Abstract
Background & Aims
Improved methods are needed to risk stratify and predict outcomes in patients with primary sclerosing cholangitis (PSC). Therefore, we sought to derive and validate a new prediction model and compare its performance to existing surrogate markers.
Methods
The model was derived using 509 subjects from a multicenter North American cohort and validated in an international multicenter cohort (n=278). Gradient boosting, a machine based learning technique, was used to create the model. The endpoint was hepatic decompensation (ascites, variceal hemorrhage or encephalopathy). Subjects with advanced PSC or cholangiocarcinoma at baseline were excluded.
Results
The PSC risk estimate tool (PREsTo) consists of 9 variables: bilirubin, albumin, serum alkaline phosphatase (SAP) times the upper limit of normal (ULN), platelets, AST, hemoglobin, sodium, patient age and the number of years since PSC was diagnosed. Validation in an independent cohort confirms PREsTo accurately predicts decompensation (C statistic 0.90, 95% confidence interval (CI) 0.84-0.95) and performed well compared to MELD score (C statistic 0.72, 95% CI 0.57-0.84), Mayo PSC risk score (C statistic 0.85, 95% CI 0.77-0.92) and SAP < 1.5x ULN (C statistic 0.65, 95% CI 0.55-0.73). PREsTo continued to be accurate among individuals with a bilirubin < 2.0 mg/dL (C statistic 0.90, 95% CI 0.82-0.96) and when the score was re-applied at a later course in the disease (C statistic 0.82, 95% CI 0.64-0.95).
Conclusions
PREsTo accurately predicts hepatic decompensation in PSC and exceeds the performance among other widely available, noninvasive prognostic scoring systems.
Keywords: Primary sclerosing cholangitis, prognosis, machine learning
Introduction
Primary sclerosing cholangitis (PSC) is a chronic cholestatic liver disorder characterized by inflammation and fibrosis in the intra and or extrahepatic ducts that is commonly associated with inflammatory bowel disease (IBD), particularly ulcerative colitis (UC).1 Advanced fibrosis and manifestations of portal hypertension can develop and are an important source of morbidity and mortality.2 Moreover, PSC is a premalignant condition and is associated with a higher risk of colorectal and hepatobiliary malignancies, which can arise regardless of the presence of advanced fibrosis.2, 3 To date, effective medical therapy for PSC is lacking. The reasons for this are multifactorial. This is partly due to the rarity of the disease, which is heterogeneous and has a low event rate that is difficult to predict. In turn, this makes clinical trials challenging. To address this, improved biomarkers are necessary to risk stratify patients in clinical trials and serve as surrogate endpoints.
A number of biomarkers have been examined to predict outcomes in PSC. Serum alkaline phosphatase (SAP) has been one of the most widely examined.4-9 A cutoff at or near 1.5 × the upper limit of normal (ULN) has been reported in multiple studies as having prognostic relevance.5, 6, 8 However, many patients with a SAP >1.5 × ULN, 62% in one study with 9 year follow up, do not experience liver related complications and individuals can have advanced liver disease with a normal SAP.8,10 The Mayo PSC risk score utilizes the patients age, bilirubin, albumin, AST and a prior history of variceal bleeding, was developed to predict short term mortality. However it has not been validated to predict long term outcomes or other clinically relevant events. Moreover, improvements in the Mayo PSC risk score and SAP failed to correlate with outcomes in a randomized controlled trial that examined high dose ursodeoxycholic acid.11, 12 Liver histology is less likely to fluctuate when compared to any single laboratory test and has been shown to predict survival.13, 14 However, it is an invasive test that is prone to sampling error and histologic changes, particularly fibrosis, may be slow to change.15 To mitigate this, the enhanced liver fibrosis (ELF) panel was developed and has been shown to predict transplant free survival.16 Elastography techniques appear to hold promise to predict clinically relevant events and prospective validation studies are ongoing.3, 17, 18 All of these prognostic variables have their own limitations and were examined using traditional statistical methods. Machine learning (ML) techniques such as gradient boosting machines (GBM) have several advantages over traditional modeling and are increasingly being examined for medical applications.19-21 ML is an application of artificial intelligence and is a method that can examine large complex data sets to generate predictive models. In addition, it is an alternative approach to conventional methods such as logistic or cox proportional hazard regression. ML has demonstrated an improved performance compared to logistic regression in a variety of clinical scenarios.20-23 Examples of ML include neural networks, support vector machines, random forest and GBM.20, 24, 25 To date, ML has not been applied in the study of cholestatic liver disease outcomes.
There has been a growing international clarion call among patients, experts in the field, industry and regulatory agencies to discover and validate better methods of predicting PSC related complications and apply them in prospective clinical trials.26 Consequently, we assembled a large, international cohort of PSC patients to create and validate a novel PSC risk score and compare its performance to existing prognostic markers using a novel approach- GBM, a ML technique.
Methods
Patient Population
Patients with cholangiographic evidence of PSC were included in the study. Exclusion criteria included: i) small duct PSC; ii) another concurrent liver disease including overlap with autoimmune hepatitis; iii) laboratory tests were unavailable; iv) cholangiocarcinoma (CCA) at baseline; v) model for end stage liver disease (MELD) score greater than 14 at baseline; vi) prior liver transplantation; or vii) presence of portal hypertension (varices, ascites, encephalopathy, splenomegaly or platelets less than 150 × 109/L) at baseline. Hence subjects with advanced PSC (i.e., those with manifest signs of portal hypertension or MELD score greater than 14, which has been the historic cutoff where individuals would begin to receive a survival benefit from transplant) were excluded.27 The rationale for this approach was to develop a tool that could be applied in the context of clinical trials. Individuals with advanced disease are less likely to be eligible for clinical trials and are less likely to benefit from pharmacotherapy in the current era.
The derivation and validation cohorts were assembled from a variety of sources (Figure 1). Patients with PSC seen at Mayo Clinic Rochester, Florida or Arizona who were enrolled in the study group entitled, PSC Resource of Genetic Risk, Environment and Synergy Studies (PROGRESS) were utilized in the derivation cohort. The international validation cohort consisted of patients obtained from 3 sources: i) North American patients with PSC who contacted (KNL) to participate in the PROGRESS registry who were not seen at one of the participating PROGRESS sites; ii) a randomly selected group of patients diagnosed with PSC and seen at Mayo Rochester who were not enrolled in PROGRESS; and iii) patients with PSC seen at medical centers in Norway. The Norwegian PSC patients were identified through a retrospective data set among PSC patients seen at the Oslo University Hospital Rikshospitalet. This data set was supplemented with laboratory and clinical records retrieved from 31 local hospitals prior to and after patients were referred to the Oslo University Hospital. Norwegian PSC patients were also identified through a prospective cohort assembled at Haukeland University Hospital in Bergen, Norway. Lastly, a cohort of 116 PSC patients who were not included in the derivation or validation cohorts who underwent a magnetic resonance elastography (MRE) at Mayo Clinic Rochester were included in a supplementary analysis.
Figure 1.
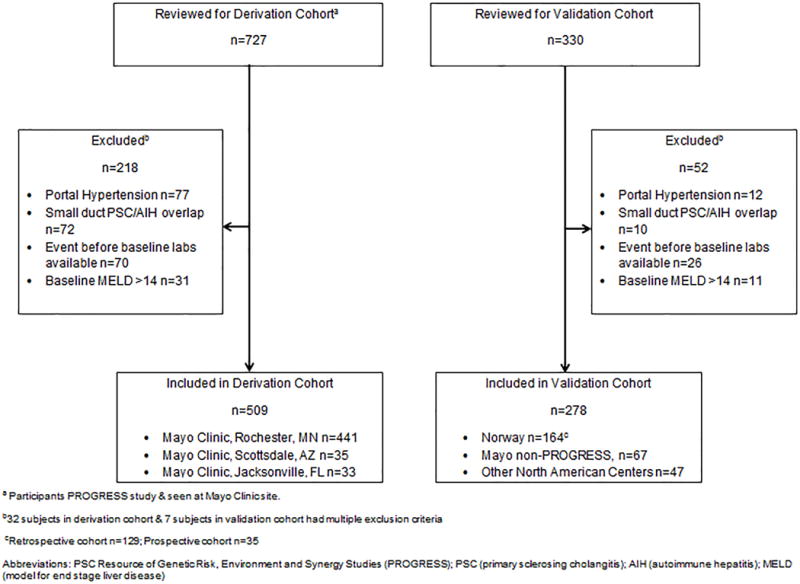
Patients Included
Data Collection & Key Definitions
The electronic and paper medical records were reviewed in detail and all pertinent clinical and laboratory data was abstracted on a standardized template. Recognizing that individuals with PSC may receive their care at multiple institutions, outside medical records (including laboratory tests) were requested for all patients and relevant data was abstracted to maximize the phenotypic depth of the cohort.
Large duct PSC was diagnosed using standard criteria.28 A biopsy was required to establish a diagnosis of small duct PSC or concomitant autoimmune hepatitis.28, 29 When biopsies were available, the fibrosis stage was graded based on the Batts-Ludwig criteria.30 The presence of a dominant stricture was determined by endoscopist or JEE using standard criteria when possible.28 A diagnosis of CCA was established if there was a mass with typical radiographic features of biliary cancer on cross-sectional imaging or a positive cytology or biopsy.31 The MELD score, Mayo PSC risk score and SAP × ULN times at baseline were abstracted along with values at later time points when possible.12 A SAP < 1.5 × ULN following a diagnosis of PSC has been associated with an improved prognosis in PSC regardless of the presence of a dominant stricture.5, 6, 8 Individuals with at least 3 SAP measurements that were < 1.5 × ULN over the course of at least one year were abstracted as having a SAP <1.5 × ULN.
The baseline was defined as the time where laboratory tests were first available following a diagnosis of PSC. The model was derived to predict a composite outcome of hepatic decompensation (i.e., variceal hemorrhage, ascites, hepatic encephalopathy, whichever occurred earliest). Individuals were censored at the time of liver transplant, when a diagnosis of CCA was established or the time of their last clinical encounter (whichever was earlier). Decompensation was selected as the primary outcome for the following reasons: i) it is an objective marker of disease severity; ii) while transplant free survival is an important endpoint, indications for transplant, severity of disease at the time of transplant and timing of transplantation can vary substantially; iii) it is plausible that biomarker(s) predictive of complications stemming from portal hypertension would not be as accurate in the prediction of liver transplantation for pruritus, recurrent cholangitis or CCA due to differences in the etiopathogenesis and the timing of transplant for these indications across multiple centers. However, liver transplant for non-malignant PSC complications or PSC related death not associated with malignancy was treated as a secondary endpoint. In this analysis, subjects were censored at the time of CCA diagnosis or their last clinical encounter (whichever was earlier).
Statistical Analyses
Continuous variables were expressed as median, interquartile range (IQR) unless otherwise specified and compared using the Kruskal-Wallis rank sum test while categorical variables were compared via the Pearson’s Chi-squared test. The model, PSC risk estimate tool (PREsTo), was created to predict endpoints within a 5 year time window. Hence if an endpoint occurred beyond that time, it was not counted as an event in the primary analysis (rather the individual would have been censored at their last follow up within the 5 year window). We applied the model at baseline and 2 years post baseline using the derivation and validation cohorts to evaluate its performance.
GBM, a ML technique, was used to create the model with the generalized boosted model package (gbm) available in the R software environment.32 GBM is an established technique for addressing regression and classification problems by producing a prediction model in the form of an ensemble of weak prediction models, typically decision trees. GBM builds the model in a step-wise fashion combining information from multiple decision trees (Supplementary Figure 1) that are iteratively built in such a way that each iteration focuses increasingly on the portions of the data that are most ill-fitting. In other words, GBM uses a series of small decision tree, each containing several of the variables from the total variable pool of a study. Unlike linear regression, GBM models the data using recursive partitioning whereby the decision tree splits the data into smaller groups (partitions) using a cutpoint (example, age > 50 years or ≤ 50 years). The resulting groups are then split again using another decision or cutpoint. After the initial decision tree is created the model has residuals (or what is unexplained by the first tree alone). GBM fits the subsequent decision tree(s) based on those residuals to improve the models predictive performance (i.e., the model learns from the earlier decision trees). This process is repeated hundreds or thousands of times (for example, in developing PREsTo, this procedure was repeated 2,500 times). Each decision tree may have different variables. Variables that have the strongest predictive power are used in more decision trees and earlier in the model building process.
Individually, each decision tree has a relatively weak predictive performance. However, when all the decision trees are combined in the final model the predictive performance is greatly enhanced. See Supplementary Figure 1 for an illustrative example.
The chief advantage of this method is that it naturally incorporates interactions between variables, is not susceptible to extreme values and handles missing values without the need to impute data. The number of decision trees included in the model (number of iterations), the depth of the decision trees and the size of the shrinkage parameter were determined by 5-fold cross-validation minimizing the Cox partial deviance. This is the recommended approach to prevent overfitting of the model. In order to assess all of the risk scores using the same subject, multiple imputation was run using the default settings of the R package “mice” using all the covariates in the gbm model.33 For numeric variables, the default imputation approach is predictive mean matching and for unordered categorical variables the default is polytomous regression. The average predictions over five instances of the imputed data were then reported. Discrimination, the ability of a risk score to accurately rank individuals from low to high risk, was assessed by calculating Harrell’s C-statistic and 95% confidence intervals were created using bootstrapping.34 Calibration, the ability accurately predict the absolute risk level, was assessed comparing observed and expected values in subjects with low, medium, and high predicted risk. The 3 risk groups were determined by the tertiles of the model’s risk score distribution. Cox models were used to examine the impact of each of the risk factors univariately.
The general formula for obtaining a risk estimate from a Cox model is: score=1−S0(t)^exp(ΣβiXi) where S0(t) is the baseline event-free rate at follow-up time t (e.g., 5 years), βi is the estimated regression coefficient and Xi is the value of the ith risk factor. With a GBM model there are no regression coefficients, however model predictions can be obtained using a series of rules (Supplementary Table 1). In this table, each row represents a set of rules. Model predictions are obtained by adding up column “y” over all the rows where a subject’s values meet the specified criteria (e.g. total bilirubin < 2.65 and sodium < 136.5). The 5 year baseline event-free rate for this model is 0.3836.
Results
Clinical Characteristics of Derivation and Validation Cohorts
One-thousand and fifty-seven subjects were reviewed for this study. Ultimately, 787 individuals were included (derivation n=509; validation n=278) (Figure 1). The median (IQR) of follow up for the derivation and validation cohort was 6.09 (2.82-13.10) and 4.21 (2.31-8.35) years. Markers of PSC disease severity (platelets, bilirubin, MELD, Mayo PSC risk score and SAP) were similar between the cohorts Table 1.
Table 1.
Baseline Features
| Derivation | Validation | ||
|---|---|---|---|
| Variable | (n=509) | (n=278) | P-value |
| Age (years) | 43.10 (30.10-54.30) | 41.70 (30.60-54.20) | 0.43 |
| Male | 61.70% (314/509) | 69.10% (192/278) | 0.05 |
| PSC Age Diagnosis (years) | 40.00 (27.60-51.40) | 35.30 (26.50-47.20) | 0.02 |
| PSC Duration (years) | 0.49 (0.12-3.06) | 1.44 (0.12-6.18) | <0.01 |
| IBD Present | 80.13% (363/453) | 83.09% (231/278) | 0.10 |
| •Ulcerative colitis | 66.89% (303/453) | 67.60% (188/278) | |
| •Crohn’s disease | 5.74% (26/453) | 10.10% (28/278) | |
| • Indeterminate colitis | 7.51% (34/453) | 5.40% (15/278) | |
| IBD Age Diagnosis | 26.60 (18.30-43.20) | 26.9 0 (16.90-42.00) | 0.46 |
| IBD Duration (years) | 9.69 (2.93-20.80) | 8.44 (1.69-18.40) | 0.42 |
| Platelets × 109/L | 258(220-318) | 273 (215-358) | 0.17 |
| Sodium mmol/L | 140 (138-141) | 140 (139-142) | 0.02 |
| Creatinine mg/dL | 0.90 (0.80-1.10) | 0.8 1 (0.70-0.92) | <0.001 |
| Albumin g/dL | 4.10 (3.70-4.40) | 4. 20 (3.90-4.50) | <0.001 |
| SAP × ULN | 1.82 (1.03-3.41) | 2.05 (1.15-3.50) | 0.26 |
| SAP > 1.5x ULN | 56.20% (213/379) | 62. 27% (148/236) | 0.13 |
| AST U/L | 58 (33-92) | 57 (36- 100) | 0.52 |
| Total Bilirubin mg/dL | 0.80 (0.50-1.30) | 0.80 (0.53 -1.35) | 0.62 |
| MELDa | 7.34 (6.43-8.87) | 7.50 (6.43 -9.36) | 0.15 |
| Mayo PSC Risk Scoreb | −0.09 (−0.71-0.69) | − 0.11 (−0.71-0.59) | 0.39 |
Available at baseline (349/509) in derivation and (215/278) in validation cohorts
Available at baseline (465/509) in derivation and (246/278) in validation cohorts Continuous variables reported as median (interquartile range)
Abbreviations: PSC (primary sclerosing cholangitis); IBD (inflammatory bowel disease); SAP (serum alkaline phosphatase); ULN (upper limit of normal); AST (aspartate aminotransferase); MELD (model for end-stage liver disease).
Decompensation within 5 years of study entry occurred among 37 subjects in the derivation cohort and 21 individuals in the validation cohort. During this same time period, 30 individuals developed CCA (derivation n=23; validation n=7) and 51 underwent a transplant (derivation n=19; validation n=32) (Table 2). Among individuals without a prior history of decompensation, transplant or cholangiocarcinoma, 2 individuals died (both in derivation cohort and both from cholangitis).
Table 2.
Outcomes in Derivation and Validation Cohorts
| Variable | Derivation (n=509) |
Validation (n=278) |
|---|---|---|
| Decompensation (0-5 yrs) | 7.27% (37/509) | 7.55% (21/278) |
| • Ascites | 5.70% (29/509) | 6.47% (18/278) |
| • Variceal Hemorrhage | 1.38% (7/509) | 0.72% (2/278) |
| • Hepatic Encephalopathy | 0.20% (1/509) | 0.36% (1/278) |
| Censoring Events (0-5 yrs) | 92.73% (472/509) | 92.44% (257/278) |
| • Liver transplantationa | 3.73% (19/509) | 11.51% (32/278) |
| • Confirmed CCA | 4.51% (23/509) | 2.51% (7/278) |
| • Last Clinical Encounter | 84.50% (430/509) | 78.42% (218/278) |
| Decompensation (2-7 yrs)b | 7.93% (13/164) | 5.10% (5/98) |
| • Ascites | 4.27% (7/164) | 3.03% (3/99) |
| • Variceal Hemorrhage | 3.04% (5/164) | 1.01% (1/99) |
| • Hepatic Encephalopathy | 0.61% (1/164) | 1.01% (1/99) |
| Censoring Events (2-7 yrs) | 92.07% (151/164) | 94.90% (93/98) |
| • Liver transplantationc | 5.49% (9/164) | 8.16% (8/98) |
| • Confirmed CCA | 3.05% (5/164) | 1.02% (1/98) |
| • Last Clinical Encounter | 83.54% (137/164) | 85.71% (84/98) |
Abbreviations: PSC (primary sclerosing cholangitis); CCA (cholangiocarcinoma).
Events that developed first are shown.
Derivation transplant indications (PSC symptoms not associated with portal hypertension n=18; concern for biliary neoplasia n=1); Validation transplant indications (PSC symptoms not associated with portal hypertension n=29; concern for biliary neoplasia n=3).
After re-applying our inclusion/exclusion criteria at year 2, the derivation cohort included 164 subjects & validation cohort included 98 subjects.
Derivation transplant indications (PSC symptoms not associated with portal hypertension n=9); Validation transplant indications (PSC symptoms not associated with portal hypertension n=8).
Creation & Calibration of PREsTo Score
Using GBM we investigated 19 potential model covariates (Supplementary Table 2) and utilized 2,500 decision trees sequentially, with each successive decision tree fitting on the residuals left over from previous decision trees. The parameter decision tree depth was optimized at 3, corresponding to three-way interactions, and the shrinkage parameter was optimized at 0.001. Covariates with a relative influence greater than 4 were included in the final model created to predict the 5 year probability of decompensation (Figure 2, Supplementary Table 2). The median (IQR) PREsTo score among those in the derivation cohort was 4.45% (3.06%-9.68%). In the derivation cohort, PREsTo accurately predicts the 5 year probability of decompensation (C statistic 0.96, 95% CI 0.93-98) and was well-calibrated in its ability to predict the 5 year risk of decompensation among patients at a low-high risk for developing an endpoint (Table 3, Figure 3a).
Figure 2.
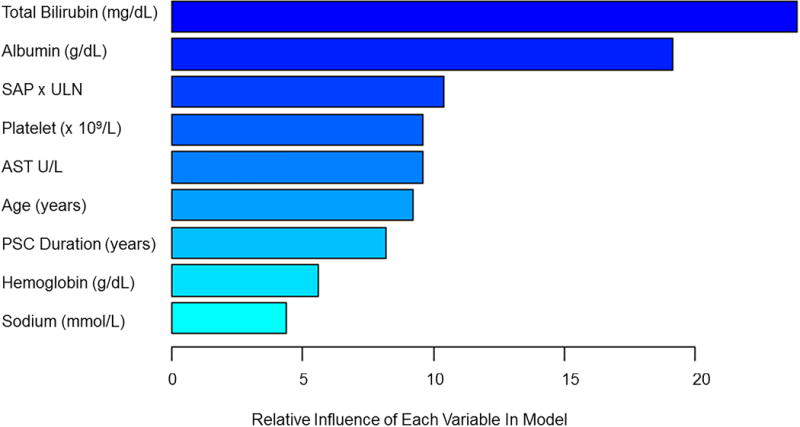
Variables Included in Model and Their Relative Importance
Table 3.
Performance of PREsTo & Other Prognostic Markers in Derivation & Validation Cohorts
| Prognostic Marker | C-statistic (95% CI) |
|---|---|
| Derivation Cohort-Secondary Endpoint | |
| PREsTo | 0.96 (0.93-0.98) |
| MELD Score | 0.73 (0.63-0.82) |
| Mayo PSC Risk | 0.84 (0.76-0.90) |
| SAP <1.5 × ULN | 0.64 (0.56-0.70) |
| Validation Cohort-Secondary Endpoint | |
| PREsTo | 0.90 (0.85-0.95) |
| MELD Score | 0.72 (0.57-0.84) |
| Mayo PSC Risk | 0.85 (0.77-0.92) |
| SAP <1.5 × ULN | 0.65 (0.55-0.73) |
Abbreviations: PREsTo (PSC risk estimate tool); CI (confidence interval); SAP × ULN (serum alkaline phosphatase × upper limit of normal); PSC (primary sclerosing cholangitis); MELD (model end stage liver disease)
Figure 3.
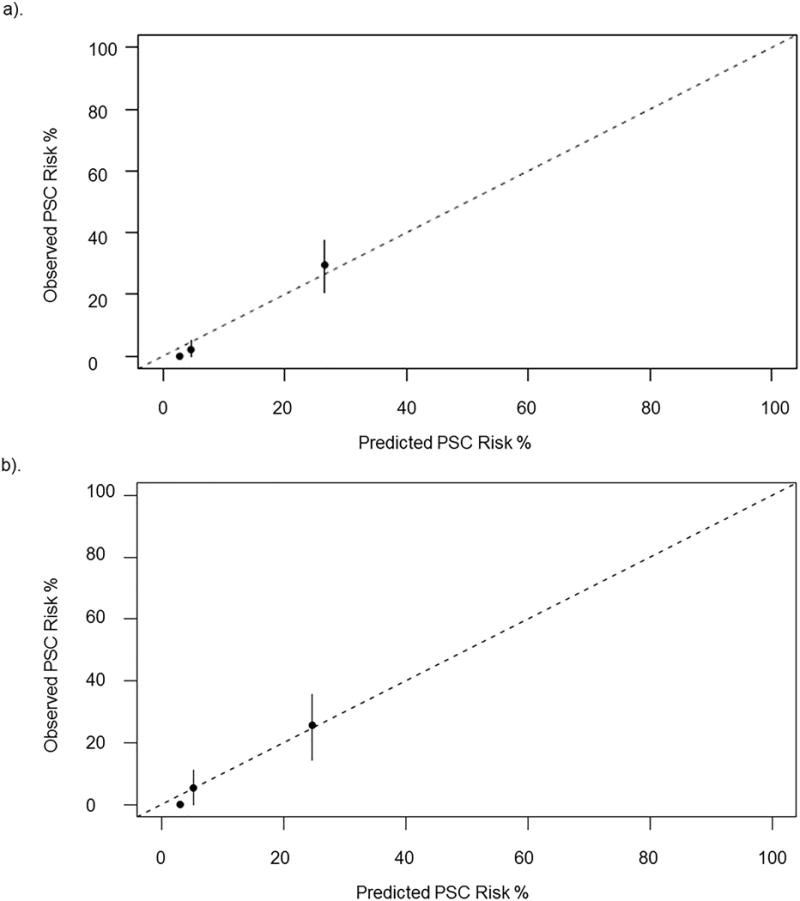
Model Calibration
a). Calibration in derivation cohort. b). Calibration in validation cohort
Utilizing an online calculator (www.web address will be inserted after acceptance of the present model), PREsTo would predict a 5 year probability of decompensation of 19% in a 41 year old patient with PSC diagnosed 2 years ago and the following labs: total bilirubin 1.0 mg/dL, SAP 150 U/L (ULN 115 U/L), albumin 3.0 mg/dL, AST 69 U/L, Platelets 204 × 109/L, Sodium 134 mmol/L and hemoglobin 14 g/dL.
Validation & Comparative Performance of PREsTo
The median (IQR) PREsTo score among those in the validation cohort was 5.1% (3.5%-9.8%). In the validation cohort, PREsTo was well-calibrated (Figure 3b) across individuals at low-high risk of decompensation and highly predictive of the 5 year risk of decompensation (C statistic 0.90, 95% CI 0.84-0.95), Table 3. Furthermore, it performed well when compared to the MELD score, Mayo PSC risk score and SAP<1.5× ULN (Table 3). Last, we also found PREsTo predictive of a secondary composite endpoint of decompensation, liver transplant for non-malignant indication or death from PSC related cause (excluding malignancy) in the derivation and validation cohorts (respectively): C statistic 0.89, 95% CI 0.85-0.92 and C statistic 0.76, 95% CI 0.69-0.83.
In several exploratory analyses, it did not appear that adding PREsTo to either liver stiffness as measured by MRE (n=213) or the fibrosis stage from liver biopsies (n=51) had a significant impact on the models performance to predict decompensation. Moreover, PREsTo alone performed similarly when compared to these other biomarkers (Supplementary Table 3). There was a strong correlation between PREsTo and liver stiffness as measured by MRE (r=0.68).
A subgroup of 114 individuals had ELF scores obtained within 3 months of their baseline PREsTo score. In this exploratory subgroup, the respective performances of PREsTo and ELF in the prediction of hepatic decompensation was C statistic 0.90, 95% CI 0.78-0.98 and C statistic 0.75, 95% CI 0.55-0.92, p=0.05, respectively. The correlation between PREsTo and ELF was moderate (r=0.43).
Performance of PREsTo at Different Time Points & Phenotypes
We examined the performance of PREsTo at different time points and disease phenotypes. We re-applied our inclusion/exclusion criteria 2 years post baseline and reassessed the models performance among 164 subjects in the derivation and 98 subjects in the validation cohorts (Table 2). This was done to ensure the model performed well at different times in the disease course and act as another mechanism to replicate the validity of our findings. Indeed, the model continued to have an excellent performance when it was applied at a later time for the prediction of decompensation (derivation cohort: C statistic 0.82, 95% CI 0.64-0.95; validation cohort: C statistic 0.89, 95% CI 0.77-1.00). PREsTo also has the ability to predict long term outcomes beyond 5 years. For example, the model’s ability to predict the primary endpoint at 10 years remained excellent: C statistic 0.86, 95% CI 0.78-0.93).
We performed several sensitivity analyses to investigate the impact of disease specific factors on the performance of the model. Indeed, the performance was maintained among those who were recently diagnosed with PSC and among those with longstanding PSC (Supplementary Table 4). The presence of an elevated bilirubin is generally regarded as a marker of disease severity but can be transiently elevated due to a biliary obstruction.26 Furthermore, the presence of jaundice often excludes individuals from participation in clinical trials. Indeed, PREsTo continued to perform well among those with a total bilirubin 2 mg/dL or less (C statistic 0.94, 95% CI 0.90-0.96) and those with a total bilirubin of 2 mg/dL or greater (C statistic 0.90, 95% CI 0.83-0.95), Supplementary Table 4. Similarly, the presence or absence of a dominant stricture, sex, presence or absence of IBD and intrahepatic disease distribution versus intra and extrahepatic disease distribution, presence of symptoms at PSC diagnosis or having normal SAP did not appear to influence the models performance (Supplementary Table 4).
Discussion
Using state of the art ML techniques, we examined a large international cohort to create and independently validate a novel model (PREsTo) that predicts the 5 year risk of hepatic decompensation among those with PSC. This model utilizes readily available clinical data, is noninvasive, inexpensive and has an excellent performance when compared to existing prognostic markers. PREsTo continued to be highly predictive when it was applied at a later point in the disease course and among various PSC subgroups that are commonly encountered in clinical practice. Moreover, the model accurately predicted 10 year outcomes.
Making direct comparisons between PREsTo and other prognostic markers published in other studies is challenging due to variations in the patients examined, endpoints and statistical methodology. However, it is notable that PREsTo was able to correctly predict the development of hepatic decompensation in 9 out of 10 patients and was well calibrated to predict the endpoint among low- high risk patients among an independently validated cohort. While the Mayo PSC risk score also performed well in this study (C statistic 0.84-0.85) it has not always correlated with outcomes in clinical trials.11 SAP <1.5 × ULN performed relatively poor in this study (C statistic 0.64-0.65). In another study, subjects with a SAP 1.3x ULN at baseline and 1 year later had a C statistic that ranged from 0.50-0.70 for transplant free survival.5 The incorporation of multiple laboratory and clinical variables in PREsTo may be one reason why it has a superior performance when compared to any single laboratory test. The ELF panel was able to discriminate between those with and without an endpoint (death or transplant) with a C statistic of 0.79 in a European validation cohort.16 Similarly, a liver stiffness of 9.5kPa, as measured by transient elastography, had a C statistic of 0.77 and spleen length had a C statistic of 0.80 for the prediction of liver-related death, hepatic decompensation or liver transplant.35 In the present study, we conducted several exploratory subgroup analyses which illustrated that the stage of liver fibrosis (C statistic 0.88) and liver stiffness measured by MRE (C statistic 0.93) were comparable to PREsTo.
PSC is a heterogeneous disorder in both its presentation and outcomes.36, 37 The presence of IBD, extrahepatic and intrahepatic disease (compared to intrahepatic alone), presence of a dominant stricture, symptoms at the time of diagnosis, jaundice or an elevated SAP have been associated with a worse prognosis.13, 37-39 Given the prognostic implications, we examined the performance of PREsTo in these key subgroups. Our findings illustrate that PREsTo continues to perform well (C statistic 0.90 or greater) in these subgroups (Supplementary Table 4). The duration of time since a diagnosis of PSC was established was an important variable in the model. However, PSC has a subclinical disease course and may be diagnosed at variable times during the natural history of the disease (i.e., development of symptoms or elevated liver tests with or without symptoms). Consequently, we performed subgroup analyses to determine the performance of PREsTo among those based on the presence of symptoms or elevations in SAP at the time of diagnosis. Indeed, PREsTo remained well-calibrated regardless of symptoms or elevations in SAP at the time of diagnosis (Supplementary Table 4).
ML has evolved within the past decade due to advances in computational power and cheaper data storage. Amazon, Google, NetFlix and others in a variety of interfaces have used ML with their customers and employees. Applying ML techniques to big data in digestive diseases is just beginning. To our knowledge, this is the first published effort to utilize GBM, a ML technique, to predict outcomes in liver disease. GBM utilizes ensemble learning where many simple learning algorithms or “decision trees” are used jointly to achieve a more optimized prediction than what is possible from any of the individual learning algorithms.40 In other words, the computer uses boosting and ensemble learning to learn from the errors in initial algorithms, modifies them to create a large pool of algorithms that together are more accurate.19 While logistic regression utilizes a single model comprised of independent variables with linear combinations, GBM uses many models and predictor variables that can have complex relationships with the outcome of interest (nonlinear, interacting variables, outliers or missing variables).6 GBM has several advantages when compared to its ML counterparts: it is less prone to overfitting; it can accommodate both continuous and categorical variables; it can estimate error rates; and it can rank the variables relative importance.21, 25 GBM has been employed in several clinical applications such as the prediction of outcomes following lower gastrointestinal bleeding, cardiovascular risk and fracture risk with good results.20, 21, 25 Hence, this technique holds promise for future studies beyond the realm of cholestatic liver disease.
Our study has several limitations. First, the participants in this study were largely seen at academic medical centers and may not be representative of the entire PSC population. Second, detailed imaging covariates, histology and measurements of liver stiffness were not available for all subjects. Consequently, we could not examine them for inclusion into the model. We tried to mitigate this by performing several exploratory analyses among subjects that had a liver biopsy or MRE. However, future studies will be needed. Indeed, it may be efficacious to have a combination of variables (biochemical, radiographic, genomic and other -omics) in a larger prognostic scoring system, which may mitigate the heterogeneous nature of PSC and individual fluctuations in laboratory tests overtime. It should also be noted that our model was created and validated among individuals who did not have markers of advanced PSC at baseline. Hence, it is uncertain how PREsTo would perform in clinical practice if applied to a different PSC population with more advanced disease. However, our aim was to create a model that could be directly applicable for the use in clinical trials where subjects with advanced PSC are excluded.
Improved methods to risk stratify PSC patients and predict outcomes are a significant unmet need in this patient population. Using ML, we have created and validated a novel prognostic score, PREsTo, that accurately predicts hepatic decompensation in comparison to other existing prognostic markers. These findings suggest PREsTo should be incorporated as an exploratory endpoint in future PSC clinical trials and be considered as a method of patient stratification. ML methods such as GBM are promising techniques for the study of liver disease in the 21st century.
Supplementary Material
Acknowledgments
Grant Support: Supported by NIH grant RO1 DK 084960 and the Chris M. Carlos and Catharine Nicole Jockisch Carlos Endowment in PSC.
Abbreviations
- PSC
primary sclerosing cholangitis
- IBD
inflammatory bowel disease
- UC
ulcerative colitis
- SAP
serum alkaline phosphatase
- ULN
upper limit of normal
- ELF
enhanced liver fibrosis
- ML
machine learning
- GBM
gradient boosting machines
- CCA
cholangiocarcinoma
- MELD
model for end stage liver disease
- PROGRESS
PSC resource of genetic risk, environment and synergy studies)
- PREsTo
PSC risk estimate tool
- MRE
magnetic resonance elastography
Footnotes
Disclosures: The authors have nothing to disclose
Transcript Profile: None.
Writing Assistance: Not utilized.
Author Contributions: All authors were involved in the drafting and critical revision of the manuscript.
References
- 1.Eaton JE, Talwalkar JA, Lazaridis KN, et al. Pathogenesis of primary sclerosing cholangitis and advances in diagnosis and management. Gastroenterology. 2013;145:521–36. doi: 10.1053/j.gastro.2013.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boonstra K, Weersma RK, van Erpecum KJ, et al. Population-based epidemiology, malignancy risk, and outcome of primary sclerosing cholangitis. Hepatology. 2013;58:2045–55. doi: 10.1002/hep.26565. [DOI] [PubMed] [Google Scholar]
- 3.Eaton JE, Dzyubak B, Venkatesh SK, et al. Performance of magnetic resonance elastography in primary sclerosing cholangitis. J Gastroenterol Hepatol. 2016;31:1184–90. doi: 10.1111/jgh.13263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hilscher M, Enders FB, Carey EJ, et al. Alkaline phosphatase normalization is a biomarker of improved survival in primary sclerosing cholangitis. Ann Hepatol. 2016;15:246–53. doi: 10.5604/16652681.1193721. [DOI] [PubMed] [Google Scholar]
- 5.de Vries EM, Wang J, Leeflang MM, et al. Alkaline phosphatase at diagnosis of primary sclerosing cholangitis and 1 year later: evaluation of prognostic value. Liver Int. 2016;36:1867–1875. doi: 10.1111/liv.13110. [DOI] [PubMed] [Google Scholar]
- 6.Rupp C, Rossler A, Halibasic E, et al. Reduction in alkaline phosphatase is associated with longer survival in primary sclerosing cholangitis, independent of dominant stenosis. Aliment Pharmacol Ther. 2014;40:1292–301. doi: 10.1111/apt.12979. [DOI] [PubMed] [Google Scholar]
- 7.Lindstrom L, Hultcrantz R, Boberg KM, et al. Association between reduced levels of alkaline phosphatase and survival times of patients with primary sclerosing cholangitis. Clin Gastroenterol Hepatol. 2013;11:841–6. doi: 10.1016/j.cgh.2012.12.032. [DOI] [PubMed] [Google Scholar]
- 8.Al Mamari S, Djordjevic J, Halliday JS, et al. Improvement of serum alkaline phosphatase to <1.5 upper limit of normal predicts better outcome and reduced risk of cholangiocarcinoma in primary sclerosing cholangitis. J Hepatol. 2013;58:329–34. doi: 10.1016/j.jhep.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 9.Stanich PP, Bjornsson E, Gossard AA, et al. Alkaline phosphatase normalization is associated with better prognosis in primary sclerosing cholangitis. Dig Liver Dis. 2011;43:309–13. doi: 10.1016/j.dld.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balasubramaniam K, Wiesner RH, LaRusso NF. Primary sclerosing cholangitis with normal serum alkaline phosphatase activity. Gastroenterology. 1988;95:1395–8. doi: 10.1016/0016-5085(88)90378-2. [DOI] [PubMed] [Google Scholar]
- 11.Lindor KD, Kowdley KV, Luketic VA, et al. High-dose ursodeoxycholic acid for the treatment of primary sclerosing cholangitis. Hepatology. 2009;50:808–14. doi: 10.1002/hep.23082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim WR, Therneau TM, Wiesner RH, et al. A revised natural history model for primary sclerosing cholangitis. Mayo Clin Proc. 2000;75:688–94. doi: 10.4065/75.7.688. [DOI] [PubMed] [Google Scholar]
- 13.Wiesner RH, Grambsch PM, Dickson ER, et al. Primary sclerosing cholangitis: natural history, prognostic factors and survival analysis. Hepatology. 1989;10:430–6. doi: 10.1002/hep.1840100406. [DOI] [PubMed] [Google Scholar]
- 14.de Vries EM, Verheij J, Hubscher SG, et al. Applicability and prognostic value of histologic scoring systems in primary sclerosing cholangitis. J Hepatol. 2015;63:1212–9. doi: 10.1016/j.jhep.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Olsson R, Hagerstrand I, Broome U, et al. Sampling variability of percutaneous liver biopsy in primary sclerosing cholangitis. J Clin Pathol. 1995;48:933–5. doi: 10.1136/jcp.48.10.933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vesterhus M, Hov JR, Holm A, et al. Enhanced liver fibrosis score predicts transplant-free survival in primary sclerosing cholangitis. Hepatology. 2015;62:188–97. doi: 10.1002/hep.27825. [DOI] [PubMed] [Google Scholar]
- 17.Corpechot C, Gaouar F, El Naggar A, et al. Baseline values and changes in liver stiffness measured by transient elastography are associated with severity of fibrosis and outcomes of patients with primary sclerosing cholangitis. Gastroenterology. 2014;146:970–9. doi: 10.1053/j.gastro.2013.12.030. quiz e15-6. [DOI] [PubMed] [Google Scholar]
- 18.Ehlken H, Wroblewski R, Corpechot C, et al. Validation of Transient Elastography and Comparison with Spleen Length Measurement for Staging of Fibrosis and Clinical Prognosis in Primary Sclerosing Cholangitis. PLoS One. 2016;11:e0164224. doi: 10.1371/journal.pone.0164224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Natekin A, Knoll A. Gradient boosting machines, a tutorial. Front Neurorobot. 2013;7:21. doi: 10.3389/fnbot.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weng SF, Reps J, Kai J, et al. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS One. 2017;12:e0174944. doi: 10.1371/journal.pone.0174944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ayaru L, Ypsilantis PP, Nanapragasam A, et al. Prediction of Outcome in Acute Lower Gastrointestinal Bleeding Using Gradient Boosting. PLoS One. 2015;10:e0132485. doi: 10.1371/journal.pone.0132485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casanova R, Saldana S, Chew EY, et al. Application of random forests methods to diabetic retinopathy classification analyses. PLoS One. 2014;9:e98587. doi: 10.1371/journal.pone.0098587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maroco J, Silva D, Rodrigues A, et al. Data mining methods in the prediction of Dementia: A real-data comparison of the accuracy, sensitivity and specificity of linear discriminant analysis, logistic regression, neural networks, support vector machines, classification trees and random forests. BMC Res Notes. 2011;4:299. doi: 10.1186/1756-0500-4-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kourou K, Exarchos TP, Exarchos KP, et al. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atkinson EJ, Therneau TM, Melton LJ, 3rd, et al. Assessing fracture risk using gradient boosting machine (GBM) models. J Bone Miner Res. 2012;27:1397–404. doi: 10.1002/jbmr.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ponsioen CY, Chapman RW, Chazouilleres O, et al. Surrogate endpoints for clinical trials in primary sclerosing cholangitis: Review and results from an International PSC Study Group consensus process. Hepatology. 2016;63:1357–67. doi: 10.1002/hep.28256. [DOI] [PubMed] [Google Scholar]
- 27.Merion RM, Schaubel DE, Dykstra DM, et al. The survival benefit of liver transplantation. Am J Transplant. 2005;5:307–13. doi: 10.1111/j.1600-6143.2004.00703.x. [DOI] [PubMed] [Google Scholar]
- 28.Chapman R, Fevery J, Kalloo A, et al. Diagnosis and management of primary sclerosing cholangitis. Hepatology. 2010;51:660–78. doi: 10.1002/hep.23294. [DOI] [PubMed] [Google Scholar]
- 29.Manns MP, Czaja AJ, Gorham JD, et al. Diagnosis and management of autoimmune hepatitis. Hepatology. 2010;51:2193–213. doi: 10.1002/hep.23584. [DOI] [PubMed] [Google Scholar]
- 30.Goodman ZD. Grading and staging systems for inflammation and fibrosis in chronic liver diseases. J Hepatol. 2007;47:598–607. doi: 10.1016/j.jhep.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 31.Rizvi S, Eaton JE, Gores GJ. Primary Sclerosing Cholangitis as a Premalignant Biliary Tract Disease: Surveillance and Management. Clin Gastroenterol Hepatol. 2015;13:2152–65. doi: 10.1016/j.cgh.2015.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridgeway G. Generalized boosted models: a guide to the gbm package. 2007 [Google Scholar]
- 33.van Buuren S, Groothuis-Oudshoorn K. Multivariate Imputation by Chained Equations in R. Journal of Statistical Software, Articles. 2011;45:1–67. [Google Scholar]
- 34.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–87. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 35.Ehlken H, Wroblewski R, Corpechot C, et al. Spleen size for the prediction of clinical outcome in patients with primary sclerosing cholangitis. Gut. 2016;65:1230–2. doi: 10.1136/gutjnl-2016-311452. [DOI] [PubMed] [Google Scholar]
- 36.Eaton JE, McCauley BM, Atkinson EJ, et al. Variations in Primary Sclerosing Cholangitis Across the Age Spectrum. J Gastroenterol Hepatol. 2017 doi: 10.1111/jgh.13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weismuller TJ, Trivedi PJ, Bergquist A, et al. Patient Age, Sex, and Inflammatory Bowel Disease Phenotype Associate With Course of Primary Sclerosing Cholangitis. Gastroenterology. 2017 doi: 10.1053/j.gastro.2017.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Broome U, Olsson R, Loof L, et al. Natural history and prognostic factors in 305 Swedish patients with primary sclerosing cholangitis. Gut. 1996;38:610–5. doi: 10.1136/gut.38.4.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chapman RW, Williamson KD. Are Dominant Strictures in Primary Sclerosing Cholangitis a Risk Factor for Cholangiocarcinoma? Current Hepatology Reports. 2017;16:124–129. doi: 10.1007/s11901-017-0341-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedman JH. Greedy function approximation: A gradient boosting machine. Ann Statist. 2001;29:1189–1232. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


