Abstract
The gut microbiota represent a highly complex ecosystem comprised of approximately 1000 species that forms a mutualistic relationship with the human host. A critical attribute of the microbiota is high species diversity, which provides system robustness through overlapping and redundant metabolic capabilities. The gradual loss of bacterial diversity has been associated with a broad array of gut pathologies and diseases including malnutrition, obesity, diabetes and inflammatory bowel disease. We formulated an in silico community model of the gut microbiota by combining genome-scale metabolic reconstructions of 28 representative species to explore the relationship between species diversity and community growth. While the individual species offered a broad range of metabolic capabilities, communities optimized for maximal growth on simulated Western and high-fiber diets had low diversities and imbalances in short-chain fatty acid (SCFA) synthesis characterized by acetate overproduction. Community flux variability analysis performed with the 28-species model and a reduced 20-species model suggested that enhanced species diversity and more balanced SCFA production were achievable at suboptimal growth rates. We developed a simple method for constraining species abundances to sample the growth-diversity tradeoff and used the 20-species model to show that tradeoff curves for Western and high-fiber diets resembled Pareto-optimal surfaces. Compared to maximal growth solutions, suboptimal growth solutions were characterized by higher species diversity, more balanced SCFA synthesis and lower exchange rates of crossfed metabolites between more species. We hypothesized that modulation of crossfeeding relationships through host-microbiota interactions could be an important means for maintaining species diversity and suggest that community metabolic modeling approaches that allow multiobjective optimization of growth and diversity are needed for more realistic simulation of complex communities.
Author summary
The gut microbiota serve a critical role in maintaining a healthy state in the human host. The gut contains approximately 1,000 bacterial species that provide a wide range of metabolic capabilities including the breakdown of dietary compounds and the synthesis of useful metabolites. The robust function of the gut community is intimately connected to its diversity, both with respect to the number of species and the relative abundance of those species. The gradual loss of diversity is a key element of microbiota dysbiosis, which has been correlated with a wide range of health problems including inflammatory bowel disease. To investigate species diversity in the gut microbiota, we developed an in silico community model by combining genome-scale metabolic reconstructions of 28 representative species from the most abundant genera in the human gut. Our model predicted that maximal community growth produced low species diversity and no synthesis of the health-promoting metabolite butyrate. After reducing the community model to 20 species, we showed that suboptimal community growth allowed much higher species diversity and butyrate synthesis more consistent with in vivo studies. The model predicted that increased diversity could be achieved through modulation of metabolite crossfeeding relationships between species, an experimentally testable hypothesis.
Introduction
The gut microbiota comprise a highly complex ecosystem that has been characterized as an additional organ within the human host [1, 2]. The microbiota form a mutualistic relationship with the host, with saccharolytic species enzymatically degrading complex carbohydrates into fermentable sugars and fermentative species converting sugars and other available nutrients into a variety of absorbable metabolites [3, 4]. A particularly important function of the microbiota is to ferment dietary fiber into the short-chain fatty acids (SCFAs) acetate, butyrate and propionate [5, 6]. While significant variations are possible depending on diet, the molar ratio of these three SCFAs is approximately 60:20:20 [7]. SCFAs are consumed by host colonocytes as a primary energy source, with butyrate being the preferred SCFA but acetate probably supplying more energy to its higher concentration in vivo.
The gut microbiota consist of approximately 1,000 species [8] and 7,000 unique strains [2] in a typical human host. The two dominant phyla in healthy humans are Firmicutes and Bacteroidetes, which comprise more than 90% of the community [9, 10]. Other important but much less abundant phyla are Proteobacteria, Actinobacteria, Euryarchaeota and Verrucomicrobia as well as Eukaryota such as fungi [11, 12]. Metagenomic studies have shown wide variations in bacterial composition in healthy humans [13, 14], demonstrating that microbiota composition is an individual characteristic and an inadequate measure for assessing gut health across patient populations.
A hallmark of healthy gut communities is high diversity [15, 16], both in terms of the species present and the relative abundance of these species [17, 18]. An integrated gene catalog developed from 1,267 sequenced samples and comprising almost 10 million genes provides detailed information on the number of genes from over 700 gut genera [19]. The high diversity of the gut microbiota is demonstrated by 75 genera having relative gene counts (genes counts in that genus divided by total gene counts) of at least 0.1%. Numerous studies have shown a strong correlation between bacterial diversity and health/disease states, with long-term loss of diversity a key characteristic of dysbiosis [20–22]. The loss of bacterial diversity has been implicated in broad range of diseases including Clostridium difficile infections [23], inflammatory bowel and Crohn’s diseases [24, 25], obesity [26], diabetes [27], cardiovascular disease [28], rheumatoid arthritis [29], colorectal cancer [30], cystic fibrosis [31] and depression [32].
Genome-scale metabolic modeling has emerged as an important tool for computationally interrogating the metabolism of microbial communities. A number of alternative methods for combining metabolic reconstructions of single species into community models are now available. These methods are invariably based on growth rate maximization, either with regard to the species individually [33, 34] or the community as a whole [35–37]. Compared to other techniques, the recently developed SteadyCom method represents an important advance by performing community flux balance analysis (FBA) to determine the relative abundance of each species for maximal community growth while ensuring that all metabolites are properly balanced within each species and the community [38]. While the usual FBA objective of maximal growth has been experimentally demonstrated for individual bacteria such as Escherichia coli [39, 40], limited data is available to support the adoption of maximal growth as a community objective [41–43]. Indeed this objective has the potential to favor the fastest growing species and produce communities with low diversity that are inconsistent with healthy gut communities observed in vivo. In this study, we utilized the SteadyCom method to investigate the tradeoff between community growth and species diversity for in silico communities comprised of 20 and 28 representative gut species.
Materials and methods
Gut microbiota models
Semi-curated genome-scale metabolic reconstructions for representative species within the 28 most abundant genera [19] in the human gut were obtained from the Virtual Metabolic Human database (vmh.uni.lu) [44]. These models represented the five major bacterial phyla (Actinobacteria, 2 species; Bacteroidetes, 4 species; Firmicutes, 15 species; Fusobacteria, 1 species; Proteobacteria, 6 species), including 10 species from the highly prevalent Firmicutes order Clostridia (Table 1). The 28 genera covered almost 85% of reference genes by occurrence frequency according to a recent integrated catalog of the human gut microbiota [19]. Each species was constrained according to either a Western or high-fiber diet [44] and assigned a non-growth ATP maintenance (ATPM) value of 10 mmol/gDW/h, which is within the range reported for curated bacterial reconstructions. Because the species biomass equations were not curated [44], all species models used the same growth-dependent maintenance energy. The community metabolic model was constructed from the single-species reconstructions using the createCommModel function provided within the SteadyCom suite of MATLAB tools [38]. The community model accounted for 22,203 genes, 26,867 metabolites and 35,031 reactions within the 28 species as well as 354 uptake and 354 secretion reactions for the extracellular space.
Table 1. Properties of the 28 genome-scale metabolic reconstructions comprising the gut microbiota model.
| Number+ | Species* | Genes | Metabolites | Reactions |
|---|---|---|---|---|
| 1/1 | Bacteroides thetaiotaomicron | 841 | 1032 | 1370 |
| 2/2 | Clostridium clostridioforme | 1277 | 990 | 1352 |
| 3/3 | Faecalibacterium prausnitzii | 619 | 825 | 1008 |
| 4/4 | Eubacterium rectale | 700 | 941 | 1194 |
| 5/5 | Blautia wexlerae | 854 | 1006 | 1248 |
| 6/6 | Streptococcus salivarius | 604 | 868 | 1156 |
| 7/7 | Ruminococcus callidus | 564 | 912 | 1138 |
| 8/8 | Collinsella tanakaei | 691 | 959 | 1239 |
| 9/9 | Escherichia coli | 1193 | 1152 | 1795 |
| 10/10 | Roseburia inulinivorans | 663 | 903 | 1097 |
| 11/- | Lactobacillus mucosae | 582 | 852 | 1080 |
| 12/11 | Prevotella ruminicola | 695 | 916 | 1119 |
| 13/- | Alistipes finegoldii | 667 | 882 | 1139 |
| 14/- | Bifidobacterium longum | 529 | 729 | 890 |
| 15/12 | Enterobacter cloacae | 1330 | 1223 | 1776 |
| 16/13 | Klebsiella pneumoniae | 1395 | 1186 | 1801 |
| 17/14 | Coprococcus catus | 824 | 969 | 1211 |
| 18/15 | Veillonella atypica | 663 | 895 | 1120 |
| 19/- | Parabacteroides distasonis | 797 | 1049 | 1357 |
| 20/16 | Fusobacterium varium | 805 | 963 | 1228 |
| 21/17 | Haemophilus parainfluenzae | 656 | 973 | 1257 |
| 22/- | Pseudoflavonifractor capillosus | 678 | 885 | 1070 |
| 23/18 | Dorea longicatena | 689 | 851 | 1017 |
| 24/19 | Citrobacter amalonaticus | 1317 | 1218 | 1811 |
| 25/- | Phascolarctobacterium succinatutens | 560 | 832 | 1018 |
| 26/20 | Desulfovibrio desulfuricans | 717 | 1022 | 1279 |
| 27/- | Megasphaera elsdenii | 649 | 931 | 1172 |
| 28/- | Acidaminococcus fermentans | 644 | 903 | 1089 |
| Total 28 species model | 22203 | 26867 | 35031 | |
| Total 20 species model | 17097 | 19804 | 26216 |
*underlined species are not included in the 20 species community
+species numbers for 28 species/20 species communities
The uptake reactions in the extracellular space of the community model were constrained with the chosen diet as the union of all the maximum nutrient uptake rate constraints from the 28 species models. An Excel file with community uptake constraints for the four models (20 and 28 species; Western and high-fiber diets) is available in the Supporting Information (S1 File). With the exceptions noted below, the maximum nutrient uptake rates of each species were set equal to values defined by the chosen diet for that species. This approach ensured that each species would produce the same single-species growth rate with SteadyCom as obtained with standard flux balance analysis (FBA). Crossfeeding of all 21 amino acids and eight common metabolic byproducts (acetate, CO2, ethanol, formate, H2, D-lactate, L-lactate, succinate) was promoted by increasing the maximum nutrient uptake rates of these nutrients to 10 mmol/gDW/h. A single value was used due to the lack of metabolite- and species-dependent data for byproduct uptakes in the literature. While these constraints had no effect on single-species metabolism due to the extracellular constraints, they allowed species with different single-species growth rates to coexist through crossfeeding. The SCFAs butyrate and propionate were not allowed to be consumed under the assumption that crossfeeding of these two SCFAs was negligible compared to utilization by the host [45].
Community simulations
The community metabolic models were solved with the SteadyComCplex function within SteadyCom [38]. The IBM ILOG Cplex solver was used for linear program (LP) solution. SteadyCom performs community FBA by computing the relative abundance of each species for maximal community growth while ensuring that all metabolites are properly balanced within each species and the community. SteadyCom provides the capability to constrain the species abundances to explore various features of community behavior. Most simulations were performed with the default abundance constraints (lower bound zero, upper bound unity), which is referred to as the unconstrained case. Community flux variability analysis (FVA) was performed with respect to the species abundances using the SteadyComFVACplex function within SteadyCom. FVA showed that all maximal growth communities had unique species abundances. Using community FVA results for the 28-species community, species which could only coexist at 70% or less of the maximal community growth rate were eliminated to yield a 20-species community (Table 1). The 20-species model was used to explore the tradeoffs between community growth and diversity by constraining the species abundances with upper bounds computed from community FVA results obtained at different percentages of the maximal growth rate (see below).
Simulation results were analyzed with respect to the community growth rate, species abundances and diversity in the community, and the number and type of crossfeeding relationships. The growth rate and species abundances were direct outputs of SteadyCom. Species diversity was quantified using the Inverse Simpson equitability index [46–48] that accounted both for the number of participating species (e.g. richness) and the abundance of each species (e.g. evenness),
| (1) |
where N is the total number of species and pi ∈ [0, 1] is the relative abundance of species i, another direct output of SteadyCom. The diversity measure Dcom varied from if the community has a single participating member to unity if the all species participated and had the equal abundances. While the community models were built from metabolic reconstructions of particular species, the species name and associated genus were used interchangeably since the modeling goal was to achieve diversity in the genera.
To investigate the tradeoff between community growth and species diversity, maximum abundances in the 20-species community model were constrained using FVA results as follows,
| (2) |
where is the maximum abundance of species j calculated at a specified percentage of the maximal growth rate using FVA, r is a uniform random number in the closed interval [0 1] and is the upper abundance bound of species j. The maximum abundance of species j was divided by the sum of the maximum abundances of all species such that the scaled maximum abundances summed to unity. The bounds were randomized to more completely sample the growth-diversity tradeoff and were generated subject to the constraint to avoid producing structurally infeasible LP problems. The multiplier 2 was introduced because r was uniformly distributed with expected value 0.5. As compared to completely random bounds, Eq (2) allowed species with higher FVA abundances to have higher upper bounds on average. These bounds tended to constrain the solution such that higher diversity than the optimal solution was obtained, especially when FVA solutions at lower growth rates (e.g. 60% of maximal) were used. Eq (2) is heuristic in the sense that the calculated upper bounds do not ensure Pareto optimality [49, 50] of the resulting solutions in the growth-diversity space. More sophisticated methods that allow the calculation of the maximal diversity at a given growth rate would be required for this purpose. For each diet, a total of 900 cases were performed at FVA growth rates 60–99% of the maximal value, with more cases run at lower growth rates (150 cases at 60% and 65%, 125 cases at 70% and 75%, 100 cases at 80%, 85% and 90%, 25 cases at 95% and 99%) to adequately sample higher diversities. When was close to unity, SteadyCom often returned a solution where was outside the default tolerance of 10−4 specified within SteadyCom. These solutions were discarded to maintain the same accuracy of all suboptimal growth solution, substantially reducing the total number of cases (e.g. 900 to 595 for the Western diet).
Results
Single species provide a broad range of metabolic capabilities
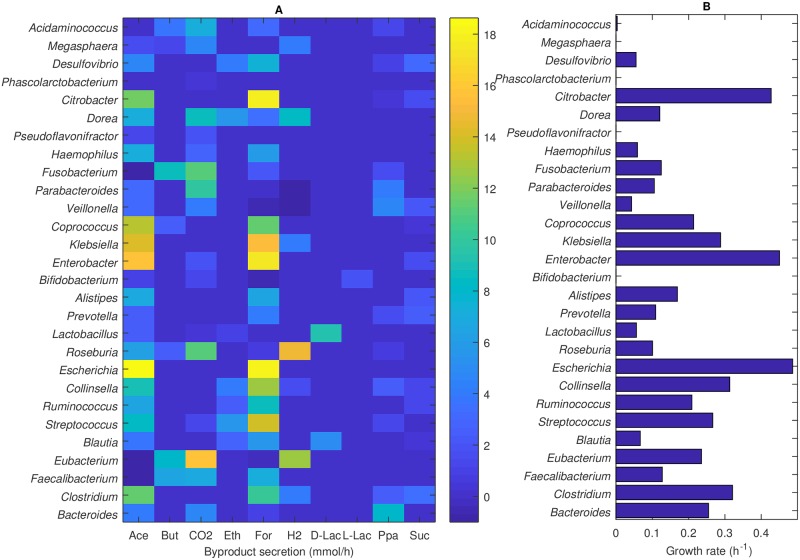
First the community model was constrained to investigate the metabolic capabilities of each species individually on the in silico Western diet. These single-species simulations were performed within SteadyCom by constraining the abundance of all other species to zero. The growth rate and the secretion rates of ten primary metabolic byproducts were determined for each species (Fig 1). The 28 species exhibited a wide range of growth rates, including three species (Escherichia, Enterobacter, Citrobacter) with growth rates exceeding 0.4 h−1 and four species (Bifidobacterium, Pseudoflavonifractor, Phascolarctobacterium, Megasphaera) with growth rates of zero for the ATP maintenance value of 10 mmol/gDW/h. While species with high individual growth rates were expected to have a competitive advantage in the in silico community, slower growing species had the possibility of coexisting by increasing their growth rates through metabolite crossfeeding. With regard to SCFA synthesis, 24 species secreted acetate, 7 species secreted butyrate including 3 major butyrate producers (Faecalibacterium, Eubacterium, Fusobacterium), and 13 species secreted propionate including 3 major propionate producers (Bacteroides, Veillonella, Parabacteroides). While the SCFA synthesis capabilities of the genera Bacteroides, Faecalibacterium and Eubacterium are well documented [51, 52], the other SCFA predictions also appear to be consistent with experimental studies [53, 54].
Fig 1. (A) Byproduct synthesis rates and (B) growth rates of the 28 species on an in silico Western diet.
The species are listed by their genera. The byproducts listed are acetate (ace), butyrate (but), carbon dioxide (CO2), ethanol (eth), formate (for), hydrogen (H2), D-lactate (D-lac), L-lactate (L-lac). propionate (ppa) and succinate (suc).
Optimal community growth leads to low species diversity
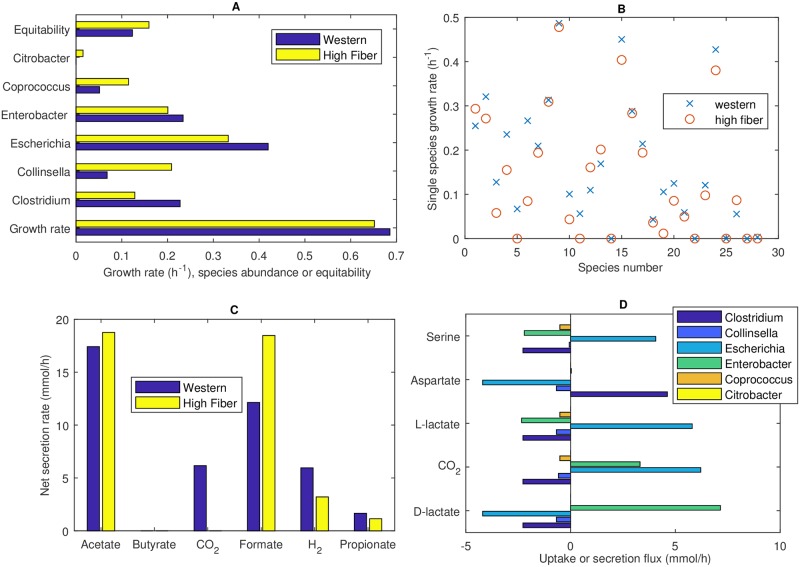
SteadyCom was used to determine the optimal growth rate and species abundances of the 28-member community on Western and high-fiber diets. The growth rate on each diet was similar (0.69 h−1 Western diet, 0.65 h−1 high-fiber diet) (Fig 2A) and appeared to be consistent with limited data available for in vivo gut community growth rates [55, 56]. Each community consisted of a small number of species, with only five species for the Western diet and six species for the high-fiber diet having non-zero abundances. The abundances of the 28 species correlated strongly with their single-species growth rates (Fig 2B; P < 10−4 for either diet), as would be expected from a community modeling methodology based on growth maximization. The dominant species included generally beneficial commensals from the genera Clostridium [57], Collinsella [58] and Coprococcus [59] but also represented several genera associated with inflammatory bowel disease (IBD) pathogenesis, including Escherichia [60], Enterobacter [61] and Citrobacter [62]. As a result of dominance by a few species, both communities exhibited low diversity, which has been correlated with a wide variety of gut pathologies [20–22]. Only slightly higher diversity was achieved for the Western diet when the ATP maintenance value of each species was tuned to the extent possible to achieve a uniform single-species growth rate of 0.2 h−1 (S1 Fig), demonstrating that the domination of particular species was partially attributable to their ability to more effectively exploit crossfed metabolites for growth.
Fig 2. Optimized 28-species communities for in silico Western and high-fiber diets.
(A) Growth rate (h−1), non-zero species abundances and equitability measure Dcom. (B) Single-species growth rates (h−1) with species numbers corresponding to Table 1. (C) Net synthesis rates of major metabolic byproducts including the three SCFAs acetate, butyrate and propionate. The byproducts ethanol, D-lactate, L-lactate and succinate are not shown because their net secretion rates were zero. (D) Uptake and secretion fluxes of the five metabolites most significantly crossfed between the participating species for the Western diet.
The community model was formulated to allow crossfeeding of all 21 amino acids and eight metabolic byproducts. Ethanol, D-lactate, L-lactate and succinate were crossfed to the extent that their net secretion rates (difference between the sum of the species synthesis and uptake rates) on either diet were zero (not shown). Both diets produced relatively high formate and acetate net secretion rates (Fig 2C), while the rates of CO2, H2, propionate were comparatively low and no butyrate was produced. Predicted ratios of the acetate:butyrate:propionate rates of 91:0:9 and 94:0:6 for the Western and high-fiber diets, respectively, were inconsistent with reported in vivo SFCA levels, which commonly are in the range of 60:20:20 [7]. The SCFA imbalance predicted for both diets was attributable to acetate production by all participating species, a lack of propionate producers (only Clostridium and Collinsella), and the absence of acetate consumers and butyrate producers. The five most significantly cross-fed metabolites were predicted to be the amino acids aspartate and serine and the byproducts D-lactate, L-lactate and CO2 (Fig 2D). Clostridium, Escherichia and Enterobacter formed a mutualistic three-species subcommunity with both large uptake and secretion rates of the five metabolites. By contrast, Collinsella and Coprococcus exhibited commensal interactions by solely consuming the secretion products while Citrobacter did not participate in the crossfeeding of these metabolites.
Suboptimal community growth offers the potential for enhanced species diversity
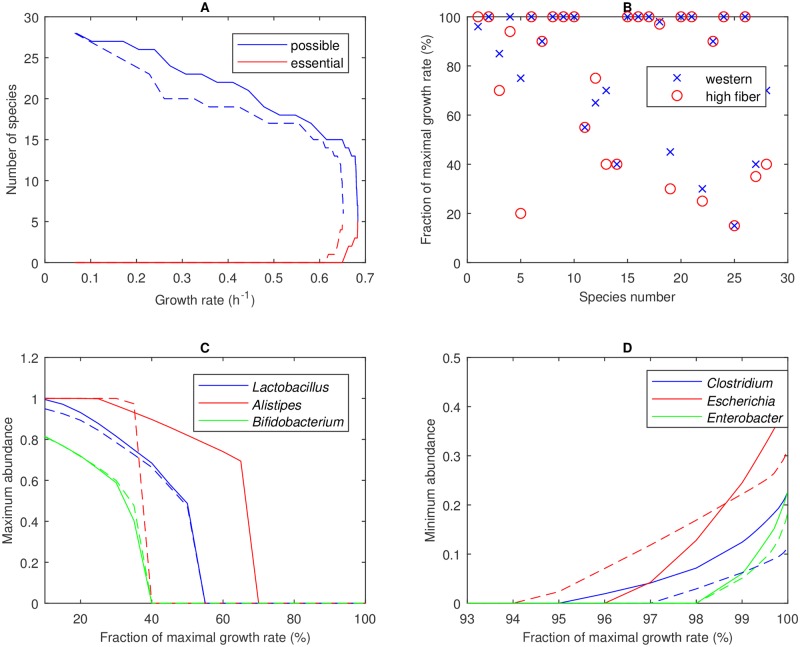
In silico communities optimized for maximal growth exhibited a lack of species diversity and SCFA imbalance characterized by low butyrate levels, both of which are strongly correlated to gut disease [5, 6, 20, 53]. To explore species diversity and SCFA synthesis at suboptimal growth rates, SteadyCom was used to perform community flux variability analysis (FVA) with respect to the species abundances. For growth rates between 10% and 99.99% of the maximal value, the number of species abundances that could be maximized to exceed 1% of the community abundances (possible species) or could be minimized to be exceed 1% of the community abundances (essential species) were determined. Uniqueness of the maximal growth communities was indicated by the convergence of the number of possible and essential species to a single value at the maximal growth rate (Fig 3A). FVA produced unique species abundances for all maximal growth communities S1 Folder. While the Western diet could only support five species at the maximal growth rate, the possible community size increased to 13 species at 99% and to 18 species at 80% of the maximal growth rate. Nine species were predicted to be incapable of coexistence at growth rates greater than 70% of the maximal value (Fig 3B), including saccharolytic Alistipes and the common probiotics Lactobacillus and Bifidobacterium (Fig 3C). No species were essential until 96% of the maximal growth rate and all five species that comprised the optimized community were not essential until 99.92% (Fig 3D). Similar results were obtained with the high-fiber diet. These results suggest that substantially enhanced diversity may be achievable at even marginally suboptimal growth rates.
Fig 3. Community FVA performed for the 28-species community with Western (solid lines) and high-fiber (dashed lines) diets.
(A) Number of possible species, defined as a species for which its abundance could be maximized to exceed 1%, and essential species, defined as a species for which its abundance could be minimized to be exceed 1%. (B) Community growth rate at which the abundance of each species could no longer exceed 1% when maximized with species numbers corresponding to Table 1. (C) Three representative species that could not coexist below 70% of the maximal growth rate. (D) Three representative species that had to be present in the community at high growth rates.
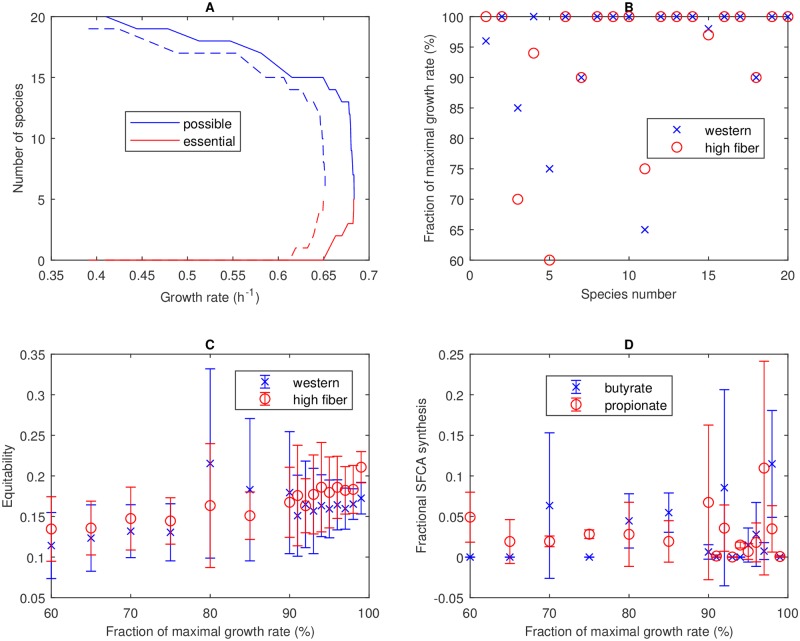
Based on the results in Fig 3B, the 28-species community was reduced to a 20-species community by removing eight species that could only coexist at growth rates less than 70% of the maximal value on both diets (Table 1). These species belonged to the genera Lactobacillus, Alistipes, Bifidobacterium, Parabacteroides, Pseudoflavonifractor, Phascolarctobacterium, Megasphaera and Acidaminococcus. This reduction in community size allowed a more efficient exploration of species diversity at growth rates above 70% of the maximal value, where the eliminated species were ensured not to coexist. Community FVA performed with the 20-species community suggested that 18 species could coexist at 80% and 15 species could coexist at 95% of the maximal growth rate on the Western diet (Fig 4A). Similar results were obtained for the high-fiber diet. By construction, all species could coexist at 70% of the maximal growth rate for at least one diet (Fig 4B).
Fig 4. Community FVA performed for the 20-species community with Western and high-fiber diets.
(A) Number of possible species and essential species for Western (solid lines) and high-fiber (dashed lines) diets. (B) Growth rate at which the abundance of each species could no longer exceed 1%. (C) Equitability measure Dcom calculated from the 40 FVA solutions at each growth rate. Symbols represent the mean and error bars represent the standard deviation. (D) Fractional butyrate and propionate synthesis, defined as the synthesis rate of the SCFA divided by the synthesis rate of all SCFAs (acetate, butyrate and propionate), calculated from the 40 FVA solutions at each growth rate for the Western diet. Symbols represent the mean and error bars represent the standard deviation.
We hypothesized that the tradeoff between community growth and species diversity could be described by a Pareto optimal surface [49, 50, 63–65] with increased diversity achieved only at the expense of reduced growth. However, community FVA did not necessarily provide a means to sample this tradeoff surface since diversity is not considered as part of the analysis. To investigate this issue, the 40 FVA solutions (20 solutions each for species abundance minimization and maximization) generated at each growth rate were used to compute the equitability measure Dcom. The average Dcom value for all growth rates and both diets was in the small range 0.11–0.22 (Fig 4C). Of the 2,720 FVA solutions tested, the largest Dcom was 0.43 and only nine cases produced Dcom > 0.35. In other words, the FVA solutions proved inadequate for generating high diversity, which was not surprising given that FVA solutions were computed by minimizing or maximizing a particular species abundance. This lack of diversity again translated to SCFA imbalance, with the average butyrate and propionate fractions over all 1,360 cases for the Western diet being 2% and 3%, respectively (Fig 4D).
Modulation of metabolite crossfeeding establishes tradeoff between growth and diversity
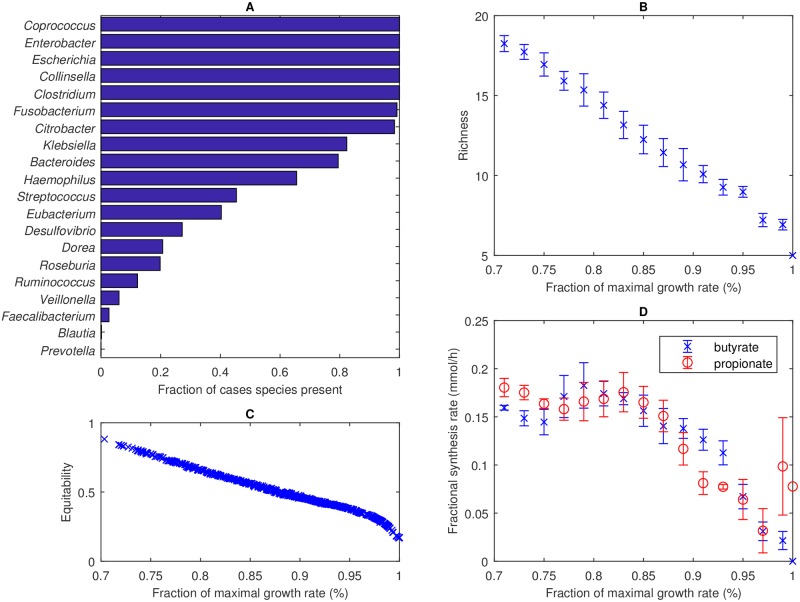
While useful for generating bounds on achievable species diversity and SCFA production, community FVA did not provide a direct means to investigate the tradeoff between community growth and diversity. However, we found that this tradeoff could be explored effectively by using FVA solutions at a particular growth rate as inputs to Eq 2 for calculation of an upper bound on each species abundance. When FVA solutions at low growth rates (e.g. 60% of maximal) were used for constraint calculation, the resulting solutions tended to have relatively high diversity and low growth. Using 575 simulation cases performed with the 20-species community and the Western diet, the fraction of cases in which the abundance of each species exceeded 1% was calculated as a measure of species fitness over a wide range of growth rates (Fig 5A). The five species that comprised the community at the optimal growth rate were present in all 575 communities. Citrobacter and the high butyrate producer Fusobacterium were present in over 98% of communities, while Klebsiella and the high propionate producer Bacteroides were present in at least 80% of communities. By contrast, the high butyrate producer Faecalibacterium and and high propionate producer Veillonella were present in no more than 6% of communities. Similar results were obtained with the high-fiber diet (S2A Fig).
Fig 5. Optimized 20-species communities for the Western diet with species abundance bounds calculated from community FVA solutions.
(A) Fraction of the 575 simulation cases for which the species abundance exceeded 1%. (B) Binned community richness, defined as the number of species with abundances greater than 1%. Results of the 575 cases were collected into 15 bins centered at growth rates ranging from 0.71 to 0.99 of the maximal value. The symbols represent the mean and error bars represent the standard deviation in each bin. (C) Equitability measure Dcom calculated for all 575 cases. (D) Binned fractional butyrate and propionate synthesis.
Results of the 575 Western diet cases were collected into 15 bins in the growth rate space. While the maximal growth solution consisted of only five species with non-zero abundances, the richness of suboptimal solutions was found to routinely exceed ten species (Fig 5B) with a maximum richness of 19 species achieved at 70.3% of the maximal growth rate. The binned richnesses showed small variabilities, partially due to the use of FVA solutions with zero maximum abundance values for some species. Similar richness trends were observed for the high-fiber diet, with 61% and 18% of the 568 simulation cases having richnesses of at least 10 and 15 species, respectively (S2B Fig).
The 575 Western diet cases produced a remarkably simple tradeoff between community growth and species diversity represented by a line (R2 = 0.996) for growth rates less than 97% of the optimal value (Fig 5C). We hesitate to refer this curve as “Pareto optimal” because our computational procedure does not ensure Pareto optimality of the calculated points. Regardless, this curve clearly showed that species diversity could only be achieved at the expense of community growth and visa versa. A very similar growth-equitability curve was generated with the high-fiber diet (S2C Fig).
Enhanced species diversity at suboptimal growth rates tended to produce more favorable ratios of SCFA net synthesis rates (Fig 5D). For the Western diet, the bin centered at 0.79 produced average butyrate and propionate fractions of 18% and 17%, respectively, which appeared to be more consistent than the maximal growth solution with the 20% values commonly reported for in vivo levels of these two SCFAs. The binned SCFA fractions showed small variabilities, consistent with predicted richness variations. Less favorable SCFA synthesis rates were predicted for the high-fiber diet (S2D Fig). While the propionate fractional rate averaged 18% over the range of 70–95% of the maximal growth rate, the butyrate fractional rate never averaged more 12% in any bin and reached a single-case maximum of 13%. Lower butyrate synthesis compared to the Western diet was attributed to reduced participation of the high butyrate producers Fusobacterium, Eubacterium and Faecalibacterium in the simulated communities.
While our method of imposing calculated bounds on the species abundances proved effective for investigating the growth-diversity tradeoff, such a direct mechanism for modulating species abundances is not biologically plausible. Several mechanisms for tuning community composition have been widely studied, including spatial structuring of the participating species in multispecies biofilms [66, 67] and the modulation of metabolite crossfeeding between species [68–70]. Because SteadyCom is based on the assumption of a homogeneous environment, the only mechanism available to tune community composition is modulation of nutrient uptake rates, including the uptake rates of crossfed metabolites. To further investigate how crossfeeding was modulated to achieve high diversity at suboptimal growth rates, we compared the optimal solution for the Western diet to the results in Fig 5 for the simulation cases binned at 79% of the optimal growth rate.
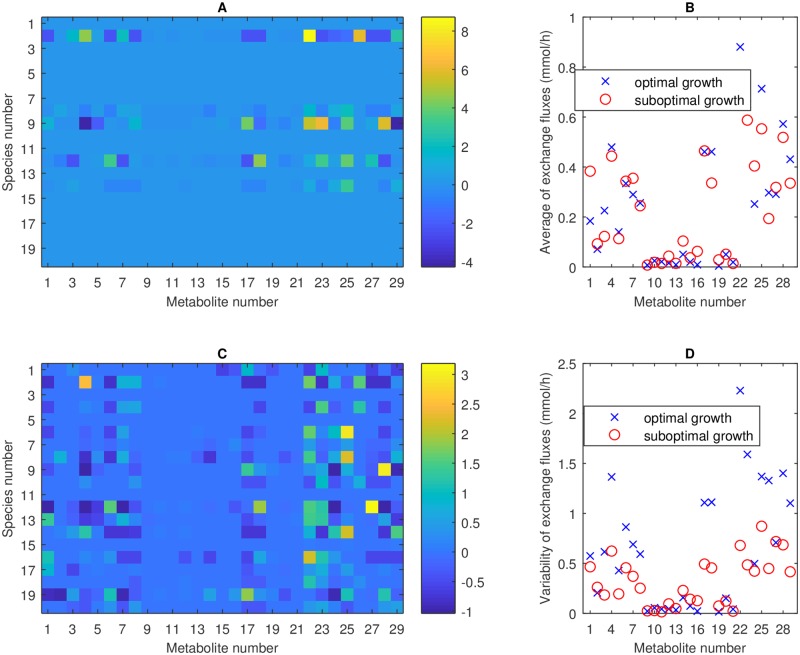
A heatmap of uptake/secretion rates of the 29 crossfed metabolites for each of the 20 species shows that the optimal solution was characterized by relatively high crossfeeding rates between a small number (five) of participating species (Fig 6A). Crossfed metabolites with the largest uptake/secretion rates were aspartate, serine, CO2, D-lactate, L-lactate (metabolites 4, 17, 23, 27, 28; see also Fig 2D). Comparable results for suboptimal growth at 79% of the optimal growth rate were generated by averaging the metabolite uptake/secretion rates across the 34 simulation cases within this bin. By contrast to the optimal case, suboptimal growth was characterized by relatively low crossfeeding rates between a large number (15.4 on average) of participating species (Fig 6B).
Fig 6. Metabolite crossfeeding for the 20-species community and the Western diet for the maximal growth case and 34 suboptimal growth cases binned around 79% of the maximal growth rate.
(A) Maximal growth heatmap of uptake (negative) and secretion (positive) rates in mmol/h for each crossfed metabolite (numbered in S1 Table) and each species (numbered in Table 1). (B) Suboptimal growth heatmap of uptake and secretion rates in mmol/h for each crossfed metabolite and each species averaged across 34 cases. (C) Absolute value of the exchange rate of each crossfed metabolite averaged across the 20 species and the 34 cases (for suboptimal growth). (D) Standard deviation of the exchange rates associated with Fig 6C.
To better understand the modulation of crossfeeding rates between optimal and suboptimal growth, we averaged the absolute values of the exchange (uptake and secretion) rates of each crossfed metabolite across the 20 species and the 34 simulation cases (for suboptimal growth). While individual crossfed metabolites differed with respect to their average exchange rates, the overall utilization of the 29 metabolites was similar (Fig 6C) with the average exchange rate across all metabolites being 0.25 mmol/h for maximal growth and 0.24 mmol/h for the suboptimal growth cases. By contrast, the standard deviation of the exchange rate across all metabolites was 0.64 mmol/h for maximal growth and 0.32 mmol/h for the suboptimal growth cases. Similar results were obtained for the high-fiber diet (S3 Fig). This analysis reinforced a central theme of this in silico study: optimal growth resulted in large metabolite exchange rates between few species while suboptimal growth was characterized by reduced metabolite exchange rates between many species.
Discussion
Metabolic modeling of the human gut microbiota has emerged as an important in silico tool for investigating community growth and composition as well as species interactions. While yielding useful insights into community behavior, previous gut microbiota models [38, 71–75] have been limited with respect to the number of species included and the metabolic interactions allowed. To our knowledge, the gut community models developed in this study represent the most complete descriptions to date both with respect to the number of species (e.g. 28 bacteria) and model size (e.g. 26,867 metabolites, 35,031 reactions). Our ability to generate and solve such large community models is directly attributable to the availability of gut bacteria reconstructions in the Virtual Metabolic Human database (https://vmh.uni.lu) [44] and the computational efficiency of the SteadyCom method [38]. The model database allowed the species to be chosen based on the most abundant genera in the gut [19] rather than by the availability of curated reconstructions, which remains limited to a few dozen species. The tradeoff for this unprecedented diversity of modeled species was that the reconstructions were only semi-curated and could not be expected to have the fidelity of fully curated models. Given the qualitative nature of our study focused on examining community growth and diversity, this limitation was deemed acceptable. Rather than requiring a priori specification of particular interactions between species, SteadyCom allowed arbitrary crossfeeding of secreted metabolites between all species. We limited crossfeeding to the 21 amino acids and 8 common byproducts, yielding 21,924 possible crossfeeding relationships for the 28-species community.
The 28 species included in the first gut community offered a wide diversity of metabolic capabilities, including the synthesis of short-chain fatty acids (SCFAs) used by host colonocytes as a primary energy source. As observed in healthy gut communities [5, 6, 76], SCFA synthesis was diversified across the community, with 24 species, 7 species and 13 species secreting acetate, butyrate and propionate, respectively. However, the maximal growth communities determined with SteadyCom consisted of only five species for the Western diet and six species for the high-fiber diet. The optimized communities were enriched in genera known to be overrepresented in inflammatory bowel disease, namely Escherichia [60], Enterobacter [61] and Citrobacter [62]. The communities also exhibited large imbalances in SCFA production, with over 90% of SCFA synthesis yielding acetate and no butyrate secreted, another hallmark of IBD [20–22]. Our simulation results suggest that maximal community growth unchecked by the host may evolve to disease states such as IBD.
We used SteadyCom to perform community flux variability analysis (FVA) as a means to determine limits on achievable species diversity and SCFA synthesis. FVA suggested that suboptimal community growth rates offered the potential for substantially enhanced diversity with 20 of the 28 species capable of coexisting at 70% of the optimal growth rate for at least one diet. Based on these results, we generated a reduced 20-species community by eliminating the eight species capable of coexisting only at growth rates less than 70% of the maximal values. The eight species eliminated represented several genera known to be beneficial for gut health, most notably Lactobacillus [77], Alistipes and [78], Bifidobacterium [79]. Due to their low predicted growth rates compared to other community members, these species may need to establish favorable metabolic niches along the intestine to robustly coexist [80–82]. While outside the scope this study due to the homogeneous assumption underlying SteadyCom, the effect of such spatial gradients would be interesting topic for future research.
FVA performed with the reduced 20-species community further demonstrated the potential for achieving high species richness (defined as the number of species with abundances of at least 1%) and equitability (see Eq 1) as well as balanced SCFA production at suboptimal growth rates. We developed a simple randomized method for using the FVA results to constrain species abundances in SteadyCom to achieve suboptimal growth rates and sample the growth-diversity tradeoff surface. A remarkably simple linear relationship between the community growth rate and species equitability was predicted, with high levels of diversity (richness ≥ 12 species, equitability ≥ 0.55) achievable at growth rates below 85% of the maximal value. Increased species diversity resulted in more balanced SCFA synthesis, with butyrate comprising 14–18% of total production. These predictions are consistent with known characteristics of healthy gut communities, suggesting that simulated suboptimal growth represents a “healthy” state and simulated maximal growth represents a “dysbiosis” state such as IBD.
We further analyzed our suboptimal growth solutions to determine how SteadyCom achieved enhanced diversity and how the host might promote diversity in vivo. In addition to modulating the uptake of available nutrient across species, SteadyCom tuned the secretion/uptake rates of crossfed amino acids and metabolic byproducts to enhance the growth rates of otherwise slower growing species. Compared to maximal growth, suboptimal solutions were characterized by lower secretion and uptake rates of crossfed metabolites between a larger number of species. These results suggest modulation of crossfeeding relationships is one possible mechanism available to the host for promoting diversity at the expense of growth. From a theoretical perspective, host-microbiota metabolic interactions might be viewed as a type of bilevel optimization problem with the microbiota attempting to achieve maximal community growth and the host modulating the gut environment to maximize species diversity. Gut diseases such as IBD might result from the host “losing the battle” due to inflexibilities resulting from poor diet and/or sudden loss of diversity due to antibiotic treatment. While a few metabolic modeling methods that address diversity have been proposed [83–85], additional modeling tools that directly address the growth-diversity tradeoff in microbial communities are needed.
As has been reported in numerous in vivo studies [86–89], we expected the in silico high-fiber diet to promote species diversity and enhance butyrate synthesis as compared to the Western diet. Instead our model predicted that diet had little effect on the growth-diversity tradeoff and high fiber actually resulted in reduced butyrate levels. These discrepancies could reflect insufficient numbers of fiber-degrading and butyrate-producing species in our simulated communities. Indeed only four members (Bacteroides, Prevotella, Alistipes, Desulfovibrio) of the 28-species community exhibited faster single-species growth rates on the high-fiber diet, and two of these species were removed to generate the 20-species community. The communities contained only three major butyrate producers, with one species (Faecalibacterium) being among the least competitive members of the 20-species community. The computational efficiency of SteadyCom allows the construction of larger community models with more representation of fiber-degrading and butyrate-producing species.
Supporting information
(A) Growth rate (h−1), non-zero species abundances and equitability measure Dcom. (B) Single-species growth rates (h−1) with species numbers corresponding to Table 1. (C) Net synthesis rates of major metabolic byproducts including the three SCFAs acetate, butyrate and propionate. The byproducts ethanol, D-lactate, L-lactate and succinate are not shown because their net secretion rates were zero. (D) Uptake and secretion fluxes of the five metabolites most significantly crossfed between the participating species.
(TIF)
(A) Fraction of the 568 simulation cases for which the species abundance exceeded 1%. (B) Binned community richness, defined as the number of species with abundances greater than 1%. Results of the 568 cases were collected into 15 bins centered at growth rates ranging from 0.71 to 0.99 of the maximal value. The symbols represent the mean and error bars represent the standard deviation in each bin. (C) Equitability measure Dcom calculated for all 568 cases. (D) Binned fractional butyrate and propionate synthesis.
(TIF)
(A) Maximal growth heatmap of uptake (negative) and secretion (positive) rates in mmol/h for each crossfed metabolite (numbered in S1 Table) and each species (numbered in Table 1). (B) Suboptimal growth heatmap of uptake and secretion rates in mmol/h for each crossfed metabolite and each species averaged across 36 cases. (C) Absolute value of the exchange rate of each crossfed metabolite averaged across the 20 species and the 36 cases (for suboptimal growth). (D) Standard deviation of the exchange rates associated with S3 FigC.
(TIF)
(PDF)
Community uptake constraints for the four models (20 and 28 species; Western and high-fiber diets).
(XLSX)
Matlab codes and data files used for generating results.
(ZIP)
Acknowledgments
The authors wish to thank Prof. Ines Thiele and Dr. Almut Heinken (University of Luxembourg) for providing access to the genome-scale metabolic reconstructions. The authors also wish to thank Prof. Costas Maranas and Dr. Siu Hung (Joshua) Chan (Pennsylvania State University) for assistance with SteadyCom.
Data Availability
SteadyCom is freely available at http://www.maranasgroup.com/software.htm. The MATLAB simulation codes used in this study are included as supplementary materials available from the journal website.
Funding Statement
The authors wish to acknowledge National Institutes of Health (Award U01EB019416) and National Science Foundation (Award CBET 1511346) for partial financial support of this research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funds from these grants were used to cover the costs to publish in open access.
References
- 1. Sommer F, Bäckhed F. The gut microbiota–masters of host development and physiology. Nature Reviews Microbiology. 2013;11(4):227 10.1038/nrmicro2974 [DOI] [PubMed] [Google Scholar]
- 2. Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: an integrative view. Cell. 2012;148(6):1258–1270. 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thompson JA, Oliveira RA, Xavier KB. Chemical conversations in the gut microbiota. Gut microbes. 2016;7(2):163–170. 10.1080/19490976.2016.1145374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell host & microbe. 2011;10(4):336–347. 10.1016/j.chom.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Byrne C, Chambers E, Morrison D, Frost G. The role of short chain fatty acids in appetite regulation and energy homeostasis. International journal of obesity. 2015;39(9):1331–1338. 10.1038/ijo.2015.84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7(3):189–200. 10.1080/19490976.2015.1134082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. Journal of lipid research. 2013;54(9):2325–2340. 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shreiner AB, Kao JY, Young VB. The gut microbiome in health and in disease. Current opinion in gastroenterology. 2015;31(1):69 10.1097/MOG.0000000000000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krajmalnik-Brown R, Ilhan ZE, Kang DW, DiBaise JK. Effects of gut microbes on nutrient absorption and energy regulation. Nutrition in Clinical Practice. 2012;27(2):201–214. 10.1177/0884533611436116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Warner D, George C. Erythrocyte sedimentation rate and related factors in end-stage renal failure. Nephron. 1991;57(2):248–248. 10.1159/000186266 [DOI] [PubMed] [Google Scholar]
- 11. Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nature Reviews Microbiology. 2011;9(4):279 10.1038/nrmicro2540 [DOI] [PubMed] [Google Scholar]
- 12. Donaldson GP, Lee SM, Mazmanian SK. Gut biogeography of the bacterial microbiota. Nature Reviews Microbiology. 2016;14(1):20 10.1038/nrmicro3552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, et al. Genomic variation landscape of the human gut microbiome. Nature. 2013;493(7430):45 10.1038/nature11711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome medicine. 2016;8(1):51 10.1186/s13073-016-0307-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Greenhalgh K, Meyer KM, Aagaard KM, Wilmes P. The human gut microbiome in health: establishment and resilience of microbiota over a lifetime. Environmental microbiology. 2016;18(7):2103–2116. 10.1111/1462-2920.13318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2015; p. gutjnl–2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207 10.1038/nature11234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, et al. An integrated catalog of reference genes in the human gut microbiome. Nature biotechnology. 2014;32(8):834 10.1038/nbt.2942 [DOI] [PubMed] [Google Scholar]
- 20. Lin L, Zhang J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC immunology. 2017;18(1):2 10.1186/s12865-016-0187-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thursby E, Juge N. Introduction to the human gut microbiota. Biochemical Journal. 2017;474(11):1823–1836. 10.1042/BCJ20160510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Selber-Hnatiw S, Rukundo B, Ahmadi M, Akoubi H, Al-Bizri H, Aliu AF, et al. Human gut microbiota: toward an ecology of disease. Frontiers in microbiology. 2017;8:1265 10.3389/fmicb.2017.01265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ross CL, Spinler JK, Savidge TC. Structural and functional changes within the gut microbiota and susceptibility to Clostridium difficile infection. Anaerobe. 2016;41:37–43. 10.1016/j.anaerobe.2016.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ahmed I, Roy BC, Khan SA, Septer S, Umar S. Microbiome, Metabolome and Inflammatory Bowel Disease. Microorganisms. 2016;4(2):20 10.3390/microorganisms4020020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wright EK, Kamm MA, Teo SM, Inouye M, Wagner J, Kirkwood CD. Recent advances in characterizing the gastrointestinal microbiome in Crohn’s disease: a systematic review. Inflammatory bowel diseases. 2015;21(6):1219–1228. 10.1097/MIB.0000000000000382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dahiya DK, Puniya M, Shandilya UK, Dhewa T, Kumar N, Kumar S, et al. Gut microbiota modulation and its relationship with obesity using prebiotic fibers and probiotics: a review. Frontiers in microbiology. 2017;8:563 10.3389/fmicb.2017.00563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang WW, Kitai T, Hazen SL. Gut microbiota in cardiovascular health and disease. Circulation research. 2017;120(7):1183–1196. 10.1161/CIRCRESAHA.117.309715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Al Khodor S, Reichert B, Shatat IF. The Microbiome and Blood Pressure: Can Microbes Regulate Our Blood Pressure? Frontiers in pediatrics. 2017;5:138 10.3389/fped.2017.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Forbes JD, Van Domselaar G, Bernstein CN. The gut microbiota in immune-mediated inflammatory diseases. Frontiers in microbiology. 2016;7:1081 10.3389/fmicb.2016.01081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dulal S, Keku TO. Gut microbiome and colorectal adenomas. Cancer journal (Sudbury, Mass). 2014;20(3):225 10.1097/PPO.0000000000000050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Burke D, Fouhy F, Harrison M, Rea MC, Cotter PD, O’Sullivan O, et al. The altered gut microbiota in adults with cystic fibrosis. BMC microbiology. 2017;17(1):58 10.1186/s12866-017-0968-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dash S, Clarke G, Berk M, Jacka FN. The gut microbiome and diet in psychiatry: focus on depression. Current opinion in psychiatry. 2015;28(1):1–6. 10.1097/YCO.0000000000000117 [DOI] [PubMed] [Google Scholar]
- 33. Stolyar S, Van Dien S, Hillesland KL, Pinel N, Lie TJ, Leigh JA, et al. Metabolic modeling of a mutualistic microbial community. Molecular systems biology. 2007;3(1):92 10.1038/msb4100131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Heinken A, Thiele I. Systematic prediction of health-relevant human-microbial co-metabolism through a computational framework. Gut Microbes. 2015;6(2):120–130. 10.1080/19490976.2015.1023494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Khandelwal RA, Olivier BG, Röling WF, Teusink B, Bruggeman FJ. Community flux balance analysis for microbial consortia at balanced growth. PloS one. 2013;8(5):e64567 10.1371/journal.pone.0064567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shoaie S, Ghaffari P, Kovatcheva-Datchary P, Mardinoglu A, Sen P, Pujos-Guillot E, et al. Quantifying diet-induced metabolic changes of the human gut microbiome. Cell metabolism. 2015;22(2):320–331. 10.1016/j.cmet.2015.07.001 [DOI] [PubMed] [Google Scholar]
- 37. Zomorrodi AR, Maranas CD. OptCom: a multi-level optimization framework for the metabolic modeling and analysis of microbial communities. PLoS computational biology. 2012;8(2):e1002363 10.1371/journal.pcbi.1002363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chan SHJ, Simons MN, Maranas CD. SteadyCom: Predicting microbial abundances while ensuring community stability. PLoS computational biology. 2017;13(5):e1005539 10.1371/journal.pcbi.1005539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ibarra RU, Edwards JS, Palsson BO. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420(6912):186 10.1038/nature01149 [DOI] [PubMed] [Google Scholar]
- 40. Fong SS, Marciniak JY, Palsson BØ. Description and interpretation of adaptive evolution of Escherichia coli K-12 MG1655 by using a genome-scale in silico metabolic model. Journal of bacteriology. 2003;185(21):6400–6408. 10.1128/JB.185.21.6400-6408.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gottstein W, Olivier BG, Bruggeman FJ, Teusink B. Constraint-based stoichiometric modelling from single organisms to microbial communities. Journal of the Royal Society Interface. 2016;13(124):20160627 10.1098/rsif.2016.0627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perez-Garcia O, Lear G, Singhal N. Metabolic network modeling of microbial interactions in natural and engineered environmental systems. Frontiers in microbiology. 2016;7:673 10.3389/fmicb.2016.00673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Biggs MB, Medlock GL, Kolling GL, Papin JA. Metabolic network modeling of microbial communities. Wiley Interdisciplinary Reviews: Systems Biology and Medicine. 2015;7(5):317–334. 10.1002/wsbm.1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Magnúsdóttir S, Heinken A, Kutt L, Ravcheev DA, Bauer E, Noronha A, et al. Generation of genome-scale metabolic reconstructions for 773 members of the human gut microbiota. Nature biotechnology. 2017;35(1):81 10.1038/nbt.3703 [DOI] [PubMed] [Google Scholar]
- 45. Kettle H, Louis P, Holtrop G, Duncan SH, Flint HJ. Modelling the emergent dynamics and major metabolites of the human colonic microbiota. Environmental microbiology. 2015;17(5):1615–1630. 10.1111/1462-2920.12599 [DOI] [PubMed] [Google Scholar]
- 46. Li K, Bihan M, Yooseph S, Methe BA. Analyses of the microbial diversity across the human microbiome. PloS one. 2012;7(6):e32118 10.1371/journal.pone.0032118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johnson KVA, Burnet PW. Microbiome: Should we diversify from diversity? Gut microbes. 2016;7(6):455–458. 10.1080/19490976.2016.1241933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hill TC, Walsh KA, Harris JA, Moffett BF. Using ecological diversity measures with bacterial communities. FEMS microbiology ecology. 2003;43(1):1–11. 10.1111/j.1574-6941.2003.tb01040.x [DOI] [PubMed] [Google Scholar]
- 49. Byrne D, Dumitriu A, Segrè D. Comparative multi-goal tradeoffs in systems engineering of microbial metabolism. BMC systems biology. 2012;6(1):127 10.1186/1752-0509-6-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Towbin BD, Korem Y, Bren A, Doron S, Sorek R, Alon U. Optimality and sub-optimality in a bacterial growth law. Nature communications. 2017;8:14123 10.1038/ncomms14123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rios-Covian D, Salazar N, Gueimonde M, de los Reyes-Gavilan CG. Shaping the metabolism of intestinal Bacteroides population through diet to improve human health. Frontiers in microbiology. 2017;8:376 10.3389/fmicb.2017.00376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Frontiers in microbiology. 2016;7:979 10.3389/fmicb.2016.00979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. MBio. 2014;5(2):e00889–14. 10.1128/mBio.00889-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Durant J, Nisbet D, Ricke S. Comparison of Batch Culture Growth and Fermentation of a PoultryVeillonellaIsolate and SelectedVeillonellaSpecies Grown in a Defined Medium. Anaerobe. 1997;3(6):391–397. 10.1006/anae.1997.0129 [DOI] [PubMed] [Google Scholar]
- 55. Korem T, Zeevi D, Suez J, Weinberger A, Avnit-Sagi T, Pompan-Lotan M, et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science. 2015;349(6252):1101–1106. 10.1126/science.aac4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vandeputte D, Falony G, Vieira-Silva S, Tito RY, Joossens M, Raes J. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut. 2015; p. gutjnl–2015. 10.1136/gutjnl-2015-309618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fite A, Macfarlane S, Furrie E, Bahrami B, Cummings JH, Steinke DT, et al. Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. Journal of clinical microbiology. 2013;51(3):849–856. 10.1128/JCM.02574-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pascal V, Pozuelo M, Borruel N, Casellas F, Campos D, Santiago A, et al. A microbial signature for Crohn’s disease. Gut. 2017; p. gutjnl–2016. 10.1136/gutjnl-2016-313235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Shaw KA, Bertha M, Hofmekler T, Chopra P, Vatanen T, Srivatsa A, et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome medicine. 2016;8(1):75 10.1186/s13073-016-0331-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rhodes JM. The role of Escherichia coli in inflammatory bowel disease. Gut. 2007;56(5):610–612. 10.1136/gut.2006.111872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kaakoush NO, Day AS, Huinao KD, Leach ST, Lemberg DA, Dowd SE, et al. Microbial dysbiosis in pediatric patients with Crohn’s disease. Journal of clinical microbiology. 2012;50(10):3258–3266. 10.1128/JCM.01396-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Saleh M, Elson CO. Experimental inflammatory bowel disease: insights into the host-microbiota dialog. Immunity. 2011;34(3):293–302. 10.1016/j.immuni.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Higuera C, Villaverde AF, Banga JR, Ross J, Morán F. Multi-criteria optimization of regulation in metabolic networks. PLoS one. 2012;7(7):e41122 10.1371/journal.pone.0041122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shoval O, Sheftel H, Shinar G, Hart Y, Ramote O, Mayo A, et al. Evolutionary trade-offs, Pareto optimality, and the geometry of phenotype space. Science. 2012; p. 1217405. [DOI] [PubMed] [Google Scholar]
- 65. Rabbers I, van Heerden JH, Nordholt N, Bachmann H, Teusink B, Bruggeman FJ. Metabolism at evolutionary optimal States. Metabolites. 2015;5(2):311–343. 10.3390/metabo5020311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nature reviews microbiology. 2004;2(2):95 10.1038/nrmicro821 [DOI] [PubMed] [Google Scholar]
- 67. Tolker-Nielsen T, Molin S. Spatial organization of microbial biofilm communities. Microbial ecology. 2000;40(2):75–84. [DOI] [PubMed] [Google Scholar]
- 68. El Aidy S, Van den Abbeele P, Van de Wiele T, Louis P, Kleerebezem M. Intestinal colonization: how key microbial players become established in this dynamic process. Bioessays. 2013;35(10):913–923. 10.1002/bies.201300073 [DOI] [PubMed] [Google Scholar]
- 69. Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environmental microbiology. 2007;9(5):1101–1111. 10.1111/j.1462-2920.2007.01281.x [DOI] [PubMed] [Google Scholar]
- 70. Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environmental microbiology. 2017;19(1):29–41. 10.1111/1462-2920.13589 [DOI] [PubMed] [Google Scholar]
- 71. Bauer E, Thiele I. From Network Analysis to Functional Metabolic Modeling of the Human Gut Microbiota. MSystems. 2018;3(3):e00209–17. 10.1128/mSystems.00209-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. van der Ark KC, van Heck RG, Dos Santos VAM, Belzer C, de Vos WM. More than just a gut feeling: constraint-based genome-scale metabolic models for predicting functions of human intestinal microbes. Microbiome. 2017;5(1):78 10.1186/s40168-017-0299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bauer E, Zimmermann J, Baldini F, Thiele I, Kaleta C. BacArena: Individual-based metabolic modeling of heterogeneous microbes in complex communities. PLoS computational biology. 2017;13(5):e1005544 10.1371/journal.pcbi.1005544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Mendes-Soares H, Mundy M, Soares LM, Chia N. MMinte: an application for predicting metabolic interactions among the microbial species in a community. BMC bioinformatics. 2016;17(1):343 10.1186/s12859-016-1230-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shoaie S, Karlsson F, Mardinoglu A, Nookaew I, Bordel S, Nielsen J. Understanding the interactions between bacteria in the human gut through metabolic modeling. Scientific reports. 2013;3:2532 10.1038/srep02532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, et al. Gut microbiota functions: metabolism of nutrients and other food components. European journal of nutrition. 2017; p. 1–24. 10.1007/s00394-017-1445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mikelsaar M, Sepp E, Štšepetova J, Songisepp E, Mändar R. Biodiversity of intestinal lactic acid bacteria in the healthy population In: Advances in Microbiology, Infectious Diseases and Public Health. Springer; 2016. p. 1–64. [DOI] [PubMed] [Google Scholar]
- 78. Dziarski R, Park SY, Kashyap DR, Dowd SE, Gupta D. Pglyrp-regulated gut microflora Prevotella falsenii, Parabacteroides distasonis and Bacteroides eggerthii enhance and Alistipes finegoldii attenuates colitis in mice. PloS one. 2016;11(1):e0146162 10.1371/journal.pone.0146162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bottacini F, van Sinderen D, Ventura M. Omics of bifidobacteria: research and insights into their health-promoting activities. Biochemical Journal. 2017;474(24):4137–4152. 10.1042/BCJ20160756 [DOI] [PubMed] [Google Scholar]
- 80. Corcoran B, Stanton C, Fitzgerald G, Ross R. Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Applied and environmental microbiology. 2005;71(6):3060–3067. 10.1128/AEM.71.6.3060-3067.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Maldonado-Gómez MX, Martínez I, Bottacini F, O’Callaghan A, Ventura M, van Sinderen D, et al. Stable engraftment of Bifidobacterium longum AH1206 in the human gut depends on individualized features of the resident microbiome. Cell host & microbe. 2016;20(4):515–526. 10.1016/j.chom.2016.09.001 [DOI] [PubMed] [Google Scholar]
- 82. Pereira FC, Berry D. Microbial nutrient niches in the gut. Environmental microbiology. 2017;19(4):1366–1378. 10.1111/1462-2920.13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Budinich M, Bourdon J, Larhlimi A, Eveillard D. A multi-objective constraint-based approach for modeling genome-scale microbial ecosystems. PloS one. 2017;12(2):e0171744 10.1371/journal.pone.0171744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Van Hoek MJ, Merks RM. Emergence of microbial diversity due to cross-feeding interactions in a spatial model of gut microbial metabolism. BMC systems biology. 2017;11(1):56 10.1186/s12918-017-0430-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Tzamali E, Poirazi P, Tollis IG, Reczko M. A computational exploration of bacterial metabolic diversity identifying metabolic interactions and growth-efficient strain communities. BMC systems biology. 2011;5(1):167 10.1186/1752-0509-5-167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Requena T, Martinez-Cuesta MC, Peláez C. Diet and microbiota linked in health and disease. Food & Function. 2018;. [DOI] [PubMed]
- 87. Simpson HL, Campbell BJ. dietary fibre–microbiota interactions. Alimentary pharmacology & therapeutics. 2015;42(2):158–179. 10.1111/apt.13248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Havenaar R. Intestinal health functions of colonic microbial metabolites: a review. Beneficial microbes. 2011;2(2):103–114. 10.3920/BM2011.0003 [DOI] [PubMed] [Google Scholar]
- 89. McNabney SM, Henagan TM. Short Chain Fatty Acids in the Colon and Peripheral Tissues: A Focus on Butyrate, Colon Cancer, Obesity and Insulin Resistance. Nutrients. 2017;9(12):1348 10.3390/nu9121348 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Growth rate (h−1), non-zero species abundances and equitability measure Dcom. (B) Single-species growth rates (h−1) with species numbers corresponding to Table 1. (C) Net synthesis rates of major metabolic byproducts including the three SCFAs acetate, butyrate and propionate. The byproducts ethanol, D-lactate, L-lactate and succinate are not shown because their net secretion rates were zero. (D) Uptake and secretion fluxes of the five metabolites most significantly crossfed between the participating species.
(TIF)
(A) Fraction of the 568 simulation cases for which the species abundance exceeded 1%. (B) Binned community richness, defined as the number of species with abundances greater than 1%. Results of the 568 cases were collected into 15 bins centered at growth rates ranging from 0.71 to 0.99 of the maximal value. The symbols represent the mean and error bars represent the standard deviation in each bin. (C) Equitability measure Dcom calculated for all 568 cases. (D) Binned fractional butyrate and propionate synthesis.
(TIF)
(A) Maximal growth heatmap of uptake (negative) and secretion (positive) rates in mmol/h for each crossfed metabolite (numbered in S1 Table) and each species (numbered in Table 1). (B) Suboptimal growth heatmap of uptake and secretion rates in mmol/h for each crossfed metabolite and each species averaged across 36 cases. (C) Absolute value of the exchange rate of each crossfed metabolite averaged across the 20 species and the 36 cases (for suboptimal growth). (D) Standard deviation of the exchange rates associated with S3 FigC.
(TIF)
(PDF)
Community uptake constraints for the four models (20 and 28 species; Western and high-fiber diets).
(XLSX)
Matlab codes and data files used for generating results.
(ZIP)
Data Availability Statement
SteadyCom is freely available at http://www.maranasgroup.com/software.htm. The MATLAB simulation codes used in this study are included as supplementary materials available from the journal website.