Abstract
Recent development in statistical methodology for personalized treatment decision has utilized high-dimensional regression to take into account a large number of patients’ covariates and described personalized treatment decision through interactions between treatment and covariates. While a subset of interaction terms can be obtained by existing variable selection methods to indicate relevant covariates for making treatment decision, there often lacks statistical interpretation of the results. This paper proposes an asymptotically unbiased estimator based on Lasso solution for the interaction coefficients. We derive the limiting distribution of the estimator when baseline function of the regression model is unknown and possibly misspecified. Confidence intervals and p-values are derived to infer the effects of the patients’ covariates in making treatment decision. We confirm the accuracy of the proposed method and its robustness against misspecified function in simulation and apply the method to STAR*D study for major depression disorder.
Keywords: Large p Small n, Model Misspecification, Optimal Treatment Regime, Robust Regression
1. Introduction
Precision medicine aims to design optimal treatment for each individual according to their specific conditions in the hope of lowering medical cost and improving efficacy of treatments. Existing methods can be broadly partitioned into regression-based or classification-based approaches. Popular regression-based approaches include Q-learning [21, 11, 3, 6, 7, 17] and A-learning [13, 10, 8, 16, 15]. Q-learning models the conditional mean of the outcome given covariates and treatment while A-learning directly models the interaction between treatment and covariates that is sufficient for treatment decisions. Other regression-based methods have been developed in [18], [12], [19], etc. In contrast, classification-based approaches, also known as policy search or value search, estimate the marginal mean of the outcome for every treatment regime within a pre-specified class and then take the maximizer as the estimated optimal regime; see, for example, [13], [22], [23], [25], [26]. As emerging technology makes it possible to gather an extraordinarily large number of prognostic factors for each individual, such as genetic information and clinical measurements, regression-based and classification-based methods have been extended to handle high-dimensional data [12, 25, 26, 19]
This paper focuses on regression-based approaches and plans to address the current limitation that the selected interaction effects often lack statistical interpretation when the number of covariates exceeds the sample size. Consider the following semi-parametric model. Denote X as the n × p design matrix, where n is the number of patients and p is the number of patients’ covariates. Let be the design matrix with an additional column of 1’s and the i-th row of . Denote Yi as the outcome and Ai = {0,1} the treatment assignment of the i-th patient. Assume
| (1) |
where μ(Xi) is an unspecified baseline function and β0 is the vector of unknown coefficients for the interactions between treatment and patients’ covariates. We are interested in inference on β0 and optimal treatment decisions based on the estimated β0.
We propose an asymptotically unbiased estimator for β0 and derive its limiting distribution when p > n. Consequently, confidence intervals and p-values of the interaction coefficients can be calculated. Because the baseline function is unknown and possibly misspecified, we investigate the robustness of the estimator to misspecified μ(Xi).
The proposed estimator for β0 and the p-value calculated from data can be utilized to make significance-based optimal treatment decision. We illustrate the efficiency of the new treatment regime in a real application to STAR*D study for major depression disorder.
2. Method and Theory
2.1. An A-learning framework
We adopt the robust regression approach of [16] and transform the interaction from to , where π(Xi) = E(Ai|Xi) is the propensity score. Because E(Ai −π|Xi) = 0, the transformed interaction is orthogonal to the baseline function μ(Xi) given Xi. By doing so, we can protect the estimation of β0 from the effect of baseline function misspecification [8, 16]. For simplicity of presentation, we consider a completely randomized study with prespecified propensity score, i.e. A = {0,1} and π(Xi) = P(Ai = 1|Xi) = π. Let , then (1) is equivalent to
The true μπ(·) is unknown. Let be an estimator of μπ(·) that does not necessarily converge to the true μπ(·). Assume uniformly for some function . Because E(Ai − π|Xi) = 0, then and are orthogonal, which implies that
| (2) |
where D(A,π) = diag{A1 − π,…,An − π}. We apply the Lasso to regress on and obtain
| (3) |
It is well-known that the limiting distribution of Lasso solution is difficult to derive. The misspecified baseline function adds another layer of difficulty [2].
2.2. Unbiased estimation with misspecified baseline function
We propose a de-sparsified estimator for β0 motivated by [24] and [20]:
| (4) |
where is an estimator of the precision matrix of .
We show that this estimator is asymptotically unbiased and derive its limiting distribution as follows. Decompose into three terms:
Where , and is the sample covariance matrix of .
Consequently, for an arbitrary q × (p + 1) matrix, H, we can decompose into three terms. The reason to study is to explore the asymptotic joint distribution of q arbitrary linear contrasts of , which includes the limiting distribution of a single as a special case. We derive the limiting distribution of by showing that , and .
We first show a preliminary result on the lasso solution with misspecified baseline function. As is an important component in Δ1, this preliminary result helps to derive the asymptotic properties of . Consider the following assumptions, where C denotes a generic constant, possibly varying from place to place:
Condition 1: The random error i of the true model (1) are independent and identically distributed with and .
Condition 2: There exists a such that for any ,
and
Condition 3: Denote as the jth column of . for all 1 ≤ j ≤ p + 1.
Condition 4: Define Σ as the covariance matrix of . Σ has smallest eigenvalue .
Condition 5: The number of important interaction terms satisfies that .
Lemma 2.1. Consider model (1). Assume Conditions 1 – 5. The lasso solution from (3) with satisfies
| (5) |
Next, we show that and are at the order of op (1). Similar to [20], we apply lasso for nodewise regression to construct . Details of the construction are presented in Section A.1. Consider additional assumptions:
Condition 6: The prespecified matrix Hq×(p+1) satisfies
||H||∞ ≤ C and
ht = |{j ≠ t: Ht,j ≠ 0}| ≤ C for any t ∈ {1,…,q}.
Condition 7: The precision matrix Θ of satisfies
||Θ||∞ ≤ C and
sj = |{k ≠ j: Θj,k ≠ 0}| ≤ C for any j ∈ {1,…,p}.
Lemma 2.2. Consider model (1). Assume Conditions 1 – 7. Obtain by (3) with and by nodewise regression with for any j ∈ {1,…,p}. Then
| (6) |
| (7) |
The remaining component of is . We show that converges to a multivariate normal distribution. Since η does not involve , less conditions are needed in the following lemma.
Lemma 2.3. Consider model (1). Assume Conditions 1– 3 and 6 – 7. Obtain by nodewise regression with for any j ∈ {1,…,p}. Then
| (8) |
where G = HΘVΘTHT is a q×q nonnegative matrix, , , and ||G||∞ < ∞.
We summarize the above results in the following theorem.
Theorem 2.4. Consider model (1). Assume Conditions 1 – 7. Obtain by (3) with and by nodewise regression with for any j ∈ {1,…,p}. Then
where G is defined in (8).
2.3. Some examples for
In real applications, we need to choose an estimator for the baseline function. The only requirement for is Condition 2, which is very general. Here we discuss two examples for .
Example 1: . In this case, Condition 2 (a) holds trivially.
Condition 2 (b) is implied by
Condition 8: .
The following corollary presents the limiting distribution of in this case. Proof of this corollary is straightforward and, thus, omitted.
Corollary 2.5. Consider model (1). Implement . Assume
Conditions 1, 3 – 7, and 8. Obtain by (3) with and by nodewise regression with for any j ∈ {1,…,p}. Then,
where G is defined in (8).
Example 2: , where is obtained by
| (9) |
Note that the solution from (9) is equivalent to the solution from (3) (detail of the proof is included in the proof of Corollary 2.6). In this case, we replace Condition 2 with Condition 9: Define ; γ∗ satisfies
and
Corollary 2.6. Consider model (1). Implement and obtain and from (9) with Assume Conditions 1, 3 – 7, and 9.
Obtain by nodewise regression with for any j ∈ {1,…,p}.
Then,
where G is defined in (8).
2.4. Interval estimation and p-value
The theoretical result on the asymptotic normality of the de-sparsified estimator can be utilized to construct confidence intervals for the coefficients of interest and to perform hypothesis testing. The variance-covariance matrix G can be approximated by where
Therefore, a pointwise (1 − α) confidence interval of β0,j can be constructed as
| (10) |
where , and Φ(·) is the c.d.f of N(0,1).
One can also calculate the asymptotic p-value for testing
for a given j ∈ {1,…,p}. A non-zero β0,j means that variant Xj is relevant in making treatment decision for a patient.
3. Simulation
We consider three simulation settings with different baseline functions:
Case 1: ,
Case 2: ,
Case 3: , where X1 is the first covariate of X.
We simulate the random error from N(0,1) and the design matrix X from multivariate normal MN(0,Σ), where Σ = AR(0.5). The parameters in the baseline functions is set as γ = (1,1,0,···,0)T, and the parameter of interest β0 has s nonzero elements with values 1 or 1.5. The treatment main effect is set at zero.
We apply our de-sparsified estimator : in (4) with and derive confidence intervals by (10). To evaluate the finite-sample performance of our method, we report the following measures. Denote CIj as the 95% confidence interval for β0,j and Strue = {j ∈ {2,…p + 1}: β0,j ≠ 06}.
MAB( ): the mean absolute bias of for relevant variables, calculated as .
MAB( ): the mean absolute bias of lasso solution for relevant variables, calculated as .
Coverage for noise variables: the empirical value of .
Coverage for relevant variables: the empirical value of .
Length of CI for noise variables: the empirical value of .
Length of CI for relevant variables: the empirical value of .
Tables 1–2 summarize the performance measures for case 1–3 from 100 simulations with n = 200, p = 300 and s = 5 or 10. It is shown that the mean absolute bias for relevant variables of our estimator is significantly smaller than that of the lasso solution in all settings. The coverages of the confidence intervals are fairly consistent for all cases 1–3, concurring with the theoretical results on the robustness of the method in Corollary 2.6. We notice that the coverage of confidence intervals for noise variables is very close to the nominal level of 0.95, but that for the relevant variants is lower than 0.95. Similar phenomena have been observed for the original de-sparsified estimator in [20] and [4]. This phenomenon indicates that even though the bias of the original Lasso estimator has been corrected asymptotically by the de-sparsifying procedure, the non-zero coefficients can still be under-estimated with finite sample. Further, given that the implemented , it agrees with our expectation that the lengths of the confidence intervals are shortest for Case 1 when the true baseline function is linear and increases in Case 2 and 3 with nonlinear baseline functions.
Table 1.
Coverage of confidence interval and mean absolute bias (MAB) of with p = 300 and s = 5. Standard errors are in parenthesis.
| Intensity=1 | Intensity=1.5 | |||||
|---|---|---|---|---|---|---|
| Case 1 | Case 2 | Case 3 | Case 1 | Case 2 | Case 3 | |
| 0.17(0.06) | 0.40(0.14) | 0.30(0.10) | 0.17(0.06) | 0.41(0.15) | 0.30(0.11) | |
| 0.23(0.06) | 0.50(0.14) | 0.39(0.11) | 0.23(0.06) | 0.53(0.18) | 0.40(0.13) | |
| Coverage for noise variables | 0.95(0.01) | 0.95(0.01) | 0.95(0.03) | 0.95(0.01) | 0.94(0.05) | 0.95(0.01) |
| Coverage for relevant variables | 0.88(0.13) | 0.89(0.15) | 0.85(0.18) | 0.88(0.13) | 0.86(0.20) | 0.86(0.17) |
| Length of CI for noise variables | 0.65(0.04) | 1.50(0.17) | 1.21(0.91) | 0.65(0.04) | 1.50(0.17) | 1.12(0.12) |
| Length of CI for relevant variables |
0.65(0.04) | 1.49(0.16) | 1.20(0.92) | 0.65(0.04) | 1.49(0.16) | 1.11(0.11) |
Table 2.
Coverage of confidence interval and mean absolute bias (MAB) of with p = 300 and s = 10.
| Intensity=1 | Intensity=1.5 | |||||
|---|---|---|---|---|---|---|
| Case 1 | Case 2 | Case 3 | Case 1 | Case 2 | Case 3 | |
| 0.19(0.04) | 0.42(0.10) | 0.33(0.08) | 0.19(0.04) | 0.46(0.27) | 0.43(0.96) | |
| 0.24(0.06) | 0.51(0.10) | 0.40(0.09) | 0.24(0.06) | 0.57(0.27) | 0.52(0.96) | |
| Coverage for noise variables | 0.95(0.03) | 0.94(0.04) | 0.95(0.03) | 0.94(0.05) | 0.94(0.04) | 0.94(0.05) |
| Coverage for relevant variables | 0.86(0.14) | 0.86(0.16) | 0.84(0.14) | 0.84(0.18) | 0.84(0.16) | 0.82(0.18) |
| Length of CI for noise variables | 0.69(0.05) | 1.58(0.18) | 1.19(0.13) | 0.69(0.05) | 1.64(0.46) | 1.23(0.47) |
| Length of CI for relevant variables |
0.69(0.05) | 1.58(0.18) | 1.18(0.13) | 0.69(0.05) | 1.63(0.45) | 1.22(0.47) |
4. Application to STAR*D study
We consider the dataset from multi-site, randomized, multi-step STAR*D (Sequenced Treatment Alternatives to Relieve Depression) study for Major depressive disorder (MDD), which is a chronic and recurrent common disorder [5, 14]. In STAR*D study, patients received citalopram (CIT) which is a selective serotonin reuptake inhibitor antidepressant at Level 1. Patients who have had unsatisfactory outcome at level 1 are included in level 2 to randomly receive one of the two treatment switch options: sertraline (SER) and bupropion (BUP). Patients who received treatment at level 2 but have not showed sufficient improvement will be randomized at level 2A.
Our data contains 319 patients who received treatment switch options at level 2. Among them, 48% of the patients received BUP and 52% received SER. Except for treatment indicator, 308 covariate variables of the patients are included. The outcome of interest is the Quick Inventory of Depressive SymptomatologySelf-report QIDS-SR16. Similar to existing studies on STAR*D [15], we transform the outcome to be the negative of the original outcome. We are interested in making optimal treatment decision between SER and BUR to maximize the mean outcome.
We apply the de-sparsified estimator in (4) with and derive the p-value based on the limiting distribution in Corollary 2.6. The top-ranked p-values that are less than 0.05 are presented in Table 3 with the corresponding covariates.
Table 3.
Top-ranked p-values and the corresponding covariates that are most relevant for making treatment decision.
| Index | Covariate | p-value | se of | |
|---|---|---|---|---|
| 1 | Qccur_r_rate | 0.0056 | −1.84 | 0.66 |
| 2 | URNONE | 0.0079 | 2.35 | 0.88 |
| 3 | NVTRM | 0.0097 | −1.45 | 0.56 |
| 4 | IMPWR | 0.010 | −1.47 | 0.57 |
| 5 | hWL | 0.011 | −1.58 | 0.62 |
| 6 | GLT2W | 0.013 | −1.52 | 0.61 |
| 7 | DSMTD | 0.017 | −1.50 | 0.63 |
| 8 | IMSPY | 0.019 | −1.36 | 0.58 |
| 9 | hMNIN | 0.020 | 1.40 | 0.60 |
| 10 | EARNG | 0.028 | −1.43 | 0.65 |
| 11 | URPN | 0.029 | −1.23 | 0.56 |
| 12 | PETLK | 0.039 | −1.18 | 0.57 |
| 13 | NVCRD | 0.041 | 1.17 | 0.57 |
| 14 | EMSTU | 0.046 | −1.39 | 0.69 |
The meanings of the notations in Table 3 are as follows. Qccur r rate: QIDSC score changing rates. URNONE: no symptoms in patients’ urination category. NVTRM: tremors. IMPWR: indicating whether patients thought they have special powers. hWL: hRS Weight loss. DSMTD: recurrent thoughts of death, recurrent suicidal ideation, or suicide attempt. hMNIN: hRS Middle insomnia. EMSTU: did you worry a lot that you might do something to make people think that you were stupid or foolish?
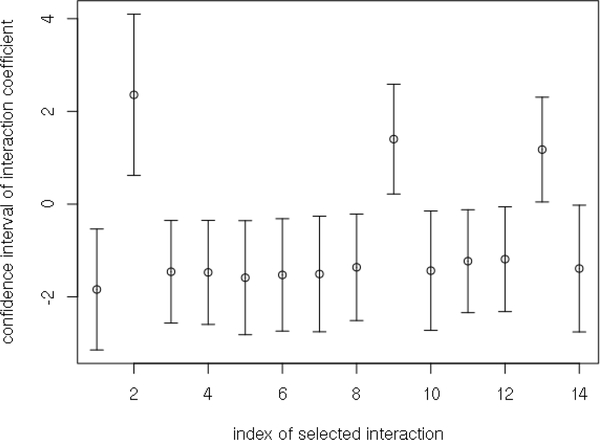
We have also derived the 95% confidence intervals by (10) for the interaction effects between all covariates and treatment options. The confidence intervals for the top 14 interaction effects are presented in Figure 1.
Fig 1.
The 95% confidence intervals of the top 14 interaction effects
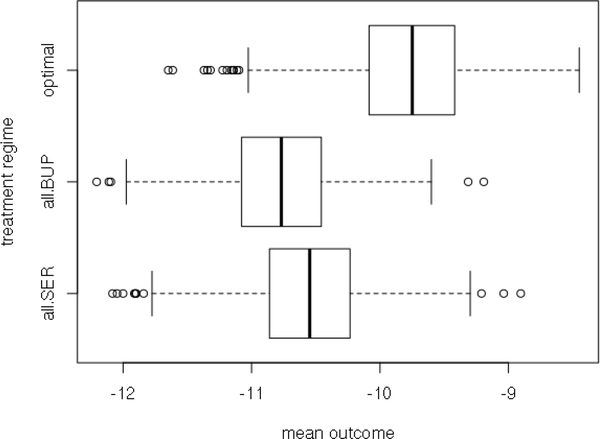
We also apply cross validation to evaluate the effects of the selected variables in making optimal treatment decision. Specifically, we randomly divide the data in half into the training dataset and the testing dataset. On the training dataset, we use the 14 selected variables to compute the least squares estimator for interaction coefficients and formulate the estimated optimal treatment regime. On the corresponding testing dataset, we use the estimated value function derived by the inverse probability weighted estimator [23] to measure the performance of the estimated optimal treatment regime. For comparison, we also compute the estimated value functions for the regimes of assigning only SER and only BUP. We conducted 1000 data splittings and present the boxplot of the estimated value functions of all the treatment regimes in Figure 2. It shows that our estimated optimal treatment regime generally gives larger estimated value functions than those of assigning only SER and only BUP.
Fig 2.
Boxplot of the estimated value functions from cross validation testing samples. ’optimal’ is for the estimated optimal treatment regime; ’all.BUP’ is for the regime of assigning only BUP; ’all.SER’ is for the regime of assigning only SER.
5. Discussion
In this paper, we have considered randomized studies with a pre-specified propensity score. The proposed method can be extended to observation studies with high-dimensional covariates, where the propensity score model needs to be correctly specified from data. Related work on propensity score estimation can be found in [16]. However, the derivation of the limiting distribution of our desparsified estimator for the treatment-covariates interaction coefficients would be much more involved and requires further investigation.
Appendix A: Appendix
A.1. Construction of
We apply lasso for nodewise regression to obtain a matrix such that is close to I as in [9]. Let denote the matrix obtained by removing the jth column of . For each j = 1,…,p + 1, let
with components ,k = 1,…,p + 1 and k ≠ j. Further, define
and
A.2. A preliminary lemma
Define
| (11) |
Lemma A.1. Assume conditions 1 – 3, we have
Proof of Lemma A.1:
| (12) |
For the first term of (12), it is clear that by condition 1,
Further, by the moment inequality in Chapter 6.2.2 of [1],
where the second inequality is by conditions 1 and 3. Then, by Markov inequality,
| (13) |
Next, consider the second term of (12). It is easy to see that since ,
Then, similar arguments as those leading to (13) combined with conditions 2 (b) and 3 gives
| (14) |
Finally, by condition 2 (a), the third term of (12)
| (15) |
A.3. Proof of Lemma 2.1
By the construction of , we have
Recall the definition of in (11), then
The first term of the righ-hand side
where the order of is derived in Lemma A.1.
The rest of the proof is similar to the proof of the second claim of Lemma 2 in [2], where it is shown that given conditions 3 – 5, the compatibility condition holds with probability tending to 1. Then using Lemma A.1 and similar arguments in Section 6.2.2 of [1], the inequality in (5) holds.
A.4. Proof of Lemma 2.2
Consider (6) first. Recall
Rewrite as
Since and by condition 3, by
Multivariate Central Limit Theorem, the (p + 1)-vector
On the other hand, condition 7 combined with similar arguments as in the proof of the first claim of Lemma 2 in [2] gives Summarizing the above with condition 6 and Slutsky’s Theorem gives
| (16) |
(6) follows from (16) and condition 2 (a).
Next, consider (7).
By condition 6, Similar arguments as those leading to inequality (10) in [20], combined with conditions 3 and 7, result in . Combing the above with Lemma 2.2 gives (7).
A.5. Proof of Lemma 2.3
We first show . By definition of η, we have
The first term of the above is equal to 0 because by condition 1. The second term also equals to 0 since .
Next, rewrite as
where
The second moment of Wi is
The first term
The second term
Note that by condition 1. This combined with conditions 2 (b) and 3 gives ||V||∞ < ∞. Consequently, ||HΘVΘTHT|| < ∞ is implied by conditions 6 – 7.
On the other hand, condition 7 combined with similar arguments as in the proof of the first claim of Lemma 2 in [2] gives . By Multivariate Central Limit Theorem and Slutsky’s Theorem,
Result in (8) follows.
A.6. Proof of Corollary 2.6
After obtaining and from (9), implement . Define
First, we show that By the construction of β∗,
| (17) |
On the other hand, the construction of implies
where the second inequality is by (17). Therefore, by the uniqueness of the solution of convex optimization, .
Secondly, we show that
| (18) |
Given the fact that and are orthogonal and from condition 1, definition of γ∗ implies
Define . Then conditions 1 and 9 (b) imply
| (19) |
By the second claim of Lemma 2 of [2], (19) combined with conditions 3 – 5 and 9 (a) gives (18).
Next, it is easy to see that condition 2 (a) is implied by (18) and condition 3, and condition 2 (b) holds trivially given condition 9 (b). Therefore, condition 2 is satisfied and the rest of the proof is the same as the proof of Theorem 2.4.
Footnotes
MSC 2010 subject classifications: Primary 62J05, 62F35; secondary 62P10.
References
- [1].Bühlmann P and van de Geer S (2011). Statistics for high-dimensional data - methods, theory and applications. Springer. [Google Scholar]
- [2].Bühlmann P and van de Geer S (2015). High-dimensional inference in misspecified linear models. Electronic Journal of Statistics 9, 1449–1473. [Google Scholar]
- [3].Chakraborty B, Murphy SA, and Strecher VJ (2010). Inference for non-regular parameters in optimal dynamic treatment regimes. Statistical Methods in Medical Research 19(3), 317–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Dezeure R, Bühlmann P, Meier L, and Meinshausen N (2015). Highdimensional inference: Confidence intervals, p-values and r-software hdi. Statistical Science 30(4), 533–558. [Google Scholar]
- [5].Fava M, Rush AJ, Trivedi MH, Nierenberg AA, Thase ME, Sackeim HA, Quitkin FM, Wisniewski S, Lavori PW, Rosenbaum JF, et al. (2003). Background and rationale for the sequenced treatment alternatives to relieve depression (star* d) study. Psychiatric Clinics of North America 26(2), 457–494. [DOI] [PubMed] [Google Scholar]
- [6].Goldberg Y, Song R, and Kosorok MR (2013). Adaptive q-learning. Institute of Mathematical Statistics Collections 9, 150–162. [Google Scholar]
- [7].Laber EB, Linn KA, and Stefanski LA (2014). Interactive model building for q-learning. Biometrika 101, 831–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lu W, Zhang HH, and Zeng D (2013). Variable selection for optimal treatment decision. Statistical Methods in Medical Research 22(5), 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Meinshausen N and Buhlmann P (2006). High-dimensional graphs and variable selection with the Lasso. The Annals of Statistics 34(3), 1436–1462. [Google Scholar]
- [10].Murphy S (2003). Optimal dynamic treatment regimes. Journal of The Royal Statistical Society: Series B 65, 331–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Murphy S (2005). A generalization error for q-learning. Journal of Machine Learning Research 6, 1073–1097. [PMC free article] [PubMed] [Google Scholar]
- [12].Qian M and Murphy SA (2011). Performance guarantees for individualized treatment rules. The Annals of Statistics 39, 1180–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Robins J, Hernan MA, and Brumback B (2000). Marginal structural models and causal inference in epidemiology. Epidemiology 11, 550–560. [DOI] [PubMed] [Google Scholar]
- [14].Rush AJ, Fava M, Wisniewski SR, Lavori PW, Trivedi MH, Sackeim HA, Thase ME, Nierenberg AA, Quitkin FM, Kashner TM, et al. (2004). Sequenced treatment alternatives to relieve depression (star* d): rationale and design. Controlled Clinical Trials 25(1), 119–142. [DOI] [PubMed] [Google Scholar]
- [15].Shi C, Fan A, Song R, and Lu W (2018). High-dimensional a-learning for optimal dynamic treatment regimes. The Annals of Statistics 46(3), 925–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Shi C, Song R, and Lu W (2016). Robust learning for optimal treatment decision with np-dimensionality. Electronic Journal of Statistics 10, 2894–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Song R, Wang W, Zeng D, and Kosorok MR (2015). Penalized qlearning for dynamic treatment regimens. Statistica Sinica 25, 901–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Su X, Tsai C-L, Wang H, Nickerson DM, and Li B (2009). Subgroup analysis via recursive partitioning. Journal of Machine Learning Research 10, 141–158. [Google Scholar]
- [19].Tian L, Alizadeh AA, Gentles AJ, and Tibshirani R (2014). A simple method for estimating interactions between a treatment and a large number of covariates. Journal of the American Statistical Association 109, 1517–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].van de Geer S, Bühlmann P, Ritov Y, and Dezeure R (2014). On asymptotically optimal confidence regions and tests for high-dimensional models. The Annals of Statistics 42(3), 1166–1202. [Google Scholar]
- [21].Watkins CJCH and Dayan P (1992). Q-learning. Machine Learning 8, 279–292. [Google Scholar]
- [22].Zhang B, Tsiatis AA, Davidian M, Zhang M, and Laber EB (2012). Estimating optimal treatment regimes from a classification perspective. Stat 1, 103–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhang B, Tsiatis AA, Laber EB, and Davidian M (2012). A robust method for estimating optimal treatment regimes. Biometrics 68, 1010–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang C and Zhang SS (2014). Confidence intervals for low dimensional parameters in high dimensional linear models. Journal of the Royal Statistical Society. Series B 76(1), 217–242. [Google Scholar]
- [25].Zhao Y, Zeng D, Rush AJ, and Kosorok MR (2012). Estimating individualized treatment rules using outcome weighted learning. Journal of the American Statistical Association 107, 1106–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhao YQ, Zeng D, Laber EB, Song R, Yuan M, and Kosorok MR (2015). Doubly robust learning for estimating individualized treatment with censored data. Biometrika 102, 151–168. [DOI] [PMC free article] [PubMed] [Google Scholar]