Abstract
Environmental signals mediated via the aryl hydrocarbon receptor (AHR) shape the developing immune system and influence immune function. Developmental exposure to AHR binding chemicals causes persistent changes in CD4+ and CD8+ T cell responses later in life, including dampened clonal expansion and differentiation during influenza A virus (IAV) infection. Naïve T cells require activation by dendritic cells (DCs), and AHR ligands modulate the function of DCs from adult organisms. Yet, the consequences of developmental AHR activation by exogenous ligands on DCs later in life has not been examined. We report here that early life activation of AHR durably reduces the ability of DC to activate naïve IAV-specific CD8+ T cells; however, activation of naïve CD4+ T cells was not impaired. Also, DCs from developmentally exposed offspring migrated more poorly than DCs from control dams in both in vivo and ex vivo assessments of DC migration. Conditional knockout mice, which lack Ahr in CD11c lineage cells, suggest that dampened DC emigration is intrinsic to DCs. Yet, levels of chemokine receptor 7 (CCR7), a key regulator of DC trafficking, were generally unaffected. Gene expression analyses reveal changes in Lrp1, Itgam, and Fcgr1 expression, and point to alterations in genes that regulate DC migration and antigen processing and presentation as being among pathways disrupted by inappropriate AHR signaling during development. These studies establish that AHR activation during development causes long-lasting changes to DCs, and provide new information regarding how early life environmental cues shape immune function later in life.
Introduction
The immune system develops during gestation and following birth. The environment experienced in utero and during early postnatal life can influence the immune system, leading to durable changes that influence health later in life. For instance, reports from human population studies show that prenatal and early postnatal exposure to certain pollutants correlates with immune dysregulation later in life [1–4]. Research in animal models further supports the idea that immune function at maturity is influenced by early life exposures [5–9], and that the developing immune system is more sensitive than the mature immune system to enduring modulation by environmental factors [10–12]. While these studies reveal links between developmental exposures and life-long changes in immune function, how early life exposures shape the immune system is not fully understood.
Recent reports show that activation of the aryl hydrocarbon receptor (AHR) during development changes immune responses later in life, suggesting that this environment-sensing transcriptional regulator is one means via which the early life environment influences immune function. The AHR binds many synthetic and naturally derived chemicals that are commonly found in environment, including pollutants, dietary substances, and byproducts of microorganisms [13, 14]. The AHR is widely expressed in most tissues and cells, including immune cells, and is emerging as a key regulator of immune cell development, differentiation, and function [15, 16]. One group of exogenous AHR ligands to which humans are regularly exposed are dioxins and polychlorinated biphenyls (PCBs). These anthropogenic chemicals bioaccumulate in the food chain, and exposure is primarily via the diet [17, 18]. Fetuses and neonates are exposed as well, as dioxins and PCBs cross the placenta, and are found in breast milk, cord, and infant blood [19–22]. Moreover, several different human cohort studies show strong associations between early life exposure to these AHR-binding pollutants and altered immune function, including more severe or frequent respiratory infections and decreased antibody responses to vaccinations [23–28]. In mice, early life exposure to a prototype environmental AHR ligand, 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD), causes durable alterations in T cell responses later in life. For example, developmentally exposed adult offspring have significantly fewer virus-specific CD8+ cytotoxic T lymphocytes (CTL) and conventional CD4+ T helper cells after infection with influenza A virus [7, 8, 29, 30].
In order for naïve T cells to become activated, proliferate, and differentiate into appropriately armed effector cells, they must receive an integrated set of signals from dendritic cells (DCs). The AHR is emerging as an important regulator of DC function, although this evidence is from studies of direct exposure (i.e., not developmental exposure) to AHR ligands [31–39]. However, whether triggering of the AHR during development changes the function of DCs later in life has not been investigated. In the present study, we determined whether maternal exposure to a representative environmental AHR ligand modifies DC function in adult offspring, including their ability to stimulate naïve T cells, their frequency and distribution in vivo, and their migration. We used targeted transcriptome analyses to investigate intrinsic differences in gene expression in DCs from developmentally exposed mice. Our results show that AHR activation during development induces long-lasting changes in DC functions. Given the pivotal role that DCs play in establishing and maintaining T cell responses, the ability of early life AHR activation to affect DC functional properties later in life has many implications as we seek to understand AHR-mediated regulation of the immune system, and to understand how the early life environmental cues shape the way the immune system is poised to respond.
Results
AHR activation during development reduces the ability of DCs to stimulate naïve CD8+ T cells, but not naïve CD4+ T cells
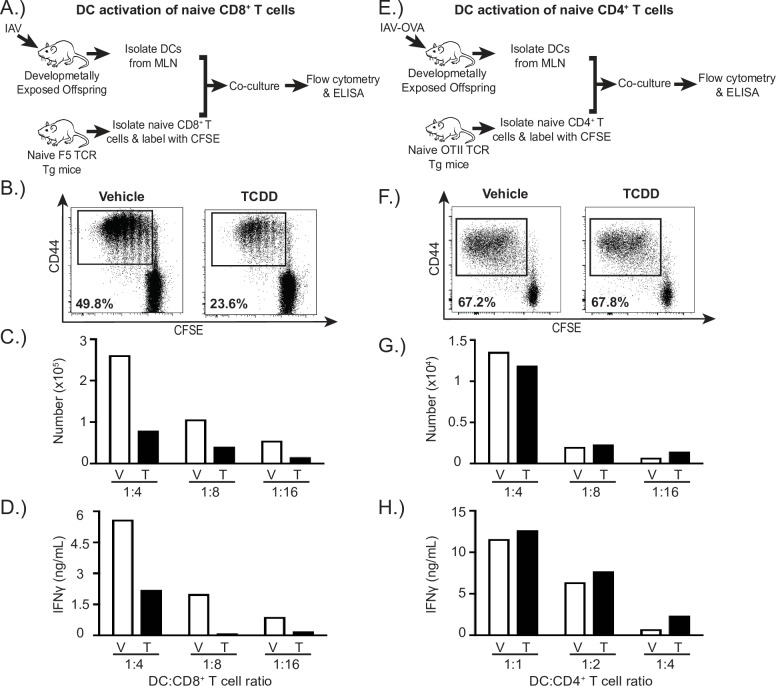
During a primary immune challenge, naïve T cells require activation signals from DCs. Given that developmental exposure to environmental AHR ligands, such as TCDD, durably disrupts CD8+ and CD4+ T cell responses [7, 8, 29, 40], we sought to determine whether early life AHR activation affects the ability of DCs to activate naïve T cells using an established assay of APC function [35, 38]. Specifically, after respiratory infection we isolated DCs from the lung-draining mediastinal lymph nodes (MLN), the principle site of naïve T cell activation during IAV infection. We used an equivalent number of DCs from adult infected offspring of vehicle or TCDD exposed dams to activate naïve, CFSE-labeled TCR transgenic T cells ex vivo. To measure activation of CD8+ T cells, DCs were isolated from the MLN of developmentally exposed, infected adult offspring and co-cultured with naïve CD8+ T cells from naïve, untreated F5 TCR transgenic mice (Fig 1A). The TCR on CD8+ T cells of F5 mice recognizes a specific immunodominant peptide from IAV nucleoprotein (NP366-374) [41]. Compared to DCs isolated from infected offspring of vehicle-treated dams, DCs from adult offspring of TCDD-treated dams had a two-fold reduction in the percentage of activated and proliferated (CD44hiCFSEdecay) F5 CD8+ T cells (Fig 1B). The number of CD44hiCFSEdecayCD8+ T cells was also reduced when F5 CD8+ T cells were cultured with DCs from offspring of TCDD treated dams (Fig 1C). To examine CD8+ T cell differentiation, we measured IFNγ levels. CD8+ T cells cultured with DCs from offspring of TCDD-treated dams secreted roughly half as much IFNγ as CD8+ T cells stimulated by DCs from vehicle controls (Fig 1D). These data indicate that AHR activation during development reduced the ability of DCs to act as optimally effective APCs for naïve CD8+ T cells, as they stimulated lower levels of CD8+ T cell proliferation and differentiation. That is, activation of the AHR early in life, via direct treatment of the dam and vertical exposure to the fetus and neonate, leads to perturbation in the ability of DCs to stimulate naïve CD8+ T cells in adult offspring.
Fig 1. DCs from developmentally exposed mice have an impaired ability to stimulate naïve virus-specific CD8+ T cells but not naïve CD4+ T cells.
(A) Adult offspring of dams treated with vehicle (V) or TCDD (T) were infected (i.n.) with IAV (Memphis/102/72). On day 3 post infection, MLNs were pooled from animals of the same group (≥ 30 mice/group), and DCs enriched with immunomagnetic separation. Naïve (CD44lo) CD8+ T cells were isolated from spleens of untreated and uninfected F5 TCR transgenic mice, and labeled ex vivo with CFSE. Serially diluted DCs were and co-cultured with CFSE-labeled naïve F5 CD8+ T cells (2x105 T cells/well) in a range from 1:4–1:16 DCs:T cells. After 3 days in culture, cells were collected and stained for flow cytometric analysis. (B) The dot plots indicate the percentage of proliferating (CD44hiCFSEdecay) F5 CD8+ T cells after culture with DCs from vehicle or TCDD exposed offspring (1:4 DC:T cell ratio). F5 CD8+ cells were identified as Vβ11+CD8+ cells. (C) The bar graph shows the number of proliferating F5 CD8+ T cells. (D) The graph depicts IFNγ levels in supernatants at diminishing DC:T cell ratios. (E) Adult offspring were infected (i.n.) with HKx31/OVAII (≥ 30 mice/group). On day 3 post infection, MLNs were pooled from animals of the same group (≥ 30 mice/group), and DCs were enriched from the pool of MLN cells. Naïve (CD44lo) CD4+ T cells were isolated from spleens of untreated and uninfected OTII TCR transgenic mice and labeled with CFSE. DCs were co-cultured with CFSE labeled naïve OTII CD4+ T cells (2x105 T cells/well) for 4 days in a range from 1:4–1:16 DCs:T cells. (F) The dot plots indicate the percentage of proliferating (CD44hiCFSEdecay) OTII CD4+ T cells after culture with DCs from vehicle or TCDD exposed offspring (1:4 DC:T cell ratio). OTII CD4+ cells were identified as Vβ5+CD4+ cells. (G) Bar graphs show the number of proliferating OTII CD4+ T cells stimulated by DCs. (H) Bar graph shows IFNγ levels in supernatant at indicated DC:T cell ratios. Data are from one representative experiment, except IFNγ levels in DC:OTII CD4+ T cell co-cultures, which show the combined data from two experiments. Each experiment was independently repeated at least 2 times with similar results. Underlying data can be found in S1 Data.
CD4+ T cells are also important for host defenses against IAV and other respiratory pathogens [42]. Therefore, we investigated whether developmental exposure affected the ability of DCs to stimulate naïve CD4+ T cells. We used a similar approach to the ex vivo system with naïve CD8+ T cells, with two changes. We isolated CD4+ T cells from naïve, untreated OTII TCR transgenic mice, whose CD4+ T cells bear TCRs that recognize a peptide fragment of ovalbumin (OVA323-339), and isolated DCs from developmentally exposed mice infected with a transgenic IAV that expresses this peptide: x31/OVAII [43, 44]. After infection, DCs from MLN of developmentally exposed adult offspring were co-cultured with naïve CFSE-labeled naïve CD4+ T cells from OTII TCR transgenic mice (Fig 1E). Regardless of whether the offspring were from vehicle or TCDD treated dams, DCs stimulated a similar percentage and number of OTII CD4+ T cells to become activated and proliferate (Fig 1F and 1G). Furthermore, these CD4+ T cells produced similar levels of IFNγ (Fig 1H). Thus, in contrast to an impaired ability to stimulate CD8+ T cells, early life AHR activation does not appear to affect the ability of DCs to serve as APCs for naïve CD4+ T cells in the context of IAV infection.
In vivo distribution of DCs
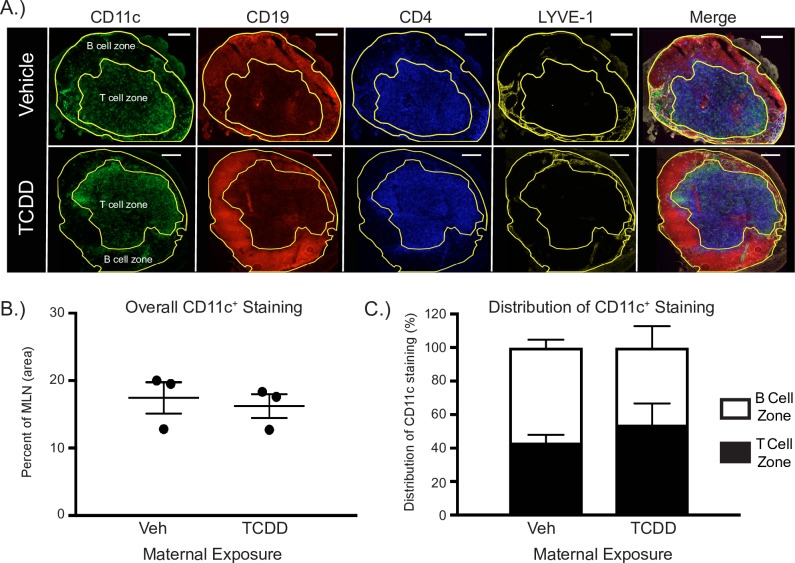
In addition to affecting their ability to activate naive T cells, early life exposure could alter the distribution of DC within lymph nodes, which could contribute to poorer T cell responses during in vivo immune challenge. Therefore, we evaluated whether developmental exposure affects the in vivo distribution of DCs using immunohistochemistry and enumerated distinct DC subsets using analytical flow cytometry. One of the main sites where DCs act as APCs for naïve T cells is the T cell zone in lymph nodes. We utilized immunofluorescent microscopy to visualize the distribution of DCs in the MLN 3 days following IAV infection (Fig 2A). The percentage of the MLN cross-sectional area that was positive for CD11c+ staining was not different between adult offspring from vehicle or TCDD treated dams (Fig 2B). Moreover, the relative distribution of CD11c+ staining in T and B cell zones was equivalent between treatment groups (Fig 2C). Specifically, 43% and 54% of CD11c staining was in T cell zones of MLNs from vehicle and TCDD groups, respectively. These results suggest that, at this macro-level, developmental activation of AHR does not change the overall distribution of DCs between the T cell and B cell zones of the MLN during early stages of the response to IAV infection; a time of peak DC interaction with naïve T cells.
Fig 2. Developmental activation of the AHR does not alter the overall distribution of DCs in T or B cell zones in the MLN.
(A) Representative fluorescent immunohistochemistry images from mature male offspring of dams exposed to vehicle (V; top row) or TCDD (T; bottom row) 3 days after infection with IAV (HKx31). Serial frozen sections were stained with anti-CD11c (DCs, green), anti-CD19 (B cells, red), anti-CD4 (CD4+ T cells, blue), and anti-LYVE-1 (lymphatic vessels, yellow) are shown, along with the merged image. The B cell zone and T cell zones were defined using CD19 or CD4, respectively. The outer boundary of the tissue section was defined using LYVE-1. Scale bar, 200 μm. (B) The graph shows the average percentage (± SEM) of CD11c staining throughout the MLN cross-sections analyzed. (C) The stacked bar graph shows the average percentage (± SEM) of positive CD11c staining in the B cell zone (white bars) and T cell zone (black bars) in MLN of vehicle (V) or TCDD (T) offspring. 3–8 sections analyzed/per mouse, from 3 different mice per group. Underlying data can be found in S1 Data.
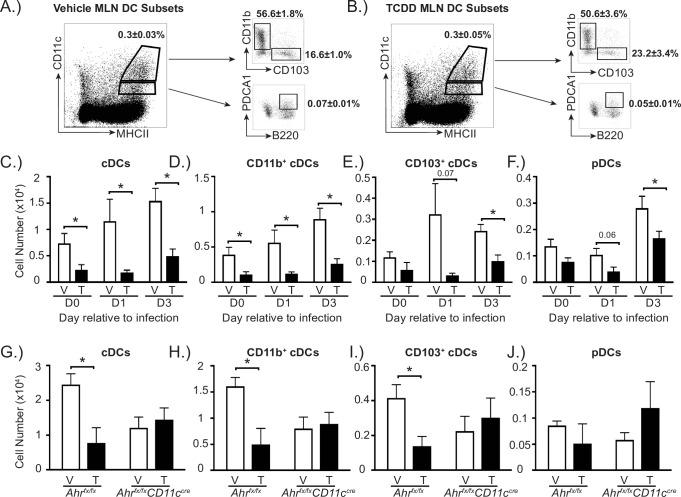
Activation of the AHR during development could also alter the number of distinct DC subsets within the MLN of the offspring. DCs are a heterogeneous population of cells, which are broadly categorized into conventional DCs (cDCs) and plasmacytoid DCs (pDCs) [45–47]. cDCs can be further divided into different subtypes. In the lung and lung-draining lymph nodes, two of the main types of cDCs are CD11b+ and CD103+ DCs. Both of these cDC subsets emigrate from the lung during respiratory antigen challenge, and present antigen to naïve T cells in the lung-draining lymph nodes [35, 38, 48–51]. We used multiple parameter analytical flow cytometry to identify and enumerate pDCs, total cDCs, CD11b+ cDCs and CD103+ cDCs, in adult offspring before and during IAV infection. Representative FACS plots depict the gating strategy to define DCs, and show the proportion of cDCs, CD11b+ cDCs, CD103+ cDCs, and pDCs in MLNs of mature offspring of vehicle (Fig 3A) and TCDD-treated dams (Fig 3B). Prior to IAV infection, there were significantly fewer cDCs, CD11b+ cDCs in particular, in the MLN of offspring of the TCDD-treated dams (Fig 3C and 3D). One day after IAV infection, we observed a reduction in number of total cDCs, CD11b+ cDC and CD103+ cDC subsets, as well as pDCs (Fig 3C–3F). Three days after infection, the number of cDCs and pDCs remained significantly decreased in adult offspring that were developmentally exposed compared to those from control dams (Fig 3C–3F). We also observed a reduced percentage of CD103+ cDCs and increase in the proportion of CD11b+ cDCs one day after IAV infection, but these differences were not observed in uninfected offspring or 3 days after infection (S1 Table). In contrast to the MLN, in the lung there were no statistically significant differences in the percentage or number of cDCs or pDCs between treatment groups prior to or after infection (S1 Fig and S1 Table).
Fig 3. Activation of AHR during development reduces DCs in the MLN in an AHR-dependent manner.
Mice were exposed to vehicle or TCDD during development. At maturity, developmentally exposed offspring were unchallenged, or infected with IAV (HKx31). The percentage and number of DC subsets in the MLN were determined by flow cytometry. (A, B) Following gating to exclude doublets and auto-fluorescent cells, DCs were identified as follows: conventional DCs (cDCs; CD11chiMHCIIhi cells) and plasmacytoid DCs (pDCs; CD11cloMHCIIhi PDCA1+CD45R+ cells). Conventional DCs were further subdivided into two populations: CD11b+ cDCs (CD11b+CD103-CD11chiMHCIIhi cells) and CD103+ cDCs (CD103+CD11b-CD11chiMHCIIhi cells). Representative dot plots depict the gating used to define cDCs, CD11b+ cDCs, CD103+ cDCs and pDCs in the MLN of vehicle (A) and TCDD (B) exposed offspring. The dot plots indicate the average percentage (±SEM) of the indicated DC subset 3 days after infection. (C-F) The bar graphs show the number (±SEM) of the indicated DC population in MLN from naïve (day 0) or infected mice. (G-J) Bar graphs show the number (±SEM) of DC in the MLN 3 days after IAV infection in Ahrfx/fx or Ahrfx/fxCD11ccre mice from V or T dams treated with 10 μg/kg BW TCDD on GD14 and PND2. At each point in time, all wildtype offspring within a group were from a separate dam, n = 6–9 mice per group per day. Some Ahrfx/fx or Ahrfx/fxCD11ccre mice offspring within a group were from the same dam, n = 3–5 mice per group, because of limited number of offspring. Day 0 data are representative of 4 independent experiments, day 1 data are representative of 3 independent experiments, day 3 data are representative of 6 independent experiments with similar results. The experiment using Ahrfx/fxCD11ccre mice was performed once. An * indicates p ≤ 0.05. Underlying data can be found in S1 Data.
To determine whether the decreased number of DCs in the MLN was dependent on expression of AHR in CD11c+ cells, we used Ahrfx/fxCD11ccre conditional knockout mice [38]. Specifically, Ahrfx/fx dams, which express a functional AHR protein, were mated with male Ahrfx/fxCD11ccre mice. Impregnated dams were dosed with 10 μg TCDD/kg body weight or vehicle control. This increased dose was used because Ahrfx/fx mice express an allelic variant of the AHR, Ahrd/d, which encodes a protein with 10 times lower affinity for TCDD than the Ahrb/b expressed by B6 mice [52]. Using adult Ahrfx/fx and Ahrfx/fxCD11ccre offspring of vehicle and TCDD-treated dams, we determined the number of DC subsets in the MLN 3 days after IAV infection (Fig 3G–3J). Maternal TCDD treatment significantly reduced number of all cDCs and of CD11b+cDCs and CD103+ cDCs, but not pDCs, in Ahrfx/fx offspring, compared to Ahrfx/fx offspring of vehicle treated dams (Fig 3G–3J). In contrast, this reduction was not observed in Ahrfx/fxCD11ccre littermates that were developmentally exposed to TCDD (Fig 3G–3J). Additionally, while the number of cDCs appears diminished in vehicle control Ahrfx/fxCD11ccre offspring compared to their vehicle Ahrfx/fx littermates, the difference in these values was not statistically significant. Overall, these findings suggest that AHR activation during development decreases the number of DCs in the lung-draining lymph node via a mechanism that, at least in part, requires AHR expression in cells of the CD11c lineage.
Developmental AHR activation reduces DC migration
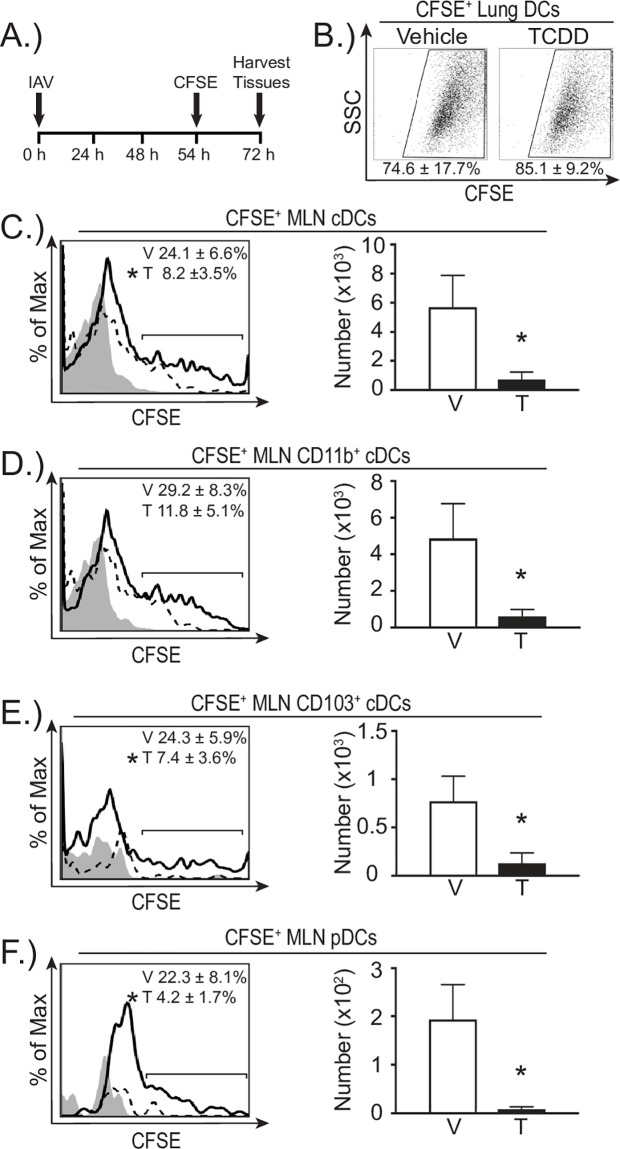
The decreased number of DCs in the MLN of offspring that were developmentally exposed to TCDD could reflect that developmental exposure increases DC death and/or affects DC migration. Apoptotic and dead DCs were identified using flow cytometry. Regardless of whether we examined DCs prior to or after infection, we did not observe any statistically significant differences in apoptotic or dead DCs from the MLN or lungs of offspring from TCDD-treated dams compared to vehicle offspring (S2 Fig). In contrast, we found evidence that developmental exposure affects DC migration. To directly measure the ability of DCs to emigrate from the infected lung to the MLN, we fluorescently labeled cells in the respiratory tract with CFSE, and enumerated CFSE+ DCs in the MLN 3 days after infection [35, 53] (Fig 4A). The percentage of DCs in the lung that were CFSE+ was not different between offspring of dams treated with vehicle or TCDD (Fig 4B). Thus, developmental exposure did not affect the labeling of DCs by intranasally instilled CFSE. However, early life activation of the AHR reduced the percentage of CFSE+ DCs in the MLN compared to CFSE+DCs in MLNs of infected offspring of vehicle treated dams (Fig 4C–4F, histograms). Furthermore, this reduction spanned all DC subsets, and there were significantly fewer CFSE+ cDCs, CD11b+, CD103+, and pDCs in the MLN 3 days after infection in offspring of TCDD-treated dams (Fig 4C–4F, bar graphs). These findings indicate that AHR activation during development leads to DCs with a diminished capacity to emigrate from the lung to lymph node during infection.
Fig 4. In vivo DC trafficking from the lung to MLN is reduced in offspring of TCDD treated dams.
(A) At maturity, developmentally exposed offspring were infected with IAV (HKx31). CFSE (8mM) diluted in PBS was instilled (i.n.) 54 h after infection. Mice were sacrificed 18 h after CFSE treatment and cells were stained for flow cytometry. (B) Representative FACS plots show CFSE+ DCs in lungs of vehicle or TCDD exposed mice. The average percentage (±SEM) of DCs that were CFSE+ is shown below plots. (C-F) CFSE+ DCs in the MLN of TCDD (T) or vehicle (V) exposed adult offspring were enumerated 72 h after infection. Representative histograms show the CFSE+ staining of each DC subset (vehicle, solid line; TCDD, dotted line; gray depicts the CFSE FMO control). The average percentage (±SEM) of each DC subset that was CFSE+ is indicated on each plot. The bar graphs show the average number of the indicated DC subset that was CFSE+. Error bars depict ± SEM. An * indicates p ≤ 0.05. All offspring within a group are from a separate dam (n = 3–7 per group). Data are from one experiment that is representative of two independent experiments. Underlying data can be found in S1 Data.
To further test whether developmental exposure blunts DC migration properties, we used a well-established in vitro DC migration assay. Specifically, we generated DCs from the bone marrow of naïve adult offspring of dams treated with either vehicle or TCDD (Fig 5A), and measured their ability to migrate towards a concentration gradient of CCL21 in a Transwell system. Consistent with prior reports, maternal treatment with this low dose of TCDD did not alter the total number of bone marrow cells obtained [29], nor did it affect the number of immature BMDCs generated after day 8 in culture, nor the number of mature BMDCs obtained following LPS treatment (Fig 5B–5D). However, fewer DCs from the TCDD-exposed group migrated from the upper toward the lower chamber, containing CCL21 (Fig 5E). Area under the curve (AUC) analyses further support that the proportion of migrated DCs was significantly reduced for BMDCs generated from animals that were developmentally exposed to TCDD (Fig 5F). These results further support that early life activation of the AHR results in DCs with a reduced ability to migrate towards an optimal stimulus.
Fig 5. In vitro BMDC migration is reduced by developmental AHR activation.
(A) Bone marrow dendritic cells (BMDC) were generated from bone marrow cells of uninfected offspring of vehicle and TCDD treated dams. (B-D) The bar graphs depict the number of bone marrow cells (B), the number of immature BMDC (C), and the number of mature CD11chiMHCIIhi BMDCs (D) from adult offspring in each group. (E, F) Migration towards a gradient of CCL21 was determined using a Transwell system. (E) The graph depicts the average number of BMDCs collected from the bottom well at each CCL21 concentration. p-values are listed above data points. (F) The bar graph shows the average area under the curve (AUC) for DC migration in the two treatment groups. Error bars depict ± SEM. An * indicates p ≤ 0.05. BMDCs from all offspring within a group are from a separate dam (n = 12 mice per group). Data were combined from two independently conducted experiments. Underlying data can be found in S1 Data.
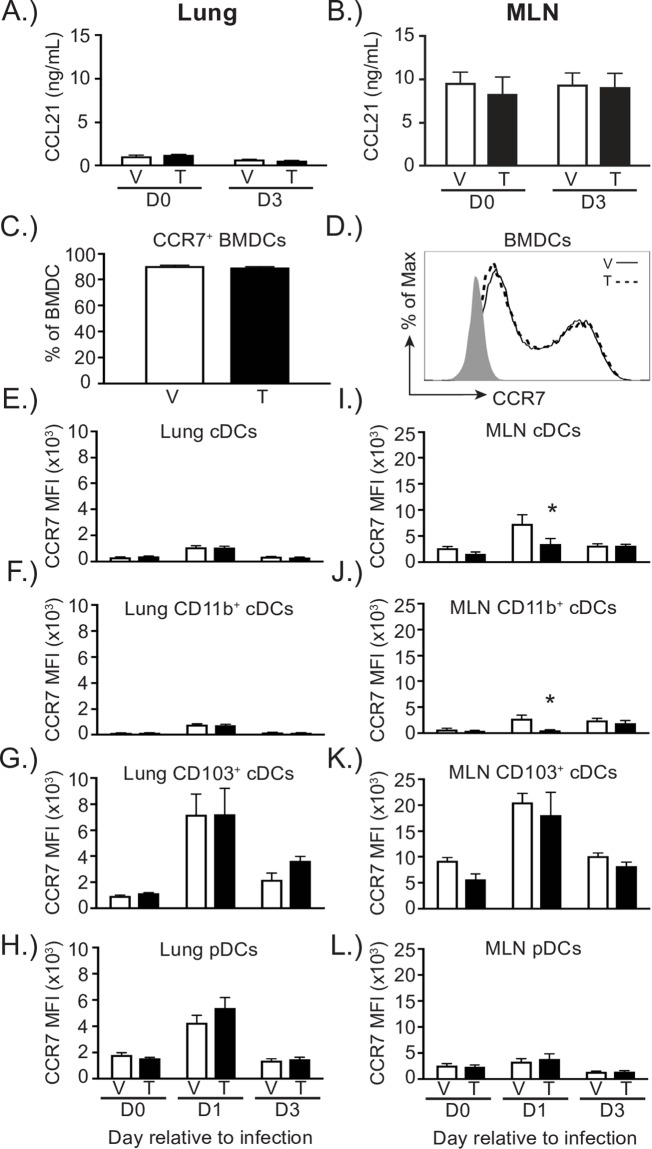
Given that major regulators of leukocyte migration are CCL21 and chemokine receptor 7 (CCR7), the receptor that CCL21 binds [54–56], we next examined whether developmental exposure decreased in vivo levels of CCL21, or diminished CCR7 expression on DCs. Using ELISAs to measure CCL21 in tissue homogenates, the levels of CCL21 were about nine times higher in the MLN than the lung (Fig 6A and 6B). However, prior to and three days after infection, the levels of CCL21 in the lung and MLN were equivalent between developmental exposure groups (Fig 6A and 6B). Using BMDCs from developmentally exposed offspring, there was no difference in the percentage of BMDCs that were CCR7+ or in the mean fluorescence intensity (MFI) of CCR7 on BMDCs from these two exposure groups (Fig 6C and 6D). Similarly, in the lung the relative level of expression of CCR7 on DCs, examined using MFI, was not reduced on any of the DC subsets examined (Fig 6E–6H). Moreover, there were no differences in the percentage or number of CCR7-expressing DC subsets in the lung (S1 Table). In contrast, cDCs, specifically CD11b+ cDCs, in the MLN of developmentally exposed offspring had significantly reduced CCR7 levels 1 day after infection (Fig 6I–6L). In addition to, the proportion of all cDCs and of CD11b+ cDCs that were CCR7+ on day 1 after infection in the MLN was also reduced (S1 Table). However, this decrease in CCR7 expression was transient, as the MFI and percentage of CCR7+ DCs in the MLN was not different between the two groups prior to infection or 3 days after infection.
Fig 6. CCL21 production is not affected by early life activation of the AHR, while CCR7 levels are moderately changed on cDC in the MLN.
(A-B) CCL21 levels in lung and MLN homogenates were measured by ELISA. The bar graph shows the levels of CCL21 in ng/mL from the lung (A) and MLN (B) of uninfected (d0) or infected (d3; HKx31) offspring. (C-L) Single cell suspensions from BMDC, lung, and MLN were stained for flow cytometric analysis, and DC subsets are defined as in Fig 3 with the addition of an anti-CCR7 antibody. (C) The bar graph shows the percentage of LPS stimulated BMDC that are CCR7+. (D) The histogram shows mean fluorescence intensity of CCR7 on either V (solid line) or T (dotted line) exposed LPS stimulated BMDCs. Solid histogram represents FMO control. (E-H) The bar graphs show the geometric mean fluorescence intensity (MFI) of CCR7 on DC subsets in the lung prior to (day 0) or after infection (HKx31; days 1–3). (I-L) The bar graphs show the MFI of CCR7 on the indicated DC subsets in the MLN prior to or after infection (HKx31; day 0 or days 1–3, respectively). Error bars depict ± SEM. An * indicates p ≤ 0.05. All offspring within a group are from a separate dam (n = 6–9 mice per group per day). Lung CCL21 data are representative of one experiment, and MLN CCL21 data are representative of two independent experiments. CCR7 data are from a single experiment that is representative of two independent experiments. Underlying data can be found in S1 Data.
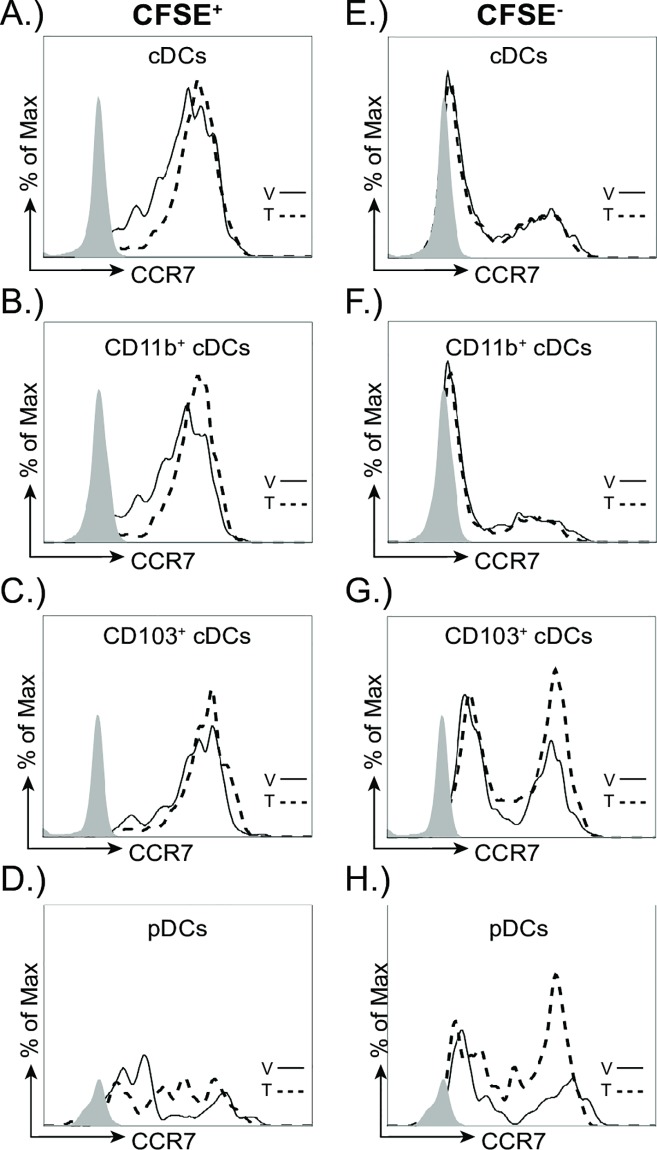
To further determine whether developmental exposure affects CCR7 levels specifically on the DCs that trafficked to the MLN from the infected lung, we compared CCR7 levels on CFSE+ and CFSE- DCs in the MLN (Fig 7). Overall, the expression of CCR7 was higher on CFSE+ cDC subsets (Fig 7A–7D) compared to CFSE- cDCs (Fig 7E–7H). Also, a greater proportion of the CFSE+ cDCs were CCR7+, which further suggests that CCR7 is higher on cDCs in the MLN that emigrated from the lung. However, between the treatment groups, the MFI of CCR7 was not significantly different on any of the DC subsets examined (Table 1). These data suggest that early life AHR activation does not change CCR7 levels on DCs that have emigrated from the lung to the MLN.
Fig 7. CCR7 levels on resident or recently migrated DC are not changed with early life exposure to TCDD in the MLN.
At maturity, developmentally exposed offspring were infected with H3N2 IAV (HKx31). CFSE (8mM) was instilled (i.n.) 54 h after infection. Mice were sacrificed 18 h after CFSE treatment and single cell suspensions were made from MLN. Vehicle (V, solid line), TCDD (T, dotted line). Grey histograms indicate the fluorescence minus one (FMO) control. (A-D) Representative histograms depict CCR7 levels on recently migrated (CFSE+) DC subsets in the MLN 3 days after IAV infection. (E-H) Representative histograms depict CCR7 levels three days after infection on resident CFSEˉ DC subsets in MLNs of vehicle or TCDD exposed offspring. Error bars depict ± SEM. All offspring within a group are from a separate dam (n = 3–9 mice per group per day). Data are representative of at least two experiments with similar results.
Table 1. Mean fluorescence intensity (MFI) of CCR7.
| DC type | CFSE+ | CFSE- | ||
|---|---|---|---|---|
| Vehicle | TCDD | Vehicle | TCDD | |
| cDCs | 2710 ± 125 | 2784 ± 390 | 1029 ± 226 | 991 ± 96 |
| CD11b+ | 2527 ± 242 | 2269 ± 468 | 857 ± 317 | 700 ± 78 |
| CD103+ | 4531 ± 584 | 5516 ± 444 | 1899 ± 217 | 2474 ± 152 |
| pDCs | 1975 ± 1068 | 1893 ± 475 | 2186 ± 622 | 1692 ± 216 |
Table shows the MFI (± SEM) of CCR7 on each CFSE+ and CFSEˉ DC subset in the MLN three days after IAV infection. All offspring within a group are from a separate dam (n = 3–9 mice per group per day). Data are representative of at least two experiments with similar results.
Developmental activation of AHR alters gene expression in DCs
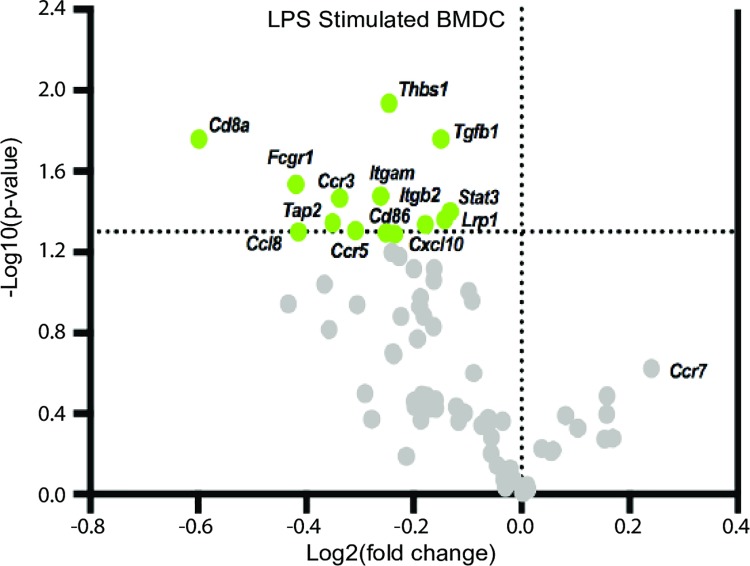
Given that early life AHR activation reduced the capacity of DCs to stimulate naïve CD8+ T cells and also affected DC trafficking, we next wanted to identify DC-specific functional pathways that are altered by developmental AHR activation. To uncover cellular pathways in DCs affected by developmental exposure, we utilized a PCR array that measured 84 genes involved in DC function. In mature BMDCs from offspring of dams that were treated with vehicle or TCDD, 71 of 84 genes were expressed. Of these genes, 14 were differentially expressed in BMDCs from offspring of TCDD-treated dams (Fig 8), while the remaining 58 genes were not significantly different between the two exposure groups. Interestingly, all 14 of the differentially expressed genes (DEGs) were down-regulated in the TCDD group, compared to BMDCs from the control group (Table 2). The 14 DEGs encode proteins that contribute to five aspects of DC function: antigen processing and presentation, cellular adhesion and migration, chemokines, cellular survival and differentiation, and signal transduction.
Fig 8. Developmental activation of the AHR down-regulates BMDC genes involved in pathways critical for DC function.
Mature BMDC were generated from adult offspring (8–10 weeks old) of B6 dams treated with vehicle control or TCDD. The volcano plot shows -log10(p-value) vs. log2(fold change) of genes assayed by PCR array in LPS stimulated BMDC. Log2(fold change) calculated relative to LPS stimulated BMDCs from offspring developmentally exposed to vehicle. The horizontal dotted line indicates significance at -log10(0.05). Genes were considered significantly different when p ≤ 0.05. All offspring within a group are from a separate dam (n = 6 mice per group). Underlying data can be found in S1 Data.
Table 2. Significantly differentially expressed genes in BMDC.
| Gene ID | aLog2(fold change) | bp-value | Pathway |
|---|---|---|---|
| CD8a | -0.60 | 0.02 | Antigen processing and presentation |
| FCGR1 | -0.42 | 0.03 | Antigen processing and presentation |
| LRP1 | -0.14 | 0.04 | Antigen processing and presentation |
| TAP2 | -0.35 | 0.05 | Antigen processing and presentation |
| CD86 | -0.24 | 0.05 | Antigen processing and presentation |
| THBS1 | -0.25 | 0.01 | Cellular adhesion and migration |
| ITGAM | -0.26 | 0.03 | Cellular adhesion and migration |
| CCR3 | -0.34 | 0.03 | Cellular adhesion and migration |
| ITGB2 | -0.18 | 0.05 | Cellular adhesion and migration |
| CCR5 | -0.31 | 0.05 | Cellular adhesion and migration |
| TGFB1 | -0.15 | 0.02 | Cellular survival, proliferation, and differentiation |
| CCL8 | -0.41 | 0.05 | Chemokines |
| CXCL10 | -0.25 | 0.05 | Chemokines |
| STAT3 | -0.13 | 0.04 | Signal transduction |
The table shows the log2(fold change), p-value, and associated pathway of significantly differentially expressed genes in BMDC. Bolded genes represent genes that were similarly observed in MLN DC (Table 3). All offspring within a group are from a separate dam (n = 6 mice per group).
aValues calculated relative to Vehicle LPS BMDC
bAll expression changes were statistically significant with p ≤ 0.05
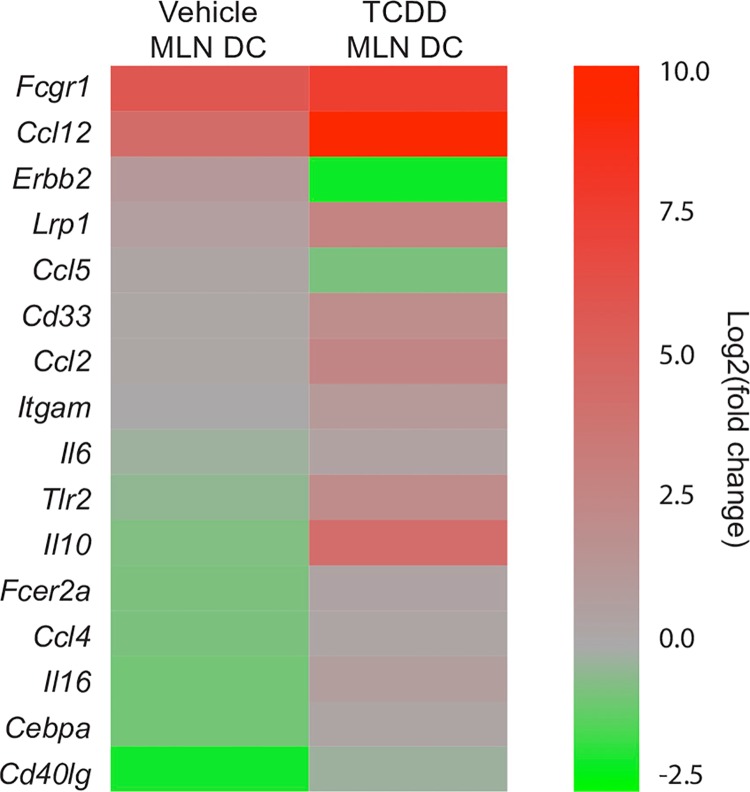
Using the same PCR array system, we compared gene expression in DCs from MLN of developmentally exposed mice. In MLN-derived DCs, there were 16 DEGs, while 63 genes were not significantly different between the two exposure groups and 5 genes were not detected (Fig 9). The number of DEGs was similar in BMDCs and MLN-derived DCs. Yet, in contrast to DEGs in BMDCs, the direction of change among the DEGs from MLN-derived DCs was mostly increased (Fig 9). The differences in the change in direction may reflect differences in the timing of DC stimulation relative to gene expression assessment and differences in DC activation in vitro versus in vivo. Despite these differences, the DEGs in DCs from MLN were similar to BMDCs with regard to the functional pathways affected. Similar to BMDCs, pathways changed by developmental exposure include antigen processing and presentation, cellular adhesion and migration, cellular survival and differentiation, chemokines, cytokines, and signal transduction pathways (Table 3). Thus, although only three DEGs were the same in MLN DCs and BMDCs (Lrp1, Fcgr1 and Itgam), the overall pathways affected by developmental exposure were similar in DCs from the MLN and bone marrow.
Fig 9. Genes in MLN DC are up-regulated after early life AHR activation.
CD11c+ cells (DC) were enriched from the MLNs of adult offspring from dams that were exposed to vehicle control or TCDD. The heat map shows significantly differentially expressed genes (DEGs) in DC from vehicle or TCDD exposed offspring 3 days after IAV infection (HKx31) based on the average log2(fold change) relative to naïve DCs from 3 replicate samples per group and are ordered from most significantly up-regulated to most significantly down-regulated in the vehicle group. Genes were considered significantly different between vehicle and TCDD groups when p ≤ 0.05. All offspring within a group are from a separate dam (n = 12 mice per replicate, 3 independent replicates per group). Underlying data can be found in S1 Data.
Table 3. Significantly differentially expressed genes in MLN DC.
| Gene ID | aLog2(fold change) from naïve DCs | bp-value | Pathway | |
|---|---|---|---|---|
| aVehicle | aTCDD | |||
| TLR2 | -0.43 | 2.09 | 0.00 | Antigen processing and presentation |
| CD40lg | -1.96 | -0.22 | 0.01 | Antigen processing and presentation |
| FCER2a | -0.76 | 0.45 | 0.03 | Antigen processing and presentation |
| LRP1 | 0.71 | 2.70 | 0.04 | Antigen processing and presentation |
| FCGR1 | 5.83 | 7.58 | 0.04 | Antigen processing and presentation |
| CD33 | 0.19 | 2.00 | 0.05 | Antigen processing and presentation |
| ITGAM | 0.09 | 1.11 | 0.02 | Cellular adhesion and migration |
| CEBPa | -0.89 | 0.24 | 0.04 | Cellular survival, proliferation, differentiation |
| CCL2 | 0.13 | 2.50 | 0.01 | Chemokines |
| CCL12 | 4.31 | 9.31 | 0.01 | Chemokines |
| CCL4 | -0.78 | 0.26 | 0.02 | Chemokines |
| CCL5 | 0.30 | -0.77 | 0.03 | Chemokines |
| IL6 | -0.89 | 0.81 | 0.00 | Cytokines |
| IL10 | -0.70 | 4.27 | 0.01 | Cytokines |
| IL16 | -0.23 | 0.51 | 0.02 | Cytokines |
| ERBB2 | 1.15 | -2.02 | 0.00 | Signal transduction |
The table shows the log2(fold change) from naïve vehicle or TCDD DCs, p-value, and associated pathway of significant DEGs in DC. Bolded genes represent genes that were similarly observed in BMDC (Table 2).
aValues calculated relative to respective naïve MLN DC treatment group
bAll expression changes were statistically significant with p ≤ 0.05
In addition to the genes in the PCR array, we measured Ccr7, Ido1 and Cyp1a1 expression. There were no statistically significant differences in the expression of Ccr7 in BMDCs or MLN DCs from the TCDD group compared to DCs from the vehicle exposed offspring (Fig 8 and S2 Table). This corresponds with the general lack of durable differences in CCR7 protein expression on DCs in offspring of vehicle and TCDD treated dams (Fig 6). Cyp1a1 and Ido1 represent two genes that are upregulated by direct exposure to AHR agonists. The expression levels of Ido1 in BMDCs or MLN DCs were not different between treatment groups (S2 Table). Consistent with prior reports [7], we did not detect Cyp1a1 expression in MLN DCs from either exposure group (S2 Table). This suggests that the DEGs in cells from adult offspring are not direct AHR target genes, but instead reflect altered transcriptional regulation in DCs that is triggered by AHR activation earlier in life.
Discussion
AHR activation during development has a lasting impact on adaptive immune responses in the offspring, modulating T cell responses in several model systems, including viral infection. Yet, our understanding of how early life AHR activation specifically affects functional properties of many types of leukocytes is scant. For instance, although DCs are critical for the initiation of adaptive immunity, the consequences of AHR activation during development on DCs has not been established. Given that direct AHR signaling modifies DC function, it is logical to hypothesize that exposure to exogenous AHR ligands during development could affect this important leukocyte lineage later in life. In this report, we show that developmental activation of the AHR influences some aspects of DC function, such as their ability to stimulate naïve CD8+ T cells and their ability to migrate, while leaving other DC characteristics without perceptible alteration. Thus, inappropriate AHR signaling early in life can lead to changes to immune defense mechanisms via durable modifications to DCs.
One critical function of DCs is the stimulation of naïve T cells during an immune challenge. Despite their central role, only a handful of studies have examined whether DCs are sensitive to perturbation by early life exposures, and just a few of these directly assessed APC function [57–62]. For instance, DCs from allergen-naïve offspring of asthmatic mothers exhibited an increased ability to stimulate proliferation of CD4+ T cells in vitro [57]. In another study, developmental exposure of mice to the antibiotic neomycin resulted in a reduction in the ability of a mixture of macrophages and DCs to stimulate antigen-specific CD4+ and CD8+ T cells [61]. Thus, the early life environment can influence APC function in offspring. Our findings add to this knowledge base by showing that developmental exposure to an AHR ligand affects this important property of DCs.
Direct AHR activation influences the ability of DCs to activate naive T cells [34, 35, 38, 63, 64]. Therefore, it is not surprising that developmental exposure to an AHR ligand influences this central DC function. However, a notable difference between direct versus developmental exposure may be that developmental AHR activation leads to differential effects on the capacity of DCs to stimulate naïve CD4+ versus CD8+ T cells. Here, we show that in a head-to-head comparison, triggering AHR during development leads to DCs that have a reduced capacity to activate virus-specific naïve CD8+ T cells, but there were no differences in their ability to activate naïve CD4+ T cells during IAV infection. AHR activation during development, such as via maternal exposure to TCDD, disrupts activation of both naïve CD4+ and CD8+ T cells in vivo [7, 8]. The TCR transgenic T cells used to measure DC APC function were from untreated and uninfected mice, allowing us to interrogate the potential DC contribution to this altered T cell response. Two in vivo studies of direct AHR activation showed that DCs from TCDD-exposed mice were equally capable of stimulating OVA-specific CD4+ T cell responses, highlighting a difference between direct and developmental activation of AHR [33, 36]. However, activation of CD8+ T cells was not examined in these studies. Moreover, an impact on DC-CD4+ T cell activation has not been consistently observed with direct exposure. For example, in another study, splenic DCs from mice treated directly with TCDD showed enhanced proliferation of OVA-specific CD4+ T cells in an ex vivo assay [63]. In contrast, direct treatment of DCs with the AHR agonist, 2-(1′H-indole-3′-carbonyl)-thiazole-4-carboxylic acid methyl ester (ITE) reduced the proliferation of antigen-specific CD4+ T cells in vitro [34]. While less well studied in comparison to CD4+ T cells, direct activation of the AHR also affects their ability to stimulate CD8+ T cells. For example, fewer IAV-specific CD8+ T cells were activated by DCs isolated from TCDD treated mice [35, 38]. Thus, AHR signaling in the mature immune system, by direct treatment with an AHR ligand, affects the capacity of DCs to stimulate CD4+ and CD8+ T cells, although whether AHR activation enhances or diminishes T cell activation appears to be dependent on the context in which DCs were evaluated, while developmental AHR signaling affects the capacity of DCs to stimulate CD8+ T cells but not CD4+ T cells in the context of IAV infection.
While the mechanisms that contribute to the differential ability of DCs to activate CD4+ and CD8+ T cells following AHR activation during development are not yet known, our study provides important insight. For instance, this difference may reflect functional disparities among DC subsets, as distinct DC subsets (e.g., CD103+ vs CD11b+) may play a more dominant role in CD4+ vs. CD8+ T cell activation at distinct points in time after respiratory antigen challenge [38, 48–50]. Also, a unique function of some DC subsets, such as migratory CD103+ cDCs, is that they can present exogenously derived peptides with MHCI through a process called cross-presentation. Cross presentation is particularly important for initiating CD8+ T cell responses during viral infections and anti-tumor responses [65, 66]. There is some circumstantial evidence that direct (i.e. non-developmental) exposure to AHR agonists affects cross-presentation. Specifically, AHR activation reduced the ability of highly purified CD103+ DCs from IAV-infected mice to stimulate naïve virus-specific CD8+ T cells [38]. Further support that triggering AHR during development influences DC cross-presentation later in life comes from examination of gene expression in DCs from offspring of treated dams. DEGs in DCs include Tap2, Fcgr1 and Lrp1. Tap2 encodes transporter associated with antigen processing 2 (TAP2), which is directly associated with cross-presentation by DCs [66]. Fcgr1 encodes the high affinity immunoglobulin gamma Fc receptor I (FCGR1), which mediates internalization of antigen-IgG complexes and DC cross presentation [67, 68]. Lrp1 encodes a scavenger receptor called low density lipoprotein receptor-related protein 1 or LRP1, which aids in the internalization of antigen-heat shock protein complexes [69–71]. Both FCGR1 and LRP1 play a role in cross-presentation of exogenous antigen in the context of MHCI on DCs [67–72]. Thus, disruption of cross-presentation is a mechanism by which early life activation of the AHR could dampen DC:CD8+ T cell interactions. Moreover, this provides a potential explanation for how AHR activation during development affects the ability of DCs to activate naïve CD8+, but not CD4+ T cells, following infection. In addition to cross presentation, inappropriate triggering of the AHR during development may also affect other mechanisms that govern DC and T cell interactions, such as the frequency and duration of DC:T cell contacts, antigen uptake, availability, and the balance between immunostimulatory and regulatory DC functions [73–76].
Another key finding is that AHR activation during development reduced DC migration later in life. Studies of direct AHR activation point to an emerging role for the AHR in the control of leukocyte migration. For example, direct exposure to TCDD reduced bone marrow cell migration in vivo [77]. In other studies, in vivo DC migration from the lung to the MLN during IAV infection was reduced upon AHR activation [35]. AHR signaling also affects neutrophil trafficking, although it may do so indirectly, via signaling in non-hematopoietic cells, and the direction of change depends upon the stimuli [30, 78–81]. Also, although AHR signaling affects neutrophil recruitment, it does not affect the number of neutrophils in peripheral tissues in the absence of antigen challenge [78–80]. Similarly, we did not observe differences in the number of DCs in lungs before or after infection. Yet, when cued to emigrate, such as in vivo during infection or ex vivo in response to a chemokine gradient, fewer DCs migrated. Although we observed fewer DCs emigrating from the lung to the MLN upon infection, it is possible that rather than failing to migrate, the DCs are migrating improperly and accumulating in a different anatomical site. Thus, while early life AHR activation influences DC migration properties, it likely does so in conjunction with, or in the context of, other signals to the DCs.
Although recent studies have linked direct AHR activation to increases in CCR7 expression levels [32, 82], developmental exposure led to only modest and transient decreases in CCR7 levels on cDCs in the MLN, and did not alter CCR7 expression levels on BMDCs or lung DCs. Thus, it is likely that other factors that regulate DC migration are affected by developmental AHR activation. For instance, signals that promote cellular cytoskeletal rearrangement or adhesion could be altered as a result of early life AHR activation in DC. Candidates revealed by gene expression analyses include molecular regulators of cell adhesion and migration, such as Lrp1 and Itgam (CD11b). CD11b is one of four β2 integrins involved in cell signaling, migration, and in DC-T cell interactions [83]. In addition to playing a role in cross-presentation, LRP1 signaling can influence cell migration [84–87], and LRP1 directly associates with CD11b and modulates cellular adhesion [84, 88]. Interestingly, differential expression of Lrp1 and Itgam was observed in both BMDCs and in DCs from the MLN of developmentally exposed mice. This further suggests a possible central role for AHR-mediated changes in these genes, or in the upstream regulators of the expression of these genes. An important consideration in the interpretation of gene expression analyses in DCs isolated from the MLN is that these results are skewed by preferentially reflecting gene expression in the DC populations that have already migrated to the lymph nodes. This limitation is mitigated by our observation that in developmentally exposed mice, decreased DCs number reflects a reduction in all subsets examined.
The changes observed in DCs appear to be inherent to the DC lineage, and suggest that triggering AHR during development causes long-lasting programming of DCs. Evidence includes the differences in ex vivo APC function, diminished DC migration, including abrogation of the reduced DC number in MLNs of developmentally exposed mice lacking Ahr in the CD11c lineage. Given that the lifecycle of DCs is several days to a few weeks [89–91], the DCs evaluated at maturity are not present in the fetus and neonate. That is, the DCs interrogated at maturity are not directly exposed to the exogenous AHR ligand given to the dams. Instead, it is likely that DC progenitor cells, such as a monocyte-dendritic cell progenitors (MDP) or common dendritic cell progenitors (CDP), are affected by early life AHR activation. Changes in DC function observed in developmentally exposed offspring may reflect alterations in epigenetic programming in the DC lineage. Epigenetic regulatory mechanisms, such as DNA methylation, influence gene expression and cellular function [92]. Although not extensively studied, developmental exposure to other environmental factors, such as being born to an asthmatic mother, alters DNA methylation marks in DCs of the offspring [57, 58]. Furthermore, early life activation of the AHR modifies DNA methylation patterns and gene expression in CD8+ T cells of adult offspring [7]. Thus, it is possible that inappropriate AHR activation during development alters DNA methylation, and potentially other epigenetic regulatory mechanisms, in the DC lineage, leading to durable changes in DC functions later in life.
The research reported here focuses on how developmental exposure to a representative AHR agonist influences the response of DCs during acute, primary IAV infection. DCs are fundamental to appropriate immune responses to many pathogens, to vaccines, and to establishing anti-tumor immunity. Thus, the significance of these initial findings extends beyond IAV infection, and suggest developmental exposure to AHR-binding substances affects DC responses in other contexts. More broadly, these data support the idea that the early life environment shapes DC function in an enduring manner, influencing immune function into adulthood. Given that DCs are critical regulators of adaptive immune responses during may different types of immune challenges, this work reveals that AHR-mediated events influence DCs may be an important factor in many communicable, and non-communicable, diseases.
Materials and methods
Mice
C57Bl/6 (B6) and B6.Cg-Tg (Itgax-cre) 1-1Reiz/J (CD11ccre) [93] mice (5–6 weeks old) were purchased from the National Cancer Institute (Frederick, MD) or The Jackson Laboratory (Bar Harbor, ME). Dr. Christopher Bradfield (University of Wisconsin, Madison, WI) provided breeding stock of Ahrfx/fx mice. Breeding stock of F5 T cell receptor (TCR) transgenic mice (F5 mice) were provided by Dr. Demetrius Moskophidis (Medical College of Georgia, Augusta, GA) and Dr. Dimitris Kioussis (National Institute for Medical Research, London, UK). The TCR on CD8+ T cells of F5 mice recognizes amino acids 366–374 of the nucleoprotein (NP366-374) of influenza virus A/Memphis/102/72 in the context of H-2Db [41]. Breeding stock of OTII TCR transgenic mice (OTII mice) were provided by Dr. Minsoo Kim (University of Rochester, Rochester, NY). The TCR on CD4+ T cells of OTII mice recognizes amino acids 323–339 of chicken ovalbumin in the context of I-Ab [94]. The phenotype of F5 and OTII mice is determined using flow cytometry [41, 94]. Conditional knockout Ahrfx/fxCD11ccre mice were generated by mating male CD11ccre mice with female Ahrfx/fx mice. DNA from tail biopsies was obtained by digestion with Proteinase K (Invitrogen, Carlsbad, CA) and Direct PCR Lysis Reagent (Viagen Biotech, Los Angeles, CA). Genotyping was performed by PCR using the following primers: Ahrfx/fx OL4064 (5’- CAG TGG GAA TAA GGC AAG AGT GA– 3’) and OL4088 (5’- GGT ACA AGT GCA CAT GCC TGC– 3’); CD11c Cre transgene forward (5’–ACT TGG CAG CTG TCT CCA AG– 3’) and CD11c Cre transgene reverse (5’–GCG AAC ATC TTC AGG TTC TG– 3’). All mice are housed in microisolator cages in a specific pathogen-free facility at the University of Rochester Medical Center, and are provided food and water ad libitum. Animals were sacrificed using anesthetic overdose, followed by a secondary method. All animal treatments were conducted with approval of Institutional Animal Care and Use Committee and Institutional Biosafety Committee of the University of Rochester.
Developmental exposure
Nulliparous female B6 mice were housed with B6 males, and checked daily for the presence of a vaginal plug; designated day 0 of gestation (GD0). Impregnated B6 mice were singly housed and treated with 1 μg TCDD/kg body weight or the peanut oil vehicle control (Vehicle) by gavage on GD 0, 7, and 14 and 2 days after parturition (PND2). This dose of TCDD is not overtly toxic to the dam or pups [7, 8, 29, 30]. In experiments where Ahrfx/fxCD11ccre mice were used, female Ahrfx/fx mice were mated with male Ahrfx/fxCD11ccre mice to generate Ahrfx/fx and Ahrfx/fxCD11ccre offspring. Ahrfx/fx dams were dosed with 10 μg TCDD/kg body weight or vehicle control on GD14 and PND2. This increased dose was used because Ahrfx/fx mice express an allelic variant of the AHR, Ahrd/d, which encodes a protein with 10 times lower affinity for TCDD than the Ahrb/b expressed by B6 mice [52, 80, 95]. The timing of dosing of impregnated Ahrfx/fx mice was altered due to pup death that occurred when TCDD was administered during early pregnancy (i.e., dams were not treated on GD0 and GD7). For the dosing solution, TCDD (≥99% purity; Cambridge Isotope Laboratories, Woburn, MA) was dissolved in anisole and diluted in peanut oil. The vehicle control consisted of peanut oil containing an equivalent concentration of anisole (0.01%). No culling of litters was performed, and offspring were weaned at 20–21 days of age.
Viral infection
Adult offspring (8–12 weeks of age) of TCDD- or vehicle-treated dams were anesthetized by intraperitoneal (i.p.) injection of avertin (2,2,2-tribromoethanol; Sigma Aldrich, Milwaukee, WI) and inoculated intranasally (i.n.) with 25 μl sterile PBS containing 120 hemagglutinating units (HAU) of influenza virus strain A/HK/x31 (x31; H3N2), 1x107 plaque forming units (PFU) of A/Memphis/102/72 (Mem/102; H3N2), or 8.25x105 PFU of A/HK/x31/OVAII (x31/OVAII; H3N2; [44]). This work was conducted with prior review and approval of the Institutional Biosafety Committee of the University of Rochester.
Isolation of immune cells
Single-cell suspensions of mediastinal lymph node (MLN), lung, bone marrow, and spleen cells were obtained as previously described [29, 35, 96]. Briefly, MLNs were disrupted between the frosted ends of 2 microscope slides, and digested with collagenase-containing media for 25 min at 37°C in 5% CO2 (RPMI 1640 medium containing 1 mg/mL collagenase A (Worthington Biochemical, Lakewood, NJ), 30 μg/mL DNase I (Roche), 2.5% FBS, and 10 mM HEPES). Lungs were perfused with a solution of 0.6 mM EDTA (Invitrogen, Carlsbad, CA) in 1X phosphate buffered saline (PBS; Lonza, Walkersville, MD). Immune cells from the airways and interstitial spaces of the lung were obtained by digesting lungs with RPMI containing collagenase and DNAse I. Bone marrow (BM) cells were collected from the long bones suspended in complete RPMI (cRPMI) media (RPMI 1640 containing 10% FBS, 100 U/mL penicillin/streptomycin, 2 mM Glutamax-I (L-alanyl-L-glutamine dipeptide), 1 mM sodium pyruvate, and 1X non-essential amino acids; Gibco, Grand Island, NY). Naïve F5 CD8+ T cells and naïve OTII CD4+ T cells were enriched from spleens of untreated and uninfected mice using the MagCelect Mouse Naïve CD8+ T Cell Isolation Kit, or MagCelect Mouse Naïve CD4+ T Cell Isolation Kit, respectively (R&D Systems, Minneapolis, MD). The purity of naïve CD8+ T cells (Vβ11+CD8+CD44lo) and naïve CD4+ T cells (Vβ5+CD4+CD44lo) was determined by flow cytometry, and was >95%.
Flow cytometry
Cells were incubated with anti-mouse CD16/32, and stained with previously determined optimal concentrations of fluorochrome-conjugated mAbs. To identify DCs, antibodies against MHC class II (M5/114.15.2; I-A/I-E), CD11c (N418), CD11b (M1/70), CD103 (M290), B220 (RA2-6B2), PDCA-1 (ebio927). In some experiments, fluorochrome-conjugated antibodies for annexin V, live/dead, or CCR7 (4B12) were added. To identify T cells, antibodies against CD8 (53.67), Vβ11 (RR3–15), CD4 (GK1.5), Vβ5 (MR9-4), and CD44 (IM7) were used. Antibodies were purchased from eBioscience, BD Biosciences or BioLegend. Fluorescence minus one (FMO) controls were used to define gating parameters. Doublet discrimination and autofluorescent cell exclusion was included in the gating strategy. Data acquisition was performed using LSR-II cytometers (BD BioSciences) and data analyses were performed using FlowJo software (Tree Star, Ashland, OR).
Ex vivo assay of APC function
DCs were isolated from pooled MLN of Mem/102- or X31/OVAII-infected adult offspring (>30 mice per group) and enriched using a Mouse CD11c Microbead Kit (Miltenyi Biotec, Auburn, CA) [35, 38]. On the same day, naïve T cells from spleens of F5 or OTII mice were isolated using MagCelect Mouse Naïve CD8+ T Cell Isolation Kit or Naïve CD4+ T Cell Isolation Kits, respectively (R&D Systems, Minneapolis, MD), and labeled with 2 μM carboxyfluorescein diacetate succinimidyl ester (CFSE; Invitrogen, Carlsbad, CA) [97]. CFSE-labeled naïve F5 CD8+ or CFSE-labeled OTII CD4+ T cells (2x105) were cultured in 96-well plates with serially diluted CD11c+ MLN cells. After 3 (F5) or 4 (OTII) days in culture, cells were collected and stained with antibodies to Vβ11 and CD8, or Vβ5 and CD4, along with CD44 for flow cytometric analysis. Activated and proliferated T cells were defined based on up-regulation of CD44 and loss of CFSE staining (CFSEdecayCD44hi) [38].
Fluorescent immunohistochemistry
MLNs were snap frozen in OCT (Sakura Finetek, Netherlands). Using a cryostat, whole lymph nodes were cut into 10 μm thick sections, and 3–4 sections were placed onto coated slides. To determine the middle section, the total number of sections for each MLN was divided in half. The slide containing this section, and the slides with serial sections directly before and after it were stained, giving a total of 6–9 stained MLN sections per sample. Tissues were fixed with 2% paraformaldehyde (PFA), washed with PAB (1X phosphate buffer saline (PBS), containing 1% bovine serum albumin (BSA), and 0.1% sodium azide). Fixed slides were stained with a cocktail of the following fluorochrome-conjugated antibodies: CD11c PE (N418; 0.004 μg/μl; eBioscience), CD19 APC (1D3; 0.004 μg/μl; eBioscience), CD4 PerCP (RM4-5; 0.006 μg/μl; BD Biosciences), and LYVE-1 AF488 (ALY7; 0.01 μg/μl; eBioscience). An anti-mouse CD16/CD32 (93; eBioscience) antibody was included to reduce nonspecific binding to Fc receptors. Fluorescent images were captured using a conventional fluorescence microscope with an Olympus DP80 camera and automated stage (Olympus Corporation, Waltham, MA). Olympus cellSens software was used to create montage fluorescent images of each MLN. Image analysis was performed on raw images using an open source software distribution of ImageJ called Fiji (https://fiji.sc/) [98]. Determination of T cell zones and B cell zones was performed using Fiji’s polygon tool, by drawing demarcations based on anti-CD19 and anti-CD4 staining. Images were thresholded to determine the percentage of CD11c positive area across the entire surface area, and in each compartment.
In vivo DC migration
CFSE was diluted in sterile endotoxin-free PBS to 8 mM. Mice were anesthetized and diluted CFSE was instilled (i.n.) 54 h after infection with IAV (HKx31), to label all cells in the respiratory tract [35, 53]. After 18 h of CFSE treatment (3 d after infection), mice were sacrificed and MLN cells were stained with antibodies against MHCII and CD11c to identify DCs that had migrated from the lung (CFSE+CD11c+MHCII+).
BMDC generation and ex vivo migration assay
To generate bone marrow derived DCs (BMDCs), bone marrow cells (2x106 cells/well) were cultured in cRPMI supplemented with 25 ng/mL mouse GM-CSF and 10 ng/mL mouse IL-4 (Peprotech, Rocky Hill, NJ) for 8 days [99, 100]. On day 8, immature BMDCs were harvested and resuspended in cRPMI, and treated for 24 h with LPS (1 μg/mL) to create mature DCs [99, 101]. After 24 h, mature BMDCs were enumerated, and used in migration assays, stained for flow cytometry, or had RNA extracted. Migration towards a gradient (0–1000 ng/mL) of CCL21 (Peprotech, Rocky Hill, NJ) was determined by plating 2x105 BMDCs in the top wells of 8 μm pore, 96-well Transwell plates (Corning, Corning, NY) and enumerating cells in the bottom well after 2 h incubation at 37°C.
Cytokine and chemokine analysis
Culture supernatants were collected from wells of DC:T cell co-cultures and the concentration of IFNγ was measured using a sandwich ELISA (BD Biosciences). CCL21 concentrations were measured in lung and MLN homogenates using a pre-fabricated CCL21 ELISA kit (R&D Systems, Minneapolis, MD) per the manufacturers protocol.
Real-time PCR analysis
Total RNA was isolated from LPS stimulated BMDC with an RNeasy Mini Kit (Qiagen, Germantown, MD) and cDNA generated using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA). RNA was isolated from CD11c+ DC enriched from MLNs using an RNeasy Plus Kit (Qiagen, Germantown, MD) and cDNA conversion was performed using a NuGEN WT-Ovation PicoSL Kit (NuGEN Technologies, San Carlos, CA). A mouse Dendritic and Antigen Presenting Cell RT2 Profiler PCR Array System was used (Qiagen, Germantown, MD; https://www.qiagen.com/us/shop/pcr/primer-sets/rt2-profiler-pcr-arrays/?catno=PAMM-406Z#geneglobe) following the manufacturer’s instructions. Additional gene-specific amplification was achieved using the following primers: mouse Ccr7 (forward 5’GGA AAA TGA CAA GGA GAG CCA3’, reverse 5’GAG ACA AGA ACC AAA AGC ACAG3’; exon 1–2, IDT), mouse Ido1 (forward 5’GCA TAA GAC AGA ATA GGA GGCA3’, reverse 5’GGT ACA TCA CCA TGG CGT AT3’; IDT), mouse Cyp1a1 (forward 5’TTT GGA GCT GGG TTT GAC AC3’, reverse 5’CTG CCA ATC ACT GTG TCT A3’; IDT), and mouse L13 as internal housekeeping gene (forward 5’CTA CAG TGA GAT ACC ACA CCA AG3’, reverse 5’TGG ACT TGT TTC GCC TCC TC’; IDT). Real-time RT PCR was performed using a Bio-Rad CFX96 Touch detection system with RT2 SYBR Green qPCR Master Mix (Qiagen, Germantown, MD) or iQ SYBR Green Supermix (Bio-Rad, Hercules, CA). Changes in gene expression were determined using the 2-ΔΔCT method [102].
Statistical analyses
The dam was defined as the statistical unit for all experiments, because the dams, not her offspring, were directly treated with TCDD or vehicle control. For most experiments, experimental groups were comprised of 6–9 adult male offspring from separate dams. For some ex vivo assays, pooling of mice from within the same exposure group was required to yield ample biological material. Statistical analyses were performed using JMP software (SAS Institute, Cary, NC). A two-way ANOVA, followed by a Tukey HSD post hoc test, was used to compare differences between multiple independent variables (e.g. multiple genotypes, over time, or across different concentrations). Differences between means of vehicle or TCDD groups at a single point in time were evaluated using a Student's t-test. Means were considered significantly different when p-values were less than or equal to 0.05. Error bars on all graphs represent the standard error of the mean (SEM).
Supporting information
DCs were evaluated prior to and up to 3 days after infection with IAV (HKx31). Flow cytometry was used to identify DC subsets as follows: conventional DCs (cDCs; CD11chiMHCIIhi cells), CD11b+ cDCs (CD11chiMHCIIhiCD11b+CD103- cells), CD103+ cDCs (CD11chiMHCIIhiCD103+CD11b- cells), and plasmacytoid DCs (pDCs; CD11cloMHCIIhi PDCA1+CD45R+ cells). (A, B) Representative dot plots depict the gating used to define cDCs, CD11b+ cDCs, CD103+ cDCs and pDCs in the lungs of vehicle (V) and TCDD (T) exposed offspring after gating to exclude doublets, dead cells, and autofluorescent cells. The percentage on the plot indicates the average percentage of the indicated DC subset 3 days after infection. cDC and pDC percentages are of all immune cells in the lung, whereas CD11b+ DCs and CD103+ DCs indicate the proportion of CD11chiMHCIIhi cells (cDCs). (C-F) The bar graphs show the number (±SEM) of the indicated DC population in the lung from naïve (day 0) or infected mice. At each point in time, all offspring within a group were from a separate dam, n = 6–9 mice per group per day. Day 0 data are representative of 4 independent experiments, day 1 data are representative of 3 independent experiments, day 3 data are representative of 6 independent experiments with similar results. Underlying data can be found in S1 Data.
(DOCX)
DCs were evaluated prior to and 3 days after infection with IAV (HKx31). Flow cytometry was used to identify DC subsets with the addition of annexin V and live/dead stains to detect apoptotic and dead cells. Specifically, annexin V binds phosphatidyl serine residues on the outer leaflet of exposed plasma membranes and live/dead covalently binds intracellular amines from cells with compromised membranes; the detection of cells double positive for these markers indicate dead cells. The bar graphs show the number (±SEM) of DC subsets that were double positive for Annexin V+LiveDead+ in the lung (A-C) and MLN (D-F) from naïve (day 0) or infected mice (day 3). At each point in time, all offspring within a group were from a separate dam, n = 6–9 mice per group per day. Underlying data can be found in S1 Data.
(DOCX)
Flow cytometry was used to identify DC subsets prior to and up to 3 days after infection with IAV (HKx31) as follows: conventional DCs (cDCs; CD11chi MHCIIhi cells), CD11b+ cDCs (CD11chiMHCIIhi CD11b+CD103- cells), CD103+ cDCs (CD11chiMHCIIhi CD103+CD11b- cells), and plasmacytoid DCs (pDCs; CD11cloMHCIIhi PDCA1+CD45R+ cells). In separate experiments, cells were further defined by expression of CCR7. Previous gating excluded doublets and autofluorescent cells. Percentage and number of DC subsets are indicated in the table. aPercentage of all immune cells in the lung. bPercentage of MLN cells. cPercentage of cDCs. For CCR7+ DC subsets, percentages are of cDC, CD11b+, CD103+, or pDC that were positive for CCR7. For CCR7+ DC subsets, all numbers are x103. All values ± SEM. An * indicates significance compared to vehicle (p ≤ 0.05).
(DOCX)
Mature BMDC were generated from bone marrow of naïve or DCs were enriched from the MLNs of IAV infected adult offspring from dams that were exposed to vehicle control or TCDD. The table shows the fold change of Ccr7, Ido1, and Cyp1a1 in DCs relative to their respective vehicle (BMDC) or uninfected (MLN DC) controls. Changes in gene expression were determined using the 2-ΔΔCT method. All offspring within a group are from a separate dam (BMDC, n = 6 mice per group; MLN DC, n = 12 mice per replicate, 3 replicates per group).
(DOCX)
(XLSX)
Acknowledgments
Dr. Christopher Bradfield for mice, Dr. David Topham for X31-OVA IAV, Dr. Minsoo Kim for OTII mice, Dr. Lisbeth Boule for experimental assistance, Dr. Timothy Bushnell and Mr. Matt Cochran at the UR Flow Cytometry Core, and Dr. John Ashton and Ms. Michelle Zanche at the UR Genomics Research Core.
Abbreviations
- AHR
aryl hydrocarbon receptor
- APC
antigen presenting cell
- BM
bone marrow
- BMDC
bone marrow derived dendritic cell
- CCL
C-C motif chemokine ligand
- CCL12
C-C motif chemokine ligand 12
- CCL2
C-C motif chemokine ligand 2
- CCL4
C-C motif chemokine ligand 4
- CCL5
C-C motif chemokine ligand 5
- CCL8
C-C motif chemokine ligand 8
- CCR
C-C motif chemokine receptor
- CCR3
C-C motif chemokine receptor 3
- CCR5
C-C motif chemokine receptor 5
- CCR7
chemokine receptor 7
- CD
cluster of differentiation
- CD33
cluster of differentiation 33
- CD40lg
cluster of differentiation 40 ligand
- CD86
cluster of differentiation 86
- CD8a
cluster of differentiation 8a
- cDC
conventional dendritic cell
- CDP
common dendritic cell progenitor
- CEBPa
CCAAT/enhancer-binding protein alpha
- CFSE
carboxyfluorescein diacetate succinimidyl ester
- cKO
conditional knockout
- CTL
cytotoxic T lymphocyte
- CXCL10
C-X-C motif chemokine 10
- DC
dendritic cell
- DEG
differentially expressed gene
- ELISA
enzyme-linked immunosorbent assay
- ERBB2
erb-b2 receptor tyrosine kinase 2
- FCER2a
Fc epsilon receptor 2 alpha
- FCGR1
immunoglobulin gamma Fc receptor I
- FMO
fluorescence minus one
- GD
gestational day
- HAU
hemagglutinating unit
- IAV
influenza A virus
- IFNγ
interferon gamma
- IL10
interleukin 10
- IL16
interleukin 16
- IL6
interleukin 6
- ITGAM
integrin alpha M
- ITGB2
integrin subunit beta 2
- LN
lymph node
- LPS
lipopolysaccharide
- LRP1
low density lipoprotein receptor-related protein 1
- MDP
monocyte-dendritic cell progenitor
- MFI
mean fluorescence intensity
- MLN
mediastinal lymph node
- PCB
polychlorinated biphenyl
- pDC
plasmacytoid dendritic cell
- PFU
plaque forming unit
- PND
postnatal day
- SEM
standard error of the mean
- STAT3
signal transducer and activator of transcription 3
- TAP2
transporter associated with antigen processing 2
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- TCR
T cell receptor
- TGFB1
transforming growth factor beta 1
- THBS1
thrombospondin 1
- TLR2
toll-like receptor 2
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The National Institute of Environmental Health Sciences (grant numbers T32ES07026, R01ES017250, R01ES023260, P30ES01247) and National Institute of General Medical Sciences (R25GM064133) supported this work. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Grandjean P, Andersen EW, Budtz-Jorgensen E, Nielsen F, Molbak K, Weihe P, et al. Serum vaccine antibody concentrations in children exposed to perfluorinated compounds. JAMA. 2012;307(4):391–397. 10.1001/jama.2011.2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dietert RR. Developmental immunotoxicity, perinatal programming, and noncommunicable diseases: Focus on human studies. Adv Med. 2014;2014:867805 10.1155/2014/867805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao J, Xu X, Hylkema MN, Zeng EY, Sly PD, Suk WA, et al. Early-life exposure to widespread environmental toxicants and health risk: A focus on the immune and respiratory systems. Ann Glob Health. 2016;82(1):119–131. 10.1016/j.aogh.2016.01.023 [DOI] [PubMed] [Google Scholar]
- 4.Zhou A, Chang H, Huo W, Zhang B, Hu J, Xia W, et al. Prenatal exposure to bisphenol A and risk of allergic diseases in early life. Pediatr Res. 2017;81(6):851–856. 10.1038/pr.2017.20 [DOI] [PubMed] [Google Scholar]
- 5.Ng SP, Zelikoff JT. The effects of prenatal exposure of mice to cigarette smoke on offspring immune parameters. J Toxicol Environ Health A. 2008;71(7):445–453. 10.1080/15287390701839281 [DOI] [PubMed] [Google Scholar]
- 6.Bauer SM, Roy A, Emo J, Chapman TJ, Georas SN, Lawrence BP. The effects of maternal exposure to bisphenol A on allergic lung inflammation into adulthood. Toxicol Sci. 2012;130(1):82–93. 10.1093/toxsci/kfs227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Winans B, Nagari A, Chae M, Post CM, Ko CI, Puga A, et al. Linking the aryl hydrocarbon receptor with altered DNA methylation patterns and developmentally induced aberrant antiviral CD8+ T cell responses. J Immunol. 2015;194(9):4446–4457. 10.4049/jimmunol.1402044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boule LA, Winans B, Lawrence BP. Effects of developmental activation of the AhR on CD4+ T-cell responses to influenza virus infection in adult mice. Environ Health Perspect. 2014;122(11):1201–1208. 10.1289/ehp.1408110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilbert KM, Bai S, Barnette D, Blossom SJ. Exposure cessation during adulthood did not prevent immunotoxicity caused by developmental exposure to low-level trichloroethylene in drinking water. Toxicol Sci. 2017;157(2):429–437. 10.1093/toxsci/kfx061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luebke RW, Chen DH, Dietert R, Yang Y, King M, Luster MI. The comparative immunotoxicity of five selected compounds following developmental or adult exposure. J Toxicol Environ Health B Crit Rev. 2006;9(1):1–26. 10.1080/15287390500194326 [DOI] [PubMed] [Google Scholar]
- 11.Winans B, Humble MC, Lawrence BP. Environmental toxicants and the developing immune system: a missing link in the global battle against infectious disease? Reprod Toxicol. 2011;31(3):327–336. 10.1016/j.reprotox.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boule LA, Lawrence BP. Influence of early-life environmental exposures on immune function across the life span In: Esser C, editor. Environmental influences on the immune system. Vienna: Springer; Vienna; 2016. p. 21–54. [Google Scholar]
- 13.Nguyen LP, Bradfield CA. The search for endogenous activators of the aryl hydrocarbon receptor. Chem Res Toxicol. 2008;21(1):102–116. 10.1021/tx7001965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, et al. Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front Cell Infect Microbiol. 2018;8(13):1–22. 10.3389/fcimb.2018.00013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esser C, Rannug A. The aryl hydrocarbon receptor in barrier organ physiology, immunology, and toxicology. Pharmacol Rev. 2015;67(2):259–279. 10.1124/pr.114.009001 [DOI] [PubMed] [Google Scholar]
- 16.Gutierrez-Vazquez C, Quintana FJ. Regulation of the immune response by the aryl hydrocarbon receptor. immunity. 2018;48(1):19–33. 10.1016/j.immuni.2017.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malisch R, Kotz A. Dioxins and PCBs in feed and food—review from European perspective. Sci Total Environ. 2014;491–492:2–10. 10.1016/j.scitotenv.2014.03.022 [DOI] [PubMed] [Google Scholar]
- 18.Warenik-Bany M, Strucinski P, Piskorska-Pliszczynska J. Dioxins and PCBs in game animals: Interspecies comparison and related consumer exposure. Environ Int. 2016;89–90:21–29. 10.1016/j.envint.2016.01.007 [DOI] [PubMed] [Google Scholar]
- 19.Nau H, Bass R, Neubert D. Transfer of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) via placenta and milk, and postnatal toxicity in the mouse. Arch Toxicol. 1986;59(1):36–40. [DOI] [PubMed] [Google Scholar]
- 20.Wang SL, Lin CY, Guo YL, Lin LY, Chou WL, Chang LW. Infant exposure to polychlorinated dibenzo-p-dioxins, dibenzofurans and biphenyls (PCDD/Fs, PCBs)—correlation between prenatal and postnatal exposure. Chemosphere. 2004;54(10):1459–1473. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki G, Nakano M, Nakano S. Distribution of PCDDs/PCDFs and Co-PCBs in human maternal blood, cord blood, placenta, milk, and adipose tissue: dioxins showing high toxic equivalency factor accumulate in the placenta. Biosci Biotechnol Biochem. 2005;69(10):1836–1847. [DOI] [PubMed] [Google Scholar]
- 22.Tsukimori K, Morokuma S, Hori T, Takahashi K, Hirata T, Otera Y, et al. Characterization of placental transfer of polychlorinated dibenzo-p-dioxins, dibenzofurans and polychlorinated biphenyls in normal pregnancy. J Obstet Gynaecol Res. 2013;39(1):83–90. 10.1111/j.1447-0756.2012.01906.x [DOI] [PubMed] [Google Scholar]
- 23.Dallaire F, Dewailly É, Vézina C, Muckle G, Weber JP, Bruneau S, Ayotte P. Effect of prenatal exposure to polychlorinated biphenyls on incidence of acute respiratory infections in preschool inuit children. Environ Health Perspect. 2006;114(8):1301–1305. 10.1289/ehp.8683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glynn A, Thuvander A, Aune M, Johannisson A, Darnerud PO, Ronquist G, et al. Immune cell counts and risks of respiratory infections among infants exposed pre- and postnatally to organochlorine compounds: A prospective study. Environ Health. 2008;7:62 10.1186/1476-069X-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heilmann C, Grandjean P, Weihe P, Nielsen F, Budtz-Jorgensen E. Reduced antibody responses to vaccinations in children exposed to polychlorinated biphenyls. PLoS Med. 2006;3(8):e311 10.1371/journal.pmed.0030311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heilmann C, Budtz-Jorgensen E, Nielsen F, Heinzow B, Weihe P, Grandjean P. Serum concentrations of antibodies against vaccine toxoids in children exposed perinatally to immunotoxicants. Environ Health Perspect. 2010;118(10):1434–1438. 10.1289/ehp.1001975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hochstenbach K, van Leeuwen DM, Gmuender H, Gottschalk RW, Stolevik SB, Nygaard UC, et al. Toxicogenomic profiles in relation to maternal immunotoxic exposure and immune functionality in newborns. Toxicol Sci. 2012;129(2):315–324. 10.1093/toxsci/kfs214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jusko TA, De Roos AJ, Lee SY, Thevenet-Morrison K, Schwartz SM, Verner MA, et al. A birth cohort study of maternal and infant serum PCB-153 and DDE concentrations and responses to infant tuberculosis vaccination. Environ Health Perspect. 2016;124(6):813–821. 10.1289/ehp.1510101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vorderstrasse BA, Cundiff JA, Lawrence BP. Developmental exposure to the potent aryl hydrocarbon receptor agonist 2,3,7,8-tetrachlorodibenzo-p-dioxin impairs the cell-mediated immune response to infection with influenza a virus, but enhances elements of innate immunity. J Immunotoxicol. 2004;1(2):103–112. 10.1080/15476910490509244 [DOI] [PubMed] [Google Scholar]
- 30.Hogaboam JP, Moore AJ, Lawrence BP. The aryl hydrocarbon receptor affects distinct tissue compartments during ontogeny of the immune system. Toxicol Sci. 2008;102(1):160–170. 10.1093/toxsci/kfm283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vogel CF, Goth SR, Dong B, Pessah IN, Matsumura F. Aryl hydrocarbon receptor signaling mediates expression of indoleamine 2,3-dioxygenase. Biochem Biophys Res Commun. 2008;375(3):331–335. 10.1016/j.bbrc.2008.07.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bankoti J, Burnett A, Navarro S, Miller AK, Rase B, Shepherd DM. Effects of TCDD on the fate of naive dendritic cells. Toxicol Sci. 2010;115(2):422–434. 10.1093/toxsci/kfq063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bankoti J, Rase B, Simones T, Shepherd DM. Functional and phenotypic effects of AhR activation in inflammatory dendritic cells. Toxicol Appl Pharmacol. 2010;246(1–2):18–28. 10.1016/j.taap.2010.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quintana FJ, Murugaiyan G, Farez MF, Mitsdoerffer M, Tukpah AM, Burns EJ, et al. An endogenous aryl hydrocarbon receptor ligand acts on dendritic cells and T cells to suppress experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A. 2010;107(48):20768–20773. 10.1073/pnas.1009201107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin GB, Moore AJ, Head JL, Neumiller JJ, Lawrence BP. Aryl hydrocarbon receptor activation reduces dendritic cell function during influenza virus infection. Toxicol Sci. 2010;116(2):514–522. 10.1093/toxsci/kfq153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Simones T, Shepherd DM. Consequences of AhR activation in steady-state dendritic cells. Toxicol Sci. 2011;119(2):293–307. 10.1093/toxsci/kfq354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vogel CF, Wu D, Goth SR, Baek J, Lollies A, Domhardt R, et al. Aryl hydrocarbon receptor signaling regulates NF-kappaB RelB activation during dendritic-cell differentiation. Immunol Cell Biol. 2013;91(9):568–575. 10.1038/icb.2013.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin GB, Winans B, Martin KC, Lawrence BP. New insights into the role of the aryl hydrocarbon receptor in the function of CD11c(+) cells during respiratory viral infection. Eur J Immunol. 2014;44(6):1685–1698. 10.1002/eji.201343980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goudot C, Coillard A, Villani AC, Gueguen P, Cros A, Sarkizova S, et al. Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages. Immunity. 2017;47(3):582–596.e6. 10.1016/j.immuni.2017.08.016 [DOI] [PubMed] [Google Scholar]
- 40.Boule LA, Burke CG, Fenton BM, Thevenet-Morrison K, Jusko TA, Lawrence BP. Developmental activation of the AHR increases effector CD4+ T cells and exacerbates symptoms in autoimmune disease-prone Gnaq+/- mice. Toxicol Sci. 2015;148(2):555–566. 10.1093/toxsci/kfv203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mamalaki C, Elliott J, Norton T, Yannoutsos N, Townsend AR, Chandler P, et al. Positive and negative selection in transgenic mice expressing a T-cell receptor specific for influenza nucleoprotein and endogenous superantigen. Dev Immunol. 1993;3(3):159–174. 10.1155/1993/98015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun J, Braciale TJ. Role of T cell immunity in recovery from influenza virus infection. Curr Opin Virol. 2013;3(4):425–429. 10.1016/j.coviro.2013.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson JM, Jensen PE, Evavold BD. DO11.10 and OT-II T cells recognize a C-terminal ovalbumin 323–339 epitope. J Immunol. 2000;164(9):4706–4712. [DOI] [PubMed] [Google Scholar]
- 44.Thomas PG, Brown SA, Yue W, So J, Webby RJ, Doherty PC. An unexpected antibody response to an engineered influenza virus modifies CD8(+) T cell responses. Proc Natl Acad Sci U S A. 2006;103(8):2764–2769. 10.1073/pnas.0511185103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ueno H, Klechevsky E, Morita R, Aspord C, Cao T, Matsui T, et al. Dendritic cell subsets in health and disease. Immunol Rev. 2007;219:118–142. 10.1111/j.1600-065X.2007.00551.x [DOI] [PubMed] [Google Scholar]
- 46.Steinman RM, Idoyaga J. Features of the dendritic cell lineage. Immunol Rev. 2010;234(1):5–17. 10.1111/j.0105-2896.2009.00888.x [DOI] [PubMed] [Google Scholar]
- 47.Merad M, Sathe P, Helft J, Miller J, Mortha A. The dendritic cell lineage: Ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu Rev Immunol. 2013;31:563–604. 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ballesteros-Tato A, Leon B, Lund FE, Randall TD. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8(+) T cell responses to influenza. Nat Immunol. 2010;11(3):216–224. 10.1038/ni.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.del Rio ML, Rodriguez-Barbosa JI, Kremmer E, Forster R. CD103- and CD103+ bronchial lymph node dendritic cells are specialized in presenting and cross-presenting innocuous antigen to CD4+ and CD8+ T cells. J Immunol. 2007;178(11):6861–6866. [DOI] [PubMed] [Google Scholar]
- 50.Hao X, Kim TS, Braciale TJ. Differential response of respiratory dendritic cell subsets to influenza virus infection. J Virol. 2008;82(10):4908–4919. 10.1128/JVI.02367-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sung SS, Fu SM, Rose CE Jr., Gaskin F, Ju ST, Beaty SR. A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J Immunol. 2006;176(4):2161–2172. [DOI] [PubMed] [Google Scholar]
- 52.Poland A, Palen D, Glover E. Analysis of the four alleles of the murine aryl hydrocarbon receptor. Mol Pharmacol. 1994;46(5):915–921. [PubMed] [Google Scholar]
- 53.Legge KL, Braciale TJ. Accelerated migration of respiratory dendritic cells to the regional lymph nodes is limited to the early phase of pulmonary infection. Immunity. 2003;18(2):265–277. [DOI] [PubMed] [Google Scholar]
- 54.Forster R, Schubel A, Breitfeld D, Kremmer E, Renner-Muller I, Wolf E, et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99(1):23–33. [DOI] [PubMed] [Google Scholar]
- 55.Forster R, Braun A, Worbs T. Lymph node homing of T cells and dendritic cells via afferent lymphatics. Trends Immunol. 2012;33(6):271–280. 10.1016/j.it.2012.02.007 [DOI] [PubMed] [Google Scholar]
- 56.Braun A, Worbs T, Moschovakis GL, Halle S, Hoffmann K, Bolter J, et al. Afferent lymph-derived T cells and DCs use different chemokine receptor CCR7-dependent routes for entry into the lymph node and intranodal migration. Nat Immunol. 2011;12(9):879–887. 10.1038/ni.2085 [DOI] [PubMed] [Google Scholar]
- 57.Fedulov AV, Kobzik L. Allergy risk is mediated by dendritic cells with congenital epigenetic changes. Am J Respir Cell Mol Biol. 2011;44(3):285–292. 10.1165/rcmb.2009-0400OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mikhaylova L, Zhang Y, Kobzik L, Fedulov AV. Link between epigenomic alterations and genome-wide aberrant transcriptional response to allergen in dendritic cells conveying maternal asthma risk. PLoS One. 2013;8(8):e70387 10.1371/journal.pone.0070387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abdala-Valencia H, Berdnikovs S, Soveg FW, Cook-Mills JM. Alpha-tocopherol supplementation of allergic female mice inhibits development of CD11c+CD11b+ dendritic cells in utero and allergic inflammation in neonates. Am J Physiol Lung Cell Mol Physiol. 2014;307(6):L482–496. 10.1152/ajplung.00132.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdala-Valencia H, Soveg F, Cook-Mills JM. Gamma-tocopherol supplementation of allergic female mice augments development of CD11c+CD11b+ dendritic cells in utero and allergic inflammation in neonates. Am J Physiol Lung Cell Mol Physiol. 2016;310(8):L759–771. 10.1152/ajplung.00301.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu Y, Jin P, Peng J, Zhang X, Wong FS, Wen L. Different immunological responses to early-life antibiotic exposure affecting autoimmune diabetes development in NOD mice. J Autoimmun. 2016;72:47–56. 10.1016/j.jaut.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferrini M, Carvalho S, Cho YH, Postma B, Miranda Marques L, Pinkerton K, et al. Prenatal tobacco smoke exposure predisposes offspring mice to exacerbated allergic airway inflammation associated with altered innate effector function. Part Fibre Toxicol. 2017;14(1):30 10.1186/s12989-017-0212-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vorderstrasse BA, Dearstyne EA, Kerkvliet NI. Influence of 2,3,7,8-tetrachlorodibenzo-p-dioxin on the antigen-presenting activity of dendritic cells. Toxicol Sci. 2003;72(1):103–112. [DOI] [PubMed] [Google Scholar]
- 64.Schulz VJ, van Roest M, Bol-Schoenmakers M, van Duursen MB, van den Berg M, Pieters RH, et al. Aryl hydrocarbon receptor activation affects the dendritic cell phenotype and function during allergic sensitization. Immunobiology. 2013;218(8):1055–1062. 10.1016/j.imbio.2013.01.004 [DOI] [PubMed] [Google Scholar]
- 65.Joffre OP, Segura E, Savina A, Amigorena S. Cross-presentation by dendritic cells. Nat Rev Immunol. 2012;12(8):557–569. 10.1038/nri3254 [DOI] [PubMed] [Google Scholar]
- 66.Cruz FM, Colbert JD, Merino E, Kriegsman BA, Rock KL. The biology and underlying mechanisms of cross-presentation of exogenous antigens on MHC-I molecules. Annu Rev Immunol. 2017;35:149–176. 10.1146/annurev-immunol-041015-055254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Regnault A, Lankar D, Lacabanne V, Rodriguez A, Théry C, Rescigno M, et al. Fcγ receptor–mediated induction of dendritic cell maturation and major histocompatibility complex class I–restricted antigen presentation after immune complex internalization. J Exp Med. 1999;189(2):371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boross P, van Montfoort N, Stapels DA, van der Poel CE, Bertens C, Meeldijk J, et al. FcRgamma-chain ITAM signaling is critically required for cross-presentation of soluble antibody-antigen complexes by dendritic cells. J Immunol. 2014;193(11):5506–5514. 10.4049/jimmunol.1302012 [DOI] [PubMed] [Google Scholar]
- 69.Basu S, Binder RJ, Ramalingam T, Srivastava PK. CD91 is a common receptor for heat shock proteins gp96, hsp90, hsp70, and calreticulin. Immunity. 2001;14(3):303–313. [DOI] [PubMed] [Google Scholar]
- 70.Subramanian M, Hayes CD, Thome JJ, Thorp E, Matsushima GK, Herz J, et al. An AXL/LRP-1/RANBP9 complex mediates DC efferocytosis and antigen cross-presentation in vivo. J Clin Invest. 2014;124(3):1296–1308. 10.1172/JCI72051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kinner-Bibeau LB, Sedlacek AL, Messmer MN, Watkins SC, Binder RJ. HSPs drive dichotomous T-cell immune responses via DNA methylome remodelling in antigen presenting cells. Nat Commun. 2017;8:15648 10.1038/ncomms15648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hart JP, Gunn MD, Pizzo SV. A CD91-positive subset of CD11c+ blood dendritic cells: characterization of the APC that functions to enhance adaptive immune responses against CD91-targeted antigens. J Immunol. 2004;172(1):70–78. [DOI] [PubMed] [Google Scholar]
- 73.Mandl JN, Liou R, Klauschen F, Vrisekoop N, Monteiro JP, Yates AJ, et al. Quantification of lymph node transit times reveals differences in antigen surveillance strategies of naïve CD4+ and CD8+ T cells. Proc Natl Acad Sci U S A. 2012;109(44):18036–18041. 10.1073/pnas.1211717109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Henrickson SE, Perro M, Loughhead SM, Senman B, Stutte S, Quigley M, et al. Antigen availability determines CD8(+) T cell-dendritic cell interaction kinetics and memory fate decisions. Immunity. 2013;39(3):496–507. 10.1016/j.immuni.2013.08.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Macagno A, Napolitani G, Lanzavecchia A, Sallusto F. Duration, combination and timing: the signal integration model of dendritic cell activation. Trends Immunol. 2007;28(5):227–233. 10.1016/j.it.2007.03.008 [DOI] [PubMed] [Google Scholar]
- 76.Qian C, Cao X. Dendritic cells in the regulation of immunity and inflammation. Semin Immunol. 2017;35:3–11. 10.1016/j.smim.2017.12.002 [DOI] [PubMed] [Google Scholar]
- 77.Casado FL, Singh KP, Gasiewicz TA. Aryl hydrocarbon receptor activation in hematopoietic stem/progenitor cells alters cell function and pathway-specific gene modulation reflecting changes in cellular trafficking and migration. Mol Pharmacol. 2011;80(4):673–682. 10.1124/mol.111.071381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vorderstrasse BA, Lawrence BP. Protection against lethal challenge with Streptococcus pneumoniae is conferred by aryl hydrocarbon receptor activation but is not associated with an enhanced inflammatory response. Infect Immun. 2006;74(10):5679–5686. 10.1128/IAI.00837-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Teske S, Bohn AA, Hogaboam JP, Lawrence BP. Aryl hydrocarbon receptor targets pathways extrinsic to bone marrow cells to enhance neutrophil recruitment during influenza virus infection. Toxicol Sci. 2008;102(1):89–99. 10.1093/toxsci/kfm282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wheeler JL, Martin KC, Lawrence BP. Novel cellular targets of AhR underlie alterations in neutrophilic inflammation and inducible nitric oxide synthase expression during influenza virus infection. J Immunol. 2013;190(2):659–668. 10.4049/jimmunol.1201341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Loughman JA, Yarbrough ML, Tiemann KM, Hunstad DA. Local generation of kynurenines mediates inhibition of neutrophil chemotaxis by uropathogenic Escherichia coli. Infect Immun. 2016;84(4):1176–1183. 10.1128/IAI.01202-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aguilera-Montilla N, Chamorro S, Nieto C, Sanchez-Cabo F, Dopazo A, Fernandez-Salguero PM, et al. Aryl hydrocarbon receptor contributes to the MEK/ERK-dependent maintenance of the immature state of human dendritic cells. Blood. 2013;121(15):e108–117. 10.1182/blood-2012-07-445106 [DOI] [PubMed] [Google Scholar]
- 83.Schittenhelm L, Hilkens CM, Morrison VL. Beta2 integrins as regulators of dendritic cell, monocyte, and macrophage function. Front Immunol. 2017;8:1866 10.3389/fimmu.2017.01866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cao C, Lawrence DA, Li Y, Von Arnim CA, Herz J, Su EJ, et al. Endocytic receptor LRP together with tPA and PAI-1 coordinates Mac-1-dependent macrophage migration. EMBO J. 2006;25(9):1860–1870. 10.1038/sj.emboj.7601082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bergstrom SE, Bergdahl E, Sundqvist KG. A cytokine-controlled mechanism for integrated regulation of T-lymphocyte motility, adhesion and activation. Immunology. 2013;140(4):441–455. 10.1111/imm.12154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bergstrom SE, Uzunel M, Talme T, Bergdahl E, Sundqvist KG. Antigen-induced regulation of T-cell motility, interaction with antigen-presenting cells and activation through endogenous thrombospondin-1 and its receptors. Immunology. 2015;144(4):687–703. 10.1111/imm.12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rabiej VK, Pflanzner T, Wagner T, Goetze K, Storck SE, Eble JA, et al. Low density lipoprotein receptor-related protein 1 mediated endocytosis of beta1-integrin influences cell adhesion and cell migration. Exp Cell Res. 2016;340(1):102–115. 10.1016/j.yexcr.2015.11.020 [DOI] [PubMed] [Google Scholar]
- 88.Spijkers PP, da Costa Martins P, Westein E, Gahmberg CG, Zwaginga JJ, Lenting PJ. LDL-receptor-related protein regulates beta2-integrin-mediated leukocyte adhesion. Blood. 2005;105(1):170–177. 10.1182/blood-2004-02-0498 [DOI] [PubMed] [Google Scholar]
- 89.Chen M, Huang L, Shabier Z, Wang J. Regulation of the lifespan in dendritic cell subsets. Mol Immunol. 2007;44(10):2558–2565. 10.1016/j.molimm.2006.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen M, Wang J. Programmed cell death of dendritic cells in immune regulation. Immunol Rev. 2010;236:11–27. 10.1111/j.1600-065X.2010.00916.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhan Y, Chow KV, Soo P, Xu Z, Brady JL, Lawlor KE, et al. Plasmacytoid dendritic cells are short-lived: Reappraising the influence of migration, genetic factors and activation on estimation of lifespan. Sci Rep. 2016;6:25060 10.1038/srep25060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500. 10.1038/nrg.2016.59 [DOI] [PubMed] [Google Scholar]
- 93.Caton ML, Smith-Raska MR, Reizis B. Notch–RBP-J signaling controls the homeostasis of CD8(−) dendritic cells in the spleen. J Exp Med. 2007;204(7):1653–1664. 10.1084/jem.20062648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Barnden MJ, Allison J, Heath WR, Carbone FR. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol Cell Biol. 1998;76(1):34–40. 10.1046/j.1440-1711.1998.00709.x [DOI] [PubMed] [Google Scholar]
- 95.Walisser JA, Glover E, Pande K, Liss AL, Bradfield CA. Aryl hydrocarbon receptor-dependent liver development and hepatotoxicity are mediated by different cell types. Proc Natl Acad Sci U S A. 2005;102(49):17858–17863. 10.1073/pnas.0504757102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Neff-LaFord H, Teske S, Bushnell TP, Lawrence BP. Aryl hydrocarbon receptor activation during influenza virus infection unveils a novel pathway of IFN-gamma production by phagocytic cells. J Immunol. 2007;179(1):247–255. [DOI] [PubMed] [Google Scholar]
- 97.Quah BJC, Warren HS, Parish CR. Monitoring lymphocyte proliferation in vitro and in vivo with the intracellular fluorescent dye carboxyfluorescein diacetate succinimidyl ester. Nat Protoc. 2007;2:2049–2056. 10.1038/nprot.2007.296 [DOI] [PubMed] [Google Scholar]
- 98.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, et al. Fiji—an open source platform for biological image analysis. Nat Methods. 2012;9(7):676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lutz MB, Suri RM, Niimi M, Ogilvie AL, Kukutsch NA, Rossner S, et al. Immature dendritic cells generated with low doses of GM-CSF in the absence of IL-4 are maturation resistant and prolong allograft survival in vivo. Eur J Immunol. 2000;30(7):1813–1822. [DOI] [PubMed] [Google Scholar]
- 100.Son YI, Egawa S, Tatsumi T, Redlinger RE Jr., Kalinski P, Kanto T. A novel bulk-culture method for generating mature dendritic cells from mouse bone marrow cells. J Immunol Methods. 2002;262(1–2):145–157. [DOI] [PubMed] [Google Scholar]
- 101.Rivas-Caicedo A, Soldevila G, Fortoul TI, Castell-Rodriguez A, Flores-Romo L, Garcia-Zepeda EA. Jak3 is involved in dendritic cell maturation and CCR7-dependent migration. PLoS One. 2009;4(9):e7066 10.1371/journal.pone.0007066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DCs were evaluated prior to and up to 3 days after infection with IAV (HKx31). Flow cytometry was used to identify DC subsets as follows: conventional DCs (cDCs; CD11chiMHCIIhi cells), CD11b+ cDCs (CD11chiMHCIIhiCD11b+CD103- cells), CD103+ cDCs (CD11chiMHCIIhiCD103+CD11b- cells), and plasmacytoid DCs (pDCs; CD11cloMHCIIhi PDCA1+CD45R+ cells). (A, B) Representative dot plots depict the gating used to define cDCs, CD11b+ cDCs, CD103+ cDCs and pDCs in the lungs of vehicle (V) and TCDD (T) exposed offspring after gating to exclude doublets, dead cells, and autofluorescent cells. The percentage on the plot indicates the average percentage of the indicated DC subset 3 days after infection. cDC and pDC percentages are of all immune cells in the lung, whereas CD11b+ DCs and CD103+ DCs indicate the proportion of CD11chiMHCIIhi cells (cDCs). (C-F) The bar graphs show the number (±SEM) of the indicated DC population in the lung from naïve (day 0) or infected mice. At each point in time, all offspring within a group were from a separate dam, n = 6–9 mice per group per day. Day 0 data are representative of 4 independent experiments, day 1 data are representative of 3 independent experiments, day 3 data are representative of 6 independent experiments with similar results. Underlying data can be found in S1 Data.
(DOCX)
DCs were evaluated prior to and 3 days after infection with IAV (HKx31). Flow cytometry was used to identify DC subsets with the addition of annexin V and live/dead stains to detect apoptotic and dead cells. Specifically, annexin V binds phosphatidyl serine residues on the outer leaflet of exposed plasma membranes and live/dead covalently binds intracellular amines from cells with compromised membranes; the detection of cells double positive for these markers indicate dead cells. The bar graphs show the number (±SEM) of DC subsets that were double positive for Annexin V+LiveDead+ in the lung (A-C) and MLN (D-F) from naïve (day 0) or infected mice (day 3). At each point in time, all offspring within a group were from a separate dam, n = 6–9 mice per group per day. Underlying data can be found in S1 Data.
(DOCX)
Flow cytometry was used to identify DC subsets prior to and up to 3 days after infection with IAV (HKx31) as follows: conventional DCs (cDCs; CD11chi MHCIIhi cells), CD11b+ cDCs (CD11chiMHCIIhi CD11b+CD103- cells), CD103+ cDCs (CD11chiMHCIIhi CD103+CD11b- cells), and plasmacytoid DCs (pDCs; CD11cloMHCIIhi PDCA1+CD45R+ cells). In separate experiments, cells were further defined by expression of CCR7. Previous gating excluded doublets and autofluorescent cells. Percentage and number of DC subsets are indicated in the table. aPercentage of all immune cells in the lung. bPercentage of MLN cells. cPercentage of cDCs. For CCR7+ DC subsets, percentages are of cDC, CD11b+, CD103+, or pDC that were positive for CCR7. For CCR7+ DC subsets, all numbers are x103. All values ± SEM. An * indicates significance compared to vehicle (p ≤ 0.05).
(DOCX)
Mature BMDC were generated from bone marrow of naïve or DCs were enriched from the MLNs of IAV infected adult offspring from dams that were exposed to vehicle control or TCDD. The table shows the fold change of Ccr7, Ido1, and Cyp1a1 in DCs relative to their respective vehicle (BMDC) or uninfected (MLN DC) controls. Changes in gene expression were determined using the 2-ΔΔCT method. All offspring within a group are from a separate dam (BMDC, n = 6 mice per group; MLN DC, n = 12 mice per replicate, 3 replicates per group).
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.