Abstract
Background/Aim
Cancer-associated fibroblasts (CAFs) are important factors in the progression of hepatocellular carcinoma (HCC). But the characterization of these cells remains incomplete. This study aims to identify a panel of markers for CAFs that are associated with HCC progression.
Materials and methods
The sequencing data and clinicopathological characteristics of 366 patients were obtained from the Cancer Genome Atlas (TCGA) database (366 HCC tissues and there were 50/366 cases with corresponding normal liver tissues). In vitro validation of the markers was performed by quantitative real-time PCR using the hepatic stellate cell line LX2 induced by the HCC cell line Huh7. The activation of LX2 was confirmed by α-smooth muscle actin and fibroblast activation protein, using quantitative real-time PCR and immunofluorescence staining. In vivo detections of the 12 markers were done in 40 tissue samples (30 HCC and 10 normal).
Results
We successfully identified 12 CAF markers from TCGA data: FGF5, CXCL5, IGFL2, MMP1, ADAM32, ADAM18, IGFL1, FGF8, FGF17, FGF19, FGF4, and FGF23. The 12-marker panel was associated with the pathological and clinical progressions of HCC. All 12 markers were upregulated in vitro. In vivo expressions of these markers were paralleled with those in TCGA data.
Conclusion
A 12-marker panel of CAFs in HCC is identified, which is associated with both pathological and clinical progressions of cancer.
Keywords: hepatocellular carcinoma, cancer-associated fibroblasts, CAFs marker panel, TCGA database analysis, transcriptome profiling
Introduction
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer death in both men and women worldwide.1 There is an increased number of activated fibroblasts, also known as the cancer-associated fibroblasts (CAFs) within the HCC microenvironment, which derives mainly from the activation of hepatic stellate cells (HSCs). Multiple studies have shown that CAFs can be a poor prognostic factor in HCC.2–4 There are ample evidences that suggest the active involvement of CAFs in the progression of different cancers.5,6 Although CAFs bear the features of activated fibroblasts, they are identified merely by one or two fibroblastic markers in most studies, eg, the α-smooth muscle actin (α-SMA).4,7 Therefore, the role of CAFs needs to be further characterized by more fibroblastic markers before their involvement in HCC progression can be better studied.
Not until recently, there has been little progress in the characterization of these heterogeneous stromal cells. Increased numbers of CAFs subpopulations have been found owing to the differential expression of fibroblastic panels in these cells, which is explored, instead of a single marker.8–10 It is believed that these stromal cells may share dissimilar fibroblastic patterns in different cancers and cancer stages,4,11–13 while some markers of CAFs are known to affect many aspects of HCC progression.7,14 In this sense, defining a certain panel of the fibroblastic markers can help to identify the CAFs coevolving with HCC.
The screening for fibroblastic markers can be performed for differential expression profiles in cancer tissues to obtain a more complete panel in HCC. As an integrated source for the sequencing data of most known human cancers, the Cancer Genome Atlas (TCGA) offers a good platform for the investigation. In addition, the known markers of activated fibroblasts are categorized as mesenchymal cell markers, cytokines, chemokines, growth factors, extracellular matrix components, and related enzymes.4,7
The aim of this study was to explore and identify the marker panel of CAFs associated with tumor progression in HCC. The differential expression profiles between 1) normal liver tissues and HCC tissues and 2) early stage and advanced-stage tumors were retrieved from TCGA database. Based on these, the identified fibroblastic markers were associated with the pathological progression of HCC. The correlation of the marker panel with clinical outcomes was then investigated. The panel was validated in the in vitro model of induced CAFs, ie, the HSC cell line activated by HCC cancer cell, as described in previous study,15 and in the postsurgical specimens from HCC patients.
Materials and methods
HCC mRNA sequencing data from the public TCGA database
The HCC mRNA sequencing data were acquired in the open-source TCGA database, from the National Cancer Institute GDC data portal, USA (https://portal.gdc.cancer.gov/). We searched for the HCC mRNA sequencing data by the following preconditions: 1) transcriptome profiling; 2) gene expression quantification; 3) RNA sequencing; 4) primary cancer site defined as the liver; 5) TCGA program; 6) pathological diagnosis as the liver HCC; and 7) both genders, all races, all ages, and all follow-up status. Altogether, 371 cases were identified with 424 sequencing files (RNA sequencing data form of the Illumina HiSeq 2000 RNA Sequencing platform). After excluding cases with incomplete clinical information, we included 366 cases for further analysis. There were 50 cases with sequencing data of the normal/nontumor liver tissues.
Data integration and differential gene screening in software R
Screening the differentially expressed genes for further CAF expression profile analysis was performed in the software R. The 366 included samples were classified according to pathological stages (I, II, III, and IV). Subsequently, two different patterns of early stage and advanced-stage groups were defined: 1) pathological stage I and stage II–IV, and 2) pathological stage I–II and stage III–IV respectively. Then, the differential gene expressions were screened between 1) the normal and tumor tissues, and 2) the early stage and advanced-stage tumor tissues using the edgeR package in R3.4.3. The cutoff set as a false discovery rate was <0.05, and the genes with |logFC| of >1 were considered as significantly changed. The differential genes obtained were then used for further analysis using the pheatmap package in the software R.
CAFs expression profiles in different stages of HCC
The differential genes obtained were matched with the fibroblast profile.4,7 Subsequently, the relative expression profiles of CAFs in different HCC stages were plotted in the GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla, CA, USA).
Integration of all markers and determination of low/high expression in each panel
To evaluate the correlation between different panels and clinical outcomes, we integrated the expression of all markers in each panel for every patient. In the integration process, we first assumed the same weighted score to each marker. For example, 1/12 was applied to the expression of each marker in the 12-CAF marker panel. Thereafter, the weighed expressions of these markers were added up to become one integrated expression. Accordingly, the integrated expressions of these panels were made in each patient. The cutoff values of high and low expression were then set at 50% of the integrated expressions among all patients. Subsequently, these patients were divided into two groups in each CAF panel, ie, the low and high expression groups.
Patient specimens
Thirty fresh tumor tissue samples and 10 normal liver tissues were collected right after surgical resections (total number = 40), between the year 2010 and 2017, in The Fifth Affiliated Hospital, Sun Yat-sen University, Guangdong Province, China. These specimens were then immediately stored at −80°C. All patients received no treatments before their first surgical resections in our center. All tumor cases were pathologically confirmed HCC. The clinicopathological characteristics are shown in Table S1. The normal liver tissues were collected from trauma patients who underwent partial hepatectomy with no other known diseases. The TNM staging were determined according to the 2010 American Joint Committee on Cancer staging system. All samples were collected after obtaining written informed consent from the patients in our center, and the study was approved by the Institutional Ethical Review Board of The Fifth Affiliated Hospital, Sun Yat-sen University.
Cell lines and cell culture
The HCC cancer cell line Huh7 was obtained from Cancer Center, Sun Yat-sen University, Guangzhou, Guangdong, China.16 The HSC line LX2 (Merck Millipore, Billerica, MA, USA)17 was obtained from Fudan University, Shanghai, China. The use of the cell lines was approved by the Institutional Ethical Review Board of The Fifth Affiliated Hospital, Sun Yat-sen University. LX2 was cultured in RPMI-1640 medium (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) and Huh7 was cultured in DMEM medium (Gibco). All culture media were supplemented with 10% FBS, 50 U/mol penicillin, and 50 mg/mL streptomycin (Sigma-Aldrich Co., St Louis, MO, USA). The cells were maintained in a humidified incubator at 37°C with 5% CO2. Conditioned medium (CM) from Huh7 was collected, centrifuged at 3,000g for 20 minutes, and then filtered with 0.45 µM filters. The CM was supplied with RPMI-1640 containing 10% FBS (1:1) for the induction of LX2 cells. LX2 was induced in the CM for at least 48 hours.
Immunofluorescence staining
LX2 was plated, induced, and grown on glass coverslips up to 60% confluence. After being fixed in 4% paraformaldehyde, the cells were incubated in primary rabbit anti-α-SMA (Abcam, Ab32575, 1:500 dilutions) overnight at 4°C. Afterward, the cells were washed and incubated for 1-hour with fluorescence-conjugated secondary antibody (BA1105, 1:500 dilutions; BOSTER Biological Technology, Wu Han, China). Finally, cells were washed and stained by DAPI (BOSTER Biological Technology). Then, the cells were observed and imaged by the fluorescence microscope (ZEISS, Oberkochen, Germany).
Quantitative real-time PCR
Total RNA from cultured cells were extracted by using RNAiso Plus (Takara Bio, Da Lian, China). NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) was used to determine the concentration of extracted RNA. TaKaRa One Step RNA PCR Kit (AMV) (Takara Bio) was used for cDNA synthesis. The PowerUp SYBR Master Mix Applied Biosystems (Invitrogen, Thermo Fisher Scientific) was used for the detection of relative gene expression. Each PCR was run in at least three independent experiments (Eppendorf Realplex System, Eppendorf AG, Hamburg, Germany). The comparative Ct method (ΔCt) was used for the relative expression calculation, which was normalized using β-actin as the internal control. The primer sequences were either designed on the NCBI website (http://www.ncbi.nlm.nih.gov/tools/primer-blast) or obtained online from the Prim-erBank website (http://pga.mgh.harvard.edu/primerbank/). The primer sequences are shown in Table S2.
Statistical analysis
The SPSS statistical software (version 16.0; IBM SPSS, Armonk, NY, USA) and the GraphPad Prism software (version 6.0) were used for statistical analyses. All survival statistics (including the survival curve plotting) were done by the Kaplan–Meier survival method with log-rank test in SPSS. Cox proportional hazards regression, univariate survival analysis, and associations among different factors (chi-squared test) were performed using SPSS16.0. P-value <0.05 was considered significant.
Results
Differential expression profile in HCC tumor tissues
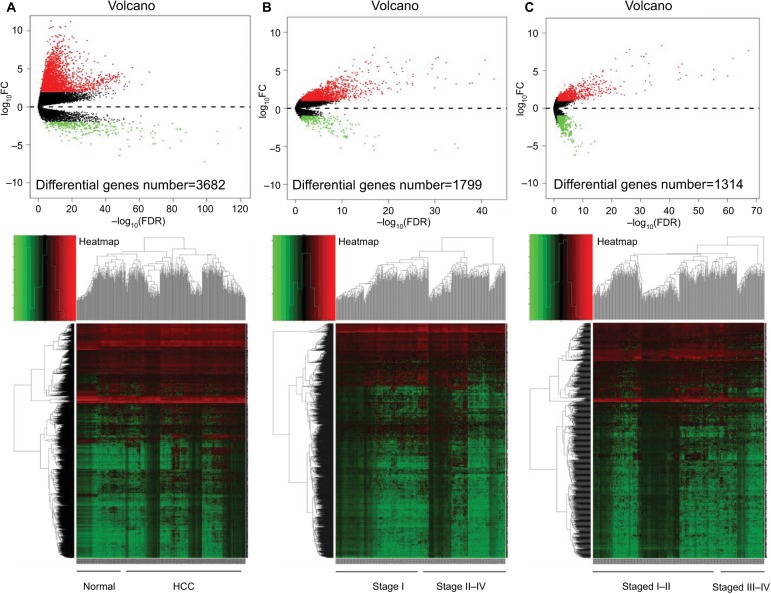
To set the background genes for further analysis, we compared the sequencing data between HCC and the adjacent nontumor samples (Figure 1A and Table S3). Meanwhile, we found the genes involved in tumor development by comparing the data in the early stage and the advanced-stage tumors. The definitions of different stages have been described in Materials and methods. Subsequent results were shown between stage I and stage II–IV tumors (Figure 1B and Table S4); stage I–II and stage III–IV tumors (Figure 1C and Table S5).
Figure 1.
Differential gene expression using the TCGA mRNA sequencing data of HCC.
Notes: Volcano map and heat map for differential gene expressions between (A) the normal (n=50 cases) and all HCC tissues (n=366 cases); (B) stage I (n=179 cases) and stage II–IV HCC tissues (n=187); (C) stage I–II (n=274 cases), and stage III–IV HCC tissues (n=92 cases).
Abbreviations: HCC, hepatocellular carcinoma; TCGA, The Cancer Genome Atlas.
Identify the CAF marker panel associated with pathological progression of HCC
Next, among the identified gene profiles, we searched for fibroblastic markers according to published articles.7 As a result, the CAF profile differentially expressed in cancer was identified between normal liver and tumor (Figure S1A and Table S6). And the stage-associated CAF profile was found between early stage and advanced-stage HCC tumors (Figure S1B and Table S7; Figure S1C and Table S8). Then we crossed the three CAF profiles to reach a specific panel associated with the progression of HCC (Tables S6 and S8). Subsequently, 12 markers were identified for the panel (Figure S1D and Table 1).
Table 1.
The markers included in different subpanels.
| Gene set | Markers |
|---|---|
|
| |
| 12-CAF marker panel | FGF5, CXCL5, IGFL2, MMP1, ADAM32, ADAM18, IGFL1, FGF8, FGF17, FGF19, FGF4, and FGF23 |
| 11-CAF marker panel | FGF5, CXCL5, IGFL2, MMP1, ADAM32, ADAM18, IGFL1, FGF8, FGF17, FGF19, and FGF4 |
| 8-Advanced CAF marker panel | FGF5, CXCL5, IGFL2, MMP1, ADAM32, ADAM18, IGFL1, and FGF8 |
| 4-Early CAF marker panel | FGF17, FGF19, FGF4, and FGF23 |
| 3-Early CAF marker panel | FGF17, FGF19, and FGF4 |
Abbreviations: ADAM, a disintegrin and metalloproteinase; CAFs, cancer-associated fibroblasts; CXCL, C-X-C motif chemokine ligand; FGF, fibroblast growth factor; IGFL, IGF-like family member; MMP, matrix metalloproteinase.
The 12 markers consisted of fibroblast growth factors (FGF5, FGF8, FGF17, FGF19, FGF4, and FGF23), matrix metalloproteinase-1 (MMP1), members from the family of ADAMs (a disintegrin and metalloproteinase, ADAM32 and ADAM18), C-X-C motif chemokine ligand-5 (CXCL5), and members of the insulin-like growth factor family of signaling molecules (IGFL2 and IGFL1).
To verify the expression patterns of the 12 markers in different HCC stages, we analyzed the expression of each gene according to the TNM staging. The relative expression profile was plotted for each marker (Figure S2). The panel was grouped into four different subpanels based on the stage expression patterns of these markers (Table 1). The association of these panels and the clinical features in HCC was subsequently explored.
Association of the CAF panels and clinical outcomes in HCC patients
Integration of all markers and determination of low/high expression of the panels have been described in the Materials and methods.
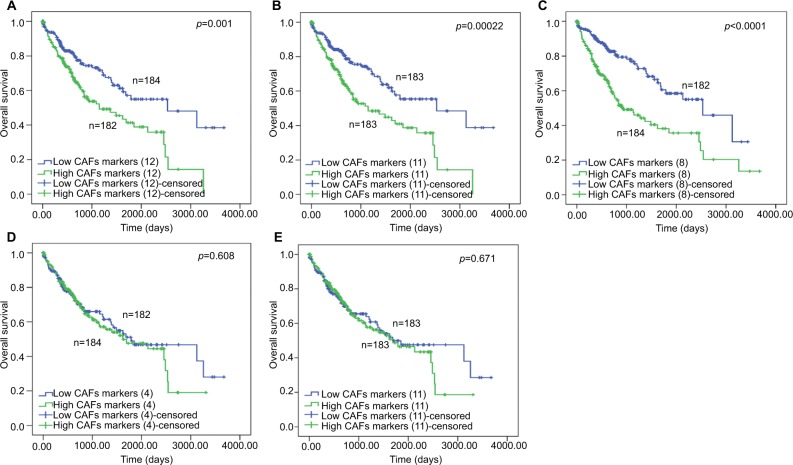
The association of different CAFs gene panels and patient clinical features is shown in Table S9. To evaluate the association between the expression of different panels and HCC patient survival, we built the overall survival curves using Kaplan–Meier method (Figure 2).
Figure 2.
The Kaplan–Meier analysis of overall survival for different CAF marker subpanels for HCC.
Notes: The overall survival curves were plotted and compared between the low expression and high expression group of each subpanels: (A) the 12-marker panel, (B) the 11-marker panel, (C) the 8-advanced panel, (D) 4-early panel, and (E) 3-early panel.
Abbreviations: CAFs, cancer-associated fibroblasts; HCC, hepatocellular carcinoma.
As a result, the high expression of the panel with advanced markers predicted worse patient survival (12-CAF marker panel, P=0.001; 11-CAF marker panel, P<0.001; 8-advanced CAF marker panel, P<0.0001; Kaplan–Meier, log-rank test). Meanwhile, the expression of the panels with only early stage markers showed no influence on HCC patient survival (4-early CAF marker panel, P=0.608; 3-early CAF marker panel, P=0.671; log-rank test).
Furthermore, the multivariate analysis showed that the high expression of the panel with only advanced-stage markers was an independent risk factor for HCC patient survival (8-advanced CAF marker, Cox regression, P<0.011) (Table 2).
Table 2.
Univariate and multivariate analysis for clinicopathological factors of HCC patients.
| Factors | OSM
|
||||
|---|---|---|---|---|---|
| Univariate
|
Multivariate
|
||||
| P-value | HR | 95% Cl
|
P-value | ||
| Lower | Upper | ||||
|
| |||||
| Age: ≤Y68 vs >Y68 | 0.890 | ||||
| Gender: male vs female | 0.244 | ||||
| Race: White vs Asian vs Black vs others | 0.428 | ||||
| Hepatitis/alcohol/other risk factors: yes vs no | 0.073 | 0.927 | |||
| TNM staging: I vs II vs III vs IV | <0.001 | 2.628 | 1.251 | 5.519 | 0.011 |
| Vascular invasion: yes vs no | 0.476 | ||||
| Tumor grade (1 vs 2 vs 3 vs 4) | 0.931 | ||||
| AFP (µg/L): ≤20 vs >20 | 0.984 | ||||
| Platelet count (×109/L): <100 vs 100–300 vs >300 | <0.001 | 0.816 | |||
| Albumin (g/L): <35 vs 35–50 vs > 50 | <0.001 | 0.610 | |||
| Total bilirubin, mg/dL (µmol/L): ≤ 1.0 (17.1) vs >1.0 (17.1) | <0.001 | 2.577 | 1.651 | 4.023 | <0.001 |
| Child–Pugh classification: A vs B vs C | 0.004 | 0.411 | |||
| 12-CAF marker combination: low vs high expression | 0.001 | 0.177 | |||
| 11-CAF marker panel: low vs high expression | <0.001 | 0.619 | |||
| 8-Advanced CAF marker panel: low vs high expression | <0.001 | 2.034 | 1.389 | 2.978 | <0.001 |
| 4-Early CAF marker panel: low vs high expression | 0.608 | ||||
| 3-Early CAF marker panel: low vs high expression | 0.671 | ||||
Note: p<0.05. Bold values indicate statistical significance.
Abbreviations: AFP, α-fetoprotein; CAFs, cancer-associated fibroblasts; OS, overall survival; HCC, hepatocellular carcinoma.
To reach a more precise conclusion, we performed stratified analysis of the 8-advanced marker panel. The stratifications performed were according to the two early marker panels (4- and 3-early CAF marker panel, Figure S3). The result showed that patients with high expression of the advanced marker panel significantly correlated with patient’s poor survival, regardless of the expression levels of the early markers (Figure S3, Table S10).
Taken together, the 12-CAF marker panel was significantly associated with the clinical progression of HCC. The subpanel analysis further confirmed that the panel consisted of markers from different tumor stages and was parallel to clinical features and patient prognosis.
In vitro confirmation of the 12-marker panel in HCC-induced CAFs model
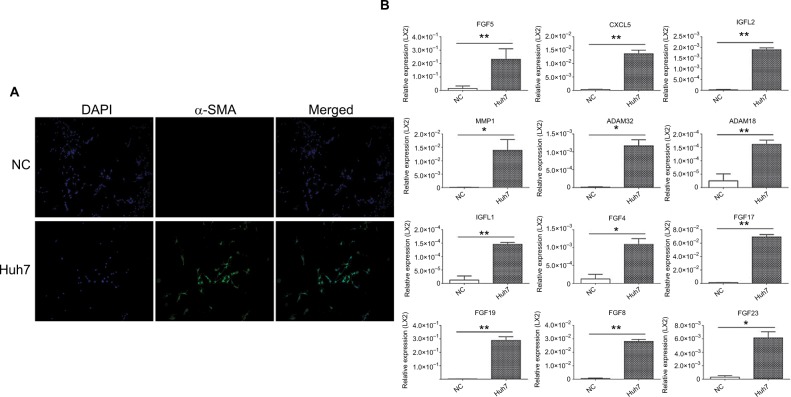
We confirmed the panel in HCC-induced CAFs model, ie, the HSCs cell line LX2 that was activated by the HCC cancer cell line Huh7. As in most HCC studies, the activation of LX2 (induced CAFs) was indicated by the increased expression of α-SMA (Figure 3A, Figure S4) and fibroblast activation protein (Figure S4). In the induced CAFs model, we validated that all 12 markers were increased in the CAFs model upon Huh7 induction, using the quantitative real-time PCR assay (Figure 3B).
Figure 3.
In vitro confirmation of the 12-marker panel in activated LX2.
Notes: (A) Immunocytofluorescence of LX2, (B) quantitative real-time PCR determination of the 12 markers in the LX2 (Huh7, LX2 activated by Huh7 conditioned cultured medium; NC, RPMI-1640 treated LX2 as the negative control; *P<0.01; **P<0.001).
Abbreviations: α-SMA, α-smooth muscle actin; NC, negative control.
In vivo validation of the 12-marker panel in patient specimens from our center
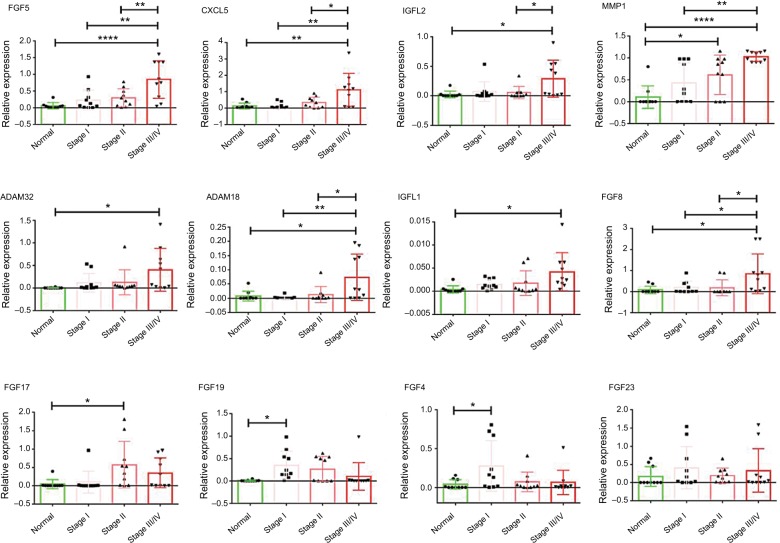
The 12 markers were further validated in the postsurgical tissues from patients in our center. In consistent with the results found in TCGA data, the eight advanced markers (FGF5, CXCL5, IGFL2, MMP1, ADAM32, ADAM18, IGFL1, and FGF8) were upregulated in stage III–IV tumors. FGF17 were significantly increased in stage II and stage III–IV HCC tumors. FGF19 and FGF4 were significantly higher in stage I tumors. The change of FGF23 was insignificant, although there was discernable upregulation in some tumor tissues (Figure 4).
Figure 4.
In vivo validation of the panel in normal liver tissues and different stages of HCC tumor tissues.
Notes: The total number of patients is 40, as for each group, n=10; *P<0.01; **P<0.001; ****P<0.00001.
Abbreviation: HCC, hepatocellular carcinoma.
Discussion
In this study, we have successfully identified 12 markers for the CAFs panel in HCC, which was associated with the pathological progression of tumor. Furthermore, subpanel analysis confirmed that the panel correlated with both the TNM stages and the clinical progression. In vitro study also verified that all 12 markers were upregulated in the induced model of CAFs. The markers were validated in the postsurgical tissues from HCC patients in our center.
The 12-marker panel offers a solution concerning the molecular phenotyping of CAFs in HCC. Although there are emerging studies for various subpopulations of CAFs, their presence in HCC still depends on the application of individual markers.7–10,13 According to the previous reports, the 12-marker panel built in this study is also based on the expression patterns of fibroblast markers.8,10 Because it is our major concern to identify the CAFs that can coevolve with HCC, the panel includes general markers not only in the cancer tissues but also associated with both the early- and advanced-stage HCC, pathologically.
The panel recognized in our study should be considered as the marker pool for the identification of several subpopulations related to the process of HCC progression. In general, patients with high expression of the 12-marker CAFs panel suffered from increased clinical events of tumor progression. And it is in consistence with the results of previous studies that increased CAF features in patients indicate poor prognosis.18–20
HSCs have been reported to be the important source for CAFs in HCC.4 Meanwhile, they show enhanced features of CAFs after receiving stimulations from cancer cells. It is also suggested that CAFs and activated HSCs may share a lot of common gene signatures.15,21–23 Therefore, the in vitro model used in our study may resemble the activated CAFs. As a validation of the specificity of the panel, all 12 markers are found to be upregulated in the in vitro CAFs model induced by HCC cancer cell line.
But it is more likely that CAFs bear changing expression of these markers across different tumor stages. As depicted in TCGA data and validated in the postsurgical tissues in our center, the 12-marker panel consists of at least two distinct subpanels, the early and advanced stage. The two subpanels are in parallel with the pathological and clinical prognostic features that correspond to different tumor stages. To recognize even more precise subpopulations by using the panel, a more stage-specific cell model is in need. One of the solutions is the primary culture of CAFs from human HCC tissues. Moreover, the relative expression of these markers should also be validated by more methods, eg, the immunohistochemistry staining in vivo and the flow cytometry in vitro. In fact, the crosstalk between cancer cells and these stromal fibroblasts remains largely unknown.4 Hopefully, exploring the 12-marker panel in future studies could benefit and boost the understanding of the cancer–stroma relationship.
There are some frequently used markers for the CAF studies in HCC, eg, the α-SMA and fibroblast-specific protein.3,11,18,24 But these markers failed to be included in the panel. It is thus highly suggested that they are not good indicators for the progression-related CAFs that we try to define here. It brings up the concern again that the study of CAFs using one or two of the fibroblastic markers is indeed not specific enough. However, these markers are sensitive to indicate the presence of activated fibroblasts in tumor tissues.7
In summary, we report a 12-marker panel of CAFs in HCC, which is associated with both pathological and clinical progressions. The panel is validated in the in vitro CAFs model and in the HCC tumor tissues in vivo. By using such a panel, it will facilitate us to identify the specific CAFs in the process of HCC development. It can also help us understand how the microenvironment can adapt to different HCC developmental stages in coming studies.
Acknowledgments
This work was supported by the Fundamental Research Funds for the Central Universities (Sun Yat-sen University, Grant 17ykpy60).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Lee JS, Yoo JE, Kim H, et al. Tumor stroma with senescence-associated secretory phenotype in steatohepatitic hepatocellular carcinoma. PLoS One. 2017;12(3):e0171922. doi: 10.1371/journal.pone.0171922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau EY, Lo J, Cheng BY, et al. Cancer-associated fibroblasts regulate tumor-initiating cell plasticity in hepatocellular carcinoma through c-Met/FRA1/HEY1 signaling. Cell Rep. 2016;15(6):1175–1189. doi: 10.1016/j.celrep.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 4.Affo S, Yu LX, Schwabe RF. The role of cancer-associated fibroblasts and fibrosis in liver cancer. Annu Rev Pathol. 2017;12:153–186. doi: 10.1146/annurev-pathol-052016-100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiong S, Wang R, Chen Q, et al. Cancer-associated fibroblasts promote stem cell-like properties of hepatocellular carcinoma cells through IL-6/STAT3/Notch signaling. Am J Cancer Res. 2018;8(2):302–316. [PMC free article] [PubMed] [Google Scholar]
- 6.Liu C, Liu L, Chen X, et al. LSD1 stimulates cancer-associated fibroblasts to drive Notch3-dependent self-renewal of liver cancer stem-like cells. Cancer Res. 2018;78(4):938–949. doi: 10.1158/0008-5472.CAN-17-1236. [DOI] [PubMed] [Google Scholar]
- 7.Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev Cancer. 2016;16(9):582–598. doi: 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 8.Costa A, Kieffer Y, Scholer-Dahirel A, et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33(3):e10, 463–479. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Su S, Chen J, Yao H, et al. CD10+GPR77+ cancer-associated fibroblasts promote cancer formation and chemoresistance by sustaining cancer stemness. Cell. 2018;172(4):e16, 841–856. doi: 10.1016/j.cell.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Öhlund D, Handly-Santana A, Biffi G, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J Exp Med. 2017;214(3):579–596. doi: 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki R, Devhare P, Ray RB, Ray R. Hepatitis C virus-induced tumor-initiating cancer stem-like cells activate stromal fibroblasts in a xenograft tumor model. Hepatology. 2017;66(6):1766–1778. doi: 10.1002/hep.29346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni WD, Yang ZT, Cui CA, Cui Y, Fang LY, Xuan YH. Tenascin-C is a potential cancer-associated fibroblasts marker and predicts poor prognosis in prostate cancer. Biochem Biophys Res Commun. 2017;486(3):607–612. doi: 10.1016/j.bbrc.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 13.Öhlund D, Elyada E, Tuveson D. Fibroblast heterogeneity in the cancer wound. J Exp Med. 2014;211(8):1503–1523. doi: 10.1084/jem.20140692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cadamuro M, Nardo G, Indraccolo S, et al. Platelet-derived growth factor-D and Rho GTPases regulate recruitment of cancer-associated fibroblasts in cholangiocarcinoma. Hepatology. 2013;58(3):1042–1053. doi: 10.1002/hep.26384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sancho-Bru P, Juez E, Moreno M, et al. Hepatocarcinoma cells stimulate the growth, migration and expression of pro-angiogenic genes in human hepatic stellate cells. Liver Int. 2010;30(1):31–41. doi: 10.1111/j.1478-3231.2009.02161.x. [DOI] [PubMed] [Google Scholar]
- 16.Liu M, Chen L, Ma NF, et al. CHD1L promotes lineage reversion of hepatocellular carcinoma through opening chromatin for key developmental transcription factors. Hepatology. 2016;63(5):1544–1559. doi: 10.1002/hep.28437. [DOI] [PubMed] [Google Scholar]
- 17.Xu L, Hui AY, Albanis E, et al. Human hepatic stellate cell lines, LX-1 and LX-2: new tools for analysis of hepatic fibrosis. Gut. 2005;54(1):142–151. doi: 10.1136/gut.2004.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang XM, Yu DM, Mccaughan GW, Gorrell MD. Fibroblast activation protein increases apoptosis, cell adhesion, and migration by the LX-2 human stellate cell line. Hepatology. 2005;42(4):935–945. doi: 10.1002/hep.20853. [DOI] [PubMed] [Google Scholar]
- 19.Ju MJ, Qiu SJ, Fan J, et al. Peritumoral activated hepatic stellate cells predict poor clinical outcome in hepatocellular carcinoma after curative resection. Am J Clin Pathol. 2009;131(4):498–510. doi: 10.1309/AJCP86PPBNGOHNNL. [DOI] [PubMed] [Google Scholar]
- 20.Mazzocca A, Fransvea E, Dituri F, Lupo L, Antonaci S, Giannelli G. Down-regulation of connective tissue growth factor by inhibition of transforming growth factor beta blocks the tumor-stroma cross-talk and tumor progression in hepatocellular carcinoma. Hepatology. 2010;51(2):523–534. doi: 10.1002/hep.23285. [DOI] [PubMed] [Google Scholar]
- 21.Bárcena C, Stefanovic M, Tutusaus A, et al. Angiogenin secretion from hepatoma cells activates hepatic stellate cells to amplify a self-sustained cycle promoting liver cancer. Sci Rep. 2015;5:7916. doi: 10.1038/srep07916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makino Y, Hikita H, Kodama T, et al. CTGF mediates tumor-stroma interactions between hepatoma cells and hepatic stellate cells to accelerate HCC progression. Cancer Res. 2018;78(17):4902–4914. doi: 10.1158/0008-5472.CAN-17-3844. [DOI] [PubMed] [Google Scholar]
- 23.Schoepp M, Ströse AJ, Haier J. Dysregulation of miRNA expression in cancer associated fibroblasts (CAFs) and its consequences on the tumor microenvironment. Cancers. 2017;9(6):E54. doi: 10.3390/cancers9060054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang DY, Goossens N, Guo J, et al. A hepatic stellate cell gene expression signature associated with outcomes in hepatitis C cirrhosis and hepatocellular carcinoma after curative resection. Gut. 2016;65(10):1754–1764. doi: 10.1136/gutjnl-2015-309655. [DOI] [PMC free article] [PubMed] [Google Scholar]