Abstract
HIV-1-associated neurocognitive impairment (HAND) is associated with blood-brain-barrier (BBB) inflammation, and inflammation involves toll-like receptors (TLRs) signaling. It is not known whether primary human brain microvascular endothelial cells (HBMEC), the major BBB component, express TLRs or whether TLRs are involved in BBB dysfunction and HAND. We demonstrate that HBMEC express TLR3, 4, 5, 7, 9, and 10, and TLR3 was the most abundant. HIV-1 and TLR3 activation increased endothelial TLR3 transcription and expression. HIV-1-positive human subjects showed significantly higher TLR3 expression in brain tissues and blood vessels, with higher TLR3 levels in subjects with HAND. HIV-1 and TLR3 activation increased endothelial IL6 expression by 6-to-127-fold (P<0.001), activated c-jun(serine-63) and SAPK/JNK(Thr183/Tyr185). HIV-1 upregulated IL6 through interleukin-1 receptor-associated-kinase(IRAK)-1/4/TAK1/JNK pathways, via ATP-dependent JNK activation. TLR3 activation upregulated IL6 through TAK1/JNK pathways, via ATP-dependent or -independent JNK activation. HIV-1 and TLR3 activation also upregulated transcription factors associated with IL6 and TAK1/JNK pathways (Jun, CEBPA, STAT1). Blocking TLR3 activation prevented HIV-1-and TLR3 ligands-induced upregulation of these transcription factors, prevented IL6 transcription and expression, c-jun and JNK activation. HIV-1 and TLR3 ligands significantly increased monocytes adhesion and migration through the BBB, and decreased endothelial claudin-5 expression. Blocking TLR3 and JNK activation prevented HIV-1- and TLR3 ligands-induced claudin-5 downregulation, monocytes adhesion and transendothelial migration. These data suggests that viral immune recognition via endothelial TLR3 is involved in endothelial inflammation and BBB dysfunction in HIV/AIDS and HAND. Our data provides novel insights into the molecular basis of these HIV-1- and TLR3-mediated effects.
Keywords: TLR3, HIV-1, blood-brain barrier, IL6, TAK1/MKK7/JNK signaling, human brain microvascular endothelial cells, claudin-5, monocytes
Background
Toll-like receptors (TLRs) are pattern-recognition receptors that detect invading pathogens via their pathogen associated molecular patterns (PAMP), and the resulting TLR/PAMP interaction triggers the innate immune system [1, 2]. Thus, TLRs play a major role in innate immunity. Thirteen different TLRs have been discovered so far, of which 11 (TLR1 to TLR11) are expressed in human cells [3, 4]. TLR1, 2, 4, 5, 6, and 10 are mostly expressed on the outer cell membranes, whereas TLR3, 7, 8, and 9 are mostly expressed on endosomal membranes [3, 4]. However, the localization of TLRs vary amongst cell types; TLR3 is often located intracellularly on endosomes in many cells, but in human astrocytes TLR3 is expressed both intracellularly and on the cell surface [5]. TLR3 can be activated by viral double-stranded RNA (dsRNA) [6], viral single-stranded RNA (ssRNA) [7], endogenous viral mRNA [8], and polyinosinic-polycytidylic acid (PIC), a synthetic analogue of viral dsRNA [9]. It has been demonstrated that viral dsRNA is a replication intermediate of several ssRNA viruses; and that dsRNA are produced during the course of replication of ssRNA viruses such as the respiratory syncytial virus, encephalomyocarditis virus and West Nile virus, and activate TLR3 [10–12].
Ligand-induced TLR activation can be beneficial since the TLR-ligand complex initiates a quick and protective innate immune response against invading pathogens [1, 2]. However, it can also be detrimental because uncontrolled and unregulated TLR activation can result in excessive inflammation, cell injury, and tissue damage [13–15]. In fact, TLR3 has been implicated in the pathogenesis of several viral infections, including HIV-1- and SIV-induced inflammation in lymph nodes and astrocytes [16, 17], HIV-1 transactivation, replication, and inflammation [18, 19].
Human brain microvascular endothelial cells (HBMEC) are a major component of the blood-brain barrier (BBB). Under normal physiological conditions, HBMEC functions as an interface between the blood and the brain parenchyma, strictly regulating influx of ion molecules and leukocytes into the central nervous system (CNS) [20, 21]. During HIV-1 infection, the BBB function is compromised, and this may enable the entry of free virus and/or HIV-infected mononuclear phagocytes into the brain, infection of resident CNS cells and inflammation, and these may contribute to the development of HIV-1 associated neurocognitive impairments (HAND) [21–24]. We previously showed that HIV-1-induced BBB dysfunction was associated with activation of pro-inflammatory cytokines and chemokines in HBMEC, and that IL6 and TLR signaling were major canonical pathways involved in this HIV-induced BBB inflammation [25, 26]. It is not known whether primary HBMEC express TLRs, and which of the 11 human TLRs may be involved in HIV-induced BBB dysfunction. In the present study, we identify the TLRs expressed in primary HBMEC. We show that HBMEC express TLR3, 4, 5, 7, 9, and 10, and TLR3 was the most abundant TLR expressed in HBMEC. Ex-vivo studies using human brain tissues confirmed these findings and showed significantly higher TLR3 transcription and expression in brain tissues of HIV-1-infected subjects, mostly on the brain blood vessels; with much higher TLR3 levels in brain tissues of subjects with HAND or HIV encephalitis (HIVE). We also showed that exposure of HBMEC to HIV-1 or the TLR3 ligand PIC increased TLR3 levels and IL6 expression via Transforming growth factor-β activated kinase-1 (TAK1) and c-Jun N-terminal kinase (JNK) Pathways. Functional studies showed that both HIV-1 and PIC significantly increased monocytes adhesion and migration through in vitro BBB models, and decreased the expression of the endothelial tight junction protein claudin-5. The TLR3/dsRNA complex inhibitor, as well as JNK inhibitors blocked HIV-1- and PIC-induced claudin-5 downregulation, monocytes adhesion and transendothelial migration. These data suggest that viral immune recognition via endothelial TLR3 is involved HIV-1-induced endothelial inflammation and BBB dysfunction; and that HIV-induced inflammation, BBB dysfunction and HAND are associated with increased TRL3 expression in the CNS and brain endothelium.
Materials and Methods
Brain endothelial cells (EC) culture
Primary HBMEC were isolated from brain tissue obtained during surgical removal of epileptogenic cerebral cortex in adult patients, under an Institutional Review Board-approved protocol at the University of Arizona as described previously [27, 28]. Routine evaluation by immunostaining for von-Willebrand factor, Ulex europaeus lectin and CD31 showed that cells were >99% pure. Freshly isolated cells were cultured in collagen-coated flasks or 6-well culture plates using Dulbecco’s Modified Eagle Medium (DMEM)/F12 (Life Technologies, Grand Island, NY, USA) containing 10% fetal bovine serum (FBS, Atlanta Biologicals, Flowery Branch, GA, USA), supplemented with 10 mmol/l L-glutamine (Life Technologies), 1% heparin (Thermo Fisher Scientific, Pittsburgh, PA, USA), 1% endothelial cell growth supplement (ECGS; BD Bioscience, San Jose, CA, USA), 1% penicillin-streptomycin (Life Technologies), 1% fungizone (MP Biomedicals, Solon, OH, USA). Cells at passage 2 to 4 were used in this study.
Human brain tissues
Brain tissues (cortex) from HIV-1-seropositive patients with or without neurocognitive impairment, and HIV-1/2-seronegative controls were obtained from the National NeuroAIDS Tissue Consortium and the Department of Pharmacology and Experimental Neuroscience brain bank. The clinical histories of all brain tissue donors are detailed in Table 1.
Table 1.
Clinical history of brain tissues donors
| HIV-1 Status |
ID | Gender/ Age (y) |
PMI (h) |
Neurocognition/ Neuropathology |
Other autopsy diagnosis |
|---|---|---|---|---|---|
| Neg | N1 | M/35 | 8.5 | Normal/None | Mild Alzheimer gliosis |
| Neg | N2 | N/A | N/A | Normal/None | N/A |
| Neg | N3 | F/38 | 5.75 | Normal/Not significant | Mild gliosis, lung and bile duct carcinoma |
| Neg | N4 | M/32 | 4.25 | Normal/Not significant | Cystic fibrosis, and multiorgan failure |
| Neg | N5 | F/46 | 4 | Normal/Not significant | Mild fibrosis, mild patchy gliosis |
| Neg | N6 | F/49 | 4.5 | Normal/Not significant | Hepatic cirrhosis, liver failure |
| Neg | N7 | M/52 | 5.25 | Normal/None | Hypertension, renal failure |
| Neg | N8 | M/72 | 3 | Normal/None | Hypertension, COPD |
| Neg | N9 | M/64 | 3.3 | Normal/None | Hepatic carcinoma |
| Neg | N10 | M/56 | 3 | Normal/Not significant | Lung adenocarcinoma, mild nonspecific cortical atrophy |
| Neg | N11 | M/67 | 3.5 | Normal/None | COPD, TB |
| Neg | N12 | M/61 | 5.75 | Normal/Not significant | Cardiomyopathy, mild gliosis |
| Pos | P1 | ?/46 | 2.75 | Normal/None | N/A |
| Pos | P2 | ?/27 | 8 | Normal/None | N/A |
| Pos | P3 | ?/37 | 5 | Normal/None | N/A |
| Pos | P4 | M/39 | 11 | Normal/None | Minimal non-diagnostic abnormalities |
| Pos | P5 | M/35 | 6.5 | Normal/None | Non-Hodgkins lymphoma, AIDS |
| Pos | P6 | M/54 | 6.5 | Normal/None | AIDS |
| Pos | P7 | M/48 | 15 | Normal/None | Liver disease |
| Pos | P8 | M/52 | 8 | Normal/None | AIDS |
| Pos | P9 | M/38 | 14 | Normal | Pneumonia, hemophilia A, AIDS |
| Pos | HAND1/D1 | M/30 | 6 | HAND/HIVE | Minimal terminal anoxia |
| Pos | HAND2/D2 | ?/50 | 21 | HAND/HIVE | N/A |
| Pos | HAND3/D3 | ?/39 | 12 | HAND/HIVE | N/A |
| Pos | HAND4/D4 | ?/40 | 12 | HAND/HIVE | N/A |
| Pos | HAND5/D5 | M/47 | 11 | HAND/HIVE | Encephalopathy, microglial nodule encephalitis, meningoencephalitis with microvascular damage |
| Pos | HAND6/D6 | M/40 | 5 | HAND/HIVE | Non-Hodgkin’s lymphoma, AIDS, lymphocytic meningitis |
| Pos | HAND7/D7 | M/44 | 4 | HAND/HIVE | Microglial nodules, astrocytosis, widespread gliosis, AIDS |
| Pos | HAND8/D8 | M/52 | 5 | HAND/HIVE | Atherosclerosis |
| Pos | HAND9/D9 | M/34 | 11.5 | HAND/HIVE | AIDS |
| Pos | HAND10/D10 | M/38 | 7 | HAND/HIVE | HIV encephalopathy / Leukoencephalopathy, AIDS |
Neg indicates HIV seronegative; Pos, HIV seropositive; HAND, HIV-associated neurocognitive disorders; HIVE, HIV encephalitis; y, years; M, male; F, female; ? or N/A, not available; and PMI, postmortem interval; AIDS, acquired immunodeficiency syndrome; COPD, chronic obstructive pulmonary disease; TB, tuberculosis.
RNA extraction and cDNA synthesis
Confluent HBMEC in six-well plates were treated with high molecular weight PIC (50 µg/ml) (InvivoGen, San Diego, CA, USA) or HIV-1ADA (multiplicity of infection, MOI: 0.001) for 2 to 24 hours. Controls consisted of untreated cells and cells treated with similar concentrations of PIC or HIV-1 in the presence of the TLR3/dsRNA Complex Inhibitor (TLR3.CI) (30 nM). Total RNA was extracted from HBMEC and human brain tissues using the Trizol reagent (Life Technologies) according to the manufacturer’s protocol. RNA was further cleaned using Total RNA cleanup kit (Qiagen, Valencia, CA, USA); RNA yield and quality were checked using a NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE) and for all samples absorbance ratio of 260/280 was ≥2. cDNA was generated from 1 µg RNA in a 20 µl reaction volume, using the Verso cDNA kit (Thermo Fisher, Waltham, MA, USA), according to the manufacturer’s instructions. Reverse transcription was carried out for 30 min at 42°C.
Reverse transcription (RT)-PCR
Two microliters of the cDNA obtained was used for RT-PCR, in a 20 µl reaction volume containing 50 U/ml Taq DNA polymerase supplied in a proprietary reaction buffer (pH 8.5) (Promega, Madison, WI, USA), 400 µM each dNTP, 3 mM MgCl2 (all from Promega), and 25 µM human TLRs (1–10) specific primers (InvivoGen) (Table 2). Positive controls consisted of PCR performed using 10 ng TLRs (1–10) cDNA provided by the manufacturer (InvivoGen). PCR was performed in a GeneAmp® PCR System 9700 (Applied Biosystems, Foster City, CA, USA), using the following conditions: 95°C for 2 min, followed by 35 cycles of 95°C for 30 sec, 60°C for 30 sec, 72°C for 2 min and 72°C for 5 min, and overnight incubation at 16°C. For endogenous control, additional PCR were performed using GAPDH-specific primers (forward primer: 5’-CGAGATCCCTCCAAAATCAA-3’; reverse primer: 5’-TGTGGTCATGAGTCCTTCCA-3’). Ten microliters of each PCR products were analyzed by electrophoresis on a 2% agarose gel containing ethidium bromide, and the images were captured using a Gel Imaging system (G:BOX, Syngene, Frederick, MD, USA).
Table 2.
TLR primers
| Gene name | Invivogen catalog# | Amplicon size (bp) |
|---|---|---|
| Human TLR1 | rtp-htlr1 | 706 |
| Human TLR2 | rtp-htlr2 | 657 |
| Human TLR3 | rtp-htlr3 | 602 |
| Human TLR4 | rtp-htlr4 | 550 |
| Human TLR5 | rtp-htlr5 | 493 |
| Human TLR6 | rtp-htlr6 | 435 |
| Human TLR7 | rtp-htlr7 | 387 |
| Human TLR8 | rtp-htlr8 | 335 |
| Human TLR9 | rtp-htlr9 | 312 |
| Human TLR10 | rtp-htlr10 | 214 |
Real-time PCR
The cDNA generated as described above was diluted (1:10) in nuclease-free water. A 10 µl reaction mixture containing 2 µl of each diluted cDNA sample, 5 µl master mix (containing the polymerase enzyme, dNTPs and MgCl2), 0.5 µl primer-probe mixture [containing 900 nM of each forward and reverse primer and 250 nM Taqman minor groove binder (MGB) probe], and 2.5 µl nuclease-free water was used for quantitative real-time PCR. Gene amplification was performed in a LightCycler® 480 II Real-Time PCR System (Roche, Indianapolis, IN) and the software monocolor hydrolysis probe universal cycling conditions (95 °C for 10 min, followed by 45 cycles of 95 °C for 10 sec, 60 °C for 30 sec, and 72 °C for 1 sec). Gene quantification was performed using the Ct method as described in the software user manual. All PCR primers and probes were obtained from Applied Biosystems and primer IDs were as follows: IL6 (Hs00985639), JUN (Hs99999141_s1), RELB (Hs00232399_m1), cAMP response element binding protein (CEBP)A (Hs00269972_s1), CEBPG (Hs01922818_s1), signal transducer and activator of transcription (STAT)1 (Hs00234829_m1), and TLR3 (HS01551078_m1). For endogenous control, each gene expression was normalized to the sample GAPDH (Hs99999905_m1).
Immunofluorescence and confocal microscopy
Sections (5 µM) were cut from each human brain tissue using a cryostat (Leica, Buffalo Grove, IL, USA), mounted on glass slides, fixed, permeabilized, and incubated for 1 hour in PBS containing 3% bovine serum albumin to block non-specific binding. Tissues were incubated overnight with antibodies to TLR3 and glucose transporter-1 (GLUT1) (Abcam, Cambridge, MA, USA), followed by staining (1 hour in the dark at room temperature) with secondary antibodies coupled with Alexa Fluor-488 or −635 (Invitrogen, Waltham, MA, USA) at 1:500 dilutions. Stained tissues were washed in PBS, mounted in Prolong Gold anti-fade reagent containing DAPI (Molecular Probes, Grand Island, NY) and examined under an Olympus FV500-IX 81 confocal laser scanning imaging system. The triple laser lines excitation were 405-nm for nucleus-stains; 484-nm for TLR3, and 635-nm for GLUT1. A fourth laser line (excitation: 543-nm; emission: 595-nm) was used to detect auto-fluorescent pigments.
Protein extraction and Western blot analysis
Following cellular treatment, protein extraction, quantification, and Western blot analysis were performed as previously described [26, 28, 29]. Briefly, cells were lysed using the mammalian cell lysis buffer CelLytic M (Sigma, St Louis, MO, USA), and protein quantified using the bicinchoninic acid assay as we previously described [26, 28, 29]. Protein (35 µg) was fractionated in a 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. Membranes were blocked for 1 hour with SuperBlock T-20 (Pierce, Rockford, IL, USA) and blotted for 2 hours or overnight with monoclonal antibodies to TLR3, claudin-5 (Abcam), JNK, phopho-JNK, c-Jun, phospho-c-Jun (all from Cell Signaling Technology, Danvers, MA, USA). These primary antibodies were used at 1:1,000 dilutions. Membranes were then washed, blotted for 1 hour with horseradish peroxidase-conjugated secondary antibody, washed again, and visualized using the enhanced chemiluminescence (Pierce) and gel doc system (Syngene, Frederick, MD, USA). After each western blot experiment, membranes were stripped using the Restore Western Blot Stripping Buffer (Pierce) and re-blotted with β-actin antibody (Abcam) to confirm equal loading.
Interleukin-6 ELISA
Interleukin (IL)-6 expression and secretion was quantified using the BD OptEIA Human IL6 ELISA kit II (BD Biosciences, CA, USA) according to the manufacturer’s protocol. Briefly, confluent HBMEC were treated with PIC (50 µl/ml), HIV-1ADA (MOI: 0.001) and/or pharmacological inhibitors (Table 3) for 48 hours. Controls consisted of untreated cells and cells treated with the TLR3/dsRNA Complex Inhibitor (TLR3.CI) (30 nM). Following cell treatment, culture supernatants were collected and further centrifuged for 5 min at 2350 g to remove any cellular debris, and 100 µl of each sample used for IL6 ELISA according to manufacturer’s instructions. Absorbance readings were obtained using SpectraMax M5 ELISA plate reader (Molecular Devices, Sunnyvale, CA, USA). For each experiment, IL6 standards provided with the kit were used to generate a standard curve and determine IL6 concentrations in each sample. Each experimental condition was tested in duplicate or triplicate.
Table 3.
Description of pharmacological inhibitors used in the study
| Inhibitor name | Catalog# / Abbreviation |
Mechanism of action | Manufacturer |
|---|---|---|---|
| TLR3/dsRNA Complex Inhibitor | TLR3.CI | Acts as a direct, competitive and high affinity inhibitor of dsRNA binding to TLR3. Selectively antagonizes TLR3 signaling without affecting other TLRs. | Calbiochem- EMD Millipore |
| NFκB Activation Inhibitor | 481406 | Inhibitor of NFκB transcriptional activation | Calbiochem- EMD Millipore |
| SR 11302 | SR11302 | Inhibitor of activator protein-1 (AP-1) transcriptional activity | Tocris Biosciences |
| JNK Inhibitor II | 420119 | Selective and reversible inhibitor of c-Jun N-terminal kinase (JNK1, JNK2, and JNK3). | Calbiochem- EMD Millipore |
| JNK Inhibitor V | 420129 | Reversible and ATP-competitive inhibitor of c-Jun N-terminal kinase (JNK1, JNK2, and JNK3). | Calbiochem- EMD Millipore |
| JNK Inhibitor III | 420130 | Specifically disrupt c-Jun/JNK complex formation and the subsequent phosphorylation and activation of c-Jun by JNK | Calbiochem- EMD Millipore |
| IRAK-1/4 inhibitor | IRAK-1/4 | Inhibitor of interleukin-1 receptor-associated kinases (IRAK)-1 and −4 | Sigma-Aldrich |
| 5Z-7-Oxozeaenol | 5ZO | ATP-competitive irreversible inhibitor of TAK1 (MEKK7), MKK7, MEK1, and ERK2. Has no activity against other MAPK | Sigma-Aldrich |
Abbreviations: TLR3: Toll-like receptor-3; JNK: c-Jun N-terminal kinase; dsRNA: double-stranded RNA; AP-1: activator protein-1; IRAK: Interleukin-1 Receptor-Associated Kinase; NFkB: nuclear factor kappaB; TAK1: Transforming growth factor-p activated kinase-1; MEKK7: MAP Kinase Kinase Kinase-7; MKK7: MAP Kinase Kinase-7; MEK1: MAP Kinase Kinase-1; ERK2: extracellular-signal-regulated kinase-2 / MAP Kinase-1
Monocyte isolation and labeling
Human monocytes were obtained from HIV-1/2 and hepatitis-B seronegative donor leukopaks, and separated by countercurrent centrifugal elutriation as previously described [26, 28, 30]. Cells were identified as >98% pure monocytes by Wright staining and CD68 immunostaining (at 1:50 dilution) (Dako, Carpentaria, CA, USA). Monocytes were used for adhesion and transendothelial migration assays within 24 hours of elutriation. Freshly elutriated monocytes were washed twice with pre-warmed (37°C) PBS, re-suspended in pre-warmed PBS containing 5 µM of 5-carboxyfluorescein diacetate, succinimidyl ester (CFDA-SE) (Life Technologies); 1 × 106 cells / ml, and incubated at 37°C, 5% CO2, for 15 min. Monocytes were then pelleted by centrifugation at 300 g for 10 minutes, re-suspended in pre-warmed media and incubated at 37°C, 5% CO2, for 30 min. CFDA-SE-labeled monocytes were washed three times with pre-warmed serum-free media before use for adhesion and migration assays.
Monocyte adhesion and migration through in vitro BBB models
Adhesion and migration experiments were performed as we previously described [26, 28, 30]. Briefly, for adhesion assays, HBMEC were plated on 96-well collagen-coated black plates, and for migration assay, HBMEC were plated on collagen-coated FluoroBlok-tinted tissue culture inserts (3-µm pore size; BD Biosciences). Confluent HBMEC were treated with PIC (50 µl/ml), HIV-1ADA (MOI: 0.001), and/or pharmacological inhibitors (Table 3) for 24 or 48 hours. Controls consisted of untreated HBMEC and cells treated with similar concentrations of pharmacological inhibitors for 24 or 48 hours. Following treatment, HBMEC were rinsed three times to remove HIV-1, PIC, or pharmacological inhibitors. For adhesion assays, HBMEC were exposed to 2.5 × 105 CFDA-SE-labeled monocytes and incubated at 37°C, 5% CO2, for 15 min. After the 15 min incubation, HBMEC were washed 3 times with PBS and the number of adherent monocytes quantified by spectrophotometry (top readings, absorbance 494 nm; emission, 517 nm), with a standard curve derived from a serial dilution of a known number of CFDA-SE-labeled monocytes. For migration assays, following treatment and rinsing of HBMEC, 2.5 × 105 CFDA-SE-labeled monocytes were added to the upper chamber of the FluoroBlok inserts and allowed to migrate for 2 hours (37°C, 5% CO2). The numbers of migrated monocytes were quantified by spectrophotometry (bottom readings, absorbance 494 nm; emission, 517 nm), with a standard curve derived from a serial dilution of a known number of CFDA SE-labeled monocytes.
Statistical analyses
Data were analyzed by t-test (two-tailed) for two-group comparisons and one- or two-way ANOVA followed by Tukey’s multiple-comparisons tests using GraphPad Prism 5.0b. (GraphPad Software, La Jolla, CA, USA). Threshold of significance level was 0.05. Data are presented as means ± standard error of the mean.
Results
Expression of TLRs in primary HBMECs
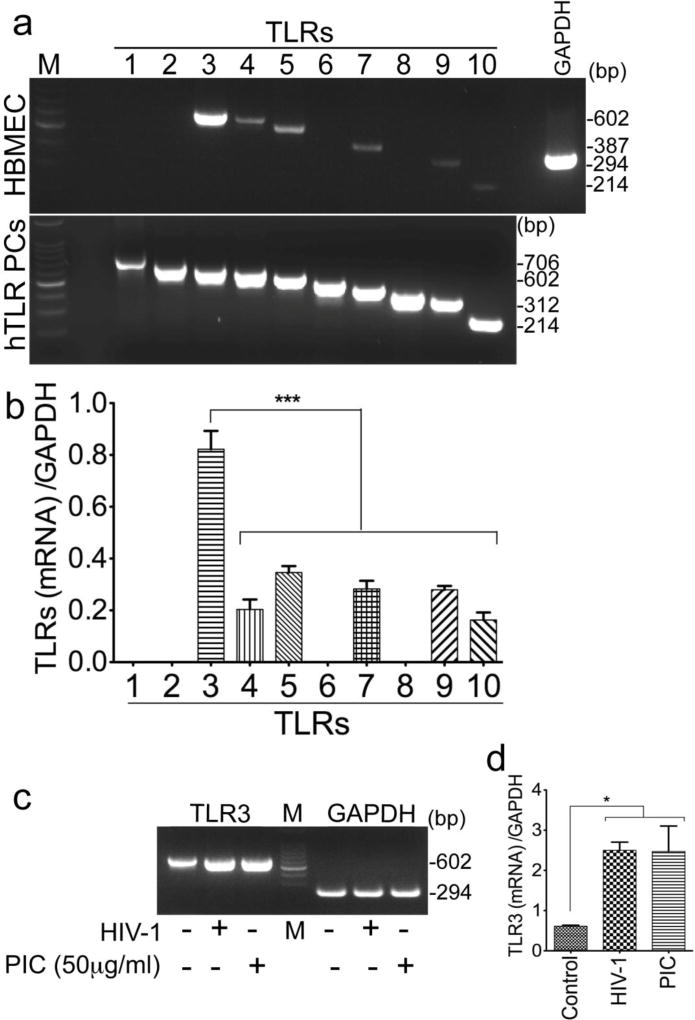
Our previous studies showed that signaling via TLRs was a major canonical pathway involved in HIV-induced activation of pro-inflammatory cytokines and chemokines in HBMEC, and BBB inflammation [25]. We performed RT-PCR to investigate the expression of TLRs in HBMEC and their role in HIV-induced BBB inflammation. We show that primary HBMEC express six TLRs (TRL3, 4, 5, 7, 9, 10) mRNA (Fig. 1a). TLR3 was the most abundant TLR expressed and no mRNA for TLR1, 2, 6, or 8 was detected (Fig. 1a). These results were confirmed by densitometry analyses of data from three independent experiments using HBMEC from three different human donors (Fig. 1b).
Fig. 1. TLR3 mRNA is the most abundant TLR transcript expressed on HBMEC, and HIV-1 and TRL3 ligands increased TLR3 transcription.
(a): Semi-quantitative RT-PCR using primers specific for TLRs 1–10 showed mRNA for TLR3, 4, 5, 7, 9, and 10 in HBMEC, and TLR3 were the most abundant. hTLR PCs: human TLRs positive controls; RT-PCR was performed using 10 ng TLRs (1–10) cDNA from InvivoGen. (b): Densitometry analyses of RT-PCR data from three independent experiments using HBMEC from three different human donors. (c): Exposure of HBMEC to HIV-1ADA (MOI: 0.001) or PIC for 24 hours increased TLR3 transcription, and densitometry analyses of data from three independent experiments using HBMEC from three different human donors, confirmed these results (d). Additional controls consisted of RT-PCR with GAPDH primers (a, c). M: molecular weight marker; bp: base pairs; *P<0.05, ***P<0.001.
Exposure of HBMEC to HIV-1 or TLR3 ligands increased TLR3 transcription and expression
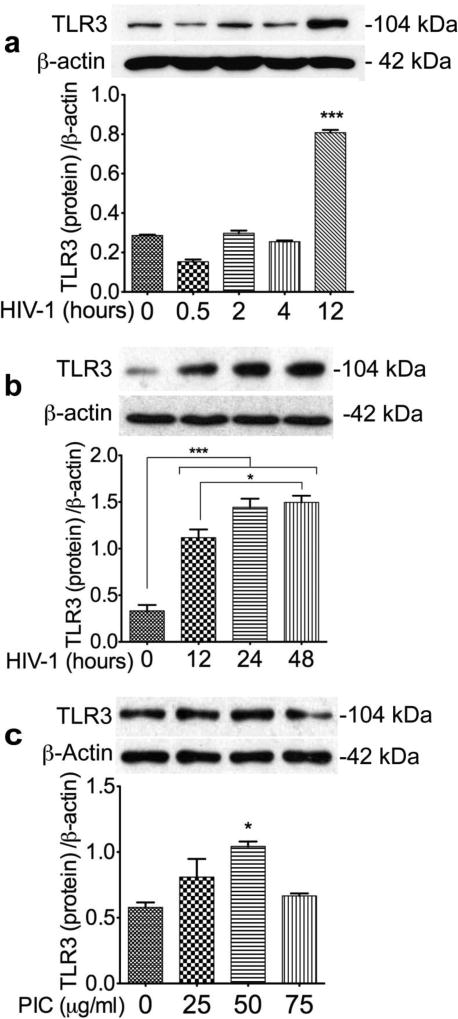
To determine whether HIV or TLR3 ligands can alter TLR3 expression on the brain endothelium, we assessed TRL3 mRNA and proteins levels in HBMEC exposed to HIV-1ADA (MOI: 0.001) or PIC (25 to 75 µg/ml) for 0.5 to 24 hours. Exposure of HBMEC to HIV-1 or the TLR3 ligand PIC increased TLR3 transcription (Fig. 1c), and densitometry analyses of data from three independent experiments using HBMEC from three different human donors, confirmed these results (Fig. 1d). Exposure of HBMEC to HIV-1 or the TLR3 ligand PIC also increased TLR3 expression (Fig. 2). Initial reduction in TLR3 expression was observed at 30 min HIV-1 exposure, but TLR3 levels gradually increased thereafter (Fig. 2a), with time-dependent increase in TLR3 expression observed from 12 to 48 hours HIV-1 exposure (Fig. 2b). Exposure of HBMEC to PIC for 24 hours increased TLR3 expression, with maximal levels observed with 25 to 50 µg/ml PIC (Fig. 2c). Densitometry analyses of data from three independent experiments using HBMEC from three different donors, confirmed that HIV-1 and PIC increased TLR3 expression (Fig. 2).
Fig. 2. HIV-1 and TRL3 ligands increased TLR3 expression in HBMEC.
(a): Exposure of HBMEC to HIV-1ADA increased TLR3 expression, with higher levels observed from 12 hours post-exposure. (b): Exposure of HBMEC to HIV-1ADA for 12 to 48 hours induced a time-dependent increase in TLR3 expression. (c): Exposure of HBMEC to PIC (for 24 hours) increased TLR3 expression, with higher levels observed with 25 to 50 µg/ml PIC. Additional controls consisted of Western blot analyses with β-actin antibodies; densitometry analyses of data from three independent experiments using HBMEC from three different human donors, confirmed that HIV-1 and PIC increased TLR3 expression (a-c). kDa: kilodalton. *P<0.05, ***P<0.001.
Increased transcription and expression of TLR3 in brain tissues of HIV-1-infected humans
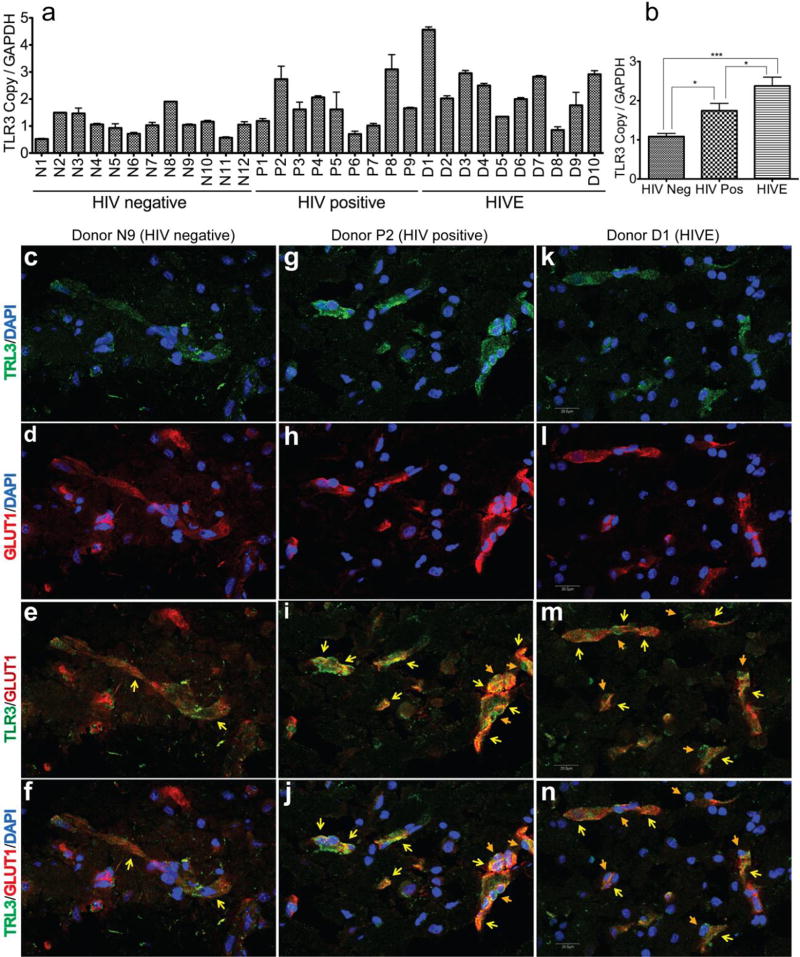
To determine whether HIV-1 infection alter TLR3 levels in the human brain, we analyzed brain tissues from 12 HIV-1/2-seronegative control subjects, 9 HIV-1-seropositive patients without evidence of HIVE or HAND, and 10 HIV-1-seropositive patients with HIVE and HAND. All brain tissues were from the cortical regions, with 28 of the 31 samples from the frontal cortex, 2 samples from the parietal cortex, and 1 sample from the temporal cortex. Table 1 shows the age, gender, clinical history, post-mortem interval (PMI) between the time of death and autopsy, and a summary of post-mortem findings for all 31 human subjects. No significant differences were detected in age and PMI between the seronegative, HIV-1-infected, or HIVE groups. For seronegative controls, HIV-1-infected, and HIVE patients, the age ranges in years (and means ± standard deviations) were respectively 32 to 72 (mean: 52±13.4), 27 to 54 (mean: 41.78±8.8), and 30 to 52 (mean: 40.6±7.76). For seronegative controls, HIV-1-infected, and HIVE patients, the PMI ranges in hours (and means ± standard deviations) were respectively 3 to 8.5 (mean: 4.6±1.6), 2.75 to 15 (mean: 8.5±4), and 4 to 21 (mean: 9.45±5.16).
Quantitative real-time PCR showed significant upregulation of TLR3 mRNA transcripts in brain tissues from HIV-1-infected humans, with the highest TLR3 mRNA levels in brain tissues of patients with HIVE/HAND (Fig. 3a,b). To further investigate the effects of HIV-1 infection on TLR3 expression in the human BBB in vivo, we analyzed brain tissue sections from seronegative controls, HIV-infected and HIVE patients by confocal microscopy. Data showed TLR3 expression on the brain blood vessels of seronegative controls (Fig. 3c–f, yellow arrows), HIV-positive (Fig. 3g–j, yellow arrows) and HIVE (Fig. 3k–n, yellow arrows) patients, with higher TLR3 expression in the blood vessels and tight junction strands of HIV+ and HIVE patients brains, as demonstrated by increased staining intensity. Brain tissues from HIV-positive and HIVE patients also showed mononuclear phagocytes expressing TLR3 crossing the blood vessels (Fig. 3, orange arrows).
Fig. 3. Increased TLR3 expression in brain tissues and CNS blood vessels of HIV-1-infected humans.
(a, b): Quantitative real-time PCR show increased TLR3 mRNA in brain tissues of HIV-1-positive individuals (HIV pos, donors P1 to P9), and infected patients with HAND or HIVE (donors D1 to D10), compared to seronegative controls (HIV Neg, donors N1 to N12). *P<0.05, ***P<0.001. (c-n): Confocal microscopy of human brain tissues sections stained with monoclonal antibodies to TLR3 (green) and the endothelial glucose transporter-1 (GLUT1) (red). Images with (c, d, f, g, h, j, k, l, n) and without (e, i, m) DAPI staining shows TLR3 expression on blood vessels, including blood vessels tight junction strands (yellow arrows). For HIV+ and HIVE patients, data also shows TLR3 expression on mononuclear phagocytes ingressing across the blood-brain barrier (orange arrows). Scale bar for all panels: 20 µM.
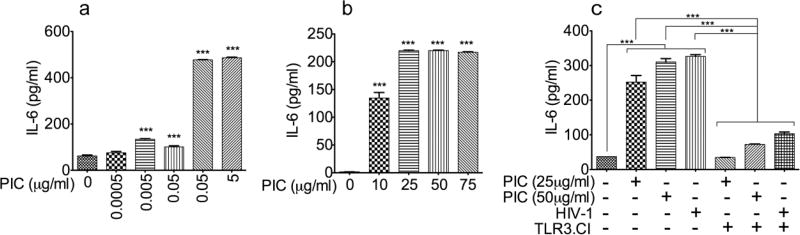
HIV-1 and TLR3 ligands upregulate IL6 expression in HBMEC
To determine whether PIC- and HIV-1-induced increase in TLR3 expression in HBMEC (Figs. 1 and 2) could be associated with increased BBB inflammation, we analyzed the effect of TLR3 ligands and HIV-1 on IL6 expression and secretion in HBMEC. Studies using HBMEC from three different human donors (Fig. 4a–c) showed that PIC at 0.5 to 75 µg/ml increased IL6 expression and secretion in primary HBMEC by 6 to 127-fold (Fig. 4a–c, P<0.001). Studies using HBMEC from the third donor showed that the TLR3/dsRNA complex inhibitor (TLR3.CI) blocked both PIC- and HIV-1-induced IL6 expression (Fig. 4c, P<0.001).
Fig. 4. HIV-1 and TLR3 ligands upregulate IL6 expression in HBMEC.
Exposure of HBMEC to PIC [0.5 ng/ml to 5 µg/ml (a) or 10 to 75 µg/ml (b)] induced IL6 expression and secretion. Experiments in panels a and b were performed using endothelial cells from two different human donors. Experiments using HBMEC from a third donor (c) showed that exposure of HBMEC to PIC (25 to 50 µg/ml) or HIV-1ADA (MOI: 0.001) induced IL6 expression and secretion; and the TLR3/dsRNA complex inhibitor (TLR3.CI) blocked both PIC- and HIV-1-induced IL6 expression (c). ***P<0.001.
HIV-1 and TLR3 ligands induced upregulation of mRNA transcripts for IL6 and transcription factors associated with the TAK1 and JNK pathways in HBMEC
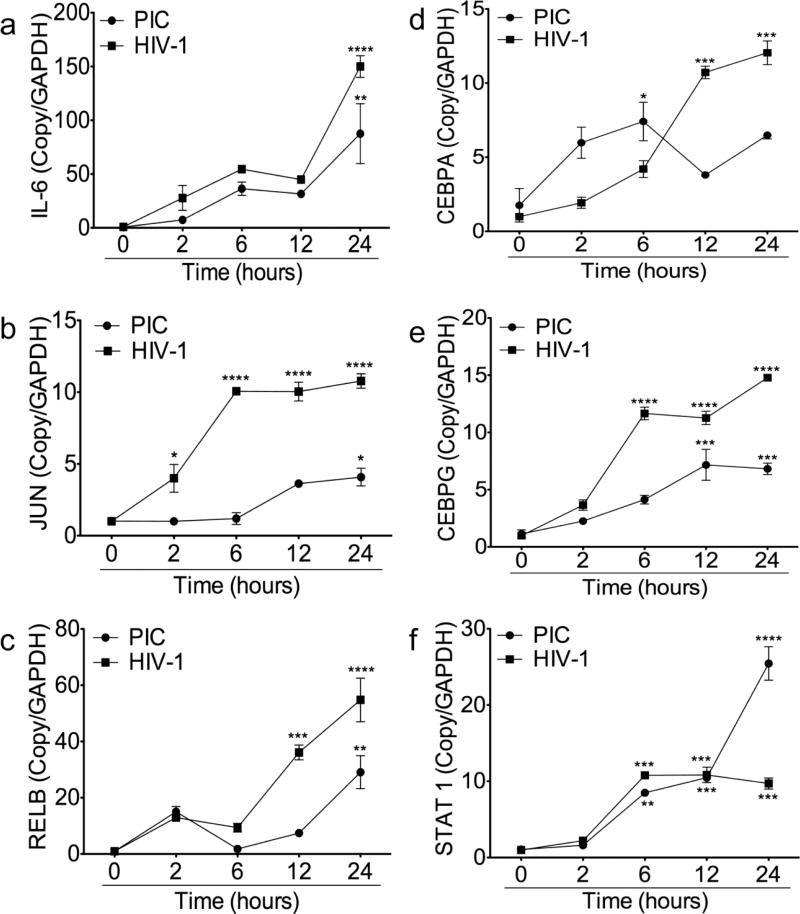
To determine whether HIV-1- and PIC-induced IL6 expression was associated with increased IL6 transcription, we used real-time PCR to quantify IL6 mRNA in HBMEC exposed to HIV-1 (MOI: 0.001) and PIC (50 µg/ml) for 2 to 24 hours. Both PIC and HIV-1 induced a time-dependent increase in IL6 transcription in HBMEC, with the highest IL6 mRNA levels observed from 24 hours HIV-1 or PIC exposure (Fig. 5a). The IL6 promoter contains several cis-acting response elements that regulate its transcription and expression. These included the activator protein-1 (AP-1), NFκB, STATs, and the cAMP response element binding protein (CREB/CEBP) [31–33]. Therefore, we analyzed the effects of HIV-1 and TLR3 ligands on the expression of these transcription factors. Compared to untreated controls both PIC and HIV-1 significantly increased JUN, RELB, CEBPA, CEBPG, and STAT1 transcription in HBMEC (Fig. 5b–f). PIC-and HIV-1-induced expression of these transcription factors was mostly time-dependent, with the highest mRNA levels observed at 24 hours exposure (Fig. 5b–f). Compared to untreated controls, exposure of HBMEC to PIC for 2 to 24 hours increased IL6, JUN, RELB, CEBPA, CEBPG, and STAT1 mRNA levels respectively by 9 to 108.6-fold (Fig. 5a), 3 to 4 fold (Fig. 5b), 1.7 to 29-fold (Fig. 5c), 2 to 4.2-fold (Fig. 5d), 1.97 to 6.26-fold (Fig. 5e), and 9 to 108.6-fold (Fig. 5f). Exposure of HBMEC to HIV-1 for 2 to 24 hours increased IL6, JUN, RELB, CEBPA, CEBPG, and STAT1 and mRNA levels respectively by 27.8 to 150-fold (Fig. 5a), 4 to 10.78 fold (Fig. 5b), 13 to 54.78-fold (Fig. 5c), 1.92 to 12-fold (Fig. 5d), 3.65 to 14.8-fold (Fig. 5e), and 27.8 to 150-fold (Fig. 5f).
Fig. 5. HIV-1 and TLR3 ligands induce transcriptional upregulation of IL6 and transcription factors associated with the TAK1 and JNK pathways in HBMEC.
Quantitative real-time PCR showed that exposure of HBMEC to PIC (50 µg/ml) or HIV-1ADA (MOI: 0.001) for 2 to 24 hours induce transcriptional upregulation of IL6 (a), JUN (b), RELB (c), CEBPA (d), CEBPG (e), and STAT1 (f). *P<0.05, **P<0.01, ***P<0.001 compared to untreated controls. These are representative data from three independent experiments using HBMEC from three different human donors.
TLR3/dsRNA complex inhibitors blocked both TLR3 ligands- and HIV-1-induced upregulation of mRNA transcripts for IL6 and transcription factors associated with the TAK1 and JNK pathways
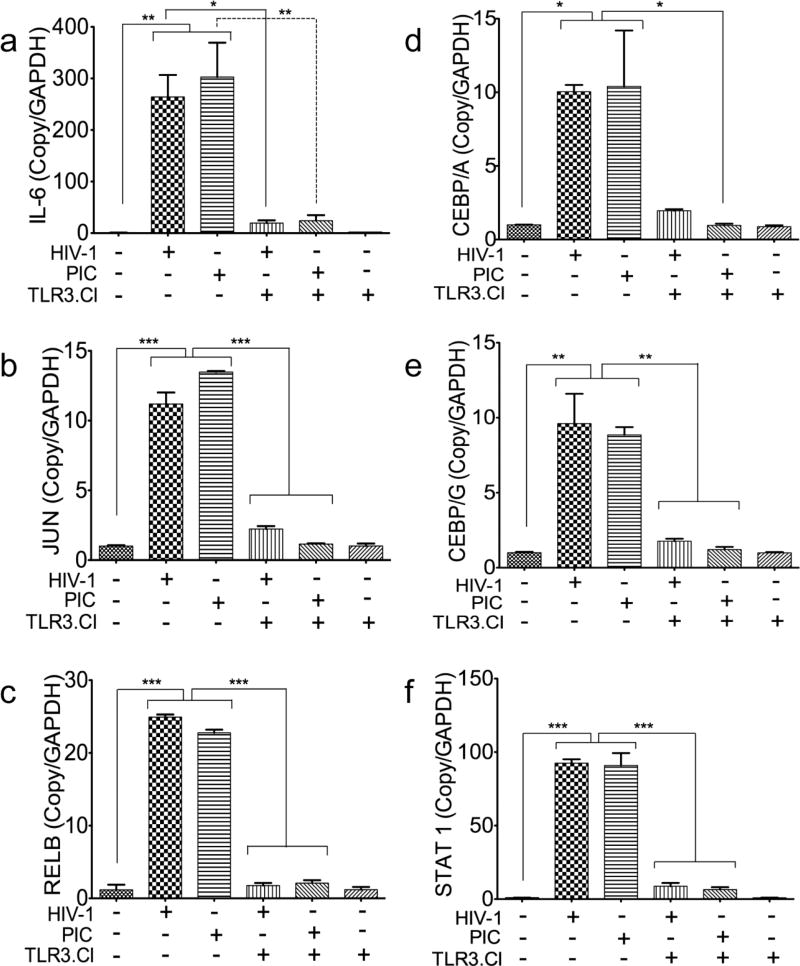
To confirm the involvement of TLR3 in HIV-1-induced upregulation of mRNA transcripts for IL6 and the transcription factors identified above (Fig. 5) we performed real-time PCR to quantify their mRNA levels in HBMEC exposed for 24 hours to HIV-1ADA (MOI: 0.001) or PIC (50 µg/ml) with and without the presence of the TLR3/dsRNA complex inhibitors (TLR3.CI). Compared to untreated controls, exposure of HBMEC to HIV-1 increased the transcription of IL6, JUN, RELB, CEBPA, CEBPG, and STAT1 respectively by 264-, 11.2-, 24.93-, 10-, 9.6-, and 92.5-fold (Fig. 6). Exposure of HBMEC to PIC increased the transcription of IL6, JUN, RELB, CEBPA, CEBPG, and STAT1 respectively by 302.8-, 13.48-, 22.78-, 10.4-, 8.86-, and 91-fold (Fig. 6). The TLR3.CI blocked both HIV-1- and PIC-induced transcription of IL6 (Fig. 6a, P<0.05 and P<0.01), JUN (Fig. 6b, P<0.001), RELB (Fig. 6c, P<0.001), CEBPA (Fig. 6d, P<0.05), CEBPG (Fig. 6e, P<0.01), and STAT1 (Fig. 6f, P<0.001).
Fig. 6. The TLR3/dsRNA complex inhibitor blocked both TLR3 ligands- and HIV-1-induced transcriptional upregulation of IL6 and transcription factors associated with the TAK1 and JNK pathways.
Quantitative real-time PCR showed that exposure of HBMEC to PIC (50 µg/ml) and HIV-1ADA (MOI: 0.001) for 24 hours induce transcriptional upregulation of IL6 (a), JUN (b), RELB (c), CEBPA (d), CEBPG (e), and STAT1 (f). The inhibitor of TLR3/dsRNA complex (TLR3.CI) blocked both PIC- and HIV-1-induced upregulation of IL6 (a), JUN (b), RELB (c), CEBPA (d), CEBPG (e), and STAT1 (f). *P<0.05, **P<0.01, ***P<0.001. These are representative data from three independent experiments using HBMEC from three different human donors.
HIV-1 and TLR3 ligands induced phosphorylation of c-Jun and JNK in HBMEC
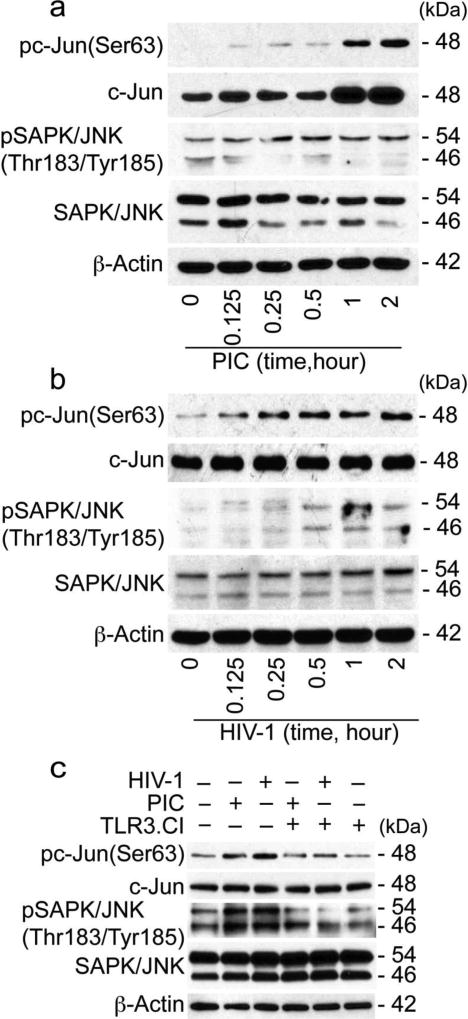
Because HIV-1 and TLR3 ligands upregulates the transcription factors associated with the JNK pathways (Figs. 5 and 6), we analyzed the effects of HIV-1 and PIC on c-Jun and JNK activation in HBMEC. Exposure of HBMEC to both PIC (Fig. 7a) and HIV-1 (Fig. 7b) induced the phosphorylation of c-Jun at serine-63 (Ser63). PIC- and HIV-1-induced c-Jun phosphorylation occurred as early as 5 min and gradually increased for up to 2 hours, with maximal phosphorylation observed at 2 hours (Fig. 7a and 7b). Both PIC (Fig. 7a) and HIV-1 (Fig. 7b) increased the phosphorylation of SAPK/JNK at Thr183/Tyr185; with PIC-induced SAPK/JNK phosphorylation occurring as early as 5 min following PIC exposure (Fig. 7a), and HIV-1-induced SAPK/JNK phosphorylation starting at 30 min following viral exposure (Fig. 7b). Repeated experiments using HBMEC from different donors confirmed these results, and showed that TLR3.CI inhibited both HIV-1- and PIC-induced phosphorylation of c-Jun, and also inhibited HIV-1- and PIC-induced phosphorylation of SAPK/JNK (Fig. 7c).
Fig. 7. HIV-1 and TLR3 ligands induced the phosphorylation of c-Jun and JNK in HBMEC.
(a): Exposure of primary HBMEC to the TLR3 ligand PIC (50 µg/ml) induced the phosphorylation of c-Jun at Ser63, with maximal phosphorylation at 2 hours; increased c-Jun, and increased the phosphorylation of SAPK/JNK at Thr183/Tyr185. (b): Exposure of primary HBMEC to HIV-1ADA (MOI: 0.001) induced the phosphorylation of c-Jun at Ser63 and SAPK/JNK at Thr183/Tyr185. (c): The inhibitor of TLR3/dsRNA complex (TLR3.CI) blocked PIC- and HIV-1-induced phosphorylation of c-Jun and SAPK/JNK. These are representative data from two independent experiments using HBMEC from two different human donors.
TAK1 and JNK pathways mediates HIV-1 or TLR3 ligands induced IL6 expression HBMEC
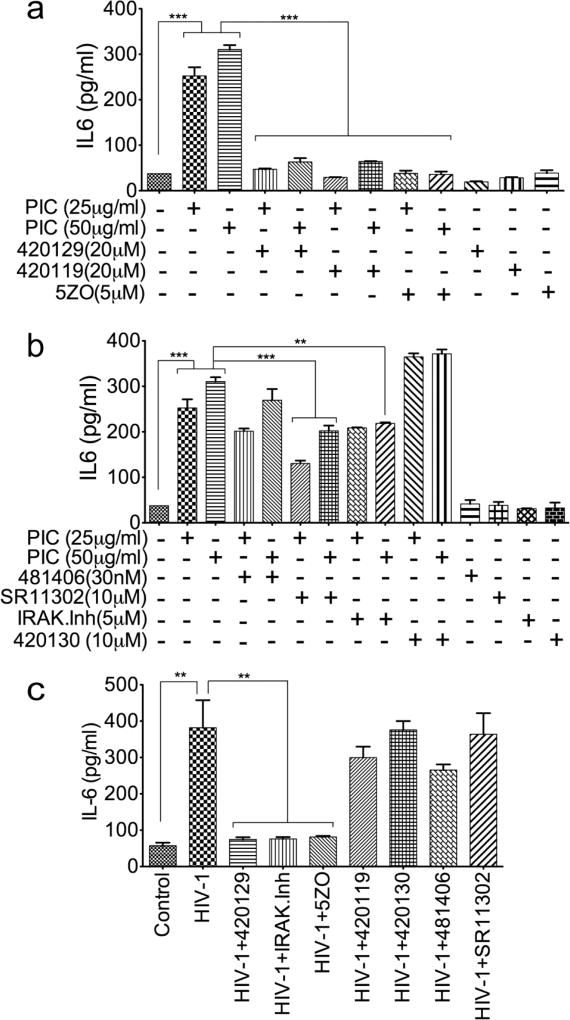
To determine whether blocking the pathways associated with transcription factors upregulated by TLR3 ligands and HIV-1 (Figs. 5 and 6) can alter PIC- and HIV-1-induced IL6 expression, we quantified IL6 expression and secretion in HBMEC exposed to PIC or HIV-1, in the presence or absence of specific inhibitors of effectors associated with those transcription factors. The ATP-competitive JNK inhibitor (420129), the JNK inhibitor 420119, and the TAK1/MKK7 inhibitor (5ZO) blocked PIC-induced IL6 expression and secretion (Fig. 8a), whereas the inhibitor of c-Jun/JNK complex (420130), the inhibitor of NFκB transcriptional activation (481406), the inhibitor of AP-1 transcription (SR11302), and the IRAK1/4 inhibitor only partially reduced or had no major effect on PIC-induced IL6 expression (Fig. 8b). The ATP-competitive JNK inhibitor (420129), the TAK1/MKK7 inhibitor (5ZO), and the IRAK1/4 inhibitor blocked HIV-1-induced IL6 expression and secretion (Fig. 8c); whereas the JNK inhibitor 420119, the inhibitor of c-Jun/JNK complex (420130), the inhibitor of NFκB transcriptional activation (481406), and the inhibitor of AP-1 transcription (SR11302) had no major effect on HIV-1-induced IL6 expression (Fig. 8c).
Fig. 8. The JNK and TAK1/MKK7 pathways mediate TLR3 ligands- and HIV-1- induced IL6 expression HBMEC.
(a) The JNK inhibitor 420119, the ATP-competitive JNK inhibitor 420129, and the TAK1/MKK7 inhibitor 5ZO blocked PIC induced IL6 expression and secretion. (b): PIC induced IL6 expression was reduced by the inhibitor of AP-1 transcriptional activity SR11302, whereas the inhibitor of NFκB transcriptional activation 481406, the IRAK1/4 inhibitor, and the inhibitor of c-Jun/JNK complex 420130 had minimal to no effect. (c): The ATP-competitive JNK inhibitor 420129, the IRAK1/4 inhibitor, and the TAK1/MKK7 inhibitor 5ZO blocked HIV-1-induced IL6 expression and secretion; whereas the JNK inhibitor 420119, the inhibitor of c-Jun/JNK complex 420130, the inhibitor of NFκB transcriptional activation 481406, and the inhibitor of AP-1 transcriptional activity SR11302, had minimal to no effect. *P<0.05, **P<0.01, ***P<0.001.
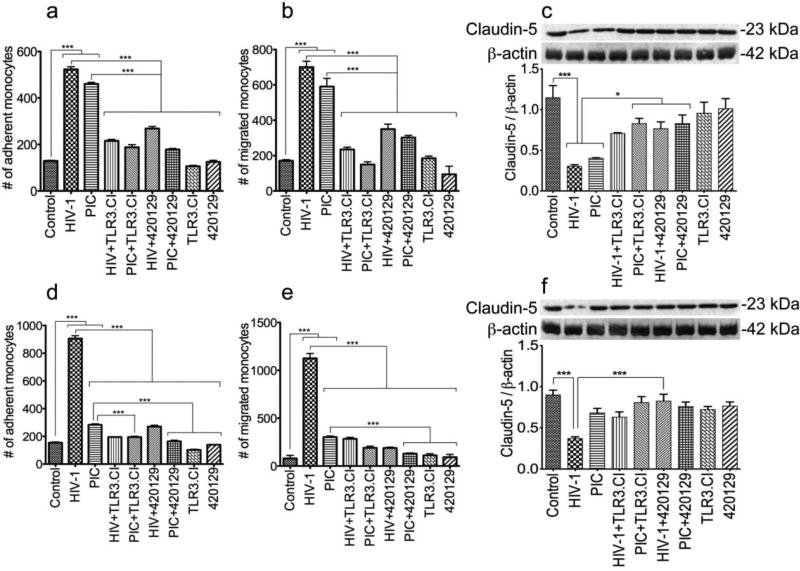
Effects of HIV-1 and TLR3 ligands on the blood-brain barrier properties and function
We previously demonstrated that HIV-1 and viral factors impair the BBB properties and function, and this was associated with decreased expression of tight junction proteins, increased adhesion and migration of human mononuclear phagocytes through the BBB [26, 29, 30]. To determine whether this involves TLR3, we assessed the effects on HIV-1 and TLR3 ligands on claudin-5 expression in primary HBMEC, and monocytes adhesion and migration through in vitro BBB models. Exposure of HBMEC to HIV-1 and PIC (50 µg/ml) for 24 hours increased monocytes adhesion (by 4- and 3.5-fold respectively, Fig. 9a) and transmigration through the BBB (by 4.1- and 3.45-fold respectively, Fig. 9b) (P<0.001), and decreased claudin-5 expression in HBMEC by 3.77- and 2.85-fold respectively (Fig.9c, P<0.001). Exposure of HBMEC to HIV-1 and PIC for 48 hours increased monocytes adhesion (by 5.86- and 1.83-fold respectively, Fig. 9d) and transmigration through the BBB (by 14- and 3.78-fold respectively, Fig. 9e) (P<0.001), and decreased claudin-5 expression in HBMEC by 2.43- and 1.32-fold respectively (Fig. 9f). PIC-induced monocyte adhesion and trans-endothelial migration decreased over time, with less adhesion and migration after 48 hours (Fig. 9d, e), compared to 24 hours PIC treatment (Fig. 9a, b). Similarly, 48 hours PIC treatment of HBMEC did not significantly altered claudin-5 expression (Fig. 9f), whereas 24 hours PIC treatment significantly decreased claudin-5 expression (Fig. 9c, P<0.001).
Fig. 9. Effects of HIV-1 and TLR3 ligands on the blood-brain barrier properties and function.
Exposure of HBMEC to HIV-1ADA (MOI: 0.001) for 24 hours (a, b, c) or 48 hours (d, e, f) increased monocytes adhesion to HBMEC by 4 to 5.86-fold (a, d), increased monocytes transmigration through the BBB by 4 to 14-fold (b, e), and decreased claudin-5 expression by 2.43 to 3.77-fold (c, f). Exposure of HBMEC to PIC (50 µg/ml) for 24 hours (a, b, c) or 48 hours (d, e, f) increased monocytes adhesion to HBMEC by 1.83 to 3.56-fold (a, d), increased monocytes transmigration through the BBB by 3.45 to 3.78-fold (b, e), and decreased claudin-5 expression by 1.32 to 2.85-fold (c, f). The TLR3/dsRNA complex inhibitor (TLR3.CI) and the ATP-competitive JNK inhibitor (420129) significantly reduced HIV-1- and PIC-induced monocytes adhesion (a, d), transendothelial migration (b, e), and claudin-5 downregulation (c, f). ***P<0.001. Controls consisted of untreated HBMEC, and HBMEC exposed only to TLR3.CI or the JNK inhibitor. These controls showed no alteration in BBB function. These are representative data from two independent experiments using HBMEC from two or three different human donors.
The TLR3.CI decreased HIV-1- induced monocytes adhesion (by 2.4 to 4.6-fold, Fig. 9a,d), BBB transmigration (by 3 to 4-fold, Fig. 9b, e) (P<0.001), and abrogated HIV-1-induced decrease in claudin-5 expression (Fig. 9c, f). TLR3.CI also decreased PIC-induced monocytes adhesion (by 1.4 to 2.4-fold, Fig. 8a, c), transmigration through the BBB (by 1.6 to 4-fold, Fig. 8b, d) (P<0.001), and abrogated PIC-induced downregulation of claudin-5 expression (Fig. 9c, f).
Data in Figs 5, 6, and 8 showed that the JNK pathways mediated HIV-1- and TLR3 ligands-induced inflammation. To determine whether this pathway was also involved in HIV-1-and PIC-induced alterations in BBB properties and function, we assessed the effects of an ATP-competitive JNK inhibitor (420129) on monocytes adhesion, BBB transmigration, and endothelial claudin-5 expression. This JNK inhibitor reduced HIV-1-induced monocytes adhesion (by 2- to 3.3-fold, Fig. 9a, d) and transmigration through the BBB (by 2- to 6-fold, Fig. 9b, e) (P<0.001). It significantly reduced HIV-1-induced decrease of claudin-5 expression (by 2.2- to 2.5-fold, Fig. 9c, f). This JNK inhibitor also reduced PIC-induced monocytes adhesion (by 1.7- to 2.5-fold, Fig. 9a, d), transmigration through the BBB (by 1.95- to 2.3-fold, Fig. 9b, e) (P<0.001), and also abrogated PIC-induced decrease in claudin-5 expression (Fig. 9c, P<0.05). Densitometry analyses of data from two independent experiments using HBMEC from two different donors, confirmed HIV-1 and PIC-induced downregulation of claudin-5 expression, as well as the protective effects of the TLR3.CI and JNK inhibitor (Fig. 9c, f).
Discussion
In HIV-infected individuals, the BBB is exposed to virions and viral products from the systemic circulation, and there is evidence that HIV-1 induced BBB injury enables viral entry into the brain, infection of resident CNS cells and HAND [21–24]. This HIV-1-induced BBB injury is associated with signaling via IL6 and TLRs [25], and the present study show that primary HBMEC, the major component of the BBB, express TLR3, 4, 5, 7, 9, and 10. We also show that TLR3 was the most abundant TLR expressed in these cells. It has also been shown that the human cerebral endothelial cell line hCMEC/D3 express mRNA for TLR2, 3, 4, and 6 [34] and that primary rat cerebral EC express mRNA for TRL2, 3, and 6 [34]. The fact that TLR2 and 6 are expressed in the hCMEC/D3 cell line and rat cerebral EC but not in primary HBMEC; and that TLR5, 7, 9, and 10 are expressed in primary HBMEC, but not in hCMEC/D3 or rat cerebral EC, shows heterogeneity in TLRs expression on blood vessels, with differential expression in primary brain EC compared to cell lines, and in EC from animals brain vessels compared to EC from human brain vessels. EC from other vascular beds also express TLRs. Human lung and dermal EC [35], as well as human choroidal and retinal EC [36] express TLR1, 2, 3, 4, 5, 6, and 9 [35, 36].
TLR3 can be activated by viral dsRNA [6], viral ssRNA [7], or by dsRNA generated by RNA polymerases as intermediates during replication of positive-sense ssRNA viruses [12, 37, 38]. HIV-1 is a positive-sense ssRNA virus; in HIV-infected subjects, the BBB is constantly exposed to virus from the peripheral blood, which could also activate TLR3. In the current study, we showed that exposure of HBMEC to HIV-1 or TLR3 ligands increased TLR3 transcription and expression. Significantly, we showed increased transcription and expression of TLR3 in brain tissues from HIV-1-infected humans, with higher TLR3 expression in blood vessels and tight junction strands of HIV+ and HIVE patients. This indicates that HIV-1 CNS infection is associated with increased TLR3 expression in the brain, and that HIV-induced encephalitis further increased TLR3 expression. Our data showing expression of TLR3 in mononuclear phagocytes on blood vessels suggest that increased TLR3 expression in HIV-1 infected subjects was associated with increased infiltration of blood mononuclear phagocytes into the CNS. These data demonstrates the involvement of TLR3 in HIV-1-induced BBB injury and HIVE, and are in agreement with other studies implicating TLR3 in HIV/AIDS pathogenesis. In fact, there is increased TLR3 expression in peripheral blood mononuclear cells of HIV-1 infected patients [39], with the highest TLR3 levels in cells from patients with advanced AIDS and higher viral load [40].
Human studies showed that increased plasma or serum IL6 levels in HIV-infected subjects is associated with higher viremia, faster progression to AIDS, worse clinical outcomes and high mortality [41–43]. We previously showed that HIV-infection and HAND was associated with increased expression of IL6 in human brain tissues and the BBB [25, 26, 28, 29]. Our current study shows that exposure of primary HBMEC to HIV-1 or TLR3 ligands increased IL6 transcription and expression. This involvement of TLR3 in HIV-mediated IL6 production and BBB inflammation was further confirmed by data showing that inhibitors of TLR3/dsRNA complex blocked both HIV-1- and PIC-induced IL6 transcription and expression. There is evidence that TLR3 activation of EC from other vascular beds also induce inflammation in humans. Exposure of human glomerular EC [44], lung EC [35, 45], dermal microvascular EC [35], choroidal and retinal EC [36], and coronary artery and umbilical vein EC [46] to PIC increased the expression of IL6 [35, 36, 44, 45], as well as IL8 and IP10 in vitro and in vivo [45, 46]; TNFα, IFNβ, CCL2, CCL5, and CXCL10 [44].
The IL6 gene has functional cis-regulatory elements that contain binding sites for NFκB, CEBP, AP-1, STATs, and CEBPs [31–33]. In the current study, we showed that in addition to increasing IL6 transcription and expression, exposure of HBMEC to HIV-1 or PIC significantly upregulated the transcription factors JUN, RELB, CEBPA, CEBPG, and STAT1. TLR3 dimerizes to form a signaling complex [47] and ligands bind to the TLR3 dimer via its N-terminal ectodomain [48]. Ligand binding results in a conformational change and recruitment of Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF), which then triggers a signaling cascade that results in expression of INF-β or proinflammatory cytokines [49]. Our data confirm the specificity of TLR3 pathways in PIC- and HIV-1-induced IL6 expression, as we demonstrate that both HIV-1- and PIC-induced IL6 expression and secretion could be blocked by an inhibitor of the TLR3/dsRNA complex (TLR3.CI) that specifically antagonize TLR3 signaling by blocking the binding of dsRNA to TLR3. TLR3.CI also blocked HIV-1- and PIC-induced upregulation of mRNA transcripts for IL6, JUN, RELB, CEBPA, CEBPG, and STAT1. c-Jun dimerizes with c-Fos to form AP-1 [50]; and c-Jun is activated through phosphorylation by the JNK pathways [50, 51]. We show that TLR3.CI also blocked both HIV-1- and PIC-induced phosphorylation of c-jun and JNK. These data further confirmed the involvement of JNK pathways downstream of TLR3 in HIV-1-induced BBB dysfunction.
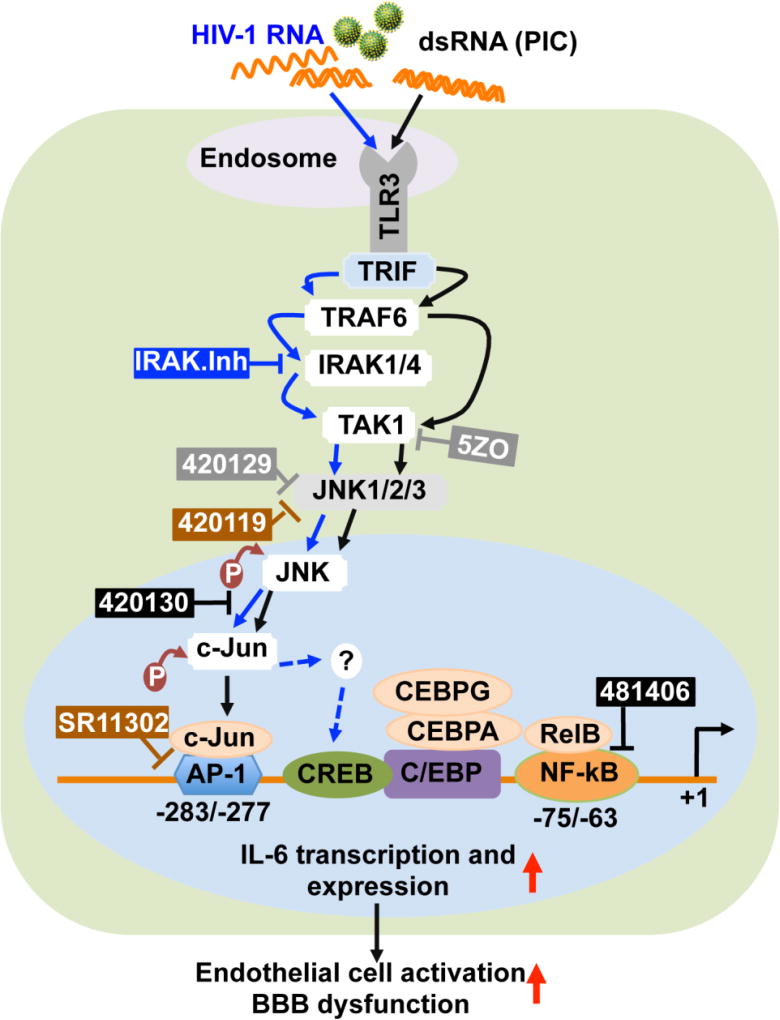
HIV-1-induced and TLR3-mediated IL6 expression in HBMEC was blocked by 5ZO, an ATP-dependent and competitive inhibitor of TAK1 and the MAP kinases MKK7, MEK1 and ERK2; and was also blocked by 420129, an ATP-dependent and competitive JNK inhibitor. In the MAPK/JNK pathways, stress signal activates small GTPases such as Rac1 and Rho; activated GTPase then stimulate MAPKKK, which phosphorylates MKK4 or MKK7 [52–54]. Following activation, MKK4/7 phosphorylates and activates JNK. Activated JNKs form dimers, translocate to the nucleus, bind to the c-Jun transcriptional domain, and phosphorylate c-Jun at serine or threonine residues [52, 54]. Thus, JNK regulates c-Jun/AP-1 activity and it has been shown that phosphorylation of c-Jun at Ser63 or Ser73 increases the transcription of downstream target genes [50–53]. This was confirmed by our present data showing that exposure of HBMEC to HIV-1 and TLR3 ligands induced Ser63 phosphorylation of c-Jun and Thr183/Tyr185 phosphorylation of JNK, and that this was associated with increased IL6 expression. HIV-1- and PIC-induced IL6 expression was blocked by TLR3.CI and by pharmacological inhibitors of JNK and TAK1/MKK7, all downstream effectors of TLR3 signaling (Fig. 10). The MAP kinases TAK1, MKK7, MEK1, and ERK2 all contain a cysteine residue in their ATP-binding site; suggesting that 5ZO and JNK inhibitors may block IL6 expression by interfering with cysteine residues in the ATP-binding region of these kinases. This would suggest that a region on these kinases ATP-binding site is involved in HIV-1-induced and TLR3-mediated endothelial inflammation and BBB dysfunction. The fact that the ATP-dependent and competitive JNK inhibitor 420129 blocked both PIC- and HIV-1-induced IL6 expression and secretion while the non-ATP-dependent JNK inhibitor 420119 only blocked PIC and had no effect on HIV-1-induced IL6 expression suggests that TLR3-mediated IL6 expression can occur via ATP-dependent or-independent activation of JNK, whereas HIV-1-induced IL6 expression on the brain endothelium occur strictly via ATP-dependent activation of JNK.
Fig. 10. Model illustrating the signaling pathways in HIV-1-induced and TLR3-mediated IL6 expression and BBB dysfunction.
Arrows with solid blue lines indicate activation and direct signaling of HIV-1. Dotted blue lines indicated potential/alternative pathways. Arrows with solid black lines indicate activation and direct signaling of TLR3 ligands. The ┴ symbol indicates pharmacological inhibitors. Grey color rectangular boxes indicate compounds that inhibited both HIV-1- and TLR3 ligands-mediated effects. The blue rectangular boxes indicate compounds that inhibited HIV-1-mediated IL6 expression, but had minimal effects PIC-mediated effects. The brown color rectangular boxes indicate compounds that blocked or significantly diminished TLR3 ligands-induced IL6 expression but had no effects on HIV-1-mediated effects. The black color boxes indicate compounds that had no effect on HIV-1 or TLR3 ligands-mediated effects.
 indicates an increase.
indicates an increase.
Functional studies showed that HIV-1 and PIC decreased claudin-5 expression in HBMEC, significantly increased monocytes adhesion and migration through the BBB, and that the ATP-dependent JNK inhibitor 420129 and TLR3.CI blocked both PIC- and HIV-1-induced monocytes adhesion and trans-endothelial migration, and abrogated PIC- and HIV-1-induced claudin-5 downregulation. This suggests that both HIV-1 and TLR3 ligands use similar TAK1-and JNK-dependent pathways upstream to induce IL6 expression in HBMEC and BBB dysfunction (Fig. 10); and that this occurs through ATP-dependent mechanisms. Inhibitors of IRAK1/4, downstream effectors of TLR signaling, blocked HIV-1-induced IL6 expression, but only partially diminished PIC-induced IL6 expression. Activated JNK phosphorylates c-Jun, which dimerizes with c-Fos to form AP-1 [50, 51]. The inhibitor of c-Jun/JNK complex formation, the inhibitor of AP-1 transcriptional activity, and the inhibitor of NFκB transcriptional activation had no major effect on HIV-1- or PIC-induced IL6 expression. This suggests that HIV-1-induced and TLR3-mediated IL6 expression & inflammation occurs principally via TAK1 and JNK pathways, not by affecting the c-Jun/JNK complex or AP1 transcriptional activity, nor by affecting NFκB transcriptional activation. Antibodies to IL6 or IL6 receptors are clinically used for the treatment of inflammatory diseases [55–57]. Our current study provides novel insights into the molecular basis for HIV-1-induced and TLR3-mediated IL6 expression in HBMEC, BBB dysfunction and CNS injury. The data suggests that targeting TLR3, TAK1, and JNK signaling pathways could provide a therapeutic approach for preventing BBB inflammation, dysfunction and CNS injury in HIV/AIDS.
Acknowledgments
This work was supported by grant from the National Institute of Health, National Institute of Mental Health RO1 MH094160. We would like to thank Dr. Joe Zhou, Director of the Microscopy Core Facility, University of Nebraska-Lincoln Center for Biotechnology for assistance with confocal imaging, and Ms. Hong Li for technical assistance.
Abbreviations
- TLR
Toll-like receptor
- PAMP
Pathogen associated molecular patterns
- HIV
Human immunodeficiency virus
- BBB
Blood-brain barrier
- HBMEC
Human brain microvascular endothelial cells
- EC
endothelial cells
- CNS
Central nervous system
- PIC
Polyinosinic-polycytidylic acid
- ssRNA
single stranded ribonucleic acid
- dsRNA
double stranded ribonucleic acid
- TLR3.CI
TLR3/dsRNA complex inhibitor
- GLUT1
Glucose transporter-1
- MOI
Multiplicity of infection
- kDa
kilo Dalton
- bp
base pairs
- RT-PCR
reverse transcription polymerase chain reaction
- GAPDH
Glyceraldehyde 3-phosphate dehydrogenase
- cDNA
Complementary DNA
- IL6
interleukin-6
- ATP
Adenosine triphosphate
- CFDA-SE
5-carboxyfluorescein diacetate, succinimidyl ester
- CREB/CEBP
cAMP response element binding protein
- STAT-1
Signal transducer and activator of transcription 1
- 5ZO
5Z-7-oxozeaenol. Table 1 and Table 3 include additional list of abbreviations
Footnotes
Authors’ contributions: B.B. carried out immunoassays, RT-PCR, real-time PCR, Western blot, adhesion and migration assays; participated in the making of Figures and Tables, data analysis and writing of the manuscript. G.D.K. conceived and designed the study, participated in the making of Figures and Tables, confocal microscopy, data analysis, and wrote the manuscript.
Conflicting interests: The authors declare that they have no conflict of interest.
References
- 1.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem Biophys Res Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 2.Beutler BA. Tlrs and innate immunity. Blood. 2009;113:1399–1407. doi: 10.1182/blood-2008-07-019307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Botos I, Segal DM, Davies DR. The structural biology of toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Jack CS, Arbour N, Manusow J, Montgrain V, Blain M, McCrea E, Shapiro A, Antel JP. Tlr signaling tailors innate immune responses in human microglia and astrocytes. J Immunol. 2005;175:4320–4330. doi: 10.4049/jimmunol.175.7.4320. [DOI] [PubMed] [Google Scholar]
- 6.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded rna and activation of nf-[kappa]b by toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Smith D, Bot S, Dellamary L, Bloom A, Bot A. Noncoding rna danger motifs bridge innate and adaptive immunity and are potent adjuvants for vaccination. The Journal of Clinical Investigation. 2002;110:1175–1184. doi: 10.1172/JCI15536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karikó K, Ni H, Capodici J, Lamphier M, Weissman D. Mrna is an endogenous ligand for toll-like receptor 3. Journal of Biological Chemistry. 2004;279:12542–12550. doi: 10.1074/jbc.M310175200. [DOI] [PubMed] [Google Scholar]
- 9.Wesch D, Beetz S, Oberg H-H, Marget M, Krengel K, Kabelitz D. Direct costimulatory effect of tlr3 ligand poly(i:C) on human γδ t lymphocytes. The Journal of Immunology. 2006;176:1348–1354. doi: 10.4049/jimmunol.176.3.1348. [DOI] [PubMed] [Google Scholar]
- 10.Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates west nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10:1366–1373. doi: 10.1038/nm1140. [DOI] [PubMed] [Google Scholar]
- 11.Vercammen E, Staal J, Beyaert R. Sensing of viral infection and activation of innate immunity by toll-like receptor 3. Clin Microbiol Rev. 2008;21:13–25. doi: 10.1128/CMR.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsumoto M, Oshiumi H, Seya T. Antiviral responses induced by the tlr3 pathway. Reviews in medical virology. 2011 doi: 10.1002/rmv.680. [DOI] [PubMed] [Google Scholar]
- 13.Hanamsagar R, Hanke ML, Kielian T. Toll-like receptor (tlr) and inflammasome actions in the central nervous system. Trends Immunol. 2012;33:333–342. doi: 10.1016/j.it.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong Y, Le Y. Toll-like receptors in inflammation of the central nervous system. Int Immunopharmacol. 2011;11:1407–1414. doi: 10.1016/j.intimp.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 15.Arroyo DS, Soria JA, Gaviglio EA, Rodriguez-Galan MC, Iribarren P. Toll-like receptors are key players in neurodegeneration. Int Immunopharmacol. 2011;11:1415–1421. doi: 10.1016/j.intimp.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanghavi SK, Reinhart TA. Increased expression of tlr3 in lymph nodes during simian immunodeficiency virus infection: Implications for inflammation and immunodeficiency. Journal of immunology. 2005;175:5314–5323. doi: 10.4049/jimmunol.175.8.5314. [DOI] [PubMed] [Google Scholar]
- 17.Kim H, Yang E, Lee J, Kim SH, Shin JS, Park JY, Choi SJ, Kim SJ, Choi IH. Double-stranded rna mediates interferon regulatory factor 3 activation and interleukin-6 production by engaging toll-like receptor 3 in human brain astrocytes. Immunology. 2008;124:480–488. doi: 10.1111/j.1365-2567.2007.02799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buitendijk M, Eszterhas SK, Howell AL. Toll-like receptor agonists are potent inhibitors of human immunodeficiency virus-type 1 replication in peripheral blood mononuclear cells. AIDS research and human retroviruses. 2014;30:457–467. doi: 10.1089/AID.2013.0199. [DOI] [PubMed] [Google Scholar]
- 19.Bhargavan B, Woollard SM, Kanmogne GD. Toll-like receptor-3 mediates hiv-1 transactivation via nfκb and jnk pathways and histone acetylation, but prolonged activation suppresses tat and hiv-1 replication. Cellular Signalling. 2016;28:7–22. doi: 10.1016/j.cellsig.2015.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ballabh P, Braun A, Nedergaard M. The blood-brain barrier: An overview: Structure, regulation, and clinical implications. Neurobiol Dis. 2004;16:1–13. doi: 10.1016/j.nbd.2003.12.016. [DOI] [PubMed] [Google Scholar]
- 21.Persidsky Y, Ramirez SH, Haorah J, Kanmogne GD. Blood-brain barrier: Structural components and function under physiologic and pathologic conditions. J Neuroimmune Pharmacol. 2006;1:223–236. doi: 10.1007/s11481-006-9025-3. [DOI] [PubMed] [Google Scholar]
- 22.Berger JR, Avison M. The blood brain barrier in hiv infection. Front Biosci. 2004;9:2680–2685. doi: 10.2741/1427. [DOI] [PubMed] [Google Scholar]
- 23.Toborek M, Lee YW, Flora G, Pu H, Andras IE, Wylegala E, Hennig B, Nath A. Mechanisms of the blood-brain barrier disruption in hiv-1 infection. Cell Mol Neurobiol. 2005;25:181–199. doi: 10.1007/s10571-004-1383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banks WA, Ercal N, Price TO. The blood-brain barrier in neuroaids. Curr HIV Res. 2006;4:259–266. doi: 10.2174/157016206777709447. [DOI] [PubMed] [Google Scholar]
- 25.Chaudhuri A, Duan F, Morsey B, Persidsky Y, Kanmogne GD. Hiv-1 activates proinflammatory and interferon-inducible genes in human brain microvascular endothelial cells: Putative mechanisms of blood-brain barrier dysfunction. J Cereb Blood Flow Metab. 2008;28:697–711. doi: 10.1038/sj.jcbfm.9600567. [DOI] [PubMed] [Google Scholar]
- 26.Chaudhuri A, Yang B, Gendelman HE, Persidsky Y, Kanmogne GD. Stat1 signaling modulates hiv-1-induced inflammatory responses and leukocyte transmigration across the blood-brain barrier. Blood. 2008;111:2062–2072. doi: 10.1182/blood-2007-05-091207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woollard SM, Bhargavan B, Yu F, Kanmogne GD. Differential effects of tat proteins derived from hiv-1 subtypes b and recombinant crf02_ag on human brain microvascular endothelial cells: Implications for blood-brain barrier dysfunction. J Cereb Blood Flow Metab. 2014;34:1047–1059. doi: 10.1038/jcbfm.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhargavan B, Kanmogne GD. Differential mechanisms of inflammation and endothelial dysfunction by hiv-1 subtype-b and recombinant crf02_ag tat proteins on human brain microvascular endothelial cells: Implications for viral neuropathogenesis. Molecular neurobiology. 2017 doi: 10.1007/s12035-017-0382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang B, Akhter S, Chaudhuri A, Kanmogne GD. Hiv-1 gp120 induces cytokine expression, leukocyte adhesion, and transmigration across the blood-brain barrier: Modulatory effects of stat1 signaling. Microvascular research. 2009;77:212–219. doi: 10.1016/j.mvr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanmogne GD, Schall K, Leibhart J, Knipe B, Gendelman HE, Persidsky Y. Hiv-1 gp120 compromises blood-brain barrier integrity and enhances monocyte migration across blood-brain barrier: Implication for viral neuropathogenesis. J Cereb Blood Flow Metab. 2007;27:123–134. doi: 10.1038/sj.jcbfm.9600330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen-Kiang S, Hsu W, Natkunam Y, Zhang X. Nuclear signaling by interleukin-6. Curr Opin Immunol. 1993;5:124–128. doi: 10.1016/0952-7915(93)90091-6. [DOI] [PubMed] [Google Scholar]
- 32.Akira S. Il-6-regulated transcription factors. Int J Biochem Cell Biol. 1997;29:1401–1418. doi: 10.1016/s1357-2725(97)00063-0. [DOI] [PubMed] [Google Scholar]
- 33.Baccam M, Woo SY, Vinson C, Bishop GA. Cd40-mediated transcriptional regulation of the il-6 gene in b lymphocytes: Involvement of nf-kappa b, ap-1, and c/ebp. J Immunol. 2003;170:3099–3108. doi: 10.4049/jimmunol.170.6.3099. [DOI] [PubMed] [Google Scholar]
- 34.Nagyoszi P, Wilhelm I, Farkas AE, Fazakas C, Dung NT, Hasko J, Krizbai IA. Expression and regulation of toll-like receptors in cerebral endothelial cells. Neurochem Int. 2010;57:556–564. doi: 10.1016/j.neuint.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Pegu A, Qin S, Fallert Junecko BA, Nisato RE, Pepper MS, Reinhart TA. Human lymphatic endothelial cells express multiple functional tlrs. J Immunol. 2008;180:3399–3405. doi: 10.4049/jimmunol.180.5.3399. [DOI] [PubMed] [Google Scholar]
- 36.Stewart EA, Wei R, Branch MJ, Sidney LE, Amoaku WM. Expression of toll-like receptors in human retinal and choroidal vascular endothelial cells. Exp Eye Res. 2015;138:114–123. doi: 10.1016/j.exer.2015.06.012. [DOI] [PubMed] [Google Scholar]
- 37.Weber F, Wagner V, Rasmussen SB, Hartmann R, Paludan SR. Double-stranded rna is produced by positive-strand rna viruses and DNA viruses but not in detectable amounts by negative-strand rna viruses. J Virol. 2006;80:5059–5064. doi: 10.1128/JVI.80.10.5059-5064.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawai T, Akira S. Toll-like receptor and rig-i-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- 39.Scagnolari C, Selvaggi C, Chiavuzzo L, Carbone T, Zaffiri L, d’Ettorre G, Girardi E, Turriziani O, Vullo V, Antonelli G. Expression levels of tlrs involved in viral recognition in pbmcs from hiv-1-infected patients failing antiretroviral therapy. Intervirology. 2009;52:107–114. doi: 10.1159/000218082. [DOI] [PubMed] [Google Scholar]
- 40.Lester RT, Yao XD, Ball TB, McKinnon LR, Kaul R, Wachihi C, Jaoko W, Plummer FA, Rosenthal KL. Toll-like receptor expression and responsiveness are increased in viraemic hiv-1 infection. AIDS. 2008;22:685–694. doi: 10.1097/QAD.0b013e3282f4de35. [DOI] [PubMed] [Google Scholar]
- 41.Bastard JP, Soulie C, Fellahi S, Haim-Boukobza S, Simon A, Katlama C, Calvez V, Marcelin AG, Capeau J. Circulating interleukin-6 levels correlate with residual hiv viraemia and markers of immune dysfunction in treatment-controlled hiv-infected patients. Antivir Ther. 2012;17:915–919. doi: 10.3851/IMP2093. [DOI] [PubMed] [Google Scholar]
- 42.Hamlyn E, Fidler S, Stohr W, Cooper DA, Tambussi G, Schechter M, Miro JM, McClure M, Weber J, Babiker A, et al. Interleukin-6 and d-dimer levels at seroconversion as predictors of hiv-1 disease progression. AIDS. 2014;28:869–874. doi: 10.1097/QAD.0000000000000155. [DOI] [PubMed] [Google Scholar]
- 43.Olwenyi OA, Naluyima P, Cham F, Quinn TC, Serwadda D, Sewankambo NK, Gray RH, Sandberg JK, Michael NL, Wabwire-Mangen F, et al. Brief report: Differential associations of interleukin 6 and intestinal fatty acid-binding protein with progressive untreated hiv-1 infection in rakai, uganda. J Acquir Immune Defic Syndr. 2016;72:15–20. doi: 10.1097/QAI.0000000000000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagele H, Allam R, Pawar RD, Anders HJ. Double-stranded rna activates type i interferon secretion in glomerular endothelial cells via retinoic acid-inducible gene (rig)-1. Nephrol Dial Transplant. 2009;24:3312–3318. doi: 10.1093/ndt/gfp339. [DOI] [PubMed] [Google Scholar]
- 45.Huang LY, Stuart C, Takeda K, D’Agnillo F, Golding B. Poly(i:C) induces human lung endothelial barrier dysfunction by disrupting tight junction expression of claudin-5. PloS one. 2016;11:e0160875. doi: 10.1371/journal.pone.0160875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimmer S, Steinmetz M, Asdonk T, Motz I, Coch C, Hartmann E, Barchet W, Wassmann S, Hartmann G, Nickenig G. Activation of endothelial toll-like receptor 3 impairs endothelial function. Circ Res. 2011;108:1358–1366. doi: 10.1161/CIRCRESAHA.111.243246. [DOI] [PubMed] [Google Scholar]
- 47.Leonard JN, Ghirlando R, Askins J, Bell JK, Margulies DH, Davies DR, Segal DM. The tlr3 signaling complex forms by cooperative receptor dimerization. Proc Natl Acad Sci U S A. 2008;105:258–263. doi: 10.1073/pnas.0710779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bell JK, Askins J, Hall PR, Davies DR, Segal DM. The dsrna binding site of human toll-like receptor 3. Proc Natl Acad Sci U S A. 2006;103:8792–8797. doi: 10.1073/pnas.0603245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lai Y, Yi G, Chen A, Bhardwaj K, Tragesser BJ, Rodrigo AV, Zlotnick A, Mukhopadhyay S, Ranjith-Kumar CT, Kao CC. Viral double-strand rna-binding proteins can enhance innate immune signaling by toll-like receptor 3. PloS one. 2011;6:e25837. doi: 10.1371/journal.pone.0025837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kallunki T, Deng T, Hibi M, Karin M. C-jun can recruit jnk to phosphorylate dimerization partners via specific docking interactions. Cell. 1996;87:929–939. doi: 10.1016/s0092-8674(00)81999-6. [DOI] [PubMed] [Google Scholar]
- 51.Gupta S, Barrett T, Whitmarsh AJ, Cavanagh J, Sluss HK, Derijard B, Davis RJ. Selective interaction of jnk protein kinase isoforms with transcription factors. The EMBO journal. 1996;15:2760–2770. [PMC free article] [PubMed] [Google Scholar]
- 52.Minden A, Karin M. Regulation and function of the jnk subgroup of map kinases. Biochim Biophys Acta. 1997;1333:F85–104. doi: 10.1016/s0304-419x(97)00018-8. [DOI] [PubMed] [Google Scholar]
- 53.Minden A, Lin A, Claret FX, Abo A, Karin M. Selective activation of the jnk signaling cascade and c-jun transcriptional activity by the small gtpases rac and cdc42hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 54.van Dam H, Castellazzi M. Distinct roles of jun : Fos and jun : Atf dimers in oncogenesis. Oncogene. 2001;20:2453–2464. doi: 10.1038/sj.onc.1204239. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka T, Narazaki M, Kishimoto T. Il-6 in inflammation, immunity, and disease. Cold Spring Harbor Perspectives in Biology. 2014;6 doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tanaka T, Narazaki M, Ogata A, Kishimoto T. A new era for the treatment of inflammatory autoimmune diseases by interleukin-6 blockade strategy. Seminars in Immunology. 2014;26:88–96. doi: 10.1016/j.smim.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 57.Rossi J-F, Lu Z-Y, Jourdan M, Klein B. Interleukin-6 as a therapeutic target. Clinical Cancer Research. 2015;21:1248–1257. doi: 10.1158/1078-0432.CCR-14-2291. [DOI] [PubMed] [Google Scholar]