Introductory paragraph
Mutations of the mitochondrial genome (mtDNA) underlie a significant portion of mitochondrial disease burden. These disorders are currently incurable and effectively untreatable, with heterogeneous penetrance, presentation and prognosis. To address the lack of effective treatment for these disorders, we exploited a recently developed mouse model that recapitulates common molecular features of heteroplasmic mtDNA disease in cardiac tissue, the m.5024C>T tRNAALA mouse. Through application of a programmable nuclease therapy approach, using systemically administered, mitochondrially targeted zinc finger-nucleases (mtZFNs) delivered by adeno-associated virus, we induced specific elimination of mutant mtDNA across the heart, coupled to a reversion of molecular and biochemical phenotypes. These findings constitute proof-of-principle that mtDNA heteroplasmy correction using programmable nucleases could provide a therapeutic route for heteroplasmic mitochondrial diseases of diverse genetic origin.
Introduction
Mitochondrial diseases are a broad group of hereditary, multi-system disorders, a substantial portion of which are transmitted through mutations of mitochondrial DNA (mtDNA) with minimum prevalence of 1 in 5,000 adults 1. Human mtDNA is a small, double-stranded, multi-copy genome present at ~ 100 – 10,000 copies per cell 2. In the disease state, mutated mtDNA often co-exists with wild-type mtDNA in heteroplasmy, and disease severity in conditions caused by heteroplasmic mtDNA mutations correlates with mutation load 3. A threshold effect, where > 60% mutant mtDNA load must be exceeded before symptoms manifest, is a definitive feature of heteroplasmic mtDNA diseases, and attempts to shift the heteroplasmic ratio below this threshold have driven much research towards treatment of these incurable and essentially untreatable disorders. One such approach relies on directed nucleolysis of mtDNA using, among other programmable genome engineering tools, mitochondrially targeted zinc finger-nucleases (mtZFNs) 4–6. Because mammalian mitochondria lack efficient DNA double-strand break (DSB) repair pathways 7, selective introduction of DSBs into mutant mtDNA leads to rapid degradation of these molecules by components of the mtDNA replisome 8. As mtDNA copy number is maintained at a cell type-specific steady-state level, selective elimination of mutant mtDNA stimulates replication of the remaining mtDNA pool, eliciting shifts in the heteroplasmic ratio.
In previous work, we have described methods for delivery of zinc finger proteins (ZFPs) to mitochondria in cultured cells 9,10 and the assembly and function of efficient mtZFN architectures, capable of producing large heteroplasmic shifts that result in phenotype rescue of patient-derived cell cultures 5,11–13. Using the first available mouse model of heteroplasmic mitochondrial disease, bearing the point mutation m.5024C>T in mitochondrial tRNAALA (mt-tRNAALA), which faithfully recapitulates key molecular features of mitochondrial disorders in cardiac tissue 14, we now demonstrate efficient manipulation of mtDNA heteroplasmy with concomitant rescue of molecular and biochemical phenotypes across the heart following delivery of mtZFNs by systemically administered adeno-associated virus (AAV).
Results
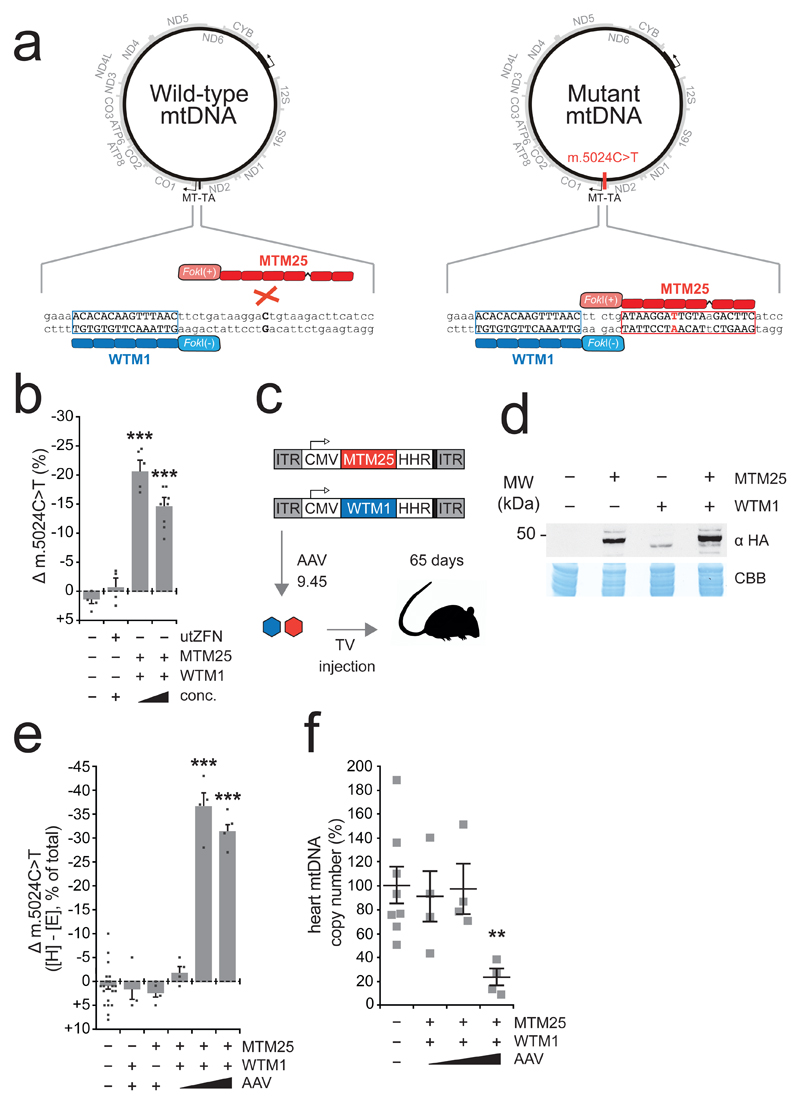
In the context of second generation tail-tail mtZFN architectures (mtZFN2G) shown to be efficacious in previous work 5,15, we set out to generate pairs of zinc finger proteins (ZFP) with single nucleotide binding specificity for m.5024C>T. As this site in the mouse mtDNA is challenging for ZFPs, a selection of targeting strategies with varying numbers of zinc finger motifs, spacer region lengths and additional linkers were employed. Assembly of candidate ZFPs yielded a library (Fig. S1A and Table S1) consisting of 24 unique ZFPs targeting the m.5024C>T site, referred to as mutant-specific monomer (MTM), and a single partner ZFP targeting an adjacent sequence on the opposite strand, referred to as wild-type-specific monomer 1 (WTM1). These constructs were subjected to several rounds of screening in mouse embryonic fibroblasts (MEFs) bearing ~ 65% m.5024C>T to assess heteroplasmy shifting activity (Fig. S1B). These screens identified consistent, specific activity of pairing MTM25/WTM1 (Fig. S1C and Fig. 1A), which produced a shift of ~ 20%, from 65% to 45% m.5024C>T in the MEF cell line as determined by pyrosequencing (Fig. 1B). We additionally confirmed exclusive mitochondrial localization of MTM25 and WTM1 in MEF cells (Fig. S2), and then selected this pair for in vivo experiments.
Figure 1. Strategy to eliminate m.5024C>T and in vivo mtDNA heteroplasmy modification.
A Illustration of mtZFN strategy. A wild-type specific monomer (WTM1), bind upstream of m.5024 in wild-type and mutant genomes; a mutant specific monomer (MTM25) binds preferentially to the mutated site. Dimerization of obligatory heterodimeric FokI domains produces DNA double-stand breaks resulting in specific depletion of mutant mtDNA. B Pyrosequencing of m.5024C>T heteroplasmy from MEFs transfected with controls or MTM25/WTM1 at differing concentrations facilitated by tetracycline-sensitive HHR 12. Change (Δ) in m.5024C>T heteroplasmy is plotted. utZFN is a mtZFN that does not have a target site in mouse mtDNA 12. n = 5 (mtZFN, low expression), 8 (mtZFN, high expression), 4 (all other conditions) biologically independent cell cultures (Table S2). Error bars indicate SD. Statistical analysis performed: two-tailed Student’s t-test. Vehicle/mtZFN low expression p = 0.000021, vehicle/mtZFN high expression p = 0.000083. Measure of center is the mean. C Scheme of in vivo experiments. MTM25 and WTM1 are encoded in separate AAV genomes, encapsidated in AAV9.45 then simultaneously administered by tail-vein (TV) injection. Animals are sacrificed at 65 days post-injection. D Western blot of total heart protein from animals injected with 5*1012 vg MTM25 and/or WTM1. Both proteins include the HA tag and are differentiated by molecular weight. This blot was performed twice with similar results. Raw data are available for this panel (Fig. S9). E Pyrosequencing of m.5024C>T heteroplasmy from ear [E] and heart [H] total DNA. Change (Δ) in m.5024C>T is plotted. n = 20 (vehicle), 3 (WTM1 only), 4 (all other conditions) animals (Table S2). Error bars indicate SEM. Statistical analysis performed: two-tailed Student’s t-test. Vehicle/intermediate dose p < 0.00001, Vehicle/high dose p < 0.00001. Measure of center is the mean. F Assessment of mtDNA copy number by qPCR. n = 8 (vehicle), 4 (all other conditions) animals (Table S2). Error bars indicate SEM. Statistical analysis performed: two-tailed Student’s t-test p = 0.007931. Measure of center is the mean.
MTM25 and WTM1 mtZFN monomers were encoded in separate viral genomes and encapsidated within the cardiac-tropic, engineered AAV9.45 serotype (Fig. 1C) 16. Following tail-vein administration of 5*1012 viral genomes (vg) per monomer per mouse, robust expression of MTM25 and WTM1 in total mouse heart tissue was detected by western blotting (Fig. 1D). Despite equal quantities of injected viral genomes, lower expression levels of WTM1 were consistently detected, possibly due to lower stability of the translated protein. Next, various doses of mtZFN-AAV9.45 were administered into mt-tRNAALA animals harbouring m.5024C>T heteroplasmy ranging from 44 % - 81 % (Table S2). As only minimal variance in heteroplasmy is observed between tissues of the m.5024C>T mouse13, mtDNA heteroplasmy is assessed by comparison of pyrosequencing data, expressed as the change (Δ) between ear punch genotype (E) determined at two weeks of age (prior to experimental intervention) and post-mortem heart genotype (H). Analysis of animals at 65 days post-injection revealed specific elimination of the m.5024C>T mutant mtDNA in mtZFN-treated mice, but not in vehicle- or single monomer-injected controls (Fig. 1E). The extent to which heteroplasmy was altered by mtZFN treatment followed a biphasic AAV dose-dependent trend, with the intermediate dose (5*1012 vg) being the most efficient in eliminating m.5024C>T mutant mtDNA (Fig. 1E). The lowest (1*1012 vg) dose did not result in heteroplasmy shifts (Fig. 1E), due to insufficient concentration of mtZFNs and mosaic transduction of the targeted tissue by AAV (Fig. S3). The highest dose (1*1013 vg) exhibited diminished heteroplasmy shifting activity compared with the intermediate dose (5*1012 vg), likely due to mitochondrial off-target effects resulting in partial mtDNA copy number depletions, which are not observed when lower doses are administered (Fig. 1F). It is unclear what effect, if any, these partial depletions of mtDNA copy number could exert over time, however this lattermost result is consistent with our previous observations 12, underscoring the importance of fine-tuning mtZFN levels in mitochondria for efficient mtDNA heteroplasmy modification. AAV9.45 transduction could not be detected in non-cardiac tissues, and no shifts in heteroplasmy were detected in the liver at 65 days post-injection, irrespective of viral dose (Fig. S3). As AAV transduction of post-mitotic tissues, particularly in short-lived mammals, is essentially permanent, a time-dependence of heteroplasmy shifting is expected. Accordingly, measurements of mtDNA heteroplasmy over time in cardiac tissue demonstrate significant increases in heteroplasmy shifting activity in the latest post-treatment time points (Fig. S4). Despite the presence of two regions with significant homology to the mtDNA target site in the nuclear genome, no evidence for off-target effects exerted by mtZFNs could be detected at these sites (Fig. S5A,B), consistent with our previous reports of exclusive mitochondrial localization of mtZFNs 5,9,10,12. Additionally, no evidence for non-homologous end-joining (NHEJ) at the target site in mtDNA could be detected, confirming previous data that mtZFN-induced DNA DSBs do not result in NHEJ activity (Fig. S5C) 12.
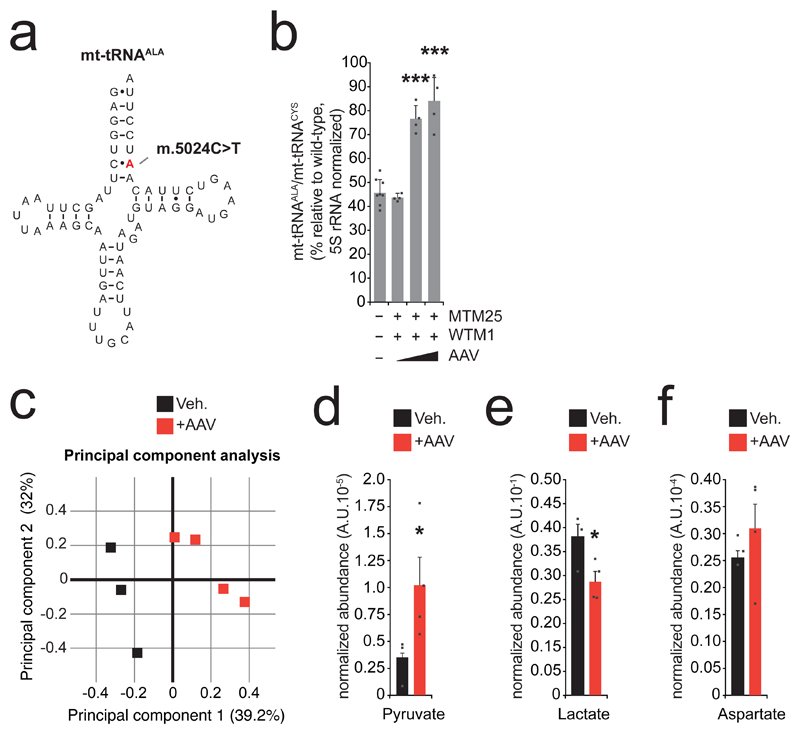
Having defined conditions within which a robust shift of m.5024C>T heteroplasmy is achieved in vivo, we next addressed disease-relevant phenotype correction in the model. A common feature of mt-tRNA mutations in mitochondrial diseases, recapitulated in the tRNAALA mouse model 14, is the instability of mt-tRNA molecules in proportion with mutant load (Fig. 2A) 17. To assess the effects of mtZFN treatment on the stability of mt-tRNAALA in the hearts of animals across the dosage range, we used high-resolution northern blotting, which revealed a significant increase in mt-tRNAALA steady-state levels (Fig. 2B and Fig. S6) that are proportional to heteroplasmy shifts detected in these mice (average m.5024C>T heteroplasmy: control 71% pre-, 73% post-treatment; low AAV dose, 73% pre-, 71% post-treatment; medium AAV dose, 73% pre-, 37% post-treatment; high AAV dose, 71% pre-, 40% post-treatment) (Fig. 1E and Table S2) and consistent with previously reported data 14. Depletions of mtDNA copy number associated with administration of high viral doses (Fig. 1F), did not appear to impact recovery of mt-tRNAALA steady-state levels following heteroplasmy shift (Fig. 2B). This agrees with previously published data that even severe mtDNA depletion does not manifest in proportional changes of mitochondrial RNA steady-state levels 18.
Figure 2. Reduction of m.5024C>T mtDNA heteroplasmy results in phenotype rescue.
A Illustration of mt-tRNAALA bearing the m.5024C>T mutation. Given the nature and position of this mutation, transcribed tRNA molecules containing the mutation mispair are unlikely to fold correctly or be aminoacylated, resulting in reduced steady-state levels of mt-tRNAALA at high levels of m.5024C>T heteroplasmy 14. B Quantification of high-resolution northern blot data from total heart RNA extracts. mt-tRNAALA and mt-tRNACYS abundance was normalized to 5S rRNA. n = 8 (vehicle), 4 (all other conditions) animals (Table S2). Error bars indicate SEM. Statistical analysis performed: two-tailed Student’s t-test. Vehicle/intermediate dose p < 0.00001, vehicle/high dose p = 0.00011. Measure of center is the mean. C Principal component analysis (PCA) plot of metabolomic data for intermediate dose AAV-treated mice and age/initial heteroplasmy-matched controls acquired by LC-MS (Table S2). n = 3 (vehicle), 4 (AAV) animals. D Total metabolite levels of pyruvate from samples measured in C. n = 3 (vehicle), 4 (AAV) animals. Error bars indicate SEM. Statistical analysis performed: one-tailed Student’s t-test. p = 0.046403. Measure of center is the mean. E Total metabolite levels of lactate from samples measured in C. n = 3 (vehicle), 4 (AAV) animals. Error bars indicate SEM. Statistical analysis performed: one-tailed Student’s t-test. p = 0.03505. Measure of center is the mean. E Total metabolite levels of aspartate from samples measured in C. Error bars indicate SEM. n = 3 (vehicle), 4 (AAV) animals. Measure of center is the mean
To assess the physiological effects of mt-tRNAALA molecular phenotype rescue, we analyzed steady-state metabolite levels in cardiac tissue from mice with high m.5024C>T mutant heteroplasmy treated with the intermediate viral titer (5*1012 vg) and heteroplasmy/age matched controls (Table S2). This analysis revealed an altered metabolic signature in mtZFN treated mice (Fig. 2C and Fig. S7), demonstrating significantly increased pyruvate levels (Fig. 2D) and significantly decreased lactate levels (Fig. 2E) in treated mice, suggestive of a diminished reliance on glycolysis, coupled to elevated aspartate levels (Fig. 2F) in treated mice, suggestive of improved mitochondrial respiration 19. These indicators of improved mitochondrial metabolism are not observed in mice treated with the highest AAV dose (Fig. S8), which also exhibit substantial copy number depletions (Fig. 1F). Due to phenotypic heterogeneity of mice bearing high levels of mtDNA heteroplasmy, changes in gross cardiac function following heteroplasmic shifts could not be assessed. Taken together, these data indicate that partial m.5024C>T heteroplasmy shift (Fig. 1E) results in recovery of mt-tRNAALA steady-state levels and rescue of mitochondrial function (Fig. 2B-F).
Discussion
Our previous reports on the use of mtZFN technology have demonstrated that these programmable nucleases can target multiple genetic lesions, producing phenotypically relevant shifts of mtDNA heteroplasmy in cellular models of mitochondrial dysfunction 5,12,13. Here, we have further demonstrated the flexibility and future potential of mtZFN technology by targeting another heteroplasmic mutation in mouse mtDNA, m.5024C>T, manipulating the heteroplasmy of this variant both in vitro and in vivo (Fig. 1), which results in molecular and physiological rescue of disease phenotypes in heart tissue (Fig. 2).
Despite the time elapsed since mtDNA mutations were first associated with human disease in the late 1980’s 20,21, effective treatments for heteroplasmic mitochondrial disease have not been forthcoming. Preventing the transmission of mtDNA mutations through mitochondrial replacement therapy/mitochondrial donation has gained traction 22,23, although given the nature of the mtDNA bottleneck 24, issues surrounding carryover of mutant mtDNA 25, heterogeneous mitochondrial disease presentation 26 and the subsequent lack of family history of mitochondrial disease in the majority of new cases, these approaches can only be of limited use. More recently, several intriguing molecular pathways to treatment of mitochondrial disease have been defined and explored by a number of groups 27, however, hopes for clinically-relevant therapy for heteroplasmic mitochondrial disease, thus far, remain unfulfilled 28. The data we describe in this letter, and those from Bacman et al. 29, constitute proof-of-principle that somatic mitochondrial genome editing using programmable nucleases, in combination with the ever-increasing collection of engineered, tissue-specific AAV serotypes, may offer a potentially universal route to treatment for heteroplasmic mitochondrial disease. Given the magnitude of in vivo heteroplasmy modification demonstrated using these tools, total amelioration of clinical symptoms and/or halting of disease progression could be expected. As such, this development has the potential to transform the prospects of many mitochondrial disease patients, and further work enabling the translation of these tools into effective medicines is vital.
Online Methods
Constructs, plasmids and viral vectors
All mtZFN architectures used were as reported for second generation mtZFN (mtZFN2G), with the exception of the ZFP domains 5,15. The MTM(n)_T2A_WTM1 m.5024C>T candidate library was cloned by insertion of the MTM ZFP domains upstream of FokI(+) between 5’ EcoRI and 3’ BamHI restriction sites. This product was then PCR amplified to include a 5’ ApaI site and remove the 3’ stop codon while also incorporating a T2A sequence and 3’ XhoI site. This fragment was then cloned into pcmCherry (Addgene 62803) using ApaI/XhoI sites. The WTM1 ZFP was separately cloned upstream of FokI(-) in the pcmCherry_3k19 vector (Addgene 104499) incorporating the 3’ hammerhead ribozyme (HHR) using 5’ EcoRI and 3’ BamHI sites, and the resulting product was PCR amplified to include 5’ XhoI and 3’ AflII sites allowing cloning downstream of MTM(n) variants. MTM25(+) and WTM1(-) monomers were cloned into separate pcmCherry and pTracer vectors as described previously 15. Vector construction of mtZFNs intended for AAV production was achieved by PCR amplification of MTM25(+)_HHR and WTM1(-)_HHR transgenes, incorporating 5’ EagI and 3’ BglII sites. These products were then cloned into rAAV2-CMV between 5’ EagI and 3’ BamHI sites. The FLAG epitope tag of WTM1(-) was replaced with a hemagglutinin (HA) tag at the same position in the WTM1(-) open reading frame by PCR. The resulting plasmids were used to generate recombinant AAV2/9.45-CMV-MTM25 and AAV2/9.45-CMV-WTM1 viral particles at the UNC Gene Therapy Center, Vector Core Facility (Chapel Hill, NC). The 3K19 hammerhead ribozyme (HHR) sequence 30 was incorporated into mtZFN-AAV9.45 constructs to allow ubiquitous expression of the transgene from CMV while limiting the expression level, allowing administration of the high viral titers required to ensure effective co-transduction of cells in the targeted tissue without inducing large mtDNA copy number depletions.
Maintenance, transfection and fluorescence activated cell sorting of cell cultures
Wild-type and m.5024C>T mouse embryonic fibroblast (MEF) cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 2 mM L-glutamine, 110 mg/L sodium pyruvate (Life Technologies) and 10% FCS (PAA Laboratories). Cells were transfected by electroporation using Nucleofector II apparatus (Lonza) using a MEF1 kit and T20 program. Fluorescence activated cell sorting (FACS) was performed as described previously 15. Control of mtZFN expression was achieved through titration of tetracycline into culture media, controlling the rate of HHR autocatalysis as described previously 12.
Use of animal models
All animal experiments were carried out in accordance with the UK Animals (Scientific Procedures) Act 1986 (PPL70/7538) and EU Directive 2010/63/EU. The C57BL/6j-tRNAALA mice used in this study were housed from one to four per cage in a temperature controlled (21°C) room with a 12 h light-dark cycle and 60% relative humidity. The experimental design included only male mice between 2 to 8 months of age harboring 44 % - 81 % m.5024C>T heteroplasmy (20 Vehicle, 7 Single Monomer, 4 per mtZFN-AAV9.45 dosage) (Table S1). Treatments of vehicle (1 x PBS, 350 mM NaCl, 5% w/v D-sorbitol) and AAVs were administered by tail vein injection.
Protein extraction and quantitation
For cultured cells, total cellular protein was extracted as described previously 12. For mouse heart tissue, 50 mg was homogenised in RIPA buffer (150 mM NaCl, 50 mM Tris pH 8, 1% (v/v) Triton X-100, 0.5% (v/v) deoxycholate, 0.1% (v/v) SDS) using a gentleMACS dissociator (Miltenyi). The resulting homogenate was centrifuged at 10,000 x g at 4C for 10 minutes, supernatant was then recovered and centrifuged at 10,000 x g at 4C for 10 minutes. Concentration of both cellular and tissue protein extracts was determined by BCA assay (Pierce).
Immunodetection of proteins
The localization of proteins by immunofluorescence in fixed MEF cells was performed as described previously 10. The following antibodies were used: rabbit anti-TOM20 (Santa Cruz Biotechnology, sc-11415, 1:200), Alexa Fluor 647 anti-rabbit (Abcam, ab150079, 1:1000), mouse anti-FLAG (Sigma, F1804, 1:1000), Alexa Fluor 594 anti-mouse (Life Technologies, R37121, 1:1000), rat anti-HA (Roche, 11867431001, 1:200), Alexa Fluor 488 anti-rat (Life Technologies, A11006). Immunofluorescence images were captured using a Zeiss LSM880 confocal microscope and processed using ImageJ.
Detection of proteins by western blotting was achieved by resolving 20-100 μg of extracted protein on SDS-PAGE 4-12% Bis-Tris Bolt gels. These were transferred to nitrocellulose using an iBlot 2 transfer cell (Life Technologies). Antibodies used for western blotting in this work: rat anti-HA (Roche, 11867431001, 1:500), goat anti-rat HRP (Santa Cruz, SC2065, 1:1000). Gels were stained for loading using Coomassie Brilliant Blue (Life Technologies).
Tissue histology and fluorescence microscopy
To evaluate GFP expression in histological sections, mouse tissues (heart, liver, brain, kidney and skeletal muscles) were snap-frozen in isopentane pre-cooled in liquid nitrogen. Eight μm-thick sections on positive-charged glass slides were fixed in 4% PFA, washed with PBS and finally mounted with Prolong Diamond Antifade Mountant with DAPI. Images were acquired using a Zeiss Axio Observer Z1 microscope LSM 880 confocal module, equipped with an Argon Ion MultiLine Laser, Solid State Diode Laser (405 nm), AOTF filter, and a Plan-Apochromat 63x/1.4 NA oil immersion objective). All settings were preserved during image acquisition for all samples. Image J was used to process the images.
DNA extraction and quantitation
DNA was extracted from both cultured cells and whole tissues using a Qiagen DNEasy Blood & Tissue kit, according to the manufacturer’s instructions. Once acquired, DNA concentrations were assessed by spectrophotometry.
Pyrosequencing and qPCR
Assessment of m.5024C>T mtDNA heteroplasmy was carried out by pyrosequencing. PCR reactions for pyrosequencing were prepared using KOD DNA polymerase (Takara) for 40 cycles using 100 ng template DNA with the following primers:
m.4,962 – 4,986 Forward
5’ ATACTAGTCCGCGAGCCTTCAAAG 3’
m.5,360 – m.5,383 Reverse
5’ [Btn] GAGGGTTCCGATATCTTTGTGATT 3’
m.5003 – m.5022 Sequencing primer
5’ AAGTTTAACTTCTGATAAGG 3’
Mitochondrial DNA copy number of mouse heart samples was determined by qPCR using PowerUp SYBR Green Master Mix according to the manufacturer’s protocol (Applied Biosystems). Samples were analysed using a 7900HT Fast Real-Time PCR System (Thermo Fisher). The following primers were used:
MT-COI Forward
5’ TGCTAGCCGCAGGCATTACT 3’
MT-COI Reverse
5’ CGGGATCAAAGAAAGTTGTGTTT 3’
RNaseP Forward
5’ GCCTACACTGGAGTCCGTGCTACT 3’
RNaseP Reverse
5’ CTGACCACACACGAGCTGGTAGAA 3’
All primers for pyrosequencing and qPCR were designed using NCBI reference sequences GRCm38.p6 and NC_005089.1 for the C57BL/6j mouse nuclear and mitochondrial genomes respectively.
Amplicon resequencing of nuclear DNA off-target sites
Two regions of the NCBI reference sequence for C57BL/6j nuclear DNA demonstrated significant homology (>75% sequence identity) with the mtZFN target site in mtDNA. Amplicons containing these sites were obtained by PCR using primers listed below:
Ch.2 Forward
5’ GGGTTCCGATATCTTTGTGATTGG 3’
Ch.2 Reverse
5’ GAGCATAAGCCATTGTTGTTCTG 3’
Ch.5 Forward
5’ GACTACCTGAGCAAGGAGTC 3’
Ch.5 Reverse
5’ CTACAGGAGATGGAGGACAC 3’
All primers were designed using NCBI reference sequence GRCm38.p6 for the C57BL/6j mouse nuclear genome. PCR amplicons were subjected to Nextera sample processing, and resulting libraries were assessed by 2 x 150-cycle paired-end sequencing using a MiSeq instrument (Illumina). Quality trimming and 3’-end adapter clipping of sequenced reads were performed simultaneously with Trim Galore! (--paired) and aligned to GRCm38 using bowtie2. Only reads that contained the entire region chr5: 60042834-60042934 or chr2: 22589909-22590009 were selected for counting with SAMtools (flagstat) and insertion/deletion count based on CIGAR string (I/D). All individual samples yielded >10,000 reads per nucleotide.
Amplicon resequencing of the mtDNA target site
The region m.4,962 - 5,383, also used for pyrosequencing analysis, was amplified by PCR using un-biotinylated primers. PCR amplicons were subjected to Nextera sample processing, and resulting libraries were assessed by 2 x 150-cycle paired-end sequencing using a MiSeq instrument (Illumina). Quality trimming and 3’-end adapter clipping of sequenced reads were performed simultaneously with Trim Galore! (--paired) and aligned to GRCm38 using bowtie2. Only reads that contained the entire region m.4,994 – 5,094 were selected for counting with SAMtools (flagstat) and insertion/deletion count based on CIGAR string (I/D). All individual samples yielded >10,000 reads per nucleotide.
RNA extraction and northern blotting
Total RNA was extracted from 25 mg of mouse heart tissue using Trizol (Ambion) by homogenization using a gentleMACS dissociator (Miltenyi). Northern blotting was performed as described previously 31. Briefly, 5 µg of total RNA was resolved on a 10 % (w/v) polyacrylamide gel containing 8 M urea. Gels were dry blotted onto a positively charged nylon membrane (Hybond-N+), with the resulting membrane cross-linked by exposure to 254 nm UV light, 120 mJ/cm2. For tRNA probes, cross-linked membranes were hybridised with radioactively labelled RNA probes T7 transcribed from PCR fragments corresponding to appropriate regions of mouse mtDNA. 5S rRNA was probed with a complementary α[32P]-end labelled DNA oligo. Membranes were exposed to a storage phosphor screen and scanned using a Typhoon phosphor imaging system (GE Healthcare). The signals were quantified using Fiji software. The following primers/oligonucleotides were used:
MT-TA Forward
5’ TAATACGACTCACTATAGGGAGACTAAGGACTGTAAGACTTCATC 3’
MT-TA Reverse
5’ GAGGTCTTAGCTTAATTAAAG 3’
MT-TC Forward
5’ TAATACGACTCACTATAGGGAGACAAGTCTTAGTAGAGATTTCTC 3’
MT-TC Reverse
5’ GGTCTTAAGGTGATATTCATG 3’
MT-TL1 Forward
5’ TAATACGACTCACTATAGGGAGACTATTAGGGAGAGGATTTGAAC 3’
MT-TL1 Reverse
5’ ATTAGGGTGGCAGAGCCAGG 3’
5S rRNA oligo:
5’ AAGCCTACAGCACCCGGTATTCCCAGGCGGTCTCCCATCCAAGTACTAACCA 3’
All primers for northern blotting were designed using NCBI reference sequences GRCm38.p6 and NC_005089.1 for the C57BL/6j mouse nuclear and mitochondrial genomes respectively.
Sample preparation and liquid chromatography coupled to mass spectrometry (LC-MS) analysis
Snap-frozen tissue specimens were cut and weighed into Precellys tubes prefilled with ceramic beads (Stretton Scientific Ltd., Derbyshire, UK). An exact volume of extraction solution (30% acetonitrile, 50% methanol and 20% water) was added to obtain 40 mg specimen per mL of extraction solution. Tissue samples were lysed using a Precellys 24 homogenizer (Stretton Scientific Ltd., Derbyshire, UK). The suspension was mixed and incubated for 15 minutes at 4°C in a Thermomixer (Eppendorf, Germany), followed by centrifugation (16,000 g, 15 min at 4°C). The supernatant was collected and transferred into autosampler glass vials, which were stored at -80°C until further analysis. Samples were randomized in order to avoid bias due to machine drift and processed blindly. LC-MS analysis was performed using a QExactive Orbitrap mass spectrometer coupled to a Dionex U3000 UHPLC system (Thermo). The liquid chromatography system was fitted with a Sequant ZIC-pHILIC column (150 mm × 2.1 mm) and guard column (20 mm × 2.1 mm) from Merck Millipore (Germany) and temperature maintained at 40°C. The mobile phase was composed of 20 mM ammonium carbonate and 0.1% ammonium hydroxide in water (solvent A), and acetonitrile (solvent B). The flow rate was set at 200 µL/min with the gradient as described previously 32. The mass spectrometer was operated in full MS and polarity switching mode. The acquired spectra were analyzed using XCalibur Qual Browser and XCalibur Quan Browser software (Thermo Scientific).
Statistical analysis
One and two-tailed Student’s t-test were used to compare independent means. Statistical analysis was performed using Prism 5 software.
Supplementary Material
Acknowledgements
This work was supported by the Medical Research Council (MC_U105697135 and MC_UU_00015/4 to M.M., MC_UU_12022/6 to C.F. and MC_UU_00015/5 to M.Z.), ERC Advanced Grant (FP7-322424 to M.Z.), NRJ-Institut de France (to M.Z.) and the Max Planck Society (to J.B.S.). P. R.-G. was supported by "Fundação para a Ciência e a Tecnologia" (PD/BD/105750/2014). We would like to acknowledge the significant contribution to model development made by Prof. Nils-Göran Larsson, which was essential to this work. We are grateful to the personnel at Phenomics Animal Care Facility for their technical support in managing our mouse colonies. We are grateful to Martin Rice, Phenomics Animal Care Facility, for technical assistance with viral administration. We thank Regina Dirksen (MPI, Cologne) for isolation and immortalization of the MEFs. All FACS experiments were performed at the NIHR BRC Cell Phenotyping Hub, Cambridge, UK, by Chris Bowman, Esther Perez, Jelena Markovic Djuric and Anna Petrunkina-Harrison.
Footnotes
Data availability statement
All NGS data generated in the present study are available from the BioProject database using accession number PRJNA479953. All other datasets and materials are available from the corresponding authors.
Author Contributions
P.A.G. designed the research, performed biochemical, in vitro and in vivo experiments, analyzed data and wrote the paper. C.V. performed in vivo experiments. M.-L.S. contributed to model characterization. A.S.H.C. and E.G. performed mass spectrometry-based metabolomic experiments and analyzed data. C.A.P. and L.V.H. performed biochemical experiments and analyzed data. B.J.M performed biochemical and immunofluorescence experiments. P.R.-G. and R.C. performed histology experiments. L.Z. designed and assembled the ZFP library. E.J.R. oversaw ZFP library preparation. M.Z. oversaw in vivo experiments. C.F. oversaw mass spectrometry-based metabolomic experiments. J.B.S. provided cell and mouse models and contributed to model characterization. M.M. oversaw the project and co-wrote the paper, with all authors’ involvement.
Competing Financial Interests Statement
E.J.R. and L.Z. are current full-time employees of Sangamo Therapeutics.
References
- 1.Gorman GS, et al. Prevalence of nuclear and mitochondrial DNA mutations related to adult mitochondrial disease. Ann Neurol. 2015;77:753–759. doi: 10.1002/ana.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wachsmuth M, Hubner A, Li M, Madea B, Stoneking M. Age-Related and Heteroplasmy-Related Variation in Human mtDNA Copy Number. Plos Genet. 2016;12:e1005939. doi: 10.1371/journal.pgen.1005939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gorman GS, et al. Mitochondrial diseases. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.80. 16080. [DOI] [PubMed] [Google Scholar]
- 4.Bacman SR, Williams SL, Pinto M, Peralta S, Moraes CT. Specific elimination of mutant mitochondrial genomes in patient-derived cells by mitoTALENs. Nat Med. 2013;19:1111–1113. doi: 10.1038/nm.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gammage PA, Rorbach J, Vincent AI, Rebar EJ, Minczuk M. Mitochondrially targeted ZFNs for selective degradation of pathogenic mitochondrial genomes bearing large-scale deletions or point mutations. EMBO Mol Med. 2014;6:458–466. doi: 10.1002/emmm.201303672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy P, et al. Selective elimination of mitochondrial mutations in the germline by genome editing. Cell. 2015;161:459–469. doi: 10.1016/j.cell.2015.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alexeyev M, Shokolenko I, Wilson G, LeDoux S. The maintenance of mitochondrial DNA integrity--critical analysis and update. Cold Spring Harb Perspect Biol. 2013;5:a012641. doi: 10.1101/cshperspect.a012641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peeva V, et al. Linear mitochondrial DNA is rapidly degraded by components of the replication machinery. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04131-w. 1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minczuk M, Papworth MA, Kolasinska P, Murphy MP, Klug A. Sequence-specific modification of mitochondrial DNA using a chimeric zinc finger methylase. Proc Natl Acad Sci U S A. 2006;103:19689–19694. doi: 10.1073/pnas.0609502103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minczuk M, Kolasinska-Zwierz P, Murphy MP, Papworth MA. Construction and testing of engineered zinc-finger proteins for sequence-specific modification of mtDNA. Nat Protoc. 2010;5:342–356. doi: 10.1038/nprot.2009.245. [DOI] [PubMed] [Google Scholar]
- 11.Minczuk M, Papworth MA, Miller JC, Murphy MP, Klug A. Development of a single-chain, quasi-dimeric zinc-finger nuclease for the selective degradation of mutated human mitochondrial DNA. Nucleic Acids Res. 2008;36:3926–3938. doi: 10.1093/nar/gkn313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gammage PA, et al. Near-complete elimination of mutant mtDNA by iterative or dynamic dose-controlled treatment with mtZFNs. Nucleic Acids Res. 2016;44:7804–7816. doi: 10.1093/nar/gkw676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaude E, et al. NADH Shuttling Couples Cytosolic Reductive Carboxylation of Glutamine with Glycolysis in Cells with Mitochondrial Dysfunction. Mol Cell. 2018;69:581–593 e587. doi: 10.1016/j.molcel.2018.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kauppila JH, et al. A Phenotype-Driven Approach to Generate Mouse Models with Pathogenic mtDNA Mutations Causing Mitochondrial Disease. Cell Rep. 2016;16:2980–2990. doi: 10.1016/j.celrep.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gammage PA, Van Haute L, Minczuk M. Engineered mtZFNs for Manipulation of Human Mitochondrial DNA Heteroplasmy. Methods Mol Biol. 2016;1351:145–162. doi: 10.1007/978-1-4939-3040-1_11. [DOI] [PubMed] [Google Scholar]
- 16.Pulicherla N, et al. Engineering liver-detargeted AAV9 vectors for cardiac and musculoskeletal gene transfer. Mol Ther. 2011;19:1070–1078. doi: 10.1038/mt.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yarham JW, Elson JL, Blakely EL, McFarland R, Taylor RW. Mitochondrial tRNA mutations and disease. Wiley Interdiscip Rev RNA. 2010;1:304–324. doi: 10.1002/wrna.27. [DOI] [PubMed] [Google Scholar]
- 18.Jazayeri M, et al. Inducible expression of a dominant negative DNA polymerase-gamma depletes mitochondrial DNA and produces a rho0 phenotype. J Biol Chem. 2003;278:9823–9830. doi: 10.1074/jbc.m211730200. [DOI] [PubMed] [Google Scholar]
- 19.Birsoy K, et al. An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell. 2015;162:540–551. doi: 10.1016/j.cell.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–719. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 21.Wallace DC, et al. Mitochondrial DNA mutation associated with Leber's hereditary optic neuropathy. Science. 1998;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 22.Craven L, et al. Pronuclear transfer in human embryos to prevent transmission of mitochondrial DNA disease. Nature. 2010;465:82–85. doi: 10.1038/nature08958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tachibana M, et al. Towards germline gene therapy of inherited mitochondrial diseases. Nature. 2013;493:627–631. doi: 10.1038/nature11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Floros VI, et al. Segregation of mitochondrial DNA heteroplasmy through a developmental genetic bottleneck in human embryos. Nat Cell Biol. 2018;20:144–151. doi: 10.1038/s41556-017-0017-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamada M, et al. Genetic Drift Can Compromise Mitochondrial Replacement by Nuclear Transfer in Human Oocytes. Cell Stem Cell. 2016;18:749–754. doi: 10.1016/j.stem.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vafai SB, Mootha VK. Mitochondrial disorders as windows into an ancient organelle. Nature. 2012;491:374–383. doi: 10.1038/nature11707. [DOI] [PubMed] [Google Scholar]
- 27.Viscomi C, Bottani E, Zeviani M. Emerging concepts in the therapy of mitochondrial disease. Biochim Biophys Acta. 2015;1847:544–557. doi: 10.1016/j.bbabio.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Pfeffer G, et al. New treatments for mitochondrial disease-no time to drop our standards. Nat Rev Neurol. 2013;9:474–481. doi: 10.1038/nrneurol.2013.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bacman SR, et al. MitoTALEN reduces mutant mtDNA load and restores tRNAAla levels in a mouse model of heteroplasmic mtDNA mutation. Nature Medicine. 2018 doi: 10.1038/s41591-018-0166-8. in press (back-to-back submission) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beilstein K, Wittmann A, Grez M, Suess B. Conditional control of mammalian gene expression by tetracycline-dependent hammerhead ribozymes. ACS Synth Biol. 2015;4:526–534. doi: 10.1021/sb500270h. [DOI] [PubMed] [Google Scholar]
- 31.Pearce SF, et al. Maturation of selected human mitochondrial tRNAs requires deadenylation. Elife. 2017;6 doi: 10.7554/eLife.27596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mackay GM, Zheng L, van den Broek NJ, Gottlieb E. Analysis of Cell Metabolism Using LC-MS and Isotope Tracers. Methods Enzymol. 2015;561:171–196. doi: 10.1016/bs.mie.2015.05.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.