Abstract
Autosomal recessive ataxias are characterised by a fundamental loss in coordination of gait with associated atrophy of the cerebellum. There is significant clinical and genetic heterogeneity amongst inherited ataxias; however, an early molecular diagnosis is essential with low-risk treatments available for some of these conditions. We describe two female siblings who presented early in life with unsteady gait and cerebellar atrophy. Whole exome sequencing revealed compound heterozygous inheritance of two pathogenic mutations (p.Leu277Pro, c.1506+1G>A) in the coenzyme Q8A gene (COQ8A), a gene central to biosynthesis of coenzyme Q (CoQ). The paternally derived p.Leu277Pro mutation is predicted to disrupt a conserved motif in the substrate-binding pocket of the protein, resulting in inhibition of CoQ10 production. The maternal c.1506+1G>A mutation destroys a canonical splice donor site in exon 12 affecting transcript processing and subsequent protein translation. Mutations in this gene can result in primary coenzyme Q10 deficiency type 4, which is characterized by childhood onset of cerebellar ataxia and exercise intolerance, both of which were observed in this sib-pair. Muscle biopsies revealed unequivocally low levels of CoQ10, and the siblings were subsequently established on a therapeutic dose of CoQ10 with distinct clinical evidence of improvement after 1 year of treatment. This case emphasises the importance of an early and accurate molecular diagnosis for suspected inherited ataxias, particularly given the availability of approved treatments for some subtypes.
Electronic supplementary material
The online version of this article (10.1007/8904_2017_73) contains supplementary material, which is available to authorized users.
Keywords: Autosomal recessive cerebellar ataxia, CoQ10, COQ8A
Introduction
Individuals with autosomal recessive cerebellar ataxia (ARCA) present with considerable clinical diversity but fundamentally have an inability to coordinate movement and balance due to cerebellar dysfunction. Individuals are typically diagnosed in childhood or as young adults (<30 years of age), with cerebellar atrophy visible by MRI (Montero et al. 2007). While categorisation of clinical entities has improved with the identification of the genetic basis for many of these conditions, there is still substantial genetic heterogeneity, making diagnosis challenging and an efficient molecular diagnosis difficult. However, the importance of a precise genetic diagnosis (enabling discrimination between the many types of autosomal recessive ataxias) has been underlined by the realisation that a minority of these disorders can be effectively treated with pharmacotherapy, resulting in significant improvement in health.
The advent of next generation sequencing technologies has tremendously enhanced the efficiency of molecular diagnosis for complex and highly heterogeneous neurodevelopmental disorders such as the inherited ataxias, providing answers for families, facilitating family planning and occasionally presenting the prospect of treatment strategies. We have applied this approach to identify the genetic basis for an ARCA caused by a deficiency in CoQ10 synthesis, resulting in a treatment strategy for a family 9 years after symptom onset.
Cases
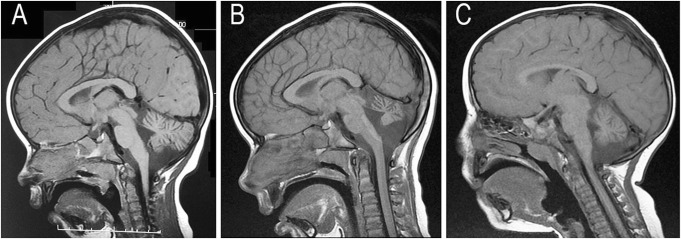
We describe a family with two girls who presented primarily with gait ataxia and cerebellar atrophy. There was no known consanguinity between the two Caucasian parents, and their family history was non-contributory. The proband (II.1) was born after an uncomplicated pregnancy and delivery at term. She sat, rolled and crawled at appropriate ages but only succeeded in walking, unsteadily, with a walker at 2 years and 6 months, and her gait remains unsteady at her current age of 11 years. She presented primarily with dysmetria (with no clonus); however there has been no discernable deterioration since her diagnosis. She is not dysmorphic, but does have reduced muscle bulk and tone. Her reflexes are normal, and she has no visual disturbance or nystagmus. MRI at 2 years and 2 months showed a small cerebellar vermis but normal height of the cerebellar hemispheres (Fig. 1a). A repeat MRI at 7 years and 9 months showed prominent cerebellar sulci, with a loss of height of the cerebellar hemispheres, and atrophy of the vermis (Fig. 1b). Both scans revealed a normal brainstem, including the pons.
Fig. 1.
T1-weighted midline sagittal MRI scans of the two siblings with COQ8A mutations. (a) Scan of the oldest sibling at 2 years and 2 months of age demonstrating a small cerebellar vermis; (b) follow-up scan of the older sibling at age 7 years and 9 months demonstrating progressive loss of volume of the cerebellar vermis and thinning of the folia consistent with an atrophic mechanism underlying her ataxic presentation; (c) scan of the younger sibling at age 2 years and 2 months demonstrating moderate volume loss in the cerebellar vermis and increased spaces between the folia
The proband’s sister (II.2) demonstrated a similar course, with truncal ataxia preventing walking until 3 years of age. At 2 years and 2 months, MRI revealed a small cerebellar vermis and increased spaces between the folia especially in the inferior cerebellum (Fig. 1c). The height of the cerebellar hemispheres was normal, and the brainstem and pons appeared normal. The proband also has a younger brother (III.3) who shows no clinical abnormalities at 3 years of age.
The metabolic workup for both girls has been extensive with no abnormalities in plasma cholesterol, plasma albumin, transferrin isoelectric focussing, plasma and urinary amino acids, urinary organic acids, cerebrospinal fluid (CSF) neurotransmitters and CSF lactate. Comparative genomic hybridisation (Agilent ISCA (v2)) revealed no significant imbalance. Liver mitochondrial enzymology was considered normal, and muscle mitochondrial enzymology revealed complex II + III to be low (reflecting the enzymatic deficiency later proposed by genetics – full mitochondrial analysis can be found in Supplementary Table 1). Electron microscopy analysis of the same muscle tissue showed no evidence of giant abnormal mitochondria. Other fine structures were unremarkable. Initial genetic investigations by Sanger sequencing ruled out mutations in APTX, and no causative triplet repeat expansions were identified in FRDA, ATXN1, ATXN2, ATXN3, CACNA1A, ATXN7, TBP, and ATN1. We subsequently performed whole exome sequencing on both affected siblings (Supplementary Methods).
Variation filtering identified the presence of two single nucleotide variants (SNVs) in COQ8A (coenzyme Q8A, previous symbol ADCK3 (AARF domain-containing kinase 3); NM_020247.4, OMIM 606980) in both affected children (Table 1). Sanger sequencing of parental DNA confirmed compound heterozygous inheritance of these variants. Mutations in COQ8A result in coenzyme Q10 deficiency, primary, 4 (COQ10D4, OMIM 612016), an autosomal recessive disorder characterized by childhood onset of cerebellar ataxia and exercise intolerance (Lagier-Tourenne et al. 2008), which is phenotypically concordant with this family’s presentation.
Table 1.
COQ8A variant annotations
| Gene | Chr | HGVS DNA ref | HGVS protein ref | Variant type | Predicted effect | Genotype |
|---|---|---|---|---|---|---|
| COQ8A | 1 | NM_020247.4:c.830T>C | NM_020247.4:p.(Leu277Pro) | Missense | Aa change | Heterozygous |
| COQ8A | 1 | NG_012825.2:c.1506+1G>A | NG_012825.2:p.(=) | Splice donor | Destroys splice site | Heterozygous |
The paternally derived SNV encodes a non-synonymous missense change from leucine to proline (c.830T>C, p.Leu277Pro) in exon 6. This amino acid is conserved in vertebrates from human to lamprey (UCSC (Kent et al. 2002), PhyloP (Pollard et al. 2010); Supplementary Figure 1) and is located in an N-terminal motif that is conserved across all members of the AARF domain-containing kinase family: the KxGK motif (positions 276–279) (Lagier-Tourenne et al. 2008). The variant has been previously observed in heterozygote state in a single European individual in the gnomAD dataset (AF = 4.48–06) (Lek et al. 2016). This variant is predicted to be damaging by mutation impact prediction algorithms (PolyPhen-2, SIFT Blink, SNPs&GO, PROVEAN, full details in Supplementary Table 2).
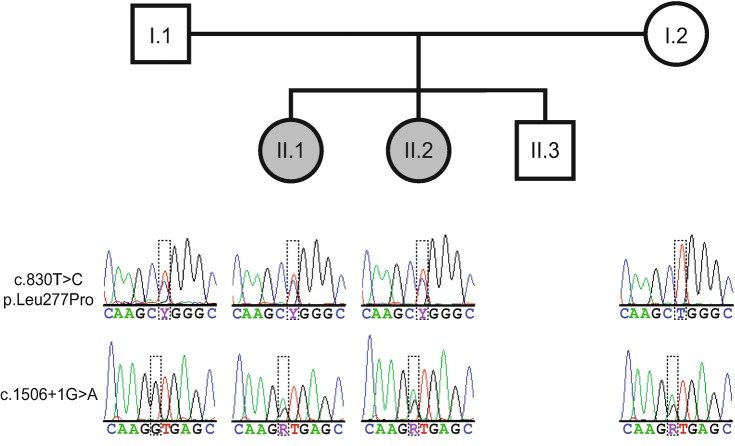
The maternally derived SNV alters the canonical splice donor site in exon 12 (c.1506+1G>A); this mutation has not been reported previously and is absent in public variant databases. BDGP splice site and ASSP programmes predict that this variant will destroy the splice donor site of exon 12, which is expected to affect removal of intron 12 from all reported protein-coding isoforms. The effect of the c.1506+1G>A splice site mutation was determined using RNA sequencing (Supplementary Methods) on blood obtained from both parents. No significant abundance differences were observed between the parents for any annotated COQ8A protein-coding transcript (200,000 vs. 190,926 FPKM, p = 0.43 for canonical transcript NM_020247.4). However, all mRNA molecules originating from the maternal c.1506+1G>A allele retained all or part of intron 12; conversely, intron 12 was correctly spliced in all mRNAs originating from the mother’s wild-type allele and both paternal alleles. RNAseq fragments derived from the maternal wild-type allele were twice as abundant as those from the c.1506+1G>A allele (52 vs. 25 fragments). Mutation haplotypes and parental inheritance were validated by PCR followed by Sanger sequencing (Fig. 2).
Fig. 2.
Family pedigree and transmission of the c.830T>C and c.1506+1G>A mutations in the COQ8A gene. Sanger sequencing electropherograms for both loci are shown below the corresponding family member in the lower part of the figure
Following the discovery of the COQ8A variants, muscle and plasma total CoQ10 were measured using high-performance liquid chromatography with electrochemical detection, similarly to Tang et al. (2001). The muscle CoQ10 was mildly reduced in II.1 (16.3 ± 3.4 nmol/g tissue, reference range 20–70 nmol/g wet tissue) when compared to biopsies from myopathy patients not suspected of CoQ10 deficiencies and previously published reference intervals (Lopez et al. 2006). Plasma CoQ10 was reported as low-normal (0.68 μmol/L, 0.52 μmol/L in II.1 and II.2, respectively; reference range 0.45–1.71 μmol/L), which is consistent with the literature on biosynthetic CoQ10 defects (Molyneux et al. 2005, 2008; Yubero et al. 2014).
The siblings underwent treatment with oral CoQ10 (20 mg/kg/day, (Blumkin et al. 2014)) and follow-up was performed at 12 months. Use of a validated clinical tool for the assessment of ataxia (Trouillas et al. 1997) was instituted to objectively measure the effect of treatment. The scale is scored 0–100 with a score of 0 signifying no ataxic symptoms and 100 indicating a maximal score. The proband’s baseline score was 40/100, and after treatment for 12 months, the score had reduced to 29/100 (full ataxia assessment is detailed in Supplementary Table 3). The parents of the child also reported an improvement in energy and classroom performance, observations that were reinforced when teachers, blinded to the deliberate omission of daily doses of CoQ10, volunteered their observations of discernible deterioration in function over the school day. The younger sibling also demonstrated an improvement in ataxia score from 49/100 to 43/100 over the same time frame as her sister.
Discussion
Autosomal recessive ataxias due to primary CoQ10 deficiency are a heterogeneous group of disorders caused by mutations in genes involved in the CoQ10 biosynthetic pathway (Desbats et al. 2015). Coenzyme Q10 acts as an electron carrier in the mitochondrial respiratory chain and serves as an antioxidant in the intracellular environment (Crane et al. 1993); hence there has been much clinical interest in its potential therapeutic benefits. Indeed, therapeutic doses of CoQ10 have been successful in ameliorating symptoms for some cases of primary CoQ10 deficiency ataxias (Mignot et al. 2013). We describe compound heterozygous mutations in COQ8A, a gene central to the CoQ10 biosynthetic pathway, in two female siblings who presented with gait ataxia and cerebellar atrophy. The older sibling had moderately decreased muscle CoQ10 and both underwent a trial of CoQ10. Remarkably, both siblings showed improvement in their ataxia scores following 12 months of treatment, with functional improvements also evident in daily classroom performance.
Causative mutations in the COQ8A gene were first reported in childhood ataxia by Lagier-Tourenne in 2001 who proposed the term autosomal recessive cerebellar ataxia 2 (ARCA2) after observing cerebellar atrophy and exercise intolerance during childhood with an associated reduction of muscle CoQ10 (Lagier-Tourenne et al. 2008). COQ8A encodes an unorthodox protein kinase-like (uPKL) protein that localises to the mitochondrial matrix and is central to CoQ biosynthesis (Khadria et al. 2014; Stefely et al. 2016). The deletion of the yeast and E. coli homologues eliminates CoQ biosynthesis in these organisms (Poon et al. 2000; Do et al. 2001), and a mouse COQ8A knockout model exhibits an ataxic phenotype and pathological signatures that align with the human condition (including degeneration of cerebellar Purkinje cells and abnormal skeletal mitochondria morphology) (Stefely et al. 2016). The structure of the protein was originally described by Stefely et al. as an UbiB protein with an atypical protein kinase-like fold containing particular features inhibitory to protein kinase activity (Stefely et al. 2015). More recently, they provide evidence that the protein encoded by COQ8A functions similar to yeast Coq8p and further argue that it is in fact an uPKL with noncanonical activities which support CoQ biosynthesis (amongst other functions) (Stefely et al. 2016).
Atypical kinase COQ8A, mitochondrial, features a long N-terminal extension, which folds into α-helices to form a KxGQ motif which appears to play a central role in CoQ biosynthesis, as mutating the region results in autophosphorylation and inhibition of CoQ production in vivo (Stefely et al. 2015, 2016). The p.Leu277Pro mutation identified in this New Zealand family is located in one of the predicted alpha helical domains (GQα2), which contribute to the KxGQ motif. The well-described tendency of proline to distort and destabilise alpha helices in aqueous environments (Khan and Vihinen 2007) suggests that the mutation affects protein stability, as demonstrated for other known pathogenic mutations in the GQα2 helix (Stefely et al. 2015).
The second mutation, c.1506+1G>A, destroys the splice donor site in exon 12 resulting in retention of intron 12 leading to premature termination of COQ8A protein synthesis, ultimately resulting in reduced CoQ10 levels. Primary cell lines from an ACRA2 patient harbouring similar mutations to the case described here (splice and missense) showed reduced protein and total CoQ10 levels and exhibited ultrastructural changes to the mitochondria (Cullen et al. 2016).
Some patients with primary CoQ10 synthesis defects respond to supplementation with high-dose oral CoQ10 (Desbats et al. 2015); however, patients with COQ8A mutations show a somewhat varied response, with many not responding well (Lamperti et al. 2003; Aure et al. 2004; Lagier-Tourenne et al. 2008; Mollet et al. 2008; Anheim et al. 2010; Gerards et al. 2010; Horvath et al. 2012; Terracciano et al. 2012; Mignot et al. 2013; Blumkin et al. 2014; Liu et al. 2014; Barca et al. 2016; Hikmat et al. 2016; Malgireddy et al. 2016), as summarised in Supplementary Table 4. There have, however, been three reported cases of improvement, two of which are later onset than the girls presented here. The latest, reported by Barca et al., showed improvement in speech and gait in a 48-year man (onset at 20 years of age) after 1 year of treatment with 400 mg/day CoQ10 (Barca et al. 2016). This individual harbours a homozygous deletion (c.1511_1512delCT) that leads to a premature truncation of the protein (p. Ala504fs). This mutation is located 5 bp from the splice site mutation described in our New Zealand sib-pair, both of which reside in the protein kinase domain. Another study by Liu et al. reported improvement in a Pakistani sib-pair (age of onset: 10 years of age), who harbour a homozygous frameshift mutation (c.1844_1845insG) in the C-terminus of mitochondrial atypical kinase COQ8A, which is predicted to extend the open reading frame by 81 amino acids (Liu et al. 2014). These siblings showed significant improvements in myoclonic movements, ataxic gait and dysarthric speech 3 months after treatment with CoQ10 at 200 mg twice a day. The third reported case of improvement describes partial improvement in motor skills balance and strength at 5 years of age with 20 mg/kg/day oral CoQ10 (Blumkin et al. 2014). When the drug was ceased at 6 years of age (pre-empted by the patient who had an accompanying psychiatric condition), their condition deteriorated. This individual and her sister (milder presentation) harbour compound heterozygous mutations in the protein kinase domain (p.P502R), near the site disrupted by the splice site mutation described here, and a previously observed deletion at c.1750_1752delACC. There are also some self-reported improvements from patients on oral CoQ10 (Mollet et al. 2008; Liu et al. 2014). The sib-pair described here provides further evidence of the therapeutic benefits of CoQ10 for this condition in some families.
This case outlines the clinical and genetic heterogeneity of autosomal recessive ataxias and highlights the importance of early and accurate diagnosis (in this instance using whole exome sequencing). This seems particularly prudent given the possible response to a low-risk treatment option that could be given prior to severe central nervous system damage.
Electronic Supplementary Material
Evolutionary conservation of the locus affected by the missense (PDF 731 kb)
■■■■ (PDF 85 kb)
■■■■ (XLSX 24 kb)
■■■■ (PDF 65 kb)
Acknowledgements
We would like to thank Kristine Boxen at the Auckland Science Analytical Services for Sanger sequencing services and the New Zealand eScience Infrastructure for high-performance computing support.
Take-Home Message
Compound heterozygous COQ8A mutations cause treatment-responsive CoQ10 deficiency ataxia.
Contributions of Individual Authors
JCJ, KL and RGS designed and conducted experiments and wrote the manuscript; WW and BS conducted experiments; JT, DRL and RH clinically confirmed research results; SPR clinically evaluated patients and conducted the trial of CoQ10 treatment; SM, PMG and RM performed muscle and plasma CoQ10 analysis.
Corresponding Author
Professor Russell Snell.
Competing Interest Statement
JCJ, WW, BS, JT, DRL, RH, SM, PMG, RM, SPR, RGS and KL declare they have no conflict of interest.
Details of Funding
JCJ is supported by a Rutherford Discovery Fellowship from the New Zealand Government and administered by the Royal Society of New Zealand; SPR is supported by Curekids NZ. The research was funded by the Neurological Foundation of New Zealand, the University of Auckland and the Minds for Minds Charitable Trust.
Details of Ethics Approval
The study was approved by the New Zealand Northern B Health and Disability Ethics Committee (ref 12/NTB/59), and parents provided written informed consent.
Patient Consent Statement
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. Informed consent was obtained from all patients for being included in the study.
Contributor Information
Russell G. Snell, Email: r.snell@auckland.ac.nz
Collaborators: Matthias Baumgartner, Marc Patterson, Shamima Rahman, Verena Peters, Eva Morava, and Johannes Zschocke
References
- Anheim M, Fleury M, Monga B, et al. Epidemiological, clinical, paraclinical and molecular study of a cohort of 102 patients affected with autosomal recessive progressive cerebellar ataxia from Alsace, Eastern France: implications for clinical management. Neurogenetics. 2010;11:1–12. doi: 10.1007/s10048-009-0196-y. [DOI] [PubMed] [Google Scholar]
- Aure K, Benoist JF, Ogier de Baulny H, Romero NB, Rigal O, Lombes A. Progression despite replacement of a myopathic form of coenzyme Q10 defect. Neurology. 2004;63:727–729. doi: 10.1212/01.WNL.0000134607.76780.B2. [DOI] [PubMed] [Google Scholar]
- Barca E, Musumeci O, Montagnese F, et al. Cerebellar ataxia and severe muscle CoQ10 deficiency in a patient with a novel mutation in ADCK3. Clin Genet. 2016;90:156–160. doi: 10.1111/cge.12742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumkin L, Leshinsky-Silver E, Zerem A, Yosovich K, Lerman-Sagie T, Lev D. Heterozygous mutations in the ADCK3 gene in siblings with cerebellar atrophy and extreme phenotypic variability. JIMD Rep. 2014;12:103–107. doi: 10.1007/8904_2013_251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crane FL, Sun IL, Sun EE. The essential functions of coenzyme Q. Clin Investig. 1993;71:S55–S59. doi: 10.1007/BF00226841. [DOI] [PubMed] [Google Scholar]
- Cullen JK, Abdul Murad N, Yeo A, et al. AarF domain containing kinase 3 (ADCK3) mutant cells display signs of oxidative stress, defects in mitochondrial homeostasis and lysosomal accumulation. PLoS One. 2016;11:e0148213. doi: 10.1371/journal.pone.0148213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbats M, Lunardi G, Doimo M, Trevisson E, Salviati L. Genetic bases and clinical manifestations of coenzyme Q10 (CoQ10) deficiency. J Inherit Metab Dis. 2015;38:145–156. doi: 10.1007/s10545-014-9749-9. [DOI] [PubMed] [Google Scholar]
- Do TQ, Hsu AY, Jonassen T, Lee PT, Clarke CF. A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in Saccharomyces cerevisiae abc1 mutants. J Biol Chem. 2001;276:18161–18168. doi: 10.1074/jbc.M100952200. [DOI] [PubMed] [Google Scholar]
- Gerards M, van den Bosch B, Calis C, et al. Nonsense mutations in CABC1/ADCK3 cause progressive cerebellar ataxia and atrophy. Mitochondrion. 2010;10:510–515. doi: 10.1016/j.mito.2010.05.008. [DOI] [PubMed] [Google Scholar]
- Hikmat O, Tzoulis C, Knappskog PM, et al. ADCK3 mutations with epilepsy, stroke-like episodes and ataxia: a POLG mimic? Eur J Neurol. 2016;23:1188–1194. doi: 10.1111/ene.13003. [DOI] [PubMed] [Google Scholar]
- Horvath R, Czermin B, Gulati S, et al. Adult-onset cerebellar ataxia due to mutations in CABC1/ADCK3. J Neurol Neurosurg Psychiatry. 2012;83:174–178. doi: 10.1136/jnnp-2011-301258. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadria AS, Mueller BK, Stefely JA, Tan CH, Pagliarini DJ, Senes A. A Gly-zipper motif mediates homodimerization of the transmembrane domain of the mitochondrial kinase ADCK3. J Am Chem Soc. 2014;136:14068–14077. doi: 10.1021/ja505017f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S, Vihinen M. Spectrum of disease-causing mutations in protein secondary structures. BMC Struct Biol. 2007;7:56. doi: 10.1186/1472-6807-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagier-Tourenne C, Tazir M, López LC, et al. ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am J Hum Genet. 2008;82:661–672. doi: 10.1016/j.ajhg.2007.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamperti C, Naini A, Hirano M, et al. Cerebellar ataxia and coenzyme Q10 deficiency. Neurology. 2003;60:1206–1208. doi: 10.1212/01.WNL.0000055089.39373.FC. [DOI] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y-T, Hersheson J, Plagnol V, et al. Autosomal-recessive cerebellar ataxia caused by a novel ADCK3 mutation that elongates the protein: clinical, genetic and biochemical characterisation. J Neurol Neurosurg Psychiatry. 2014;85:493–498. doi: 10.1136/jnnp-2013-306483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez LC, Schuelke M, Quinzii CM, et al. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am J Hum Genet. 2006;79:1125–1129. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malgireddy K, Thompson R, Torres-Russotto D. A novel CABC1/ADCK3 mutation in adult-onset cerebellar ataxia. Parkinsonism Relat Disord. 2016;33:151–152. doi: 10.1016/j.parkreldis.2016.10.010. [DOI] [PubMed] [Google Scholar]
- Mignot C, Apartis E, Durr A, et al. Phenotypic variability in ARCA2 and identification of a core ataxic phenotype with slow progression. Orphanet J Rare Dis. 2013;8:173. doi: 10.1186/1750-1172-8-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet J, Delahodde A, Serre V, et al. CABC1 gene mutations cause ubiquinone deficiency with cerebellar ataxia and seizures. Am J Hum Genet. 2008;82:623–630. doi: 10.1016/j.ajhg.2007.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneux SL, Florkowski CM, Lever M, George PM. Biological variation of coenzyme Q10. Clin Chem. 2005;51:455–457. doi: 10.1373/clinchem.2004.043653. [DOI] [PubMed] [Google Scholar]
- Molyneux SL, Young JM, Florkowski CM, Lever M, George PM. Coenzyme Q10: is there a clinical role and a case for measurement? Clin Biochem Rev. 2008;29:71–82. [PMC free article] [PubMed] [Google Scholar]
- Montero R, Pineda M, Aracil A, et al. Clinical, biochemical and molecular aspects of cerebellar ataxia and coenzyme Q10 deficiency. Cerebellum. 2007;6:118–122. doi: 10.1080/14734220601021700. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon WW, Davis DE, Ha HT, Jonassen T, Rather PN, Clarke CF. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J Bacteriol. 2000;182:5139–5146. doi: 10.1128/JB.182.18.5139-5146.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely JA, Reidenbach Andrew G, Ulbrich A, et al. Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. Mol Cell. 2015;57:83–94. doi: 10.1016/j.molcel.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefely JA, Licitra F, Laredj L, et al. Cerebellar ataxia and coenzyme Q deficiency through loss of unorthodox kinase activity. Mol Cell. 2016;63:608–620. doi: 10.1016/j.molcel.2016.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang PH, Miles MV, DeGrauw A, Hershey A, Pesce A. HPLC analysis of reduced and oxidized coenzyme Q10 in human plasma. Clin Chem. 2001;47:256–265. [PubMed] [Google Scholar]
- Terracciano A, Renaldo F, Zanni G, et al. The use of muscle biopsy in the diagnosis of undefined ataxia with cerebellar atrophy in children. Eur J Paediatr Neurol. 2012;16:248–256. doi: 10.1016/j.ejpn.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouillas P, Takayanagi T, Hallett M, et al. International cooperative ataxia rating scale for pharmacological assessment of the cerebellar syndrome. The Ataxia Neuropharmacology Committee of the World Federation of Neurology. J Neurol Sci. 1997;145:205–211. doi: 10.1016/S0022-510X(96)00231-6. [DOI] [PubMed] [Google Scholar]
- Yubero D, Montero R, Artuch R, Land JM, Heales SJR, Hargreaves IP. Biochemical diagnosis of coenzyme Q(10) deficiency. Mol Syndromol. 2014;5:147–155. doi: 10.1159/000362390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Evolutionary conservation of the locus affected by the missense (PDF 731 kb)
■■■■ (PDF 85 kb)
■■■■ (XLSX 24 kb)
■■■■ (PDF 65 kb)