Abstract
The SLC22 transporter family consists of more than two dozen members, which are expressed in the kidney, the liver, and other tissues. Evolutionary analysis indicates that SLC22 transporters fall into at least six subfamilies: OAT (organic anion transporter), OAT-like, OAT-related, OCT (organic cation transporter), OCTN (organic cation/carnitine transporter), and OCT/OCTN-related. Some—including OAT1 [SLC22A6 or NKT (novel kidney transporter)] and OAT3 (SLC22A8), as well as OCT1 (SLC22A1) and OCT2 (SLC22A2)—are widely studied drag transporters. Nevertheless, analyses of knockout mice and other data indicate that SLC22 transporters regulate key metabolic pathways and levels of signaling molecules (e.g., gut microbiome products, bile acids, tricarboxylic acid cycle intermediates, dietary flavonoids and other nutrients, prostaglandins, vitamins, short-chain fatty acids, urate, and ergothioneine), as well as uremic toxins associated with chronic kidney disease. Certain SLC22 transporters—such as URAT1 (SLC22A12) and OCTN2 (SLC22A5)—are mutated in inherited metabolic diseases. A new systems biology view of transporters is emerging. As proposed in the remote sensing and signaling hypothesis, SLC22 transporters, together with other SLC and ABC transporters, have key roles in interorgan and interorganism small-molecule communication and, together with the neuroendocrine, growth factor–cytokine, and other homeostatic systems, regulate local and whole-body homeostasis.
Keywords: homeostasis, drug transporter, drug metabolizing enzyme, OAT1, OAT3, OCT1, OCT2
INTRODUCTION
SLC22 transporters have a central role in moving small molecule endogenous metabolites, drugs, and toxins (exogenous and endogenous) between tissues and interfacing body fluids (e.g., the kidney proximal tubule, hepatocytes, the choroid plexus) (Figure 1) (1–7). Due to the pharmaceutical importance of SLC22 transporters, they are one of the best-studied SLC families from the viewpoint of drug handling. However, considerable recent data support an essential role in whole-organism physiology, including interorgan and interorganism communication.
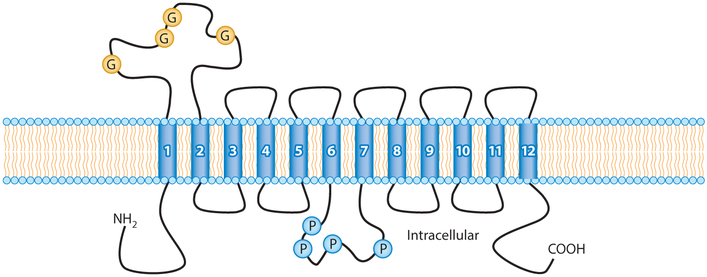
Figure 1.
SLC22 transporters in epithelial tissues. SLC22 transporters tend to have 12 transmembrane domains (numbered blue cylinders). SLC22 transporters are mostly expressed in epithelial tissues, such as the proximal tubule of the kidney, hepatocytes, cells of the choroid plexus, and the olfactory epithelium. They tend to be uptake transporters, often expressed on the basolateral (blood) side of the polarized epithelial cell, although certain transporters are expressed on the apical side. Figure adapted from Reference 146. Abbreviations: G, glycosylation sites; P, PKC phosphorylation sites.
The existence of the family, now known as SLC22, was first proposed in 1997 (8) when the gene for organic anion transporter 1 (OAT1, SLC22A6), originally designated NKT (novel kidney transporter), was first cloned (8); at that time, it was shown to be homologous to the previously identified NLT (novel liver transporter) (9), now called OAT2 (SLC22A7), and organic cation transporter 1 (OCT1, SLC22A1), which had been reported in 1994 (10). Although many reviews have generally discussed each family member in order of its numeral designation as an SLC22 (e.g., SLC22A1, SLC22A2, SLC22A3, SLC22A4) or after separation into OAT, OCT, OCTN (organic cation/carnitine transporter), and other subfamilies, this approach tends to be as much historical as based on structural or functional relatedness. As presented in Table 1, it is evident that there are many cases in which some of the most structurally related transporters—such as SLC22A6 (OAT1), SLC22A8 (OAT3), SLC22A12 (URAT1), and SCL22A20 (OAT6)—carry numerical designations that do not necessarily reflect underlying similarities. If considered in numerical order, these similarities can quickly become confusing, even to those working in the field.
Table 1.
SLC22 transporter family
| Gene symbol | Common name | Alias | Substrates | Expression |
|---|---|---|---|---|
| SLC22A1 | OCT1 | Cationic drugs Putrescine Thiamine Carnitine |
Liver Small intestine |
|
| SLC22A2 | OCT2 | Cationic drugs Acetylcholine Dopamine Creatinine |
Kidney Small intestine |
|
| SLC22A3 | OCT3 |
EMT EMTH |
Cationic drugs Epinephrine |
Heart Skeletal muscle Central nervous system |
| SLC22A4 | OCTN1 | ETT | Ergothioneine Carnitine Acetylcholine |
Kidney Intestine |
| SLC22A5 | OCTN2 | Carnitine Choline |
Skeletal muscle Kidney |
|
| SLC22A6 | OAT1 | NKT | Anionic drugs α-Ketoglutarate Indoxyl sulfate p-Cresol sulfate Urate |
Kidney Choroid plexus |
| SLC22A7 | OAT2 | NLT | Anionic drugs cGMP (cyclic guanosine monophosphate) Creatinine |
Liver Kidney |
| SLC22A8 | OAT3 | ROCT | Anionic drugs Estrone sulfate Bile acids Flavonoids Creatinine |
Kidney Brain Retina Testis |
| SLC22A9 | OAT7 | Estrone sulfate | Liver | |
| SLC22A10 | Liver | |||
| SLC22A11 | OAT4 | Urate Estrone sulfate |
Placenta Kidney Brain |
|
| SLC22A12 | URAT1 | RST | Urate | Kidney |
| SLC22A13 | OAT10 | ORCTL3 | Organic anions Urate |
Kidney Brain Colon |
| SLC22A14 | Oct12 | ORCTL4 | Kidney Colon |
|
| SLC22A15 | FLIPT1 | Kidney Brain Liver |
||
| SLC22A16 | FLIPT2 |
CT2 OCT6 |
Carnitine Drugs |
Testis Bone marrow |
| SLC22A17 | BOIT |
BOCT1 24p3R |
Brain Choroid plexus |
|
| SLC22A18 | ORCTL2 | IMPT1 | Kidney | |
| Slc22a19 | Oat5 | Estrone sulfate | ||
| SLC22A20 | OAT6 | Odorants | Olfactory (mouse) | |
| Slc22a21 | Octn3 | |||
| Slc22a22 | OatPG | Prostaglandin | ||
| SLC22A23 | Boct2 | Brain Liver |
||
| SLC22A24 | NET46 | |||
| SLC22A25 | UST6 | Liver | ||
| SLC22A31 | Larynx Prostate |
In the absence of full functional and/or structural characterization of all, or nearly all, SLC22 family members, another approach is to describe and discuss family members in terms of evolutionary relatedness, which is presumed to reflect to a significant degree the underlying structural or functional similarity (11). This has the additional advantage of placing orphan or understudied members of the SLC22 family in relation to better-studied members, suggesting possible functions for the orphan family members.
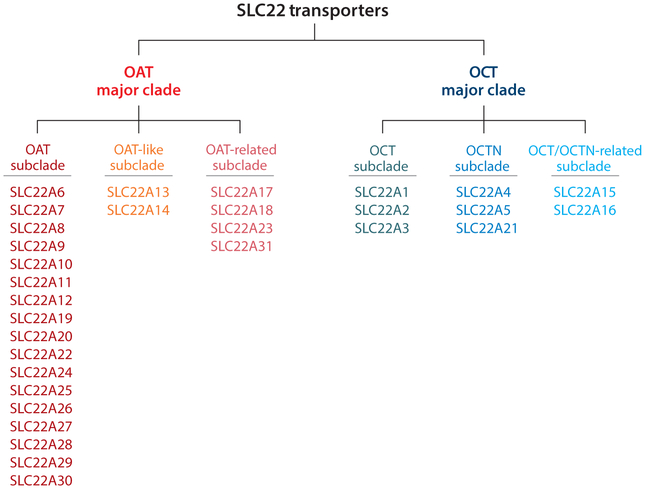
Recent evolutionary studies make it possible to present and discuss the family in this way (11), and this is the approach taken here. Thus, on the basis of evolutionary analysis, the SLC22 family can be divided into at least six subclades or subfamilies (Figure 2): (a) OAT (SLC22A6, SLC22A7, SLC22A8, SLC22A9, SLC22A10, SLC22A11, SLC22A12, SLC22A19, SLC22A20, SLC22A22, SLC22A24, SLC22A25, SLC22A26, SLC22A27, SLC22A28, SLC22A29, SLC22A30), (b) OAT-like (SLC22A13, SLC22A14), (c) OAT-related (SLC22A17, SLC22A18, SLC22A23, SLC22A31), (d) OCT (SLC22A1, SLC22A2, SLC22A3), (e) OCTN (SLC22A4, SLC22A5, SLC22A21), and (f) OCT/OCTN-related (SLC22A15, SLC22A16). Again, by focusing on these evolutionarily distinct groups of SLC22 genes, it may be easier to appreciate functional similarities and differences within and between subfamilies, as well as to identify areas worthy of further study. [Although we may use the term SLC22 transporters to refer to the family in general, it should be noted that transport capacity has not been established for each of the family members and, in at least one case, it has been argued that despite some sequence similarity, the gene SLC22A18 may not be a legitimate family member (11).]
Figure 2.
The six subfamilies of SLC22 transporters. Evolutionary analysis indicates that SLC22 transporters are highly conserved and found in fly, worm, sea urchin, and other organisms. The SLC22 family consists of two major clades: OAT (organic anion transporter) and OCT (organic cation transporter). Each of these clades can be further divided into three subclades, designated as OAT, OAT-like, OAT-related, OCT, OCTN (organic cation/carnitine transporter), and OCT/OCTN-related.
Although we will discuss the medical, pharmacological, and toxicological relevance of each transporter (to the extent that these are reasonably well established), our overall focus will be on conveying the fundamental physiological relevance of single transporters or groups of transporters in the handling of endogenous metabolites and signaling molecules. This is in accordance with recent calls for more systems biology approaches to the SLC and ABC transporters (1, 12, 13). Indeed, in certain systems biology analyses, SLC22 appears to be at the center of a transporter interaction network involving many other SLC families in different organs (13). Thus, with a particular emphasis on the role of various SLC22 transporters in remote interorgan and interorganism communication, the discussion will be framed by the remote sensing and signaling hypothesis, which is summarized below.
THE PHYSIOLOGICAL ROLE OF SLC22 TRANSPORTERS: THE REMOTE SENSING AND SIGNALING HYPOTHESIS
SLC22 transporters are highly conserved throughout evolution and are expressed in different sites (or at different levels) during prenatal development compared with adulthood, suggesting a physiological role in embryogenesis (14, 15). Even in the adult, some SLC22 transporters are highly expressed in tissues not usually associated with drug elimination [e.g., OAT6 in the olfactory epithelium, various OCTs in the central nervous system (CNS)]. When considered together, the roughly thirty SLC22 transporters have broad substrate specificity, ranging from organic anions to organic zwitterions and organic cations, as well as other molecules (16–18). Mutations in at least two transporters (SLC22A12 or URAT1; SLC22A5 or OCTN2) are associated with metabolic diseases related to urate and carnitine handling that are inherited along classic Mendelian lines (19, 20). Metabolomics data from various SLC22 transporter knockouts indicate that, in some cases, they have a central role in modulating the levels of endogenous metabolites and signaling molecules—including those derived from the activity of the gut microbiome—in the plasma, as well as other body fluids (21–26). Many of these metabolites and signaling molecules have well-described roles in cell, organ, interorgan, and interorganism communication (12, 27). Based on in vitro and other data, other transporters appear to be relatively selective transporters of urate (URAT1), prostaglandins (OATPG), odorant-type molecules (OAT6 or SLC22A20), or carnitine and ergothioneine (OCTN1) (19, 28–31). Other transported molecules (e.g., bile acids, α-ketoglutarate, and β-hydroxybutyrate) are involved in metabolite signaling via G protein-coupled receptors and epigenetic and other modifications capable of altering the transcriptional state of cells.
All of this suggests that the highly publicized role of SLC22 transporters (particularly OAT1, OAT3, OCT1, and OCT2) as drug transporters is somewhat misleading, and we must ask, “What do drug transporters really do?” Despite the type of evidence discussed above, even now the biological perspective seems to be mostly ignored, especially considering the attention that these transporters receive from the pharmaceutical industry and the US Food and Drug Administration (FDA), as well as other regulatory agencies (32–34). For these transporters are very much a part of organ and systemic physiology, as well as pathophysiology (4, 12, 35–39).
Beginning in 2006, an alternative view of the biological role of SLC22 drug transporters—and, indeed, all SLC and ABC drug transporters—began to be developed based on the endogenous functions of these widely expressed transporters of metabolites and signaling molecules, and their roles in interorgan and interorganism communication (for example, gut microbiome–gut–plasma–liver–kidney–urine, or gut microbiome–gut–plasma–blood–brain barrier–CNS–cerebrospinal fluid) (1, 12, 16, 18, 27, 30). This theory, which now seems to be supported by a wide variety of converging data—such as that surveyed above and detailed below—is now called the remote sensing and signaling hypothesis (Figure 3). It has been discussed in detail elsewhere (1, 12, 18, 27); here, we focus on its particular relevance to SLC22 transporters and use it as a framework for the remainder of this review.
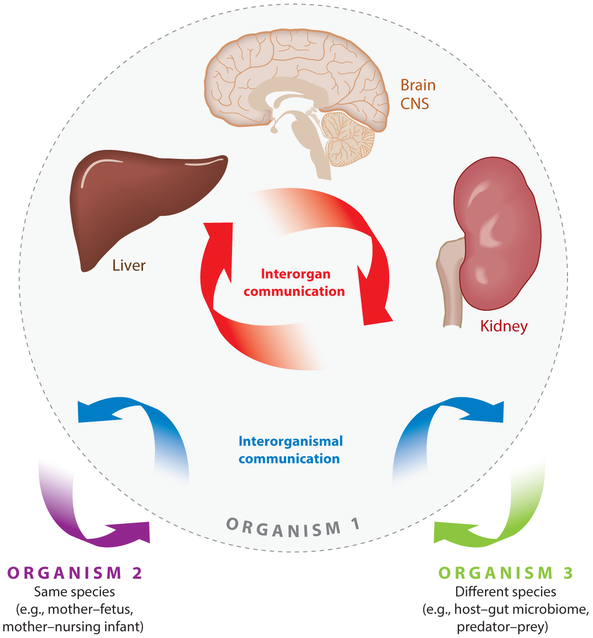
Figure 3.
The role of SLC22 transporters in remote sensing and signaling. The remote sensing and signaling hypothesis argues that SLC22 transporters and other SLC and ABC drug and nondrug transporters are part of a large interorgan and interorganism small-molecule communication network that maintains homeostasis in epithelial and nonepithelial tissues and body fluid compartments, such as blood, bile, cerebrospinal fluid, and urine. This remote sensing and signaling system in SLC and ABC transporters is hypothesized to work in parallel with the neuroendocrine and growth factor–cytokine systems and is, indeed, intertwined with them. Figure adapted from Reference 1. Abbreviation: CNS, central nervous system.
Recent systems biology analyses suggest that SLC22 might be a superhub among all SLC transporters in that it sits at, or near the center of, a larger SLC transporter network that connects to a wide variety of SLC transporters regulating various aspects of metabolism and expressed in many different organs (13). Moreover, metabolic reconstructions (explained below) of transcriptomics and metabolic data derived from Oat1 and Oat3 knockout mice indicate broad effects on many biological pathways involved in metabolism and signaling throughout the body (21, 24–26, 40). Although some SLC22 transporters (i.e., OAT1, OAT3, OCT1, and OCT2) are multispecific (for both metabolites and drugs), others have limited specificity (e.g., OAT6 for odorants, URAT1 for urate, OATPG for prostaglandins, OCTN2 for carnitine, and OCTN1 for ergothioneine). Nevertheless, multispecific OATs can also transport odorants, urate, prostaglandins, and carnitine (or its metabolites).
Early on (e.g., 2006–2009), one potential example of remote sensing and signaling—still hypothetical but circumstantially supported by transport data, expression patterns, and knockout metabolomics data—was proposed as follows (16, 29, 30). The gut microbiome produces odorants that are transported by SLC22 and other multi-, oligo-, and monospecific transporters along the gut–liver–kidney axis such that these volatile odorants enter the urine via OAT1 and OAT3 (23, 26). Once volatilized, these odorants can potentially interact with OAT6 (which has a high affinity for certain odorants), odorant receptors, or both, in the olfactory mucosa of an animal of the same or different species (e.g., mouse, cat, dog) (16, 29, 30). The expected, but unproven, behavioral consequence might be that this molecular mechanism of interorgan and interorganism communication (e.g., gut bacteria to mouse gut–liver–kidney axis to urine to cat olfactory system) affects the behavioral interactions of mice, cats, and dogs.
Although the previous, somewhat hypothetical, example provides insight into how remote sensing and signaling via SLC22 and other transporters might work through interorgan and interorganism communication, a validated, and somewhat surprising, example comes from the renal and extrarenal handling of uric acid in the setting of hyperuricemia (or gout) in the absence versus the presence of kidney disease (35, 41, 42). The human and nonhuman in vivo evidence supporting this example (described below) demonstrates how multispecific (OAT1, OAT3) and more specific (URAT1, OAT4) renal SLC22 transporters work together (and with members of other transporter families such as ABCG2 and SLC2A9) to regulate uric acid levels. In the setting of kidney disease, the function of an intestinal uric acid transporter, ABCG2, becomes considerably more important (35, 41, 42). This supports a mechanism for remote communication between the dysfunctional kidney and the intact intestine to regulate uric acid levels when the normal renal elimination of this potentially toxic organic anion is affected by declining kidney function, thereby averting more severe renal and nonrenal pathology due to high uric acid levels. It also begs the question about mechanisms of transporter-mediated remote communication between different organs in the service of maintaining homeostasis (including SLC and ABC transporters); as discussed below, potential molecular mechanisms are beginning to emerge.
The remote sensing and signaling hypothesis will not only be the framework through which we view what follows regarding SLC22 transporters, but it is also discussed as a corrective to the tremendous bias of viewing these transporters as drug transporters, which often overlooks the well-established biological roles of these and other SLC and ABC drug transporters in local as well as systemic physiology and pathophysiology involving endogenous metabolites and signaling molecules. Nevertheless, the implications of the remote sensing and signaling hypothesis are quite far-ranging—including considering how the network of multispecific, oligospecific, and monospecific transporters throughout the body integrates with other physiological systems, such as the endocrine, growth factor–cytokine, and autonomic systems to maintain homeostasis—and these implications have been discussed elsewhere in detail (1, 12, 18, 27).
CLASSIFYING SLC22 TRANSPORTERS BASED ON EVOLUTIONARY RELATEDNESS
When OAT1 was discovered as NKT in 1996–1997 (43), it was noticed that NKT was homologous to two other genes in the database that had not previously been linked: OCT1 and NLT (now OAT2). Thus, a new family was proposed, consisting of these three genes likely to be involved in organic anion transport, organic cation transport, or both (8). With the subsequent identification of more than two dozen other SLC22 transporters, SLC22 became a family that in humans and rodents comprises approximately 30 members (Table 1).
SLC22 transporters are conserved in nonmammalian model organisms, such as fly, worm, zebrafish, and sea urchin (11, 44, 45). Although it is not always possible to trace back the orthologous gene from humans or rodents to more ancient species, it is possible to determine whether the gene is a member of the SLC22 transporter family and, often, whether it is a member of the OAT or OCT clades. In some cases, it is also possible to link the ancient gene to one of the six subclades, or subfamilies, that are described below.
From an evolutionary standpoint, SLC22 divides into two major clades: the large OAT clade (23 members) and the smaller OCT clade (8 members). Each of these clades subdivides into three subclades, resulting in six groups distinguished by sequence homology through evolution, an apparent common ancestral gene, subclade-selective amino acid sequence motifs, or a combination of these (11). Below, the physiology, pharmacology, and toxicology of each group (SLC22 subclade) are described in more detail. SLC22 is one of the best-studied transporter families and has hundreds of drug, toxin, nutrient, and metabolite ligands; it is beyond the scope of this review to enumerate all of these, and the focus here is on the largely neglected area of endogenous ligands that are important in normal physiology and disease. For lists of in vitro and in vivo drugs and metabolites the reader is referred to various reviews and original knockout metabolomics papers (1–7, 17, 18, 22–24, 25, 26, 43).
OAT Group
By far, the best-studied members of the OAT group are OAT1 and OAT3 (5, 18, 46, 47). This is due to their great pharmaceutical importance, as indicated by FDA guidelines that list these two transporters among several SLC and ABC drug transporters that are to be tested for the transport of new drugs (32). As far as common drugs are concerned, it has not been easy to identify clear differences between the substrate specificity of OAT1 and OAT3, although a recent study employing machine learning methods, along with other studies, has indicated that OAT3 has a greater preference for some drugs with a slightly cationic character (16, 48, 49). However, the in vitro data on metabolite transport for OAT1 and OAT3, as well as data from metabolomics of the knockouts, are beginning to suggest major differences in endogenous substrate specificity, with OAT1 handling smaller molecules [e.g., TCA (tricarboxylic acid) cycle intermediates] and OAT3 able to handle larger, more hydrophobic molecules with multiple rings (e.g., bile acids and flavonoids) (18, 22–26).
The OAT subclade includes a number of transporters that appear to be much more selective in their substrate preference. For example, URAT1, which was originally identified in mouse as renal-specific transporter (Rst) (50), is a major renal uric acid transporter expressed in the apical membrane of the proximal tubule (51) (discussed in more detail below).
OAT2 was originally identified as NLT (9), and it was this OAT that, along with OAT1 (originally NKT) and OCT1, made it possible to propose the existence of the SLC22 transporter family based on sequence homology (8). OAT2 is highly expressed in the liver and in the kidney (52). Although it transports some drugs (as do many of the other OATs, apart from OAT1 and OAT3), among its most intriguing substrates are cyclic nucleotides [e.g., cyclic guanosine monophosphate (cGMP)] (53). Thus, it is possible that, as with MRP4 (ABCC4) (54), this drug transporter plays a part in modulating signal transduction pathways.
Another interesting OAT, from the perspectives of both substrate selectivity and expression, is OAT6 (SLC22A20) (16, 29, 30, 55, 56), which is closely related to OAT1 and OAT3 and is expressed primarily in the olfactory epithelium (30), as well as the testes (57). Given its olfactory expression pattern, it is particularly noteworthy that OAT6 interacts at relatively high affinity (compared with other OATs) with a number of well-known odorant molecules (e.g., benzoate, heptanoate) and has sequence similarity to certain odorant receptors (56). Although the role (if any) of OAT6 in olfaction remains to be fully explored, some possibilities are discussed below.
OAT-Like Group
This subclade is not well studied, but based on in vitro assays as well as human association studies (58, 59), OAT10 (SLC22A13) appears to be a urate transporter. It also appears to transport nicotinate (59).
OAT-Related Group
SLC22A17 is interesting for multiple reasons. It has high expression in the choroid plexus, a tissue that also expresses other SLC22 family members, including OAT1 and OAT3 (60). Its actual substrate is not well understood, although it has been proposed to participate in iron transport via functioning as a receptor for lipocalin (NGAL) (61). SLC22A18 is, arguably, an outlier among SLC22 transporters, not only in this subclade but also in the SLC22 family as a whole. By amino acid sequence, it appears related to the bacterial drug–proton antiporters (11). Much more work needs to be done to understand the endogenous function of transporters in this intriguing subclade.
OCT Group
OCT1, OCT2, and OCT3 have substantially different tissue distributions, although they overlap in some tissues (62, 63). From the general viewpoint of organ function, the major liver OCT is OCT1; the major kidney OCT is OCT2; and OCT3, although expressed in liver and kidney, has received attention because of its potential importance in neurotransmitter uptake in the CNS (64).
The drug substrate specificities for OCT1 and OCT2 are similar, and machine learning studies have not been able to consistently identify molecular features that predispose to being transported by OCT1 versus OCT2 (49). With respect to endogenous function, OCT1 is also capable of transporting polyamines, thiamine, and carnitine, and OCT2 is capable of interacting with dopamine, creatinine, and acetylcholine (65).
OCTN Group
OCTN1 is likely to be primarily an ergothioneine and carnitine transporter (28) and expressed in the kidney, intestine, liver, and other tissues. OCTN2, a well-established carnitine transporter, is highly expressed in the kidney, with some expression in the intestine, placenta, and other tissues (66).
OCT/OCTN-Related Group
From the viewpoint of amino acid sequence homology and evolution, SLC22A15 and SLC22A16, originally described, respectively, as FLIPT (fly-like putative transporter)-1 and FLIPT2, appear most closely related to the OCTN subclade of carnitine transporters (67). These transporters—expressed in the kidney and other tissues—-are not well studied, but there is some in vitro evidence to suggest that they, particularly FLIPT2 (SLC22A16), can interact with carnitine (68) and drugs such as bleomycin (69).
PHARMACOLOGICAL AND TOXICOLOGICAL ROLES BASED ON MOUSE SLC22 KNOCKOUTS
Of the SLC22 transporter murine knockouts, the following have been studied in detail: knockouts of Oct1 (Slc22a1), Oct2 (Slc22a2), Oct3 (SLC22a3), Octn1 (Slc22a4), Octn2 (Slc22a5), Oat1 (Slc22a6), Oat3 (Slc22a8), and Rst (Urat1, Slc22A12) (22–24, 70–77). Below are highlighted some of the key findings from the knockouts related to the pharmacological and toxicological roles of SLC22 transporters; later, the intriguing physiological insights from the knockouts are discussed.
OAT Subclade
As expected, the deletion of Oat1 results in diminished renal handling of its prototypical organic anion substrate para-amino hippurate (PAH) (22), and the deletion of Oat3 likewise results in diminished handling of prototypical organic anions (e.g., estrone sulfate) by the kidney (75).
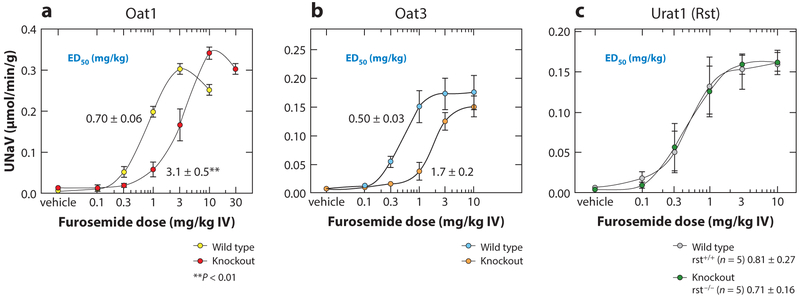
Generally, the loss of Oat1 or Oat3 has similar effects on many drugs. The loss of Oat1 or Oat3 results in a blunted natriuretic response to loop and thiazide diuretics, whereas this is not seen when Rst (Urat1) is deleted (Figure 4) (71, 78). OAT1 and OAT3 take up these albumin-bound diuretics into the cells of the proximal tubule of the kidney, whereupon they are eliminated into the lumen of the tubule. They then act on transporters in the loop of Henle and the distal convoluted tubule to inhibit net sodium reabsorption. Thus, the deletion of either of these two genes leads to a substantial loss of the effect of loop and thiazide diuretics.
Figure 4.
Loss of diuretic response in Oat knockout mice. A number of SLC22 transporters have been deleted in mice, and the pharmacological and toxicological responses of knockout mice have been studied in detail. After the deletion of either (a) Oat1 or (b) Oat3—which are known to transport diuretics in vivo—the natriuretic (urine sodium excretion) response to either loop or thiazide diuretics is blunted. This is not seen in the deletion of the closely related (c) Urat1 (Rst), which is primarily a renal urate transporter. Figure adapted from References 22 and 78. Abbreviations: ED50, half-maximal effective dose; IV, intravenous; Rst, renal specific transporter; UNaV, urinary sodium excretion.
The loss of OAT1 or OAT3 also leads to diminished renal uptake of a number of antiviral drugs (79, 80). A similar effect is seen in the choroid plexus (60). Indeed, OAT1 and OAT3 appear to be major uptake transporters for many of the antivirals used alone or in combination to treat HIV and other viral infections. The Oat3 knockouts have also been shown to have diminished renal excretion of antibiotics, including the β-lactams and fluoroquinolones (81, 82).
OAT3 is also expressed in the brain capillary endothelium (83, 84), and studies in the Oat3 knockout have shown that compounds entering the brain attain higher levels, presumably because Oat3 is unable to transport these compounds across the endothelium from the brain to the blood (85).
Mercury toxicity remains a major environmental concern, and the OATs have been implicated in transporting mercury (often conjugated to glutathione or cysteine in plasma); these conjugates act as if they are organic anions (86). The kidney is a major site of mercury toxicity due to the uptake of organic mercurial conjugates into the highly vulnerable proximal tubule cells via OATs. Consistent with this, the Oat1 knockout is well protected from damage caused by mercurials (39). Although the Oat3 knockout was not tested, the striking renal protection from mercury toxicity in the Oat1 knockout suggests that Oat1 may be relatively more important in handling organic mercurials. Oat3, however, appears to play a major part in the renal elimination of aristolochic acid, one of the presumed causative agents of Balkan endemic nephropathy (87), and the results of knockout studies are consistent with the notion that uptake via Oat3 mediates aristolochic acid nephropathy (88, 89).
The knockout of the Urat1 (Rst) gene results in altered renal handling of uric acid, which would be expected based on the human disease phenotype of the mutated URAT1 gene (71). The knockout of Oat1 and Oat3 also results in altered uric acid handling. Thus, when OAT4 and OAT10 are included, at least five SLC22 transporters appear to be important for uric acid handling. Nevertheless, the knockout phenotype with respect to uric acid for Urat1, as well as for Oat1 and Oat3, was not as great as expected, and it was suggested at the time that other transporters must be as, or more, important in urate handling (71). Subsequently, human studies indicated that among the key urate transporters are SLC2A9 and ABCG2 (90, 91), with ABCG2 becoming particularly important in the setting of renal dysfunction (35). In addition, SLC17 transporters appear to have a role in urate handling (35, 51, 92, 93).
OCT Subclade
Oct1 and Oct2 have been deleted singly and together. The Oct1 knockout has an altered uptake of prototypical organic cations (e.g., tetraethylammonium) into the liver, as well as its altered excretion into the intestinal lumen (73). In addition, there is a defect in metformin handling (76). In the Oct2 knockout, renal handling of organic cations is impaired, and this is also seen in the double knockout of Oct1 and Oct2 (74). Moreover, the double knockout kidney is less susceptible to cisplatin toxicity (72).
Consistent with Oct3’s role in transporting biogenic amines and neurotransmitters, Oct3 knockout mice demonstrate neurobehavioral abnormalities in response to particular stimuli (77). The deletion of Oct2, which can also transport neurotransmitters, has also been reported to affect behavior in mice, and the brains of these mice have altered neurotransmitter levels (70, 94).
METABOLOMICS OF MOUSE KNOCKOUTS OF SEVERAL SLC22 GENES CLARIFIES THEIR ENDOGENOUS FUNCTION
To varying degrees, targeted and untargeted metabolomics analyses have been performed, mainly on the plasma of several OCT, OAT, and OCTN knockouts. Those analyses that have provided additional insight into endogenous functions (and not extensively discussed elsewhere in this review) are highlighted here.
OAT1
From a metabolomics standpoint, a considerable amount of data about the Oat1 knockout are available (22, 23, 25). In general, metabolites involved in energy metabolism are altered, as well as odorant molecules and a number of metabolites derived from the gut microbiome that are, in the setting of kidney disease, associated with uremic toxicity (e.g., indoxyl sulfate, hippurate, p-cresol sulfate). Some of these molecules have undergone modification by phase 2 enzymes in the liver, giving rise to the notion that OATs are key players in the gut–liver–kidney axis. The metabolomics data also suggest an important role for Oatl in proximal tubule metabolism, including the TCA cycle (22).
OAT3
Current metabolomics data suggest that OAT3, even more than OAT1, is involved in the gut–liver–kidney axis. OAT3 handles dietary compounds, including flavonoids, and molecules that are derived from phase 1 and phase 2 liver metabolism (e.g., hydroxylation, sulfation, glucuronidation, acetylation). As with the Oat1 knockout, the Oat3 knockout accumulates many gut microbiome products and uremic toxins of chronic kidney disease (25, 26). There is also a suggestion that OAT3 is responsible for transporting some molecules involved in sialic acid metabolism. Taken together with the Oat1 knockout, the Oat3 metabolomics supports the concept that although the physiological roles of these two transporters overlap, OAT1 is likely more important in proximal tubule energy metabolism and OAT3 is likely more important in the flow of metabolites through the gut–liver–kidney axis (26). Furthermore, even though it has been difficult to distinguish OAT1 and OAT3 drug preferences on the basis of the physiochemical characteristics, metabolites accumulating in the Oat1 and Oat3 knockouts are distinguishable as a group on the basis of molecular weight, size, and number of rings (26).
OCT1
Metabolomics analyses of the Oct1 knockout have revealed altered levels of thiamine, and it is believed that defective transport of the latter metabolite modulates liver steatosis when the Oct1 knockout is crossed with leptin-deficient mice (95).
METABOLIC RECONSTRUCTION OF THE OAT1 KNOCKOUT FROM OMICS DATA
Genome-scale metabolic reconstructions based on transcriptomics data have been successfully applied in lower organisms to make predictions about metabolic pathways affected by perturbation, and these are increasingly being applied to mammalian models and human clinical contexts (96, 97). To create an OAT1-centered metabolic network, transcriptional changes in the Oat1 knockout mouse compared with the wild type were used to reconstruct the metabolic pathways likely to be affected by chronic loss of OAT1 function (21, 40). Many of the predictions were validated based on in vitro metabolite transport data and in vivo metabolomics data from the Oat1 knockout mouse. This information was used to construct a partly validated OAT1-centered metabolic network and pathway, and enrichment analysis implicated a number of metabolic pathways, including those involved in tryptophan metabolism, the TCA cycle, purine and pyrimidine metabolism, fatty acids, prostaglandins, amino acids, vitamins, odorants, and polyamines (see the sidebar titled Top Pathways Affected by OAT1 Loss in Knockout Mice) (40). This type of approach can potentially be used to create tentative drug transporter–centered networks for OAT3, various OATPs (organic anion transporting polypeptides; SLCO), MRPs, and other SLC and ABC transporters, and this approach has the potential to provide a general picture of how multiple drug transporters regulate endogenous metabolism involving organic anion metabolites and signaling molecules. Maps such as these not only help to explain the biology regulated or modulated by SLC and other drug transporters but also begin to explain the broader effects of one or more organic anionic drugs on metabolism beyond simple competition at the site of the transporter. Much remains to be done in this area, and the modeling techniques are rapidly improving.
TRANSCRIPTIONAL REGULATION OF SLC22 TRANSPORTERS
Coordination of interorgan crosstalk involving SLC and ABC transporters requires one or (likely) many mechanisms of integration. Apart from substrate stimulation/inducibility (98) and regulation facilitated by the molecular and cellular mechanisms mediated by hormones and genetic factors (particularly in the setting of organ dysfunction) (1, 4, 12, 18), transcriptional regulation by nuclear receptors that may bind small-molecule substrates of transporters is another potential mechanism with which to explain coordination of remote interorgan or interorganism communication. The transcriptional regulation of several SLC22 transporters has been studied in some detail in cultured cells, in ex vivo tissue preparations, and in vivo using knockouts (99–101). The hepatocyte nuclear factors, HNF4α and HNF1α, which have been shown to regulate many drug-metabolizing enzymes in the liver and other tissues, figure prominently in the regulation of several SLC22 transporters, including OAT1, OAT3, URAT1, and OCT1. This appears to be true of mature as well as developing cells and tissues. For example, by chromatin immunoprecipitation sequencing (ChIP-seq) analysis it can be demonstrated that OAT1 and OAT3 gene regulatory regions have high binding of HNF4α (101). When HNF4α, sometimes with HNF1α, is introduced into cultured cells, the messenger RNA levels of OAT1, OAT3, URAT, and OCT1 (as well as certain ABC transporters and MATEs) are increased to varying degrees (100, 101). This indicates that HNF4α and HNF1α have key roles in regulating these SLC22 transporters. Bioinformatics analyses have implicated other nuclear receptors and transcription factors in the regulation of SLC22 transporters, but these have not been analyzed in detail (101).
An endogenous ligand of HNF4α has been identified as linoleic acid (102). A number of fatty acids that bind nuclear receptors accumulate in certain SLC22 and other SLC and ABC transporter knockouts (1, 7), which raises the possibility that molecules taken up by a transporter can regulate the expression of the same or different SLC and ABC transporters—which, in turn, enables the cell to transport a variety of metabolites and signaling molecules. From the viewpoint of the remote sensing and signaling hypothesis, this is a potential mechanism for connecting nuclear receptor signaling to transport activity, presumably in the service of small molecule homeostasis regulated by the network of SLC and ABC transporters in different tissues lining different body fluid compartments, as well as the neuroendocrine system (1, 12, 18, 27).
DEVELOPMENT AND REGENERATION
SLC22 transporters are expressed during much of prenatal development and often in different sites in the developing embryo when compared with adults, including in the developing CNS, cardiovascular system, intestine, and bone (15). Indeed, this finding gave rise to the hypothesis that these drug transporters might have inherent biological roles. In the embryo, this could include transporting morphogenetic molecules (e.g., cyclic nucleotides, prostaglandins, or various activators of signaling pathways) or key metabolites (e.g., folate) into specific embryonic tissues. Or the transporters could subserve some function (e.g., signal transduction) independent of transport. In addition, the likely carnitine transporter FLIPT1 (SLC22A15) is expressed in developing brain, kidney, and lung, whereas other SLC22 transporters have high expression in the embryonic liver (67). Despite the high embryonic expression of various SLC22 transporters, single Slc22 transporter knockouts generally undergo normal development and postnatal maturation; thus, the embryonic roles of various SLC22 transporters require further investigation.
In the maturing organ—for instance, in the developing kidney—the SLC22 transporters are usually expressed at midgestation, and there is a gradual rise in the expression of transcript during development (14, 103–105). After birth, there is a rapid rise in transcript expression in the developing kidney until the late neonatal to early juvenile period, whereupon expression tends to level off during the late juvenile period and remains steady throughout adulthood (104). This has been best studied in the context of the organic anion transporters Oat1 and Oat3 in which, in addition to transcript expression, postnatal PAH transport has been analyzed in wild-type and knockout animals (104). Functional maturation of the proximal tubule, as measured by PAH transport, generally parallels transcript levels. In the developmental context, as in the adult context, HNF4α and HNF1α seem to be key mediators of the transcription of several SLC22 transporters of the OAT and OCT families (99, 101). There is also evidence from transduced mouse embryonic fibroblasts to suggest that for expression of the full complement of renal transporters, it may be necessary to suppress the liver transporter transcriptional program (100).
Regeneration is sometimes held to recapitulate aspects of development, and it is worth noting that a number of studies have examined the transcriptional and posttranscriptional changes of various OATs in the setting of acute renal injury and recovery models (106, 107). In general, there appears to be an acute drop in OAT expression followed by a later rise that sometimes overshoots basal adult expression before settling down to normal levels. Fragments of certain OATs have been detected in the urine after injury and are being studied as potential biomarkers for acute kidney injury (108).
ROLE OF SIGNALING PATHWAYS IN CELLULAR REGULATION AND FUNCTION OF SLC22 TRANSPORTERS
Over the years, a considerable amount of data have accumulated on the intersection of transporters with signaling and sorting pathways, and this is true of SLC22 transporters as well (109, 110). These transporters undergo covalent modifications, such as phosphorylation, including tyrosine phosphorylation by Src family kinases, which can affect OCT function (111). Other kinases, such as PKC, can affect trafficking (112–114). There are also strong data on the association of SLC22 transporters such as URAT1 with PDZK1 domains, which is important in membrane localization and function (115). Indeed, one of the mutations that affects human uric acid levels is caused by a defect in the association of URAT1 with PDZK1 (116).
In addition, certain SLC22 transporters, such as the olfactory transporter OAT6, have been found to have strong sequence similarity to parts of odorant receptors, which are G protein–coupled receptors (56). Although much of the similarity appears to be in what are presumed to be common odorant binding sites, this has raised the question as to whether one or more SLC22 transporters can function as a transceptor, as occurs in the case of nutrient transporters in bacteria and higher organisms. This intriguing possibility remains to be studied in detail.
Reports support the importance of SLC22A17 in acting indirectly in iron transport; apparently, this transporter is actually a receptor for NGAL, also known as LCN2R, and is involved in the endocytosis of iron-free and iron-bound lipocalin (61). Because of the broad importance of iron and lipocalin in physiology and cell differentiation, SLC22A17 provides an interesting example of how SLC22 transporters may function in unexpected ways beyond those we associate with the classic OATs, OCTs, and OCTNs.
Thus, there are many ways in which classical signaling pathways can affect SLC22 transporter sorting, localization, and function. The examples of SLC22A17 and possibly OAT6 (SLC22A20) suggest novel roles of these transporters in homeostasis that remain to be fully explored. This is particularly interesting from the perspective of the remote sensing and signaling hypothesis, which explicitly emphasizes the intersection of classical neuroendocrine and growth factor–cytokine–dependent signaling pathways with the endogenous physiological roles of SLC and ABC transporters in interorgan communication for the purpose of maintaining the homeostasis of small molecules important to organ and systemic metabolism, as well as signaling in normal and disease states.
CLINICAL SLC22-ASSOCIATED SYNDROMES
OAT Subclade
Early on, nonsynonymous single-nucleotide polymorphisms (SNPs) in the coding regions of OAT1, OAT3, and URAT1 were found to be relatively uncommon (117), but SNPs were much more common in the noncoding regions of these genes (118). Consistent with knockout and in vitro studies, SNPs in OAT1 and OAT3 have been implicated in mercury toxicity (in miners), diuretic responsiveness, and β-lactam antibiotic handling (in an East Asian population) (119–121).
URAT1, originally discovered as Rst in mice, was rediscovered in Japanese patients with exercise-induced uric acid stones (122). These patients have a mutation in URAT1. Subsequently, SNPs in URAT1 were found to be associated with hyperuricemia, although it now appears, based on a number of genome-wide association studies, that SNPs in ABCG2 and SLC2A9 are at least as important in various populations studied (123). Among SLC22 transporters, SNPs in OAT1, OAT3, and OAT4 have also been shown to have effects on uric acid levels, although these effects are much more modest than those of the aforementioned transporters (117).
OATs have been implicated in diabetic nephropathy, which appears to have reduced expression of OAT1 and OAT3 and a metabolic profile in affected patients that resembles that seen in the OAT knockout mice (124). OAT3 has also been implicated in gestational diabetes through an interesting mechanism possibly consistent with the remote sensing and signaling hypothesis (125). CMPF (3-carboxy-4-methyl-5-propyl-2-furanpropanoic acid), derived from dietary and gut microbiome sources, crosses the intestine and enters the blood, whereupon it is taken up by OAT3 in the pancreatic β-cells and, via its effect on redox state, alters insulin secretion (125). This appears to be an example of remote interorgan (if not interorganism) communication via drug transporters leading to aberrant signaling and metabolic disease.
OCTN Subclade
Systemic carnitine deficiency, an autosomal recessive disease that results in cardiomyopathy, skeletal myopathy, and metabolic abnormalities, results from a mutation in OCTN2 (126). Thus, renal reabsorption of carnitine is blocked, leading to renal wasting and lower plasma levels of carnitine, which is necessary for mitochondrial oxidation of fatty acids and generating acetyl coenzyme A (127). SNPs in OCTN1 and OCTN2 are also associated with inflammatory syndromes, including inflammatory bowel disease and rheumatoid arthritis (128–130).
TOXINS, UREMIC SOLUTES, NEUROTRANSMITTERS, AND CREATININE
OAT Subclade
OAT3 is a transporter of ochratoxin A and aristolochic acid, dietary compounds that have been implicated in Balkan endemic nephropathy, which is associated with malignancy in the lower urinary tract (87, 131). As with several other nephrotoxins (i.e., organic mercurial or cephaloridine, which is no longer used), these dietary toxins enter the proximal tubule via OAT3 (and OAT1 in the case of mercury and cephaloridine), whereupon they cause tubular damage (47, 88, 132).
Among endogenous toxins, both OAT1 and OAT3 have received a great deal of attention because they appear to be the major transporters of small molecule uremic toxins, such as indoxyl sulfate, p-cresol sulfate, CMPF, and many others (23–25, 47, 133). These molecules accumulate in the serum of the Oat1 and Oat3 knockouts. Interestingly, trimethylamine N-oxide (TMAO), a uremic toxin associated with cardiovascular morbidity (134), accumulates in the Oat3 knockout (26), although it is not clear whether it can act as a substrate for OAT3. Many of these uremic toxins are tryptophan metabolites derived from gut microbial flora; they cross the intestine and enter the blood. Some are modified by phase 1 (e.g., hydroxylation) or phase 2 (e.g., sulfation, glucuronidation) enzymes in the liver, resulting in potentially toxic compounds, such as indoxyl sulfate (23, 25, 26). This compound is one of several uremic toxins responsible for the uremic syndrome of chronic kidney disease (CKD). Uremic toxins are thought to perturb overall metabolism and result in pathology in the CNS, heart, pericardium, peripheral nerves, skeletal muscle, and other tissues. Indoxyl sulfate also enters the proximal tubule via the OATs, where it appears to accelerate the progression of CKD (133, 135). Thus, there have been efforts to diminish levels of indoxyl sulfate in CKD patients; one such effort tried oral delivery of intestinal sorbents of indole compounds (e.g., AST-120) (135, 136).
Other uremic toxins derived from the gut flora and diet are unmodified by the liver but, nonetheless, appear to enter tissues and cells affected in the uremic syndrome of CKD. Some of these, such as kynurenine, can affect signaling in many tissues, including the CNS (137); others, such as CMPF, affect metabolism (125). Tubular secretion may become more important as renal function declines (138, 139). As indicated by knockout and in vitro data, many of these compounds, such as indoxyl sulfate, are eliminated by OAT1, OAT3, or both (23, 24). Thus, the pathophysiology of uremia appears to be, in part, the result of disordered remote sensing and signaling (1, 12, 18, 25, 27). This pathophysiology involves interorganism communication (e.g., between gut microbes and host) and interorgan communication (e.g., the gut–liver–kidney axis), and it is mediated through metabolites, signaling molecules, and endogenous toxins transported in and out of tissues (e.g., in the CNS) and body fluid compartments by OATs and other multispecific and monospecific transporters.
Although it is not entirely clear which are the main renal transporters determining the secretion of creatinine, a classical marker of renal function that is sometimes listed as a uremic toxin, both OCTs and OATs have been implicated by in vitro studies, knockout mouse studies, or both (140–143).
OCT Subclade
Certain SNPs in OCT1 can alter the hepatic uptake of metformin, an antidiabetes drug, thereby diminishing its efficacy (144). As mentioned above, OCTs are also important creatinine transporters (136, 137).
OCT3 is expressed in the brain, as well as in the kidney and liver. In the brain, it is expressed in the cortex and hippocampus and believed to be important in the extraneuronal reuptake of neurotransmitters (145). Likewise, OCT2, which is expressed in the limbic system as well as in the kidney, is also capable of transporting biogenic amines (70). Because of their CNS expression and ability to transport neurotransmitters, as well as the knockout behavioral phenotypes described above, these genes are receiving attention as potential targets for the treatment of anxiety disorders and depression.
CLOSING COMMENTS ON UNDERSTANDING SLC22 TRANSPORTERS FROM THE PERSPECTIVE OF REMOTE SENSING AND SIGNALING
It is increasingly clear that SLC22 transporters have an important endogenous function and that the subdivision of the transporters into subclades, as described here, provides a novel way to connect their evolutionary history, sequence homology, and endogenous function. The SLC22 transporter family is highly connected to other SLC transporter families involved in key metabolic and signaling processes, and the remote sensing and signaling hypothesis (1, 12, 16, 18, 27) proposes that these drug and nondrug SLC and ABC transporters—expressed in diverse epithelial and endothelial cells (among others), which line body fluid compartments (e.g., urine, amniotic fluid, bile) or important tissue interfaces (e.g., the blood-brain barrier)—are part of a large, endogenous interorgan and interorganism communication network that is intertwined with traditional homeostatic networks, such as the neuroendocrine and growth factor–cytokine systems. The theory implies that the fundamental biology underlying pharmacokinetics needs to be rewritten from the perspective of endogenous physiology, and it broadly expands the role of drug transporters in human and nonhuman homeostasis.
TOP PATHWAYS AFFECTED BY OAT1 LOSS IN KNOCKOUT MICE.
This list is adapted from Reference 40.
∎TCA cycle
∎Tyrosine metabolism
∎Alanine, aspartate, and glutamate metabolism
∎Butanoate metabolism
∎Arginine and proline metabolism
∎Tryptophan metabolism
∎Nicotinate and nicotinamide metabolism
∎Valine, leucine, and isoleucine degradation
∎Nitrogen metabolism
∎Glyoxylate and dicarboxylate metabolism
∎Propanoate metabolism
∎Glycine, serine, and threonine metabolism
∎Purine metabolism
∎Pyrimidine metabolism
ACKNOWLEDGMENTS
The author thanks Dr. Kevin T. Bush for his invaluable assistance. Figure 4 was a control for the papers cited in the figure caption (22, 28) and was a collaboration with the group of Volker Vallon. This work was supported in whole or part by US National Institutes of Health grants R01-DK109392, RO1GM104098, and U54-HD090259.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Nigam SK. 2015. What do drug transporters really do? Nat. Rev. Drug Discov 14:29–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizwan AN, Burckhardt G. 2007. Organic anion transporters of the SLC22 family: biopharmaceutical, physiological, and pathological roles. Pharm. Res 24:450–70 [DOI] [PubMed] [Google Scholar]
- 3.Pelis RM, Wright SH. 2014. SLC22, SLC44, and SLC47 transporters—organic anion and cation transporters: molecular and cellular properties. Curr. Top. Membr 73:233–61 [DOI] [PubMed] [Google Scholar]
- 4.Saito H. 2010. Pathophysiological regulation of renal SLC22A organic ion transporters in acute kidney injury: pharmacological and toxicological implications. Pharmacol. Ther 125:79–91 [DOI] [PubMed] [Google Scholar]
- 5.VanWert AL, Gionfriddo MR, Sweet DH. 2010. Organic anion transporters: discovery, pharmacology, regulation and roles in pathophysiology. Biopharm. Drug Dispos 31:1–71 [DOI] [PubMed] [Google Scholar]
- 6.Wang L, Sweet DH. 2013. Renal organic anion transporters (SLC22 family): expression, regulation, roles in toxicity, and impact on injury and disease. AAPS J 15:53–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.You GF, Morris ME, eds. 2014. Drug Transporters: Molecular Characterization and Role in Drug Disposition. Hoboken, NJ: Wiley; 2nd ed. [Google Scholar]
- 8.Lopez-Nieto CE, You G, Bush KT, Barros EJ, Beier DR, Nigam SK. 1997. Molecular cloning and characterization of NKT, a gene product related to the organic cation transporter family that is almost exclusively expressed in the kidney. J. Biol. Chem 272:6471–78 [DOI] [PubMed] [Google Scholar]
- 9.Simonson G, Vincent A, Roberg K, Huang Y, Iwanij V. 1994. Molecular cloning and characterization of a novel liver-specific transport protein. J. Cell Sci 107:1065–72 [DOI] [PubMed] [Google Scholar]
- 10.Grundemann D, Gorboulev V, Gambaryan S, Veyhl M, Koepsell H. 1994. Drug excretion mediated by a new prototype of polyspecific transporter. Nature 372:549–52 [DOI] [PubMed] [Google Scholar]
- 11.Zhu C, Nigam KB, Date RC, Bush KT, Springer SA, et al. 2015. Evolutionary analysis and classification of OATs, OCTs, OCTNs, and other SLC22 transporters: structure-function implications and analysis of sequence motifs. PLOS ONE 10:e0140569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahn SY, Nigam SK. 2009. Toward a systems level understanding of organic anion and other multispecific drug transporters: a remote sensing and signaling hypothesis. Mol. Pharmacol 76:481–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cesar-Razquin A, Snijder B, Frappier-Brinton T, Isserlin R, Gyimesi G, et al. 2015A call for systematic research on solute carriers. Cell 162:478–87 [DOI] [PubMed] [Google Scholar]
- 14.Brouwer KL, Aleksunes LM, Brandys B, Giacoia GP, Knipp G, et al. 2015. Human ontogeny of drug transporters: review and recommendations of the Pediatric Transporter Working Group. Clin. Pharmacol. Ther 98:266–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pavlova A, Sakurai H, Leclercq B, Beier DR, Yu AS, Nigam SK. 2000. Developmentally regulated expression of organic ion transporters NKT (OAT1), OCT1, NLT (OAT2), and Roct. Am. J. Physiol. Renal Physiol 278:F635–43 [DOI] [PubMed] [Google Scholar]
- 16.Kaler G, Truong DM, Khandelwal A, Nagle M, Eraly SA, et al. 2007. Structural variation governs substrate specificity for organic anion transporter (OAT) homologs: potential remote sensing by OAT family members. J. Biol. Chem 282:23841–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koepsell H 2013. Polyspecific organic cation transporters and their biomedical relevance in kidney. Curr. Opin. Nephrol. Hypertens 22:533–38 [DOI] [PubMed] [Google Scholar]
- 18.Nigam SK, Bush KT, Martovetsky G, Ahn SY, Liu HC, et al. 2015. The organic anion transporter (OAT) family: a systems biology perspective. Physiol. Rev 95:83–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichida K,Hosoyamada M,Hisatome I,Enomoto A,Hikita M,et al. 2004. Clinical and molecular analysis of patients with renal hypouricemia in Japan—influence of URAT1 gene on urinary urate excretion. J. Am. Soc. Nephrol 15:164–73 [DOI] [PubMed] [Google Scholar]
- 20.Seth P, Wu X, Huang W, Leibach FH, Ganapathy V. 1999. Mutations in novel organic cation transporter (OCTN2), an organic cation/carnitine transporter, with differential effects on the organic cation transport function and the carnitine transport function. J. Biol. Chem 274:33388–92 [DOI] [PubMed] [Google Scholar]
- 21.Ahn SY, Jamshidi N, Mo ML, Wu W, Eraly SA, et al. 2011. Linkage of organic anion transporter-1 to metabolic pathways through integrated “omics”-driven network and functional analysis. J. Biol. Chem 286:31522–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eraly SA, Vallon V, Vaughn DA, Gangoiti JA, Richter K, et al. 2006. Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. J. Biol. Chem 281:5072–83 [DOI] [PubMed] [Google Scholar]
- 23.Wikoff WR, Nagle MA, Kouznetsova VL, Tsigelny IF, Nigam SK. 2011. Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1). J. Proteome Res 10:2842–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu W, Jamshidi N, Eraly SA, Liu HC, Bush KT, et al. 2013. Multispecific drug transporter Slc22a8 (Oat3) regulates multiple metabolic and signaling pathways. Drug Metab. Dispos 41:1825–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu W, Bush KT, Nigam SK. 2017. Key role for the organic anion transporters, OAT1 and OAT3, in the in vivo handling of uremic toxins and solutes. Sci. Rep 7:4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bush KT, Wu W, Lun C, Nigam SK. 2017. The drug transporter OAT3 (SLC22A8) and endogenous metabolite communication via the gut-liver-kidney axis. J. Biol. Chem 292:15789–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu W, Dnyanmote AV, Nigam SK. 2011. Remote communication through solute carriers and ATP binding cassette drug transporter pathways: an update on the Remote Sensing and Signaling Hypothesis. Mol. Pharmacol 79:795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grundemann D 2012. The ergothioneine transporter controls and indicates ergothioneine activity—a review. Prev. Med 54(Suppl.):S71–74 [DOI] [PubMed] [Google Scholar]
- 29.Kaler G, Truong DM, Sweeney DE, Logan DW, Nagle M, et al. 2006. Olfactory mucosa-expressed organic anion transporter, Oat6, manifests high affinity interactions with odorant organic anions. Biochem. Biophys. Res. Commun 351:872–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monte JC, Nagle MA, Eraly SA, Nigam SK. 2004. Identification of a novel murine organic anion transporter family member, OAT6, expressed in olfactory mucosa. Biochem. Biophys. Res. Commun 323:429–36 [DOI] [PubMed] [Google Scholar]
- 31.Shiraya K, Hirata T, Hatano R, Nagamori S, Wiriyasermkul P, et al. 2010. A novel transporter of SLC22 family specifically transports prostaglandins and co-localizes with 15-hydroxyprostaglandin dehydrogenase in renal proximal tubules. J. Biol. Chem 285:22141–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.US FDA (Food Drug Adm.). 2012. Guidance for Industry: Drug Interaction Studies—Study Design, Data Analysis, Implications for Dosing, and Labeling Recommendations. Silver Spring, MD: US FDA [Google Scholar]
- 33.Eur. Med. Agency. 2012. Guideline on the Investigation of Drug Interactions. London: Eur. Med. Agency [Google Scholar]
- 34.MHLW Res. Group. 2014. Drug Interaction Guideline for Drug Development and Labeling Recommendations. Tokyo: Minist. Health Labour Welf. [Google Scholar]
- 35.Bhatnagar V, Richard EL, Wu W, Nievergelt CM, Lipkowitz MS, et al. 2016. Analysis of ABCG2 and other urate transporters in uric acid homeostasis in chronic kidney disease: potential role of remote sensing and signaling. Clin. Kidney J 9:444–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandoni A, Hazelhoff MH, Bulacio RP, Torres AM. 2012. Expression and function of renal and hepatic organic anion transporters in extrahepatic cholestasis. World J. Gastroenterol 18:6387–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Di Giusto G, Anzai N, Endou H, Torres AM. 2008. Elimination of organic anions in response to an early stage of renal ischemia–reperfusion in the rat: role of basolateral plasma membrane transporters and cortical renal blood flow. Pharmacology 81:127–36 [DOI] [PubMed] [Google Scholar]
- 38.Di Giusto G, Anzai N, Ruiz ML, Endou H, Torres AM. 2009. Expression and function of Oat1 and Oat3 in rat kidney exposed to mercuric chloride. Arch. Toxicol 83:887–97 [DOI] [PubMed] [Google Scholar]
- 39.Torres AM, Dnyanmote AV, Bush KT, Wu W, Nigam SK. 2011. Deletion of multispecific organic anion transporter Oat1/Slc22a6 protects against mercury-induced kidney injury. J. Biol. Chem 286:26391–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu HC, Jamshidi N, Chen Y, Eraly SA, Cho SY, et al. 2016. An organic anion transporter 1 (OAT1)–centered metabolic network. J. Biol. Chem 291:19474–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu H, Klaassen C. 2008. Gender differences in mRNA expression of ATP-binding cassette efflux and bile acid transporters in kidney, liver, and intestine of 5/6 nephrectomized rats. Drug Metab. Dispos 36:16–23 [DOI] [PubMed] [Google Scholar]
- 42.Yano H, Tamura Y, Kobayashi K, Tanemoto M, Uchida S. 2014. Uric acid transporter ABCG2 is increased in the intestine of the 5/6 nephrectomy rat model of chronic kidney disease. Clin. Exp. Nephrol 18:50–55 [DOI] [PubMed] [Google Scholar]
- 43.Lopez-Nieto CE, You G, Barros EJG, Beier DR, Nigam SK. 1996. Molecular cloning and characterization of a novel transport protein with very high expression in the kidney. J. Am. Soc. Nephrol 7:1301 (Abstr.) [Google Scholar]
- 44.Eraly SA, Monte JC, Nigam SK. 2004. Novel slc22 transporter homologs in fly, worm, and human clarify the phylogeny of organic anion and cation transporters. Physiol. Genom 18:12–24 [DOI] [PubMed] [Google Scholar]
- 45.Wu W, Baker ME, Eraly SA, Bush KT, Nigam SK. 2009. Analysis of a large cluster of SLC22 transporter genes, including novel USTs, reveals species-specific amplification of subsets of family members. Physiol. Genom 38:116–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emami Riedmaier A, Nies AT, Schaeffeler E, Schwab M. 2012. Organic anion transporters and their implications in pharmacotherapy. Pharmacol. Rev 64:421–49 [DOI] [PubMed] [Google Scholar]
- 47.Nigam SK, Wu W, Bush KT, Hoenig MP, Blantz RC, Bhatnagar V. 2015. Handling of drugs, metabolites, and uremic toxins by kidney proximal tubule drug transporters. Clin. J. Am. Soc. Nephrol 10:2039–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahn SY, Eraly SA, Tsigelny I, Nigam SK. 2009. Interaction of organic cations with organic anion transporters. J. Biol. Chem 284:31422–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu HC, Goldenberg A, Chen Y, Lun C, Wu W, et al. 2016. Analysis of molecular properties of drugs interacting with SLC22 transporters OAT1, OAT3, OCT1, and OCT2: a machine-learning approach. J. Pharmacol. Exp. Ther 359:215–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mori K, Ogawa Y, Ebihara K, Aoki T, Tamura N, et al. 1997. Kidney-specific expression of a novel mouse organic cation transporter-like protein. FEBS Lett. 417:371–74 [DOI] [PubMed] [Google Scholar]
- 51.Sakurai H. 2013. Urate transporters in the genomic era. Curr. Opin. Nephrol. Hypertens 22:545–50 [DOI] [PubMed] [Google Scholar]
- 52.Shen H, Lai Y, Rodrigues AD. 2017. Organic anion transporter 2: an enigmatic human solute carrier. Drug Metab. Dispos 45:228–36 [DOI] [PubMed] [Google Scholar]
- 53.Cropp CD, Komori T, Shima JE, Urban TJ, Yee SW, et al. 2008. Organic anion transporter 2 (SLC22A7) is a facilitative transporter of cGMP. Mol. Pharmacol 73:1151–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russel FG, Koenderink JB, Masereeuw R. 2008. Multidrug resistance protein 4 (MRP4/ABCC4): a versatile efflux transporter for drugs and signalling molecules. Trends Pharmacol. Sci 29:200–7 [DOI] [PubMed] [Google Scholar]
- 55.Schnabolk GW, Youngblood GL, Sweet DH. 2006. Transport of estrone sulfate by the novel organic anion transporter Oat6 (Slc22a200). Am. J. Physiol. Renal. Physiol 291:F314–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu W, Bush KT, Liu HC, Zhu C, Abagyan R, Nigam SK. 2015. Shared ligands between organic anion transporters (OAT1 and OAT6) and odorant receptors. Drug Metab. Dispos 43:1855–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnabolk GW, Gupta B, Mulgaonkar A, Kulkarni M, Sweet DH. 2010. Organic anion transporter 6 (Slc22a20) specificity and Sertoli cell-specific expression provide new insight on potential endogenous roles. J. Pharmacol. Exp. Ther 334:927–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.AbuAli G, Grimm S. 2014. Isolation and characterization of the anticancer gene organic cation transporter like-3 (ORCTL3) In Anticancer Genes: Advances in Experimental Medicine and Biology, Vol. 818, ed. Grimm S, pp. 213–27. London: Springer-Verlag; [DOI] [PubMed] [Google Scholar]
- 59.Bahn A, Hagos Y, Reuter S, Balen D, Brzica H, et al. 2008. Identification of a new urate and high affinity nicotinate transporter, hOAT10 (SLC22A13). J. Biol. Chem 283:16332–41 [DOI] [PubMed] [Google Scholar]
- 60.Nagle MA, Wu W, Eraly SA, Nigam SK. 2013. Organic anion transport pathways in antiviral handling in choroid plexus in Oat1 (Slc22a6) and Oat3 (Slc22a8) deficient tissue. Neurosci. Lett 534:133–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Devireddy LR, Gazin C, Zhu X, Green MR. 2005. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell 123:1293–305 [DOI] [PubMed] [Google Scholar]
- 62.Ciarimboli G, Gautron S, Schlatter E, eds. 2016. Organic Cation Transporters: Integration of Physiology, Pathology and Pharmacology. Cham, Switz.: Springer [Google Scholar]
- 63.Koepsell H 2004. Polyspecific organic cation transporters: their functions and interactions with drugs. Trends Pharmacol. Sci 25:375–81 [DOI] [PubMed] [Google Scholar]
- 64.Duan H, Wang J. 2010. Selective transport of monoamine neurotransmitters by human plasma membrane monoamine transporter and organic cation transporter 3. J. Pharmacol. Exp. Ther 335:743–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koepsell H, Schmitt BM, Gorboulev V. 2003. Organic cation transporters. Rev. Physiol. Biochem. Pharmacol 150:36–90 [DOI] [PubMed] [Google Scholar]
- 66.Wu X, Prasad PD, Leibach FH, Ganapathy V. 1998. cDNA sequence, transport function, and genomic organization of human OCTN2, a new member of the organic cation transporter family. Biochem. Biophys. Res. Commun 246:589–95 [DOI] [PubMed] [Google Scholar]
- 67.Eraly SA, Nigam SK. 2002. Novel human cDNAs homologous to Drosophila Orct and mammalian carnitine transporters. Biochem. Biophys. Res. Commun 297:1159–66 [DOI] [PubMed] [Google Scholar]
- 68.Enomoto A, Wempe MF, Tsuchida H, Shin HJ, Cha SH, et al. 2002. Molecular identification of a novel carnitine transporter specific to human testis: insights into the mechanism of carnitine recognition. J. Biol. Chem 277:36262–71 [DOI] [PubMed] [Google Scholar]
- 69.Aouida M, Poulin R, Ramotar D. 2010. The human carnitine transporter SLC22A16 mediates high affinity uptake of the anticancer polyamine analogue bleomycin-A5. J. Biol. Chem 285:6275–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bacq A, Balasse L, Biala G, Guiard B, Gardier AM, et al. 2012. Organic cation transporter 2 controls brain norepinephrine and serotonin clearance and antidepressant response. Mol. Psychiatry 17:926–39 [DOI] [PubMed] [Google Scholar]
- 71.Eraly SA, Vallon V, Rieg T, Gangoiti JA, Wikoff WR, et al. 2008. Multiple organic anion transporters contribute to net renal excretion of uric acid. Physiol. Genom 33:180–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Franke RM, Kosloske AM, Lancaster CS, Filipski KK, Hu C, et al. 2010. Influence of Oct1/Oct2-deficiency on cisplatin-induced changes in urinary N-acetyl-β-d-glucosaminidase. Clin. Cancer Res 16:4198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jonker JW, Wagenaar E, Mol CA, Buitelaar M, Koepsell H, et al. 2001. Reduced hepatic uptake and intestinal excretion of organic cations in mice with a targeted disruption of the organic cation transporter 1 (Oct1 [Slc22a1]) gene. Mol. Cell. Biol 21:5471–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jonker JW, Wagenaar E, Van Eijl S, Schinkel AH. 2003. Deficiency in the organic cation transporters 1 and 2 (Oct1/Oct2 [Slc22a1/Slc22a2]) in mice abolishes renal secretion of organic cations. Mol. Cell. Biol 23:7902–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sweet DH, Miller DS, Pritchard JB, Fujiwara Y, Beier DR, Nigam SK. 2002. Impaired organic anion transport in kidney and choroid plexus of organic anion transporter 3 (Oat3 [Slc22a8]) knockout mice. J. Biol. Chem 277:26934–43 [DOI] [PubMed] [Google Scholar]
- 76.Wang DS, Jonker JW, Kato Y, Kusuhara H, Schinkel AH, Sugiyama Y. 2002. Involvement of organic cation transporter 1 in hepatic and intestinal distribution of metformin. J. Pharmacol. Exp. Ther 302:510–15 [DOI] [PubMed] [Google Scholar]
- 77.Wultsch T, Grimberg G, Schmitt A, Painsipp E, Wetzstein H, et al. 2009. Decreased anxiety in mice lacking the organic cation transporter 3. J. Neural. Transm 116:689–97 [DOI] [PubMed] [Google Scholar]
- 78.Vallon V, Rieg T, Ahn SY, Wu W, Eraly SA, Nigam SK. 2008. Overlapping in vitro and in vivo specificities of the organic anion transporters OAT1 and OAT3 for loop and thiazide diuretics. Am. J. Physiol. Renal. Physiol 294:F867–73 [DOI] [PubMed] [Google Scholar]
- 79.Nagle MA, Truong DM, Dnyanmote AV, Ahn SY, Eraly SA, et al. 2011. Analysis of three-dimensional systems for developing and mature kidneys clarifies the role of OAT1 and OAT3 in antiviral handling. J. Biol. Chem 286:243–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Truong DM, Kaler G, Khandelwal A, Swaan PW, Nigam SK. 2008Multi-level analysis of organic anion transporters 1, 3, and 6 reveals major differences in structural determinants of antiviral discrimination. J. Biol. Chem 283:8654–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vanwert AL, Bailey RM, Sweet DH. 2007. Organic anion transporter 3 (Oat3/Slc22a8) knockout mice exhibit altered clearance and distribution of penicillin G. Am. J. Physiol. Renal. Physiol 293:F1332–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vanwert AL, Srimaroeng C, Sweet DH. 2008. Organic anion transporter 3 (Oat3/Slc22a8) interacts with carboxyfluoroquinolones, and deletion increases systemic exposure to ciprofloxacin. Mol. Pharmacol 74:122–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mori S, Takanaga H, Ohtsuki S, Deguchi T, Kang YS, et al. 2003. Rat organic anion transporter 3 (rOAT3) is responsible for brain-to-blood efflux of homovanillic acid at the abluminal membrane of brain capillary endothelial cells. J. Cereb. Blood Flow Metab 23:432–40 [DOI] [PubMed] [Google Scholar]
- 84.Mori S, Ohtsuki S, Takanaga H, Kikkawa T, Kang YS, Terasaki T. 2004. Organic anion transporter 3 is involved in the brain-to-blood efflux transport of thiopurine nucleobase analogs. J. Neurochem 90: 931–41 [DOI] [PubMed] [Google Scholar]
- 85.Ose A, Ito M, Kusuhara H, Yamatsugu K, Kanai M, et al. 2009. Limited brain distribution of [3R,4R,5S]-4-acetamido-5-amino-3-(1-ethylpropoxy)-1-cyclohexene-1-carboxylate phosphate (Ro 64–0802), a pharmacologically active form of oseltamivir, by active efflux across the blood–brain barrier mediated by organic anion transporter 3 (Oat3/Slc22a8) and multidrug resistance–associated protein 4 (Mrp4/Abcc4). Drug Metab. Dispos 37:315–21 [DOI] [PubMed] [Google Scholar]
- 86.Zalups RK, Ahmad S. 2005. Handling of cysteine S-conjugates of methylmercury in MDCK cells expressing human OAT1. Kidney Int 68:1684–99 [DOI] [PubMed] [Google Scholar]
- 87.Stiborova M, Arlt VM, Schmeiser HH. 2016. Balkan endemic nephropathy: an update on its aetiology. Arch. Toxicol 90:2595–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bakhiya N, Arlt VM, Bahn A, Burckhardt G, Phillips DH, Glatt H. 2009. Molecular evidence for an involvement of organic anion transporters (OATs) in aristolochic acid nephropathy. Toxicology 264:74–79 [DOI] [PubMed] [Google Scholar]
- 89.Xue X, Gong LK, Maeda K, Luan Y, Qi XM, et al. 2011. Critical role of organic anion transporters 1 and 3 in kidney accumulation and toxicity of aristolochic acid I. Mol. Pharm 8:2183–92 [DOI] [PubMed] [Google Scholar]
- 90.Li S, Sanna S, Maschio A, Busonero F, Usala G, et al. 2007. The GLUT9 gene is associated with serum uric acid levels in Sardinia and Chianti cohorts. PLOS Genet 3:e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Woodward OM, Kottgen A, Coresh J, Boerwinkle E, Guggino WB, Kottgen M. 2009. Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. PNAS 106:10338–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Togawa N, Juge N, Miyaji T, Hiasa M, Omote H, Moriyama Y. 2015. Wide expression of type I Na+-phosphate cotransporter 3 (NPT3/SLC17A2), a membrane potential–driven organic anion transporter. Am. J. Physiol. Cell Physiol 309:C71–80 [DOI] [PubMed] [Google Scholar]
- 93.Reimer RJ. 2013. SLC17: a functionally diverse family of organic anion transporters. Mol. Asp. Med 34:350–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Farthing CA, Sweet DH.2014. Expression and function of organic cation and anion transporters (SLC22 family) in the CNS. Curr. Pharm. Des 20:1472–86 [DOI] [PubMed] [Google Scholar]
- 95.Chen L, Shu Y, Liang X, Chen EC, Yee SW, et al. 2014. OCT1 is a high-capacity thiamine transporter that regulates hepatic steatosis and is a target of metformin. PNAS 111:9983–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Aurich MK, Thiele I. 2016. Computational modeling of human metabolism and its application to systems biomedicine. Methods Mol. Biol 1386:253–81 [DOI] [PubMed] [Google Scholar]
- 97.Szallasi Z, Stelling J, Periwal V, eds. 2006. System Modeling in Cell Biology: From Concepts to Nuts and Bolts. Cambridge, MA: MIT Press [Google Scholar]
- 98.Hewitt WR, Wagner PA, Bostwick EF, Hook JB. 1977. Transport ontogeny and selective substrate stimulation as models for identification of multiple renal organic anion transport systems. J. Pharmacol. Exp. Ther 202:711–23 [PubMed] [Google Scholar]
- 99.Gallegos TF, Martovetsky G, Kouznetsova V, Bush KT, Nigam SK. 2012. Organic anion and cation SLC22 “drug” transporter (Oat1, Oat3, and Oct1) regulation during development and maturation of the kidney proximal tubule. PLOS ONE 7:e40796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Martovetsky G, Bush KT, Nigam SK. 2016. Kidney versus liver specification of SLC and ABC drug transporters, tight junction molecules, and biomarkers. Drug Metab. Dispos 44:1050–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martovetsky G, Tee JB, Nigam SK. 2013. Hepatocyte nuclear factors 4α and 1α regulate kidney developmental expression of drug-metabolizing enzymes and drug transporters. Mol. Pharmacol 84:808–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yuan X, Ta TC, Lin M, Evans JR, Dong Y, et al. 2009. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLOS ONE 4:e5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rosines E, Sampogna RV, Johkura K, Vaughn DA, Choi Y, et al. 2007. Staged in vitro reconstitution and implantation of engineered rat kidney tissue. PNAS 104:20938–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sweeney DE, Vallon V, Rieg T, Wu W, Gallegos TF, Nigam SK. 2011. Functional maturation of drug transporters in the developing, neonatal, and postnatal kidney. Mol. Pharmacol 80:147–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sweet DH, Eraly SA, Vaughn DA, Bush KT, Nigam SK. 2006. Organic anion and cation transporter expression and function during embryonic kidney development and in organ culture models. Kidney Int 69:837–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Habu Y, Yano I, Okuda M, Fukatsu A, Inui K. 2005. Restored expression and activity of organic ion transporters rOAT1, rOAT3 and rOCT2 after hyperuricemia in the rat kidney. Biochem. Pharmacol 69:993–99 [DOI] [PubMed] [Google Scholar]
- 107.Preising C, Schneider R, Bucher M, Gekle M, Sauvant C.2015. Regulation of expression of renal organic anion transporters OAT1 and OAT3 in a model of ischemia/reperfusion injury. Cell. Physiol. Biochem 37:1–13 [DOI] [PubMed] [Google Scholar]
- 108.Bulacio RP, Anzai N, Ouchi M, Torres AM. 2015. Organic anion transporter 5 (Oat5) urinary excretion is a specific biomarker of kidney injury: evaluation of urinary excretion of exosomal Oat5 after N-acetylcysteine prevention of cisplatin induced nephrotoxicity. Chem. Res. Toxicol 28:1595–602 [DOI] [PubMed] [Google Scholar]
- 109.Cocucci E, Kim JY, Bai Y, Pabla N. 2017. Role of passive diffusion, transporters, and membrane trafficking–mediated processes in cellular drug transport. Clin. Pharmacol. Ther 101:121–29 [DOI] [PubMed] [Google Scholar]
- 110.Xu D, Wang H, You G. 2016. Posttranslational regulation of organic anion transporters by ubiquitination: known and novel. Med. Res. Rev 36:964–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sprowl JA, Ong SS, Gibson AA, Hu S, Du G, et al. 2016. A phosphotyrosine switch regulates organic cation transporters. Nat. Commun 7:10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang Q, Hong M, Duan P, Pan Z, Ma J, You G. 2008. Organic anion transporter OAT1 undergoes constitutive and protein kinase C-regulated trafficking through a dynamin- and clathrin-dependent pathway. J. Biol. Chem 283:32570–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang Q, Suh W, Pan Z, You G. 2012. Short-term and long-term effects of protein kinase C on the trafficking and stability of human organic anion transporter 3. Int. J. Biochem. Mol. Biol 3:242–49 [PMC free article] [PubMed] [Google Scholar]
- 114.Xu D, Wang H, Zhang Q, You G. 2016. Nedd4-2 but not Nedd4-1 is critical for protein kinase C-regulated ubiquitination, expression, and transport activity of human organic anion transporter 1. Am. J. Physiol. Renal. Physiol 310:F821–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Anzai N, Miyazaki H, Noshiro R, Khamdang S, Chairoungdua A, et al. 2004. The multivalent PDZ domain–containing protein PDZK1 regulates transport activity of renal urate–anion exchanger URAT1 via its C terminus. J. Biol. Chem 279:45942–50 [DOI] [PubMed] [Google Scholar]
- 116.Merriman TR. 2015. An update on the genetic architecture of hyperuricemia and gout. Arthritis Res. Ther 17:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Xu G, Bhatnagar V, Wen G, Hamilton BA, Eraly SA, Nigam SK. 2005. Analyses of coding region polymorphisms in apical and basolateral human organic anion transporter (OAT) genes [OAT1 (NKT), OAT2, OAT3, OAT4, URAT (RST)]. Kidney Int 68:1491–99 [DOI] [PubMed] [Google Scholar]
- 118.Bhatnagar V, Xu G, Hamilton BA, Truong DM, Eraly SA, et al. 2006. Analyses of 5′ regulatory region polymorphisms in human SLC22A6 (OAT1) and SLC22A8 (OAT3). J. Hum. Genet 51:575–80 [DOI] [PubMed] [Google Scholar]
- 119.Engstrom K, Ameer S, Bernaudat L, Drasch G, Baeuml J, et al. 2012. Polymorphisms in genes encoding potential mercury transporters and urine mercury concentrations in populations exposed to mercury vapor from gold mining. Environ. Health Perspect 121:85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Han YF, Fan XH, Wang XJ, Sun K, Xue H,et al. 2011. Association of intergenic polymorphism of organic anion transporter 1 and 3 genes with hypertension and blood pressure response to hydrochlorothiazide. Am. J. Hypertens 24:340–46 [DOI] [PubMed] [Google Scholar]
- 121.Yee SW, Nguyen AN, Brown C, Savic RM, Zhang YC, et al. 2013. Reduced renal clearance of cefotaxime in Asians with a low-frequency polymorphism of OAT3 (SLC22A8). J. Pharm. Sci 102:3451–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tanaka M, Itoh K, Matsushita K, Matsushita K, Wakita N, et al. 2003. Two male siblings with hereditary renal hypouricemia and exercise-induced ARF. Am. J. Kidney Dis 42:1287–92 [DOI] [PubMed] [Google Scholar]
- 123.Mount DB. 2013. The kidney in hyperuricemia and gout. Curr. Opin. Nephrol. Hypertens 22:216–23 [DOI] [PubMed] [Google Scholar]
- 124.Sharma K, Karl B, Mathew AV, Gangoiti JA, Wassel CL, et al. 2013Metabolomics reveals signature of mitochondrial dysfunction in diabetic kidney disease. J. Am. Soc. Nephrol 24:1901–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Prentice KJ, Luu L, Allister EM, Liu Y, Jun LS, et al. 2014. The furan fatty acid metabolite CMPF is elevated in diabetes and induces β cell dysfunction. Cell Metab 19:653–66 [DOI] [PubMed] [Google Scholar]
- 126.Nezu J, Tamai I, Oku A, Ohashi R, Yabuuchi H, et al. 1999. Primary systemic carnitine deficiency is caused by mutations in a gene encoding sodium ion–dependent carnitine transporter. Nat. Genet 21:91–94 [DOI] [PubMed] [Google Scholar]
- 127.El-Hattab AW, Scaglia F. 2015. Disorders of carnitine biosynthesis and transport. Mol. Genet. Metab 116:107–12 [DOI] [PubMed] [Google Scholar]
- 128.Peltekova VD, Wintle RF, Rubin LA, Amos CI, Huang Q, et al. 2004. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat. Genet 11:11. [DOI] [PubMed] [Google Scholar]
- 129.Tamai I 2013. Pharmacological and pathophysiological roles of carnitine/organic cation transporters (OCTNs: SLC22A4, SLC22A5 and Slc22a21). Biopharm. Drug Dispos 34:29–44 [DOI] [PubMed] [Google Scholar]
- 130.Tokuhiro S, Yamada R, Chang X, Suzuki A, Kochi Y, et al. 2003An intronic SNP in a RUNX1 binding site of SLC22A4, encoding an organic cation transporter, is associated with rheumatoid arthritis. Nat. Genet 35:341–48 [DOI] [PubMed] [Google Scholar]
- 131.Malir F, Ostry V, Pfohl-Leszkowicz A, Malir J, Toman J. 2016. Ochratoxin A: 50 years of research. Toxins 8:191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Stefanovic V, Polenakovic M. 2009. Fifty years of research in Balkan endemic nephropathy: Where are we now? Nephron Clin. Pract 112:c51–56 [DOI] [PubMed] [Google Scholar]
- 133.Masereeuw R, Mutsaers HA, Toyohara T, Abe T, Jhawar S, et al. 2014. The kidney and uremic toxin removal: glomerulus or tubule? Semin. Nephrol 34:191–208 [DOI] [PubMed] [Google Scholar]
- 134.Tang WH, Hazen SL. 2017. Microbiome, trimethylamine N-oxide, and cardiometabolic disease. Transl. Res 179:108–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Leong SC, Sirich TL. 2016. Indoxyl sulfate—review of toxicity and therapeutic strategies. Toxins 8:358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yamaguchi J, Tanaka T, Inagi R. 2017. Effect of AST-120 in chronic kidney disease treatment: Still a controversy? Nephron 135:201–6 [DOI] [PubMed] [Google Scholar]
- 137.Kennedy PJ, Cryan JF, Dinan TG, Clarke G.2017. Kynurenine pathway metabolism and the microbiota–gut–brain axis. Neuropharmacology 112:399–412 [DOI] [PubMed] [Google Scholar]
- 138.Lowenstein J, Grantham JJ. 2016. The rebirth of interest in renal tubular function. Am. J. Physiol. Renal Physiol 310:F1351–55 [DOI] [PubMed] [Google Scholar]
- 139.Lowenstein J, Grantham JJ. 2017. Residual renal function: a paradigm shift. Kidney Int 91:561–65 [DOI] [PubMed] [Google Scholar]
- 140.Imamura Y, Murayama N, Okudaira N, Kurihara A, Okazaki O, et al. 2011. Prediction of fluoroquinolone-induced elevation in serum creatinine levels: a case of drug–endogenous substance interaction involving the inhibition of renal secretion. Clin. Pharmacol. Ther 89:81–88 [DOI] [PubMed] [Google Scholar]
- 141.Reznichenko A, Sinkeler SJ, Snieder H, van den Born J, de Borst MH, et al. 2013. SLC22A2 is associated with tubular creatinine secretion and bias of estimated GFR in renal transplantation. Physiol. Genom 45:201–9 [DOI] [PubMed] [Google Scholar]
- 142.Shen H, Liu T, Morse BL, Zhao Y, Zhang Y, et al. 2015. Characterization of organic anion transporter 2 (SLC22A7): a highly efficient transporter for creatinine and species-dependent renal tubular expression. Drug Metab. Dispos 43:984–93 [DOI] [PubMed] [Google Scholar]
- 143.Vallon V, Eraly SA, Rao SR, Gerasimova M, Rose M, et al. 2012. A role for the organic anion transporter OAT3 in renal creatinine secretion in mice. Am. J. Physiol. Renal Physiol 302:F1293–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shu Y, Brown C, Castro RA, Shi RJ, Lin ET, et al. 2008. Effect of genetic variation in the organic cation transporter 1, OCT1, on metformin pharmacokinetics. Clin. Pharmacol. Ther 83:273–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wu X, Kekuda R, Huang W, Fei YJ, Leibach FH, et al. 1998. Identity of the organic cation transporter OCT3 as the extraneuronal monoamine transporter (uptake2) and evidence for the expression of the transporter in the brain. J. Biol. Chem 273:32776–86 [DOI] [PubMed] [Google Scholar]
- 146.Eraly SA, Bush KT, Sampogna RV, Bhatnagar V, Nigam SK. 2004. The molecular pharmacology of organic anion transporters: from DNA to FDA? Mol. Pharmacol 65:479–87 [DOI] [PubMed] [Google Scholar]