with systematically reviewing the ACMG/AMP criteria to clarify and enable further quantification of individual criteria as well as harmonize the gene- and disease specifications made by the individual VCEPs. ClinGen leadership reviewed the different strategies and worked closely with the VCEPs to implement the most successful approaches. In this paper, we describe the first VCEPs and the subsequent iterative development of SOPs that ultimately coalesced into a consistent and streamlined ClinGen Expert Panel application process.
ClinGen Variant Curation Expert Panel Pilots
MYH7-Inherited Cardiomyopathies Variant Curation Expert Panel (Cardiovascular CDWG)
The MYH7/Inherited Cardiomyopathy VCEP formed under the leadership of the Cardiovascular CDWG Executive Committee, including members with appropriate expertise in cardiomyopathy and a balanced representation of nine clinicians, nine laboratory diagnosticians, and five researchers from fourteen institutions across six countries. Additional participants with expertise in statistics and bioinformatics periodically joined the group to provide guidance on systematically refining the preliminary rules for pilot testing. The full Cardiovascular Disease CDWG approved the initial specifications to the application of evidence for interpreting variants related to MYH7-associated cardiomyopathies and further refinements were made during webinars held twice per month to discuss discordances in the application of the rules and to resolve conflicts between reviewers.
The final MYH7/Inherited Cardiomyopathy guidelines were based on a pilot study of 60 MYH7 variants (Kelly et al., 2018). Internal laboratory data, such as proband count, segregation data, and de novo occurrence, were utilized for the classification of 25 of these pilot variants, emphasizing the importance of case-level data sharing. This work translated to seven variants moving from LP to P, and another five from VUS to LP, mirroring the experience of other consortia using internal data for reclassification (Furqan et al., 2017). Currently, the group is expanding the scope of its guideline specifications to include additional genes and has created a separate sub-committee (“variant curation committee”) that is solely dedicated to applying the newly created and approved rules to generate expert-panel approved variant submissions to ClinVar.
PAH-Inborn Errors of Metabolism Variant Curation Expert Panel (Metabolism CDWG)
The Inborn Errors of Metabolism (IEM) CDWG is comprised of members with balanced expertise including six clinicians (medical genetics, clinical biochemical genetics, genetic counseling), seven laboratory diagnosticians, and five researchers. The IEM CDWG initially decided to focus on genes included on newborn screening panels, using PAH as a pilot gene to establish gene-specific ACMG guidelines and their curation process. Importantly, the group collaborated with the developer of the BioPKU database (BioPKU 2018), which contains over 1000 variants found in PAH, with detailed biochemical and clinical details about the individuals in whom they were found (Blau, 2016).
The VCEP investigated different curation approaches to identify a scalable workflow. Initially, a CDWG co-chair along with one Executive Committee member completed curation of PAH variants, but this was quickly determined to be unsustainable. The review process then transitioned to include junior members of the group (e.g., junior faculty, genetics fellows, genetic counselors) paired with senior domain experts for pairwise curation and expert review. The PAH-specific guidelines included specifications to the ACMG-AMP criteria for the application PM2, PM3, PS2, PS3, PP4, and BS1 evidence tags, as described by (Zastrow et al., 2018) in this issue. A streamlined process for sustainable variant curation was later developed and finalized; junior curators are currently working independently and presenting their work and preliminary assertion for each variant to the full VCEP for review on biweekly videoconference calls.
PTEN Variant Curation Expert Panel (Hereditary Cancer CDWG)
The PTEN VCEP formed under the leadership of the Hereditary Cancer CDWG Executive Committee, and included six clinicians (clinical cancer genetics and genetic counseling), seven laboratory diagnosticians and six researchers, including experts in computational genomics and biostatistics, with significant PTEN-related expertise. Subspecialty working groups were assembled based on members’ expertise to review and present current PTEN-relevant knowledge for each of the ACMG/AMP criteria evidence types. This ensured that members represented the comprehensive background knowledge necessary for review and for making informed decisions regarding the utility and strength of criteria.
Based on presentations and discussions, the PTEN VCEP drafted and pilot-tested benign and pathogenic criteria as described by (Mester et al., 2018) in this issue. A final round of validation and testing of the specified criteria was performed using 11 variants of uncertain significance or with conflicting interpretations from ClinVar, and published variants with available functional data. Similar to other groups’ experiences, shared internal laboratory data enabled the reclassification of three variants, resolving two conflicts and reclassifying a VUS to likely benign. The PTEN VCEP will continue to conduct variant curation activities with quarterly review, and implement the curation approach developed by the PAH VCEP.
RASopathy Variant Curation Expert Panel
The RASopathy (RAS) VCEP membership represents a diverse range of expertise and qualifications including four medical geneticists, six research scientists, and seven clinical laboratory diagnosticians from three countries. The members were assigned to subgroups tasked with curating gene-specific information relevant to variant interpretation (dosage sensitivity, functional domains, hot spot analysis, functional assays etc.) and approving or disapproving of the use of certain ACMG criteria (functional domains PM1, functional studies PS3) for nine genes of interest (BRAF, HRAS, KRAS, MAP2K1, MAP2K2, PTPN11, RAF1, SHOC2, SOS1). The RAS VCEP found that grouping these well-established genes based on function and mechanism facilitated an en masse approach for generating a highly specific and robust classification framework that was effective for all variants in these genes (Gelb et al., 2018). Having sets of variants, both clearly pathogenic and benign, in the RASopathy genes also enabled additional fine-tuning of this framework. The contributions of case-level data from both diagnostic laboratories and clinicians were critical for providing the most accurate and up-to-date classification of a variant. These specifications were tested by classifying 37 well-established pathogenic variants plus an additional 66 variants in ClinVar distributed across the RASopathy genes. The RAS VCEP is currently prioritizing variants for curation and expanding the scope of its guideline specifications to additional genes associated with the RASopathies.
SHARED EXPERIENCES AND LESSONS LEARNED
As demonstrated by the pilot experiences (summarized in Table 1), the VCEPs approached the stages of their development differently, from the review and specification of how to apply the ACMG/AMP criteria to their pilot procedures for variant curation. These unique approaches generated a wealth of experiential information and guidance, including common challenges and solutions. Based on numerous ClinGen discussions and iterative improvements, these approaches formed the foundation of the streamlined and standardized ClinGen process for VCEP approval. The shared experiences in the following section emerged repeatedly during discussions of the current approval process and opportunities for improvement, sustainability, and scale-up.
Table 1 –
Summary of the pilot ClinGen Variant Curation Expert Panels (VCEPs)
| MYH7/Inherited Cardiomyopathies | PTEN | RASopathies | PAH | |
|---|---|---|---|---|
| Group Structure and Process | Core group proposed specifications for full EP review | Subgroups formed by different evidence lines. Full EP proposed and reviewed specifications | Subgroups formed by expertise and gene disease mechanism. Subgroups proposed specifications for full EP review | Core group proposed specifications for full EP review |
| Initial Guideline Optimization | Proposals refined by iterative rounds of curation and feedback | Subgroups curated literature specific to the evidence line and presented to full EP | General proposals refined by iterative rounds of curation and feedback Subgroups curated gene-specific literature to supplement use of specific rules (e.g. functional data) | Proposals refined by iterative rounds of curation and feedback |
| Consensus | Majority (66%) | Full (100%) | Majority (80%) | Full (100%) |
| Timeline | 1/2015–4/2017 | 7/2015–4/2018 | 5/2015–7/2017 | 10/2014–4/2018 |
| Curation Pilots |
60 Variants: 50 Missense 3 Deletion 7 Others |
42 Variants: 15 Benign/Likely Benign 16 Path/Likely Path 11 VUS/Conflicting |
~15 Variants per
gene for 9 genes: 5 Path 5 Non-conflicting 5 Conflicting |
15 Missense
Variants: 8 Path 7 VUS |
| Pilot Curation Method | One curator per variant | Two curators per variant | Two curators per variant | One curator per variant |
| Reviewers | Blinded double expert review (clinical and laboratory) per variant | Full EP | Triple review (clinical/research subspecialist of the gene(s) and two additional EP members) | Full EP |
| Conflict resolution | Core team with EP engagement as needed | Full EP discussion and voting | Full EP discussion and voting | Full EP discussion and voting |
| Key Lessons | Consensus determination | Recycle rule specifications that have already been successful | Subjective criteria; time intensive | Use a majority consensus approach in the future |
Group structure and membership
The VCEP group structure and composition significantly influenced the pace of guideline specification and variant curation pilots. The PTEN and RASopathies VCEPs subdivided the assessment of the eight evidence criteria based on the expertise of their memberships, while the MYH7/Inherited Cardiomyopathies and PAH VCEPs designated smaller “core” groups to draft proposals for review by the entire VCEP. In addition to senior experts in the gene- or disease-area of interest, the VCEPs required a nimble workforce, composed of dedicated, and often more junior experts and curators, to perform literature reviews, variant biocuration, and draft preliminary materials for expert review. In large part, ClinGen relies on donated effort from hundreds of working group volunteers, but it is recognized that the pace and extent of future variant curation activities will be significantly constrained if they are primarily dependent on a volunteer effort, unless this crowdsourcing approach can be greatly expanded. The VCEPs also noted a specific need to include multiple clinical laboratory diagnosticians from various institutions who have a high degree of familiarity with the ACMG/AMP guidelines based on routine interpretation of variants for clinical reporting.
Preliminary ACMG/AMP criteria specification process
Some groups found that defining the scope of work for the VCEP was challenging, but agreed that it helped to set expectations for the workload and facilitated the distribution of work. Though some VCEPs have undertaken criteria specifications for an entire disease entity (RASopathies and MYH7/Inherited Cardiomyopathies), the other VCEPs felt that this approach would be difficult for diseases that contain genes with disparate molecular mechanisms. The VCEPs also utilized similar methods to facilitate the process of specifying their guidelines, such as (1) basing their initial specifications on internal laboratory guidelines for their domain of interest, (2) presenting relevant genetic and functional information for their gene and disease (dosage sensitivity, functional domains, hotspot analysis, functional assays, etc.), (3) carrying out “bake-off” curation pilots using a small number of well-known pathogenic and benign variants to further refine the preliminary criterion specifications, and (4) conducting surveys to capture the clinical, research, and diagnostic expertise of the VCEP.
Harmonization of specifications to the ACMG/AMP guidelines
Each of the VCEPs identified a need for SVI WG standardization prior to their refinement of the ACMG/AMP guidelines. This became particularly evident as VCEPs began specifying the same criteria using different methodologies or threshold in places where generalization of the guideline might be sufficient. For example, the MYH7/Inherited Cardiomyopathies VCEP and RASopathies VCEP applied different quantitation for the number of segregations needed for each strength level for the co-segregation criterion (PP1). While some differences may be appropriate based on different rates of phenocopies and disease prevalence, it was clear that all groups needed quantitative guidance for application of the PP1 segregation criterion. Ultimately, the ongoing work of the SVI WG is expected to increase the uniformity and consistency of VCEP recommendations and reduce the need for new VCEPs to ‘reinvent the wheel’ by specifying criteria that are broadly applicable across various domains (ClinGen Sequence Variant Interpretation WG, 2018). Additionally, the ClinGen Informatics team is refining the ClinGen Variant Curation Interface (VCI) to enable VCEPs to curate using their specified rule sets. This resource is also available to users outside of the ClinGen Consortium (“ClinGen’s Curation Interfaces,” 2018).
ClinGen Variant Curation Expert Panel Application Process
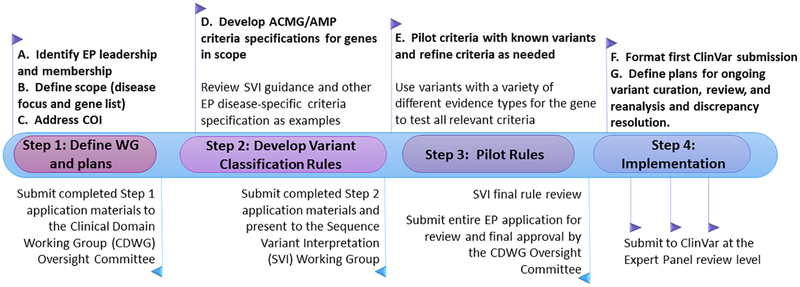
Based on the pilot experiences, a stepwise process was developed for ClinGen VCEPs to apply for the ability to submit variant assertions to ClinVar at the Expert Panel level of review (Fig. 1). Although expert panels that develop outside of the ClinGen consortium are welcome to apply for ClinVar Expert Panel status at any stage, for ClinGen-supported VCEPs the following steps are completed in a specific order and approved by the SVI WG and the ClinGen CDWG Oversight Committee. Outside groups who are interested in becoming an approved Expert Panel are encouraged to reach out early in the process, and strongly recommended to follow these steps in the same order as a ClinGen-supported VCEP.
Figure 1 -.
Stepwise process for ClinGen Expert Panel application submission and review
Step 1: Define WG and plans
Leadership and Membership
Although there is no defined minimum or maximum number of members in a VCEP, a balanced membership from a minimum of four institutions (independent affiliations) with expertise including, but not limited to, clinicians, researchers, and laboratory diagnosticians is required. VCEPs should consider reaching out and including international efforts with overlapping goals in order to capture the breadth of available knowledge in that area and ensure that the results are widely accepted.
Define Scope (disease focus and gene list)
VCEPs are recommended to focus on a single gene or a set of genes associated with a single condition or related conditions, as well as the comprehensive curation of variants in those genes. VCEPs usually choose one or several of the most common or clinically relevant genes or gene families within their area of focus and gradually enlarge the scope of the project over time.
Address Conflicts of Interest (COI)
VCEPs must declare both academic and financial COI to ensure VCEP decisions and variant classifications are devoid of bias. For example, individuals who have published on the pathogenicity of specific variants or who hold a financial interest in the outcome of a variant classification (e.g. existing clinical test specific to the variant) must abstain from final decisions about the variant’s pathogenicity.
The ClinGen COI policies are circulated for acceptance by every VCEP member and are intended to ensure transparency and consistency. COI disclosures are made publicly available on the ClinGen website.
In addition, VCEP members are asked to declare any previous, ongoing, or future competing interests, such as independent efforts to publish variant classification approaches or projects in the area. While ClinGen cannot and does not restrict members’ external activities, it is requested that these activities be disclosed to the VCEP so that decisions can be made about how to manage those activities.
Step 2: Develop Variant Classification Criteria
Develop ACMG/AMP criteria specifications for genes in scope
VCEPs begin the specification process by reviewing and incorporating guidance from the SVI WG for criteria that are broadly applicable across domains (“Sequence Variant Interpretation (SVI) Working Group,” 2018). Specification of some criteria require gene-level evidence, such as determining which functional assays are “well-established” (PS3 & BS3), identification of significant functional domains (PM1), and defining a threshold for “absence” from controls (PM2) or applying thresholds for calling benign or likely benign variants using BA1 and BS1. The membership of the VCEP is carefully selected and reviewed by the ClinGen leadership to ensure the inclusion of individuals with appropriate disease-specific expertise who can lead discussions and make determinations about these types of specifications.
In other evidence categories, VCEPs will develop rule specifications that require thorough curation of gene-specific features and literature review as well as defining evidence thresholds in the context of the gene/disease association. The SVI WG is also working on specifying criteria that can be generalized across VCEPs, such as thresholds for segregation data (PP1) and recommendations for elevating the weight of de novo (PS2/PM6) and in trans (PM3) occurrence. Specification of some criteria requires both general guidance and disease-specific information. For example, the SVI WG has established a general recommendation for determining the applicable strength of loss of function variants (PVS1) (see (Tayoun et al., 2018) this issue); however, VCEPs need to determine for which genes LOF is a relevant disease mechanism. Another example is allele frequency (BA1/BS1), where the SVI WG has provided guidance for how to go about calculating allele frequency thresholds (see Ghosh et al., 2018 this issue); however, VCEPs need to gather disease-specific information for these calculations. SVI WG guidance on these and other criteria continue to be updated and are made publicly available on the ClinGen website (https://www.clinicalgenome.org/working-groups/sequence-variant-interpretation/).
VCEPs adopt an organizational strategy for review, discussion, and decisions on developing gene/disease specifications to the ACMG/AMP guidelines. Approaches include subdividing the group and assigning a category from the guidelines to each subgroup (as exemplified by the PTEN VCEP), discussing proposed changes and specifications to the guidelines within the larger panel, and reaching consensus through voting (adopted by the MYH7/Cardiomyopathies VCEP).
Representatives from each of the VCEPs are asked to join the SVI WG conference calls to discuss difficult issues, provide feedback and recommendations, and improve harmonization across VCEPs. Once a VCEP finishes specifying the ACMG/AMP guidelines for its targeted gene or disease, it presents the draft specifications to the SVI WG, initiating iterative review and improvement steps until the ClinGen SVI gives the VCEP approval to begin their pilot.
Step 3: Pilot Gene-/Disease-specifications of Criteria
Pilot criteria with known variants and refine as needed
VCEPs pilot test their specified guidelines with relevant variants having a variety of different classifications and evidence types, with an aim to attempt testing of all criteria codes. Gene-specific criteria are applied to multiple variants that are broadly representative of each of the variant types and evidence codes used for interpretation, to provide a robust test of the classification rules. After completing the pilot, the VCEP submits its final specifications along with the variants used in the pilot and the results to the SVI WG for approval. The SVI WG provides feedback via a standardized form detailing any additional issues that need to be addressed prior to approval.
After final SVI approval of the criteria and prior to submitting pilot variant classifications to ClinVar, the Variant Curation Expert Panel submission packet is submitted to ClinVar (clinvar@ncbi.nlm.nih.gov) and the CDWG Oversight Committee for review and final approval of the VCEP.
Step 4: Implementation
Format first submission to ClinVar
After approval is received, the VCEP formats and submits their first ClinVar submission. The ClinVar variant submission template is available on ClinVar’s FTP site along with submission instructions.
Define plans for ongoing variant curation, expert review, reanalysis, and discrepancy resolution
Each VCEP then adopts an SOP for variant assessment, including protocols for preliminary curation and expert review using either ClinGen-approved methods, or another method developed by the VCEP and described in detail in the VCEP application materials. A method for conflict resolution and consensus determination for arriving at final variant classifications must also be described. In the first phase of ClinGen, the VCEPs were encouraged to experiment with different processes to determine the most effective and efficient means of curation and conflict resolution; however, standardization of best practices across ClinGen VCEPs is the eventual goal of this iterative process.
Variant curators are trained in the use of ClinGen’s Variant Curation Interface (VCI) for curating evidence, systematically applying the ACMG/AMP evidence codes, and establishing a consensus classification. Ongoing development of the VCI will add functionality such as the ability to define gene-specified criteria and generate a ClinVar submission. All variant curation activities associated with the ClinGen resource are expected to use the VCI, which will ultimately enable automated submissions of variant classifications and evidence summaries to ClinVar.
VCEPs are expected to keep their variant classifications up-to-date and expedite the reassessment of variants that have a conflicting assertion submitted to ClinVar after the VCEP’s submission. VCEPs are expected to contact the submitter of a newly submitted, medically relevant (i.e. LP vs. VUS) conflicting assertion in ClinVar and attempt to resolve or address the conflict within 3 months of notification based on discrepancy reports that are regularly issued to VCEPs. In addition, they are expected to reassess all LP and VUS classifications at least every 2 years to see if new evidence has emerged to aid in classification, or when requested by the public via the ClinGen website. VCEPs are expected to reassess LB classifications when new evidence, such as new large population datasets, becomes available.
Conclusion
A key mission of ClinGen is to provide expert assessment of the clinical significance of genomic variants based on systematic, high quality evidence review. Curation and expert review at this scale requires a massive community effort that ClinGen has organized through the development of subspecialty VCEPs, each focused on a particular group of genes or diseases. One initial metric of success will be the continual establishment of new VCEPs over time. Later, when those VCEPs are functioning in the implementation phase, we expect to see increases in the rate of expert-curated variants in ClinVar.
The main limiting factor to scaling variant curation is the ability to sustain the efforts of domain experts, biocurators, and variant analysts. As such, the development of informatics infrastructure and machine learning methods to aid in the identification and application of evidence is imperative; however, given the complexity of sequence variant interpretation, expert judgment and a large community effort are still needed to produce high quality variant classifications. Standardized volunteer training materials and proficiency assessments will make it possible for ClinGen to increasingly include the broader community in expert curation activities. The SOPs, user interfaces, and informatics infrastructure thus represent substantial work products that will facilitate the ongoing work of multiple VCEPs.
ClinGen is extending its outreach to NIH institutes, professional societies, and other organizations to enable the development and support of new VCEPs and expand existing VCEPs to accelerate the throughput of variant curation and classification. Table 2 shows the ongoing efforts across the different CDWGs. For example, the Eunice Kennedy Shriver National Institute of Child Health and Human Development has supported three new VCEPs through an innovative U24 program (RFA-HD-17–001) developed in collaboration with ClinGen. In addition, ClinGen is working with professional organizations, including the American Society of Hematology, which funded the establishment of two new VCEPs for platelet disorders and malignant blood disorders, and the Association for Clinical Genomic Science in the United Kingdom, which partnered with ClinGen to support the cardiomyopathy VCEP. ClinGen is optimistic that the systematic and comprehensive work of our existing and new expert groups will accelerate the pace of variant curation efforts, and lead to classification resolution for genomic variants with discrepant or uncertain ClinVar assertions, enabling improved diagnoses in patients.
Table 2 –
Current and ongoing ClinGen-affiliated VCEPs. †VCEPs funded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and the American Society of Hematology.
| Cardiovascular CDWG | Hereditary Cancer CDWG | Inborn Errors of Metabolism CDWG | Neurodevelopmental Diseases CDWG | Hearing Loss CDWG | Hemostasis and Thrombosis CDWG | Somatic Cancer CDWG | Not Affiliated to CDWG | |
|---|---|---|---|---|---|---|---|---|
|
Step 1 Define WG and Plans |
FBN1 (Marfan Syndrome) | Myeloid Malignancy† | Angelman/Rett-like Diseases | Platelet Disorders† | TP53 (Somatic) | |||
|
Step 2 Develop Variant Classification Rules |
KCNQ1 (Long QT) | GAA/Storage
Diseases Mitochondrial Diseases† |
Monogenic Diabetes† | |||||
|
Step 3 Pilot Rules |
Familial Hypercholesterolemia | CDH1 (Diffuse Gastric
Cancer) TP53 (Li-Fraumeni Syndrome) |
Brain Malformations† | Hereditary Hearing Loss | ||||
|
Step
4 Implementation |
MYH7/Inherited Cardiomyopathy | PTEN (PTEN Hamartoma Tumor Syndrome) | PAH (Phenylketonuria) | RASopathy |
Acknowledgments
Grant Sponsor: The National Human Genome Research Institute (NHGRI) primarily funded this work via the following cooperative agreements: 1U01HG007437, 1U41HG006834, 1U01HG007436, U41HG009649, U41HG009650.
Footnotes
Conflict of interest statement:
R.D.S. is a consultant to and holds stock options in Acer Therapeutics and Censa Pharmaceuticals. C.E. is a member of the External Strategic Advisory Board of N-of-One, and is the pro bono CMO of Family Care Path, Inc. and of PlexSeq Diagnostics/Covariance, Inc.
Several authors are clinical service providers and are employed by laboratories that offer fee-based clinical sequencing. This employment is noted in the author affiliations. {The authors declare no additional conflicts of interest beyond their employment affiliation}
References
- Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, … Rehm HL (2016). Performance of ACMG-AMP Variant-Interpretation Guidelines among Nine Laboratories in the Clinical Sequencing Exploratory Research Consortium. American Journal of Human Genetics, 98(6), 1067–1076. doi: 10.1016/j.ajhg.2016.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzariti DR, Riggs ER, Niehaus A, Rodriguez LL, Ramos EM, Kattman B, … Rehm HL (2018). Points to consider for sharing variant-level information from clinical genetic testing with ClinVar. Molecular Case Studies, 4(1). doi: 10.1101/mcs.a002345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bean LJH, & Hegde MR (2016). Gene variant databases and sharing: creating a global genomic variant database for personalized medicine. Human Mutation, 37(6), 559–563. doi: 10.1002/humu.22982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau N (2016). Genetics of phenylketonuria: then and now. Human Mutation, 37(6), 508–515. doi: 10.1002/humu.22980 [DOI] [PubMed] [Google Scholar]
- Bowdin S, Gilbert A, Bedoukian E, Carew C, Adam MP, Belmont J, … Krantz ID (2016). Recommendations for the integration of genomics into clinical practice. Genetics in Medicine, 18(11), 1075–1084. doi: 10.1038/gim.2016.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong JX, Buckingham KJ, Jhangiani SN, Boehm C, Sobreira N, Smith JD, … Bamshad MJ (2015). The genetic basis of mendelian phenotypes: discoveries, challenges, and opportunities. American Journal of Human Genetics, 97(2), 199–215. doi: 10.1016/j.ajhg.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ClinGen. (2018, March 11). ClinGen Expert Panels. Retrieved April 4, 2018, from https://www.clinicalgenome.org/expert-groups/
- ClinGen’s Curation Interfaces. (2018). Retrieved July 27, 2018, from https://www.clinicalgenome.org/resources-tools/
- ClinVar. (2017, September 26). Review Status in ClinVar. Retrieved March 29, 2018, from https://www.ncbi.nlm.nih.gov/clinvar/docs/review_status/
- Ellard S, Baple EL, Owens M, Eccles DM, Abbs S, Deans ZC, … McMullan DJ (2017). ACGS Best Practice Guidelines for Variant Classification 2017. Association for Clinical Genetic Science. [Google Scholar]
- Furqan A, Arscott P, Girolami F, Cirino AL, Michels M, Day SM, … SHaRe Consortium. (2017). Care in specialized centers and data sharing increase agreement in hypertrophic cardiomyopathy genetic test interpretation. Circulation. Cardiovascular Genetics, 10(5). doi: 10.1161/CIRCGENETICS.116.001700 [DOI] [PubMed] [Google Scholar]
- Garber KB, Vincent LM, Alexander JJ, Bean LJH, Bale S, & Hegde M (2016). Reassessment of genomic sequence variation to harmonize interpretation for personalized medicine. American Journal of Human Genetics, 99(5), 1140–1149. doi: 10.1016/j.ajhg.2016.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelb BD, Cavé H, Dillon MW, Gripp KW, Lee JA, Mason-Suares H, … Vincent LM (2018). ClinGen’s RASopathy Expert Panel consensus methods for variant interpretation. Genetics in Medicine. doi: 10.1038/gim.2018.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R, Harrison S, Rehm H, Plon S, Bisecker L, & ClinGen Sequence Variant Interpretation Working Group. (2018). Updated Recommendation for the Benign Stand Alone ACMG/AMP Criterion. Human Mutation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghouse J, Skov MW, Bigseth RS, Ahlberg G, Kanters JK, & Olesen MS (2018). Distinguishing pathogenic mutations from background genetic noise in cardiology: The use of large genome databases for genetic interpretation. Clinical Genetics, 93(3), 459–466. doi: 10.1111/cge.13066 [DOI] [PubMed] [Google Scholar]
- Han PKJ, Umstead KL, Bernhardt BA, Green RC, Joffe S, Koenig B, … Biesecker BB (2017). A taxonomy of medical uncertainties in clinical genome sequencing. Genetics in Medicine, 19(8), 918–925. doi: 10.1038/gim.2016.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison SM, Dolinsky JS, Knight Johnson AE, Pesaran T, Azzariti DR, Bale S, … Rehm HL (2017). Clinical laboratories collaborate to resolve differences in variant interpretations submitted to ClinVar. Genetics in Medicine, 19(10), 1096–1104. doi: 10.1038/gim.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly MA, Caleshu C, Morales A, Buchan J, Wolf Z, Harrison SM, … Funke B (2018). Adaptation and validation of the ACMG/AMP variant classification framework for MYH7-associated inherited cardiomyopathies: recommendations by ClinGen’s Inherited Cardiomyopathy Expert Panel. Genetics in Medicine, 20(3), 351–359. doi: 10.1038/gim.2017.218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, … Maglott DR (2016). ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Research, 44(D1), D862–8. doi: 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebo MS, Zakoor K-R, Chun K, Speevak MD, Waye JS, McCready E, … Canadian Open Genetics Repository Working Group. (2017). Data sharing as a national quality improvement program: reporting on BRCA1 and BRCA2 variant-interpretation comparisons through the Canadian Open Genetics Repository (COGR). Genetics in Medicine, 20(3), 294–302. doi: 10.1038/gim.2017.80 [DOI] [PubMed] [Google Scholar]
- Manolio TA, Chisholm RL, Ozenberger B, Roden DM, Williams MS, Wilson R, … Ginsburg GS (2013). Implementing genomic medicine in the clinic: the future is here. Genetics in Medicine, 15(4), 258–267. doi: 10.1038/gim.2012.157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mester JL, Ghosh R, Pesaran T, Huether R, Karam R, Hruska KS, … Zhang L (2018). Gene-Specific Criteria for PTEN Variant Curation: Recommendations from the ClinGen PTEN Expert Panel. Human Mutation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milko LV, Funke BH, Hershberger RE, Azzariti DR, Lee K, Riggs ER, … Rehm HL (2018). Development of Clinical Domain Working Groups for the Clinical Genome Resource (ClinGen):Lessons Learned and Plans for the Future. Genetics in Medicine. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, … ClinGen. (2015). ClinGen--the Clinical Genome Resource. The New England Journal of Medicine, 372(23), 2235–2242. doi: 10.1056/NEJMsr1406261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, … ACMG Laboratory Quality Assurance Committee. (2015). Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine, 17(5), 405–424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequence Variant Interpretation (SVI) Working Group. (2018). Retrieved July 27, 2018, from https://www.clinicalgenome.org/working-groups/sequence-variant-interpretation/
- Tayoun AA, Pesaran T, DiStefano M, Oza A, Rehm H, Biesecker L, … ClinGen Sequence Variant Interpretation Working Group. (2018). Recommendations for Interpreting the Loss of Function PVS1 ACMG/AMP Variant Criteria. Human Mutation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zastrow DB, Baudet H, Shen W, Thomas A, Shi Y, Weaver MA, … Feigenbaum A (2018). Unique Aspects of Sequence Variant Interpretation for Inborn Errors of Metabolism (IEM): The ClinGen IEM Working Group and the Phenylalanine Hydroxylase Gene. Human Mutation. [DOI] [PMC free article] [PubMed] [Google Scholar]