Abstract
Scope:
This study aimed to investigate the role of the epigenetic regulator SET domain-containing lysine methyltransferase 7 (Setd7) in regulating the antioxidant Nrf2 pathway in prostate cancer (PCa) cells and examined the effects of two phytochemicals, phenethyl isothiocyanate (PEITC) and ursolic acid (UA).
Methods and results:
Lentivirus-mediated shRNA knockdown of Setd7 in LNCaP and PC-3 cells decreased the expression of downstream Nrf2 targets, such as NAD(P)H: quinone oxidoreductase 1 (Nqo1) and glutathione S-transferase theta 2 (Gstt2). Down-regulation of Setd7 decreased soft agar colony formation ability of PCa cells. Knockdown of Setd7 increased reactive oxygen species (ROS) generation. Furthermore, Setd7 knockdown attenuated Nqo1 and Gstt2 expression in response to H2O2 challenge, whereas increased DNA damage was observed in Setd7 knockdown cells in comet assay. Interestingly, Setd7 expression could be induced by the dietary phytochemicals PEITC and UA. Chromatin immunoprecipitation (ChIP) assays showed that Setd7 knockdown decreased H3K4me1 enrichment in the Nrf2 and Gstt2 promoter regions, while PEITC and UA treatments elevated the enrichment.
Conclusion:
Taken together, these results indicate that Setd7 knockdown decreases Nrf2 and Nrf2-target genes expression and that PEITC and UA induce Setd7 expression, which activates the Nrf2/antioxidant response element (ARE) signaling pathway and protects DNA from oxidative damage.
Keywords: Nrf2, Oxidative stress, PEITC, Prostate cancer cells, Setd7
Introduction
Lysine methylation is a major posttranslational modification. At the chromatin level, a variety of histone methyltransferases are involved in multiple epigenetic regulation-associated processes, including gene transcription, gene silencing, chromatin structure establishment and maintenance, and DNA repair and replication [1]. The SET domain-containing lysine methyltransferase 7 (Setd7, also called Set7/9), one of the first identified lysine methyltransferases, targets lysine 4 on histone H3 (H3K4) and is the mono-methyltransferase associated with transcriptional activation. Methylation of H3K4 by Setd7 inhibits nucleosome remodeling and histone deacetylase (NuRD) at the histone H3 tail and blocks methylation of H3K9 by Suppressor of variegation 3–9 homolog 1 (Suv39h1). Among the epigenetic regulatory proteins, Setd7 plays multiple roles in regulating histone assembly and non-histone protein activities. Setd7 is involved in regulating a wide range of targets, including transcription factors, tumor suppressors and membrane-associated receptors [2–5]. In addition, numerous non-histone Setd7 targets, including DNMT1, PCAF, p65 and p53, have been reported as potential mediators of the effects of Setd7 [6–9]. Therefore, Setd7 primarily functions by methylating various target proteins, altering the stability and activity of the target proteins. A previous study revealed that the role of Setd7 is modulatory, subtle, and highly dependent on the cell type and/or physiological conditions [1]. However, further elucidation of the epigenetic regulation of Setd7 is needed.
Recently, epigenetic alterations, such as DNA methylation, histone modification, nucleosome remodeling, and miRNA silencing, have been implicated in oxidative stress-induced cancer initiation and progression. Previous work from our laboratory provided compelling evidence that the expression of NF-E2-related factor 2 (Nrf2) is epigenetically regulated during prostate tumor development in TRAMP mice [10]. Similar to other age-related cancers, PCa is characterized by increased intracellular oxidative stress [10]. An increasing body of evidence has demonstrated that Nrf2 is a key transcription factor in antioxidant responses and xenobiotic metabolism via modulation of a broad range of antioxidant and phase II detoxification genes [11, 12], including heme oxygenase-1 (Ho-1), NAD(P)H:quinone oxidoreductase 1 (Nqo1), UDP-glucuronosyltransferase 1A1 (Ugt1a1) and glutathione S-transferase theta 2 (Gstt2). When cells are exposed to oxidative and electrophilic stresses, this cellular protective system will be triggered through activation of the Nrf2/antioxidant response element (ARE) signaling pathway. A recent study showed that the histone methyltransferase mixed-lineage leukemia (MLL), which targets H3K4 as the trimethyltransferase, is associated with an active chromatin state. The Nrf2 and Ho-1 expression was decreased when MLL was knocked down by siRNA in 5-fluorouracil-resistant colon cancer cells [13]. However, the roles of Nrf2 in cancer cell survival and resistance to chemotherapeutics and radiotherapy are still controversial [14].
Accumulating evidence suggests that dietary consumption of blueberries, cranberries, apple peels, and cruciferous vegetables, which contain abundant levels of the triterpenoid ursolic acid (UA) and the glucosinolate phenethyl isothiocyanate (PEITC), substantially reduces the risk of PCa [15–19]. Indeed, dietary intake is considered an important factor that influences the epigenome. In this context, the Nrf2 pathway can also be activated by multiple natural chemopreventive compounds, such as UA, sulforaphane (SFN) and PEITC [20–22]. In the last few decades, studies have shown that PEITC and UA exert various biological effects, including anti-metastatic, anti-angiogenic, antioxidant, anti-inflammatory, antimicrobial and other therapeutic functions [23, 24]. Previous reports indicated that PEITC may prevent PCa through epigenetic mechanisms; for example, PEITC inhibits histone deacetylases and aberrant CpG island methylation of various genes that contribute to PCa carcinogenesis and progression [25]. Recently, studies have also found that UA can activate Nrf2, block cellular transformation of mouse epidermal cells and protect the brain and liver from cerebral ischemia and CCl4-induced damage in mice via epigenetic modifications [20, 26, 27]. However, the effect of PEITC and UA on epigenetic regulation of Setd7 has not been previously examined.
In this study, we found that PEITC and UA induced Setd7 expression. We further investigated the potential role of Setd7 in regulating the Nrf2-mediated cellular antioxidative system in PCa cells. Down-regulation of Setd7 reduced the expression of downstream Nrf2 targets and was associated with colony formation efficiency. However, down-regulation of Setd7 also increased the DNA damage induced by H2O2 and reactive oxygen species (ROS) generation. Additionally, chromatin immunoprecipitation (ChIP) assays demonstrated that the enrichment of H3K4me1 in the promoter regions of Nrf2 and Gstt2 was decreased by Setd7 knockdown.
Materials and methods
Materials
Dulbecco’s modified Eagle’s medium/nutrient mixture F-12 (DMEM/F-12), RPMI 1640 medium, fetal bovine serum (FBS), penicillin-streptomycin (10,000 U/ml), puromycin, versene and trypsin-EDTA were supplied by Gibco (Grand Island, NY, USA). PEITC, UA, hydrogen peroxide (H2O2), and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich (St. Louis, MO, USA). The NE-PER Nuclear and Cytoplasmic Extraction Reagents and Chloromethyl derivative- 2′,7′-dichlorodihydrofluorescein diacetate (CM-H2DCFDA) (General Oxidative Stress Indicator) were supplied by Thermo Fisher Scientific (Waltham, MA, USA). Antibodies were obtained from Abcam (Cambridge, MA, USA; Nrf2, Nqo1, and H3K4me1 antibodies), Cell Signaling Technology (Beverly, MA, USA; Setd7 antibody), GeneTex, Inc. (Irvine, CA, USA; Gstt2 antibody) and Santa Cruz Biotechnology (Santa Cruz, CA, USA; actin (I-19) antibody and all secondary antibodies).
Cell culture and treatment
PCa LNCaP and PC3 cell lines were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) and cultured in either RPMI 1640 or DMEM/F-12, with 10% FBS at 37 °C in a humidified 5% CO2 atmosphere. Cells were grown to approximately 80% confluence and then treated with a medium containing PEITC, UA or H2O2 at the indicated concentrations for 24 h or other indicated times. DMSO (0.1%) was used as the vehicle control.
Setd7 knockdown in LNCaP and PC-3 cells
Lentivirus-mediated short hairpin RNAs (shRNAs) were used to establish stable mock (control, sh-Mock) and Setd7 knockdown (sh-SETD7) in LNCaP and PC-3 cells. The shRNA clone sets were obtained from GeneCopoeia (Rockville, MD, USA), and lentiviral-mediated transduction was performed according to the manufacturer’s manual. After selection in RPMI 1640 and DMEM supplemented with 10% FBS and 2 mg/mL puromycin for 3 weeks, the sh-Mock and sh-SETD7 cells were further used to evaluate the role of SETD7. For determination of the proliferation rate of sh-Mock and sh-SETD7 LNCaP and PC-3 cells, the cells were seeded in 60-mm tissue culture plates at an initial density of 10,000 cells. The cells were counted, and the number was recorded after 24, 48, and 72 h of incubation using a TC20 automated cell counter (Bio-Rad, Hercules, CA, USA).
RNA isolation and quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from the treated cells using a GeneJET RNA Purification Kit (Thermo Scientific, Waltham, MA, USA). According to the manufacturer’s instructions, first-strand complementary DNA was synthesized from 1 μg of RNA using TaqMan® Reverse Transcription Reagents (Applied Biosystems, Carlsbad, CA, USA). qPCR analysis was performed in an ABI7900HT system (Applied Biosystems) with SYBR Green PCR Master Mix (Applied Biosystems) using complementary DNA as the template. The sequences of the primers used for PCR amplifications are listed in Table 1. The PCR conditions were as follows: initial denaturation for 10 min at 95 °C, followed by 40 cycles of 15 s at 95 °C, and 1 min at 60 °C. A melting curve analysis was performed to ensure the specificity of the products. The relative gene expression levels were normalized to the GAPDH level using the 2-ΔΔCT formula (ΔΔCT = ΔCT sample - ΔCT control).
Table 1.
The primers used in qPCR.
| Primer | Sequence (5’→3’) |
|---|---|
| NRF2 For | CAAAAGGAGCAAGAGAAAGCC |
| NRF2 Rev | TCTGATTTGGGAATGTGGGC |
| SETD7 For | TCACGGAGAAAAGAACGGACG |
| SETD7 Rev | ATCATCCACATAATACCCCTCCA |
| NQO1 For | TCACCGAGAGCCTAGTTCC |
| NQO1 Rev | TCATGGCATAGTTGAAGGAACG |
| GSTT2 For | CCTAGAGCTGTTTCTTGACCTG |
| GSTT2 Rev | ACTCCTTGCTCTTGTGCTG |
| GAPDH For | ACATCGCTCAGACACCATG |
| GAPDH Rev | TGTAGTTGAGGTCAATGAAGGG |
| NRF2-CHIP For | AATAGGCAGGTTTGGAGGGC |
| NRF2-CHIP Rev | CCCCATTCTCAAGACCACCC |
| GSTT2-CHIP For | TCCCAGCTGTGCGTTCATAG |
| GSTT2-CHIP Rev | GGCCAAGACTCCACCACC |
Western blotting
The cells were harvested using radioimmunoprecipitation assay (RIPA) buffer (Millipore, Billerica, MA, USA) supplemented with a protein inhibitor cocktail (Sigma, St. Louis, MO). Then, the cell lysates were sonicated at 4 °C and centrifuged at 13,000 rpm for 15 min at 4 °C. The supernatants were collected and quantified using a Pierce™ bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Rockford, IL, USA). Then, 40 μg of total protein was diluted with Laemmli’s sodium dodecyl sulfate sample buffer (Boston Bioproducts, Ashland, MA, USA) and denatured at 95 °C for 10 min. The proteins were loaded and separated on 4–15% Criterion™ Tris-HCl Precast Gels (Bio-Rad, Hercules, CA, USA) and then transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad, Hercules, CA, USA), followed by blocking with 5% bovine serum albumin (BSA) in Tris-buffered saline-0.1% Tween 20 (TBST) buffer for 1 h at room temperature. The membrane was sequentially incubated with specific primary antibodies and HRP-conjugated secondary antibodies. The protein bands were visualized using SuperSignal™ West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific, Waltham, MA, USA) with a Gel Documentation 2000 system (Bio-Rad, Hercules, CA, USA).
Comet assay
For investigation of Setd7-mediated protection against oxidant-induced DNA damage, the alkaline version of a single cell microgel electrophoresis assay (comet assay) was performed to assess the impact of H2O2. LNCaP cells were seeded in 60-mm tissue culture plates. After incubation for 24 h, the cells were harvested, washed and then treated with H2O2 at 100 μM for 15 min at 37 °C. The comet assay was performed as described by Tice et al. and Glei et al. [28, 29]. Individual cells were embedded in a thin agarose gel on a microscope slide. All cellular proteins were then removed from the cells by lysing. The DNA was allowed to unwind under alkaline conditions. Following unwinding, the DNA underwent electrophoresis, allowing the broken DNA fragments or damaged DNA to migrate away from the nucleus. After the samples were stained with the DNA-specific fluorescent dye ethidium bromide (Sigma-Aldrich, Taufkirchen, Germany), the gel was assessed for fluorescence levels in the head and tail and the length of the tail. The extent of DNA liberated from the head of the comet is directly proportional to the amount of DNA damage.
Colony formation assay
LNCaP sh-Mock and sh-SETD7 and PC-3 sh-Mock and sh-SETD7 cells were seeded in 6-well plates at an initial density of 1000 cells/well for 24 h. The cells were then treated with vehicle (0.1% DMSO) or the genotoxic compound cumene hydroperoxide (CumOOH) (Sigma-Aldrich, Taufkirchen, Germany) at 2 μM at 37 °C in a humidified 5% CO2 atmosphere for 14 days. Then, the medium was removed, and the cells were rinsed carefully with PBS. The PBS was removed, and 500 μL of 0.01% crystal violet was added. After 30 min, the crystal violet was carefully removed, and the samples were rinsed with dH2O. The plates with colonies were then dried at room temperature. The colonies were photographed using a computerized microscope system with the Nikon ACT-1 program (Version 2.20). The number of cell colonies was calculated and analyzed as the ratio of the number of LNCaP sh-SETD7 cells to LNCaP sh-Mock cells.
Measurement of ROS
Intracellular ROS generation in LNCaP cells was measured with flow cytometry following staining with CM-H2DCFDA. After incubation with UA (5 or 20 μM) or PEITC (2.5 or 5 μM) for 12 h, the cells were washed and incubated with 10 μM CM-H2DCFDA at 37 °C for 30 min. The cells were then washed three times with PBS, and the fluorescence intensity was measured using flow cytometry.
ChIP assay
ChIP assays were performed using a MAGnify™ Chromatin Immunoprecipitation System (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions. Briefly, LNCaP sh-Mock and sh-SETD7 cells were treated with UA (20 μM) or PEITC (5 μM) for 24 h. After the samples were harvested and washed twice with PBS, chromatin was cross-linked with 1% formaldehyde for 10 min at room temperature. Next, 1.25 M glycine was added to quench the excess formaldehyde. The cells were pelleted by centrifugation, resuspended in lysis buffer and sonicated to generate 200 to 500 bp DNA fragments using a Bioruptor sonicator (Diagenode Inc., Sparta, NJ, USA). The diluted chromatin solution was immunoprecipitated with 2 μg of anti-H3K4me1 antibody (Abcam, Cambridge, MA, USA) or rabbit immunoglobulin G. After a wash, cross-link reversal, DNA elution and DNA purification, the relative amount of immunoprecipitated DNA was quantified via qPCR using the primers listed in Table 1. The enrichment of the precipitated DNA was calibrated using a standard curve constructed from serial dilution of the inputs, which were treated in parallel with the precipitated samples except antibody was not added in the immunoprecipitation procedure, and the data are presented as the fold changes in the signal-to-input ratio normalized to the control.
Statistical analysis
Statistical significance was tested with a one-way ANOVA followed by Dunnett’s post hoc test between multiple experimental groups and the Student’s t-test between two experimental groups using GraphPad PRISM software. The values are presented as the mean ± SD (standard deviation). p-values less than 0.05 were considered statistically significant.
Results
Setd7 played an important role in the process of PEITC- and UA-induced Nrf2-related detoxifying/antioxidant target gene expression
To determine the dynamic changes in Setd7 and Nrf2-related detoxifying/antioxidant target gene expression after PEITC and UA treatment, we treated PCa LNCaP and PC-3 cells with UA (20 μM) or PEITC (5 μM) for different periods of time (6, 12, 24, or 36 h) or with UA (5 or 20 μM) and PEITC (2.5 or 5 μM) for 24 h. Then, the cells were harvested, and the Setd7, Nrf2, Nqo1, and Gstt2 mRNA and protein levels were analyzed using qPCR and western blotting, respectively. The results showed that UA treatment for 12 h affected Setd7, treatment for 6 h affected Nrf2, and treatment for 24 h affected both Nqo1 and Gstt2 mRNA levels. PEITC affected Setd7, Nqo1, and Gstt2 mRNA expression at the 12 h time point and Nrf2 expression at the 6 h time point (Fig. 1A). In addition, Nqo1 mRNA expression was induced by UA (20 μM) and PEITC (5 μM), while Gstt2 mRNA expression was induced by UA (20 μM) after 24 h of treatment (Fig. 1B). We also found that the Setd7 protein level was gradually increased after PEITC and UA treatment in LNCaP cells. Only Setd7 and Nqo1 protein expression was significantly increased by PEITC (5 μM) treatment at 24 h (Fig. 1C-F).
FIGURE 1.
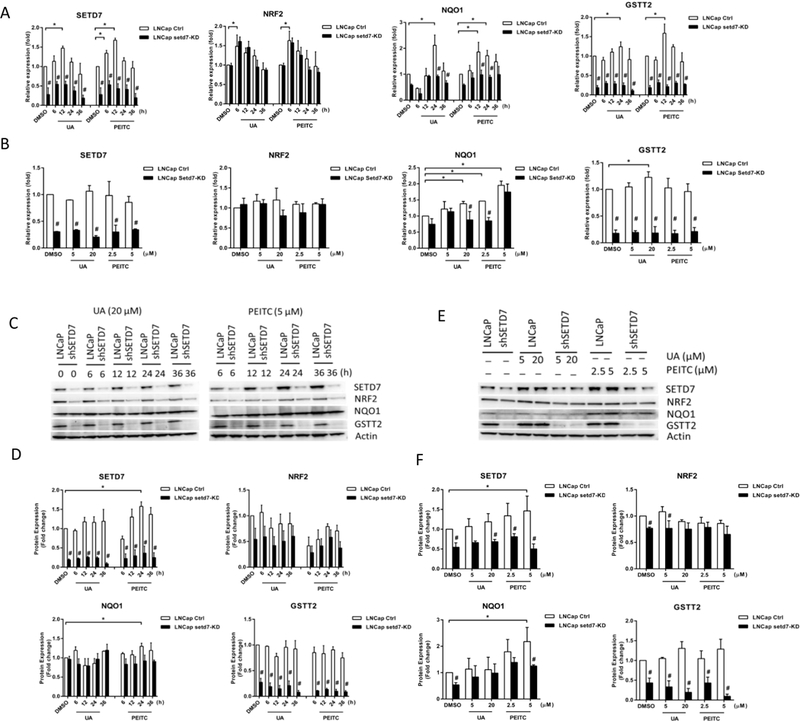
Dynamic changes in Setd7 and Nrf2-related gene expression following PEITC and UA treatment of LNCaP and LNCaP Setd7-KD cells. LNCaP cells were treated with 0.1% DMSO (Control), UA (20 μM) or PEITC (5 μM) for 6, 12, 24, or 36 h; or LNCaP cells were treated with 0.1% DMSO (Control), UA (5 or 20 μM) and PEITC (2.5 or 5 μM) for 24 h; and then, the cells were harvested and analyzed to determine the Setd7, Nrf2, Nqo1 and Gstt2 mRNA and protein levels using qPCR and western blotting. The protein amounts were normalized to the β-actin levels and are expressed as the relative fold change compared with LNCaP DMSO control cells. (A and B) The relative Setd7, Nrf2, Nqo1 and Gstt2 mRNA levels; (C and E) representative immunoblot of Setd7, Nrf2, Nqo1 and Gstt2 proteins; and (D and F) the relative Setd7, Nrf2, Nqo1 and Gstt2 protein levels. The mean values and standard deviations represent triplicate samples. The data shown are representative of three experiments. * The difference was statistically significant when p < 0.05 compared with the levels in LNCaP DMSO control cells. # p < 0.05 vs. the LNCaP cell line.
Setd7 plays a prominent role in histone and non-histone protein lysine methylation. To determine whether Setd7 is involved in the effects of PEITC and UA on the Nrf2-ARE pathway, we established sh-Mock and sh-SETD7 cells using lentiviral shRNA vectors. Decreased Setd7 mRNA and protein levels were confirmed in sh-SETD7 cells by qPCR and western blot analyses, respectively. Importantly, Setd7, Nqo1, and Gstt2 mRNA and protein expression in LNCaP cells was decreased at different time points and dosages when Setd7 was stably inhibited (Fig. 1).
The profiles in PC-3 cells were similar to those in LNCaP cells. Treatment with UA for 24 h affected Setd7 and Nqo1 and treatment for 12 h affected Nrf2 and Gstt2 mRNA levels. PEITC affected Setd7, Nrf2, and Nqo1 mRNA expression at the 12 h time point and Gstt2 expression at the 24 h time point (Fig. 2A). In addition, Setd7, Nqo1, and Gstt2 mRNA expression was induced by UA (20 μM), while Nqo1 and Gstt2 mRNA expression was induced by UA (20 μM) and PEITC (5 μM) after 24 h of treatment (Fig. 2B). We also found that the Setd7 protein level gradually increased after PEITC and UA treatment in PC-3 cells. Setd7 protein expression was induced by UA (20 μM) and PEITC (5 μM) treatment at 24 h and 36 h, while Nqo1 and Gstt2 expression was induced at 24 h and 6 h, respectively (Fig. 2C-F). Additionally, Setd7 knockdown significantly decreased the Setd7, Nqo1 and Gstt2 mRNA and Setd7, Nrf2, Nqo1 and Gstt2 protein levels in PC-3 cells (Fig. 2). Taken together, these results suggest that both the time course and the dose are important for Setd7 induction by PEITC and UA and that Setd7 plays an important role in the Nrf2-ARE signaling pathway in LNCaP and PC-3 cells.
FIGURE 2.
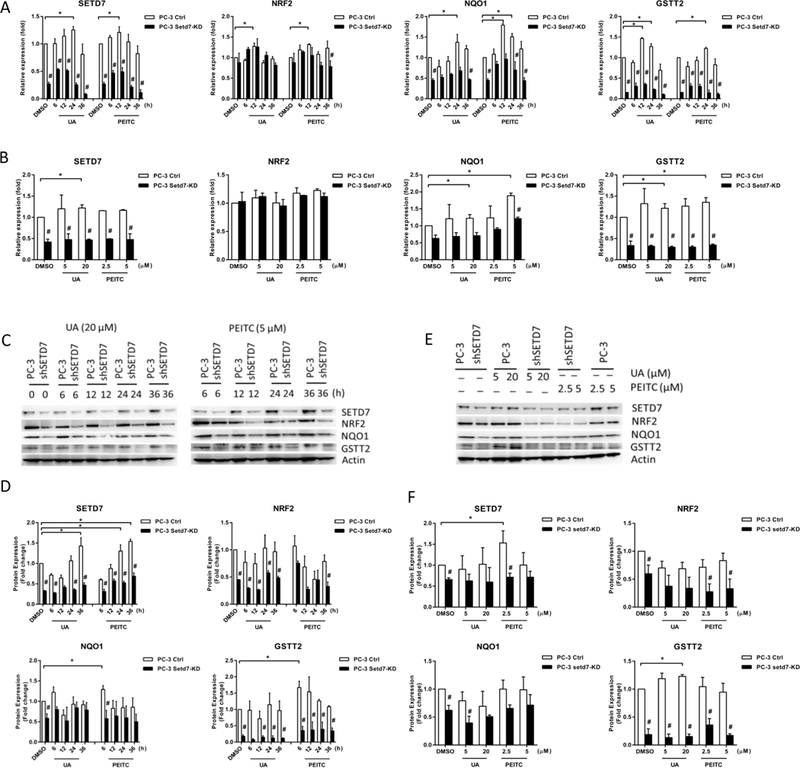
Dynamic changes in Setd7 and Nrf2-related gene expression following PEITC and UA treatment of PC-3 and PC-3 Setd7-KD cells. PC-3 cells were treated with 0.1% DMSO (Control), UA (20 μM) or PEITC (5 μM) for 6, 12, 24, or 36; or PC-3 cells were treated with 0.1% DMSO (Control), UA (5 or 20 μM) and PEITC (2.5 or 5 μM) for 24 h; and then, the cells were harvested and analyzed to determine the Setd7, Nrf2, Nqo1 and Gstt2 mRNA and protein levels using qPCR and western blotting. The protein amounts were normalized to the β-actin levels and are expressed as the relative fold change compared with PC-3 DMSO control cells. (A and B) The relative Setd7, Nrf2, Nqo1 and Gstt2 mRNA levels; (C and E) representative immunoblot of Setd7, Nrf2, Nqo1 and Gstt2 proteins; (D and F) the relative Setd7, Nrf2, Nqo1 and Gstt2 protein levels. The mean values and standard deviations represent triplicate samples. The data shown are representative of three experiments. * The difference was statistically significant when p < 0.05 compared with the results in PC-3 DMSO control cells. # p < 0.05 vs. the PC-3 cell line.
Knockdown of Setd7 reduced colony formation in LNCaP and PC-3 cells
As described above, Setd7 influences the expression of Nrf2 and its downstream genes, including the phase II enzyme Gstt2, which exhibits peroxidase activity [30]. CumOOH, a model substrate of Gstt2, has pronounced genotoxic activity. To induce oxidative DNA damage, we used CumOOH in colony formation assays. The results showed that Setd7 knockdown significantly decreased the number and size of colonies formed by LNCaP and PC-3 cells. In the control groups, Setd7 knockdown significantly decreased colony formation by approximately 80% compared with that of the sh-Mock cells. Meanwhile, the CumOOH-mediated inhibition of colony formation was substantially reduced by 80% in LNCaP and 70% in PC-3 cells. In contrast, in LNCaP sh-Mock and LNCaP sh-SETD7 cells, CumOOH significantly suppressed colony formation by 70% compared with that of the control treatment cells, and the suppression was 40% in PC-3 sh-SETD7 cells (Fig. 3). Based on these results, we concluded that Setd7 protected cells from DNA damage induced by genotoxic CumOOH.
FIGURE 3.
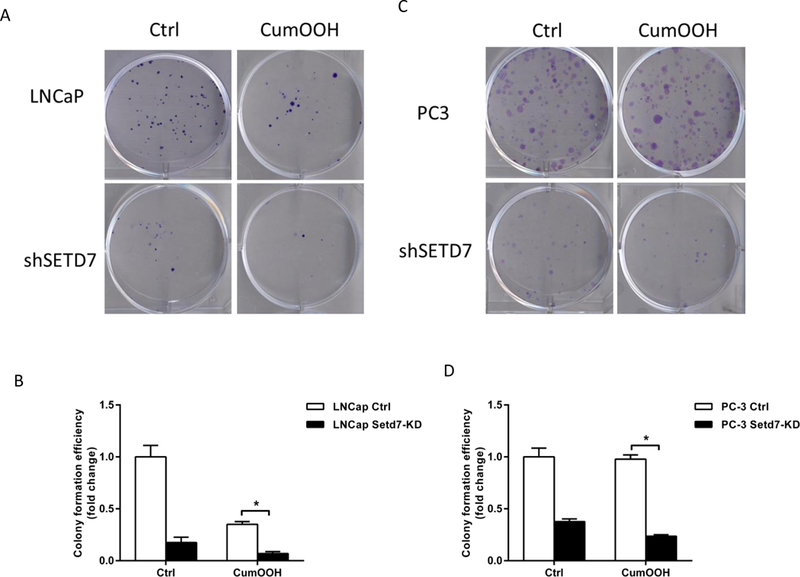
Setd7 knockdown reduced colony formation efficiency in PCa cells. After treatment with 2 μM CumOOH in 6-well plates for 14 days, the cells were fixed and stained with crystal violet, and the cell colonies were counted. (A and B) Representative images of the colony formation assays in LNCaP cells; (C and D) representative images of the colony formation assays in PC-3 cells. The bars represent the mean values of triplicate samples (mean ± SD). * p < 0.05.
Setd7 knockdown increased ROS generation and DNA damage
Next, we designed experiments to test ROS generation in LNCaP and LNCaP Setd7-KD cells following PEITC and UA treatment. Intracellular ROS generation in DMSO-, UA- and PEITC-treated LNCaP and LNCaP Setd7-KD cells was assessed with flow cytometry following staining with CM-H2DCFDA. The results showed that in the DMSO group and the UA and PEITC treatment groups, knockdown of Setd7 significantly increased ROS production (Fig. 4). This further confirmed that Setd7 offered some protection to DNA integrity in LNCaP cells.
FIGURE 4.
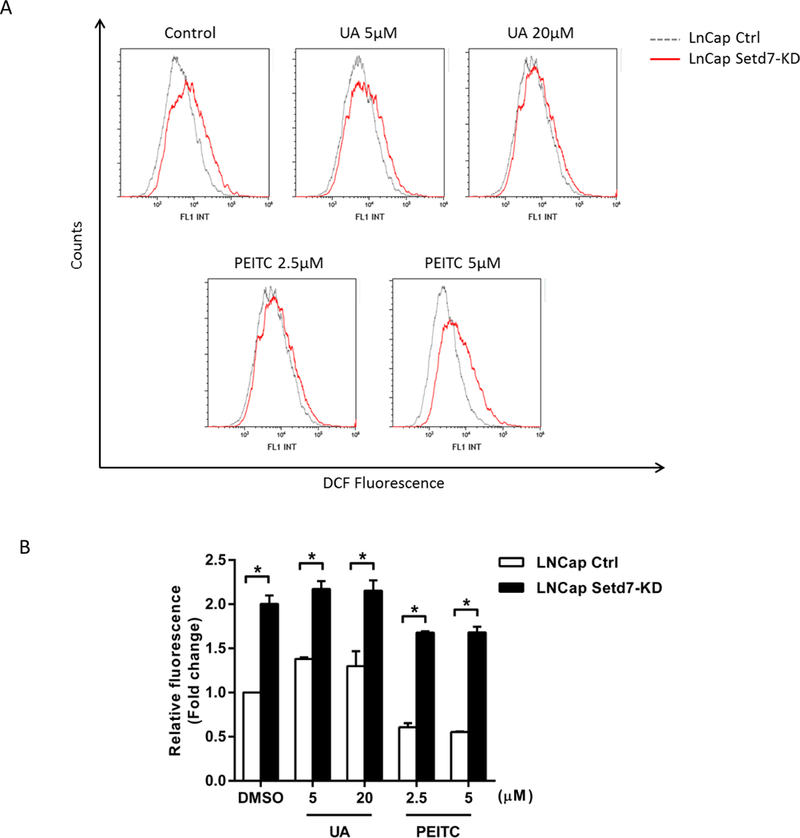
Setd7 down-regulation increased ROS generation. (A) LNCaP and LNCaP Setd7-KD cells were treated with the indicated concentrations of PEITC and UA for 12 h, incubated with 10 μM CM-H2DCFDA for a total of 30 min, and then analyzed via flow cytometry. (B) Fold changes in DCF fluorescence in LNCaP and LNCaP Setd7-KD cells. The data presented in panel A are representative of three independent experiments, and the statistical results from these experiments are presented in panel B (* p < 0.05, significantly different compared with the LNCaP cell line).
To further characterize the roles of Setd7 in the Nrf2-ARE signaling pathway, we stimulated cells with the oxidant H2O2. The Setd7, Nrf2, Nqo1 and Gstt2 mRNA expression levels were investigated using qPCR. In LNCaP sh-Mock cells, higher H2O2 concentration upregulated Setd7, Nrf2, Nqo1 and Gstt2 mRNA levels. Meanwhile, Setd7 knockdown repressed basal and H2O2-induced Setd7, Nqo1 and Gstt2 expression (Fig. 5A). Nrf2 is an essential regulator of cytoprotective detoxifying/antioxidant enzymes, and accordingly, the results showed that Setd7 played an important role in oxidative stress conditions at least in part by augmenting detoxifying/antioxidant enzymes, which was mediated by enhanced Nrf2 expression.
FIGURE 5.
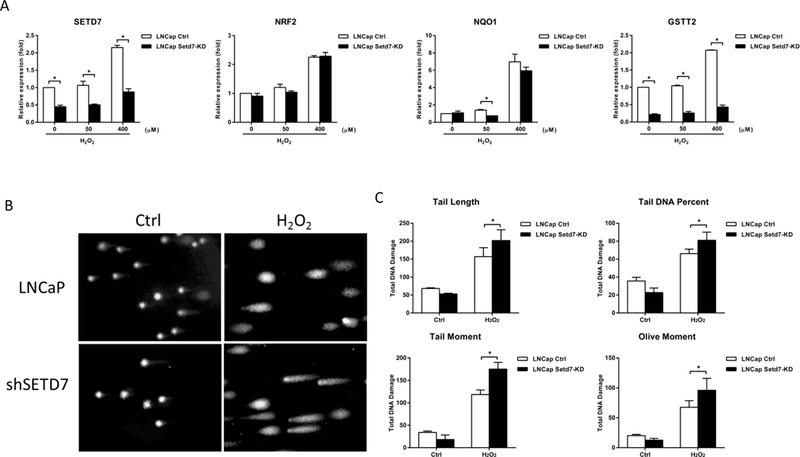
Setd7 knockdown increased the DNA damage following oxidative stress induced by H2O2. (A) LNCaP cells were treated with H2O2 at 50 and 400 μM, and then, the cells were harvested and analyzed to determine the Setd7, Nrf2, Nqo1 and Gstt2 mRNA levels using qPCR; (B) the levels of DNA damage in LNCaP cells induced by 100 μM H2O2 were measured with an alkaline comet assay. Representative images of the comet assays to assess DNA integrity; (C) parameters tested for DNA damage. The mean values and standard deviations represent triplicate samples. The data shown are representative of three experiments. * The difference was statistically significant when p < 0.05 compared with the control cell results.
Next, a comet assay was performed to further confirm the DNA-protective role of Setd7. H2O2 can induce oxidative stress and cause DNA damage in cells. The LNCaP sh-Mock and sh-SETD7 cells were exposed to H2O2, and DNA integrity was measured using the comet assay. In the present study, cells treated with 100 μM H2O2 exhibited a significant increase in DNA damage compared to cells in the control group. Setd7 knockdown significantly augmented the DNA fragmentation induced by H2O2. The results of the tail length, tail DNA percentage, tail moment and olive moment analyses showed that Setd7 knockdown significantly increased DNA damage by approximately 30%, 20%, 40%, and 30%, respectively, compared with that in the sh-Mock cells (Fig. 5B). These findings indicate that Setd7 is a key enzyme in many cellular processes, including DNA protection and epigenetic modification.
Setd7 knockdown decreased H3K4me1 recruitment to the Nrf2 and Gstt2 promoter regions
To confirm that Setd7 regulates Nrf2 and Gstt2 expression through mono-methylation of H3K4, we used ChIP assays to further examine the Nrf2 and Gstt2 transcriptional regulatory mechanism and histone modification in LNCaP cells. When Setd7 expression was knocked down, the H3K4me1 enrichment on the Nrf2 and Gstt2 promoter regions was significantly reduced. In contrast, PEITC and UA treatment increased the H3K4me1 enrichment at the Nrf2 and Gstt2 promoters in LNCaP cells (Fig. 6A and B). The specificity of the ChIP assay was verified using non-specific rabbit IgG, which is not specifically amplified in qPCR, in the precipitation procedure as a negative control (data not shown). To confirm the results, we extracted and analyzed nuclear Nrf2 protein using western blots. Setd7 knockdown inhibited Nrf2 protein expression in LNCaP cells, which was consistent with our previous results (Fig. 6C and D). These data suggest that Nrf2 and Gstt2 may be indirectly regulated by Setd7 via H3K4 methylation. Setd7 knockdown reduced the H3K4me1 enrichment on the Nrf2 and Gstt2 promoter regions, subsequently inhibiting Nrf2 and Gstt2 expression. Thus, Gstt2 expression is not only influenced by the Nrf2 level but also by Setd7 induction.
FIGURE 6.
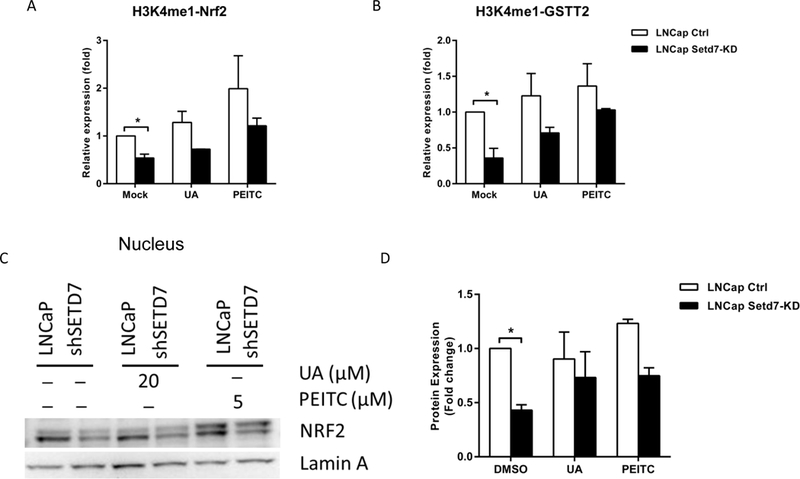
Effect of Setd7 on recruitment of the transcriptional machinery to Nrf2 and Gstt2. LNCaP and LNCaP Setd7-KD cells were treated with UA (20 μM) and PEITC (5 μM) for 24 h, and then, the cells were harvested and analyzed with ChIP assays and western blots. (A and B) ChIP assays were performed to examine H3K4me1 enrichment on the promoter region of Nrf2 and Gstt2 in LNCaP cells. The immunoprecipitated DNA was used as a template for qPCR, and the enrichment was quantified as the proportion of its self-input. Relative fold change was then calculated by normalizing the ratio to that of the mock control; (C) western blots were performed to analyze the nuclear Nrf2 expression in LNCaP cells with or without UA and PEITC treatment; (D) Relative expression level of Nrf2 was measured by comparing with the expression of Lamin A control in each individual experiment. * p < 0.05 indicates significant differences between the LNCaP cells and LNCaP Setd7-KD cells.
Discussion
Our present study showed that Setd7 expression in LNCaP and PC-3 cells can be induced by the phytochemicals PEITC and UA, which regulate the Nrf2/ARE signaling pathway and protect DNA from oxidative damage. Furthermore, we elucidated the detailed molecular mechanism by which Setd7 modulates key proteins in these pathways.
Setd7 specifically methylates H3K4 and enhances transcriptional activation. Methylation of H3K4 by Setd7 inhibits the association between the histone deacetylase NuRD and the H3 tail. In addition, H3K4 methylation precludes Suv39h1-mediated methylation at K9 of H3 (H3K9). H3K4 and H3K9 methylation dynamically compete with each other and have differential effects on subsequent histone acetylation by p300 [2, 3].In contrast, Setd7-mediated methylation of non-histone proteins, especially transcription factors, is associated with gene activation or repression through modulation of protein stability in a site- and context-specific manner. This process has been shown to be crucial for multiple cellular responses, including the response to stress stimuli [8, 9, 31, 32]. Recent in vivo and in vitro studies have demonstrated that Setd7-mediated changes in lysine methylation on histones have no effect on the p53-mediated cell cycle [32–35]. Using lentivirus shRNA-mediated Setd7 knockdown in PCa LNCaP and PC-3 cells, our study showed that Setd7 regulates the expression of downstream genes targeted by the transcription factor Nrf2. These results suggest that the histone methyltransferase activity of Setd7 may be important for these effects. We found that Setd7 knockdown significantly down-regulated Nqo1 and Gstt2 mRNA and protein expression.
Setd7 influences chromatin conformation and transcription activity in various ways. For example, Setd7 methylates the tumor suppressor protein p53 and estrogen receptor α, activating them [9, 36]. In contrast, RelA/p65, E2F1 and DNMT1 methylation renders them unstable and dysfunctional [6, 31, 37, 38]. In addition, Setd7 enhances androgen receptor (AR)-mediated transactivation but does not affect AR protein levels [39, 40]. Setd7 activates farnesoid X receptor (FXR) transcriptional activity without altering protein stability [7]. Meanwhile, Setd7-mediated protein methylation can also influence protein-protein and protein-RNA interactions [41]. Given the evidence indicating a critical role of Setd7 methylation in Nrf2 activity, we hypothesized that Setd7 stabilizes the Nrf2 protein level, while the transcriptional activity is not affected. Based on our results, in PCa cells, Setd7 knockdown significantly decreased the Nrf2 protein level rather than the mRNA level. The ChIP assay results indicated that Nrf2 and Gstt2 expression was suppressed through regulation of H3K4me1 by Setd7. Gstt2 expression may not only be influenced by Nrf2 but also by H3K4 methylation. The expression of the transcription factor Nrf2 may be modulated by multiple factors at different levels. However, we cannot eliminate the possibility that Setd7 directly methylates the lysine residues of Nrf2. He et al. hypothesized that Setd7 methylation may alter the interaction between Nrf2 and Keap1, leading to a feedback mechanism by binding to the ARE region in the Nrf2 promoter [42]. There is some evidence showing that Nrf2 is directly acetylated by CREB-binding protein, leading to an increase in promoter-specific DNA binding of Nrf2. In contrast, heterologous sirtuin 1 (SIRT1) can decrease Nrf2 acetylation associated with Nrf2-dependent gene transcription. The acetylation and deacetylation of Nrf2 indicate that there are novel regulatory mechanisms modulating the Nrf2-dependent antioxidant response [43, 44]. Nevertheless, the above hypotheses require more studies for confirmation, especially whether Nrf2 can be directly methylated by Setd7 and whether other proteins participate in this process.
In this study, the mRNA and protein expression of the Nrf2 downstream target genes Nqo1 and Gstt2 were both inhibited when Setd7 was down-regulated. Petermann et al. reported that Gstt2 protects cells from oxidative DNA damage induced by the synthetic hydroperoxide CumOOH, indicating that Gstt2 has anti-genotoxic function [45]. Therefore, we addressed the question of whether Setd7 protects against genotoxic stress by stabilizing Gstt2 expression. To analyze this, we challenged LNCaP and PC-3 cells with the Gstt2 substrate CumOOH. The colony formation assay results clearly demonstrated that Setd7 knockdown significantly reduced the colony number and size compared to those of the control cells, indicating that Setd7 can protect PCa cells against genotoxic damage at least in part through expression of the phase II gene Gstt2. Furthermore, to confirm the function of Setd7, we used H2O2, which causes oxidative DNA damage [46]. The comet assay analysis of LNCaP cells showed that the DNA damage induced by H2O2 was increased when Setd7 expression was inhibited. Similarly, the qPCR results showed that Setd7 knockdown significantly repressed the Gstt2 mRNA expression under H2O2 oxidative stress. Notably, this observation is consistent with the results from the UA- and PEITC-treated LNCaP and PC-3 cells. In addition, the flow cytometry results showed that Setd7 knockdown markedly increased intracellular ROS generation. Our results suggest that elevated Setd7 levels may protect PCa cells from oxidative stress caused by CumOOH, H2O2 and endogenously produced peroxides.
Interestingly, our data showed that the phytochemicals PEITC and UA could induce Setd7 expression in LNCaP and PC-3 cells. A growing body of evidence suggests that PEITC and UA have both in vitro and in vivo therapeutic functions, including antioxidant, anti-inflammatory and anticancer activities, and can prevent PCa through epigenetic regulation. Recently, many studies have attempted to elucidate the PEITC- and UA-mediated epigenetic modifications. PEITC and UA have been reported to reduce the expression of epigenetic modification enzymes, such as the histone deacetylases, in mouse epidermal JB6 P+ and LNCaP cells [20, 25]. Similarly, UA can activate Nrf2 and its downstream cytoprotective detoxifying/antioxidant enzymes (Gst and Nqo1) in a cigarette smoke extract (CSE)-induced bronchial epithelial cell culture model [27, 47]. The epigenetic effects of UA can potentially contribute to the prevention of skin cancer [20]. PEITC can selectively increase H3K4 methylation [25], and can suppress prostate cancer cell invasiveness through epigenetic mechanisms including microRNAs [48]. We found that Setd7 induction could account for the activated Nrf2/ARE signaling pathway and the elevated H3K4 methylation caused by PEITC and UA. Accordingly, inhibition of Setd7 expression led to significant repression of Nrf2 and its downstream target genes Nqo1 and Gstt2. In addition, Setd7 knockdown abrogated the effects of PEITC and UA when combined with these two compounds. Although in the last few decades, great progress has been made in identifying and characterizing the effects of PEITC and UA on PCa chemoprevention, there is still much to be learned. For example, drug resistance and off-target actions of these compounds have limited their use for cancer treatment. Several reports have shown that resistance to PEITC- and UA-induced apoptosis can be mediated through multiple signaling pathways in PCa cells [49–52]. These results demonstrated that PEITC or UA may have the potential to be considered in future combination PCa therapy, but drug resistance continues to be a problem. In our study, we showed for the first time that Setd7 was induced by PEITC and UA, and more importantly, Setd7 regulated the Nrf2/ARE signaling pathway and protected DNA against oxidative stress induced by CumOOH and H2O2 in LNCaP and PC-3 cells. These results strongly suggest that Setd7 is a key regulator of Nrf2 function and plays an important role in antioxidant response, xenobiotic disposition, DNA protection and cell survival. Correspondingly, the effects of Setd7 may confer drug resistance to PCa cells. However, the molecular mechanism of this process has not been completely elucidated. Future studies examining the cross-talk between Setd7 and other signaling pathways are also needed.
Recently, many preclinical studies have suggested that histone methyltransferase inhibitors are prospective drug candidates for cancer therapy. Setd7 is a drug target for diseases such as type II diabetes, AIDS and hormone-dependent breast cancer [53]. Accordingly, (R)-PFI-2 has been shown to be a potent and selective inhibitor of the methyltransferase activity of Setd7 in cells [1]. Therefore, based on our new findings, Setd7 may be a promising therapeutic target for PCa. Furthermore, the epigenetic mechanism of Setd7 underlying PCa chemoprevention needs to be clearly defined, and further studies are required to address the above topics, especially the effects of Setd7 inhibitors.
Acknowledgments
This work is supported in part by institutional funds and by AT009152 from the National Center for Complementary and Integrative Health (NCCIH), R01AT007065 from NCCIH and the Office of Dietary Supplements (ODS) and CA200129 from the National Cancer Institute (NCI). The authors thank all the members in Dr. Kong’s lab for their helpful discussion of this work.
Abbreviations:
- ARE
antioxidant response element
- CumOOH
cumene hydroperoxide
- DMSO
dimethyl sulfoxide
- GSTT2
glutathione S-transferase theta 2
- H2O2
hydrogen peroxide
- HO1
heme oxygenase-1
- NQO1
NAD (P) H: quinone oxidoreductase 1
- NRF2
NF-E2-related factor 2
- PCa
Prostate cancer
- PEITC
phenethyl isothiocyanate
- qPCR
quantitative polymerase chain reaction
- SETD7
SET domain containing lysine methyltransferase 7
- SFN
sulforaphane
- UA
ursolic acid
- UGT1A1
UDP-glucuronosyltransferase 1A1
Footnotes
Conflict of interest statement
The authors declare no conflict of interest.
References
- [1].Barsyte-Lovejoy D, Li F, Oudhoff MJ, Tatlock JH, Dong A, Zeng H, Wu H, Freeman SA, Schapira M, Senisterra GA, Kuznetsova E, Marcellus R, Allali-Hassani A, Kennedy S, Lambert JP, Couzens AL, Aman A, Gingras AC, Al-Awar R, Fish PV, Gerstenberger BS, Roberts L, Benn CL, Grimley RL, Braam MJ, Rossi FM, Sudol M, Brown PJ, Bunnage ME, Owen DR, Zaph C, Vedadi M, Arrowsmith CH, (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proceedings of the National Academy of Sciences of the United States of America 2014, 111, 12853–12858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nishioka K, Chuikov S, Sarma K, Erdjument-Bromage H, Allis CD, Tempst P, Reinberg D, Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes & development 2002, 16, 479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y, Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Molecular cell 2001, 8, 1207–1217. [DOI] [PubMed] [Google Scholar]
- [4].Otwinowski Z, Minor W, Processing of X-ray diffraction data collected in oscillation mode. Method Enzymol 1997, 276, 307–326. [DOI] [PubMed] [Google Scholar]
- [5].Vagin A, Teplyakov A, MOLREP: an automated program for molecular replacement. J Appl Crystallogr 1997, 30, 1022–1025. [Google Scholar]
- [6].Esteve PO, Chin HG, Benner J, Feehery GR, Samaranayake M, Horwitz GA, Jacobsen SE, Pradhan S, Regulation of DNMT1 stability through SET7-mediated lysine methylation in mammalian cells. Proceedings of the National Academy of Sciences of the United States of America 2009, 106, 5076–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Balasubramaniyan N, Ananthanarayanan M, Suchy FJ, Direct methylation of FXR by Set7/9, a lysine methyltransferase, regulates the expression of FXR target genes. American journal of physiology Gastrointestinal and liver physiology 2012, 302, G937–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ea CK, Baltimore D, Regulation of NF-kappaB activity through lysine monomethylation of p65. Proceedings of the National Academy of Sciences of the United States of America 2009, 106, 18972–18977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, McKinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, Reinberg D, Regulation of p53 activity through lysine methylation. Nature 2004, 432, 353–360. [DOI] [PubMed] [Google Scholar]
- [10].Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, Foster BA, Kan YW, Kong AN, Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PloS one 2010, 5, e8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y, An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochemical and biophysical research communications 1997, 236, 313–322. [DOI] [PubMed] [Google Scholar]
- [12].McMahon M, Itoh K, Yamamoto M, Chanas SA, Henderson CJ, McLellan LI, Wolf CR, Cavin C, Hayes JD, The Cap’n’Collar basic leucine zipper transcription factor Nrf2 (NF-E2 p45-related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer research 2001, 61, 3299–3307. [PubMed] [Google Scholar]
- [13].Kang KA, Piao MJ, Ryu YS, Kang HK, Chang WY, Keum YS, Hyun JW, Interaction of DNA demethylase and histone methyltransferase upregulates Nrf2 in 5-fluorouracil-resistant colon cancer cells. Oncotarget 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lau A, Villeneuve NF, Sun Z, Wong PK, Zhang DD, Dual roles of Nrf2 in cancer. Pharmacological research 2008, 58, 262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ikeda Y, Murakami A, Ohigashi H, Ursolic acid: an anti- and pro-inflammatory triterpenoid. Molecular nutrition & food research 2008, 52, 26–42. [DOI] [PubMed] [Google Scholar]
- [16].Kolonel LN, Hankin JH, Whittemore AS, Wu AH, Gallagher RP, Wilkens LR, John EM, Howe GR, Dreon DM, West DW, Paffenbarger RS Jr., Vegetables, fruits, legumes and prostate cancer: a multiethnic case-control study. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 2000, 9, 795–804. [PubMed] [Google Scholar]
- [17].Abdull Razis AF, Noor NM, Cruciferous vegetables: dietary phytochemicals for cancer prevention. Asian Pacific journal of cancer prevention : APJCP 2013, 14, 1565–1570. [DOI] [PubMed] [Google Scholar]
- [18].Shanmugam MK, Dai X, Kumar AP, Tan BK, Sethi G, Bishayee A, Ursolic acid in cancer prevention and treatment: molecular targets, pharmacokinetics and clinical studies. Biochemical pharmacology 2013, 85, 1579–1587. [DOI] [PubMed] [Google Scholar]
- [19].Stoewsand GS, Bioactive organosulfur phytochemicals in Brassica oleracea vegetables--a review. Food and chemical toxicology : an international journal published for the British Industrial Biological Research Association 1995, 33, 537–543. [DOI] [PubMed] [Google Scholar]
- [20].Kim H, Ramirez CN, Su ZY, Kong AN, Epigenetic modifications of triterpenoid ursolic acid in activating Nrf2 and blocking cellular transformation of mouse epidermal cells. The Journal of nutritional biochemistry 2016, 33, 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Krajka-Kuzniak V, Szaefer H, Bartoszek A, Baer-Dubowska W, Modulation of rat hepatic and kidney phase II enzymes by cabbage juices: comparison with the effects of indole-3-carbinol and phenethyl isothiocyanate. The British journal of nutrition 2011, 105, 816–826. [DOI] [PubMed] [Google Scholar]
- [22].Saw CL, Cintron M, Wu TY, Guo Y, Huang Y, Jeong WS, Kong AN, Pharmacodynamics of dietary phytochemical indoles I3C and DIM: Induction of Nrf2-mediated phase II drug metabolizing and antioxidant genes and synergism with isothiocyanates. Biopharmaceutics & drug disposition 2011, 32, 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kashyap D, Tuli HS, Sharma AK, Ursolic acid (UA): A metabolite with promising therapeutic potential. Life sciences 2016, 146, 201–213. [DOI] [PubMed] [Google Scholar]
- [24].Gupta P, Wright SE, Kim SH, Srivastava SK, Phenethyl isothiocyanate: a comprehensive review of anti-cancer mechanisms. Biochimica et biophysica acta 2014, 1846, 405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang LG, Chiao JW, Prostate cancer chemopreventive activity of phenethyl isothiocyanate through epigenetic regulation (review). International journal of oncology 2010, 37, 533–539. [DOI] [PubMed] [Google Scholar]
- [26].Li L, Zhang X, Cui L, Wang L, Liu H, Ji H, Du Y, Ursolic acid promotes the neuroprotection by activating Nrf2 pathway after cerebral ischemia in mice. Brain research 2013, 1497, 32–39. [DOI] [PubMed] [Google Scholar]
- [27].Ma JQ, Ding J, Zhang L, Liu CM, Protective effects of ursolic acid in an experimental model of liver fibrosis through Nrf2/ARE pathway. Clinics and research in hepatology and gastroenterology 2015, 39, 188–197. [DOI] [PubMed] [Google Scholar]
- [28].Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayashi H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF, Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environmental and molecular mutagenesis 2000, 35, 206–221. [DOI] [PubMed] [Google Scholar]
- [29].Glei M, Hofmann T, Kuster K, Hollmann J, Lindhauer MG, Pool-Zobel BL, Both wheat (Triticum aestivum) bran arabinoxylans and gut flora-mediated fermentation products protect human colon cells from genotoxic activities of 4-hydroxynonenal and hydrogen peroxide. Journal of agricultural and food chemistry 2006, 54, 2088–2095. [DOI] [PubMed] [Google Scholar]
- [30].Hurst R, Bao Y, Jemth P, Mannervik B, Williamson G, Phospholipid hydroperoxide glutathione peroxidase activity of human glutathione transferases. The Biochemical journal 1998, 332 ( Pt 1), 97–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Yang XD, Huang B, Li M, Lamb A, Kelleher NL, Chen LF, Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. The EMBO journal 2009, 28, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ivanov GS, Ivanova T, Kurash J, Ivanov A, Chuikov S, Gizatullin F, Herrera-Medina EM, Rauscher F 3rd, Reinberg D, Barlev NA, Methylation-acetylation interplay activates p53 in response to DNA damage. Molecular and cellular biology 2007, 27, 6756–6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lehnertz B, Rogalski JC, Schulze FM, Yi L, Lin S, Kast J, Rossi FM, p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Molecular cell 2011, 43, 673–680. [DOI] [PubMed] [Google Scholar]
- [34].Campaner S, Spreafico F, Burgold T, Doni M, Rosato U, Amati B, Testa G, The methyltransferase Set7/9 (Setd7) is dispensable for the p53-mediated DNA damage response in vivo. Molecular cell 2011, 43, 681–688. [DOI] [PubMed] [Google Scholar]
- [35].Kurash JK, Lei H, Shen Q, Marston WL, Granda BW, Fan H, Wall D, Li E, Gaudet F, Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Molecular cell 2008, 29, 392–400. [DOI] [PubMed] [Google Scholar]
- [36].Subramanian K, Jia D, Kapoor-Vazirani P, Powell DR, Collins RE, Sharma D, Peng J, Cheng X, Vertino PM, Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Molecular cell 2008, 30, 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Link PA, Gangisetty O, James SR, Woloszynska-Read A, Tachibana M, Shinkai Y, Karpf AR, Distinct roles for histone methyltransferases G9a and GLP in cancer germ-line antigen gene regulation in human cancer cells and murine embryonic stem cells. Molecular cancer research : MCR 2009, 7, 851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kontaki H, Talianidis I, Lysine methylation regulates E2F1-induced cell death. Molecular cell 2010, 39, 152–160. [DOI] [PubMed] [Google Scholar]
- [39].Ko S, Ahn J, Song CS, Kim S, Knapczyk-Stwora K, Chatterjee B, Lysine methylation and functional modulation of androgen receptor by Set9 methyltransferase. Molecular endocrinology 2011, 25, 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gaughan L, Stockley J, Wang N, McCracken SR, Treumann A, Armstrong K, Shaheen F, Watt K, McEwan IJ, Wang C, Pestell RG, Robson CN, Regulation of the androgen receptor by SET9-mediated methylation. Nucleic acids research 2011, 39, 1266–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhang X, Huang Y, Shi X, Emerging roles of lysine methylation on non-histone proteins. Cellular and molecular life sciences : CMLS 2015, 72, 4257–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].He S, Owen DR, Jelinsky SA, Lin LL, Lysine Methyltransferase SETD7 (SET7/9) Regulates ROS Signaling through mitochondria and NFE2L2/ARE pathway. Scientific reports 2015, 5, 14368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kawai Y, Garduno L, Theodore M, Yang J, Arinze IJ, Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. The Journal of biological chemistry 2011, 286, 7629–7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Sun Z, Chin YE, Zhang DD, Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response. Molecular and cellular biology 2009, 29, 2658–2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Petermann A, Miene C, Schulz-Raffelt G, Palige K, Holzer J, Glei M, Bohmer FD, GSTT2, a phase II gene induced by apple polyphenols, protects colon epithelial cells against genotoxic damage. Molecular nutrition & food research 2009, 53, 1245–1253. [DOI] [PubMed] [Google Scholar]
- [46].Sohal RS, Weindruch R, Oxidative stress, caloric restriction, and aging. Science 1996, 273, 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liu W, Tan X, Shu L, Sun H, Song J, Jin P, Yu S, Sun M, Jia X, Ursolic acid inhibits cigarette smoke extract-induced human bronchial epithelial cell injury and prevents development of lung cancer. Molecules 2012, 17, 9104–9115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zhang C, Shu L, Kim H, Khor TO, Wu R, Li W, Kong AN, Phenethyl isothiocyanate (PEITC) suppresses prostate cancer cell invasion epigenetically through regulating microRNA-194. Molecular nutrition & food research 2016, 60, 1427–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Limami Y, Pinon A, Leger DY, Pinault E, Delage C, Beneytout JL, Simon A, Liagre B, The P2Y2/Src/p38/COX-2 pathway is involved in the resistance to ursolic acid-induced apoptosis in colorectal and prostate cancer cells. Biochimie 2012, 94, 1754–1763. [DOI] [PubMed] [Google Scholar]
- [50].Shin SW, Kim SY, Park JW, Autophagy inhibition enhances ursolic acid-induced apoptosis in PC3 cells. Biochimica et biophysica acta 2012, 1823, 451–457. [DOI] [PubMed] [Google Scholar]
- [51].Xiao D, Johnson CS, Trump DL, Singh SV, Proteasome-mediated degradation of cell division cycle 25C and cyclin-dependent kinase 1 in phenethyl isothiocyanate-induced G2-M-phase cell cycle arrest in PC-3 human prostate cancer cells. Molecular cancer therapeutics 2004, 3, 567–575. [PubMed] [Google Scholar]
- [52].Mukherjee S, Bhattacharya RK, Roy M, Targeting protein kinase C (PKC) and telomerase by phenethyl isothiocyanate (PEITC) sensitizes PC-3 cells towards chemotherapeutic drug-induced apoptosis. Journal of environmental pathology, toxicology and oncology : official organ of the International Society for Environmental Toxicology and Cancer 2009, 28, 269–282. [DOI] [PubMed] [Google Scholar]
- [53].Wagner T, Jung M, New lysine methyltransferase drug targets in cancer. Nature biotechnology 2012, 30, 622–623. [DOI] [PubMed] [Google Scholar]


