Abstract
Background and Aims:
Longitudinal electronic health record (EHR) data offer a large-scale, untapped source of phenotypic information on harmful alcohol use. Using established, alcohol-associated variants in the gene that encodes the enzyme alcohol dehydrogenase 1B (ADH1B) as criterion standards, we compared the individual and combined validity of three longitudinal EHR-based phenotypes of harmful alcohol use: Alcohol Use Disorders Identification Test-Consumption (AUDIT-C) trajectories; mean age-adjusted AUDIT-C; and diagnoses of Alcohol Use Disorder (AUD).
Design:
With longitudinal EHR data from the Million Veteran Program (MVP) linked to genetic data, we used two population-specific polymorphisms in ADH1B that are strongly associated with AUD in African Americans (AAs) and European Americans (EAs): rs2066702 (Arg369Cys, AAs) and rs1229984 (Arg48His, EAs) as criterion measures.
Setting:
United States Department of Veterans Affairs Healthcare System
Participants:
167,721 veterans (57,677 AAs and 110,044 EAs; 92% male, mean age of 63 years). Data collected from October 1, 2007–May 1, 2017.
Measurements:
Using all AUDIT-C scores and AUD diagnostic codes recorded in the EHR, we calculated age-adjusted mean AUDIT-C values, longitudinal statistical trajectories of AUDIT-C scores, and ICD-9/10 diagnostic groupings for AUD.
Findings:
19,793 AAs (34.3%) had one or two minor alleles at rs2066702 (minor allele frequency (MAF)=0.190) and 6,933 EAs (6.3%) had one or two minor alleles at rs1229984 (MAF=0.032). In both populations, trajectories and age-adjusted mean AUDIT-C were correlated (r=0.90) but, when considered separately, highest score (8+ vs 0) of age adjusted mean AUDIT-C demonstrated a stronger association with the ADH1B variants (aOR 0.54 in AAs and 0.37 in AAs) than did highest trajectory (aOR 0.71 in AAs and 0.53 in EAs); combining AUDIT-C metrics did not improve discrimination. When age-adjusted mean AUDIT-C score and AUD diagnoses were considered together, age-adjusted mean AUDIT-C (8+ vs. 0) was associated with lower odds of having the ADH1B minor allele than were AUD diagnostic codes: aOR = 0.59 vs. 0.86 in AAs and 0.48 vs. 0.68 in EAs. These independent associations combine to yield an even lower aOR of 0.51 for AAs and 0.33 for EAs.
Conclusions:
The age-adjusted mean AUDIT-C score is more strongly associated with genetic polymorphisms of known risk for alcohol use disorder than are longitudinal trajectories of AUDIT-C or AUD diagnostic codes. AUD diagnostic codes modestly enhance this association.
Keywords: Alcohol use disorder diagnostic codes, AUDIT-C, ADH1B, Arg369Cys, Arg48His, African American, European American, electronic health record data, trajectory analyses
Introduction
Alcohol use disorder (AUD) is a complex trait that is about 50% heritable (1). However, with the exception of genetic variants in ADH1B, few variants contributing to the risk of the disorder have been identified. The relatively small sample size of most published genome-wide association studies (GWASs) of AUD has provided limited statistical power to identify the many variants of small effect that likely contribute to the disorder. Linkage and candidate gene studies, and more recently GWASs, have consistently identified risk loci at the genes encoding several of the alcohol-metabolizing enzymes (2). In a discovery and replication sample of 16,087 subjects (3), we confirmed previously identified risk loci mapped to the alcohol-metabolizing enzyme genes ADH1B in African Americans (AAs; Arg369Cys or rs2066702, P = 6.33 × 10−17) and European Americans (EAs; Arg48His or rs1229984, P = 1.17 × 10−31) and identified numerous novel additional associations across the ADH region on chromosome 4. Despite the presence of AUD risk loci in the ADH gene region and elsewhere, these account for only a small percentage of the estimated heritability of the disorder. Large-scale studies [e.g., those using electronic health records (EHRs)] could identify additional risk loci, but they require a valid, widely available, and generalizable phenotype.
Many studies have used International Classification of Disease (ICD) diagnostic codes as phenotypes for harmful alcohol use, though these designations may be insensitive or non-specific (4, 5). The Alcohol Use Disorders Identification Test-Consumption (AUDIT-C), a 3-item self-reported measure of the quantity and frequency of alcohol consumption, is a practical, valid screening test for harmful drinking and risk of a current AUD (6). Since 2007, the AUDIT-C has been a required annual screening test in primary care in the U.S. Department of Veterans Affairs (VA) and is widely applicable to other medical settings.
A recent GWAS was conducted using the full 10-item AUDIT in 20,328 EA research participants from 23andMe (7). Although this was the largest study to date using the AUDIT as a phenotype for GWAS, it was restricted to a single AUDIT measure and included no AAs. The lack of genome-wide significant results in this study may have resulted from the generally low AUDIT scores in the relatively high socioeconomic status sample, the use of a single measure rather than a longitudinal phenotype, and lack of data for the key variant, ADH1B*rs1229984, which was filtered out of that sample due to problems with genotyping and imputation (Abraham A. Palmer, personal communication, November 29, 2017).
We previously developed and validated a longitudinal phenotype for harmful alcohol use using statistical trajectories of AUDIT-C (8). In a sample of 1,851 Veterans with or without HIV infection, we compared longitudinal AUDIT-C metrics on the magnitude of association with phosphatidylethanol (PEth), a direct alcohol biomarker measured in blood. In a subsample of AAs, we also used the frequency of the minor allele rs2066702, a single nucleotide polymorphism in ADH1B, which encodes the enzyme alcohol dehydrogenase 1B as a criterion measure. AUDIT-C trajectories were more strongly associated with both PEth and rs2066702 than either the highest AUDIT-C score or the score closest in time to the PEth blood draw. These findings demonstrated the value of a longitudinal measure to characterize alcohol use, as it is typically collected in general medical settings.
Although the prior study (8) was an important initial effort, it was limited to a small sample of AA Veterans with HIV infection, and demographically similar uninfected controls. That study did not consider age-adjusted mean AUDIT-C score, a simpler and more generalizable metric, nor did it evaluate the ADH1B polymorphism more commonly seen in EAs (rs1229984). Herein, we report the strength of association between the ADH1B polymorphisms—rs2066702 in AAs and rs1229984 in EAs—with previously validated AUDIT-C trajectories in the larger, more generalizable Million Veteran Program (MVP) sample (9). We also compared these associations to those using age-adjusted mean AUDIT-C score. Because ICD codes are increasingly used for phenome-wide association studies (PheWASs) (10–13), we also compared AUDIT-C metrics to ICD codes for AUD. The goal of these comparisons was to use longitudinal EHR data to determine the most informative phenotype for efforts at gene finding for harmful drinking.
Methods
Design
Using longitudinal EHR data linked to genome-wide association study (GWAS) data in the Million Veteran Program, we used two population-specific missense single nucleotide polymorphisms (SNPs) in ADH1B as criterion measures to determine whether longitudinal trajectories of AUDIT-C, age-adjusted mean AUDIT-C, or administrative codes for AUD provided the best phenotype for genetic discovery. We also considered whether additional information was gained by combining metrics.
Participants
The MVP has been described in detail elsewhere (9). Briefly, it is an observational cohort study and mega-biobank supported by the VA. Data are collected from participants using questionnaires, the VA EHR, and a blood sample for genomic and other testing. We used AUDIT-C data collected from October 1, 2007 (when first available from EHR extracts) to May 1, 2017 and lifetime ICD diagnostic codes for AUD.
Measures
AUDIT-C:
The stem asks about any alcohol consumption over the past year. The three questions are: 1) How often do you have a drink containing alcohol? 2) How many standard drinks containing alcohol do you have on a typical day? and 3) How often do you have six or more drinks on one occasion? Responses to each question are assigned 0–4 points, yielding a total score of 0–12. AUDIT-C screening thresholds that maximize sensitivity and specificity for detecting harmful alcohol use are ≥4 in men and ≥3 in women, with alcohol-related harm in both sexes increasing as AUDIT-C scores increase (14, 15).
ICD Diagnostic Codes for Alcohol Use Disorder (AUD):
We used ICD-9/10 codes to identify subjects diagnosed with AUD (either alcohol abuse or dependence) codes. The codes are based on the work of Piette et al. (16) and are included in on-line supplementary material. Participants were considered to have a diagnosis of AUD if they had at least one inpatient or two outpatient alcohol-related ICD-9 or ICD-10 codes. This requirement has been shown to improve the specificity of ICD codes compared to chart review (17). Of note, we chose not to include codes consistent with alcohol-related conditions (e.g., alcoholic cirrhosis), as we wanted to create a phenotype of drinking behavior, not its complications, to which different genetic variants may contribute.
Criterion Standard:
Variation in ADH1B is the best supported molecular genetic risk factor for harmful alcohol use among both EAs and AAs (18). The minor allele (369Cys) of the ADH1B single nucleotide polymorphism (SNP) rs2066702 is associated with a reduced risk of AUD in AAs (3), whereas the minor allele (48His) of rs1229984 is associated with a reduced risk of AUD in EAs (3).
Quality Control
We limited analyses to the two largest population groups—EAs and AAs—to provide adequate statistical power. We removed related individuals (closer than second degree) and those with evidence of admixture or divergent ancestry, discordant or missing sex, missing age, genome-wide heterozygosity rates more than 3 standard deviations above the mean, and individuals younger than 22 or older than 90 years of age.
To avoid confounding the analysis of the relations between the functional variant in each population and AUDIT-C scores, we removed individuals with the ADH1B minor allele that is uncommon in that ancestry group (i.e., 449 EAs with the minor allele of rs2066702 and 1,457 AAs with the minor allele of rs1229984). That is, an AA with the minor allele of rs1229984 could be protected from heavy drinking (as would an EA who is more likely to carry that allele).
Because trajectory analysis, one of the analytic methods used herein, requires repeated measures, we only included individuals with two or more AUDIT-C scores obtained at different ages. This approach yielded an analytic sample of 167,721 participants (57,677 AAs and 110,044 EAs). The sample was predominantly male (92%), with a mean age of 63 years at sampling and a median of seven AUDIT-C scores per subject (25th and 75th quartiles: 5 and 8).
ADH1B genotypes were obtained from the MVP GWAS, which used a custom Affymetrix Axiom Biobank Array, with 686,693 markers. The custom array is enriched for exomic SNPs, has SNPs validated for medical and psychiatric traits, and was augmented with biomarkers of specific interest to the VA population—including enrichment for African-American and Hispanic ancestry markers and validated markers for common diseases (9). The standardized Affymetrix best practice workflow was used to produce genotype data (19). Both rs2066702 and rs1229984 were directly genotyped on the array. For rs2066702 (Arg369Cys), the major (C) allele encodes an arginine and the minor (T) allele encodes a cysteine. For rs1229984 (Arg48His), the major (G) allele encodes an arginine and the minor (A) allele encodes a histidine. The genotype distribution for rs1229984 in EAs was consistent with Hardy-Weinberg Equilibrium expectations (HWEE) (χ2 (1) = 2.04, p=0.15) after exclusion of a subpopulation with a high MAF that was identified as outlying on principal components (PC) analysis. In AAs, there was no significant departure from HWEE considering the large sample size (χ2 (1) = 4.21, p=0.04).
Analyses
Using all available longitudinal EHR AUDIT-C measurements, we calculated two AUDIT-C metrics: the age-adjusted mean AUDIT-C value and a four-level trajectory of AUDIT-C scores for each participant. As previously reported (8), using the SAS 9.2 procedure, TRAJ (20), we estimated four-level longitudinal statistical trajectories of AUDIT-C (infrequent, lower risk, potentially high risk, and consistently high risk), using age as the time scale to account for decreasing alcohol use with age. The procedure calculates each participant’s probability of belonging to each trajectory and assigns him or her to the trajectory with the highest probability of membership. Age was used as the time scale, thus each individual could only contribute one AUDIT-C score for each age (i.e., year). Participants’ modal AUDIT-C scores were used for each age at which they received the AUDIT-C assessment. The analysis used a zero-inflated Poisson model (20). The median probability of membership calculated for all four groups was >90%. Trajectory analysis allows for missing longitudinal data, thus no individuals or observations were excluded due to missingness.
The mean AUDIT-C was age adjusted for comparison with the trajectories. To compute the age-adjusted mean AUDIT-C score, we used age 50 as the reference point and created weights to down-weight scores for individuals younger than 50 and up-weight scores for individuals older than 50. To create the weights, we first calculated the mean AUDIT-C score at each age (e.g., average score of all 22-year-olds, average score of all 23-year-olds). The mean score at age 50 was then divided by each age-specific mean to create the weights (e.g., mean at age 50/mean at age 22=weight for age 22). Then, the subject-specific AUDIT-C scores were multiplied by the appropriate weight (e.g., an AUDIT-C score for an individual at age 30 was multiplied by the corresponding weight for age 30). The weighted scores for a participant were summed and divided by the sum of the weights used for that participant to calculate the age-adjusted mean AUDIT-C score. The mean scores were rounded to the nearest whole from 0–12 and higher scores were collapsed due to their relatively small sample sizes. This resulted in an eight-level ordinal measure of age-adjusted mean AUDIT-C scores: 0, 1, 2, 3, 4, 5, 6 or 7, and 8+.
In subsequent analyses, we first considered covariance among the AUDIT-C metrics (mean and trajectory) and AUD diagnostic codes. We then evaluated the association of each of these measures (age-adjusted mean AUDIT-C, AUDIT-C trajectories, and AUD diagnostic codes) with rs2066702 (in AAs) and rs1229984 (in EAs) using ordered logit models, in which the allele count, i.e., 0, 1, or 2 minor alleles, was used as the outcome variable and the phenotype as the predictor in regression analyses. Of note, this approach is opposite to convention in which the phenotype is the outcome. Our goal was to compare the association between several candidate phenotype variables with an established genotype, adjusting for covariates. We are modeling association, not causation. In this context the role of dependent and independent variables is irrelevant. By modeling allele count as the outcome and candidate phenotypes as predictors adjusting for age, sex, and 10 PCs we were able to more directly compare independent associations between the candidate phenotypes and the established AHD1B variants relevant to each ancestry in a standardized manner. We used different SNPs for EAs and AAs because populations often differ in the frequency of alleles at polymorphic sites and a polymorphism in a gene in one population may not be present or may be very uncommon in another population. Two different single nucleotide polymorphisms in ADH1B are present in EAs and AAs and both are associated with alcohol dependence in the respective populations. However, the prevalence of the minor allele of each is too low in the opposite population (e.g., in AAs, rs1229984 has a minor allele frequency that is nearly zero) for analyses in the opposite populations to be meaningful. Ordered logit regression was modeled using the SAS 9.2 procedure, GENMOD.
Analyses in both populations were adjusted for ancestry PCs, sex, and age. To characterize the underlying genetic architecture of the samples using PCs, we first applied two filters to all markers: linkage disequilibrium pruning (using PLINK (20) option –indep, with threshold > 1%) and minor allele frequency (MAF) screening (MAF < 1%). We conducted PC analysis separately within the EAs and AAs using FlashPCA (21). The first 10 PCs were used in all subsequent analyses to correct for residual population stratification.
Results
AAs were younger than EAs (mean 58 vs. 66 years; p<0.001) and less likely to be male (87% vs. 94%; p<0.001) (Table 1). AAs were also more likely than EAs to have an age-adjusted mean AUDIT-C score below 2 (65% vs. 55%; p<0.001) and to be assigned to the lowest AUDIT-C trajectory characterized by infrequent drinking (38% vs. 27%; p<0.001). However, AAs were much more likely to receive an AUD diagnosis (31% vs. 14%; p<0.001).
Table 1.
Characteristics of Participants
| All (n=167,721) | African Americans (n=57,677) | European Americans (n=110,044) | |
|---|---|---|---|
| Male | 91.5% | 87.3% | 93.8% |
| Agea, mean (SD) | 63.3 (11.5) | 57.7 (11.9) | 66.2 (11.3) |
| AUDIT-C Trajectory | |||
| 1 | 30.8% | 37.9% | 27.1% |
| 2 | 32.5% | 34.1% | 31.6% |
| 3 | 29.6% | 23.1% | 33.0% |
| 4 | 7.2% | 4.9% | 8.4% |
| Age-Adjusted mean AUDIT-C | |||
| 0 | 33.7% | 39.9% | 30.4% |
| 1 | 24.4% | 25.0% | 24.1% |
| 2 | 16.6% | 15.3% | 9.4% |
| 3 | 12.1% | 9.4% | 13.6% |
| 4 | 7.7% | 5.1% | 9.0% |
| 5 | 2.7% | 2.5% | 2.8% |
| 6 or 7 | 2.0% | 2.1% | 2.0% |
| 8+ | 0.8% | 0.7% | 0.8% |
| Alcohol Use Disorder (AUD) Codesb | 19.9% | 30.8% | 14.2% |
| rs1229984 (heterozygous) | -- | -- | 6.2% |
| rs1229984 (homozygous) | -- | -- | 0.1% |
| rs2066702 (heterozygous) | -- | 30.6% | -- |
| rs2066702 (homozygous) | -- | 3.8% | -- |
SD=standard deviation; AUDIT-C=Alcohol Use Disorders Identification Test-Consumption.
Age at time of MVP enrollment
Lifetime diagnosis
Among the 57,677 AAs, 19,793 (34.3%) had one or two minor alleles at rs2066702 (minor allele frequency (MAF)=0.190). Among the 110,044 EAs, 6,933 (6.3%) had one or two minor alleles at rs1229984 (MAF=0.032). These minor allele frequencies are similar to those reported previously for AAs (0.19) (22) and EAs (0.03–0.06) (22–24).
Association among Candidate Phenotypes
As expected, the longitudinal AUDIT-C metrics (age-adjusted mean AUDIT-C and AUDIT-C trajectories) were highly correlated (r=0.90), which did not differ by population group. The proportion of individuals with an AUD increased monotonically with increasing mean AUDIT-C scores, though the correlation differed by population group (AAs: r=0.38, EAs: r=0.26; p<0.001). Among individuals with a mean AUDIT-C score of 4 or higher, more than twice the percentage of AAs as EAs (73% vs. 36%; p<0.001) had an AUD diagnosis in the EHR (Figure 1a). Similarly, nearly twice the percentage of AAs as EAs assigned to the highest AUDIT-C trajectory had an AUD diagnostic code in the EHR (83% vs. 44%; p<0.001) (Figure 1b).
Figure 1a.
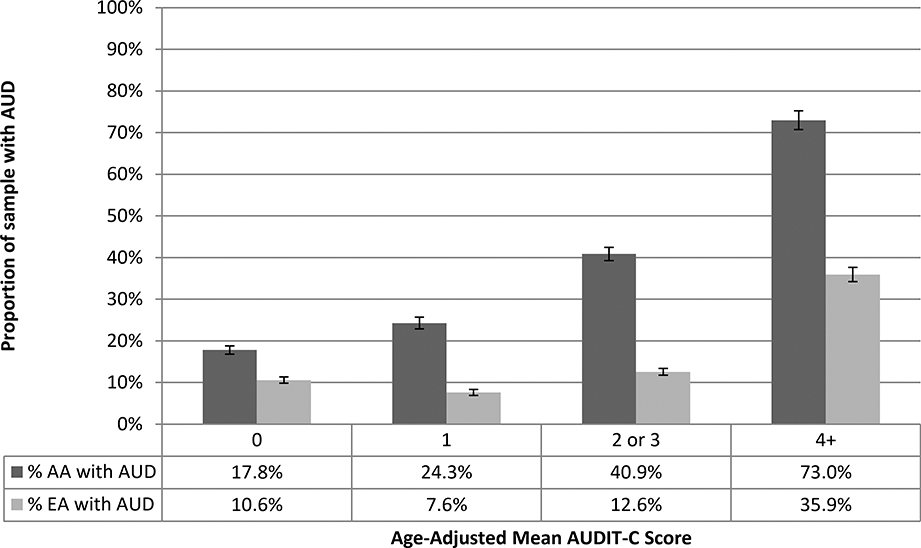
Proportion of Sample with AUD Codes by Age-Adjusted Mean AUDIT-C and Population Group
Figure 1b.
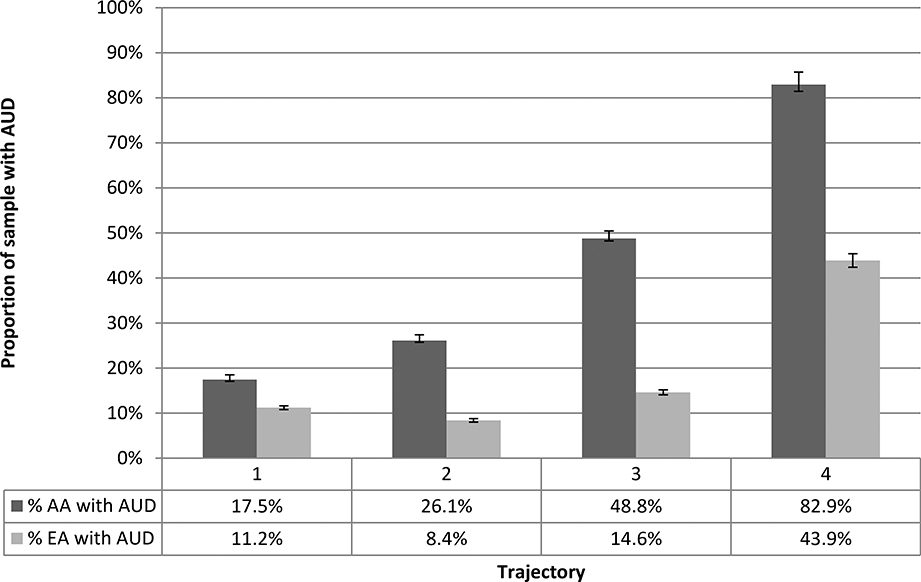
Proportion of Sample with AUD Codes by AUDIT-C Trajectories and Population Group
Association of Candidate Phenotypes with rs2066702 in African Americans
In unadjusted analyses in AAs, all three candidate phenotypes were associated with rs2066702 (χ2 p-values all < 10−21) (upper half of Table 2, Figure 2a-c). In ordered logit models, adjusting for the first 10 PCs, sex, and age, the association was strongest for age-adjusted mean AUDIT-C score, treated as an ordinal variable (p=5.12 × 10−29). After accounting for age-adjusted mean AUDIT-C score, the addition of AUDIT-C trajectories did not improve the model fit (both pseudo R2=0.014; likelihood ratio test statistic (LRT)=0.57, degrees of freedom (df)=3, p=0.90). However, the addition of AUD diagnostic codes improved discrimination (LRT=49.5, df=1, p=1.98 × 10−12) (Table 2a and b and Figure 3a).
Table 2a.
Bivariate and Adjusted Associations of Candidate Phenotypes with ADH1B Polymorphisms by Population Group
| Bivariate Association | Ordered Logit Model | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Pearson’s χ2 | Test of Trend | ||||||||
| χ2 | p-value | χ2 | p-value | LR test | df | Pseudo R2 | p-value | ||
| Association of rs2066702 with Candidate Phenotypes Among African Americans | |||||||||
| Age-adjusted mean AUDIT-Ca | 166 | 5.13 e-28 | 109 | 1.11 e-16 | 149.52 | 7 | 0.0143 | 5.12 e-29 | |
| AUDIT-C trajectoriesb | 112 | 7.60 e-22 | 68 | 9.06 e-13 | 102.40 | 3 | 0.0138 | 4.73 e-22 | |
| AUD codesc | 104 | 2.13 e-23 | 100 | 2.09 e-22 | 108.61 | 1 | 0.0138 | 1.98 e-25 | |
| Age-adjusted mean AUDIT-C + AUDIT-C trajectoriesd | n/a | -- | n/a | -- | 150.09 | 10 | 0.0143 | 3.56 e-27 | |
| Age-adjusted mean AUDIT-C + AUD codesd | n/a | -- | n/a | -- | 198.97 | 8 | 0.0149 | 1.05 e-38 | |
| Association of rs1229984 with Candidate Phenotypes Among European Americans | |||||||||
| Age-adjusted mean AUDIT-Ca | 268 | 5.40 e-49 | 159 | 2.13 e-36 | 244.37 | 7 | 0.0557 | 4.38 e-49 | |
| AUDIT-C trajectoriesb | 205 | 1.53 e-41 | 68 | 1.62 e-16 | 197.56 | 3 | 0.0548 | 1.42 e-42 | |
| AUD codesc | 183 | 1.85 e-40 | 178 | 1.58 e-40 | 158.79 | 1 | 0.0541 | 2.08 e-36 | |
| Age-adjusted mean AUDIT-C + AUDIT-C trajectoriesd | n/a | -- | n/a | -- | 250.45 | 10 | 0.0558 | 4.36 e-48 | |
| Age-adjusted mean AUDIT-C + AUD codesd | n/a | -- | n/a | -- | 321.91 | 8 | 0.0572 | 8.88 e-65 | |
AUDIT-C=Alcohol Use Disorders Identification Test-Consumption; AUD=alcohol use disorder; LR=likelihood ratio; df=degrees of freedom.
8-level ordinal measure
4-group trajectory
Based on ICD-9/10 codes
The combination of AUDIT-C measures and AUD codes does not permit bivariate associations; n/a=not applicable
Figures 2a-2f.
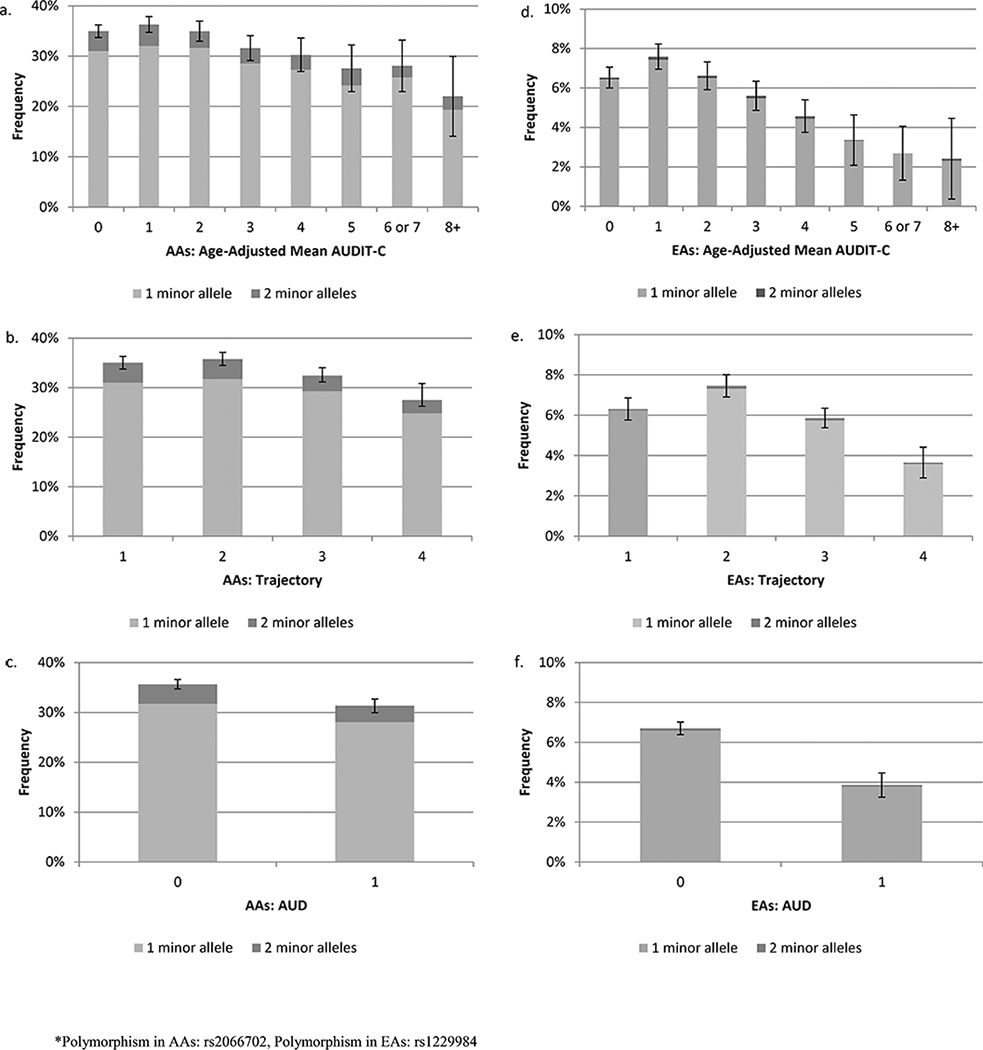
Proportion of Sample with 1 or 2 Copies of the Minor Allele by Candidate Phenotype and Population Group
Figure 2a. Age-Adjusted Mean AUDIT-C in African Americans Figure 2b. AUDIT-C Trajectories in African Americans
Figure 2c. AUD Diagnostic Codes in African Americans
Figure 2d. Age-Adjusted Mean AUDIT-C in European Americans
Figure 2e.AUDIT-C Trajectories in European Americans
Figure 2f. AUD Diagnostic Codes in European Americans
Table 2b.
Adjusted Odds Ratios for Association of ADH1B Variant Dosage and Candidate Phenotypes by Population Group
| Candidate Phenotype | African Americans | European Americans |
|---|---|---|
| Cumulative Dosage*(rs2066702) | Cumulative Dosage*(rs1229984) | |
| aOR (95% CI) | aOR (95% CI) | |
| Age-adjusted mean AUDIT-C | ||
| 0 | 1.00 | 1.00 |
| 1 | 1.07 (1.02, 1.12) | 1.06 (1.00, 1.13) |
| 2 | 0.99 (0.94, 1.05) | 0.90 (0.84, 0.97) |
| 3 | 0.86 (0.80, 0.91) | 0.77 (0.71, 0.83) |
| 4 | 0.81 (0.74, 0.88) | 0.65 (0.59, 0.73) |
| 5 | 0.71 (0.63, 0.80) | 0.51 (0.41, 0.62) |
| 6 or 7 | 0.73 (0.64, 0.83) | 0.39 (0.30, 0.51) |
| 8+ | 0.54 (0.42, 0.68) | 0.37 (0.24, 0.57) |
| AUDIT-C trajectories | ||
| 1 | 1.00 | 1.00 |
| 2 | 1.03 (1.00, 1.08) | 1.07 (1.01, 1.14) |
| 3 | 0.89 (0.85, 0.93) | 0.82 (0.77, 0.88) |
| 4 | 0.71 (0.65, 0.77) | 0.53 (0.47, 0.60) |
| AUD Codes | ||
| 0 | 1.00 | 1.00 |
| 1 | 0.82 (0.79, 0.85) | 0.59 (0.54, 0.64) |
| Age-adjusted mean AUDIT-C + AUDIT-C trajectories | ||
| Age-adjusted mean AUDIT-C | ||
| 0 | 1.00 | 1.00 |
| 1 | 1.08 (0.99, 1.18) | 0.94 (0.83, 1.05) |
| 2 | 0.99 (0.89, 1.10) | 0.79 (0.67, 0.93) |
| 3 | 0.85 (0.74, 0.97) | 0.67 (0.56, 0.81) |
| 4 | 0.80 (0.69, 0.92) | 0.58 (0.47, 0.71) |
| 5 | 0.68 (0.56, 0.83) | 0.45 (0.33, 0.61) |
| 6 or 7 | 0.70 (0.55, 0.88) | 0.35 (0.24, 0.50) |
| 8+ | 0.51 (0.37, 0.70) | 0.33 (0.20, 0.55) |
| Trajectories | ||
| 1 | 1.00 | 1.00 |
| 2 | 0.99 (0.91, 1.07) | 1.16 (1.03, 1.31) |
| 3 | 1.01 (0.90, 1.14) | 1.17 (0.98, 1.39) |
| 4 | 1.05 (0.85, 1.29) | 1.13 (0.88, 1.46) |
| Age-adjusted mean AUDIT-C + AUD Codes | ||
| Age-adjusted mean AUDIT-C | ||
| 0 | 1.00 | 1.00 |
| 1 | 1.08 (1.03, 1.13) | 1.05 (0.98, 1.12) |
| 2 | 1.02 (0.97, 1.07) | 0.90 (0.84, 0.97) |
| 3 | 0.90 (0.84, 0.96) | 0.78 (0.71, 0.84) |
| 4 | 0.86 (0.79, 0.94) | 0.68 (0.61, 0.75) |
| 5 | 0.77 (0.68, 0.87) | 0.57 (0.47, 0.70) |
| 6 or 7 | 0.80 (0.70, 0.92) | 0.47 (0.36, 0.62) |
| 8+ | 0.59 (0.47, 0.75) | 0.48 (0.31, 0.74) |
| AUD Codes | ||
| 0 | 1.00 | 1.00 |
| 1 | 0.86 (0.83, 0.90) | 0.68 (0.62, 0.74) |
aOR=adjusted odds ratio; 95% CI=95% Wald confidence interval; AUDIT-C=Alcohol Use Disorders Identification Test-Consumption; AUD=alcohol use disorder.
Reference category: 0 or 1 minor alleles and 0 minor alleles
All models adjusted for top 10 principal components, age, and sex.
Figure 3a.
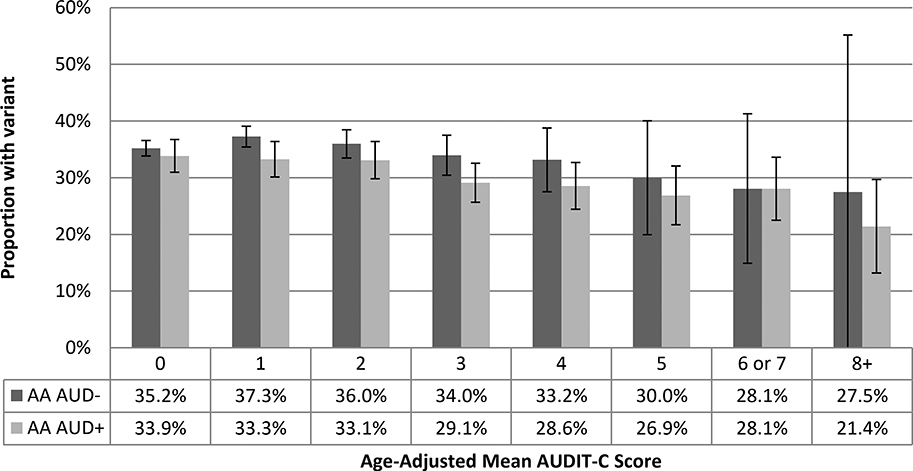
Proportion of African-American Sample with Minor Allele at rs2066702 by Age-Adjusted Mean AUDIT-C Score and AUD Codes (n=57,677)
These findings indicate that the combination of age-adjusted mean AUDIT-C score and AUD diagnostic codes yields more information among AAs. Comparing AAs with an age-adjusted mean AUDIT-C of 8 to those with a score of 0, the adjusted odds of having the ADH1B minor allele was 0.59 (95% CI 0.47, 0.75) (Table 2b). Having an AUD independently reduced the odds by 0.86 (95% CI 0.83, 0.90). Having both an AUDIT-C score of 8+ with an AUD diagnosis compared to an AUDIT-C of 0 and no AUD diagnosis would reduce the aOR of having the rs2066702 minor allele to 0.51 (data not otherwise shown).
Association of Candidate Phenotypes with rs1229984 in European Americans
In unadjusted analyses in EAs, all three candidate phenotypes were associated with rs1229984 (χ2 p-values all < 10−40) (Table 2a and b, Figure 2d-f ). In ordered logit models adjusted for the first 10 PCs, sex, and age, the association was again strongest for age-adjusted mean AUDIT-C score, treated as an ordinal variable (p=4.38 × 10−49). The addition of AUDIT-C trajectories to the age-adjusted mean AUDIT-C score model did not significantly improve model fit (both pseudo R2=0.056, LRT=6.08; df=3; p=0.11). The addition of AUD diagnostic codes, however, improved discrimination (pseudo R2=0.057, LRT=77.54, df=1, p=1.30 e-18), again showing that mean AUDIT-C score and AUD diagnostic codes can be combined to obtain more information about alcohol-related risk. Comparing EAs with an age-adjusted mean AUDIT-C of 8 to those with a score of 0, the adjusted odds of having the rs1229984 minor allele was 0.48 (95% CI 0.31, 0.74) (Table 2b). Having an AUD independently reduced the odds by 0.68 (95% CI 0.62, 0.74). Having both an AUDIT-C score of 8+ and an AUD diagnosis compared to an AUDIT-C of 0 and no AUD diagnosis would reduce the aOR of the rs1229984 minor allele to 0.33 (data not otherwise shown).
Discussion
In an analysis of longitudinal VA EHR data, we used the prevalence of the minor alleles of two functional SNPs in ADH1B—which have been shown in multiple studies to be associated with the risk of AUD ((3, 8, 18)—as a criterion to identify the best phenotype for harmful alcohol use. Because the VA EHR includes both longitudinal AUDIT-C scores and AUD codes, we compared age-adjusted mean AUDIT-C scores, AUDIT-C trajectories, and AUD codes. We found that the association with the ADH1B SNPs was strongest for age-adjusted mean AUDIT-C score in both AAs and EAs. After adjustment for the age-adjusted mean AUDIT-C score, AUD diagnostic codes demonstrated a modest, independent association in both populations.
These findings confirm our prior report of an association between longitudinal AUDIT-C scores recorded in the VA EHR and a protective ADH1B variant in AAs (rs2066702) (8), and we extended this work in several ways. First, we demonstrated that longitudinal AUDIT-C metrics are also associated with a protective ADH1B variant in EAs (rs1229984). Second, in both populations, we showed that a computationally simpler longitudinal metric—age-adjusted mean AUDIT-C score—is more strongly associated with the ADH1B variants than AUDIT-C trajectories. This finding is likely due to the fact that drinking patterns are well established by middle age (23, 24). Nonetheless, adjusting the mean AUDIT-C score for age is important, because drinking decreases after middle age (25). In contrast, trajectories would likely perform better than age-adjusted means for behaviors that change non-linearly over time. Finally, by design, AUDIT-C metrics and AUD diagnostic codes measure different aspects of harmful alcohol use. AUDIT-C is a screening tool that uses a past-year timeframe while AUD diagnostic codes reflect established lifetime diagnoses. Although longitudinal AUDIT-C scores showed more protective associations with ADH1B variants, AUD codes added independent information.
In both populations, the age-adjusted mean AUDIT-C scores (0 vs. 8+) were more strongly associated with the absence of the ADH1B minor allele than were AUD diagnostic codes: aOR = 0.59 vs. 0.86 in AAs and 0.48 vs. 0.68 in EAs. Because these effects were independent, combining an AUDIT-C score of 8+ with an AUD diagnosis compared to an AUDIT-C of 0 and no AUD yielded an aOR of 0.51 for AAs and 0.33 for EAs. When both AUDIT-C and AUD data are available, determining whether the added complexity of using both measures is justified will depend on the intended application.
Our work substantially extends the current literature on behavioral phenotypes extracted from EHR data for use in GWAS and other genetic studies. The most common approach to defining a harmful alcohol use phenotype has been the diagnosis of AUD based on the Diagnostic and Statistical Manual of Mental Disorders (DSM). Although DSM diagnoses of AUD have clinical utility, they are time consuming and costly to obtain, which limits their availability for large-scale genetic studies. The use of the ICD administrative codes as a proxy for clinical diagnosis is common for many medical conditions (12), but ICD codes are insensitive and non-specific measures of complex behaviors such as harmful alcohol use (4, 26). Importantly, our results suggest that these codes are applied differentially by race, with EAs being substantially less likely to have the code applied than AAs [despite AAs, until recently, having a lower frequency of AUD in the general population (25)]. These limitations may explain why longitudinal AUDIT-C metrics were more strongly associated with the ADH1B variants than were diagnostic codes and the inclusion of the latter only modestly improved the association. The large size of our sample made it possible to demonstrate these effects in both populations.
The utility of longitudinal, repeated AUDIT-C scores as a phenotype for harmful alcohol use may depend on the type of EHR data employed, the length of the observation period, and the statistical approach. In a prior publication, we demonstrated that a single AUDIT-C value did not have as strong a protective association with the ADH1B variants as did repeated longitudinal AUDIT-C measures incorporated into trajectories (8). This was true even when the highest AUDIT-C value was selected. In this study, our sample had an average of 7 annual AUDIT-C measurements. It provided evidence that a highly generalizable phenotype based on quantity-frequency data is valid. Because we age adjust the mean AUDIT-C, our finding could be directly reproduced in other samples with differing age distributions. Because the replication of genetic associations using independent data is an essential component of genetic discovery, having a simple, highly discriminating and generalizable phenotype is a major advance for such studies.
Our study has some limitations. First, harmful drinking is a complex trait that is likely influenced by multiple genetic variants reflecting multiple mechanisms. We have shown that the age-adjusted mean AUDIT-C score is more strongly associated with variation in ADH1B than the other phenotypes evaluated. However, it is an empirical question as to whether this phenotype would yield an advantage in detecting other genetic variants that could contribute to risk of harmful drinking via other (e.g., pharmacodynamic) mechanisms. We are in the process of examining the utility of the age-adjusted mean AUDIT-C score as an ordinal trait in a GWAS using the MVP sample. Second, because the VA EHR only includes the first three items of the AUDIT (AUDIT-C), we were not able to compare it against the full AUDIT. Third, the MVP sample is predominantly male, over 50 years of age, and in health care. Our study may not generalize well to women or younger adults. Of note, older male veterans do not drink more than age- and sex-matched comparators. According to the National Surveys on Drug Use and Health 2002–2012, self-reported heavy episodic drinking and AUD did not differ by veteran vs. civilian status among men age 50–64 years and 65 and over in the community (27). Both veterans and civilians receiving health care are more likely to endorse harmful drinking (28). Our EHR AUDIT-C data on middle-aged and older men are likely to be similar to other EHR data on this population. Thus, our findings likely do generalize to middle-aged and older men in health care.
Summary and Conclusions
The age-adjusted mean AUDIT-C score was more strongly associated with genetic polymorphisms of known risk for AUD than were trajectories of AUDIT-C or AUD diagnostic codes. Both our recent publication (8) from the Veterans Aging Cohort Study sample and the findings from the current study in the MVP sample show that it is feasible to use EHR data to derive a phenotype for harmful alcohol use that has considerable potential utility for gene discovery.
Supplementary Material
Figure 3b.
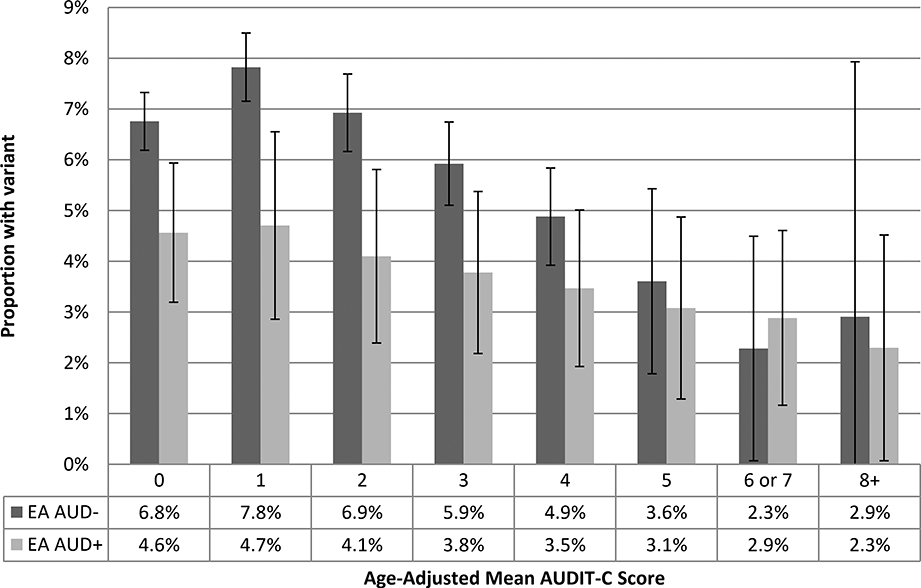
Figure 3b. Proportion of European Americans with Minor Allele at rs1229984 by Age-Adjusted Mean AUDIT-C Score and AUD Codes (n=110,044)
Acknowledgments
Financial support: This work was supported by NIAAA U24 AA020794, U01 AA020790, U10 AA013566 (completed), and VHA i01 BX003341.
Footnotes
Disclosure: Dr. Kranzler has been an advisory board member, consultant, or CME speaker for Alkermes, Indivior and Lundbeck. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which was supported in the last three years by AbbVie, Alkermes, Ethypharm, Indivior, Lilly, Lundbeck, Otsuka, Pfizer, Arbor, and Amygdala Neurosciences.
References
- 1.Verhulst B, Neale MC, Kendler KS. The heritability of alcohol use disorders: a meta-analysis of twin and adoption studies. Psychol Med. 2015;45(5):1061–72. Epub 2014/08/30. doi: 10.1017/s0033291714002165. PubMed PMID: ; PMCID: PMC4345133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Foroud T, Edenberg HJ, Crabbe JC. Genetic research: who is at risk for alcoholism. Alcohol Res Health. 2010;33(1–2):64–75. Epub 2010/01/01. PubMed PMID: ; PMCID: 3887503. [PMC free article] [PubMed] [Google Scholar]
- 3.Gelernter J, Kranzler HR, Sherva R, Almasy L, Koesterer R, Smith AH, Anton R, Preuss UW, Ridinger M, Rujescu D, Wodarz N, Zill P, Zhao H, Farrer LA. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Molecular psychiatry. 2014;19(1):41–9. Epub 2013/10/30. doi: 10.1038/mp.2013.145. PubMed PMID: ; PMCID: 4165335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGinnis K, Justice AC, Saitz R, Kraemer K, Bryant K, Fiellin D. Measures of unhealthy alcohol use among HIV infected and uninfected patients in the Veterans Aging Cohort Study. Satellite at the 33rd Annual Research Society on Alcoholism Scientific Meeting 2010. [Google Scholar]
- 5.McGinnis KA, Justice AC, Kraemer KL, Saitz R, Bryant KJ, Fiellin DA. Comparing alcohol screening measures among HIV-infected and -uninfected men. Alcohol Clin Exp Res. 2013;37(3):435–42. Epub 2012/10/10. doi: 10.1111/j.1530-0277.2012.01937.x [doi]. PubMed PMID: ; PMCID: 4492202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bush K, Kivlahan DR, McDonell MB, Fihn SD, Bradley KA. The AUDIT alcohol consumption questions (AUDIT-C): an effective brief screening test for problem drinking. Ambulatory Care Quality Improvement Project (ACQUIP). Alcohol Use Disorders Identification Test. Arch Intern Med. 1998;158(16). doi: http://www.ncbi.nlm.nih.gov/pubmed/9738608. [DOI] [PubMed] [Google Scholar]
- 7.Sanchez-Roige S, Fontanillas P, Elson SL, Gray JC, de Wit H, Davis LK, MacKillop J, Palmer AA. Genome-wide association study of alcohol use disorder identification test (AUDIT) scores in 20 328 research participants of European ancestry. Addict Biol. 2017. Epub 2017/10/24. doi: 10.1111/adb.12574. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Justice AC, McGinnis KA, Tate JP, Xu K, Becker WC, Zhao H, Gelernter J, Kranzler HR. Validating Harmful Alcohol Use as a Phenotype for Genetic Discovery Using Phosphatidylethanol and a Polymorphism in ADH1B. Alcohol Clin Exp Res. 2017;41(5):998–1003. Epub 2017/03/16. doi: 10.1111/acer.13373. PubMed PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaziano JM, Concato J, Brophy M, Fiore L, Pyarajan S, Breeling J, Whitbourne S, Deen J, Shannon C, Humphries D, Guarino P, Aslan M, Anderson D, LaFleur R, Hammond T, Schaa K, Moser J, Huang G, Muralidhar S, Przygodzki R, O’Leary TJ. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J Clin Epidemiol. 2016;70:214–23. Epub 2015/10/07. doi: 10.1016/j.jclinepi.2015.09.016. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 10.Denny JC, Bastarache L, Roden DM. Phenome-Wide Association Studies as a Tool to Advance Precision Medicine. Annu Rev Genomics Hum Genet. 2016;17:353–73. Epub 2016/05/06. doi: 10.1146/annurev-genom-090314-024956. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Ghosh J, Bejan CA, Gunter CA, Gupta S, Kho A, Liebovitz D, Sun J, Denny J, Malin B. Building bridges across electronic health record systems through inferred phenotypic topics. J Biomed Inform. 2015;55:82–93. Epub 2015/04/07. doi: 10.1016/j.jbi.2015.03.011. PubMed PMID: ; PMCID: Pmc4464930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics (Oxford, England). 2010;26(9):1205–10. Epub 2010/03/26. doi: 10.1093/bioinformatics/btq126. PubMed PMID: ; PMCID: 2859132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leader JB, Pendergrass SA, Verma A, Carey DJ, Hartzel DN, Ritchie MD, Kirchner HL. Contrasting Association Results between Existing PheWAS Phenotype Definition Methods and Five Validated Electronic Phenotypes. AMIA Annu Symp Proc. 2015;2015:824–32. Epub 2015/01/01. PubMed PMID: ; PMCID: Pmc4765620. [PMC free article] [PubMed] [Google Scholar]
- 14.Bradley KA, DeBenedetti AF, Volk RJ, Williams EC, Frank D, Kivlahan DR. AUDIT-C as a brief screen for alcohol misuse in primary care. Alcohol Clin Exp Res. 2007;31(7):1208–17. Epub 2007/04/25. doi: 10.1111/j.1530-0277.2007.00403.x. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 15.Bush K, Kivlahan DR, McDonell MB, Fihn SD, Bradley KA. The AUDIT alcohol consumption questions (AUDIT-C): an effective brief screening test for problem drinking. Ambulatory Care Quality Improvement Project (ACQUIP). Alcohol Use Disorders Identification Test. Arch Intern Med. 1998;158(16):1789–95. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 16.Piette J, B PG, Moos R First-time admissions with alcohol-related medical problems: A 10-year follow-up of a national sample of alcoholic patients. Journal of studies on alcohol. 1998;59:89–96. [DOI] [PubMed] [Google Scholar]
- 17.Justice AC, Lasky E, McGinnis KA, Skanderson M, Conigliaro J, Fultz SL, Crothers K, Rabeneck L, Rodriguez-Barradas M, Weissman SB, Bryant K. Medical disease and alcohol use among veterans with human immunodeficiency infection: A comparison of disease measurement strategies. Med Care. 2006;44(8 Suppl 2):S52–S60. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 18.Hart AB, Kranzler HR. Alcohol Dependence Genetics: Lessons Learned From Genome-Wide Association Studies (GWAS) and Post-GWAS Analyses. Alcohol Clin Exp Res. 2015;39(8):1312–27. Epub 2015/06/26. doi: 10.1111/acer.12792. PubMed PMID: ; PMCID: 4515198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Affymetrix. Axiom genotyping solution data analysis guide In: Affymetrix, editor. Rev 4 ed2016. [Google Scholar]
- 20.Jones BL, Nagin DS, Roeder K. A SAS Procedure Based on Mixture Models for Estimating Developmental Trajectories. Sociol Methods Res 2001;29(3):374–93. [Google Scholar]
- 21.Abraham G, Qiu Y, Inouye M. FlashPCA2: principal component analysis of Biobank-scale genotype datasets. Bioinformatics (Oxford, England). 2017;33(17):2776–8. Epub 2017/05/06. doi: 10.1093/bioinformatics/btx299. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 22.Xu K, Kranzler HR, Sherva R, Sartor CE, Almasy L, Koesterer R, Zhao H, Farrer LA, Gelernter J. Genomewide Association Study for Maximum Number of Alcoholic Drinks in European Americans and African Americans. Alcohol Clin Exp Res. 2015;39(7):1137–47. Epub 2015/06/04. doi: 10.1111/acer.12751. PubMed PMID: ; PMCID: PMC4706077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Platt A, Sloan FA, Costanzo P. Alcohol-consumption trajectories and associated characteristics among adults older than age 50. JStudAlcohol Drugs. 2010;71(2):169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob T, Blonigen DM, Upah R, Justice A. Lifetime drinking trajectories among veterans in treatment for HIV. Alcohol ClinExpRes. 2013;37(7):1179–87. doi: 10.1111/acer.12071 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grant BF, Chou SP, Saha TD, Pickering RP, Kerridge BT, Ruan WJ, Huang B, Jung J, Zhang H, Fan A, Hasin DS. Prevalence of 12-Month Alcohol Use, High-Risk Drinking, and DSM-IV Alcohol Use Disorder in the United States, 2001–2002 to 2012–2013: Results From the National Epidemiologic Survey on Alcohol and Related Conditions. JAMA Psychiatry. 2017;74(9):911–23. Epub 2017/08/10. doi: 10.1001/jamapsychiatry.2017.2161. PubMed PMID: ; PMCID: PMC5710229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGinnis KA, Justice AC, Kraemer KL, Saitz R, Bryant KJ, Fiellin DA. Comparing alcohol screening measures among HIV-infected and -uninfected men. Alcohol ClinExpRes. 2013;37(3):435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoggatt KJ, Lehavot K, Krenek M, Schweizer CA, Simpson T. Prevalence of substance misuse among US veterans in the general population. The American journal on addictions. 2017;26(4):357–65. Epub 2017/04/04. doi: 10.1111/ajad.12534. [DOI] [PubMed] [Google Scholar]
- 28.Roson B, Monte R, Gamallo R, Puerta R, Zapatero A, Fernandez-Sola J, Pastor I, Giron JA, Laso J. Prevalence and routine assessment of unhealthy alcohol use in hospitalized patients. European journal of internal medicine. 2010;21(5):458–64. Epub 2010/09/08. doi: 10.1016/j.ejim.2010.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


