Abstract
The precise timing of progesterone signaling through its cognate receptor, the progesterone receptor (PGR), is critical for the establishment and maintenance of pregnancy. Loss of PGR expression in the murine uterine epithelium during the preimplantation period is a marker for uterine receptivity and embryo attachment. We hypothesized that the decrease in progesterone receptor A (PGRA) expression is necessary for successful embryo implantation. To test this hypothesis, a mouse model constitutively expressing PGRA (mPgrALsL/+) was generated. Expression of PGRA in all uterine compartments (Pgrcre) or uterine epithelium (Wnt7acre) resulted in infertility with defects in embryo attachment and stromal decidualization. Expression of critical PGRA target genes, indian hedgehog, and amphiregulin (Areg), was maintained through the window of receptivity while the estrogen receptor target gene, the leukemia inhibitory factor (Lif), a key regulator of embryo receptivity, was decreased. Transcriptomic and cistromic analyses of the mouse uterus at day 4.5 of pregnancy identified an altered group of genes regulating molecular transport in the control of fluid and ion levels within the uterine interstitial space. Additionally, LIF and its cognate receptor, the leukemia inhibitory factor receptor (LIFR), exhibited PGR-binding events in regions upstream of the transcriptional start sites, suggesting PGRA is inhibiting transcription at these loci. Therefore, downregulation of the PGRA isoform at the window of receptivity is necessary for the attenuation of hedgehog signaling, transcriptional activation of LIF signaling, and modulation of solutes and fluid, producing a receptive environment for the attaching embryo.
Keywords: progesterone receptor, implantation, uterine receptivity, progesterone signaling, mouse models
Summary Sentence
Expression of PGRA at the window of receptivity transcriptionally represses LIF signaling and aberrantly regulates hedgehog and solute signaling rendering the uterus unreceptive to the implanting embryo.
Introduction
In the United States, 15% of women suffer from infertility or the inability to conceive after 1 year [1]. Additionally, pregnancy loss was estimated to occur in 70% of all pregnancies, 60% occurring within the first 6 weeks [2]. This dramatic pregnancy loss indicates a critical point during early gestation controlling the maintenance and progression of pregnancy. This critical event is embryo implantation, occurring during a narrow time frame in which the uterus is prepared to receive the implanting embryo, known as the window of receptivity [3].
The uterus is a complex endocrine organ comprised of two major compartments: the dual outer muscle layer, known as the myometrium, and the inner glandular endometrium containing the stroma, endometrial glands, and the inner uterine epithelium. The uterus responds to the secretion of the ovarian steroid hormones, estrogen (E2) and progesterone (P4), which bind to their cognate receptors, the estrogen receptor (ER, ESR1) and progesterone receptor (PGR), respectively [4]. Within the cytoplasm, hormone-bound receptors dimerize and enter the nucleus to activate or inhibit their respective target genes. The PGR is comprised of two major isoforms, the progesterone receptor A (PGRA) isoform which is the dominant mouse isoform expressed in the entirety of the uterus, and the progesterone receptor B (PGRB) isoform which is limited in expression in the uterus [5].
Through the use of genetically engineered mouse models, we and others have been able to assess the function of the steroid hormone receptors during pregnancy [6,7]. The PGR was identified to be necessary for pregnancy, as the total PGR knockout mice or PGRKO mice exhibit defects in ovulation, embryo attachment, and uterine stromal differentiation and proliferation (a process known as decidualization) [8,9]. Furthermore, studies on knockout mouse models for both PGR isoforms revealed that the PGRA isoform is required for fertility while PGRB is essential for mammary gland development [10,11].
PGR expression in the uterine epithelium at days 2–3 of pregnancy is critical for the inhibition of estrogen-induced epithelial proliferation [12] and activation of P4 target genes, such as indian hedgehog (Ihh), required for embryo implantation [3]. Progesterone receptor message levels decrease in the luminal epithelium by day 5, while dramatically increasing in stromal expression by day 5 for the promotion of uterine decidualization [13]. The decrease in epithelial PGR expression is conjectured to define the narrow window in which the uterus is receptive to the implantation of the embryo [3]. However, the functional significance of decreased PGR expression has never been examined.
In order to test whether decreased expression of PGR in the uterine epithelium at the window of receptivity is necessary for proper embryo implantation, a mouse model constitutively expressing the PGRA isoform in a tissue-specific manner was generated. This overexpression model was made utilizing a Lox-STOP-Lox construct targeted to the ROSA-26 allele. When used in combination with the whole uterine Pgrcre [14] or the epithelial-specific Wnt7acre [15] Cre recombinase mouse models, constitutive expression of the PGRA isoform results in complete infertility with failure to undergo embryo implantation or uterine decidualization. Furthermore, at the time of implantation, Ihh was significantly upregulated while the critical cytokine necessary for embryo attachment [16], leukemia inhibitory factor (Lif), was severely attenuated independent of E2 signaling. Transcriptomic and cistromic analyses were utilized to further understand the function of PGRA at the window of implantation. This is the first study providing functional evidence for the necessity of suppressed P4 signaling in the success of embryo implantation.
Methods
Generation of the progesterone receptor A overexpression mouse
The cloning system utilized to generate the PGRA expression mouse model was previously described [17]. The murine cDNA for the PGRA isoform was cloned into a shuttle vector containing a transcription start site, flag and myc-tag epitopes, and polyadenylation sequence. The shuttle vector sequence was genetically recombineered into a base vector containing a chicken β-actin ubiquitous promoter, STOP cassette flanked by LoxP sites, and homology arms to the ROSA26 locus. DNA was purified and then electroporated into mouse embryonic stem cells.
The Embryonic Stem Cell Core at Baylor College of Medicine performed the ES cell injection and expansion of the purified DNA into AB2.2 mouse embryonic stem cells. The PGRA construct was targeted to the ROSA26 locus via regions of homology graphically described in Supplemental Figure S2A. Products positive for recombination were identified via Southern blot analysis (Supplemental Figure S2B). The Genetically Engineered Mouse Core at Baylor College of Medicine injected the positive embryonic stem cells into C57BL/6J strain blastocysts and transferred them to female recipients to generate chimeras. Generic ROSA26 locus primers previously described were utilized to identify the presence of the PGRA expression allele [17].
Mouse husbandry and fertility trial
Mice were cared for according to the protocol within the Institution of Animal Care and Use Committee at Baylor College of Medicine and the Animal Care and Use Committee at the National Institute of Environmental Health Sciences. Mice exhibiting the mPgrALsL/+ allele were bred to Wnt7acre/+ or Pgrcre/+ mice to generate Wnt7acre/+mPgrALsL/+ and PRcre/+mPgrALsL/+ mice, respectively. For the 6-month breeding trial, female Wnt7acre/+mPgrALsL/+ or Pgrcre/+mPgrALsL/+ mice were bred to C57BL/6J males for 6 months. The number of litters and pups and dates of delivery were recorded.
Assessment of embryo attachment
Wnt7acre/+mPgrALsL/+ and Pgrcre/+mPgrALsL/+ mice were mated to B6D2F1 intact male mice, and the presence of the copulatory plug was recorded. Mice were sacrificed in the morning, 5 days after the presence of the coital plug. Uteri and ovaries were immediately fixed overnight in 4% paraformaldehyde and dehydrated in 70% ethanol. The tissue was subsequently processed in increasing gradients of ethanol and xylene and embedded longitudinally flat into paraffin wax. After serial sectioning of the entire reproductive tract, every fifth slide was stained in hematoxylin and eosin (H&E). Presence of implanted embryos or floating embryos was recorded and summarized for Wnt7acre/+mPgrALsL/+ and Pgrcre/+mPgrALsL/+ mice versus wildtype mice.
Artificial decidual response
To artificially assess the ability of the stroma to undergo differentiation and proliferation at the time of embryo attachment, mice were ovariectomized and treated with exogenous hormones to mimic pregnancy before applying a manual stimulus to a single uterine horn (protocol outlined previously [9]). After ovariectomy and 2 weeks of rest, mice were administered daily E2 (100 ng/mouse) injections for 3 days. After 2 days of rest, mice were treated with E2 (6.7 ng/mouse) and P4 (1 mg/mouse) for 3 days. On the third day, mice were administered a single injection of 0.025 mL of sesame oil to the right uterine horn. Mice were administered E2 and P4 for 5 more days and sacrificed on the fifth day. Uterine wet weights for the stimulated and control horns were recorded. Weight ratios were calculated by dividing stimulated horn weight by unstimulated horn weight.
Superovulation assay
Superovulation was induced in female mice by i.p. injection of 5 international units (IU) of pregnant mare's serum gonadotropin (EMD Millipore, Billerica, MA), followed by 5 IU of human chorionic gonadotropin (Pregnyl, Merck & Co., Inc., Whitehouse Station, NJ) 48 h later and placed with wildtype male mice. The mice were sacrificed 24 h later by cervical dislocation while under the anesthetic, Avertin (2,2–tribromoethyl alcohol, Sigma–Aldrich, St. Louis, MO) and oocytes were flushed from the oviducts and counted.
Immunohistochemistry
Tissues were fixed in 4% paraformaledehyde and embedded in paraffin wax. Embedded tissues were sectioned at 5 μm and baked for 20 min at 60°C. Upon cooling, slides were dewaxed in Xylenes with a decreasing gradient of pure ethanol. For H&E staining, tissues were adequately stained with H&E and were then dehydrated before coverslips were applied. For immunohistochemistry, antigen retrieval was performed according to manufacturer's instructions (Vector Labs Antigen Unmasking Solution H-3300). Endogenous peroxide was blocked using 3% hydrogen peroxide diluted in methanol. The tissue was blocked before application of primary antibody overnight (PGR: Dako A0098 (1:1000), Myc-tag: Cell Signal 71D10 (1:150), phospho-histone H3: EMD Millipore 06–570 (1:2000), ESR1: Dako M7047 (1:2000), FOXA2: Cell Signal 8186 (1:400), PTCH1: Novus Biologicals NBP1-71662 (1:500)). Secondary antibody was diluted in 1% bovine serum albumin (BSA) at a concentration of 1:200 when required. The ABC reagent was applied to tissues according to manufacturer's instructions (Vector Labs ABC PK-6100). Signal was developed using Vector Labs DAB Immpact staining according to manufacturer's instructions (Vector Labs SK-4105). Tissue was counterstained with hematoxylin and dehydrated before applying coverslips.
RNA isolation
Frozen tissue was homogenized in TRIzol reagent (Thermo Fisher). Isolation of RNA was performed using chloroform and precipitated using isopropanol with resuspension in water. For RNA prepared for microarray, TRIzol reagent was utilized followed by the aqueous phase isolation using 1-Bromo-3-chloropropane and a second aqueous phase isolation using chloroform. Pure ethanol was applied to the aqueous layer, and the total solution was administered to the Qiagen RNEasy RNA mini prep kit column. The column was washed and the RNA was isolated using manufacturer's instructions (Qiagen).
Reverse transcriptase PCR and quantitative real-time PCR
Reverse transcription of RNA into cDNA was performed using M-MLV reverse transcriptase (Thermo Fisher) according to manufacturer's instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed using Taqman Master Mix (Life Technologies) or SYBR Green Master Mix (Roche Diagnostics). Life Technologies Taqman primers were utilized, and SYBR primers were constructed based on Primer Bank sequence predictions using Sigma-Aldrich synthesized oligonucleotides (Supplemental Table S1). Delta delta Ct values were calculated using 18S control amplification results to acquire relative mRNA levels per sample.
In vitro cell culture
PGRA expressing Ishikawa cells were grown in Dulbecco's Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12) with 10% Fetal Bovine Serum (FBS). Upon reaching 70% confluency, the media was changed to phenol-red free DMEM/F12 with 5% charcoal stripped FBS and left to incubate overnight. The next morning, media was changed to phenol-red free DMEM/12 with 5% charcoal stripped FBS containing either vehicle or 10−7 M R5020. After 6 h of incubation, the media was aspirated and the cells were washed in phosphate-buffered saline (PBS). TRIzol reagent was applied to the cells, and the cells were mechanically scraped to harvest the maximum amount of RNA. RNA was purified via chloroform extraction and isopropanol precipitation. RNA was dissolved in RNAse-free water and reverse transcriptase polymerase chain reaction (PCR) was used to isolate cDNA. Quantitative real-time PCR was implemented to assess P4 and E2 target genes.
RNA microarray
RNA quality was assessed using the Agilent 2100 Bioanalyzer (Agilent Technologies). Microarrays were performed by the Genomic and RNA Profiling Core at Baylor College of Medicine. After library amplification and labeling, individual cDNA samples were hybridized to the Agilent G3 Mouse GE 8×60k array according to manufacturer's instructions (Agilent Technologies). Array data were analyzed using Bioconductor for quantile normalization. Genes with an unadjusted p-value ≤ 0.01, and an absolute fold change >1.3 were identified as differentially regulated.
Chromatin immunoprecipitation and sequencing
Whole uterine tissue was flash frozen and sent to Active Motif for Factor Path chromatin immunoprecipitation and sequencing (ChIP-seq) analysis. The tissue was fixed and then promptly sheared into small fragments before immunoprecipitation with the Active Motif PGR antibody. Bound DNA was purified and sequenced using the Illumina sequencing platform to generate a purified, amplified library of PGR-bound sequence regions. Sequence intervals were mapped to genes using Model-based Analysis of ChIP-Seq (MACS) for genomic location by the Active Motif Company. The Active Motif Company also performed validation of the PGR-binding events using chromatin immunoprecipitation with quantitative polymerase chain reaction (ChIP-qPCR) through measurement of the amount of binding events per cell compared to binding events occurring in an untranslated region.
Data analysis
Microarray data were analyzed using Ingenuity Pathway Analysis software and the public Database for Annotation, Visualization, and Integrated Discovery. The ChIP-Seq data were analyzed using the Cistrome Analysis Pipeline software (http://cistrome.org/ap/). The GraphPad Prism software was implemented for one-way analysis of variance (ANOVA), multiple comparison test, and Student t-test analyses for qRT-PCR and decidual wet weights. Hormone response elements were identified using HOMER de novo motif analysis (http://homer.salk.edu/homer/). Hierarchal clustering heatmaps were generated using Partek Genomics Suite 6.6 software.
Results
Spatiotemporal expression of PGR at the window of receptivity
To assess the expression level of PGR in the uterine compartments at the time of implantation, wildtype mice were naturally mated and sacrificed 3–5 days after the presence of the postcoital plug (day 0.5). On day 3.5 of pregnancy, PGR protein was identified in the stroma and luminal and glandular epithelium (Supplemental Figure S1A). By day 4.5, the day of embryo implantation, the luminal epithelium surrounding the implanting embryo demonstrated very low levels of PGR expression, while the stromal compartment exhibited prominent PGR staining (Supplemental Figure S1B). By day 5.5, the luminal epithelium in the interimplantation site demonstrated very low PGR levels, while the surrounding subepithelial stroma exhibited strong PGR staining (Supplemental Figure S1C). Therefore, PGR decreases in the luminal epithelium on day 4.5 at the time of embryo attachment.
Generation of a progesterone receptor A expression allele
A constitutively expressing PGRA allele was generated to assess the importance of the spatiotemporal expression of the PGRA isoform on the ability of the uterus to support embryo attachment and fetal development [17]. The murine PgrA cDNA was FLAG- and myc-tagged and inserted into the mouse ROSA26 locus in a Cre recombinase-induced expression vector as described in the Methods section (described in Supplemental Figure S2A). The mice resulting from this manipulation were termed mPgrALsL/+ mice. The mPgrALsL/+ mice were crossed to the Pgrcre and Wnt7acre mouse models [14,15]. The Pgrcre mouse expresses the Cre recombinase protein in all compartments of the uterus, while the Wnt7acre mouse model exhibits limited Cre recombinase expression in the epithelium only.
In order to assess the PGR levels of the Pgrcre mPgrALsL/+and Wnt7acre mPgrALsL/+mouse models as compared to wildtype mice, immunohistochemistry for PGR was performed on the uteri of pregnant female mice. Female mice were naturally mated with intact males and sacrificed 3 days after the presence of the coital plug (day 3.5). Upon staining for total PGR protein at day 3.5 in wildtype mice, PGR is expressed in all compartments of the uterus including the luminal epithelium (LE), stroma (S), and glandular epithelium (GE) (Figure 1A). In the total PGR knockout mouse (Pgr−/−), PGR is not expressed in the uterus as is expected (Figure 1B). However, mice positive for the PGRA expression allele (mPgrALsL/+) and Cre recombinase, either Pgrcre (Figure 1C) or Wnt7acre (Figure 1D), exhibited strong PGR expression in all uterine compartments. Since the PGRA cDNA was successfully tagged with a 5΄ myc-tag sequence, the expression of the PGRA allele can easily be distinguished from endogenous PGR expression by performing myc-tag immunohistochemistry. Myc-tag exhibits negative staining in wildtype and Pgr−/− tissues as is expected (Figure 1E and F). However, myc-tag is localized to all uterine compartments in the Pgrcre/+mPgrALsL/+ mice (Figure 1G) and only in the epithelium of the Wnt7acre/+mPgrALsl/+ (Figure 1H) mice. Therefore, expression of the knock-in allele is dependent on the presence of Cre recombinase in the uterine compartments.
Figure 1.
Validation of a conditional expression allele for PGRA. (A–H) Immunohistochemical images at day 3.5 for PGR (A–D) and myc-tag (E–H) in wildtype (WT) (A, E), Pgr−/- (B, F), Pgrcre/+mPgrALsL/+(C, G), and Wnt7acre/+mPgrALsL/+(D, H). GE = glandular epithelium, LE = luminal epithelium, S = stroma.
Constitutive progesterone receptor A expression at early pregnancy results in infertility
Upon confirmation of the expression of the PGRA allele during early implantation, the fertility status of the mice was assessed. Pgrcre/+mPgrALsL/+, Wnt7acre/+mPgrALsL/+, and wildtype mice were evaluated by breeding the mice to C57BL/6J males and the number of offspring generated over a 6-month time period was determined. Neither Pgrcre/+mPgrALsL/+ nor Wnt7acre/+mPgrALsL/+ female mice produced offspring during this time period (Table 1). In order to determine if the infertility was a result of the inability of the uterus to support embryo attachment, Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ mice were bred to B6D2F1 intact male mice and sacrificed on the fifth day or the day after embryo implantation. Uteri were harvested and fixed longitudinally. Serial sections of the reproductive tract were obtained and sequentially stained for PGR protein and with hematoxylin and eosin (H&E). At day 5.5, wildtype mice exhibited normal attached embryos with minimal epithelial tissue and limited stromal PGR staining surrounding the embryos (Figure 2A). However, only floating embryos were found in the Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ mice, adjacent to luminal epithelium exhibiting strong levels of PGR staining (Figure 2B and C). The number of floating and attached embryos for the wildtype, Pgrcre/+mPgrALsL/+, and Wnt7acre/+mPgrALsL/+ mice was subsequently recorded (Table 2). Wildtype mice exhibited normal attached embryos graphically displayed in Figure 2D, while constitutive expression of the PGRA isoform in the uterine epithelium identified in both Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ mice resulted in only floating embryos observed in the interluminal space (Figure 2E and F). Furthermore, the uterine epithelium failed to close down around the attaching embryo in both the Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ mice (Figure 2B–C, E–F). Therefore, epithelial PGRA expression during early pregnancy is sufficient to render mice infertile with embryo attachment failure.
Table 1.
Constitutive expression of the PGRA in the uterine epithelium or whole uterus results in complete infertility.
| n | No. of pups | No. of litters | Avg. no. of pups per litter | Avg. time btw. litters (days) | Avg. no. of litters per female | Avg. time to first litter (days) | |
|---|---|---|---|---|---|---|---|
| Wildtype | 4 | 164 | 24 | 6.83 | 27.80 | 6.00 | 24.50 |
| Pgrcre/+mPgrALsL/+ | 6 | 0 | 0 | – | – | – | – |
| Wnt7acre/+ mPgrALsL/+ | 5 | 0 | 0 | – | – | – | – |
Table reporting number of pups and litters, average time between litters, average litters per female, and average time to first litter after a 6-month breeding trial for wildtype, Pgrcre/+mPgrALsL/+, and Wnt7acre/+mPgrALsL/+ mice.
Figure 2.
Embryo attachment is prevented upon constitutive expression of PGRA in the uterine epithelium. PGR staining (A–C) and sequential hematoxylin and eosin (H&E) staining (D–F) of day 5.5 embryo attachment for wildtype (A, D), Pgrcre/+mPgrALsL/+(B, E), and Wnt7acre/+mPgrALsL/+ (C, F). Arrows demonstrate large interstitial space surrounding the floating embryo. LE = luminal epithelium, GE = glandular epithelium, S = stroma, emb = embryo.
Table 2.
Embryos fail to attach in PGRA-expressing mice.
| Floating | Attached | n | |
|---|---|---|---|
| Wildtype | 0 | 27 | 3 |
| Pgrcre/+mPgrALsL/+ | 18 | 0 | 4 |
| Wnt7acre/+mPgrALsL/+ | 20 | 0 | 4 |
Table depicting totaled floating and attached embryos from wildtype, Pgrcre/+mPgrALsL/+, and Wnt7acre/+mPgrALsL/+ mice at day 5.5 of pregnancy.
The number of floating and attached embryos reported in Table 2 demonstrates that less embryos were detected in the uterine sections of the Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ mice as compared to controls. In order to rule out a defect in ovulation, the ovary function was also assessed through a superovulation assay in Wnt7acre/+mPgrALsL/+ and control mice. The PGRA overexpressing and control mice produced comparable numbers of ova without a statistically significant difference (data not shown), suggesting that the Wnt7acre/+mPgrALsL/+ ovaries retain an ovulation capacity as competent as controls. The presence of less embryos in the uteri of these mice may be due to altered oviductal transport although no morphological alteration of the oviduct was observed.
Wnt7acre/+mPgrALsL/+ mice exhibit high levels of progesterone target genes and decreased estrogen targets independent of estrogen receptor levels
Embryo attachment initiates as a result of the coordination of a myriad of pathways functioning to progress the uterus to a receptive state [3]. Many of these target genes are regulated by E2 and P4. In the normal mouse uterus, the PGR is decreased in the epithelium at day 4.5 to allow for embryo attachment. However, in the Wnt7acre/+mPgrALsL/+ mice, PGRA is constitutively expressed through the window of implantation. We hypothesized that the persistent expression of PGRA may result in aberrant upregulation and downregulation of P4 target genes. In order to assess these changes in expression, Wnt7acre/+mPgrALsL/+ and wildtype mice were mated to vasectomized B6D2F1 mice and sacrificed 4 days after the presence of the coital plug (day 4.5 of pregnancy). Transcript levels were assessed by performing RNA isolation of whole uterine tissue. Total Pgr levels (Figure 3A) and Pgr epithelial target genes Ihh and amphiregulin (Areg) (Figure 3B and C) were upregulated while msh homeobox (MSX) genes, also identified as necessary for embryo attachment [18], were not altered (Figure 3D and E). Since attenuation of epithelial PGR and modulation of IHH levels have been shown to affect epithelial proliferation [12], the proliferation levels of the Wnt7acre/+mPgrALsL/+ mice were assessed. Staining for phosphorylated histone H3 demonstrated no significant difference of mitosis in the Wnt7acre/+mPgrALsL/+ mice compared to wildtype (Figure 4A–D). Nevertheless, overactive hedgehog signaling can prevent the development of the uterine glands and impair the decidual response [19]. The downstream receptor of IHH, patched 1 (PTCH1), was found upregulated in the epithelial and stromal compartment of the Wnt7acre/+mPgrALsL/+ mice compared to wildtype (Figure 4E–H), suggesting hedgehog signaling is upregulated in the uterine stroma. Since hedgehog signaling affects glandular development, the gland marker, forkhead box A2 (FOXA2), was assessed in order to evaluate appropriate glandular formation. Although uterine glands were present, FOXA2 levels were decreased in the Wnt7acre/+mPgrALsL/+ endometrial glands (Figure 4I–L), suggesting acute altered signaling of the Ihh pathway is regulating uterine gland differentiation.
Figure 3.
Constitutive epithelial PGRA expression results in aberrant levels of implantation target genes. (A–F). Relative message levels at day 4.5 natural pregnancy for Pgr (A), Ihh (B), Areg (C), Msx1 (D), Msx2 (E), Esr1 (F), Muc1 (G), Lif (H), Lifr (I), Scnn1a (J), and Cftr (K). Relative levels of mRNA in 6 h R5020 treated PGRA Ishikawa cells for AREG (L) and LIF (M). ‘†’ denotes significance with a p-value ≤ 0.05. Error bars represent ±SEM.
Figure 4.
Constitutive PGRA expression causes FOXA2 attenuation and PTCH1 upregulation. Immunohistochemistry for phospho Histone H3 (A–D), PTCH1 (E–H), FOXA2 (I–L), and ESR1 (M–P) in wildtype (A, B, E, F, I, J, M, N) and Wnt7acre/+mPgrALsL/+(C, D, G, H, K, L, O, P) at day 4.5 of pregnancy. Sample size for phospho Histone H3: wildtype n = 5, Wnt7acre/+mPgrALsL/+n = 6; PTCH1: wildtype n = 9, Wnt7acre/+mPgrALsL/+n = 13; FOXA2: wildtype n = 7, Wnt7acre/+mPgrALsL/+n = 11; ESR1: wildtype n = 5, Wnt7acre/+mPgrALsL/+n = 6.
In addition to assessing PGR target gene levels, estrogenic regulation was also evaluated. The expression of the estrogen regulated cytokine, Lif, its receptor, the leukemia inhibitory factor receptor (Lifr), and the glycoprotein, mucin 1 (Muc1), were assayed. Although ER itself is not significantly changed in mRNA (Figure 3F) or protein level (Figure 4M–P) compared to wildtype mice, its targets, Muc1, Lif, and Lifr exhibited significant attenuation (Figure 3G–I). Impaired Lif signaling and upregulation of Muc1 can lead to the prevention of embryo attachment [16,20]. Therefore, constitutive PGRA expression in the uterine epithelium of Wnt7acre/+mPgrALsL/+ mice results in an increase of P4 target genes including hedgehog signaling members, impaired FOXA2 expression within the glands, and a dramatic attenuation of known E2 target genes independent of ER levels.
Since the lumen of the Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ endometrium remained enlarged and did not collapse around the embryo, the expression of genes known to regulate fluid movement adjacent to the uterine epithelium was assayed. Recent data have described the critical role of solute channels, sodium channel, nonvoltage-gated 1 alpha (Scnn1a) and cystic fibrosis transmembrane conductance regulator (Cftr), in the promotion of embryo attachment through regulation of fluid and salt levels in the interluminal space [21]. Upregulation of Scnn1a and attenuation of Cftr was identified to aid in embryo attachment at the window of receptivity. Upon assaying message levels of Scnn1a and Cftr in the Wnt7acre/+mPgrALsL/+mice, neither of these genes were changed (Figure 3J and K), suggesting that the expansive luminal space may be regulated by another mechanism downstream of PGR action.
In order to determine if the impact of PGRA expression on Lif could be translated from mouse to human, human uterine epithelial cells were specifically engineered to stably express high levels of the PGRA isoform and are therefore P4 responsive [22]. Upon treatment of the Ishikawa PGRA-expressing cells with the progestin, R5020, for 6 h, quantitative real-time PCR analysis revealed high levels of AREG message and decreased transcript levels of LIF (Figure 3L and M). These data were consistent with the in vivo data, suggesting that PGRA not only promotes AREG in the epithelium but also governs the attenuation of LIF.
Progesterone receptor A overexpression blocks the stromal decidual reaction
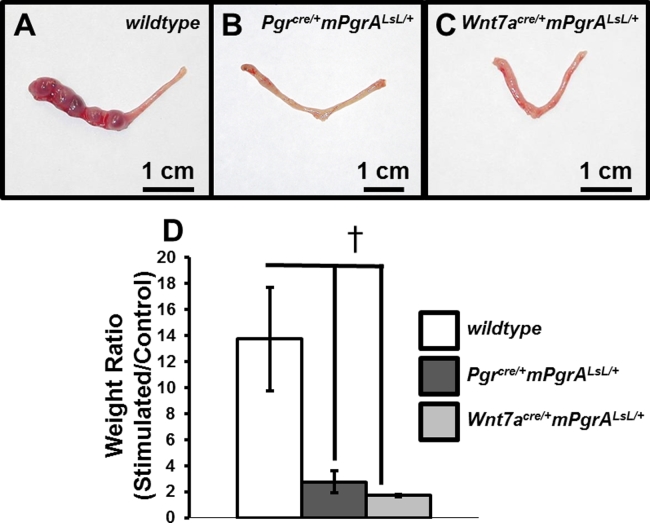
Due to the infertile phenotype of the Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ mice, the artificial decidual assay was implemented to evaluate whether the uterine stroma could mount a decidual response in the presence of a stimulus. Upon performance of the artificial decidual assay, the Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ failed to undergo a decidual response compared to wildtype (Figure 5A–C). The weight ratios of the stimulated horn divided by the control horn depicted a severe weight ratio attenuation in the Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ mice (Figure 5D). Therefore, constitutive epithelial PGRA expression prevents the stroma from eliciting a decidual response.
Figure 5.
Epithelial PGRA expression is sufficient to impair the uterine decidual response. Artificial decidual response performed on wildtype (A), Pgrcre/+mPgrALsL/+(B), and Wnt7acre/+mPgrALsL/+ (C) with the left horn stimulated by oil injection and right horn left as an unstimulated control horn. (D) Average weight ratio of stimulated horn divided by unstimulated horn for wildtype, Pgrcre/+mPgrALsL/+, and Wnt7acre/+mPgrALsL/+. ‘†’ denotes significance with a p-value ≤ 0.05. Error bars represent ±SEM.
Epithelial progesterone receptor A signaling regulates multiple pathways associated with molecular transport and cell-to-cell signaling at the window of implantation
Since the Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ mice exhibit similar phenotypes providing strong evidence for the sufficient role of elevated epithelial-specific PGRA expression in the prevention of pregnancy initiation, we focused our molecular studies on the Wnt7acre/+mPgrALsL/+ mice only. To further understand the molecular cause for infertility in the Wnt7acre/+mPgrALsL/+ mice, an RNA microarray was performed on whole uterine tissue harvested at day 4.5. A total of 889 genes were misregulated at day 4.5 with 363 genes upregulated and 526 genes downregulated. The top pathways changed in the aberrantly regulated gene list included molecular transport, small molecule biochemistry, organismal development, reproductive function, cell-to-cell signaling, and inflammatory response (Table 3) according to Ingenuity Pathway Analysis software.
Table 3.
Microarray analysis at day 4.5 in Wnt7acre/+mPgrALsL/+ mice depicts aberrant regulation of genes involved in cell signaling, adhesion, and small molecule transport.
| Categories | Molecules |
|---|---|
| Lipid metabolism, molecular transport, small molecule biochemistry | ABCC3, ABCG5, ACACA, ACAT2, ACPP, ACSS1, ADIPOR1, ADIPOR2, AHR, Akap9, AQP8, ATP7B, C3, CHST8, CLU, CSF1, CYSLTR1, DKK3, FGL1, FITM2, GAL3ST1, GATM, GNMT, H6PD, HPGDS, IL6, KLKB1, LBP, LGALS3, LGR5, LHX9, LOC102724788/PRODH, LRAT, NCEH1, PIK3CG, PLA2G10, PLA2G3, POR, PRDX6, PRF1, PRKAA2, PRKG1, PTGDR, PTGDS, PTGS1, RGS4, SAA2-SAA4, SAT1, SCTR, SERPINA6, SGK1, SGMS2, SLC18A2, SLC26A3, SMPD1, SNCA, SRGAP3, ST3GAL5, STK39, THEM5, TLR2, TSHR, TTPA, TTR, VCAM1, WNK4, XBP1 |
| Organismal development | ABCA5, ACPP, AHR, AREG, ARG1, ATP7B, AZGP1, BLNK, C3, CCNG1, CCR2, CD55, Cdkn1c, CFI, CHGB, CHST8, CLCN3, CLU, CP, CRIM1, CSF1, DCLRE1C, FAM20C, FANCC, FAT4, FCGR2B, FGF9, GADD45A, GJB1, GNMT, GUCA2A, H6PD, Hbb-b2, HP, IGFBP3, IHH, IL6, KCNQ1, LCP2, LGALS3, LGMN, MAFB, Mt1, Mug1/Mug2, MYB, MYCN, NBEAL2, NLRP6, OCLN, PCSK6, PCYT1B, PGR, PIGR, PIP, PRF1, PRKG1, PTGS1, RASGRF1, REN, RLN1, RUNX2, SCTR, SLC12A3, SLC26A3, SLC9A2, SMPD1, SOX9, SUN1, TCF15, TGFBI, TLR2, TNFSF11, TP63, TSPAN33, TYRO3, WNT7A, XBP1, YBX2 |
| Reproductive system development and function | AHR, ATP7B, C3, CATSPER1, CHST8, Mt1, Mug1/Mug2, PCSK6, PGR, PIP5K1A, PLA2G10, PLA2G3, Pld6, PRF1, PTGS1, Svs2, TEKT4, TRPM6 |
| Cell-to-cell signaling and interaction, cellular growth and proliferation | CSF1, IL6, TNFSF11 |
| Inflammatory response | AHR, ARG1, C3, CALCB, CCR2, CD55, CLU, CROCC, CSF1, CTSB, DMBT1, FANCC, FCGR2B, GADD45A, GNMT, GPX2, Havcr1, HP, HPGDS, HPX, IGFBP1, IGFBP3, IL18R1, IL6, IRAK1, KCNK2, KMO, LBP, LCP2, LECT2, LGALS3, MAPK11, MMP7, Mt1, Mug1/Mug2, NLRP6, OCLN, PGR, PIK3CG, PIP, PLA2G10, PLA2G3, PNKD, PRDX6, PRF1, PRKG1, PROS1, PSTPIP2, PTGS1, REN, SCTR, SLC9A2, Timd2, TLR2, TMEM173, TNFRSF11B, TP63, VCAM1, VTCN1, XBP1 |
| Connective tissue disorders, tissue morphology | CCR2, CTSC, FCGR2B, IHH, IL6, LECT2, PIK3CG, PSTPIP2, PTGS1, SLC39A8, TNFRSF11B, TNFSF11, TP63 |
Using Ingenuity Pathway Analysis software, the top biological functions for the Wnt7acre/+mPgrALsL/+ dataset are reported. Top biological function categories include molecular transport, small molecule biochemistry, organismal development, and cell-to-cell signaling. Genes with an unadjusted p-value ≤ 0.01 and an absolute fold change >1.3 were identified as differentially regulated.
Upon closer analysis of the microarray data, many ion and large molecule channels were differentially regulated in the dataset as shown in the heatmap in Figure 6A. Of these molecular channels, two aquaporin genes expressed in the uterus during early pregnancy [23], aquaporin 5 (Aqp5) and aquaporin 8 (Aqp8), exhibited dramatic attenuation of mRNA level in the Wnt7acre/+mPgrALsL/+ mice at day 4.5 (Figure 6B and C). With decreased aquaporin levels, fluid movement is prevented in the uterine compartments [24], providing one explanation for the large amount of fluid accumulated in the interluminal space in the Wnt7acre/+mPgrALsL/+ mice. Regarding ionic movement, potassium channel, subfamily K, member 2 (Kcnk2), a K+ importer expressed in human endometrium [25], is increased in the Wnt7acre/+mPgrALsL/+ mice (Figure 6D), implicating a low K+ environment occurring in the interstitial space. Furthermore, transient receptor potential cation channel, subfamily V, member 6 (Trpv6), a Ca+2 importer [26] expressed in the uterus at the time of attachment [27], and solute carrier family 12, member 3 (Slc12a3), a Cl− and Na+ importer [28] potentially regulated by the ovarian steroid hormones [29], are both attenuated at day 4.5 (Figure 6E and F), suggesting an overall increase in solutes in the interstitial space. Therefore, the collective misregulation of these molecular channels likely creates an interstitial environment with low K+ and high Na+, Cl−, Ca+2, with increased water levels. Thus, aberrantly upregulated epithelial PGRA levels regulate multiple molecular channels potentially resulting in an unfit environment for the attachment of the embryo.
Figure 6.
Upregulated PGRA at the window of receptivity results in the misregulation of solute transport genes. (A) Gene heatmap depicting results of the top regulated genes from the Wnt7acre/+mPgrALsL/+ dataset involved in molecular transport. Data were centered on a median of zero and color intensities were set to 1.5 and –1.5, respectively. (B–F) Relative mRNA levels for the Aqp5 (B), Aqp8 (C), Kcnk2 (D), Trpv6 (E), and Slc12a3 (F). ‘†’ denotes significance with a p-value ≤ 0.05. Error bars represent ±SEM.
Progesterone receptor A-specific ChIP-Seq defines a subset of direct targets differentially regulated at the window of implantation
To further define the microarray dataset reporting the misregulated genes at day 4.5 of natural pregnancy, a PGR ChIP-seq was performed to determine the PGRA isoform-specific cistrome. By crossing mice positive for the mPgrALsL/+ allele to the Pgr−/− [8] and Pgrcre [14] mouse models, female Pgrcre/−mPgrALsL/+ mice were generated. These mice exhibit no endogenous PGR expression [8,14]. However, presence of the ROSA26 expression allele and PGR-dependent Cre recombinase results in PGRA expression in all uterine compartments.
Pgrcre/−mPRALsL/+ mice were ovariectomized, rested for 2 weeks, and then administered P4. After 1 h, uterine horns were flash frozen and the tissue was subject to the ChIP-seq assay. The PGRA ChIP-seq demonstrated enriched binding at promoter and 5΄ untranslated regions (UTR) (Figure 7A), while motif analysis described enriched motifs for nuclear receptors, FRA1, GATA, and CEBP (Figure 7B). Therefore, PGRA is enriched at promoter regions of genes within the uterus and is found to primarily bind at locations with nuclear receptor response elements.
Figure 7.
PGRA-specific ChIP-Seq analysis exhibits enrichment at promoter and 5΄ untranslated region (UTR) and defines a unique set of PGRA-regulated genes at the window of implantation. (A) Enrichment distribution of PGRA binding on the genome compared to normal expected enrichment using the Cistrome Analysis Pipeline software. Data are equivalent to the natural log of PGRA chip binding/basal genome binding. (B) Top binding of known motifs by the PGRA isoform using HOMER motif analysis. (C) PGRA ChIP-Seq using PRcre/−mPgrALsL/+mice mapped to 15,780 genes with binding intervals within 10 kb of the transcription start site. Of these genes, 558 were differentially regulated in the Wnt7acre/+mPgrALsL/+day 4.5 microarray, graphically represented as a red circle.
In order to identify PGRA-specific target genes exhibiting differential regulation at day 4.5 in the Wnt7acre/+mPgrALsL/+ mice, the microarray dataset was overlapped with the PGRA-specific ChIP-Seq gene list. Of the 889 differentially regulated genes, 558 genes were identified as PGRA target genes (Figure 7C). Interestingly, many of the solute and molecular transport genes were identified to be directly bound by PGRA. Supplemental Figure S3 depicts PGRA-binding events at the loci of Aqp5 (A), Aqp8 (B), Slc12a3 (C), and Kcnk2 (D). Furthermore, these interactions were validated via ChIP-qPCR analysis (Supplemental Figure S3E–H). Therefore, expression of PGRA at the window of implantation directly affects the transcription of molecular channels in the uterus, resulting in an unfit environment for embryo attachment. Furthermore, LIF, the critical cytokine necessary for embryo attachment, exhibited multiple PGRA-binding events within the intronic region (Figure 8A). Additionally, the receptor of Lif, Lifr, also exhibited PGRA binding (Figure 8B). These identified PGRA-binding regions were further validated using ChIP-qPCR analysis (Figure 8C and D). Therefore, although LIF is a known ER target, these data suggest a direct inhibitory function of PGRA on the transcription of Lif and the Lifr.
Figure 8.
PGRA directly binds components of the LIF signaling pathway. Graphical description of the PGRA binding peaks in the loci of Lif (A) and Lifr (B). (C, D) ChIP-qPCR validation of the PGRA-binding events indicated with asterisks. Y-axis indicates the amount of binding events per 1000 cells. Error bars indicate the standard deviation of averaged binding occurrences. Untr = untranslated region.
Discussion
The establishment and maintenance of pregnancy within the mouse is dependent on the expression of the PGRA isoform [10]. Interestingly, PGRA message levels are functionally decreased in the uterine epithelium at the time of implantation. We were also able to confirm the reduction in PGR protein at day 4.5 within the mouse, the narrow time in which the uterus is receptive to the implanting embryo. Although this decrease in PGR expression at the window has been acknowledged for some time [13], few articles have provided an explanation for this downregulation. The generation of the mPgrALsL/+ allele and the utilization of multiple Cre recombinase models exhibiting uterine expression was instrumental in testing this phenomenon. After validating the expression pattern, we discovered that PGRA expression either in the whole uterus or just the epithelium resulted in complete infertility in a 6-month breeding trial. Furthermore, the Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ mice were able to ovulate, yet were unsuccessful at embryo attachment or closure of the epithelium around the floating embryos. Additionally, the mice were unable to mount an appropriate decidual response in the presence of hormones and a uterine stimulus. Therefore, these mice exhibited major uterine defects resulting in infertility, and constitutive PGRA epithelial expression was sufficient in causing these phenotypes.
To further understand the scope of PGRA control on the uterine epithelial environment, relative mRNA levels for critical genes involved in embryo attachment were assessed. The epithelial P4 target genes, Ihh and Areg were significantly upregulated. However, E2 target genes, Lif and Muc1, displayed attenuated RNA levels. LIF signaling at the site of attachment is required for the implantation of the embryo [16] through the activation of a complex gene network [30]. Although historically an E2 target, the Lif locus exhibited multiple PGRA-binding sites. This evidence suggests a transcriptional inhibitory role of PGRA on the Lif promoter in the epithelium. The inhibition likely occurs up until day 4.5 when epithelial PGRA decreases, allowing for the Lif locus to be transcriptionally active and responsive to the surge of E2 occurring at the time of implantation [4]. The ability for the PGRA isoform to inhibit the transcription of genes in the uterus has been described previously in both mouse and human [31,32]. This specific regulation provides evidence to the finite expression pattern of LIF and activation of the downstream signaling pathway within the narrow implantation time. Also, LIFR was previously conjectured to be inhibited by PGR during early implantation in the ewe [33]. Similar to the LIF ligand, upregulation of LIFR is dependent on the concurrent downregulation of PGR at the time of implantation. Therefore, decreased epithelial PGRA is required at the window of implantation to release the promoters of both Lif and Lifr, allowing efficient activation of LIF signaling for successful attachment of the embryo (modeled in Figure 9). Further investigations are required to validate the direct inhibition of Lif and Lifr by the PGRA isoform.
Figure 9.
Constitutive PGRA expression in the uterine epithelium inhibits LIF signaling, aberrantly promotes stromal hedgehog signaling, and causes the accumulation of fluid and solutes in the uterine lumen resulting in implantation failure. Left panel depicts normal embryo implantation occurring at the uterine epithelial surface on day 4.5 of pregnancy. The LIFR binds to LIF and activates downstream epithelial signaling promoting embryo attachment. Solute transporters for calcium (Ca+2), sodium (Na+), chlorine (Cl−), and water are increased at the epithelial surface causing a low-solute, high-water, and high-potassium (K+) level in the interstitial space. On the right panel within the Wnt7acre/+mPgrALsL/+mice, the LIFR and LIF are inhibited by high levels of epithelial PGRA. Additionally, PGRA promotes IHH, allowing for activation of hedgehog signaling within the stroma which can result in abnormal gland development and impaired decidualization. Solute and water transporters exhibit attenuation resulting in high levels of calcium (Ca+2), sodium (Na+), chlorine (Cl−), and water within the interstitial space. Potassium (K+) transporters are increased at the epithelial surface resulting in increased epithelial uptake. Therefore, high epithelial PGRA levels likely produce an unreceptive environment for embryo attachment. Epith = uterine epithelium.
Besides the direct inhibition of Lif and its receptor, there is another potential means by which PGRA expression could impact Lif message levels. Overactive hedgehog signaling has proven detrimental to decidualization and development of the uterine glands [19]. Indeed, early activation of hedgehog signaling was sufficient in preventing the formation of endometrial glands postnatally [12]. Here, we observed upregulated hedgehog signaling in the Wnt7acre/+mPgrALsL/+ mice and attenuated expression of FOXA2, a marker for glandular development and important for a successful pregnancy [34]. Therefore, the PGRA expressing mice may exhibit an altered glandular epithelial phenotype resulting in an overall decrease in glandular LIF secretions.
In depth microarray analysis of Wnt7acre/+mPgrALsL/+ uterine transcripts at day 4.5 reported 889 differentially regulated genes with top changed pathways including molecular transport, small molecule biochemistry, development, and cell-to-cell signaling. Within this dataset, over 80 differentially regulated genes were involved in the transport of molecules across an epithelial layer. To further delineate direct PGRA targets within the microarray, we performed a PGRA-specific ChIP-Seq analysis, consisting of 15,780 genes called. Upon overlap of the RNA microarray with the PGRA-specific ChIP-Seq, multiple solute channel genes were discovered to be directly bound by PGRA. PGRA was identified to directly regulate aquaporin genes, Aqp5 and Aqp8, previously reported to be critical in maintaining fluid movement in the reproductive tract [23,35]. The aquaporins are likely involved in the removal of excess fluid at day 4.5 to allow for sufficient wrapping of the embryo to the uterine epithelium. Furthermore, Kcnk2, a known K+ importer [25], was found upregulated directly by PGRA. Elevated interstitial K+ levels are essential for fertilization and have been hypothesized to be important for embryo attachment [24]. Additionally, the Ca+2 importer [26], Trpv6, and Na+ and Cl− importer [28], Slc12a3, were found attenuated in the Wnt7acre/+mPgrALsL/+ mice. Decreased expression of these importers suggests the occurrence of a high salt concentration within the interstitial space [24], which can be detrimental to embryo attachment. Additionally, previous studies have linked epithelial cell Ca+2 influx with the release of prostaglandin E2 release to allow for appropriate implantation and decidualization [36]. Since both the Pgrcre/+mPgrALsL/+ and Wnt7acre/+mPgrALsL/+ mice fail to undergo the decidual reaction, the low Ca+2 influx may play a role in the prevention of the decidual response. However, PGRA-binding intervals were only observed in the Slc12a3 locus and not in the Trpv6 locus. The resultant misregulation of these transporter levels due to high levels of PGRA create a highly fluid-filled and salt-filled (Ca+2, Na+, Cl−) environment for the attaching embryo, with a low K+ level in the interstitial space.
The high salt and fluid conditions can be detrimental for embryo attachment and decidualization through the establishment of an unfit, unreceptive uterine environment. Previously published data have provided strong evidence for the critical role of the uterine solute environment on appropriate embryo implantation [24]. In addition, K+ levels have the ability to control whole cell currents, other ion channels, cell polarization, and cell proliferation, known to affect embryo attachment [24,25]. These data suggest that under normal conditions with decreased PGRA at the window of receptivity, aquaporins, Trpv6, and Slc12a3 are upregulated and Kcnk2 is downregulated resulting in the attenuation of Na+, Ca+2, Cl−, and water molecules and increased K+ levels in the interluminal space. This low-fluid, low-solute, and high-K+ environment likely contributes to the effective wrapping of the epithelium and attainment of a receptive environment for the attaching embryo. Further investigation is required to validate the relationship between PGRA and these molecular transporters in the regulation of fluid and solute movement at the uterine epithelial surface at the time of attachment.
Appropriate P4 signaling is necessary for a successful pregnancy. Although PGRA is decreased in the uterine epithelium at day 4.5 to initiate embryo attachment, no studies have tested the significance of this attenuation. Through the utilization of multiple mouse models expressing heightened levels of the PGRA isoform, we can conclude that constitutive epithelial PGRA expression at early implantation is sufficient to render mice infertile with the inability to attach embryos or undergo the stromal decidual response. Furthermore, through the use of combined transcriptomic and cistromic analyses, this is the first article to describe the direct regulation of Lif and multiple molecular transport genes by the PGRA isoform. Increased P4 signaling due to elevated PGRA levels results in aberrant regulation of hedgehog signaling and solute and cytokine levels, creating an unreceptive environment for the implanting embryo. This mouse model and our resultant deepened understanding of P4 signaling at the window of implantation can promote the development of new therapies in order to treat women with recurring early pregnancy loss and infertility.
Supplementary Material
Supplementary data are available at BIOLRE online.
Supplemental Figure S1. PGRA is spatiotemporally expressed at the implantation window. (A–C) Immunohistochemistry for PGR in wildtype mice at natural pregnancy day 3.5 (A), day 4.5 with embryo (B), and day 5.5 (C). LE = luminal epithelium, GE = glandular epithelium, S = stroma, emb = embryo.
Supplemental Figure S2. Construction of a Cre-Inducible PGRA Expression Construct. (A) Graphical description of the conditional expression allele for the PGRA isoform targeted to the murine ROSA26 locus. CAGGS = chicken β-actin promoter, STOP = transcription stop cassette, polyA = polyadenylation sequence, 5΄ probe = southern probe annealing region. (B) Southern blot analysis for the identification of the Pgr-A knock-in allele at the ROSA26 locus.
Supplemental Figure S3. PGRA directly binds multiple solute transport genes. Graphical description of the PGRA-binding peaks in the loci of Aqp5 (A), Aqp8 (B), Slc12a3 (C), and Kcnk2 (D). (E–H) ChIP-qPCR validation of the PGRA-binding events indicated with asterisks. Y-axis indicates the amount of binding events per 1000 cells. Error bars indicate the standard deviation of averaged binding occurrences. Untr = untranslated region.
Supplemental Table S1. Complete list of SYBR primer sequences and Applied Biosystems Taqman Probe catalog numbers.
Acknowledgments
The authors thank Sungnam Cho, Jie Yang, Yiqun Zhang, Nyssa Adams, and Janet DeMayo for technical assistance. This work was impossible without the support of the advanced technology cores at Baylor College of Medicine including the Embryonic Stem Cell Core, the Genetically Engineered Mouse Core, and the Genomic and RNA Profiling Core.
Supplementary data
Supplementary data are available at BIOLRE online.
Supplemental Figure S1. PGRA is spatiotemporally expressed at the implantation window. (A–C) Immunohistochemistry for PGR in wildtype mice at natural pregnancy day 3.5 (A), day 4.5 with embryo (B), and day 5.5 (C). LE = luminal epithelium, GE = glandular epithelium, S = stroma, emb = embryo.
Supplemental Figure S2. Construction of a Cre-Inducible PGRA Expression Construct. (A) Graphical description of the conditional expression allele for the PGRA isoform targeted to the murine ROSA26 locus. CAGGS = chicken β-actin promoter, STOP = transcription stop cassette, polyA = polyadenylation sequence, 5΄ probe = southern probe annealing region. (B) Southern blot analysis for the identification of the Pgr-A knock-in allele at the ROSA26 locus.
Supplemental Figure S3. PGRA directly binds multiple solute transport genes. Graphical description of the PGRA-binding peaks in the loci of Aqp5 (A), Aqp8 (B), Slc12a3 (C), and Kcnk2 (D). (E–H) ChIP-qPCR validation of the PGRA-binding events indicated with asterisks. Y-axis indicates the amount of binding events per 1000 cells. Error bars indicate the standard deviation of averaged binding occurrences. Untr = untranslated region.
Supplemental Table S1. Complete list of SYBR primer sequences and Applied Biosystems Taqman Probe catalog numbers.
References
- 1. Chandra A, Martinez GM, Mosher WD, Abma JC, Jones J. Fertility, family planning, and reproductive health of U.S. women: data from the 2002 National Survey of Family Growth. Vital Health Stat 23, 2005:1–160. [PubMed] [Google Scholar]
- 2. Macklon NS, Geraedts JP, Fauser BC. Conception to ongoing pregnancy: the 'black box' of early pregnancy loss. Hum Reprod Update, 2002, 8:333–343. [DOI] [PubMed] [Google Scholar]
- 3. Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med, 2012, 18:1754–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang H, Dey SK. Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet, 2006, 7:185–199. [DOI] [PubMed] [Google Scholar]
- 5. Mote PA, Arnett-Mansfield RL, Gava N, deFazio A, Mulac-Jericevic B, Conneely OM, Clarke CL. Overlapping and distinct expression of progesterone receptors A and B in mouse uterus and mammary gland during the estrous cycle. Endocrinology, 2006, 147:5503–5512. [DOI] [PubMed] [Google Scholar]
- 6. Wetendorf M, DeMayo FJ. Progesterone receptor signaling in the initiation of pregnancy and preservation of a healthy uterus. Int J Dev Biol, 2014, 58:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walker VR, Korach KS. Estrogen receptor knockout mice as a model for endocrine research. ILAR J, 2004, 45:455–461. [DOI] [PubMed] [Google Scholar]
- 8. Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA Jr, Shyamala G, Conneely OM, O’Malley BW. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev, 1995, 9:2266–2278. [DOI] [PubMed] [Google Scholar]
- 9. Finn CA, Martin L. Endocrine control of the timing of endometrial sensitivity to a decidual stimulus. Biol Reprod, 1972, 7:82–86. [DOI] [PubMed] [Google Scholar]
- 10. Mulac-Jericevic B, Mullinax RA, DeMayo FJ, Lydon JP, Conneely OM. Subgroup of reproductive functions of progesterone mediated by progesterone receptor-B isoform. Science, 2000, 289:1751–1754. [DOI] [PubMed] [Google Scholar]
- 11. Mulac-Jericevic B, Lydon JP, DeMayo FJ, Conneely OM. Defective mammary gland morphogenesis in mice lacking the progesterone receptor B isoform. Proc Natl Acad Sci USA, 2003, 100:9744–9749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franco HL, Rubel CA, Large MJ, Wetendorf M, Fernandez-Valdivia R, Jeong JW, Spencer TE, Behringer RR, Lydon JP, Demayo FJ. Epithelial progesterone receptor exhibits pleiotropic roles in uterine development and function. FASEB J, 2012, 26:1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tan J, Paria BC, Dey SK, Das SK. Differential uterine expression of estrogen and progesterone receptors correlates with uterine preparation for implantation and decidualization in the mouse. Endocrinology, 1999, 140:5310–5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soyal SM, Mukherjee A, Lee KY, Li J, Li H, DeMayo FJ, Lydon JP. Cre-mediated recombination in cell lineages that express the progesterone receptor. Genesis, 2005, 41:58–66. [DOI] [PubMed] [Google Scholar]
- 15. Winuthayanon W, Hewitt SC, Orvis GD, Behringer RR, Korach KS. Uterine epithelial estrogen receptor alpha is dispensable for proliferation but essential for complete biological and biochemical responses. Proc Natl Acad Sci USA, 2010, 107:19272–19277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stewart CL, Kaspar P, Brunet LJ, Bhatt H, Gadi I, Kontgen F, Abbondanzo SJ. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature, 1992, 359:76–79. [DOI] [PubMed] [Google Scholar]
- 17. Wu SP, Lee DK, Demayo FJ, Tsai SY, Tsai MJ. Generation of ES cells for conditional expression of nuclear receptors and coregulators in vivo. Mol Endocrinol, 2010, 24:1297–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Daikoku T, Cha J, Sun X, Tranguch S, Xie H, Fujita T, Hirota Y, Lydon J, DeMayo F, Maxson R, Dey SK. Conditional deletion of Msx homeobox genes in the uterus inhibits blastocyst implantation by altering uterine receptivity. Dev Cell, 2011, 21:1014–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franco HL, Lee KY, Rubel CA, Creighton CJ, White LD, Broaddus RR, Lewis MT, Lydon JP, Jeong JW, DeMayo FJ. Constitutive activation of smoothened leads to female infertility and altered uterine differentiation in the mouse. Biol Reprod, 2010, 82:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carson DD, DeSouza MM, Kardon R, Zhou X, Lagow E, Julian J. Mucin expression and function in the female reproductive tract. Hum Reprod Update, 1998, 4:459–464. [DOI] [PubMed] [Google Scholar]
- 21. Yang JZ, Ajonuma LC, Tsang LL, Lam SY, Rowlands DK, Ho LS, Zhou CX, Chung YW, Chan HC. Differential expression and localization of CFTR and ENaC in mouse endometrium during pre-implantation. Cell Biol Int, 2004, 28:433–439. [DOI] [PubMed] [Google Scholar]
- 22. Smid-Koopman E, Kuhne LC, Hanekamp EE, Gielen SC, De Ruiter PE, Grootegoed JA, Helmerhorst TJ, Burger CW, Brinkmann AO, Huikeshoven FJ, Blok LJ. Progesterone-induced inhibition of growth and differential regulation of gene expression in PRA- and/or PRB-expressing endometrial cancer cell lines. J Soc Gynecol Investig, 2005, 12:285–292. [DOI] [PubMed] [Google Scholar]
- 23. Richard C, Gao J, Brown N, Reese J. Aquaporin water channel genes are differentially expressed and regulated by ovarian steroids during the periimplantation period in the mouse. Endocrinology, 2003, 144:1533–1541. [DOI] [PubMed] [Google Scholar]
- 24. Liu XM, Zhang D, Wang TT, Sheng JZ, Huang HF. Ion/water channels for embryo implantation barrier. Physiology (Bethesda), 2014, 29:186–195. [DOI] [PubMed] [Google Scholar]
- 25. Patel SK, Jackson L, Warren AY, Arya P, Shaw RW, Khan RN. A role for two-pore potassium (K2P) channels in endometrial epithelial function. J Cell Mol Med, 2013, 17:134–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Peng JB, Brown EM, Hediger MA. Epithelial Ca2+ entry channels: transcellular Ca2+ transport and beyond. J Physiol, 2003, 551:729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee GS, Jeung EB. Uterine TRPV6 expression during the estrous cycle and pregnancy in a mouse model. Am J Physiol Endocrinol Metab, 2007, 293:E132–E138. [DOI] [PubMed] [Google Scholar]
- 28. Wang L, Dong C, Xi YG, Su X. Thiazide-sensitive Na+-Cl- cotransporter: genetic polymorphisms and human diseases. Acta Biochim Biophys Sin (Shanghai), 2015, 47:325–334. [DOI] [PubMed] [Google Scholar]
- 29. Rojas-Vega L, Reyes-Castro LA, Ramirez V, Bautista-Perez R, Rafael C, Castaneda-Bueno M, Meade P, de Los Heros P, Arroyo-Garza I, Bernard V, Binart N, Bobadilla NA et al. , Ovarian hormones and prolactin increase renal NaCl cotransporter phosphorylation. Am J Physiol Renal Physiol, 2015, 308:F799–F808. [DOI] [PubMed] [Google Scholar]
- 30. Rosario GX, Hondo E, Jeong JW, Mutalif R, Ye X, Yee LX, Stewart CL. The LIF-mediated molecular signature regulating murine embryo implantation. Biol Reprod, 2014, 91:66, 1–18. [DOI] [PubMed] [Google Scholar]
- 31. Brayman MJ, Julian J, Mulac-Jericevic B, Conneely OM, Edwards DP, Carson DD. Progesterone receptor isoforms A and B differentially regulate MUC1 expression in uterine epithelial cells. Mol Endocrinol, 2006, 20:2278–2291. [DOI] [PubMed] [Google Scholar]
- 32. Vegeto E, Shahbaz MM, Wen DX, Goldman ME, O’Malley BW, McDonnell DP. Human progesterone receptor A form is a cell- and promoter-specific repressor of human progesterone receptor B function. Mol Endocrinol, 1993, 7:1244–1255. [DOI] [PubMed] [Google Scholar]
- 33. Song G, Satterfield MC, Kim J, Bazer FW, Spencer TE. Progesterone and interferon tau regulate leukemia inhibitory factor receptor and IL6ST in the ovine uterus during early pregnancy. Reproduction, 2009, 137:553–565. [DOI] [PubMed] [Google Scholar]
- 34. Jeong JW, Kwak I, Lee KY, Kim TH, Large MJ, Stewart CL, Kaestner KH, Lydon JP, DeMayo FJ. Foxa2 is essential for mouse endometrial gland development and fertility. Biol Reprod, 2010, 83:396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lindsay LA, Murphy CR. Aquaporins are upregulated in glandular epithelium at the time of implantation in the rat. J Mol Histol, 2007, 38:87–95. [DOI] [PubMed] [Google Scholar]
- 36. Ruan YC, Guo JH, Liu X, Zhang R, Tsang LL, Dong JD, Chen H, Yu MK, Jiang X, Zhang XH, Fok KL, Chung YW et al. , Activation of the epithelial Na+ channel triggers prostaglandin E(2) release and production required for embryo implantation. Nat Med, 2012, 18:1112–1117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at BIOLRE online.
Supplemental Figure S1. PGRA is spatiotemporally expressed at the implantation window. (A–C) Immunohistochemistry for PGR in wildtype mice at natural pregnancy day 3.5 (A), day 4.5 with embryo (B), and day 5.5 (C). LE = luminal epithelium, GE = glandular epithelium, S = stroma, emb = embryo.
Supplemental Figure S2. Construction of a Cre-Inducible PGRA Expression Construct. (A) Graphical description of the conditional expression allele for the PGRA isoform targeted to the murine ROSA26 locus. CAGGS = chicken β-actin promoter, STOP = transcription stop cassette, polyA = polyadenylation sequence, 5΄ probe = southern probe annealing region. (B) Southern blot analysis for the identification of the Pgr-A knock-in allele at the ROSA26 locus.
Supplemental Figure S3. PGRA directly binds multiple solute transport genes. Graphical description of the PGRA-binding peaks in the loci of Aqp5 (A), Aqp8 (B), Slc12a3 (C), and Kcnk2 (D). (E–H) ChIP-qPCR validation of the PGRA-binding events indicated with asterisks. Y-axis indicates the amount of binding events per 1000 cells. Error bars indicate the standard deviation of averaged binding occurrences. Untr = untranslated region.
Supplemental Table S1. Complete list of SYBR primer sequences and Applied Biosystems Taqman Probe catalog numbers.