Abstract
Myogenesis is regulated by the coordinated expression of muscle regulatory factors, a family of transcription factors that includes MYOD, MYF5, myogenin and MRF4. Muscle regulatory factors are basic helix-loop-helix transcription factors that heterodimerize with E proteins to bind the regulatory regions of target genes. Their activity can be inhibited by members of the Inhibitor of DNA binding and differentiation (ID) family, which bind E-proteins with high affinity, thereby preventing muscle regulatory factor-dependent transcriptional responses. CCAAT/Enhancer Binding protein beta (C/EBPβ) is a transcription factor expressed in myogenic precursor cells that acts to inhibit myogenic differentiation, though the mechanism remains poorly understood. We identify Id3 as a novel C/EBPβ target gene that inhibits myogenic differentiation. Overexpression of C/EBPβ stimulates Id3 mRNA and protein expression, and is required for C/EBPβ-mediated inhibition of myogenic differentiation. Misexpression of C/EBPβ in myogenic precursors, such as in models of cancer cachexia, prevents the differentiation of myogenic precursors and we show that loss of Id3 rescues differentiation under these conditions, suggesting that the stimulation of Id3 expression by C/EBPβ is an important mechanism by which C/EBPβ inhibits myogenic differentiation.
Introduction
Skeletal muscle tissue retains the ability to regenerate throughout the lifespan of an organism and this ability is dependent on the presence of a small population of skeletal muscle stem cells called satellite cells. Normally quiescent, in response to injury satellite cells can become activated to proliferate, differentiate and fuse to repair damaged muscle1,2. Myogenic differentiation is controlled by a group of four basic helix-loop-helix (bHLH) transcription factors called muscle regulatory factors (MRFs) whose coordinated expression drives muscle-specific gene expression3,4. MYF5 and MYOD are expressed during activation and proliferation of satellite cells and their expression is followed by myogenin (MYOG) and MRF4 which drive terminal differentiation5. To exert their actions, MRFs heterodimerize with E-proteins to bind the regulatory regions of their target genes.
The activity of the myogenic regulatory factors can be inhibited by members of the inhibitor of DNA binding and differentiation (ID) family6–11. The ID proteins, encoded by four genes (Id1-4), are members of the HLH superfamily; however, unlike bHLH transcription factors, they lack a DNA-binding domain and thus form inactive heterodimers with other bHLH members6,7. ID proteins bind strongly to E-proteins preventing their dimerization with MRFs and thereby inhibiting the activation of MRF target genes during myogenesis6–9. ID1, ID2 and ID3 can inhibit myogenic differentiation while ID4 is without effect9,12–14.
CCAAT/Enhancer Binding proteins (C/EBPs) are a family of six bzip transcription factors that are involved in the regulation of cellular functions such as apoptosis, differentiation and cell growth15–17. C/EBPβ plays a significant role in the regulation of mesenchymal stem cell fate including repression of osteoblastogenesis and stimulation of adipogenesis17–20. In addition to these roles, C/EBPβ is expressed in quiescent and proliferating skeletal muscle satellite cells and is rapidly decreased upon activation by the ubiquitin-proteasome system to allow for myogenic differentiation21,22. Overexpression of C/EBPβ in myoblasts blocks their differentiation, while loss of C/EBPβ expression promotes precocious differentiation and larger myotubes in vitro22. In vivo, loss of C/EBPβ in satellite cells results in larger myofibers and enhanced regenerative capacity after a single injury22. However, in this model, muscle regeneration was defective after a second injury due to impaired self-renewal and a reduction in the satellite cell pool supporting repair23. Although the role of C/EBPβ in satellite cell function has been elucidated, the specific mechanism by which C/EBPβ inhibits myogenic differentiation remains unknown.
C/EBPβ expression can interfere with the transcriptional activity of MYOD and has been shown to regulate the expression of Id1 and Id2 in the immune system24–26. In this study, we examined a potential role for ID proteins in the C/EBPβ-mediated inhibition of myogenic differentiation. We find that C/EBPβ stimulates Id3 and Id1 expression in differentiating myoblasts, but appears to only regulate Id3 transcription directly. Knockdown of Id3 or Id1 rescues myogenic differentiation in cells overexpressing C/EBPβ, though neither could rescue fusion. Further, Id3 is the most highly expressed Id family member in myoblasts, and only knockdown of ID3 could enhance myogenic differentiation in control cells.
Results
ID proteins are expressed in proliferating myoblasts and are downregulated with differentiation
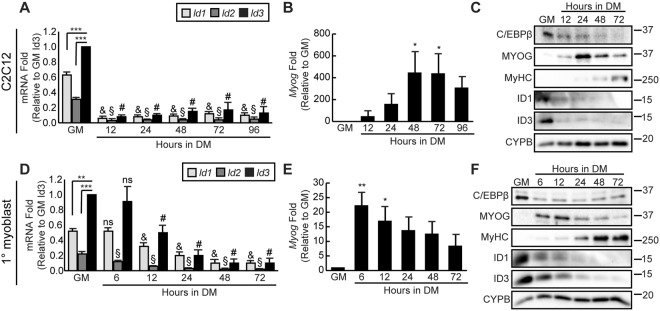
ID1, ID2 and ID3 are known inhibitors of myogenic differentiation9,12–14. Their expression patterns in proliferating myoblasts and during myogenic differentiation were evaluated in the myoblast cell line C2C12 and in primary myoblasts isolated from mouse hindlimb muscle. In C2C12 cells, Id1, Id2, and Id3 mRNAs were all detected in proliferating cells cultured in growth medium (Fig. 1A). All were rapidly and substantially downregulated 12 hours after induction to differentiate (Fig. 1A) and remained low up to 96 hours in differentiation medium. By contrast, myogenin (Myog) expression, a marker of terminal myogenic differentiation that is not expressed in proliferating myoblasts, was significantly induced after 48 hours in differentiation medium confirming differentiation (Fig. 1B). Assessment of protein expression patterns revealed that in accordance with their anti-myogenic roles, ID1, ID3 and C/EBPβ protein expression was highest in growth medium. Induction to differentiate caused a rapid downregulation of ID1 and ID3, evident 12 hours after switching to differentiation medium (Fig. 1C). C/EBPβ expression was also downregulated by 12 hours in differentiation medium, in accordance with published observations21,22. By contrast, myogenin expression was detected as early as 12 hours following induction to differentiate and levels peaked at the 24 hour time point (Fig. 1C). The structural protein, myosin heavy chain, was detected beginning at 24 hours after induction and continued to increase over the course of the experiment. ID2 protein levels were not examined as Id2 mRNA was weakly expressed in comparison to Id1 and Id3 and a reliable antibody was not found (Fig. 1A).
Figure 1.
ID protein expression is regulated during myogenic differentiation. (A) RT-qPCR analysis of Id1, Id2 and Id3 expression in C2C12 myoblasts in growth medium (GM) and after induction to differentiate (DM). Data is presented as the fold change relative to Id3 expression in GM. Bars are the mean ± SEM, n = 4, ***p < 0.001 for comparisons in GM. &p < 0.0001 relative to Id1 expression in GM. §p < 0.0001 relative to Id2 expression in GM. #p < 0.0001 relative to Id3 expression in GM. (B) Myog mRNA expression in cells cultured as in (A) presented as the fold change relative to GM condition. (C) C/EBPβ, myogenin (MYOG), myosin heavy chain (MyHC), ID1 and ID3 protein expression in C2C12 myoblasts cultured as in (A). Cyclophilin B (CYPB) is a loading control. (D) RT-qPCR analysis of Id1, Id2 and Id3 expression in proliferating (GM) and differentiating primary myoblasts (6–72 h). Data is presented as the fold change relative to Id3 expression in GM. n = 3, **p < 0.01, ***p < 0.001 for comparisons in GM. &p < 0.001 relative to Id1 expression in GM. §p < 0.001 relative to Id2 expression in GM. #p < 0.001 relative to Id3 expression in GM. ns is not statistically significant relative to GM values. (E) Myog mRNA expression in primary myoblasts cultured as in (D) and presented as the fold change relative to its expression in GM. (F) C/EBPβ, myogenin (MYOG), myosin heavy chain (MyHC), ID1 and ID3 protein expression in primary myoblasts cultured as in (D).
While C2C12 myoblasts are a useful cell line to study myogenic differentiation, they are polyploid and have inactivation of the p19/Arf locus27 and thus do not always recapitulate events in vivo. Therefore we assessed ID expression in primary myoblasts. Consistent with our findings in C2C12 myoblasts, Id1, Id2, and Id3 were all downregulated as early as 12 hours after induction to differentiate, with Id2 expressed at lower levels (Fig. 1D). The time course of myogenic differentiation is condensed in primary myoblasts as compared to C2C12 cells with the expression of myogenin mRNA and protein levels upregulated 6 hours after the addition of differentiation media (Fig. 1E,F). ID1, ID3 and C/EBPβ protein, while high in growth medium, were rapidly downregulated 6 hours after induction of differentiation (Fig. 1F). Consistent with differentiation, myosin heavy chain expression increased after myogenin expression and following downregulation of ID1 and ID3 expression (Fig. 1F).
C/EBPβ regulates the expression of Id3
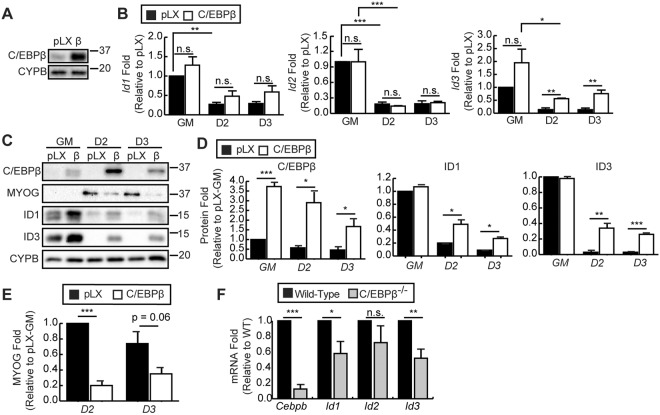
Given that C/EBPβ can inhibit myogenic differentiation and can regulate the expression of Id1 and Id2 in the immune system24–26, we examined Id gene expression in C2C12 myoblasts retrovirally transduced to express C/EBPβ (β) with an empty virus (pLXSN) under growth and differentiation conditions (Fig. 2). Overexpression of C/EBPβ was confirmed by western blot in proliferating cells (Fig. 2A). The expression of Id1, Id2 and Id3 in these cells was examined under growth and differentiation conditions (Fig. 2B). In proliferating cells, none of the ID genes were significantly upregulated. However, in differentiating cells, only Id3 mRNA expression was increased in cells overexpressing C/EBPβ as compared to empty virus controls (Fig. 2B). However, western blot analysis revealed upregulation of ID1 and ID3 protein in differentiating C/EBPβ-overexpressing cells (Fig. 2C,D). Increased ID protein expression in C/EBPβ-overexpressing myoblasts correlated with a reduction in myogenin protein expression as compared to controls (Fig. 2C,E).
Figure 2.
C/EBPβ regulates ID3 and ID1 expression. (A) C/EBPβ expression in proliferating C2C12 myoblasts that were retrovirally transduced with empty virus (pLX) or to express C/EBPβ (β). Cyclophilin B (CYPB) is a loading control. (B) Id1, Id2 and Id3 mRNA expression in cells transduced as in (A) in growth medium (GM) or cultured in differentiation medium for two (D2) or three days (D3) (n = 4). (C) C/EBPβ, myogenin (MYOG), ID1 and ID3 protein expression in C2C12 myoblasts transduced and cultured as in (B). (D) Quantification of C/EBPβ, ID1 and ID3 expression from (D) presented as fold change relative to pLX GM conditions after normalization to CYPB expression (n = 4). (E) Quantification of MYOG expression from (C) presented as fold change relative to pLXSN after two days (D2) of differentiation (n = 4). (F) Cebpb, Id1, Id2 and Id3 expression in proliferating primary myoblasts isolated from Cebpbfl/flPax7wt/wt (wild-type) and Cebpbfl/flPax7CreER/wt (C/EBPβ−/−) mice (n = 5). Bars are the mean ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, n.s. is not significant (two-tailed Student’s t-test or two-way ANOVA as required).
Conversely, in primary myoblasts isolated from a conditional null mouse where Cebpb is excised in muscle precursor cells (Cebpbfl/flPax7CreER/wt (C/EBPβ−/−)), Id1 and Id3 mRNA expression were reduced as compared to non-Cre expressing littermate controls (Cebpbfl/flPax7wt/wt (wild-type)) (Fig. 2F). The expression of Id2 was not significantly affected by knockdown of Cebpb expression. Taken together, these data suggest that C/EBPβ regulates Id1 and Id3 expression in myoblasts.
Loss of Id3 rescues C/EBPβ-mediated inhibition of myogenic differentiation
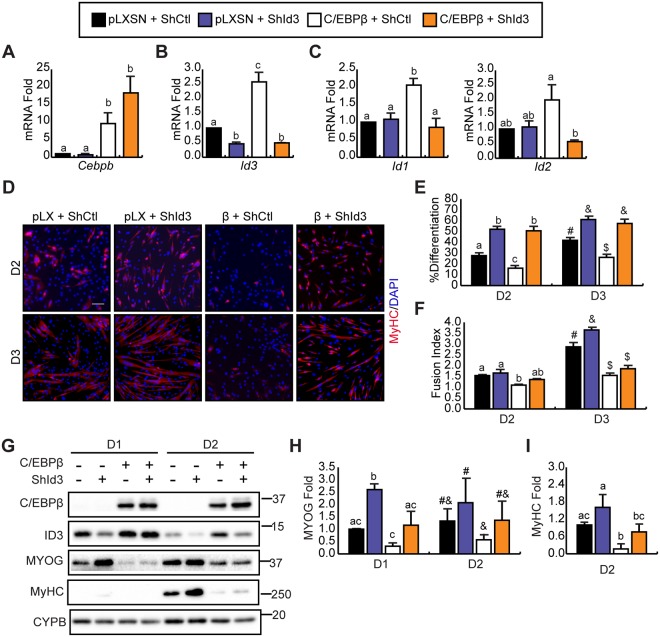
Given that Id3 is the most abundantly expressed ID family member in myoblasts and was most upregulated by C/EBPβ (Figs 1A,D, 2B–D), we first examined a potential role for ID3 in the inhibition of myogenic differentiation by C/EBPβ. C/EBPβ-overexpressing myoblasts and empty virus controls were retrovirally transduced with shRNA targeting Id3 mRNA (shId3) or with non-silencing shRNA (shCtl) and induced to differentiate. RT-qPCR confirmed both the overexpression of Cebpb and knockdown of Id3 (Fig. 3A,B). While Id3 mRNA expression was reduced approximately 50% in pLXSN-controls by the Id3-targetting shRNA, the expression of Id1 and Id2 were not affected (Fig. 3B,C). Overexpression of C/EBPβ increased Id3 expression in cultures transduced with the shCtl construct, and this was reduced approximately 80% by the shId3 construct (Fig. 3B). Id1 expression was also significantly increased by C/EBPβ overexpression, but returned to baseline levels with addition of the shId3 construct (Fig. 3C). Given that the shId3 construct does not affect Id1 and or Id2 expression in the absence of C/EBPβ-overexpression, the reduction in Id1 expression could be a consequence of enhanced differentiation, which is accompanied by downregulation of ID expression. To assess the efficacy of myogenic differentiation in these cultures, myosin heavy chain (MyHC) immunostaining was performed (Fig. 3D). Knockdown of Id3 in empty vector controls increased the percentage of nuclei found in MyHC-expressing cells after 2 and three days in differentiation medium (Fig. 3D,E). Consistent with our previous findings22, the differentiation index (% nuclei in MyHC+ cells/total nuclei) was reduced when C/EBPβ was overexpressed and this inhibition was rescued when Id3 was knocked down (Fig. 3D,E), restoring the differentiation index to the level of controls at both time points. However, loss of Id3 failed to rescue fusion (the average number of myonuclei/MyHC + cells) in C/EBPβ-overexpressing cells (Fig. 3D,F) indicating that C/EBPβ has a role in negatively regulating cell fusion independently of ID3. Western blot analysis revealed that the knockdown of Id3 expression in empty vector control cells (pLXSN) increased the expression of myogenin after one day in differentiating medium by ~2.5 fold (Fig. 3G,H). Additionally, while myogenin expression is decreased approximately 60% in C/EBPβ-overexpressing cells on day one of differentiation, this was rescued to the levels of pLXSN controls with loss of Id3 (Fig. 3G,H). On day two of differentiation, myogenin expression was no longer statistically different with knockdown of Id3, resulting in a normalization of its expression across experimental conditions at this time point (Fig. 3H). However, the loss of Id3 enhanced the expression of MyHC in pLXSN control cells and restored the defective MyHC expression observed in C/EBPβ-overexpressing cells on day two of differentiation (Fig. 3G,I).
Figure 3.
Inhibition of Id3 expression rescues C/EBPβ-mediated inhibition of myogenic differentiation. C2C12 myoblasts expressing C/EBPβ or control plasmid (pLXSN) were retrovirally transduced to express a control shRNA (ShCtl) or a shRNA directed against Id3 (ShId3) to create pooled stable cell lines. RT-qPCR analysis of (A) Cebpb, (B) Id3, and (C) Id1 and Id2 expression in myoblasts cultured in differentiation medium for two days. Data presented relative to pLXSN + ShCtl (n = 3). (D) MyHC (red) immunostaining of differentiating cells from (A) two (D2) and three (D3) days after induction. Nuclei are counterstained with DAPI (blue). Scale bar = 50 μm. (E) Differentiation index (# of MyHC+ cells relative to the total nuclei) from (D) (n = 3). (F) Fusion index (average # of nuclei per MyHC+ cell) (n = 3). (G) C/EBPβ, ID3, Myogenin (MYOG), myosin heavy chain (MyHC) protein expression in pooled stable myoblasts differentiated for one (D1) or two days (D2). Cyclophilin B (CYPB) is used as a loading control. (H) Quantification of MYOG protein expression from (G), normalized to CYPB expression and presented as fold change relative to pLXSN + ShCtl cells after one day (D1) of differentiation (n = 3). (I) Quantification of MyHC western blot from (G), normalized to CYPB expression and presented as fold change relative to pLXSN + ShCtl cells after two days (D2) of differentiation (n = 4). Bars are the mean ± SEM. Means with different letters or symbols are significantly different from one another, p < 0.05 (ANOVA, Tukey post-hoc test).
Since ID1 protein levels are affected by overexpression of C/EBPβ, we assessed whether knockdown Id1 could also rescue C/EBPβ-mediated inhibition of myogenic differentiation (Fig. S1). C2C12 myoblasts overexpressing C/EBPβ and their empty vector controls (pLXSN) were transduced to express a shRNA targeting Id1 (shId1) or a non-targeting control (shCtl) (Fig. S1A). After 3 days of differentiation, myosin heavy chain immunostaining revealed that, in contrast to knockdown of Id3, knockdown of Id1 did not enhance myogenic differentiation in pLXSN control cells as measured by the differentiation and fusion indices (Fig. S1B–D). However, the differentiation index of C/EBPβ-overexpressing cells was restored to normal levels with knockdown of Id1. Similar to shId3 knockdown, the knockdown of Id1 failed to rescue the defective fusion index in C/EBPβ-overexpressing cells. These data highlight the dominant role of ID3 over the other ID family members, at least in normal cells, as only knockdown of Id3 in control cells enhanced differentiation and fusion.
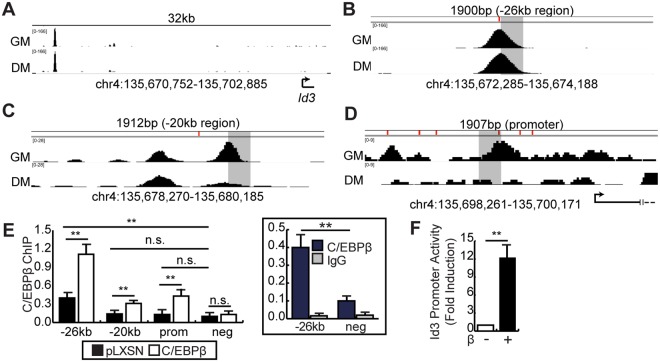
To determine if C/EBPβ directly regulates transcription of the Id3 gene, we used published ChIP-seq signal tracks from proliferating and differentiating C2C12 myoblasts (GSE36024) to identify C/EBPβ occupancy peaks at the promoter and two additional upstream regions (Fig. 4A–D). In silico analysis of the regulatory region of Id3 revealed that six putative C/EBP binding sites are located within −1 kb of the transcription start side (TSS), three of which are found within −300 bp of the TSS (Fig. 4D). We also found C/EBPβ motifs at approximately −26 kb and −20 kb from the TSS (Fig. 4B,C). ChIP-qPCR analysis in differentiating myoblasts retrovirally transduced with empty virus (pLXSN) or to overexpress C/EBPβ (β) confirmed the binding of C/EBPβ at the −26 kb upstream region (Fig. 4E and inset). C/EBPβ occupancy of the −20 kb and promoter region of Id3 in empty virus control cultures was not significantly different from that of a negative region (Fig. 4E). However, in C/EBPβ-overexpressing cells, C/EBPβ recruitment at all Id3 regulatory regions examined was enriched as compared to pLXSN (Fig. 4E) suggesting that overexpression, which occurs in vivo with sarcopenia and cachexia28,29, can force C/EBPβ onto elements that are not normally occupied. To confirm specificity, we repeated the ChIP in primary myoblasts isolated from a conditional null mouse where Cebpb is excised in muscle precursor cells (Cebpbfl/flPax7CreER/wt (C/EBPβ−/−)) (Fig. S2A). In this experiments, C/EBPβ recruitment to the −26kb region of the Id3 gene was significantly reduced in C/EBPβ−/− cells when compared to WT controls, while other regions which did not show significant C/EBPβ recruitment were unaffected (Figs S2A, 4E). To confirm the direct regulation of Id3 by C/EBPβ, we examined the ability of C/EBPβ to activate the Id3 promoter in a luciferase reporter assay30. In cells transiently expressing C/EBPβ, Id3 promoter activity was increased ~12 fold indicating that Id3 is a novel direct transcriptional target of C/EBPβ in myoblasts (Fig. 4F).
Figure 4.
C/EBPβ is a direct transcriptional regulator of Id3. (A) Id3 regulatory region (Chr4:135,670,752-135,702,885) with C/EBPβ (GSE36024) binding peaks indicated in proliferating (GM) and differentiating (DM) C2C12 myoblasts. C/EBPβ (GSE36024) binding peaks at (B) the −26kb upstream region of Id3 (Chr4:135,672,285-135,674,188), (C) the −20kb upstream region of Id3 (Chr4:135,678,270-135,680,185) and (D) the promoter region of Id3 (Chr4:135,698,261-135,700,171). C/EBP motifs are indicated by vertical dash marks above each histogram and the targeted region by qPCR is highlighted in grey. (E) qPCR-ChIP analysis of C/EBPβ recruitment to the Id3 regulatory regions in C2C12 cells retrovirally transduced with empty virus (pLXSN) or to express C/EBPβ and differentiated for three days. qPCR-ChIP data is shown as copy number as compared to a standard curve of 10% input of each condition. (n = 4). (Boxed) qPCR-ChIP data of C/EBPβ occupancy in pLXSN cells at the −26 kb region and the negative region presented with IgG control and shown as copy numbers in relation to the 10% input of each condition (n = 4). (F) Id3 promoter activity in a transient transcription assay where the Id3 promoter (−935 to + 13) drives expression of luciferase. Proliferating C2C12 cells were transiently transfected with the reporter construct and to express C/EBPβ and luciferase activity measured and shown relative to non-C/EBPβ expressing controls (n = 4). Bars are the mean ± SEM, **p < 0.01, n.s. is not significant (two-tailed Student’s t-test).
Given that ID1 protein levels are increased with C/EBPβ overexpression, we examined C/EBPβ recruitment to the Id1 promoter region. In contrast to the Id3 promoter, where overexpression of C/EBPβ increased its recruitment, the Id1 promoter was no different than a chromatin region devoid of C/EBPβ peaks (Fig. S2B).
Loss of Id3 rescues myogenic differentiation in cancer cachexia
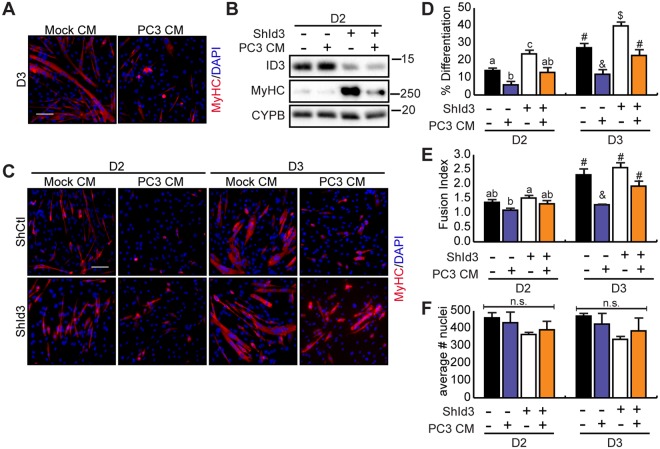
We have previously shown that exposure of myoblasts to conditioned medium from cachexia-inducing tumours, can upregulate C/EBPβ expression and inhibit myogenic differentiation. Indeed, loss of C/EBPβ rescues myoblast differentiation in this model system28. Given that ID3 is downstream of C/EBPβ and loss of ID3 expression increases myogenin expression and promotes myogenic differentiation (Fig. 3), we asked if the knockdown of ID3 could rescue differentiation in an in vitro cancer cachexia model31. C2C12 myoblasts were grown in conditioned media (CM) from a cachexia-inducing prostate cancer cell line (PC3) or proliferating C2C12 cells (mock) for 48 hours prior to induction to differentiate. Differentiation was then induced in the absence of conditioned medium for up to 3 days. Inhibition of myogenic differentiation in cells pre-treated with PC3-conditioned media was confirmed by MyHC staining (Fig. 5A). We next asked if knockdown of ID3 could rescue the defective differentiation seen in myoblasts pre-treated with PC3-conditioned media. Under differentiation conditions, knockdown of ID3 stimulated myosin heavy chain protein expression in control cells and in cells pre-cultured in PC3-conditioned media as compared to control shRNA (Fig. 5B). Indeed, pre-treatment with PC3-conditioned medium inhibited differentiation in shCtl cells but knockdown of ID3 rescued this defect in differentiation, most prominently on day 3 (Fig. 5C,D). Furthermore, while pre-treatment with PC3 conditioned medium reduced myotube size in shCtl cells on day 3, this effect was rescued in cultures lacking ID3, suggesting that the blockade in myogenic differentiation observed in cancer cachexia is mediated through misexpression of ID3 (Fig. 5C,E). Pre-treatment with PC3-conditionned medium did not influence cell density in these experiments, nor did knockdown of Id3 (Fig. 5F).
Figure 5.
Loss of ID3 rescues myogenic differentiation in an in vitro model of cachexia. (A) Representative images of myosin heavy chain (red) immunostaining of C2C12 myoblasts that grown in C2C12-conditioned media (Mock CM) or PC3-conditioned media (PC3 CM) for 48 h then differentiated in fresh medium for three days. Nuclei are counterstained with DAPI (blue). Scale bar = 50 μm. (B) ID3 and myosin heavy chain (MyHC) protein expression in C2C12 cells retrovirally transduced to express a control shRNA or a shRNA directed against Id3 (ShId3). Myoblasts were treated as in (A) and collected as differentiating cells (D2). (C) Myosin heavy chain immunostaining of C2C12 cells transduced as in (B) and treated and cultured as in (A). (D,E) Differentiation index (# of nuclei in MyHC-expressing cells/ total nuclei) and fusion index (average # of nuclei per MyHC + cell) for (C) (n = 4). (F) Average number of nuclei per field of view in cultures from (C). Data information: Bars are the mean ± SEM. Means with different letters or symbols are significantly different from one another, p < 0.05 (ANOVA, Bonferroni’s multiple comparison test).
Discussion
Herein, we demonstrate that C/EBPβ can upregulate Id1 and Id3 but not Id2 expression in myoblasts. We find that Id3 is the most abundant Id family member expressed in myoblasts and the stimulation of Id3 expression by ectopic C/EBPβ is more pronounced than that of Id1. We demonstrate that C/EBPβ binds the regulatory region of Id3 (promoter and two upstream regions) and is a direct target of C/EBPβ in myoblasts. Moreover, the knockdown of Id3 rescues the inhibition of myogenic differentiation in C/EBPβ-overexpressing cells, placing Id3 as a major effector of C/EBPβ action in this system.
While C/EBPβ is a known transcriptional regulator of both Id1 and Id2 in the immune system24–26, changes in C/EBPβ expression did not affect Id2 expression in myogenic cells, suggesting cell-type specific regulation of these genes by C/EBPβ. Interestingly, Pax7 is also a direct target of C/EBPβ22 and PAX7 has been shown to also regulate Id3 expression30, suggesting that C/EBPβ can regulate Id3 expression directly as well as indirectly through PAX7.
While all ID proteins bind E-proteins with high affinity and thus can interfere with the actions of bHLH transcription factors, in muscle, their effects are not entirely overlapping. For example, ID1 and ID2 appear to inhibit MYOD and MYF5 action but not that of MYOG or MRF4, while ID3 showed a weak inhibition of all MRFs32 suggesting that ID3 can function both in early and late myogenic differentiation if it is expressed, as occurs in cells overexpressing C/EBPβ and in our model of cancer cachexia. While the overexpression of ID2 but not ID3 in myogenic cells has been shown to inhibit the induction of myogenin expression12,14 we find that knockdown of Id3 increases myogenin protein expression even in the presence of ectopic C/EBPβ. Further, we have shown that C/EBPβ can reduce MYOD activity, an upstream regulator of myogenin22 though whether this effect is mediated directly by ID3 remains to be investigated. As such, our findings suggest that the misexpression of ID3 could therefore have a more potent impact of the progression of differentiation by targeting the late regulators of differentiation.
The shRNA used to knockdown ID3 expression is specific and failed to decrease the mRNA expression of Id1 or Id2 in control cultures. However, in the presence of ectopic C/EBPβ, the expression of the Id3-targeting shRNA did reduce Id1 mRNA, suggesting that the loss of ID3 in these cells restores the myogenic differentiation program including the downregulation of ID1 expression. Indeed, knockdown of Id1 in C2C12 cells overexpressing C/EBPβ was able to rescue differentiation but not fusion, but was without effect in control cells. While these findings would suggest that ID3 is the dominant ID protein in myoblasts under normal physiological conditions, in disease states where C/EBPβ expression is induced in myoblasts, for example cachexia, both Id1 and Id3 upregulation are likely to contribute. While we did not see recruitment of C/EBPβ to the Id1 promoter, this analysis did not include possible enhancer regions. Another condition that is associated with poor muscle regeneration and differentiation is sarcopenia, the loss of muscle mass that accompanies aging. It has been reported that the expression of Cebpb in primary myoblasts increases with mouse age (3-week, 3-month and 18-month of age)33, and Cebpb is considered part of a molecular signature of muscle aging29. Interestingly, analysis of the raw microarray data set from Price, Von Maltzahn et al.33, revealed that along with upregulation of Cebpb in aging primary myoblasts, there is upregulation of Id1 and Id3, but not Id2, consistent with our findings. These findings suggest that ID3 is an interesting therapeutic target to restore differentiation function in aged myoblasts.
Despite restoring myogenic marker expression, knockdown of Id3 was not able to restore fusion, suggesting that C/EBPβ acts to limit myotube size in a mechanism that is independent of ID3. In cancer cachexia however, where treatment of proliferating myoblasts with conditioned medium from the PC3 tumour is known to transiently upregulate C/EBPβ expression and to inhibit myogenic differentiation28, knockdown of ID3 rescues both differentiation and fusion. Indeed, given the design of the experiment with pre-treatment causing differentiation defects, it is possible that at the time of fusion, the cells have recovered from the cachectic insult. Examination of this in co-culture experiments would clarify the role for ID3 in this context.
Experimental Procedures
Cell Culture
Mouse primary myoblasts were isolated and cultured as previously described22. Briefly, hindlimb muscles of adult mice (aged 6–8 weeks) were dissected and digested with collagenase/Dispase (Roche). Isolated cells were then pre-plated to remove fibroblasts and primary myoblasts were maintained on matrigel-coated plates in Dulbecco modified eagle medium DMEM (Wisent) supplemented with 20% fetal bovine serum (FBS) (Wisent), 10% horse serum (HS) (Sigma), 10 ng/ml basic fibroblast growth factor (bFGF) and 2 ng/ml hepatocyte growth factor (HGF) (Peprotech). To induce differentiation, confluent cultures were switched to differentiation media (DMEM, 2% FBS and 10% HS). Primary myoblasts from Cebpbfl/flPax7wt/wt (WT) or Cebpbfl/flPax7CreER/wt (cKO) mice isolated as previously described22, were treated with 2 µM 4-OH tamoxifen (Sigma) for 48 h to induce excision of Cebpb in Cre-expressing cells. C2C12 myoblasts (ATCC) were grown in DMEM supplemented with 10% FBS (GM, growth media) and differentiation was induced by switching confluent cells to DMEM supplemented with 2% HS. PC-3 human prostate cancer cells (ATCC) were maintained in Roswell Park memorial institute medium (RPMI 1640) (Sigma) supplemented with 10% FBS. For in vitro cachexia experiments, conditioned media was collected from 90% confluent PC-3 cells after two days and added to C2C12 growth media (1:1 ratio) with fresh medium31. After two days in conditioned media, C2C12s were switched to fresh differentiation media.
All animal experiments were approved by the University of Ottawa Animal Care Committee and all procedures were performed in accordance with the regulations set out by the Canadian Council on Animal Care.
Retroviral Infection
Replication incompetent retroviruses were generated in Phoenix Ampho packaging cells (ATCC) as previously described22 using pLXSN-C/EBPβ plasmid. Following retroviral infection of C2C12 myoblasts with empty virus (pLXSN) or virus to express C/EBPβ (pLXSN-C/EBPβ), pooled stable cells were selected based on neomycin (Wisent) resistance. Knockdown of Id3 was accomplished by retrovirally transducing C2C12 myoblasts with a sequence targeting Id3 (pRS-shId3, Origene, TR501033), Id1 (pRS-shId1, Origene, TR511687) or a non-targeting control sequence (shCtl, Origene) and stable cells generated based on puromycin (Wisent) resistance.
Immunofluorescence
Cells were fixed in ice-cold 100% methanol and then permeabilized with PBS containing 0.5% Triton X-100 (Bioshop). Cells were incubated in primary antibody (MF20, DSHB) overnight at 4 °C followed by secondary antibody (anti-mouse Cy3, Jackson ImmunoResearch) incubation. Nuclei were counterstained with 0.5 μg/ml DAPI. Images were captured using a Zeiss AxioObserver D1 microscope (Zeiss) at the 10X objective. ImageJ software was used for the quantitative analysis of MyHC stained cells. Representative images were cropped using Adobe Photoshop.
Western Analysis
Whole cell extracts were prepared from primary myoblasts or C2C12 myoblasts. Equal amount of proteins were resolved on a 15% SDS-PAGE gel and then transferred to a PVDF membrane (Bio-Rad). Membranes were then probed with the following primary antibodies: C/EBPβ (E299, ab32358), ID1 (Biocheck, BCH-1/37-2), ID3 (Biocheck, BCH-4/17-3), myogenin (F5D, DSHB), MyHC (MF20, DSHB) and Cyclophilin B (ab16045). The ChemiDocTM MP system (Bio-Rad) was used to detect chemiluminescence. Resultant images were cropped in Adobe Photoshop.
Real-time quantitative PCR
RNA was isolated using RNeasy kit (Qiagen) as per manufacturer’s protocol. The remaining DNA was digested with DNase (Ambion) and DNA-digested RNA was used to make cDNA using iScript™ cDNA Synthesis Kit (Bio-Rad) following the manufacturer’s protocol. RT-qPCR reactions were performed using iTaqTM Universal SYBR® Green Supermix (Bio-Rad) on the CFX96 platform (Bio-Rad). Relative transcript expression was calculated using the ΔΔCt method after normalizing each to 18 S rRNA. The primer sequences used in this study are as followed: Cebpb-F: TCGAACCCGCGGACTGCAAG Cebpb-R: CGACGACGACGTGGACAGGC Id1-F: GAGGCGGCATGTGTTCCA Id1-R: CTCTGGAGGCTGAAAGGTGG Id2-F: GGACTCGCATCCCACTATCG Id2-R: GATGCCTGCAAGGACAGGAT Id3-F: AGCTCACTCCGGAACTTGTG Id3-R: GTTCAGTCCTTCTCTCGGGC Myog-F: ATCGCGCTCCTCCTGGTTGA Myog-R: CTGGGGACCCCTGAGCATTG 18s-F: CGCCGCTAGAGGTGAAATC 18s-R: CCAGTCGGCATCGTTTATGG
Chromatin Immunoprecipitation
C2C12 myoblasts were crosslinked with 1% formaldehyde for 30 min then sonicated for 30 cycles (30 sec ON/OFF) using Diagenode Bioruptor®. Equal amounts of chromatin was used to perform Immunoprecipitation (ChIP) analysis as previously described23 using C/EBPβ (C-19, Santa Cruz Biotech, Sc-150) or rabbit polyclonal IgG (Invitrogen). Data are presented as copy numbers as compared to a standard curve that was generated using 10% input of each sample. The primer sequences used are as followed: Id3_−26kb (chr4: 135, 672, 285-135, 674, 188) F:GGCTGTTCGTTGACCTTGTTT R: AGGGAATCGTGACGGTTGG, Id3_−20 kb (chr4: 135, 678, 270-135, 680, 185) F: TTCGAAAGGCTTCCGGGCTAA R: TCCCTGCGACCCAAAGCTTAC, Id3_promoter (chr4: 135, 698, 261-135, 700, 171) F: AGTTCTCGGTGGAAACGGTC R:CTAGGCGCTGAGATTGCAGA, Id1_promoter (chr2: 152, 736, 188-152, 736, 308) F: TTTGAACGTTCTGAACCCGC R: GGCTGAGAACAGAGTGTGGG, Negative region (chr11: 71, 360, 398-71, 360, 930) F: TCCCAGCTCACAGGCTAGAA R: AATGCAGAGCAGAAGGGGTC.
Luciferase Assay
The −934/ + 13 bp Id3-luciferase reporter construct was kindly provided by Dr. Andrew Lasser30. Briefly, C2C12 cells were transiently transfected with the Id3-luc reporter construct and a constitutively active RSV-β-galactosidase reporter in the presence or absence of mammalian expression plasmid for C/EBPβ using FuGENE HD (Promega). Cells were collected 48 h post-transfection to assess luciferase activity using the Dual-Luciferase Reporter Assay kit (Promega) according to manufacturer’s instructions. Luciferase activity was measured using a Monolight 2010 luminometer (Analytical Luminescence laboratory) and corrected for transfection efficiency with β-gal activity.
Statistical Analysis
Two means were compared by Student’s t-test for a minimum of three biological repeats. For multiple means, one-way or two-way ANOVA was conducted with appropriate post hoc test. *p < 0.05, **p < 0.01, ***p < 0.001. Other comparisons are explained in the relevant legends. For multiple comparisons, means indicated by different letters or symbols are statistically different from one another with a p < 0.05 as a minimum cutoff.
Electronic supplementary material
Acknowledgements
This work was supported by grant from the Canadian Institutes of Health Research (CIHR) to NWB. HA is supported by a graduate scholarship from King Saud University, Saudi Arabia. NLT had a scholarship from the University of Ottawa Brain and Mind Research Institute (uOBMRI). The Pax7-CreER mouse was kindly provided by Dr. Charles Keller at Oregon Health & Science University (Portland, OR), and the Cebpbfl/fl mouse was a kind gift from Dr. Esta Sternck at the Center for Cancer Research at the National Institutes of Health. The pGL3-Id3 was a kind gift from Dr. Andrew Lassar at Harvard Medical School (Boston, MA).
Author Contributions
H.A. contributed to the conception and design, collection, assembly, analysis and interpretation of data, the manuscript writing and final approval of the manuscript. N.L.T. contributed to the collection, assembly and the analysis of the data, the manuscript writing and final approval of the manuscript. N.W.B. contributed to the conception and design, analysis and interpretation of data, financial support, the manuscript writing, and the final approval of manuscript.
Data Availability
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files)
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34871-0.
References
- 1.Bischoff R. Regeneration of single skeletal muscle fibers in vitro. Anat. Rec. 1975;182:215–235. doi: 10.1002/ar.1091820207. [DOI] [PubMed] [Google Scholar]
- 2.Mauro A. Satellite cell of skeletal muscle fibers. J Biophys Biochem Cytol. 1961;9:493–495. doi: 10.1083/jcb.9.2.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weintraub H, et al. The myoD gene family: nodal point during specification of the muscle cell lineage. Science. 1991;251:761–766. doi: 10.1126/science.1846704. [DOI] [PubMed] [Google Scholar]
- 4.Asfour HA, Allouh MZ, Said RS. Myogenic regulatory factors: The orchestrators of myogenesis after 30 years of discovery. Exp. Biol. Med. 2018;243:118–128. doi: 10.1177/1535370217749494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornelison DD, Wold BJ. Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev Biol. 1997;191:270–283. doi: 10.1006/dbio.1997.8721. [DOI] [PubMed] [Google Scholar]
- 6.Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: A negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. doi: 10.1016/0092-8674(90)90214-Y. [DOI] [PubMed] [Google Scholar]
- 7.Sun XH, Copeland NG, Jenkins NA, Baltimore D. Id proteins Id1 and Id2 selectively inhibit DNA binding by one class of helix-loop-helix proteins. Mol. Cell. Biol. 1991;11:5603–11. doi: 10.1128/MCB.11.11.5603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Christy BA, et al. An Id-related helix-loop-helix protein encoded by a growth factor-inducible gene. Proc. Natl. Acad. Sci. USA. 1991;88:1815–9. doi: 10.1073/pnas.88.5.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jen Y, Weinttaub H, Benezra R. Overexpression of Id protein inhibits the muscle differentiation program: In vivo association of Id with E2A proteins. Genes Dev. 1992;6:1466–1479. doi: 10.1101/gad.6.8.1466. [DOI] [PubMed] [Google Scholar]
- 10.Davis RL, Cheng P-F, Lassar AB, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-V. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama S, Asahara H. The myogenic transcriptional network. Cell. Mol. Life Sci. 2011;68:1843–1849. doi: 10.1007/s00018-011-0629-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melnikova I, Christy B. Muscle Cell Differentiation Is Inhibited by the Helix-Loop-Helix Protein Id3. Cell growth Differ. 1996;7:1067–1079. [PubMed] [Google Scholar]
- 13.Atherton GT, Norton JD, Travers H. Regulation of Cell Differntiation in C2C12 Myoblasts by the Id3 Helix-Loop-Helix Protein. Cell growth Differ. 1996;7:1059–1066. [PubMed] [Google Scholar]
- 14.Melnikova IN, Bounpheng M, Schatteman GC, Gilliam D, Christy B. a. Differential biological activities of mammalian Id proteins in muscle cells. Exp. Cell Res. 1999;247:94–104. doi: 10.1006/excr.1998.4330. [DOI] [PubMed] [Google Scholar]
- 15.Tsukada J, Yoshida Y, Kominato Y, Auron PE. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54:6–19. doi: 10.1016/j.cyto.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 16.Ramji DPD, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/bj20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao Z, Umek RM, McKnight SL. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991;5:1538–1552. doi: 10.1101/gad.5.9.1538. [DOI] [PubMed] [Google Scholar]
- 18.Wiper-Bergeron N, St-Louis C, Lee JM. CCAAT/Enhancer binding protein beta abrogates retinoic acid-induced osteoblast differentiation via repression of Runx2 transcription. Mol. Endocrinol. 2007;21:2124–2135. doi: 10.1210/me.2006-0452. [DOI] [PubMed] [Google Scholar]
- 19.Darlington GJ, Ross SE, MacDougald OA. The role of C/EBP genes in adipocyte differentiation. J Biol Chem. 1998;273:30057–30060. doi: 10.1074/jbc.273.46.30057. [DOI] [PubMed] [Google Scholar]
- 20.Wiper-Bergeron N, et al. Glucocorticoid-stimulated preadipocyte differentiation is mediated through acetylation of C/EBPbeta by GCN5. Proc. Natl. Acad. Sci. USA. 2007;104:2703–2708. doi: 10.1073/pnas.0607378104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu D, Lala-Tabbert N, Lee H, Wiper-Bergeron N. Mdm2 promotes myogenesis through the ubiquitination and degradation of CCAAT/Enhancer-binding protein β. J. Biol. Chem. 2015;290:10200–10207. doi: 10.1074/jbc.M115.638577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchildon F, et al. CCAAT/enhancer binding protein beta is expressed in satellite cells and controls myogenesis. Stem Cells. 2012;30:2619–2630. doi: 10.1002/stem.1248. [DOI] [PubMed] [Google Scholar]
- 23.Lala-Tabbert N, AlSudais H, Marchildon F, Fu D, Wiper-Bergeron N. CCAAT/enhancer binding protein beta is required for satellite cell self-renewal. Skelet. Muscle. 2016;6:40. doi: 10.1186/s13395-016-0112-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saisanit S, Sun XH. Regulation of the pro-B-cell-specific enhancer of the Id1 gene involves the C/EBP family of proteins. Mol Cell Biol. 1997;17:844–850. doi: 10.1128/MCB.17.2.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu M, Nie L, Kim SH, Sun XH. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPβ. EMBO J. 2003;22:893–904. doi: 10.1093/emboj/cdg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karaya K, et al. Regulation of Id2 expression by CCAAT/enhancer binding protein beta. Nucleic Acids Res. 2005;33:1924–1934. doi: 10.1093/nar/gki339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pajcini KV, Corbel SY, Sage J, Pomerantz JH, Blau HM. Transient inactivation of Rb and ARF yields regenerative cells from postmitotic mammalian muscle. Cell Stem Cell. 2010;7:198–213. doi: 10.1016/j.stem.2010.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchildon F, Lamarche É, Lala-Tabbert N, St-Louis C, Wiper-Bergeron N. Expression of CCAAT/Enhancer Binding Protein Beta in Muscle Satellite Cells Inhibits Myogenesis in Cancer Cachexia. PLoS One. 2015;10:e0145583. doi: 10.1371/journal.pone.0145583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giresi PG, et al. Identification of a molecular signature of sarcopenia. Physiol Genomics. 2005;21:253–263. doi: 10.1152/physiolgenomics.00249.2004. [DOI] [PubMed] [Google Scholar]
- 30.Kumar D, Shadrach JL, Wagers AJ, Lassar AB. Id3 Is a Direct Transcriptional Target of Pax7 in Quiescent Satellite Cells. Mol. Biol. Cell. 2009;20:3170–3177. doi: 10.1091/mbc.e08-12-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang Z, Clemens PR. Cellular caspase-8-like inhibitory protein (cFLIP) prevents inhibition of muscle cell differentiation induced by cancer cells. Faseb J. 2006;20:2570–2572. doi: 10.1096/fj.06-6347fje. [DOI] [PubMed] [Google Scholar]
- 32.Langlands K, Yin X, Anandi G, Prochownik EV. Differential interactions of Id proteins with basic helix loop helix transcription factors. J. Biol. Chem. 1997;272:19785–19793. doi: 10.1074/jbc.272.32.19785. [DOI] [PubMed] [Google Scholar]
- 33.Price FD, et al. Inhibition of JAK-STAT signaling stimulates adult satellite cell function. Nat. Med. 2014;20:1174–1181. doi: 10.1038/nm.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its Supplementary Information files)