Nearly a century ago, a Swiss pediatrician, Guido Fanconi, described a hereditary condition of panmyelopathy accompanying aging, cancer and others, now better known as Fanconi Anemia (FA) [1]. Similar clinical symptoms shown in each group of FA patients define a shared signaling network, which was defected in FA-patients owing to germline mutations in an FA gene. There are, at least, twenty-two FA gene-encoded proteins (FANC-A/B/C/D1/D2/E/F/G/I/J/L/M/N/O/P/Q/R/S/T/U/V/W), working together with ATM, ATR, and/or others to protect humans from a variety of diseases [1].
FANCD2 sits at the center of this signaling network serving as an important “successor” to transduce upstream FA signaling upon genotoxicants and also as a critical “savior” for completing downstream FA signaling-transduction, involving all DNA-repair mechanisms known as far [2]. While fathoming FANCD2 functional importance, we discovered an unrecognized form of FANCD2, namely FANCD2-V2 in contrast to the long-known one, FANCD2-V1 [3]. Both share more than 95% identity at the amino acid level, but their biological functions are distinct from each other. The usage of the alternative polyadenylation site (APS) [4] explained part of their expression patterns and, thus, distinct functions. The proximal polyadenylation confers a higher level of FANCD2-V2 expression in normal cells/tissues. In contrast, FANCD2-V1 is elevated in transformed cells/tissues, attributed to the use of the distal APS [3]. How is the proximal or distal site of polyadenylation chosen?
Polyadenylation is an essential mRNA processing step for all genes in eukaryotes, which defines an end of the transcript. The alternate polyadenylation site (APS) will create different transcripts by inclusion or exclusion of certain RNA sequences [4]. The usage of an APS depends on both cis and trans-elements, among which methylated DNA at the area of an APS appears to be a plausible cause of an APS usage, considering roles of DNA methylation in regulating gene expression at promoter regions [5]. We, therefore, focused on the relation of DNA methylation at the vicinity of an APS (Figure 1A top-panel) with the utilization of an APS. We found that blocking DNA methyltransferases reduced the relative level of FANCD2-V2 expression (Figure 1A-bottom left & middle, a higher ratio of V1/V2), as well as the intensity of DNA methylation nearby the proximal polyadenylation site, as evidenced by methylated DNA immunoprecipitation (MeDIP) (Figure 1A-bottom right). Importantly, the association between FANCD2-V2 expression and the degree of DNA methylation nearby proximal APS was extended to human tumor samples tested (n > 2000) (Figure 1(b,c)), although the levels of FANCD2-V1 or V2 mRNA expressions were referenced from previous experiments (Figure 1B) or not yet tested (Figure 1C). Taken together [(Figure 1a-c), [3]], these results show that both ratios of V1/V2 and Me-D/Me-P are positively associated with tumor stages/grades, and the relatively high level of DNA methylation at the proximal APS area promotes the use of the proximal APS and the production of FANCD2-V2 transcript (Figure 1D).
Figure 1.
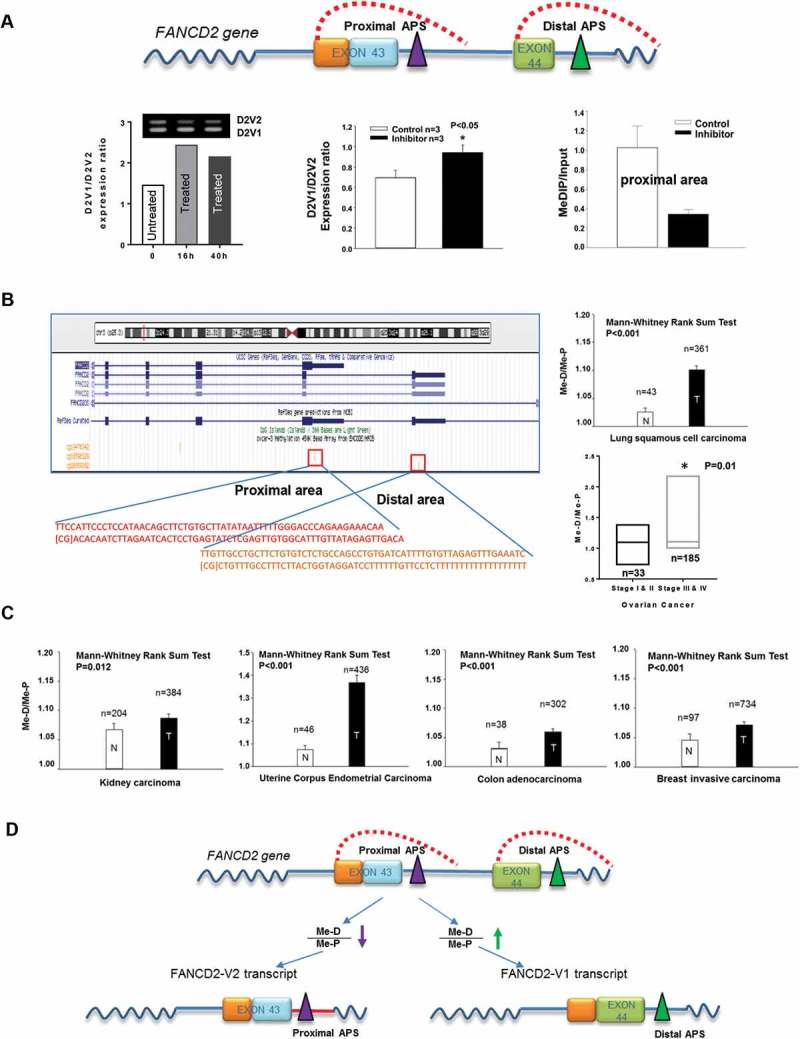
DNA Methylation at the Vicinity of an APS Influences the Usage of the Given APS. (A) The relatively high level of DNA methylation at the area of the proximal APS promotes the expression of FANCD2-V2. Top panel: Outlines of two APS areas in FANCD2 gene. FANCD2-V2 expression is a result of the use of proximal APS; while the usage of distal APS generates FANCD2-V1 transcript. Bottom left: HEK293 cells treated with DNA methyltransferase inhibitor show a relatively low level of FANCD2-V2 expression in one-time experiment via RT-PCR. The bars were plotted with the corresponding-agarose-band intensities. (V1/V2, a higher ratio ≥ V2 is relatively low). Bottom middle: In a different time of experiment, cells treated with or without the inhibitor were used for detection of FANCD2-V1 or V2 expression. Treated cells once again showed a relatively low level of FANCD2-V2 expression (real-time PCR). (V1/V2, a higher ratio = > V2 is relatively low). Bottom right: Antibodies targeting methylated DNA (MeDIP) pulled down a lower amount of DNA fragments in the vicinity of proximal APS in methyltransferase-inhibitor treated cells compared to the non-treated cells. Together, a relatively low level of DNA methylation at the proximal area (treated versus non-treated cells) is correlated significantly with FANCD2-V2 expression at a relatively low level. (B) The relative levels of human tissue DNA-methylation at proximal and distal areas of FANCD2 gene support the conclusion drawn from cell line studies via analyzing publicly available data sets (TCGA-lung and GSE72021). Left: Schematic representation of DNA methylation probes in the vicinity of proximal or distal APS. Right: The ratio (Me-D/Me-P) of DNA methylation at the distal area (Me-D) over that at the proximal area (Me-P) is statistically significant and positively associated with lung or ovarian cancer malignancy. Importantly, the ratio of V1/V2 mRNA expression we tested before in lung and ovarian tissues showed the same significant association [3] [a higher V1/V2 expression ratio in tumor tissues (T) or high-grade/stage tumors, compared to the corresponding normal tissues (N) or low-grade/stage tumors [3]]. Together, this in-vivo association supports the data generated from human cell lines (A). (C) Additional four types of human tissue samples (TCGA) show a similar association for the relative levels of DNA methylation at proximal and distal areas with the degrees of human malignancy. The ratio (Me-D/Me-P) of DNA methylation at the distal area (Me-D) over that at the proximal area (Me-P) is positively associated with human malignancy [the ratio is lower in normal tissues (N) than in tumor tissues (T)]. (D) The working hypothesis for the use of the proximal or distal APS in FANCD2 gene, leading to the expression of FANCD2-V2 or V1, respectively. In normal tissues or low-grade tumors, FANCD2-V2 expression is relatively high, which can be attributed to the proximal area of DNA methylation that is relatively high compared to the distal area (Me-D/Me-P). In contrast, the level of FANCD2-V1 expression is relatively high in human malignant tissues or high-grade/stage tumors, as a result of the relatively low level of DNA methylation in the area of the proximal APS (Me-D/Me-P).
DNA methylation is a major epigenetic mechanism, along with histone modifications, that plays an important role in many molecular and cellular events, including gene silencing, X-chromosome inactivation, genetic imprinting or cellular development. Abnormal hypermethylation of CpG Islands (CGIs: high-density CpG regions) and genome-wide hypo-methylation have often been associated with cancer [5]. In addition to promoter CGIs, shores (up to 4kb from CGIs) or open sea regions (far away from CGIs) have also shown a cancer- and tissue-specific methylation [6]. Many DNA repair genes with hypermethylated promoters and reduced or absent expression were found to be in a variety of human cancer [7]. As such, MGMT, MSH2 and WRN were hypermethylated in colon cancer with frequencies of 55%, 13% and 29% respectively. Hypermethylated promoter in MGHT or MLH1 was found in head and neck cancer with a sequence of 54% or 33% accordingly. Among FA genes, the hypermethylated promoter for FANCB or FANCF was reported, and the latter was a cause of ovarian tumor resistance developed to Cisplatin when de-hypermethylation occurred. Whether or how DNA methylation affects the center player-FANCD2 expression is rarely studied. Here, our study first documented sea-region DNA-methylation in affecting the usage of an APS in FANCD2 gene, providing a novel key to unlock in-depth insights into FA-signaling tumor-suppression functions in both FA and non-FA cells [1,2]. Additionally, corresponding translational studies over the appreciation of FA signaling remain to be pigeonholed, although evidently indicated for establishing better therapeutic strategies [8]. We believe Dr. Fanconi never imagined such when he reported the FA case. Similarly, what we have envisioned would be far less than that resulting from FA signaling in future, such as Me-D/Me-P (Figure 1D) would be turned to be a powerful biomarker in determining tumor malignancy in assisting early cancer diagnosis or grading/staging human tumors.
Funding Statement
This work was supported by the National Cancer Institute [R01CA136532]; National Cancer Institute [R01CA188251].
Acknowledgments
We thank previous lab members (Drs. Han and Shen) for their part of technical assistance. We apologize for the information used without citations owing to the number limitation for references.
References
- [1].Che R, Zhang J, Nepal M, et al. Multifaceted fanconi anemia Signaling. Trends in Genetics: TIG. 2018;34:171–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nepal M, Che R, Zhang J, et al. Fanconi anemia signaling and cancer. Trends Cancer. 2017;3:840–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Han B, Shen Y, Zhang P, et al. Overlooked FANCD2 variant encodes a promising, portent tumor suppressor, and alternative polyadenylation contributes to its expression. Oncotarget. 2017;8:22490–22500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Elkon R, Ugalde AP, Agami R.. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet. 2013;14:496–506. [DOI] [PubMed] [Google Scholar]
- [5].Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. [DOI] [PubMed] [Google Scholar]
- [6].Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Jin B, Robertson KD. DNA methyltransferases, DNA damage repair, and cancer. Adv Exp Med Biol. 2013;754:3–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhang J, Zhao D, Park HK, et al. FAVL elevation in human tumors disrupts Fanconi anemia pathway signaling and promotes genomic instability and tumor growth. J Clin Invest. 2010;120:1524–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]


