Abstract
Background and Aims
We aimed to investigate the underlying mechanism of action of risankizumab, a monoclonal antibody targeting the IL-23 p19 subunit, previously reported to induce clinical and endoscopic remission in a randomised phase II study in patients with active Crohn’s disease.
Methods
Ileum and colon biopsies obtained at screening and Week 12 from a subgroup of patients [n = 106] in the risankizumab phase II study were analysed by transcriptome-wide RNA-Seq profiling. Univariate associations were assessed using linear modelling.
Results
By Week 12, risankizumab significantly decreased [p < 0.005] the expression of 1880 and 765 genes in the colon [false-discovery rate = 0.02] and ileum [false-discovery rate = 0.05], respectively. These genes were associated with the IL-23/IL-17 axis, Th1 pathway, innate immunity, and tissue turnover. Colonic transcriptomic profiles following risankizumab treatment reflected the transcriptomic changes observed in patients achieving endoscopic response and remission at Week 12 and were significantly different from placebo [p < 0.005]. The colonic transcriptomic profile, significantly modulated by risankizumab at Week 12, was indicative of suppression of pathways associated with epithelial biology. Furthermore, pathways associated with Crohn’s disease modulated by risankizumab treatment included second messenger-mediated signalling, immune response, lymphocyte and leucocyte activation, lymphocyte differentiation and cell–cell adhesion.
Conclusions
Endoscopic remission and response observed with risankizumab in patients with active Crohn’s disease was associated with significant transcriptomic changes in the colon, compared with placebo. Differentiated expression of genes associated with the IL-23/IL-17 axis was observed in the colon and ileum 12 weeks after risankizumab treatment.
Keywords: Clinical trials, biomarkers, endoscopy
1. Introduction
Crohn’s disease [CD] is a chronic inflammatory bowel disease that is characterised by ulceration and transmural inflammation.1 IL-23 is believed to be a key mediator in the pathogenesis of CD.2–5 The Th17/IL-23 pathway contains at least 22 proteins, and 10 of the encoding genes reside in loci that are associated with inflammatory bowel disease: IL23R, TYK2, RORC, IL21, IL12B, CCR6, JAK2, IFNG, SMAD3, and STAT3.6 It has been hypothesised that the binding of IL-23 by the IL-23 receptor [IL-23R] results in activation of Janus kinase 2 [JAK2] and tyrosine kinase 2 [Tyk2] and subsequent phosphorylation of signal transducer and activator of transcription 1 [STAT1], STAT3, and STAT4, leading to the transcription of other pro-inflammatory cytokines.7 This increased transcription of pro-inflammatory cytokines is thought to result in the differentiation of CD4+ T cells into pro-inflammatory Th17 cells and the generation of an inappropriate immune response to commensal bacteria or luminal antigens in the gut.8 IL-23 promotes differentiation and also proliferation of Th17 cells through the IL-23R on Th17 cells, and patients with CD have elevated levels of IL-17A- and IL-22-producing T cells compared with healthy controls.9 Further increased levels of peripheral Th17 cells have been observed in late but not early CD.10
Selective inhibition of IL-23 with risankizumab, a monoclonal antibody targeting the IL-23 p19 subunit,11 in randomised, double-blind, phase I [placebo-controlled] and randomised, double-blind, phase II [active-controlled] clinical trials was associated with substantial and durable clinical improvement in patients with moderate-to-severe plaque psoriasis.12,13 More recently, risankizumab was more effective than placebo in inducing clinical remission, endoscopic response, and remission in patients with active CD.14 Furthermore, risankizumab was shown to significantly reduce levels of biomarkers of disease activity in faeces and serum, such as faecal calprotectin and IL-22, and C-reactive protein in the serum, after 12 weeks of treatment.14 Therefore, selective blockade of IL-23 via inhibition of p19 might be a viable therapeutic approach in CD.
The objective of the current biopsy sub-study was to gain an understanding of the mechanism of action of risankizumab, based on analyses of changes in the transcriptomic profile in the colon and ileum of patients with active CD in response to treatment. Association of these changes with endoscopic (CD Endoscopic Index of Severity [CDEIS]) response and remission, as well as faecal calprotectin [FCP] levels was determined. We also gained insights into the effects of blockade of IL-23 on pathways related to inflammatory bowel disease identified from a published meta-analysis of multiple gene expression datasets.15
2. Materials and Methods
2.1. Study population and design
Eligible patients in the phase II study [ClinicalTrials.gov number NCT02031276] had a diagnosis of CD for at least 3 months and, at screening, had moderate-to-severe CD defined as a CD Activity Index [CDAI] of 220–450, with mucosal ulcers in the ileum and/or colon, and a CDEIS ≥7 [≥4 for patients with isolated ileitis] on ileocolonoscopy scored by a blinded central endoscopy reader [previously described in Feagan et al.14]. Patients either naïve to or experienced with one or more tumour necrosis factor [TNF] antagonists were included in the study. Patients were randomly assigned [1:1:1] to receive either of two regimens of risankizumab [200 mg or 600 mg] or placebo at Weeks 0, 4, and 8, over 12 weeks.
2.2. Tissue biopsy collection and processing
In the phase II study, biopsies were collected from 53 patients treated with risankizumab and 26 patients treated with placebo. Sites were instructed to collect 6–8 biopsy samples from the region in the colon and/or ileum with the most severe inflammation at baseline, and to obtain a second set of biopsies from the same location at Week 12. Multiple biopsy samples were collected in phosphate-buffered saline solution and pooled, with separate tubes for samples from ileum and samples from the colon, before the addition of RNAlater stabilisation reagent for analysis. RNA was purified after homogenisation using a Qiagen RNeasy Fibrous Tissue Mini Kit [Qiagen, Hilden, Germany] according manufacturer’s instructions. Determination of RNA concentration and purity was performed by absorbance measurements using a NanoDrop spectrophotometer. For RNA sequencing, a polyA-based TruSeq V2 RNA sample preparation kit v2 [Illumina Inc., San Diego, CA] was used according to manufacturer’s instructions on available ileum and colon biopsies from patients.
2.3. Transcriptome analysis of colon and ileum biopsies
Global transcriptome-wide sequencing of RNA from colon and ileum biopsy samples from the 79 patients described earlier was performed using the Illumina Hi-Seq 2000 [Illumina Inc., San Diego, CA, USA]. Sequenced reads were mapped to human genome version hg19 using STAR aligner.16 Gene expression intensities and respective read counts were derived using cufflinks17 based on Ensembl release 70.18 An initial prefiltering step to remove transcripts with low expression was performed. A count of at least five was considered expressed. Based on the minimum library size of approximately 15 million, a cut-off of 0.3 counts per million was used and only transcripts with at least 10 in all location-treatment-visit groupings were selected. The samples were normalised using the trimmed mean of M-values [TMM] method.
The analyses to detect differential gene expression were performed at the univariate level. A repeated-measures linear regression model was used, with location of sample [ileum or colon], treatment [combined risankizumab doses or placebo], visit [baseline, Week 12], and location-treatment-visit interaction as fixed effects and patient as a random effect. The appropriate contrasts were then calculated for the fold changes [FDs] between risankizumab and placebo in the ileum and the colon. The R-packages limma with voom transformation were used for analyses. To account for correlation between subjects, the duplicate correlation function was used with subject-location as a blocking factor;19,20p-values of <0.005 were considered significant. The Benjamini–Hochberg procedure was applied to compute the false-discovery rate [FDR] and adjusted p-values. The approach used was to filter on p-values and state the FDR.
A one-tailed Fisher’s exact test was applied to test for over-representation of differentially expressed genes [absolute FC ≥2, p < 0.005] in the MSigDB Hallmark gene set21 and four selected MetaBase™ pathways,22 namely immune response IL-12, immune response IL-17, immune response IL-22, and immune response IL-23 signalling pathways. Gene sets with p < 0.01 were considered to be significantly enriched in deregulated genes.
To compare genes expressed in the colon and modulated by risankizumab with genes dysregulated in patients with CD versus normal healthy controls, data provided by Granlund et al.15 were used. Genes significantly deregulated in CD versus normal healthy controls [CD—N, corrected p ≤ 0.05, reported in the Supplementary Table S2 of Granlund et al.] were compared with genes significantly deregulated in the colon 12 weeks after treatment with risankizumab [p < 0.005].
2.4. Assessment of miRNAs in colon and faeces
Global transcriptome-wide sequencing of miRNA from 40 patients with colonic or ileocolonic CD was achieved using the CleanTag Small RNA Library Prep Kit protocol [TriLink BioTechnologies, San Diego, CA, USA], according to the manufacturer’s instructions, and the Illumina HiSeq 2000 [Illumina Inc., San Diego, CA, USA]. In addition, faecal miRNAs from 14 patients with matching colon biopsies were analysed using a NanoString’s human V3 CodeSet [based on miRBase v21] [NanoString Technologies, Seattle, WA, USA] that contains more than approximately 700 human miRNAs. In brief, total RNA was mixed with pairs of capture and reporter probes and hybridised on the nCounter Prep Station, and purified complexes were quantified on the nCounter Digital Analyzer and analysed by nSolver software [v1.1; NanoString Technologies, Seattle, WA]. Sequenced reads were adapter-trimmed and mapped to the human genome version hg19 using STAR aligner.16 Read counts were obtained using subread’s featureCounts,24 based on miRBase v19 annotation.25
Data were normalised using the TMM method described by Robinson and Oshlack,25 and the limma package19 was used to derive Log2FCs and corresponding FDR-adjusted p-values. Briefly, data were voom-transformed,20,26 correlations between baseline and Week 12 measurements per patient were estimated by the duplicateCorrelation function, a linear model was fitted using the lmFit-function and, finally, moderated t-statistics were computed.27 For the NanoString data analysis, the background threshold for each lane was set to the mean count level of the negative controls plus two standard deviations. Next, the positive normalisation factor was calculated as the mean of all positive controls divided by the mean of positive controls for the respective lane. Finally, miRNAs count levels were normalised to the geometric mean of the top 100 expressed miRNAs. The average of the geometric means of the top 100 expressed miRNAs for all lanes divided by the geometric mean of the respective lane was used as the normalisation factor per lane.
2.5. Assessment of faecal calprotectin and faecal lactoferrin and correlation with CDEIS
Calprotectin was assessed in faecal samples collected at baseline and Week 12 using the Bühlmann ELISA test [Schönenbuch, Switzerland]. Lactoferrin was assessed in faecal samples collected at baseline and Week 12 using the IBD-SCAN® [TECHLAB, Blacksburg, VA]. Spearman’s correlation coefficients were calculated for the correlations between the percent change from baseline in faecal calprotectin and faecal lactoferrin levels with change from baseline in CDEIS scores.
2.6. Ethics approval
The study protocol was approved by the institutional review board or ethics committee at each participating centre and conducted in accordance with applicable regulations and guidelines governing clinical study conduct, and with the ethical principles that have their origin in the Declaration of Helsinki. All patients who participated in the study provided written informed consent.
3. Results
3.1. Baseline patient characteristics
The baseline characteristics of the biopsy sub-population in the phase II study are described in Table 1. The biopsy sub-population contained a higher percentage of women than men. There were no major differences between the treatment groups in disease duration or CDEIS scores, although the clinical pattern did vary somewhat. There was a higher percentage of patients classified as ileocolonic than as colon or ileum in the biopsy sub-population only. The majority of patients were TNF-antagonist experienced.
Table 1.
Baseline characteristics of the biopsy sub-population in the phase II study.
| Placebo | Risankizumab | ||
|---|---|---|---|
| 200 mg | 600 mg | ||
| Patients with biopsies, n | 32 | 37 | 37 |
| Male, n [%] | 11 [34] | 14 [38] | 14 [38] |
| Age, years | 36 [14] | 38 [13] | 38 [12] |
| Disease duration, years | 12 [10] | 14 [9] | 13 [10] |
| Clinical disease location, n [%] | |||
| Ileum | 4 [13] | 6 [16] | 8 [22] |
| Ileocolonic | 13 [41] | 23 [62] | 14 [38] |
| Colonic | 15 [47] | 8 [22] | 14 [38] |
| Missing | 0 | 0 | 1 [3] |
| CDAI | 316 [93] | 317 [80] | 297 [63] |
| CDEIS | 13 [7] | 14 [6] | 14 [6] |
| CRP, mg/L | 27.4 [37.0] | 22.1 [24.1] | 18.5 [21.9] |
| Calprotectin, μg/g | 3006 [3672] | 2975 [5087] | 3087 [4899] |
| Previous TNF antagonist use, n [%] | 30 [94] | 35 [95] | 34 [92] |
| Concomitant corticosteroids or IM, or both, n [%] | |||
| Corticosteroid only | 6 [19] | 7 [19] | 9 [24] |
| Corticosteroid and IM | 4 [13] | 2 [5] | 2 [5] |
| IM only | 6 [19] | 7 [19] | 5 [14] |
| None | 16 [50] | 21 [57] | 21 [57] |
Patients could have had biopsies taken from colon only, colon and ileum, or ileum only. Data are mean (standard deviation [SD]) unless indicated otherwise.
CDAI, Crohn’s Disease Activity Index; CDEIS, Crohn’s Disease Endoscopic Index of Severity; CRP, C-reactive protein; IM, immunomodulator; SD, standard deviation; TNF, tumour necrosis factor.
3.2. Transcriptomic changes induced by risankizumab in the colon versus the ileum
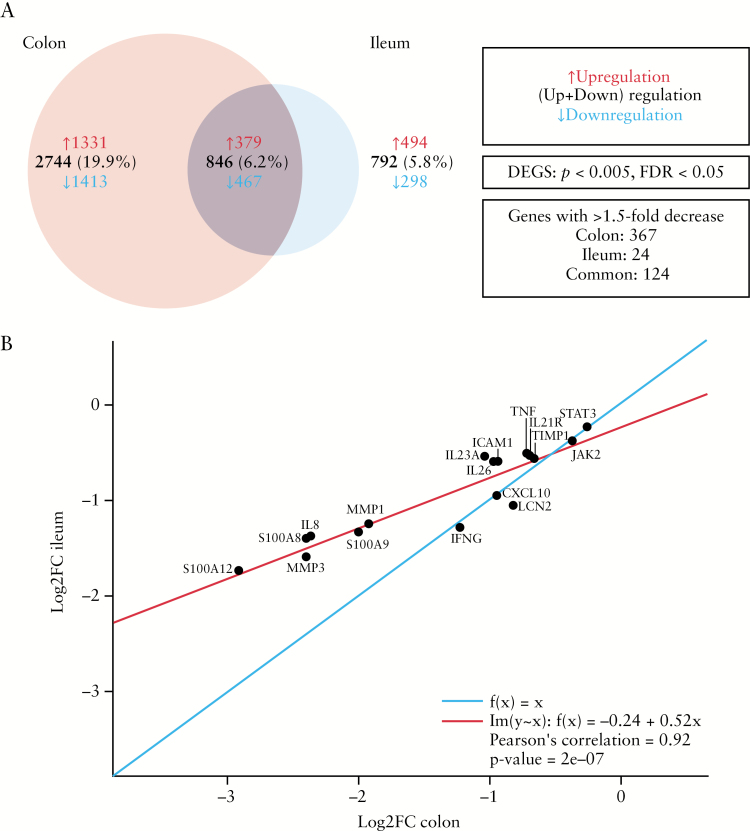
A total of 277 RNA-Seq samples [baseline and Week 12] were included in the analysis. There were 53 patients on risankizumab and 26 on placebo with at least one colon sample at baseline, and 56 patients on risankizumab and 22 on placebo with at least one ileum sample at baseline. Comparison of genes expressed in the colon and modulated by risankizumab with genes dysregulated in patients with CD versus normal healthy controls, by Granlund et al.,15 has shown an overlap in 1063 genes upregulated in CD versus normal healthy controls and downregulated upon risankizumab treatment, as well as 933 genes upregulated by risankizumab treatment and downregulated in CD versus normal healthy controls [Supplementary Figure S1a and b, available as Supplementary data at ECCO-JCC online]. Overall, there were significant decreases [p < 0.005] in expression of 1880 genes in the colon [FDR = 0.02] versus 765 genes in the ileum [FDR = 0.05] from baseline to Week 12 with risankizumab treatment [pooled 200-mg and 600-mg doses; Figure 1A]. Of these reductions in expression, there were 491 genes with >1.5-fold decrease from baseline to Week 12 in the colon compared with 148 with >1.5-fold decrease from baseline to Week 12 in the ileum.
Figure 1.
[A] Venn diagram showing numbers of common and unique genes modulated in the colon and ileum 12 weeks following treatment with risankizumab. [B] Correlation plot of Log2FC in expression of select genes associated with the IL-23/IL-17 axis and Crohn’s disease [CD] by means of RNA-Seq for colon and ileum after treatment with risankizumab. The red line represents the regression line of best fit [Pearson's correlation = 0.746] and the blue line represents a regression line of 1. DEG, differentially expressed gene; FC, fold change; FDR, false-discovery rate.
Significant changes in the expression of genes both unique and common to the colon and the ileum were associated with risankizumab treatment. Most genes that were decreased in the colon and ileum were associated with the pathogenesis of CD and the Th17 pathway. Treatment with risankizumab resulted in reduction in expression of known inflammatory genes associated with CD, including S-100A8, S-100A9, IL8, MMP1, IFNG, LCN2 [lipocalin], TIMP1, TNF, and STAT3 in the colon/ileum, and larger reductions [log FC < -1.5 for ileum and colon] in the expression of S-100A12 and MMP3 [Figure 1B]. No significant changes [p ≤ 0.05] in transcriptomic profiles were observed with placebo in the colon [FDR = 1.0] or ileum [FDR = 0.74].
3.3. Pathway analyses of genes modulated by risankizumab in the colon versus the ileum
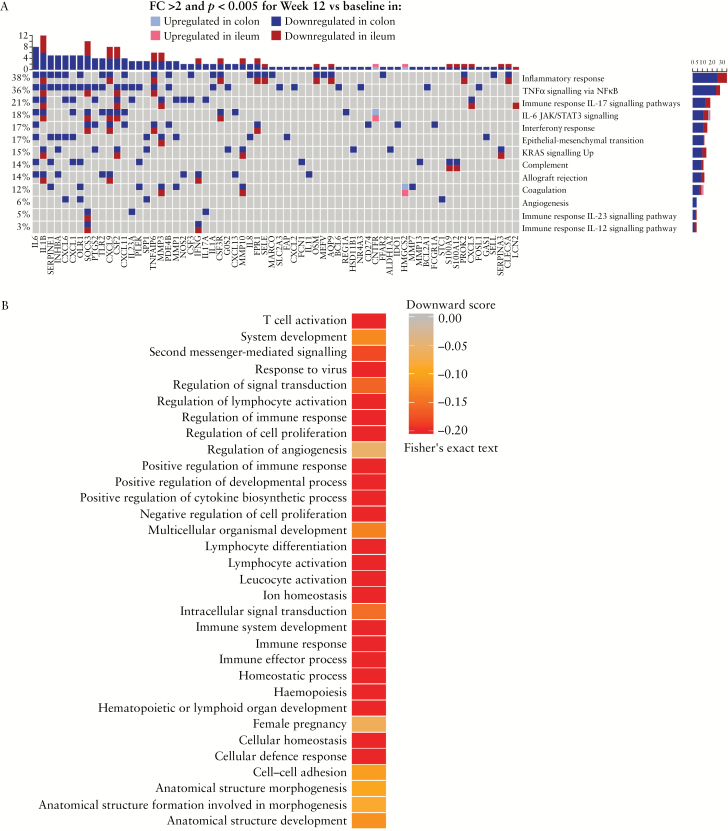
We assessed the effect of risankizumab on select biological states, processes, and signalling pathways, considering genes with absolute FCs of ≥2 [p < 0.005] modulated in the colon and ileum from baseline to Week 12 [Figure 2A]. Risankizumab treatment modulated genes associated with inflammatory response, several cytokine signalling pathways including IL-17, IL-6 JAK/STAT3, IFNγ, and IL-12 signalling, genes upregulated by KRAS activation, and TNFα signalling via NFκB in both the colon and the ileum [Figure 2A]. Apart from IL-12 signalling, in all of these gene sets or pathways more genes were differentially expressed in the colon compared with the ileum. Select genes associated with the Th17/IL-23 pathway modulated by risankizumab versus placebo in the colon are shown in Supplementary Table S1, available as Supplementary data at ECCO-JCC online.
Figure 2.
[A] Comparison of expression of hallmark pathways [Broad Institute] and selected Metabase™ signalling pathways modulated by risankizumab in the colon versus the ileum; and [B] expression of pathways relevant to CD pathogenesis using gene sets and algorithm from Granlund et al.15 CD, Crohn’s disease; IFN, interferon; JAK, Janus kinase; NES, normalised enrichment score33; STAT, signal transducer and activator of transcription; TNF, tumour necrosis factor; UV, ultraviolet.
Other gene sets encoding IL-23 signalling, components of the complement system, angiogenesis, upregulated during transplant rejection [allograft rejection], and epithelial–mesenchymal transition were only significantly enriched by genes deregulated in the colon based on the chosen cut-offs.
Differential effects of risankizumab on the colon transcriptomic profile in patients with active CD has shown modulation of disease-relevant gene ontogeny [GO] pathways, identified from a published meta-analysis of CD and UC Affymetrix datasets [Figure 2B].15 Risankizumab more strongly decreased expression of genes associated with pathways linked to second messenger-mediated signalling, lymphocyte differentiation, lymphocyte activation, leucocyte activation, immune response, and immune effector response compared with pathways associated with cell–cell adhesion, regulation of angiogenesis, anatomical structure, and development.
3.4. Transcriptomic profiles in risankizumab-treated patients versus placebo and association with endoscopic response and remission
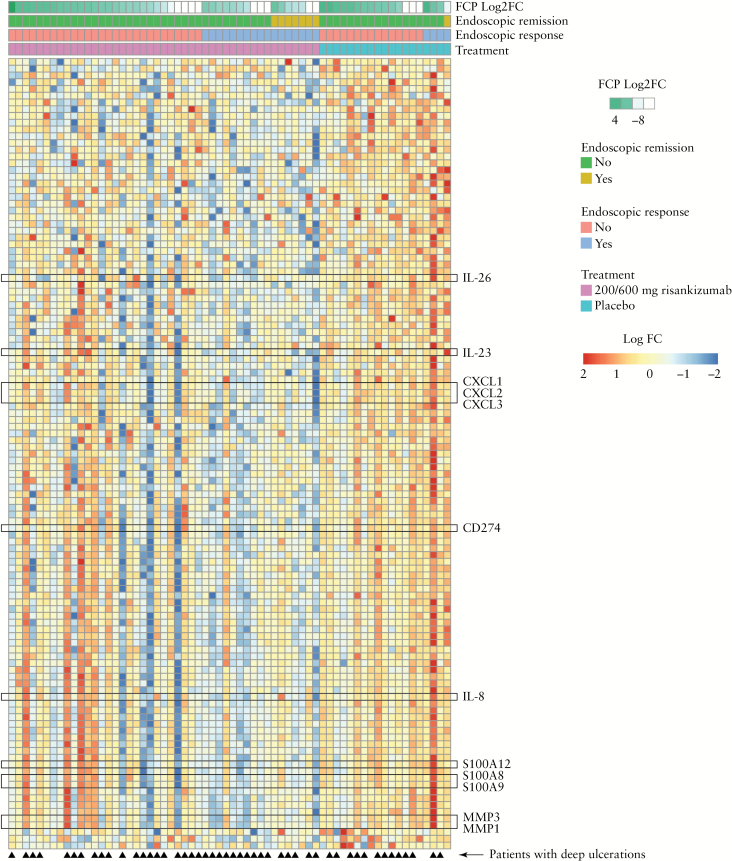
Treatment with risankizumab resulted in significant decreases in the expression of 115 genes [FC > 0.6; p < 0.005, FDR = 0.37] in the colon but not the ileum, compared with placebo [Figure 3 and Supplementary Figure S2, available as Supplementary data at ECCO-JCC online]. The number of genes downregulated from baseline to Week 12 with the 600-mg risankizumab dose was 805 [p = 0.05, FDR = 0.5] compared with 801 that were upregulated from baseline to Week 12 [p = 0.05, FDR = 0.5]. Although a significant number of genes were downregulated from baseline to Week 12 [843, p = 0.05] by the 200-mg risankizumab dose and 344 genes were upregulated, this was associated with a high FDR [0.67], making it difficult to interpret the results. Many of the genes with decreased expression were associated with the IL-23 pathway [IL26, IL23A, CD274, CCL20] and CD [CXCL1, CXCL2, CXCL3, MMP3, MMP1, IL8, IL6, TNFa, S-100A8, S-100A9, S-100A12]. Comparing the risankizumab doses, there were 152 genes that were downregulated (p < 0.05, FC > 1.5) and 33 that were upregulated [p < 0.05, FC > 1.5] by both the 200-mg and 600-mg risankizumab doses, compared with placebo. The genes decreased in expression between the two dose groups included CCL18, SLC12A3, CXCL2, CDF3, IL26, IL6, MMP3, CLEC4D, S-100A8, and S-100A12. Changes in the expression of select genes in the IL-23 pathway and associated with CD, as assessed by RNA-Seq, were also confirmed by TaqMan polymerase chain reaction [PCR; see Supplementary Figure S3, available as Supplementary data at ECCO-JCC online]. Risankizumab-treated patients who achieved endoscopic response or remission showed reduction in the expression of select genes in colon biopsies compared with patients who did not achieve endoscopic response or remission from baseline to Week 12 [Figure 3]. Furthermore, there were select patients treated with risankizumab who were classified as not achieving endoscopic response, but appeared to show transcriptomic profiles similar to those with endoscopic response at Week 12. By contrast, patients receiving placebo showed increases in gene expression compared with those receiving risankizumab, reflecting the overall transcriptomic profile. In patients with the most severe endoscopic lesions [deep ulcerations] at baseline, a larger proportion of risankizumab-treated patients achieved endoscopic response at Week 12 compared with placebo, and these patients showed a transcriptomic pattern indicating reductions in the expression of key genes associated with active CD [Figure 3]. Higher proportions of patients who had deep ulcerations at baseline in any segment of the colon achieved endoscopic response at Week 12 in the combined 200-mg and 600-mg risankizumab dose groups compared with patients with deep ulcerations in the ileum [Table 2].
Figure 3.
Heat map showing changes in transcriptomic profile in colon biopsies from patients receiving risankizumab and placebo at Week 12. Individual patient transcriptomic profiles are categorised by Log2-fold change in FCP, endoscopic remission, endoscopic response, and treatment received. White boxes for FCP indicate missing values [n = 12 patients]. The heat map with the full listing of genes is provided in Supplementary Figure 2, available as Supplementary data at ECCO-JCC online. FCP, faecal calprotectin.
Table 2.
Proportion of patients who achieved endoscopic response at Week 12 in patients with deep ulcerations at baseline in segments with biopsies.
| Location of deep ulceration, n [%] | Endoscopic response | Placebo | Pooled risankizumab [200 mg and 600 mg] |
|---|---|---|---|
| Colona | Yes/no | 1 [14.3]/7 [85.7] | 13 [44.8]/16 [55.2] |
| Ileum | Yes/no | 0 [0.0]/5 [100] | 5 [25]/15 [75] |
aIncludes rectum, sigmoid and left colon, right colon, transverse colon.
CDEIS, Crohn’s Disease Endoscopic Index of Severity.
3.5. Association of changes in faecal biomarkers with transcriptomic profiles
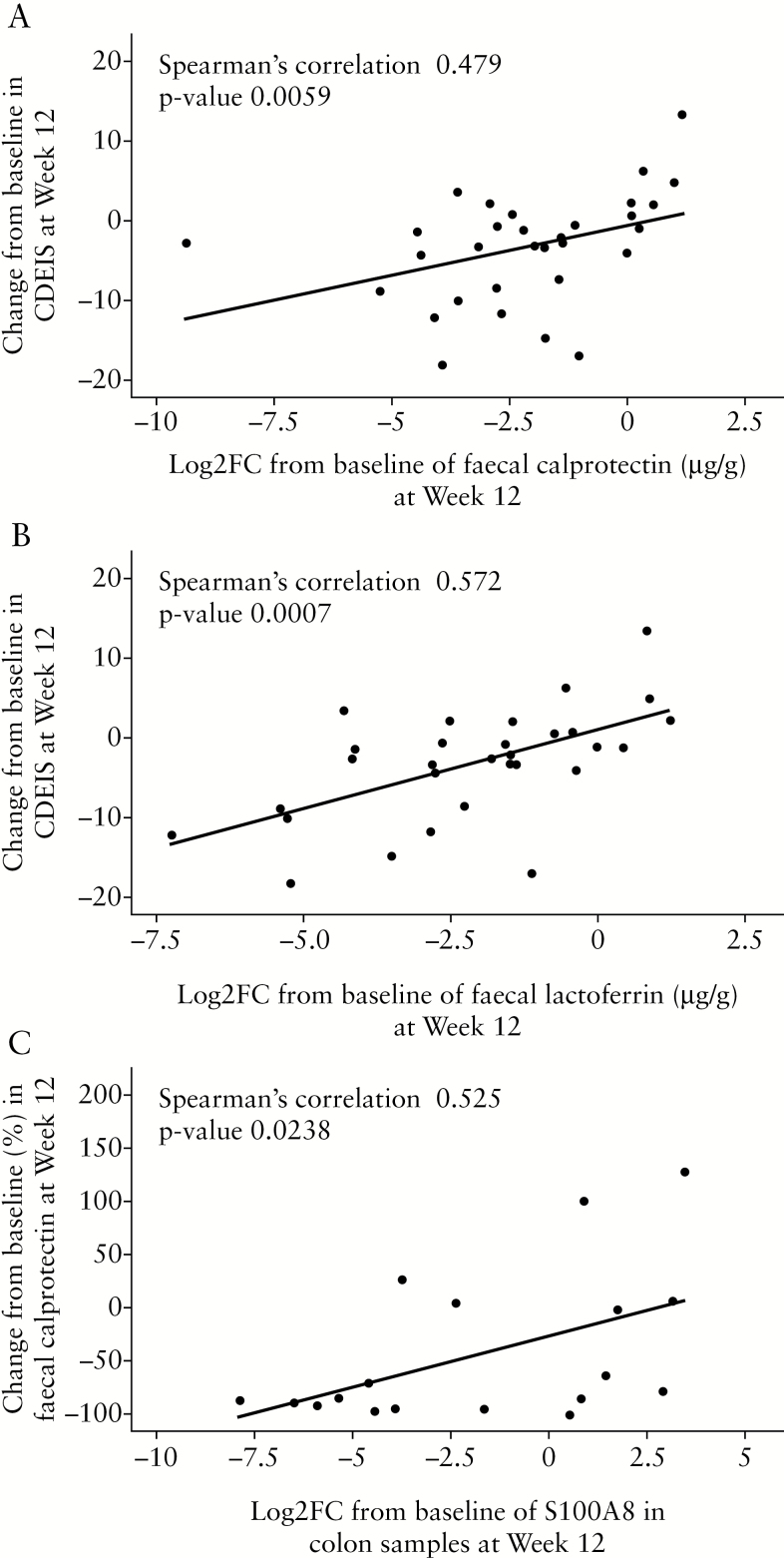
Significant reductions [baseline to Week 12] in faecal levels of calprotectin [74.4% vs 2.5%]14 and lactoferrin [69% vs 18.4%, unpublished findings] proteins were observed in patients with CD treated with 600 mg risankizumab compared with placebo. There were significant correlations between changes in faecal calprotectin [r = 0.479, p = 0.0059] or changes in lactoferrin levels [r = 0.572, p = 0.0007] and changes in CDEIS scores from baseline to Week 12 in patients treated with 600 mg risankizumab [Figure 4A and B]. A correlation was observed between the expression of calprotectin at the protein level in faeces and the reduction in the expression of S-100A8 transcript in colon biopsies in patients treated with 600 mg risankizumab [r = 0.525, p = 0.0238] and associated changes in the transcriptomic profile, as reflected in Figure 4C. Furthermore, mucosal transcriptions of S-100A8 and S-100A9 [calprotectin] were decreased in patients with reductions in protein levels of faecal biomarkers [calprotectin/lactoferrin].
Figure 4.
Correlation of change from baseline to Week 12 in CDEIS score with [A] Log2-fold change [FC] from baseline in faecal calprotectin; and [B] Log2FC from baseline in faecal lactoferrin in the colon in the 600-mg risankizumab dose group. [C] Correlation of changes from baseline to Week 12 in faecal calprotectin levels with S-100A8 transcript expression in the colon in the 600-mg risankizumab dose group. CDEIS, Crohn’s Disease Endoscopic Index of Severity.
3.6. Association of changes in tissue and faecal miRNAs
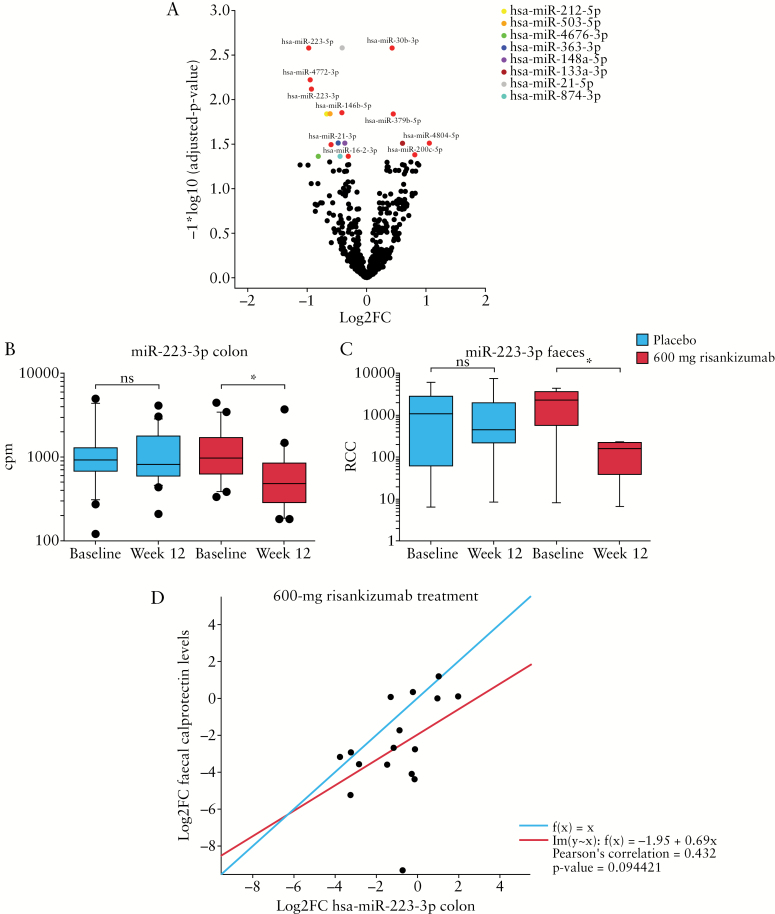
Small RNA sequencing of colon biopsies comparing baseline and Week 12 samples identified 18 significant differentially expressed miRNAs [FDR-adjusted p < 0.05] in patients treated with 600 mg risankizumab [Figure 5A]. Of these, 13 miRNAs were downregulated and five miRNAs were upregulated by risankizumab. Comparing matching colon and faecal samples from 14 patients, a strong correlation was observed for miR-223-3p. However, none of the identified miRNAs was significantly differentially expressed in both the colon and faeces in a standard differential expression analysis by limma and nSolver, respectively [data not shown]. Downregulated levels of miR-223-3p at Week 12 were observed only in risankizumab-treated patients [Figure 5B and C]. This result was not associated with endoscopic response or remission. Changes in levels of miR-223-3p, however, correlated with changes in faecal calprotectin levels in the 600-mg risankizumab dose group [Figure 5D].
Figure 5.
[A] Differentially expressed miRNAs in colon biopsies 12 weeks after treatment with 600 mg risankizumab [Log2FC from baseline] versus baseline. Coloured dots represent each of the differentially expressed miRNAs. Levels of miR-223-3p in [B] colon and [C] faeces in patients receiving placebo and 600 mg risankizumab at baseline and Week 12. Whiskers represent the 10th and 90th percentiles of expression. Datasets for figures [B] and [C] were analysed for statistical significance using two-tailed unpaired Student's t-test; *p < 0.05. [D] Correlation of Log2FC in faecal calprotectin levels with Log2FC in miR-223-3p expression in colon biopsy tissue in the 600-mg risankizumab dose group. The red line represents the regression line of best fit [Pearson's correlation = 0.432] and the blue line represents a regression line of 1. Cpm, counts per million; FC, fold change; ns, not significant; RCC, reporter code count.
4. Discussion
To our knowledge, this is the first study to characterise the effects of a selective anti-IL-23 antibody on the transcriptomic profile in the colon and ileum of patients with endoscopically active CD [despite most patients having had previous treatment with a TNF antagonist], which was significantly altered following 12 weeks of therapy with risankizumab. In this biopsy sub-population from the phase II study, patients treated with risankizumab who achieved endoscopic response or remission showed a differentiated transcriptomic profile in colon tissue compared with patients who did not achieve endoscopic response or remission at Week 12. A small number of risankizumab-treated patients classified as endoscopic non-responders had a similar transcriptomic profile in the colon as patients classified as endoscopic responders, suggesting that molecular changes in the tissue may precede endoscopic changes. These results may also represent small differences in the CDEIS scores between patients classified as endoscopic responders versus non-responders.
Greater changes in the number of genes expressed were observed in the colon compared with the ileum. Accordingly, there were several gene sets and pathways that were more significantly enriched by differentially expressed genes for the colon compared with the ileum. A recent study dissecting molecular pathways in the tissue of patients with CD, compared with non-inflammatory bowel disease patients, has shown differences in pathways related to interferon and interleukin signalling.28 This is supportive of the relevance of our results from the enrichment analysis showing that risankizumab modulated gene sets associated with inflammatory response, IL-17, IL-6 JAK/STAT3, IFNγ signalling, and key pathogenic processes that occur in CD more strongly in the colon than the ileum. Previous studies have shown that patients who are refractory to anti-TNF therapies maintain differentially expressed genes in the colon29 and the ileum,30 consistent with our results.
A limitation of this study was that biopsies were not collected from uninvolved mucosa, and thus treatment effects in healthy mucosa could not be assessed. However, a comparison of the changes in the transcriptomic profile by risankizumab in our study, with the changes in the transcriptomic profile dysregulated in patients with CD versus normal healthy controls previously reported by Granlund et al.,15 showed an overlap in 1063 genes upregulated in patients with CD versus healthy subjects and downregulated upon treatment with risankizumab. This overlap suggests that the genes evaluated do represent genuine effects of risankizumab treatment.
It is clear from the meta-analysis by Granlund et al.,15 focused on gene regulation and processes that were stably correlated with CD and ulcerative colitis disease, that a dominance of Th1 and Th17 pathways is observed. There was consistency in the normalised enrichment score across the most highly ranked categories and studies, implying that similar disease processes occur in both CD and ulcerative colitis. The most prominent pathways were those related to inflammation, as it is associated with innate and adaptive immunity, cell proliferation, as it is related to epithelial cell regeneration in response to tissue damage, and angiogenesis. We were able to build on this analysis to better understand the changes in the transcriptomic profiles in the colon and ileum upon treatment with risankizumab. Overall, many identified pathways were modulated by risankizumab treatment including second messenger-mediated signalling, immune response, lymphocyte/leucocyte activation, lymphocyte differentiation, and cell–cell adhesion.
In the subset of patients with biopsies, a higher proportion of patients with deep ulcerations in the colon, compared with those with deep ulcerations in the ileum at baseline, achieved endoscopic response at Week 12 following risankizumab treatment. A similar transcriptomic profile in the colon was observed in patients with deep ulcerations at baseline and endoscopic response versus those without deep ulcerations. The changes observed in the transcriptomic profile in the colon following 12 weeks of risankizumab treatment were not only associated with endoscopic responses and remission scores, but also with changes in faecal calprotectin levels. Furthermore, risankizumab-treated individuals with reductions in levels of faecal protein biomarkers [calprotectin and lactoferrin] showed decreases in the expression of S-100A8/A9 [calprotectin] genes in the colon and ileum. Increased expression levels of S-100A8/A9 genes and associated proteins released from neutrophils and macrophages have generally been observed in colon and ileum tissues and faeces of patients with CD, and correlate with elevated levels of Th17-associated cytokines.31 Treatment with risankizumab decreased expression of genes and proteins associated with the Th17 pathway, including IL-23, IL-26, and IL-17A, and these changes were associated with reductions in S-100A8/A9 in patients with CD.
Increased levels of miR-223-3p have been observed in the serum of patients with CD compared with healthy controls.32 IL-23 can induce the expression of miR-223-3p in intestinal tissue in patients with CD, and the associated downregulation of Claudin-8 [mRNA target] can result in a disruption of the homeostasis of the intestinal barrier.32 Here, we have also shown that 18 miRNAs were deregulated in the colon of patients treated with risankizumab versus placebo at Week 12. miR-223-3p was among the greatest downregulated miRNAs in the colon and in the faeces.
In this study, we were able to link changes in the expression of select genes in the colon tissue with changes in select miRNAs and changes in faecal protein biomarkers. Furthermore, these changes in tissue biomarkers correlated with endoscopic response/remission. Larger transcriptomic changes from baseline to Week 12 were observed in colon tissue compared with ileum tissue, showing a distinct profile. These results underscore the value of deep characterisation of biomarkers in the inflamed intestine of patients with endoscopically active CD [despite most patients having had previous treatment with a TNF antagonist] in response to new therapies, in order to generate hypotheses for differentiation from other treatments and potentially personalise care and optimise treatment outcomes.
Funding
This study was supported by Boehringer Ingelheim.
Conflict of Interest
SV, PB, RV, RS, WOB, SJP, and JSF report being employed by Boehringer Ingelheim; KMG, JWD, and KW report being employed by AbbVie Inc.; AS reports no conflicts of interests. JP reports personal fees from Boehringer Ingelheim during the conduct of the study; and personal fees for advisory boards from AbbVie, Amgen, Genentech Roche, Janssen, MSD, Novartis, Oppilan, Pfizer, Takeda, Theravance, and TiGenix, outside the submitted work.
Supplementary Material
Acknowledgments
Medical writing support was provided by Leigh Church of SuccinctChoice Medical Communications [London, UK] and funded by Boehringer Ingelheim.
Author Contributions
All authors made a substantial contribution to the concept and design of the work and the analysis and interpretation of the data. All authors made critical revisions for important intellectual content and they all approved the final version of the article and are accountable for its content.
References
- 1. Baumgart DC, Sandborn WJ. Crohn’s disease. Lancet 2012;380:1590–605. [DOI] [PubMed] [Google Scholar]
- 2. Miossec P, Kolls JK. Targeting IL-17 and TH17 cells in chronic inflammation. Nat Rev Drug Discov 2012;11:763–76. [DOI] [PubMed] [Google Scholar]
- 3. Neurath MF. IL-23: a master regulator in Crohn disease. Nat Med 2007;13:26–8. [DOI] [PubMed] [Google Scholar]
- 4. Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 2006;314:1461–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Geremia A, Arancibia-Cárcamo CV, Fleming MP, et al. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med 2011;208:1127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fransen K, van Sommeren S, Westra HJ, et al. Correlation of genetic risk and messenger RNA expression in a Th17/IL23 pathway analysis in inflammatory bowel disease. Inflamm Bowel Dis 2014;20:777–82. [DOI] [PubMed] [Google Scholar]
- 7. Parham C, Chirica M, Timans J, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol 2002;168:5699–708. [DOI] [PubMed] [Google Scholar]
- 8. Abraham C, Cho J. Interleukin-23/Th17 pathways and inflammatory bowel disease. Inflamm Bowel Dis 2009;15:1090–100. [DOI] [PubMed] [Google Scholar]
- 9. Dige A, Støy S, Rasmussen TK, et al. Increased levels of circulating Th17 cells in quiescent versus active Crohn’s disease. J Crohns Colitis 2013;7:248–55. [DOI] [PubMed] [Google Scholar]
- 10. Veny M, Esteller M, Ricart E, Piqué JM, Panés J, Salas A. Late Crohn’s disease patients present an increase in peripheral Th17 cells and cytokine production compared with early patients. Aliment Pharmacol Ther 2010;31:561–72. [DOI] [PubMed] [Google Scholar]
- 11. Singh S, Kroe-Barrett RR, Canada KA, et al. Selective targeting of the IL23 pathway: generation and characterization of a novel high-affinity humanized anti-IL23A antibody. MAbs 2015;7:778–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krueger JG, Ferris LK, Menter A, et al. Anti-IL-23A mAb BI 655066 for treatment of moderate-to-severe psoriasis: safety, efficacy, pharmacokinetics, and biomarker results of a single-rising-dose, randomized, double-blind, placebo-controlled trial. J Allergy Clin Immunol 2015;136:116–24.e7. [DOI] [PubMed] [Google Scholar]
- 13. Papp KA, Blauvelt A, Bukhalo M, et al. Risankizumab versus ustekinumab for moderate-to-severe plaque psoriasis. N Engl J Med 2017;376:1551–60. [DOI] [PubMed] [Google Scholar]
- 14. Feagan BG, Sandborn WJ, D’Haens G, et al. Induction therapy with the selective interleukin-23 inhibitor risankizumab in patients with moderate-to-severe Crohn’s disease: a randomised, double-blind, placebo-controlled phase 2 study. Lancet 2017;389:1699–709. [DOI] [PubMed] [Google Scholar]
- 15. Granlund Av, Flatberg A, Østvik AE, et al. Whole genome gene expression meta-analysis of inflammatory bowel disease colon mucosa demonstrates lack of major differences between Crohn’s disease and ulcerative colitis. PLoS One 2013;8:e56818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aken BL, Ayling S, Barrell D, et al. The Ensembl gene annotation system. Database 2016;2016:baw093. doi:10.1093/database/baw093. [DOI] [PMC free article] [PubMed]
- 19. Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu R, Holik AZ, Su S, et al. Why weight? Modelling sample and observational level variability improves power in RNA-seq analyses. Nucleic Acids Res 2015;43:e97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database [MSigDB] hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vermeire S, Schreiber S, Petryka R, et al. Clinical remission in patients with moderate-to-severe Crohn’s disease treated with filgotinib [the FITZROY study]: results from a phase 2, double-blind, randomised, placebo-controlled trial. Lancet 2017;389:266–75. [DOI] [PubMed] [Google Scholar]
- 23. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 24. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 2014;15:R29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat 2016;10:946–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weiser M, Simon JM, Kochar B, et al. Molecular classification of Crohn’s disease reveals two clinically relevant subtypes. Gut 2016;67:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Leal RF, Planell N, Kajekar R, et al. Identification of inflammatory mediators in patients with Crohn’s disease unresponsive to anti-TNFα therapy. Gut 2015;64:233–42. [DOI] [PubMed] [Google Scholar]
- 30. Arijs I, De Hertogh G, Machiels K, et al. Mucosal gene expression of cell adhesion molecules, chemokines, and chemokine receptors in patients with inflammatory bowel disease before and after infliximab treatment. Am J Gastroenterol 2011;106:748–61. [DOI] [PubMed] [Google Scholar]
- 31. Bourgonje AR, von Martels JZH, de Vos P, Faber KN, Dijkstra G. Increased fecal calprotectin levels in Crohn’s disease correlate with elevated serum Th1- and Th17-associated cytokines. PLoS One 2018;13:e0193202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang H, Chao K, Ng SC, et al. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome Biol 2016;17:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.