SUMMARY
Homeostasis in adult tissues depends on the precise regulation of stem cells and their surrounding microenvironments, or niches. Here, we show that the cell cycle inhibitor and tumor suppressor Retinoblastoma (RB) is a critical regulator of niche cells in the Drosophila testis. The testis contains a single niche, composed of somatic hub cells, that signals to adjacent germline and somatic stem cells. Hub cells are normally quiescent, but knockdown of the RB homolog Rbf in these cells causes them to proliferate and convert to somatic stem cells. Over time, mutant hub cell clusters enlarge and split apart, forming ectopic hubs surrounded by active stem cells. Furthermore, we show that Rbf’s ability to restrict niche number depends on the transcription factors E2F and Escargot and the adhesion molecule E-cadherin. Together this work reveals how precise modulation of niche cells, not only the stem cells they support, can drive regeneration and disease.
In Brief
Greenspan and Matunis find that the tumor suppressor Retinoblastoma is required in niche cells to maintain quiescence, cell fate, and niche number. Loss of Retinoblastoma causes niche cell divisions, conversion to somatic stem cells, and ectopic niche formation through niche fission, suggesting that mutations in niche cells may drive disease.
Graphical Abstract

INTRODUCTION
Stem cells maintain homeostasis within many adult tissues by producing both new stem cells (self-renewal) and daughter cells that differentiate (Greenspan et al., 2015). Signals from the surrounding microenvironment in which the stem cells reside, called the niche, are vital for promoting stem cell maintenance (Greenspan et al., 2015; Ohlstein et al., 2004). Understanding how niches regulate stem cells is key to using the regenerative capacity of stem cells for therapeutic purposes after damage. In addition, mis-regulation of cell signaling within stem cell niches can lead to tumor growth and cancer metastases (Dagogo-Jack and Shaw, 2018), underscoring the need for better understanding niche function.
The Drosophila testis provides an ideal model system to study stem cell regulation in vivo because it contains a well-defined niche in which cell types are easily identified and manipulated genetically. A major component of this niche is a cluster of quiescent somatic hub cells that signal to the attached germline stem cells (GSCs) and somatic cyst stem cells (CySCs) (Figure 1A) (Hardy et al., 1979; Kiger et al., 2001). Damage to this niche triggers an unexpected degree of cellular plasticity. Recently we found that genetic ablation of all CySCs induces hub cells to exit quiescence and begin mitotic divisions (Hétié et al., 2014). Surprisingly, this also leads to the cell fate conversion of hub cells to CySCs. This change in cell fate is accompanied by the formation of new niches throughout the testis, characterized by the presence of multiple hubs, each supporting active stem cells. However, it is still not known if hub cell quiescence and fate must be actively maintained. In addition, the molecular regulators and cellular behaviors that drive these phenotypes have not been characterized.
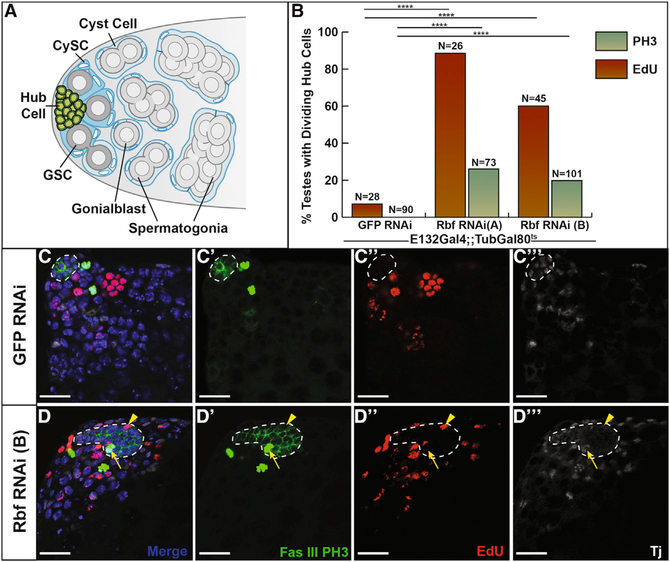
Figure 1. Hub Cells Lose Quiescence upon Rbf Knockdown.
(A) Schematic of the Drosophila testis stem cell niche, which contains a specialized microenvironment consisting of somatic hub cells (green) that signal to the attached germline stem cells (GSCs; dark gray) and somatic cyst stem cells (CySCs; dark blue). Differentiating spermatogonia (light gray) are enveloped by cyst cells (light blue) and are displaced from the testis apex.
(B) Bar graph showing the percentage of testes containing dividing hub cells as measured by either EdU incorporation indicating cells in S phase (red bars) orPH3 staining indicating cells in mitosis(green bars).Two independent Rbf RNAi lines, labeled A and B accordingly, were expressed by E132ts to control knockdown of Rbf specifically in the hub. Testes expressing either RNAi line showed a significant difference in EdU incorporation and PH3 staining in hub cells compared with E132ts > GFP RNAi controls.
(C and D) Single confocal sections through the testis apex immunostained for EdU (S phase cells; red), Fas III (hub; membranous green), PH3 (mitotic cells; nuclear green), Tj (cyst lineage; white), and DAPI (nuclei; blue). Flies were shifted to 29°C for 7 days to induce either GFP RNAi (C) or Rbf RNAi (D) knockdown. See also Figure S1.
(C–Cʹʹ) Control testis shows no EdU incorporation or PH3 staining within cells of the hub cell cluster (white outline). Merged (C), FasIII and PH3 only (Cʹ), EdU only (Cʹʹ), and Tj only (Cʹʹʹ) channels are shown.
(D–Dʹʹʹ) Loss of Rbf in hub cells using Rbf RNAi leads to hub cell divisions as seen by EdU incorporation (yellow arrowhead) and PH3 staining (yellow arrow) within the hub (white outlines). Merged (D), FasIII and PH3 only (Dʹ), EdU only (Dʹʹ), and Tj only (Dʹʹ) channels are shown.
The Drosophila Retinoblastoma homolog Retinoblastomafamily protein (Rbf) is a key cell cycle regulator that inhibits cell cycle progression by binding and repressing the cell cycle activator E2F and whose activity is controlled through phosphorylation from Cyclins (Sutcliffe et al., 2003). Rbf is known to negatively regulate cell proliferation in many cell types and is a tumor suppressor gene that is downregulated in many cancers (Cheung and Rando, 2013; Dyson, 2016). Here, we show that Rbf is required to autonomously maintain quiescence in terminally differentiated hub cells and is necessary for hub cell fate. Loss of Rbf in hub cells leads to the activation of E2F driving hub cell proliferation and conversion of hub cells to cyst lineage cells. Rbf also has unexpected effects on this model stem cell system: extended Rbf loss in hub cells causes ectopic niche formation within the testis. Live imaging and lineage tracing reveal that this process is driven by fission of the original hub. This leads to an increase in the stem cell population via two means: autonomously through the direct conversion of hub cells to stem cells and non-autonomously through the expansion of the niche. Together our results suggest a mechanism whereby cell cycle factors regulate hub cell quiescence and plasticity and demonstrate how the modulation of niche cells, as opposed to the stem cells they support, can drive regeneration and disease.
RESULTS
Rbf Is Required to Maintain Hub Cell Quiescence
Because hub cell proliferation is one of the first phenotypes observed upon recovery from CySC ablation, we hypothesized that cell cycle inhibitors might play an active role in intrinsically maintaining hub cell quiescence. Retinoblastoma (RB) is a known cell cycle regulator in many mammalian stem cell populations. Loss of RB in frequently dividing spermatogonial stem cells leads to a depletion of the stem cell pool, while loss of RB in the quiescent support cells (Sertoli cells) that contribute to the testis niche leads to Sertoli cell proliferation and dysfunction, causing a failure in sperm development (Hu et al., 2013; Rotgers et al., 2014). In the Drosophila testis, Rbf is normally expressed broadly and is detected in all of the cells within the testis apex. This includes the terminally differentiated hub cells, germ cells, and cyst lineage cells (Figures S1A and S1A‘). To determine if this cell cycle inhibitor is necessary for the maintenance of hub cell quiescence, we used the E132Gal4-UAS-TubGal80ts system (hereafter E132ts) to conditionally knock down Rbf specifically in adult hub cells. A complete loss of Rbf protein specifically within the hub, confirmed by immunostaining adult testes with anti-Rbf antisera (Figures S1B–S1Cʹ), was observed in E132ts > Rbf RNAi flies raised at the permissive temperature of 18°C and shifted to 29°C for 7 days to induce RNAi knockdown. To determine if hub cells lacking Rbf can proliferate, we immunostained testes for the mitotic marker phosphohistone H3 (PH3). PH3-positive hub cells were seen in 26% (n = 19 of 73) and 20% (n = 20 of 101) of testes with Rbf knocked down in the hub (p < 0.0001, Rbf RNAi A and B, respectively) (Figures 1B–1D) compared with E132ts > GFP RNAi control testes, in which we never saw PH3positive hub cells (n = 0 of 90). To extend these findings, hub cells lacking Rbf were assessed for incorporation of the thymidine analog ethynyl deoxyuridine (EdU), which labels cells undergoing DNA replication. EdU-positive hub cells were seen in 88% (n = 23 of 26) and 60% (n = 27 of 45) of testes expressing two independent Rbf RNAi lines for 7 days (Figures 1B–1D), confirming that these hub cells are progressing through the cell cycle. We rarely saw EdU-positive hub cells in E132ts > GFP RNAi control testes (n = 2 of 28). In comparison, low levels of S phase labeling have been reported in other studies of wild-type testes (Voog et al., 2008), suggesting that sometimes hub cells may undergo replication. However, ploidy measurements of hub cells (Figure S1I) suggest that most hub cells are normally diploid. We conclude that in contrast to wild-type hub cells, which are quiescent, hub cells lacking Rbf enter both S and M phase of the cell cycle.
Because hub cells divide in hubs lacking Rbf, we expected the hub cell clusters to increase in size. To assess this, hub volume was measured using three-dimensional (3D) reconstructions of testes expressing a bright cytoplasmic GFP transgene in hub cells.Within 3 days,hub-specific knockdown of Rbf led to a significantly larger hub volume than control testes expressing only GFP (Figures S1D, S1E, and S1H). Hub volumes continued to increase at 7 and 14 days post-induction (Figure S1H). Additionally, hub cells appeared to spread along the basement membrane of the tissue and protrude into the center of the testis, creating a neck that connected the two sections in some testes, while hubs in control testes remained unchanged (Figures S1F and S1G). Altogether these data indicate that Rbf is required in hub cells to actively maintain their quiescent state, and loss of Rbf leads to hub cell proliferation and subsequent enlargement of the hub.
Loss of Rbf in Hub Cells Causes Conversion to Cyst Lineage Cells
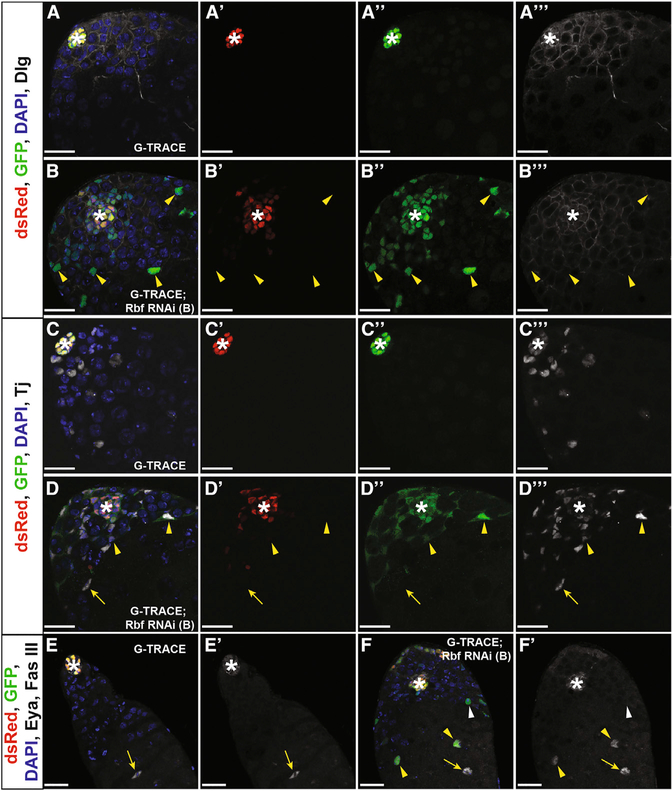
Because CySC ablation causes loss of hub cell quiescence and drives hub cell-to-CySC conversion (Hétié et al., 2014), we next asked if loss of Rbf could also change hub cell identity. To assess the fate of hub cells upon Rbf knockdown, the Gal4 technique for real-time and clonal expression (G-TRACE) (Evans et al., 2009) was used to track hub cells lacking Rbf over time. This lineage tracing system uses a red fluorescent protein (RFP) to mark current Gal4 expression and a GFP to track cells that originate from the Gal4-expressing cells. When combined with a hub-specific Gal4 driver, hub cells expressing this system will appear yellow, while cells derived from hub cells that have changed fate will lose RFP expression and retain GFP expression, appearing green. E132ts > G-TRACE, Rbf RNAi flies were shifted to 29°C for 7 days to simultaneously induce marking and Rbf knockdown specifically in hub cells. Testes from these flies frequently had green cells dispersed throughout the niche outside of the hub cell cluster (78% [n = 70 of 90 testes]), indicating that they had lost their hub cell identity (Figures 2B and2D). In comparison, control testes expressing G-TRACE alone contained yellow hub cells (RFP+GFP+) that remained confined to the apical tip of the testis (Figures 2A and 2C). As expected on the basis of prior studies using the G-TRACE system in the Drosophila testis (Voog et al., 2014), a few green cells were occasionally seen outside of the hub cluster in control testes (25% [n = 21 of 85 testes]). This background marking could be due to incomplete repression of Gal4 by Gal80 during part of development or due to effects of the hub-Gal4 driver itself. Importantly, significantly more testes contained green (hub-derived) cells outside the original hub cluster when Rbf was knocked down in hub cells (p < 0.0001; Table S1). This indicates that Rbf is required to maintain hub cell fate.
Figure 2. Hub Cells Convert to Cyst Lineage Cells upon Rbf Knockdown.
(A–F) Single confocal sections through the testis apex of E132ts > G-TRACE control testes (A, C, and E) and E132ts > G-TRACE; Rbf RNAi experimental testes (B, D, and F). Marking and RNAi induction occurred for 7 days (A–D) or 14 days (E and F) at 29°C. Testes are immunostained for dsRed (Gal4-expressing hub cells; red), GFP (hub cells and cells derived from hub cells; green), DAPI (nuclei; blue), and either Dlg (cell membranes; white, A and B), Tj (cyst lineage; white, C and D), or Fas III and Eya (hub membrane and late cyst cells, respectively; white, E and F). Merged and single channels are shown.
(A–Aʹʹʹ) Hub cells (white asterisk) are seen in a confined area at the apical tip of the control testis. These cells express the hub-GAL4 driver, allowing continuous expression of both RFP and GFP, causing cells to appear yellow. Merged (A), dsRed only (Aʹ), GFP only (Aʹʹ), and Dlg only (Aʹʹʹ) channels are shown.
(B–Bʹʹʹ) Rbfknockdown causes hub cellconversion. Cells within the main hub cluster (white asterisk) still express the hub-GAL4 driver and appear yellow. Cells originating from the hub that have migrated away from the main hub cluster have lost their hub cell identity (yellow arrowheads) and appear only green. Merged (B), dsRed only (Bʹ), GFP only (Bʹʹ), and Dlg only (Bʹʹʹ) channels are shown.
(CCʹʹʹ) Cyst lineage cells (Tj; white) do not express either RFP or GFP and thus are not derived from converting hub cells in control testes. Merged (C), dsRed only (Cʹ), GFP only (Cʹʹ), and Tj only (Cʹʹʹ) channels are shown.
(D–Dʹʹʹ) Upon Rbf knockdown, hub-derived traced cells (RFP−GFP+) express the cyst lineage marker Tj (yellow arrowheads), indicating that these cells have changed their cell fate. Tj-positive cells not derived from the hub (RFP−GFP−) are also seen (yellow arrow). See also Figure S3. Merged (D), dsRed only (Dʹ), GFP only (Dʹʹ), and Tj only (Dʹʹʹ) channels are shown.
(E and Eʹ) Late cyst cells are unmarked (RFP−GFP−), express Eya (nuclear white; yellow arrow), and are seen far away from the hub (white asterisk) in control testes. Merged (E) and Eya and FasII only (Eʹ) channels are shown.
(F and Fʹ) Upon Rbf knockdown, some converted hub cells (RFP−GFP+) that are far away from the hub (white asterisk) are seen expressing the late cyst marker Eya (nuclear white; yellow arrowheads), indicating that they can progress through cyst lineage differentiation. Unmarked (RFP−GFP−) late cyst cells (Eya+) are also seen (yellow arrow). Some marked cells (RFP−GFP+) do not express Eya (white arrowhead), suggesting that these cells have lost hub fate but have not yet acquired later cyst cell fate. Merged (F) and Eya and FasII only (Fʹ) channels are shown.
Scale bars represent 20 μm. See also Figures S2–S4, Table S1, and Videos S1, S2, and S3.
To verify that Rbf knockdown causes hub cells to leave the hub cluster and lose their identity, a second lineage-tracing reporter was used with a different drug inducible hub-specific driver (GS2295-Gal4). Using this system, 21% (n = 8 of 38) of testes lacking Rbf in hub cells showed conversion of hub cells (as indicated by GFP-marked cells outside of the hub cell cluster) by 21 days post-induction (Figures S2B and S2D). In contrast, GFP-marked cells were not detected outside of the hub in testes from lineage traced controls (n = 0 of 64 testes) (Figures S2A and S2C; Table S1). Together with the G-TRACE lineage-tracing, these data indicate that Rbf is necessary to maintain hub cell identity.
To determine the identity of lineage-traced cells arising upon hub-specific Rbf knockdown, we immunostained testes with hub, cyst lineage, and germ cell markers. Traced cells close to the hub cluster expressed the CySC marker Zinc finger homeodomain 1 (Zfh1; Figure S2B), suggesting that these cells converted into somatic stem cells. These cells also expressed the general cyst lineage marker Traffic Jam (TJ; Figures 2D and S2D) but lacked the hub-specific marker Fasciclin III (Fas III; Figures 2F, S2B, and S2D). Traced cells were seen flanking germ cells but did not express the germ cell marker Vasa (Figures S3A and S3B), suggesting that they had converted from hub cells to cyst cells. When Rbf was continuously knocked down in hub cells for a longer period of time (14 days), traced cells were seen away from the hub cluster undergoing further differentiation, as indicated by expression of the late cyst lineage marker Eyes Absent (Eya; Figures 2E and 2F). These cells also maintained their male sex identity, as staining for the female somatic cell markers Castor and Cut showed no expression in testes with Rbf knocked down in the hub (data not shown). This indicates that upon Rbf loss, a subset of hub cells convert to CySCs and progress through the cyst lineage differentiation process.
However, converted hub cells did show some aberrant behaviors compared with normal cyst cells. In wild-type testes, the only proliferating somatic cells are the CySCs next to the hub (Figures S3C and S4A) (Gönczy and DiNardo, 1996; Inaba et al., 2011). In contrast, traced hub cells were seen proliferating both next to and more than 2 cell diameters away from the hub (Figure S3D). This suggests that Rbf may also be required for cyst cell quiescence in the adult testis, as has been recently shown to occur in the Drosophila larval testis (Dominado et al., 2016). Therefore we speculated that Rbf levels might not be restored in converted cyst cells fast enough to keep them quiescent. To directly address whether Rbf promotes adult cyst cell quiescence, Rbf was knocked down specifically in cyst cells but not CySCs using the EyaGal4 driver (Ma et al., 2014). Loss of Rbf specifically in cyst cells led to an expansion of Tj-positive cells throughout the testis, and these cells were capable of dividing away from the hub, compared with controls, which only showed proliferating somatic cells within 2 cell diameters from the hub (Figures S4A and S4B). Rbf knockdown in cyst cells also led to a non-autonomous expansion of undifferentiated germ cells throughout the adult testis, as indicated by the TJ-negative DAPI bright nuclei (Figures S4A and S4B), consistent with previous reports in the larval testis (Dominado et al., 2016). Interestingly, the CySC marker Zfh1 remained restricted to the apex in testes lacking Rbf in the cyst cells (Figures S4C and S4D), suggesting that dividing cyst cells did not revert to a stem cell-like state. Altogether, these data suggest that Rbf is required in cyst cells to maintain their quiescence but not their differentiation state, demonstrating a contrast to its role in hub cells.
Live Imaging Reveals Dynamics of Hub Cell Conversion In Vivo
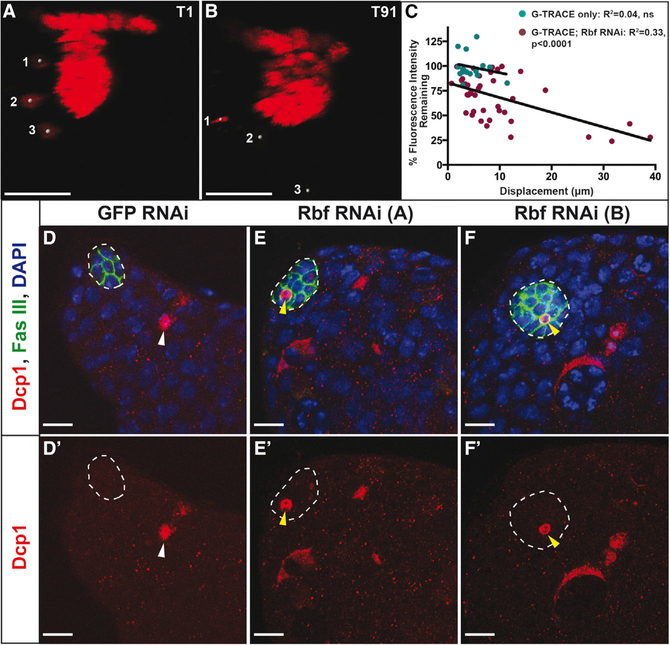
The G-TRACE lineage-tracing system (Evans et al., 2009) combined with our established techniques for live imaging (Greenspan and Matunis, 2017; Sheng and Matunis, 2011) affords us the opportunity to visualize how and from where within the hub cell fate conversion occurs in vivo. Testes co-expressing Rbf RNAi and the G-TRACE system in hub cells for 10 days were imaged live for 12–18 hr. As we expected on the basis of our fixed images, hub cells were seen dividing in most testes (n = 18 of 27 testes; Video S1) within different regions of the hub cluster, including the portion along the basement membrane, within the protrusion, and in the neck connecting these two sections. However, cell division did not correlate with those cells that converted to a cyst lineage fate, as daughter cells could be seen remaining in the hub cluster throughout the duration of the video after division (Video S1). Live imaging further revealed that these hub cells are very dynamic. Hub cells could be seen moving between different regions of the hub cluster (n = 18 of 27 testes; data not shown) as well as leaving the hub cell cluster entirely (n = 15 of 27 testes), mostly from the basement membrane but occasionally from the neck and protrusion (Video S1). Because a change in hub identity is indicated by loss of red fluorescence signal, we asked whether the cells that migrated away from the hub cluster lost their hub cell fate. To answer this question, a subset of hub cells were analyzed to determine their loss of fluorescence over time (Figures 3A and3B; Video S3). Hub cells with Rbf knocked down could be seen exhibiting three different behaviors throughout the course of the video. Some cells maintained their RFP expression and stayed within the hub cluster, some cells started to lose their RFP expression but stayed within the hub cluster, and a third category lost most of their RFP expression and migrated away from the main hub cluster (Figure 3C). Linear regression analysis showed a significant correlation between a hub cell’s displacement and its loss of RFP expression, meaning that cells that migrated the farthest from their original position tended to lose the most fluorescence (Figure 3C). In contrast to hub cells lacking Rbf, hub cells in control testes expressing the G-TRACE system alone were never seen dividing or leaving the main hub cluster during live imaging (n = 0 of 6 testes; Video S2). In addition, no correlation was seen between RFP expression levels and displacement among hub cells expressing G-TRACE alone in control testes, because these cells did not lose RFP expression or migrate far from their original position (Figure 3C; Video S2). Together these live imaging data reveal that upon Rbf loss some hub cells start to lose their hub identity before migrating away, further supporting the conclusion that Rbf is required for hub cell fate.
Figure 3. Rbf Knockdown in the Hub Causes Migration of Converting Hub Cells and Hub Cell Death.
(A and B) Still frames from live images of E132ts > G-TRACE; Rbf RNAi testes shifted for 10 days at 29°C. Testes were imaged live for approximately 15 hr, with images acquired every 10 min. Images show red channel only.
(A) Time frame 1 (T1) shows three hub cells marked by gray circles that have already left the main hub cluster.
(B) By the last time frame (T91), the hub cell marked 1 has undergone cell death, as seen through the condensation of its DNA. The other two marked hub cells (2 and 3) have traveled farther from the main hub cluster and have lost red fluorescent expression, indicating that they are no longer hub cells.
(C) Dot plot depicting the displacement from T1 to T91 versus the percentage of red fluorescence expression remaining at T91 compared with T1. Green dots show individual hub cells from control testes expressing G-TRACE only (n = 15), while red dots show individual hub cells from testes expressing G-TRACE and Rbf RNAi in the hub (n = 42). Linear regression analysis shows a significant correlation (R2 = 0.33, p < 0.0001) between the displacement of a hub cell and the amount of fluorescence it loses in testes in which Rbf has been knocked down. Control testes show no such correlation (R2= 0.04, ns). See also Videos S2 and S3.
(D–F) Single confocal sections through the testis apex of testes fixed and immunostained for Dcp1 (dying cells; red), Fas III (hub; membranous green), and DAPI (nuclei; blue). Hubs are outlined in white. E132ts flies expressing either GFP RNAi (D) or Rbf RNAi (E and F) were shifted to 29°C for 7 days to induce RNAi knockdown.
(D and Dʹ) Dying cells can be seen within control testes (white arrowhead) but are never seen within the hub. Merged (D) and Dcp1 only (Dʹ) channels are shown.
(E–Fʹ) Rbf knockdown in hub cells using two independent RNAi lines denoted A (E and Eʹ) and B (F and Fʹ) causes hub cell apoptosis, as indicated by shrinking hub cells that are marked with Dcp1 throughout the entire cell (yellow arrowheads). Merged (E and F) and Dcp1 only (Eʹ and Fʹ) channels are shown.
Scale bars represent 15 μm (A and B) and 10 μm (D–F).
During our live imaging of hubs lacking Rbf, we occasionally observed cells dying within the hub or being extruded from the hub first, then shrinking, eventually losing fluorescence (Figures 3A and 3B; Video S3). This, together with the previous findings that Retinoblastoma is known to inhibit apoptosis in certain tissues (DeGregori et al., 1997; Dyson, 2016; Tanaka-Matakatsu et al., 2009), prompted us to assay for expression of the cleaved caspase Dcp1, which marks dying somatic cells in the testis (Hasan et al., 2015; Yacobi-Sharon et al., 2013). Dcp1-positive hub cells were occasionally seen when Rbf was specifically knocked down in hub cells (n = 9 of 44 and n = 3 of 53 testes for Rbf RNAi A and B, respectively; Figures 3D–3F) but undetectable in hubs of GFP RNAi control testes (n = 0 of 61 testes). This indicates that Rbf is required not only to maintain hub cell quiescence and fate but also to prevent hub cell apoptosis. However, the level of apoptosis that occurs upon Rbf knockdown must be minimal compared with the level of cell proliferation, because overall hub size increases with time upon Rbf knockdown.
Rbf Acts through E2F to Maintain Hub Cell Quiescence and Fate
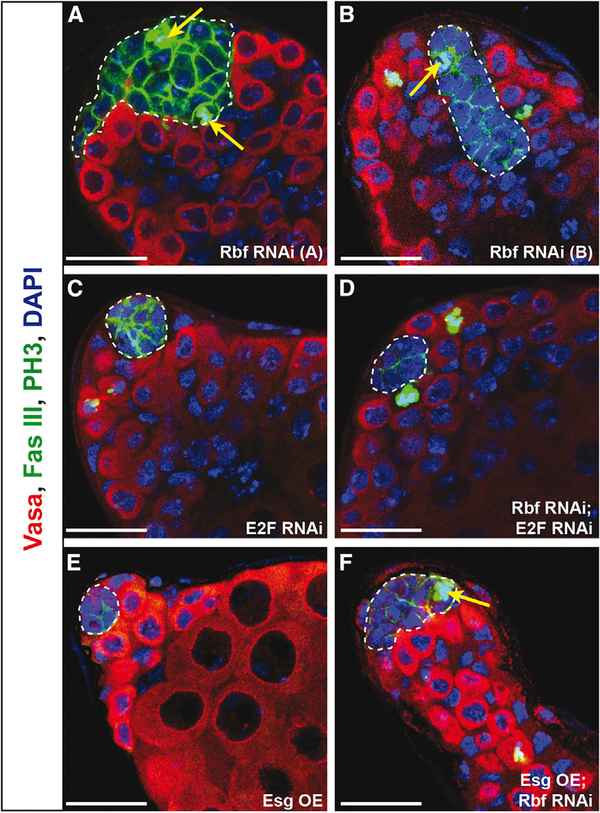
Rbf is known to inhibit the cell cycle by binding to the transcriptional activator E2F and preventing its activity. Upon hyper-phosphorylation by Cyclin D/Cdk4, Rbf releases E2F, allowing it to transcribe its target genes, many of which drive cell cycle progression (Dyson, 2016). However, Rbf is also known to have E2F-independent roles (Korenjak et al., 2012). Because knockdown of Rbf alone leads to cell cycle activation, we hypothesized that knockdown of Rbf and E2F simultaneously in hub cells could be sufficient to prevent hub cell proliferation. To ask whether Rbf acts through E2F to regulate hub cell quiescence and fate, we simultaneously removed Rbf and E2F specifically in hub cells for 14 days and assessed their proliferation status using the mitotic marker PH3. Testes lacking both Rbf and E2F in hub cells showed a complete rescue of hub size, and proliferating hub cells were undetectable (n = 0 of 42 testes; Figure 4D; Table 1). For comparison, Rbf knockdown alone yielded dividing hub cells in 52% of testes (n = 29 of 56; Figure 4A; Table 1). To ensure that this rescue was specific to E2F, flies expressing Rbf RNAi and a generic RNAi (GFP RNAi) still showed proliferating hub cells similar to Rbf RNAi alone (n = 4 of 15; Table 1). As expected, E2F knockdown alone showed no hub cell proliferation phenotype, because hub cells are normally quiescent (Figure 4C; Table 1). Together these data indicate that Rbf acts through E2F to maintain hub cell quiescence. Because testes lacking both Rbf and E2F in hub cells contain hubs that are phenotypically indistinguishable from wild-type hubs, this suggests that loss of E2F is sufficient to suppress both hub cell proliferation and the hub-to-cyst lineage conversion phenotype induced by Rbf knockdown alone. This is because if conversion were still occurring without proliferation, we would expect a complete loss of hub cells over time, which is not seen.
Figure 4. E2F Knockdown but Not Esg Overexpression Suppresses Hub Cell Proliferation Caused by Rbf Knockdown.
(A–F) Single confocal sections through the testis apex immunostained for Vasa (germ cells; red), Fas III (hub; membranous green), PH3 (mitotic cells; nuclear green), and DAPI (nuclei; blue). Hubs are outlined in white. Flies were shifted to 29°C for 14 days to induce RNAi knockdown using the E132ts driver.
(A and B) Testes with Rbf knocked down in the hub using two independent RNAi lines denoted A (A) and B (B) have extensive hub cell proliferation, indicated by PH3-marked hub cells (yellow arrows), leading to an enlargement of the hub.
(C and E) Knockdown of E2F (C) or overexpression of Esg (E) individually in the hub does not induce hub cell divisions, as indicated by the absence of PH3 in hub cells. Note that the hub size also appears normal.
(D) Knockdown of both E2F and Rbf in the hub suppresses hub cell proliferation as indicated by an absence of PH3 in the hub. Hub size appears normal. (F) Overexpression of Esg in the hub does not suppress the proliferation phenotype caused by Rbf knockdown. An enlarged hub with PH3-marked hub cells (yellow arrow) is still detected.
Scale bars represent 20 μm. See also Table 1.
Table 1.
Rescue of Rbf Knockdown Phenotypes
| Genotype E132Gal4;; | % Testes with | % Testes with |
|---|---|---|
| TubGal80ts | PH3+Hub Cells | Ectopic Hubs |
| UAS-GFP RNAi | 0% (n = 0/30) | 0% (n = 0/61) |
| UAS-Rbf RNAi (A) | 52% (n = 29/56)a,**** | 18% (n = 15/85)a,*** |
| UAS-Rbf RNAi (A); UAS-GFP RNAi |
27% (n = 4/15)a,* | 26% (n = 5/19)a,*** |
| UAS-Rbf RNAi (B) | 22% (n = 8/83)a,** | 25% (n = 29/117)a,**** |
| UAS-E2F RNAi | 0% (n = 0/53)a (ns) | 0% (n = 0/53)a (ns) |
| UAS-Rbf RNAi (A);UAS-E2F RNAi | 0% (n = 0/42)b,** | 0% (n = 0/42)b,** |
| UAS-esg; UAS-LacZ | 0% (n = 0/35)a (ns) | 0% (n = 0/35)a (ns) |
| UAS-esg; UAS-RbfRNAi (B) | 35% (n = 17/48)c,(ns) | 2% (n = 1/48)c,*** |
| UAS-shg (for Ecad overexpression) | 0% (n = 0/98)a(ns) | 0% (n = 0/98)a (ns) |
| UAS-Rbf RNAi (A);UAS-shg | 26% (n = 19/73)b(ns) | 3% (n = 2/73)b,** |
Fisher’s Exact Test
p < 0.05
p < 0.01
p < 0.001
p < 0.0001, ns, not significant.
Compared with UAS-GFP RNAi.
Compared with UAS-Rbf RNAi (A); UAS-GFP RNAi.
Compared with UAS-Rbf RNAi (B).
Esg Does Not Rescue Hub Cell Quiescence upon Rbf Knockdown
To identify pathway components that could be acting downstream of E2F, we considered the snail family transcription factor Escargot (Esg), because loss of Esg in hub cells causes hub cell-to-cyst lineage conversion (Voog et al., 2014) but not excessive hub cell proliferation. Instead, this conversion causes a loss of hub cells over time, leading to testes that completely lack hubs. In contrast, testes lacking Rbf in hub cells develop increasingly larger hubs with time as well as hub cell-to-cyst lineage conversion. Thus we hypothesized that Esg is working downstream of or in a parallel pathway to Rbf/E2F to regulate quiescence. To test this hypothesis we overexpressed Esg specifically in hub cells while simultaneously knocking down Rbf in the hub and assayed testes for hub cell proliferation. Mitotic (PH3-marked) hub cells were detected in 35% of testes expressing Esg and Rbf RNAi simultaneously (n = 17 of 48; Figure 4F), which was not statistically different from the 22% seen with Rbf RNAi alone (n = 18 of 83; Figure 4B; Table 1). Furthermore, Esg overexpression alone did not cause hub cells to divide (n = 0 of 35; Figure 4E; Table 1). This indicates that overexpression of Esg is not sufficient to induce hub cell proliferation or suppress divisions induced by loss of Rbf and thus suggests that Esg does not play a role in regulating hub cell quiescence.
Loss of Rbf in Hub Cells Results in Ectopic Niche Formation
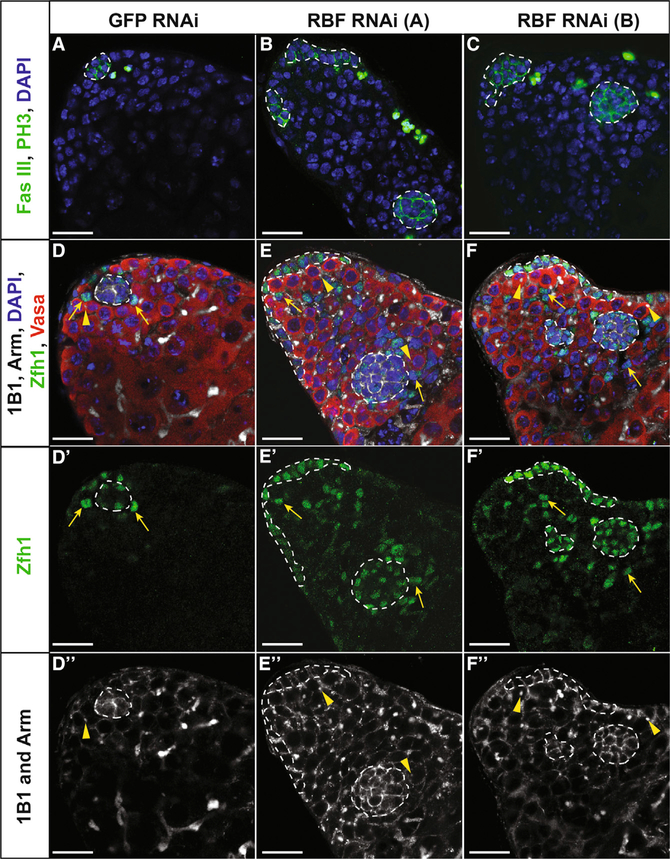
Although more than half of testes that have undergone CySC ablation are able to recover their cyst cells after damage, many (42%) of these recovering testes contain multiple niches, rather than the single niche found in wild-type testes (He´ tie´ et al., 2014). Because mis-regulated niche formation may underlie cancer metastases (Greaves et al., 2006) and Retinoblastoma is known to be downregulated in many forms of cancer (Dyson, 2016), we asked if knockdown of Rbf in hub cells for 14 days is sufficient to drive ectopic niche formation. Ectopic hubs were identified as clusters of Fas III-positive cells no longer connected to any other cluster of hub cells. Ectopic hubs were detected in 18% (n = 15 of 85) and 25% (n = 29 of 117) of testes with Rbf knocked down in the hub compared with GFP RNAi controls, which always contained a single hub (n = 0 of 61) (Figures 5A–5C; Table 1). Ectopic hubs expressed multiple hub markers, including Fas III, Armadillo, Hedgehog, and Unpaired (Figures 5A–5F and S5A–S5D). In addition, ectopic hubs were surrounded by both GSCs, as indicated by Vasa-positive cells with a dot fusome, and cyst stem cells, as demonstrated by Zfh1 staining (Figures 5D–ablation (Hétié et al., 2014). This suggests that Rbf is necessary to prevent ectopic hub formation and that loss of Rbf can cause multiple functional niches to form within the testis.
Figure 5. Loss of Rbf in Hub Cells Causes Ectopic Niche Formation.
(A–F) Single confocal sections through the testis apex of E132ts > GFP RNAi control testes (A and D) or E132ts > Rbf RNAi testes (B, C, E, and F). RNAi was induced for 14 days at 29°C. (A–C) Testes immunostained with Fas III (hub; membranous green), PH3 (mitotic cells; nuclear green), and DAPI (nuclei; blue). Control testis (A) contains only a single hub (white outline), while those testes expressing either Rbf RNAi A (B) or Rbf RNAi B (C) in hub cells contain multiple hubs. (D–F) Testes immunostained with Armadillo (hub; membranous white), 1B1 (fusome; white), Zfh1 (CySCs; green), Vasa (germ cells; red), and DAPI (nuclei; blue).
(D–Dʹʹ) Control testis with a single hub surrounded by both CySCs (yellow arrows) and GSCs represented by red cells with a dot fusome (yellow arrowhead). Merged (D), Zfh1 only (Dʹ), and 1B1 and Arm only (Dʹʹ) channels are shown.
(E–Fʹʹ) Testes with Rbf knocked down in the hub using two independent RNAi lines denoted A (E–Eʹʹ) and B (F–Fʹʹ) contain multiple hubs (white outlines) scattered throughout the testis apex, each with CySCs (yellow arrows) and GSCs (yellow arrowheads). Merged (E and F), Zfh1 only (Eʹ and Fʹ), and 1B1 and Arm only (Eʹʹand Fʹʹ) channels are shown.
Scale bars represent 20 μm. See also Figure S5 and Video S4.
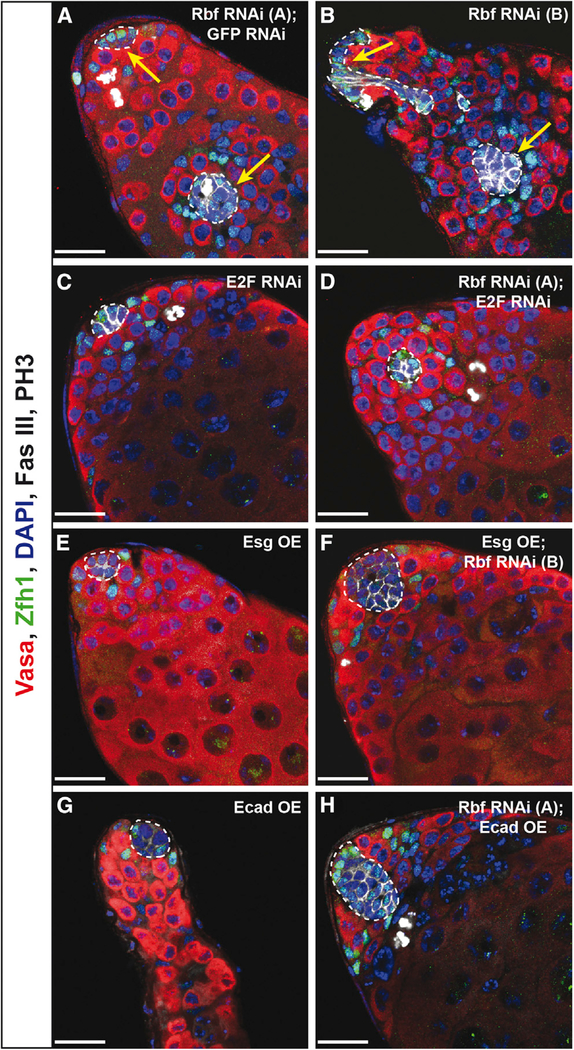
Knockdown of E2F, Overexpression of Esg, or Overexpression of Ecad Prevents Ectopic Hub Formation
Because Rbf is required to prevent ectopic hub formation, we next wanted to ask whether the same downstream components that influence quiescence and changes in fate also regulate ectopic hub formation. Therefore we simultaneously removed Rbf and E2F specifically in hub cells by RNAi knockdown for 14 days (E132ts > Rbf RNAi, E2F RNAi) and assayed for ectopic hubs using the hub marker Fas III. We saw a complete rescue of the ectopic hub phenotype (n = 0 of 42 testes with ectopic hubs; Figure 6D; Table 1) compared with Rbf knockdown alone or Rbf knockdown combined with a generic GFP RNAi (n = 15 of 85 or n = 5 of 19, respectively; Figure 6A; Table 1). As expected, E2F knockdown alone did not cause ectopic hub formation (n = 0 of 53 testes with ectopic hubs; Figure 6C; Table 1). This suggests that Rbf acts through E2F to prevent ectopic hub formation. Because Esg may work downstream of Rbf/E2F to regulate hub cell-to-cyst stem cell conversion, we next asked if Esg plays a role in the formation of multiple hubs. We saw a significant rescue in ectopic hub formation when Esg was overexpressed simultaneously with Rbf knockdown in hub cells (n = 1 of 48; Figures 6B and 6F; Table 1). This rescue was surprising, because loss of Esg alone in hub cells is not sufficient to cause ectopic hub formation (Voog et al., 2014, and data not shown). As expected, ectopic hubs were never seen when Esg was overexpressed in hub cells (n = 0 of 35 testes with ectopic hubs; Figure 6E; Table 1), because Esg is normally expressed in the hub (Hétié et al., 2014; Voog et al., 2014). Together these data indicate that both loss of Esg, which has been shown to regulate cell adhesion, and activation of E2F targets, such as those that stimulate cell proliferation, are necessary for the formation of ectopic niches.
Figure 6. E2F Knockdown, Esg Overexpression, or Ecad Overexpression Suppresses Ectopic Hub Formation upon Rbf Knockdown.
(A–H) Single confocal sections through the testis apex immunostained for Vasa (germ cells; red), Zfh1 (CySCs; green), Fas III (hub; membranous white), PH3 (mitotic cells; nuclear white), and DAPI (nuclei; blue). Hubs are outlined in white. RNAi was induced for 14 days at 29°C using the E132ts driver. Only a single hub at the apical end of the testis is detected when E2F is knocked down (C), Esg is overexpressed (E), or Ecad is overexpressed (G) individually in hub cells. In contrast, induction of Rbf RNAi (A and B) in hub cells can induce ectopic hub formation, as indicated by multiple Fas III-positive hub clusters that are not connected (white outlines with yellow arrows). Expression of two RNAis simultaneously does not dilute the strength of the knockdown as ectopic hubs are still seen when Rbf and GFP are knocked down in conjunction (A). E2F knockdown (D), Esg overexpression (F), or Ecad overexpression (H) is sufficient to prevent multiple hubs from forming upon Rbf knockdown, as only a single hub (white outline) is seen in these testes. Note that upon Rbf knockdown, E2F knockdown in the hub (D) restores normal hub size, while overexpression of Esg or Ecad (F and H, respectively) does not. These testes contain a single enlarged hub.
Because the adherens junction molecule E-cadherin (Ecad) is positively regulated by Esg in the Drosophila trachea (TanakaMatakatsu et al., 1996) and a loss of Ecad is seen when Esg is knocked down in the hub (Voog et al., 2014), we wanted to determine if Ecad expression was altered in hub cells upon Rbf knockdown. To determine Ecad expression levels, testes with Rbf knocked down in hub cells for 7 days were immunostained with Ecad antisera and protein levels assessed by fluorescence intensity. A significant reduction in the levels of Ecad expression was seen throughout the entire hub in testes in which Rbf had been knocked down specifically in hub cells compared with GFP RNAi controls (Figure S6), suggesting that Rbf can regulate levels of Ecad-mediated adhesion. To next test the role that cell adhesion plays in the formation of ectopic hubs, Ecad was overexpressed simultaneously with Rbf RNAi knockdown in hub cells for 14 days. This yielded a significant suppression of ectopic hub formation (n = 2 of 73; Figure 6H; Table 1). These testes still contained enlarged hubs with dividing hub cells (Table 1), suggesting that proliferation alone is not sufficient to drive ectopic hub formation. As expected, multiple hubs were never seen when Ecad alone was overexpressed in the hub (Figure 6G; Table 1). These data further support the model that adhesion must be downregulated in hub cells upon Rbf knockdown in order for ectopic niches to form and that upregulating adhesion molecules is sufficient to suppress their formation.
Ectopic Niches Form through Splitting of the Original Hub
The formation of new stem cell niches may be useful for regenerating tissue after injury, but its mis-regulation could lead to cancer (Greaves et al., 2006). Because this is challenging to study in vivo, we wanted to better characterize the cellular aspects of ectopic hub formation upon Rbf knockdown. Theoretically, new niches could occur in two ways: the original hub could split apart (niche fission), as seen in intestinal crypts (Withers and Elkind, 1970), or individual cells could move out of the hub and establish new niches (seeding). To see if either possibility was occurring, Drosophila testes expressing GFP specifically in the hub were imaged live for 12–18 hr after 13 days of Rbf knockdown. Clusters of hub cells were seen both spreading across the periphery of the testis along the basement membrane and protruding toward the center of the testis, as previously seen in our fixed images (Figure S1G). This dichotomy in behavior sometimes drove clusters of hub cells far enough apart to occasionally cause a single cluster of hub cells to split in to two clusters (Video S4). Fixing and immunostaining testes for the hub marker Fas III after live imaging confirmed that the membranes of these multiple hub cell clusters were no longer connected (data not shown), verifying that ectopic hubs can arise from fission of the original hub. Although this does not rule out the seeding mechanism, live imaging of testes expressing Rbf RNAi and the G-TRACE system in hub cells for 10 days (described above) did not show re-expression of the hub cell driver in cells leaving the hub, as indicated by loss of RFP expression. This suggests that converted hub cells and their progeny are most likely not involved in the formation of new hubs. Altogether these data support a model that ectopic niches form because of fission of an enlarged hub and not the seeding of cells leaving the hub.
DISCUSSION
In many tissues, stem cells have the ability to self-renew through either asymmetric or symmetric divisions. However, upon damage, other mechanisms may be required to replenish lost stem cells, as demonstrated in the crypt base columnar stem cells of the mouse intestinal niche (Beumer and Clevers, 2016). In the Drosophila testis, two additional modes of regeneration have been discovered. Dedifferentiation of spermatogonia allows GSCs to be replenished after loss (Brawley and Matunis, 2004), while direct conversion of niche cells to cyst stem cells regenerates the somatic stem cell population after ablation (Hétié et al., 2014). These studies provide vital insight into cellular plasticity, helping advance regenerative therapies for tissue repair. However, un-regulated cellular regeneration can cause cells to over-proliferate, leading to cancer. Thus understanding the mechanisms that underlie such processes is key to providing tissue regeneration therapies without inducing undesirable and unintended side effects.
The tumor suppressor Retinoblastoma (RB) is mutated in many forms of cancer, driving cellular over-proliferation (Burkhart and Sage, 2008). Although Retinoblastoma is best known for its role in cell cycle control, many studies have shown additional functions for Retinoblastoma in regulating endoreplication, apoptosis, genome stability, and cellular differentiation (Calo et al., 2010; Cayirlioglu et al., 2003; Coschi et al., 2014; DeGregori et al., 1997; Dyson, 2016; Tanaka-Matakatsu et al., 2009; Thomas et al., 2001). Although we have previously shown that overexpression of Cyclin D/Cdk4 is sufficient to induce hub cell divisions and cell fate conversion in the Drosophila testis (Hétié et al., 2014), here, we show that Retinoblastoma’s homolog, the Retinoblastoma-family protein (Rbf) is required to actively promote hub cell quiescence and identity. Therefore, we have identified a molecule that acts under normal conditions to maintain the terminally differentiated state of hub cells. Loss of Rbf in the hub causes hub cells to proliferate and convert to CySCs, which we show is mediated through the activity of Rbf’s canonical binding partner E2F. Upon Rbf knockdown in the hub, some cells maintain their hub cell fate, as shown by the appearance of hub cells upon prolonged Rbf knockdown, whereas other cells convert to a cyst lineage fate. Why this is the case is an intriguing question for future studies. Because hub cells start to lose hub their identity while still within the hub cluster (Figures 3A–3C; Videos S1 and S3), it is most likely not autocrine signaling from hub cells that maintains their identity. One possibility could be that the rate of proliferation is greater than the rate of conversion, allowing some hub cells to remain hub cells while others do not. And perhaps in the case of Esg knockdown in hub cells (Voog et al., 2014), the rate of proliferation is too slow for any hub cells to be maintained so all convert to cyst lineage cells. Another possibility could be that the hub cells are more heterogeneous than initially thought, and only a subset of hub cells are capable of maintaining their identity. Little is currently known about hub cell heterogeneity, but in situ hybridization to whole testes for transcripts from the magu gene suggests this possibility (Zheng et al., 2011). Single-cell sequencing of hub cells would provide great insight into the diversity of this cell population.
In addition to being required in hub cells, we show that Rbf is required to regulate somatic cyst cell quiescence but not stem cell fate. Loss of Rbf in cyst cells causes them to proliferate away from the hub but does not cause these cells to express stem cell markers. This suggests that even within the same tissue, Rbf can have different functions within varying somatic cell types. This is surprising because both hub cells and cyst cells are derived from a common pool of somatic gonadal precursor cells in the Drosophila embryo (Le Bras and Van Doren, 2006).
Besides regulating niche cell quiescence and fate, here, we show that Rbf plays a role in regulating niche number in the Drosophila testis. Prolonged loss of Rbf in hub cells causes hub cells to form functional ectopic niches. Multiple direct or indirect downstream targets of E2F play a role in controlling this process, as overexpression of either the transcription factor Esg or the adhesion molecule Ecad is sufficient to suppress ectopic hub formation but not hub cell proliferation. This leads to the intriguing question of what is minimally required to build a new niche. Although niche fission is known to occur within the mammalian intestinal crypt during postnatal development (Maskens and Dujardin-Loits, 1981), regeneration after certain damage (Cairnie and Millen, 1975; Wright and Al-Nafussi, 1982), and upon mutation of the Wnt signaling pathway regulator APC (Wasan et al., 1998), little is known about the mechanisms that drive these processes. Our fixed and live imaging of Rbf knockdown in hub cells of the Drosophila testis suggests that a combination of niche cell proliferation with a loss of cell-cell adhesion can lead to the splitting of the hub causing niche fission. Because crypt fission may play a role in the propagation of cancerous cells within the mammalian intestine (Greaves et al., 2006), better understanding the mechanisms that drive this process will provide vital therapeutic targets.
Great heterogeneity is often observed among the cells that comprise a single human tumor. This can occur from genomic instability within a cell population (Carter et al., 2006) or environmental cues driving vast changes in cellular morphology, proliferation, gene expression, and other cellular processes (Dagogo-Jack and Shaw, 2018; Marusyk and Polyak, 2010). Here, we show that loss of the tumor suppressor gene Rbf in the hub cells of the Drosophila testis can lead to an excess of many different cell types within the tissue through non-autonomous and autonomous means. Increasing niche cell number allows more germline and cyst stem cells to be maintained, presumably because of an increase in the physical size of the niche, niche signals, or both. In addition, Rbf knockdown in the hub leads to the increase of cyst lineage cells because of the conversion of hub cells to CySCs. Together this causes an increase in stem cell number, which consequently leads to an increase in early differentiated daughters, thus driving an expansion of different cell populations within the tissue. Furthermore, the formation of ectopic niches within the testis leads to stem cells’ residing in aberrant locations, similar to cancer metastases. This demonstrates how a mutation in a particular cell type (in this case, specifically within niche cells) can drive many different phenotypes. Thus understanding the cell or origin for many cancers may provide better insight into how certain cancers progress. Future studies dissecting whether quiescent stromal support cells contribute to cancer more generally and whether Rbf family members act in multiple niches will further illuminate the role of niche cells, not just the stem cells they support, in tissue regeneration and disease progression.
STAR⋆METHODS
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti–Fasciclin III(Drosophila) | DSHB | Cat#: 7G10; RRID: AB_528238 |
| Mouse monoclonal anti-phospho-HistoneH3 (Ser10) (6G3) | Cell Signaling Technology | Cat#: 9706S; RRID: AB_331748 |
| Guinea Pig polyclonal anti-Traffic Jam | Laboratory of D. Godt (Li et al., 2003) | N/A |
| Rabbit polyclonal anti-Vasa (d-260) | Santa Cruz Biotechnology | Cat#: SC-30210; RRID: AB_793874 |
| Mouse monoclonal anti-Rbf | Laboratory of N. Dyson (Frolov et al., 2003) | N/A |
| Chick polyclonal anti-GFP | Abcam | Cat#: ab13970; RRID: AB_300798 |
| Rabbit polyclonal anti-DsRed | Clontech | Cat#: 632496; RRID: AB_10013483 |
| Goat polyclonal anti-DsRed (C-20) | Santa Cruz Biotechnology | Cat#: SC-33354; RRID: AB_639922 |
| Mouse monoclonal anti-Discs large(Drosophila) | DSHB | Cat#: 4F3; RRID: AB_528203 |
| Mouse monoclonal anti-Eyes Absent | DSHB | Cat#: eya10H6; RRID: AB_528232 |
| Mouse monoclonal anti-Armadillo | DSHB | Cat#: N27A1; RRID: AB_528089 |
| Guinea Pig polyclonal anti-Zfh1 | Laboratory of E. Matunis | N/A |
| Rabbit polyclonal anti-cleaved DrosophilaDcp1 (Asp216) | Cell Signaling Technology | Cat#: 9578; RRID: AB_2721060 |
| Mouse monoclonal anti-hu-li tai shao | DSHB | Cat#: 1B1; RRID: AB_528070 |
| Mouse monoclonal anti-b-Galactosidase | Promega | Cat#: Z378A; RRID: AB_2313752 |
| Rabbit polyclonal anti-Stat92E | Laboratory of Erika Bach (Flaherty et al., 2010) | N/A |
| Rat monoclonal anti-DE-cadherin | DSHB | Cat#:DCAD2; RRID: AB_528120 |
| Rabbit polyclonal anti-Castor | Laboratory of Denise Montell | N/A |
| Mouse monoclonal anti-Cut | DSHB | Cat#: 2B10; RRID: AB_528186 |
| Goat anti-chicken IgY (H+L) secondary antibody, Alexa Fluor 488 conjugate | ThermoFisher Scientific | Cat#: A11039; RRID: AB_2534096 |
| Donkey anti-chicken IgY (H+L), highly cross absorbed, CF 488A | Millipore/Sigma (formerly Sigma-Aldrich) | Cat#: SAB4600031; RRID: AB_2721061 |
| Goat anti-Mouse IgG (H+L) secondary antibody, Alexa Fluor 405 conjugate | ThermoFisher Scientific | Cat#: A31553; RRID: AB_221604 |
| Goat anti-Mouse IgG (H+L) secondary antibody, Alexa Fluor 488 conjugate | ThermoFisher Scientific | Cat#: A11029; RRID: AB_2534088 |
| Goat anti-Mouse IgG (H+L) secondary antibody, Alexa Fluor 568 conjugate | ThermoFisher Scientific | Cat#: A11004; RRID: AB_2534072 |
| Goat anti-Mouse IgG (H+L) secondary antibody, Alexa Fluor 633 conjugate | ThermoFisher Scientific | Cat#: A21050; RRID: AB_2535718 |
| Goat anti-Guinea Pig IgG (H+L) secondary antibody, Alexa Fluor 488 conjugate | ThermoFisher Scientific | Cat#: A11073; RRID: AB_2534117 |
| Goat anti-Guinea Pig IgG (H+L) secondary antibody, Alexa Fluor 568 conjugate | ThermoFisher Scientific | Cat#: A11075; RRID: AB_2534119 |
| Goat anti-Guinea Pig IgG (H+L) secondary antibody, Alexa Fluor 633 conjugate | ThermoFisher Scientific | Cat#: A21105; RRID: AB_2535757 |
| Goat anti-Rabbit IgG (H+L) secondary antibody, Alexa Fluor 568 conjugate | ThermoFisher Scientific | Cat#: A11011; RRID: AB_143157 |
| Donkey anti-Rabbit IgG Antibody Cy5 CON | Millipore/Sigma (formerly Sigma-Aldrich) | Cat#: AP182SA6; RRID: AB_2721062 |
| Donkey anti-Goat IgG (H+L) secondary antibody, Alexa Fluor 568 conjugate | ThermoFisher Scientific | Cat#: A11057; RRID: AB_2534104 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 4,6-diamidino-2-phenylindole (DAPI) | Millapore/Sigma (formerly Sigma-Aldrich) | Cat#: 10236276001; CAS: 28718–90-3 |
| MIFEPRISTONE-11beta-(4-Dimethy amino) phenyl-17beta-hydroxy-17-(1-propynyl) estra-4,9-dien-3-one; RU-38486; RU-486 | Millapore/Sigma (formerly Sigma-Aldrich) | Cat#: M8046; CAS: 84371–65-3 |
| Insulin from Bovine Pancreas powder | Millapore/Sigma (formerly Sigma-Aldrich) | Cat#: I6634; CAS: 11070–73-8 |
| Fetal Bovine Serum heat inactivatedUS-FBS | Millapore/Sigma (formerly Sigma-Aldrich) | Cat#: F4135 |
| Penicillin-Streptomycin (10,000 U/mL) | ThermoFisher Scientific | Cat#: 15140122; CAS: 69–57-8 (Penicillin) and 3810–74-0 (Sreptomycin) |
| 16% Paraformaldehyde (formaldehyde) aqueous solution | Electron Microscopy Sciences | Cat#: 15710; CAS: 50–00-0 |
| Goat Serum | Millapore/Sigma (formerly Sigma-Aldrich) | Cat#: G9023 |
| Donkey Serum | Millapore/Sigma (formerly Sigma-Aldrich) | Cat#: D9663; |
| Green food color | McCormick | N/A |
| Schneider’s Drosophila Medium | ThermoFisher Scientific | Cat#: 21720024 |
| Shields and Sang M3 Insect Medium | Millapore/Sigma (formerly Sigma-Aldrich) | Cat#: S3652 |
| Vectashield antifade mounting medium | Vector Laboratories | Cat#: H-1000 |
| Poly-L-lysine Hydrobromide Mol Wt 30000 | Millapore/Sigma (formerly Sigma-Aldrich) | Cat#: P2636; CAS: 25988–63-0 |
| Critical Commercial Assays | ||
| Click-iT EdU Alexa Fluor 555 Imaging Kit | ThermoFisher Scientific | Cat#: C10338 |
| Experimental Models: Organisms/Strains | ||
| D. melanogaster: P{w[+mW.hs] = GawB} E132, w[*](also called upd-Gal4) | Bloomington Drosophila Stock Center | BDSC: 26796; FlyBase: FBti0002638 |
| D. melanogaster: w[*]; P{w[+mW.hs] = Switch2}GSG2295 | Laboratory of H. Keshishian (Nicholson et al., 2008) | BDSC: 40266 |
| Flybase: FBti0115018 | ||
| D. melanogaster: Hh-LacZ/TM3,Sb | Laboratory of K. Vani (Mohler et al., 1995) | N/A |
| D. melanogaster: EyaA3-Gal4 | Laboratory of S. DiNardo (Leatherman and Dinardo, 2008) | N/A |
| D. melanogaster: w[*]; P{w[+mC] = tubPGAL80[ts]}2/TM2 | Bloomington Drosophila Stock Center | BDSC: 7017; FlyBase: FBst0007017 |
| D. melanogaster: w[*]; P{w[+mC] = UAS-FLP.D}JD2 | Bloomington Drosophila Stock Center | BDSC: 4540; FlyBase: FBst0004540 |
| D. melanogaster: RNAi of GFP: w[1118];P{w[+mC] = UAS-GFP.dsRNA.R}142 | Bloomington Drosophila Stock Center | BDSC: 9330; FlyBase: FBst0009330 |
| D. melanogaster: RNAi of Rbf: y[1] sc[*] v[1]; P{y[+t7.7] v[+t1.8] = TRiP.HMS03004} attP2/TM3, Sb[1] | Bloomington Drosophila Stock Center | BDSC: 36744; FlyBase: FBst0036744 |
| D. melanogaster: RNAi of Rbf: y[1] sc[*] v[1];P{y[+t7.7] v[+t1.8] = TRiP.GL01293}attP40/CyO | Bloomington Drosophila Stock Center | BDSC: 41863; FlyBase: FBst0041863 |
| D. melanogaster: GTRACE: w[*];P{w[+mC] = UAS-RedStinger}4,P{w[+mC] = UAS-FLP.D}JD1, P{w[+mC] =Ubi-p63E(FRT.STOP)Stinger}9F6/CyO | Bloomington Drosophila Stock Center | BDSC: 28280; FlyBase: FBst0028280 |
| D. melanogaster: RNAi of E2F: y[1] v[1];P{y[+t7.7] v[+t1.8] = TRiP.JF02718}attP2 | Bloomington Drosophila Stock Center | BDSC: 27564; FlyBase: FBst0027564 |
| D. melanogaster: y[1] w[*]; PBac{y[+mDint2] w[+mC] = 20XUAS-6XGFP}VK00018/CyO,P{Wee-P.ph0}Bacc[Wee-P20] | Bloomington Drosophila Stock Center | BDSC: 52261; FlyBase: FBst0052261 |
| D. melanogaster: ywhsFlp; UAS-esg/CyO;UAS-LacZ/TM2 | Laboratory of A. Tomlinson (Kumar et al.,2015) | N/A |
| D. melanogaster: hs-FLP; tub > CD2 > Gal4,UAS-GFP/CyO | Laboratory of B. Ohlstein | N/A |
| D. melanogaster: E132Gal4;;TubGal80ts | Laboratory of E. Matunis | N/A |
| D. melanogaster: EyaA3Gal4;TubGal80ts | Laboratory of E. Matunis | N/A |
| D. melanogaster: UAS-GTRACE/SM6B;UAS-Rbf RNAi/TM6B: P{w[+mC] = UAS-RedStinger}4, P{w[+mC] = UAS-FLP.D}JD1, P{w[+mC] = Ubi-p63E(FRT.STOP)Stinger}9F6/SM6B; P{y[+t7.7] v[+t1.8] =TRiP.HMS03004}attP2/TM6B | This Paper | N/A |
| D. melanogaster: GS2295-Gal4/Sm6B;UAS-Rbf RNAi/TM6B: w[*]; P{w[+mW.hs] =Switch2}GSG2295/SM6B; P{y[+t7.7] v[+t1.8] = TRiP.HMS03004}attP2/TM6B | This Paper | N/A |
| D. melanogaster: tub > CD2 > Gal4, UAS-GFP/SM6B; UAS-Flp/TM6B: tub > CD2 >Gal4, UAS-GFP/SM6B; P{w[+mC] = UAS-FLP.D}JD2/TM6B | This Paper | N/A |
| D. melanogaster: UAS-Rbf RNAi/SM6B;UAS-E2F RNAi/TM6B: P{y[+t7.7] v[+t1.8] = TRiP.GL01293}attP40/SM6B; P{y[+t7.7] v[+t1.8] = TRiP.JF02718}attP2/TM6B | This Paper | N/A |
| D. melanogaster: UAS-esg/SM6B; UASRbf RNAi/TM6B: UAS-esg/SM6B; ]; P{y[+t7.7] v[+t1.8] = TRiP.HMS03004} attP2/TM6B | This Paper | N/A |
| D. melanogaster: UAS-GFP RNAi/SM6B;UAS-Rbf RNAi/TM6B: P{w[+mC] = UASGFP.dsRNA.R}142/SM6B; P{y[+t7.7] v[+t1.8] = TRiP.HMS03004}attP2/TM6B | This Paper | N/A |
| D. melanogaster: UAS-Rbf RNAi/SM6B; Hh-LacZ/TM6B: P{y[+t7.7] v[+t1.8] =TRiP.GL01293}attP40/SM6B; HH-LacZ/TM6B | This Paper | N/A |
| D. melanogaster: 20XUAS-6XGFP/SM6B; UAS-Rbf RNAi/TM6B: PBac{y[+mDint2] w[+mC] = 20XUAS-6XGFP}VK00018/CyO,P{Wee-P.ph0}Bacc[Wee-P20]/SM6B; P{y[+t7.7] v[+t1.8] = TRiP.HMS03004} attP2/TM6B | This Paper | N/A |
| D. melanogaster: w; Frt40A; UAS-shg | Laboratory of J.P. Vincent (Sanson et al., 1996) | N/A |
| D. melanogaster: UAS-Rbf RNAi/SM6B; UAS-shg/TM6B: P{y[+t7.7] v[+t1.8] =TRiP.GL01293}attP40/SM6B; UAS-shg/TM6B | This Paper | N/A |
| Software and Algorithms | ||
| Fiji | (Schindelin et al., 2012) | https://www.fiji.sc/ |
| Zeiss LSM | Carl Zeiss Microscopy | https://www.zeiss.com/microscopy/us/downloads/lsm-5-series.html |
| Zen | Carl Zeiss Microscopy | https://www.zeiss.com/microscopy/int/products/microscope-software/zen.html |
| Imaris version 7.6.5 | Bitplane | http://www.bitplane.com/imaris/imaris |
| Prism 6 | GraphPad | http://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| 35mm glass-bottom Petri dish with 10mm microwell | MatTek Corporation | Cat#: P35G-1.5–10-C |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Erika Matunis (matunis@jhmi.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila stocks and their use in this study
E132Gal4/Y;;TubGal80ts/ TubGal80ts (combined from BDSC: 26796 and 7017) – used to drive UAS-transgene expression specifically in hub cells in a temporal manner (Figure 1, 2, 3, 4, 5, 6, S1, S3, S5, S6, Table 1, S1, and Video S1, S2, S3, S4).
EyaA3Gal4/EyaA3Gal4;TubGal80ts/ TubGal80ts (combined from EyaA3Gal4 from S. DiNardo and BDSC: 7017) – used to drive UAS-transgene expression specifically in cyst cells in a temporal manner (Figure S4).
;;UAS-GFP RNAi/ UAS-GFP RNAi (BDSC: 9330) – used as a control RNAi in measuring hub cell proliferation and ectopic hub formation (Figure 1, 3, 5, S1, S4, S5, S6, and Table 1).
;UAS-Rbf RNAi/CyO; (BDSC: 41863) – used in conjunction with different Gal4 drivers to knockdown Rbf in specific cell types (Figure 1, 3, 4, 5, S1, S5, and Table 1).
;;UAS-Rbf RNAi/TM3,Sb (BDSC 36744) - used in conjunction with different Gal4 drivers to knockdown Rbf in specific cell types (Figure 1, 3, 4, 5, 6, S1, S4, S5, S6, and Table 1).
;UAS-GTRACE/CyO; (BDSC: 28280) – used for lineage tracing and live imaging in control hubs (Figure 2, S3, Table S1, and Video S2).
;UAS-GTRACE/SM6B; UAS-Rbf RNAi/TM6B (combined from BDSC: 28280 and 36744) – used for lineage tracing and live imaging in hubs with Rbf knocked down (Figure 2, S3, Table S1, and Video S1, S3).
; GS2295-Gal4/CyO; (BDSC: 40266) and;tub > CD2 > GAL4, UAS-GFP/SM6B; UAS-Flp/TM6B (combined from hsflp; tub > CD2 > GAL4, UAS-GFP/CyO from B. Ohlstein laboratory and BDSC: 4540) – used as an alternative tool to lineage trace control hub cells (Figure S2 and Table S1).
; GS2295-Gal4/Sm6B; UAS-Rbf RNAi/TM6B (combined from BDSC: 40266 and BDSC: 36744) and;tub > CD2 > GAL4, UAS-GFP/ SM6B; UAS-Flp/TM6B (combined from hsflp; tub > CD2 > GAL4, UAS-GFP/CyO from B. Ohlstein laboratory and BDSC: 4540) – used as an alternative tool to lineage trace hub cells with Rbf knocked down (Figure S2 and Table S1).
;;UAS-E2F RNAi / UAS-E2F RNAi (BDSC: 27564) – used to knockdown E2F in a cell specific manner (Figure 4, 6, and Table 1).
;UAS-Rbf RNAi/SM6B; UAS-E2F RNAi/TM6B (combined from BDSC: 41863 and 27564) – used to knockdown Rbf and E2F simultaneously in cells to test for phenotype rescue (Figure 4, 6, and Table 1).
;UAS-Rbf RNAi/SM6B; UAS-GFP RNAi/TM6B (combined from BDSC: 41863 and 9330) - used to test the effectiveness of driving two RNAi lines simultaneously by knocking down Rbf and GFP simultaneously (Figure 6 and Table 1).
yw hsFlp/Y; UAS-esg/CyO; UAS-LacZ/TM2 (from A. Tomlinson laboratory) – used to overexpress Escargot in cells (Figure 4, 6, and Table 1).
;UAS-esg/SM6B; UAS-Rbf RNAi/TM6B (combined from ywhsFlp/Y; UAS-esg/CyO; UAS-LacZ/TM2 from A. Tomlinson laboratory and BDSC: 36744) – used to overexpress Escargot and knockdown Rbf simultaneously to test for phenotype rescue (Figure 4, 6, and Table 1).
;UAS-Rbf RNAi/SM6B; Hh-LacZ/TM6B (combined from BDSC: 41863 and Hh-Lacz/TM3,Sb from K. Vani laboratory) – used to assess Hedgehog expression in ectopic hubs (Figure S5).
;20XUAS-6XGFP/SM6B; UAS-Rbf RNAi/TM6B (combined from BDSC: 52261 and 36744) – used to measure hub volume in testes in which Rbf has been knocked down. Used to image ectopic hub formation in real-time (Figure S1 and Video S4).
w; Frt40A; UAS-shg (from J.P. Vincent laboratory) – used to overexpress E-cadherin in cells (Figure 6, and Table 1).
;UAS-Rbf RNAi/SM6B; UAS-shg/TM6B (combined from BDSC: 41863 and UAS-shg from J.P. Vincent laboratory) – used to overexpress E-cadherin and knockdown Rbf simultaneously to test for phenotype rescue (Figure 6, and Table 1).
Drosophila husbandry
Flies were raised on a standard yeast/molasses medium (1212.5mL water, 14.7mL agar, 20.4g yeast, 81.8g cornmeal, 109.1ml molasses, 10.9mL Tegosept, 3.4mL propionic acid, 0.4mL phosphoric acid/ per tray of 100 vials) supplemented with dry yeast at 18C unless otherwise indicated. Male flies between 0–5 days of age were used for all experiments and subject to different conditions as noted within the text, figures, legends, and STAR methods.
METHOD DETAILS
Transgene Induction
Heat induction
Flies containing a temperature sensitive TubGal80 were grown at the permissive temperature 18C and shifted to the non-permissive temperature 29C for either 3, 7, or 14 days as indicated to induce transgene expression of RNAi lines or overexpression lines.
Drug induction
RU486 powder was dissolved in 200 proof ethyl alcohol to make a 50mg/mL stock solution stored at −20°C. For GS2295-Gal4 induction, 0–5 day old flies raised at 25°C were placed in vials with filter paper soaked in 125 μL of 2mg/mL of RU486 in apple juice and 2.5 μL green food coloring. Flies were transferred to fresh drug/apple juice vials daily for 3 days at 25°C. Flies were checked for greendyed guts to ensure drug consumption, then transferred to a standard yeast/molasses food for 18 days at 25°C prior to dissecting, fixing, and immunostaining.
In vivo EdU labeling (Leatherman and Dinardo, 2008)
For in vivo labeling of cells undergoing S phase, flies were dissected in Schneider’s Drosophila medium then testes still in their cuticles transferred to a glass dissection dish with 500 μL of 10 μM EdU in Schneider’s Drosophila medium for 20 minutes at room temperature. After soaking, testes were immediately transferred to a microtube with fixation solution (4% paraformaldehyde in 1X PBS with 0.1% Triton X-100) for 25 minutes at room temperature on a nutator. Testes were then washed in 1X PBX (1X PBS with 0.1% Triton X-100) followed by incubation in 250 μL of the ClickIT reaction cocktail (2.5 μL EdU buffer additive diluted in 22.5 μL of dH20, 25 μL 10X reaction buffer diluted in 190 μL of dH20 with 10 μL 100mM CuSO4, and 0.625 μL Alexa fluor azide 555) as per the kit’s instructions for 30 minutes to visualize EdU. After the reaction, testes were washed in 1X PBX, blocked, and stained as described below.
Dissection and Immunohistochemistry (Matunis et al., 1997)
Dissection
Flies were anesthetized using CO2 then dissected with the cuticle still surrounding the testes in 1X Becker Ringer’s solution (111 mM NaCl, 1.88 mM KCl, 64 μM NaH2PO4, 816 μM CaCl2, 2.38 mM NaHCO3) (Ashburner, 1989) and transferred immediately to fixation solution (4% paraformaldehyde in 1X PBS with 0.1% Triton X-100) for 22 minutes at room temperature on a nutator.
Immunohistochemistry
All washes were conducted using 1X PBX (1X PBS with 0.1% Triton X-100) except the final wash before adding mounting media in which 1X PBS was used. Testes were blocked overnight at 4°C in 1X PBX with 3% BSA, 0.02% NaN3, and 2% goat or donkey serum. Antisera was diluted in 1X PBX with 3% BSA and 0.02% NaN3. Testes were immunostained in primary antisera overnight at 4°C with the exception of mouse anti-Eyes Absent which was incubated at 4°C for three days and rabbit anti-STAT92E which was incubated at 4°C for one day and room temperature for one day. Secondary antibodies were diluted in 1X PBX containing 3% BSA and 0.02% NaN3 and testes incubated overnight at 4°C. The nuclear counterstain 4,6-diamidino-2-phenylindole (DAPI) (Millapore/Sigma) was added to most secondary antibody dilutions at a final concentration of 1 μg/mL. All testes were mounted in Vectashield antifade mounting medium (Vector Laboratories).
All polyclonal antibodies were stored in a 1:2 dilution at −20°C, and all monoclonal antibodies were stored at 4°C except for mouse anti-phosphohistone H3 and mouse anti-β-Galactosidase which were stored at −20°C in a 1:2 glycerol dilution. Antisera was used at the following final concentrations:
Mouse monoclonal anti-Fasciclin III 1:50
Mouse monoclonal anti-phosphohistone H3 1:400
Guinea Pig polyclonal anti-Traffic Jam 1:20,000
Rabbit polyclonal anti-Vasa 1:200
Mouse monoclonal anti-Rbf 1:10
Chick polyclonal anti-GFP 1:10000
Rabbit polyclonal anti-dsRed 1:1000
Goat polyclonal anti-dsRed 1:500
Mouse monoclonal anti-Discs Large 1:50
Mouse monoclonal anti-Eyes Absent 1:10
Mouse monoclonal anti-Armadillo 1:50
Guniea Pig polyclonal anti-Zfh1 1:1000
Rabbit polyclonal anti-cleaved Drosophila Dcp1 1:200
Mouse monoclonal anti-hu-li tai shao (1B1) 1:50
Mouse monoclonal anti-β-Galactosidase 1:1000
Rabbit polyclonal anti-Stat92E 1:1000
Rat monoclonal anti-DE-cadherin 1:20
Rabbit polyclonal anti-Castor 1:50
Mouse monoclonal anti-Cut 1:20
All Fluor 488 secondary antibodies were used at a final concentration of 1:400. All other secondary antibodies were used at a final concentration of 1:200.
Lineage Tracing
In the following genotypes, expression of the Gal4 driver caused permanent expression of GFP in hub cells and their descendants. Marked cells were detected by immunostaining. Testes with GFP cells outside the confines of the hub cluster that no longer expressed hub markers were considered positive for converting cells. The hub cluster was defined as either hub-Gal4 expressing cells marked by RFP for lineage tracing using the G-TRACE system, or Fasciclin III positive cells for lineage tracing using the alternative marking system. To trace hub cells after Rbf knockdown, E132Gal4/Y; UAS-GTRACE/+; TubGal80ts/UAS-Rbf RNAi flies were raised at 18C and 0–5 day old males shifted to 29°C for 7 or 14 days as indicated. E132Gal4/Y; UAS-GTRACE/+; TubGal80ts/+ flies lacking Rbf RNAi were processed in parallel to control for age and temperature. For the alternative lineage tracing method, GS2295-Gal4/ Tub > CD2 > Gal4,UAS-GFP; UAS-Flp/UAS-Rbf RNAi flies were raised at 25C and 0–5 day old males fed RU486 for 3 days then allowed to recover for 18 days at 25C as described above. GS2295-Gal4/Tub > CD2 > Gal4,UAS-GFP; UAS-Flp/+ flies lacking Rbf RNAi were processed in parallel to control for age, temperature, and drug consumption.
Extended Live Imaging (Greenspan and Matunis, 2017)
Flies were dissected in 1X Becker Ringer’s solution (111 mM NaCl, 1.88 mM KCl, 64 μM NaH2PO4, 816 μM CaCl2, 2.38 mM NaHCO3) (Ashburner, 1989) and testes completely removed from the cuticle and separated from the accessory glands. Testes were mounted on a 35mm glass bottom dish with a 10mm microwell (MatTek Corporation). Imaging dishes were coated with 200 μL of 1mg/mL of poly-Llysine dissolved in 0.1M Trizma buffer pH 8.5 for at least one hour then rinsed with sterile dH20 prior to testis mounting. Testes were cultured in either 500 μL or 1000 μL of Schneider’s Drosophila medium pH 7 or Shields and Sang M3 Insect medium supplemented with 15% fetal bovine serum (v/v), 0.5X penicillin/streptomycin, and 0.2mg/mL insulin from bovine pancreas dissolved in acidified water. Live images were acquired using a Zeiss LSM 780 microscope. Up to nine different positions per dish were imaged using a programmable xy stage. Images were acquired in 25 or 10 minutes intervals for up to 18 hours. Z stacks ranging 30 μm with 1.25 μm steps were acquired for each time point. Only testes that contained a strong fluorescent signal and minimal sample drift in the x, y, and z planes throughout the movie were included for analysis. Samples whose hub cells were too far away from the coverslip to follow individual cellular behaviors, or those testes with a lot of movement due to muscle sheath contraction were excluded. After completion of live imaging, culture media was removed from the imaging dish and testes fixed and stained directly in the dish as described above.
Microscopy and Image Analysis
Image acquisition
Fixed images were obtained using either a Zeiss LSM 5 Pascal equipped with a 63x oil immersion objective, 405nm diode, 488nm ArKr, and 543nm HeNe lasers with digital zoom or a Zeiss LSM 700 (JHU SOM microscope facility) equipped with a 63x oil immersion objective, 405nm diode, 488nm solid-state, 561nm solid-state, and 639nm diode lasers with digital zoom. A Zeiss LSM 780 microscope (JHU SOM microscope facility) equipped with a 63x oil immersion objective, 405nm diode, 488nm Ar, 561 solid-state, and 639 diode lasers with digital zoom was used to acquire all live imaging, fixed images taken from those samples imaged in real time, and those used to measure hub volume. Images were acquired using either Zeiss LSM or Zen software. All Z stacks through the testis tissue had a step size of 1.25 μm except for Z stack used in ploidy and E-cadherin fluorescence measurements in which a 0.5 μm step size was used.
Image processing
Images acquired using Zeiss LSM or Zen software were processed using Fiji or IMARIS software. Brightness for individual channels from single confocal slices was enhanced using Fiji, then each channel overlaid to form a merged image. All 3D reconstructions were created in IMARIS using acquired Z stacks and the volume tool to create a composite image. Brightness for individual channels was also enhanced within the IMARIS software.
QUANTIFICATION AND STATISTICAL ANALYSIS
Hub cell proliferation quantification
To quantify hub cell divisions, testes were subject to EdU labeling to label cells in S phase (as described above) and/or immunostained with the mitotic marker phosphohistone H3. Testes with EdU and/or PH3 marked cells within the confines of the hub cluster were considered positive for dividing hub cells. The hub cluster was defined as those cells marked by the hub membrane marker Fasciclin III. Testes that had no EdU incorporation throughout the entire tissue were excluded.
Apoptosis quantification
To quantify dying hub cells, testes were immunostained with the apoptotic marker cleaved Drosophila Dcp1, and the hub cluster indicated by the hub membrane marker Fasciclin III. Since some Dcp1 labeling could be seen in non-dying cells, dying hub cells were considered only those cells within the confines of the hub cluster in which Dcp1 labeled the entire cell. These cells tended to be smaller in size due to cell shrinking upon apoptosis activation.
Ectopic hub quantification
To quantify ectopic hubs, testes were immunostained with the hub membrane marker Fasciclin III. Z stacks were acquired to include the entire hub range. Hub clusters whose membranes were no longer connected in any Z planes were considered separate hubs. Testes with more than one hub cluster were considered positive for ectopic hubs.
Hub volume measurement
To determine hub volume upon Rbf knockdown, Z stacks of fixed testes whose hub cells expressed a bright cytosplasmic GFP and Rbf RNAi for 3, 7, or 14 days as indicated were acquired to include the entire hub range. Sibling flies that did not express Rbf RNAi were used to control for age and temperature. A surface rendering of the entire hub was generated using the surface tool with default parameters within IMARIS software. To generate the surface rendering, only a segment of the testis with the hub, as indicated by the GFP channel, was included. A surface area detail level of 0.25 μm was utilized with an absolute intensity that contained at least 10 voxels. The GFP fluorescence intensity threshold was adjusted for each surface rendering so that the entire hub was included within the object but bounded by Fasciclin III membrane staining. GFP expression outside the Fasciclin III bounds was considered to be converting hub cells and was excluded from hub volume measurements. Once the surface rendering was created, the volume could be automatically measured using the statistics tab.
Live cell tracking and fluorescence intensity measurements
To track cell behavior over the course of the movie, individual hub cells were randomly selected from 5 GTRACE only control movies and 7 GTRACE; Rbf RNAi movies where flies had been shifted to 29°C for 10 days prior to live imaging. Only movies acquired with 16 bits at 10min time point intervals were utilized for fluorescence quantification and testes that had too much sample drift in the x, y, or z planes were excluded. Cells were tracked manually using the Spots tool within IMARIS software. Hub cell diameter was set at 4 μm for the XY plane and 4 μm for the Z plane. Cells were tracked using the red channel only, by manually clicking on the cell at the first and last time point. Fluorescence intensity and the X, Y and Z coordinates were automatically measured for the first and last time points using the Spots function. For each cell, percent of red fluorescence intensity remaining was calculated using the following formula:
% loss = 1 + (fluorescence at last time point – fluorescence at first time point)/ fluorescence at first time point. Displacement was calculated using the following formula: . Since some movies were longer in time than others each measurement was calculated for the last common time point T91.
E-cadherin fluorescence intensity measurements
To measure levels of E-cadherin expression, testes were immunostained with E-cadherin and DAPI. Z stacks with a 0.5 μm step size were acquired to include the entire hub range. Fluorescence signal was acquired at the same gain in the linear range for all samples. Fluorescence intensity was measured using FIJI software. All stacks containing the hub were merged into a single summed slice and Ecadherin fluorescence intensity was measured by drawing an object around the entire hub and using the measure feature. The corrected total cell fluorescence (CTCF) was then calculated by taking the integrated density and subtracting the area times the mean of 3 fluorescence background readings. The CTCF for E-cadherin fluorescence was then normalized over the CTCF for the DAPI channel, which was calculated in the same manner. The normalized E-cadherin fluorescence measurements were compared for testes expressing Rbf RNAi in their hub cells to those expressing GFP RNAi in their hub cells for 7 days.
Ploidy Measurements
To determine the normal ploidy of hub cells, CTCF measurements were calculated, as described above, for the DAPI channel of control testes (testes from flies expressing GFP RNAi in their hub cells for 7 days). The CTCF measurement for the entire hub was then divided by the number of hub cells within each hub to determine the CTCF DAPI reading per hub cell. CTCF DAPI measurements of hub cells were then divided by the mean CTCF DAPI measurement of all sperm cells measured to determine ploidy. To measure DAPI fluorescence of sperm cells, seminal vesicles were opened up to allow sperm to be mounted on the same slide as testes. Z stacks with a 0.5 μm step size were acquired to include many sperm. Stacks containing many sperm were merged into a single summed slice. To measure the DAPI fluorescence of multiple sperm at a time a ROI map was generated in FIJI with a threshold of 4626 65535. Only entire sperm that were in the field of view were measured. CTCF DAPI measurements were then calculated for each sperm.
Statistical Analysis
For all quantifications, n represents the number of testes analyzed. Statistical significance was expressed as P values and determined using a Fisher’s exact test for most measurements except hub volume and E-cadherin fluorescence measurements in which an unpaired t test with Welch’s correction was used. Linear regression analysis was used to compare displacement of hub cells over time to the percent of red fluorescence remaining. All statistical tests were run using PRISM 6 software. (*) denotes p < 0.05, (**) denotes p < 0.01, (***) denotes p < 0.001, and (****) denotes p < 0.0001 and (ns) denotes values that were not significant. Error bars represent standard deviation.
Bar graphs and tables indicate the percent of testes with a certain phenotype out of all the testes analyzed with that genotype. Scatterplots show raw data of the entire distribution of values observed with the average reported as a horizontal black line and bars representing the standard deviation above and below the average. XY graph shows individual hub cells for each genotype plotted as displacement versus percent red fluorescence remaining. Linear regression lines were generated for each genotype based on the calculated R2 values.
Supplementary Material
Highlights.
Loss of Retinoblastoma in the hub causes hub cell division and conversion to CySCs.
Retinoblastoma acts through E2F to maintain hub quiescence, fate, and niche number.
Extended loss causes ectopic niche formation through fission of the original hub.
Overexpression of Esg or E-cadherin can suppress ectopic niche formation.
ACKNOWLEDGMENTS
We thank S. DiNardo, D. Drummond-Barbosa, B. Edgar, B. Ohlstein, and the Bloomington Drosophila Stock Center for flies; E. Bach, N. Dyson, D. Godt, and the Developmental Studies Hybridoma Bank for antibodies; and S. Hasan, M. Akeju, and M. de Cuevas for comments. This work was supported by NIH grants R01GM120107, R01HD052937 (to E.L.M.), and F31HD085748 (to L.J.G.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures, one table, and four videos and can be found with this article online at https://doi.org/10.1016/j.celrep.2018. 08.083.
REFERENCES
- Ashburner M (1989). Drosophila: A Laboratory Manual (Cold Spring Harbor Laboratory; ), p. 376. [Google Scholar]
- Beumer J, and Clevers H (2016). Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development 143, 3639–3649. [DOI] [PubMed] [Google Scholar]
- Brawley C, and Matunis E (2004). Regeneration of male germline stem cells by spermatogonial dedifferentiation in vivo. Science 304, 1331–1334. [DOI] [PubMed] [Google Scholar]
- Burkhart DL, and Sage J (2008). Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 8, 671–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairnie AB, and Millen BH (1975). Fission of crypts in the small intestine of the irradiated mouse. Cell Tissue Kinet. 8, 189–196. [DOI] [PubMed] [Google Scholar]
- Calo E, Quintero-Estades JA, Danielian PS, Nedelcu S, Berman SD, and Lees JA (2010). Rb regulates fate choice and lineage commitment in vivo. Nature 466, 1110–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Eklund AC, Kohane IS, Harris LN, and Szallasi Z (2006). A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet 38, 1043–1048. [DOI] [PubMed] [Google Scholar]
- Cayirlioglu P, Ward WO, Silver Key SC, and Duronio RJ (2003). Transcriptional repressor functions of Drosophila E2F1 and E2F2 cooperate to inhibit genomic DNA synthesis in ovarian follicle cells. Mol. Cell. Biol 23, 2123–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, and Rando TA (2013). Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol 14, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschi CH, Ishak CA, Gallo D, Marshall A, Talluri S, Wang J, Cecchini MJ, Martens AL, Percy V, Welch I, et al. (2014). Haploinsufficiency of an RB-E2F1-Condensin II complex leads to aberrant replication and aneuploidy. Cancer Discov. 4, 840–853. [DOI] [PubMed] [Google Scholar]
- Dagogo-Jack I, and Shaw AT (2018). Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol 15, 81–94. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Leone G, Miron A, Jakoi L, and Nevins JR (1997). Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. U S A 94, 7245–7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominado N, La Marca JE, Siddall NA, Heaney J, Tran M, Cai Y, Yu F, Wang H, Somers WG, Quinn LM, and Hime GR (2016). Rbf regulates Drosophila spermatogenesis via control of somatic stem and progenitor cell fate in the larval testis. Stem Cell Reports 7, 1152–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson NJ (2016). RB1: a prototype tumor suppressor and an enigma. Genes Dev. 30, 1492–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CJ, Olson JM, Ngo KT, Kim E, Lee NE, Kuoy E, Patananan AN, Sitz D, Tran P, Do MT, et al. (2009). G-TRACE: rapid Gal4-based cell lineage analysis in Drosophila. Nat. Methods 6, 603–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaherty MS, Salis P, Evans CJ, Ekas LA, Marouf A, Zavadil J, Banerjee U, and Bach EA (2010). chinmo is a functional effector of the JAK/STAT pathway that regulates eye development, tumor formation, and stem cell selfrenewal in Drosophila. Dev. Cell 18, 556–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Stevaux O, Moon NS, Dimova D, Kwon EJ, Morris EJ, and Dyson NJ (2003). G1 cyclin-dependent kinases are insufficient to reverse dE2F2-mediated repression. Genes Dev. 17, 723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gönczy P, and DiNardo S (1996). The germ line regulates somatic cyst cell proliferation and fate during Drosophila spermatogenesis. Development 122, 2437–2447. [DOI] [PubMed] [Google Scholar]
- Greaves LC, Preston SL, Tadrous PJ, Taylor RW, Barron MJ, Oukrif D, Leedham SJ, Deheragoda M, Sasieni P, Novelli MR, et al. (2006). Mitochondrial DNA mutations are established in human colonic stem cells, and mutated clones expand by crypt fission. Proc. Natl. Acad. Sci. U S A 103, 714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan LJ, and Matunis EL (2017). Live imaging of the Drosophila testis stem cell niche. Methods Mol. Biol 1463, 63–74. [DOI] [PubMed] [Google Scholar]
- Greenspan LJ, de Cuevas M, and Matunis E (2015). Genetics of gonadal stem cell renewal. Annu. Rev. Cell Dev. Biol 31, 291–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy RW, Tokuyasu KT, Lindsley DL, and Garavito M (1979). The germinal proliferation center in the testis of Drosophila melanogaster. J. Ultrastruct. Res 69, 180–190. [DOI] [PubMed] [Google Scholar]
- Hasan S, Hétié P, and Matunis EL (2015). Niche signaling promotes stem cell survival in the Drosophila testis via the JAK-STAT target DIAP1. Dev. Biol 404, 27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hétié P, de Cuevas M, and Matunis E (2014). Conversion of quiescent niche cells to somatic stem cells causes ectopic niche formation in the Drosophila testis. Cell Rep. 7, 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y-C, de Rooij DG, and Page DC (2013). Tumor suppressor gene Rb is required for self-renewal of spermatogonial stem cells in mice. Proc. Natl. Acad. Sci. U S A 110, 12685–12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba M, Yuan H, and Yamashita YM (2011). String (Cdc25) regulates stem cell maintenance, proliferation and aging in Drosophila testis. Development 138, 5079–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiger AA, Jones DL, Schulz C, Rogers MB, and Fuller MT (2001). Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science 294, 2542–2545. [DOI] [PubMed] [Google Scholar]
- Korenjak M, Anderssen E, Ramaswamy S, Whetstine JR, and Dyson NJ (2012). RBF binding to both canonical E2F targets and noncanonical targets depends on functional dE2F/dDP complexes. Mol. Cell. Biol 32, 4375–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SR, Patel H, and Tomlinson A (2015). Wingless mediated apoptosis: How cone cells direct the death of peripheral ommatidia in the developing Drosophila eye. Dev. Biol 407, 183–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras S, and Van Doren M (2006). Development of the male germline stem cell niche in Drosophila. Dev. Biol 294, 92–103. [DOI] [PubMed] [Google Scholar]
- Leatherman JL, and Dinardo S (2008). Zfh-1 controls somatic stem cell selfrenewal in the Drosophila testis and nonautonomously influences germline stem cell self-renewal. Cell Stem Cell 3, 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MA, Alls JD, Avancini RM, Koo K, and Godt D (2003). The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat. Cell Biol 5, 994–1000. [DOI] [PubMed] [Google Scholar]
- Ma Q, Wawersik M, and Matunis EL (2014). The Jak-STAT target Chinmo prevents sex transformation of adult stem cells in the Drosophila testis niche. Dev. Cell 31, 474–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marusyk A, and Polyak K (2010). Tumor heterogeneity: causes and consequences. Biochim. Biophys. Acta 1805, 105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maskens AP, and Dujardin-Loits RM (1981). Kinetics of tissue proliferation in colorectal mucosa during post-natal growth. Cell Tissue Kinet. 14, 467–477. [DOI] [PubMed] [Google Scholar]
- Matunis E, Tran J, Go¨ nczy P, Caldwell K, and DiNardo S (1997). punt and schnurri regulate a somatically derived signal that restricts proliferation of committed progenitors in the germline. Development 124, 4383–4391. [DOI] [PubMed] [Google Scholar]
- Mohler J, Mahaffey JW, Deutsch E, and Vani K (1995). Control of Drosophila head segment identity by the bZIP homeotic gene cnc. Development 121, 237–247. [DOI] [PubMed] [Google Scholar]
- Nicholson L, Singh GK, Osterwalder T, Roman GW, Davis RL, and Keshishian H (2008). Spatial and temporal control of gene expression in Drosophila using the inducible GeneSwitch GAL4 system. I. Screen for larval nervous system drivers. Genetics 178, 215–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlstein B, Kai T, Decotto E, and Spradling A (2004). The stem cell niche: theme and variations. Curr. Opin. Cell Biol 16, 693–699. [DOI] [PubMed] [Google Scholar]
- Rotgers E, Rivero-Müller A, Nurmio M, Parvinen M, Guillou F, Huhtaniemi I, Kotaja N, Bourguiba-Hachemi S, and Toppari J (2014). Retinoblastoma protein (RB) interacts with E2F3 to control terminal differentiation of Sertoli cells. Cell Death Dis. 5, e1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson B, White P, and Vincent JP (1996). Uncoupling cadherin-based adhesion from wingless signalling in Drosophila. Nature 383, 627–630. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng XR, and Matunis E (2011). Live imaging of the Drosophila spermatogonial stem cell niche reveals novel mechanisms regulating germline stem cell output. Development 138, 3367–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JE, Korenjak M, and Brehm A (2003). Tumour suppressors—a fly’s perspective. Eur. J. Cancer 39, 1355–1362. [DOI] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Uemura T, Oda H, Takeichi M, and Hayashi S (1996). Cadherin-mediated cell adhesion and cell motility in Drosophila trachea regulated by the transcription factor Escargot. Development 122, 3697–3705. [DOI] [PubMed] [Google Scholar]
- Tanaka-Matakatsu M, Xu J, Cheng L, and Du W (2009). Regulation of apoptosis of rbf mutant cells during Drosophila development. Dev. Biol 326, 347–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DM, Carty SA, Piscopo DM, Lee JS, Wang WF, Forrester WC, and Hinds PW (2001). The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell 8, 303–316. [DOI] [PubMed] [Google Scholar]
- Voog J, D’Alterio C, and Jones DL (2008). Multipotent somatic stem cells contribute to the stem cell niche in the Drosophila testis. Nature 454, 1132–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voog J, Sandall SL, Hime GR, Resende LPF, Loza-Coll M, Aslanian A, Yates JR 3rd, Hunter T, Fuller MT, and Jones DL (2014). Escargot restricts niche cell to stem cell conversion in the Drosophila testis. Cell Rep. 7, 722–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasan HS, Park HS, Liu KC, Mandir NK, Winnett A, Sasieni P, Bodmer WF, Goodlad RA, and Wright NA (1998). APC in the regulation of intestinal crypt fission. J. Pathol 185, 246–255. [DOI] [PubMed] [Google Scholar]
- Withers HR, and Elkind MM (1970). Microcolony survival assay for cells of mouse intestinal mucosa exposed to radiation. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med 17, 261–267. [DOI] [PubMed] [Google Scholar]
- Wright NA, and Al-Nafussi A (1982). The kinetics of villus cell populations in the mouse small intestine. II. Studies on growth control after death of proliferative cells induced by cytosine arabinoside, with special reference to negative feedback mechanisms. Cell Tissue Kinet. 15, 611–621. [PubMed] [Google Scholar]
- Yacobi-Sharon K, Namdar Y, and Arama E (2013). Alternative germ cell death pathway in Drosophila involves HtrA2/Omi, lysosomes, and a caspase-9 counterpart. Dev. Cell 25, 29–42. [DOI] [PubMed] [Google Scholar]
- Zheng Q, Wang Y, Vargas E, and DiNardo S (2011). magu is required for germline stem cell self-renewal through BMP signaling in the Drosophila testis. Dev. Biol 357, 202–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.