Abstract
Arabidopsis ABA3 is an enzyme involved in the synthesis of the sulfurated form of the molybdenum (Mo) cofactor (MoCo), which is required for the enzymatic activity of so-called Mo enzymes such as aldehyde oxidase (AO) and xanthine dehydrogenase (XDH). It has been reported that AO and XDH are essential for the biosynthesis of the bioactive compounds, ABA and allantoin, respectively. However, aba3 mutants often exhibit pleiotropic phenotypes that are not explained by defects in ABA and/or allantoin biosynthesis, leading us to hypothesize that ABA3 regulates additional metabolic pathways. To reveal the currently unidentified functions of ABA3 we compared transcriptome and metabolome of the Arabidopsis aba3 mutant with those of wild type and a typical ABA-deficient mutant aba2. We found that endogenous levels of anthocyanins, members of the flavonoid group, were significantly lower in the aba3 mutant than in the wild type or the aba2 mutant under oxidative stress. In contrast, mutants defective in the AO and XDH holoenzymes accumulated significantly higher levels of anthocyanins when compared with aba3 mutant under the same conditions. Our findings shed light on a key role of ABA3 in the ABA- and allantoin-independent accumulation of anthocyanins during stress responses.
Introduction
Molybdenum (Mo) is an essential metal for nearly all organisms because it is incorporated into pterin-based cofactors (Mo cofactor; MoCo) and functions as the catalytic center of enzymes involved in fundamental metabolic processes1. Deficiencies in MoCo result in severe pleiotropic phenotypes in both plants and animals2–6, indicating that MoCo, and hence Mo enzymes, play important roles in a wide range of biochemical and physiological processes.
The main pathway of MoCo biosynthesis is conserved across kingdoms1. It starts with the production of metal-binding pterin (MPT) from GTP and then Mo is inserted into MPT to form MoCo7,8. Among more than 40 Mo enzymes identified to date, four types of enzymes, nitrate reductase (NR)9, sulfite oxidase (SO)10, xanthine oxidase/dehydrogenase (XO/XDH)11, and aldehyde oxidase (AO)12, have been well characterized in plants. More recently, orthologous genes for mitochondrial amidoxime reducing component (mARC), which was identified as a Mo-containing enzyme in pig liver, were found in the Arabidopsis genome13,14. Based on the chemical structures of MoCos, these Mo enzymes are classified into two groups: the NR/SO/mARC family, whose Mo center forms a covalent bond with a strictly conserved cysteine residue, and the XDH/AO family, whose Mo forms a double bond with a terminal sulfur using a free cysteine as the sulfur donor. The modification reaction allowing MoCo to be utilized for AO and XDH is catalyzed by MoCo sulfurase (MOCOS or MCSU) (Fig. 1a).
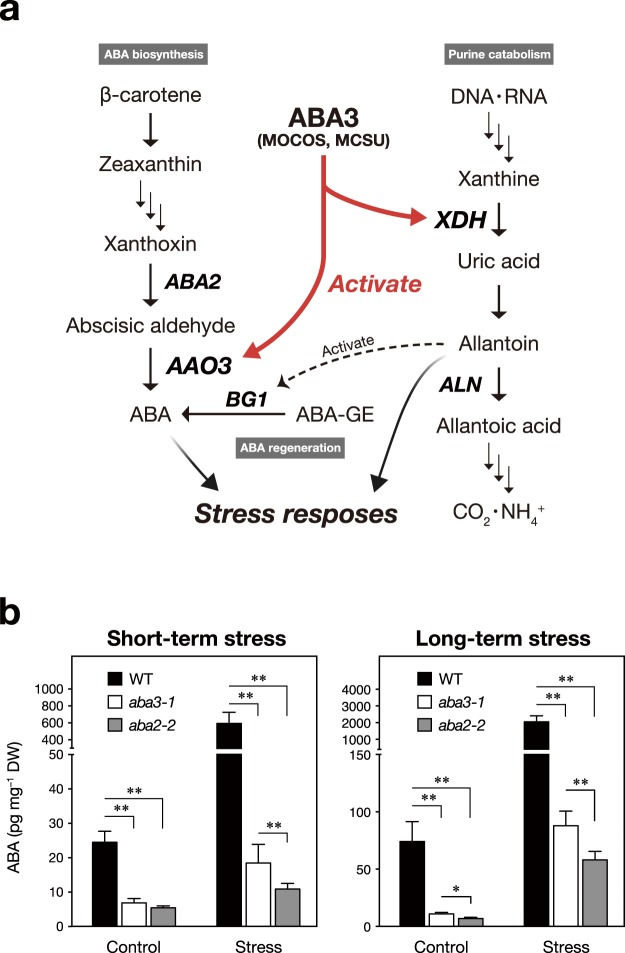
Figure 1.
Physiological functions of the Arabidopsis ABA3 enzyme in stress responses. (a) A schematic illustration of the ABA3-regulated metabolic pathways. (b) Endogenous ABA levels in wild-type (WT), aba3-1 and aba2-2 plants under short-term and long-term stress conditions. Values are means (±SD) of five biological replicates (*P < 0.01, **P < 0.001 by Tukey’s multiple comparison test).
It has been suggested that AO plays important roles in the detoxification of xenobiotics, such as drugs and pollutants, in animals15–17. In contrast, the involvement of plant AOs in response to environmental changes, including various stresses, has been well documented. For example, it was predicted that the de novo biosynthesis of the phytohormone ABA requires AOs from observations that mutants lacking AO activities, due to MoCo deficiencies, contained reduced endogenous ABA levels18,19. In Arabidopsis, the ABA-deficient aba3 mutant was originally isolated based on its ability to germinate in the presence of the gibberellin (GA) biosynthesis inhibitor paclobutrazol20, and subsequent studies showed that the causal gene (ABA3) encodes MOCOS21,22. There are four genes encoding AOs in Arabidopsis (AAO1–4)23. It has been demonstrated that the AAO3 enzyme catalyzes the oxidation of abscisic aldehyde to produce ABA24,25, and a mutation in the AAO3 gene resulted in reduction of endogenous ABA levels with increased transpiration rates and lower resistance to water shortages24,25. In contrast, the AAO1 enzyme was reported to be involved in the metabolism of an indolic compound, camalexin, which is thought to contribute to defense responses against pathogen infection26. It has also been reported recently that the AAO4 enzyme plays an important role in silique senescence, although the substrate(s) of the enzyme is unknown27.
Studies on leguminous plants such as soybean revealed that XDH is an essential enzyme in the catabolism of purine compounds synthesized from ammonium produced by root nodule bacteria to form nitrogen-rich metabolites, such as allantoin and allantoic acid, that can be translocated to sink tissues and utilized as major nitrogen sources28. RNA interference targeting of two Arabidopsis XDH genes (AtXDH1 and AtXDH2) caused typical symptoms of nitrogen deficiency, including growth retardation, early senescence, and reduced fertility, indicating that purine catabolism mediated by XDH also plays important roles in nitrogen recycling in non-leguminous plants29. It has also been reported that XDH plays important roles in response to prolonged dark periods30, water deficit31 and pathogen attack32. These functions of XDH can be explained partly by the activities of allantoin to scavenge reactive oxygen species (ROS)30,33 and/or to activate the β-glucosidase BGLU18 enzyme that catalyzes the hydrolysis of inactive ABA conjugates (ABA-glucose ester) to produce free bioactive ABA34,35.
Consistent with the roles of Mo enzymes in multiple aspects of plant life, mutants defective in MOCOS, such as Arabidopsis aba3, show pleiotropic phenotypes36. Interestingly, it is also true, however, that the phenotypes observed in the mutants are not always explained by the loss of AO and/or XDH functions. For example, the expression of several stress responsive genes was differentially regulated between aba3 and another ABA-deficient mutant, aba121. It has also been reported that the aba3 mutant is impaired in chloroplast protein import37. These observations suggest that the MOCOS enzyme has uncharacterized functions. Since MOCOS is essential for activating several Mo enzymes, we hypothesized that it might be involved in the metabolism of currently unidentified bioactive compounds.
In this study, we conducted comparative analyses of the transcriptome and metabolome using the Arabidopsis aba3 mutant and a typical ABA-deficient mutant aba2. ABA2 encodes a short-chain dehydrogenase/reductase (SDR) which catalysis the conversion of xanthoxin to abscisic aldehyde. Like ABA3, ABA2 is encoded by singe gene in Arabidopsis. However, the functions of ABA2 other than ABA biosynthesis have not been proposed. We revealed that anthocyanin biosynthesis was differentially regulated between the two mutants. Possible roles of MOCOS in ABA- and allantoin-independent stress responses are discussed.
Results
ABA-dependent and -independent transcriptional responses to osmotic stress mediated by Arabidopsis MOCOS ABA3
It has been reported that the Arabidopsis MOCOS (ABA3) enzyme plays important roles in abiotic stress responses21. In this study, we explored currently unidentified functions of ABA3 under stress. Mutations in the ABA3 gene reduce endogenous ABA content, as do mutations in other ABA biosynthesis genes, such as ABA1 and ABA220,38. ABA1 (zeaxanthin epoxidase) and ABA2 [xanthoxin dehydrogenase, also referred to as short-chain dehydrogenase/reductase (SDR)], as well as ABA3, are each encoded by a single gene. ABA1 is involved in the xanthophyll cycle in addition to ABA biosynthesis, whereas additional functions of ABA2 other than ABA biosynthesis have not been reported. Thus, we compared genome-wide gene expression between aba3 (aba3-1), aba2 (aba2-2) and wild-type (WT) plants to distinguish genes that are regulated by ABA3, both dependent and independent of ABA. A public microarray database (eFP browser, http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi 39); indicated that ABA3 mRNA levels were rapidly increased within 3 h of treatment with polyethylene glycol (PEG) solution, which induces osmotic stress, and the levels were sustained for up to 24 h (Supplementary Fig. S2). We therefore conducted microarray analysis with seedlings that were immersed in PEG solution (25%, c. −1.25 MPa) for 3 h to monitor the initial stages of the response, which we termed short-term response (Fig. 1b and Supplementary Fig. S3). In addition, to determine the roles of ABA3 during long-term stress responses, we analyzed the transcriptome after incubation of seedlings on low water potential (−1.2 MPa) agar plates40 for 8 h (Fig. 1b and Supplementary Fig. S3): it has been reported that endogenous ABA content and expression of ABA-responsive genes attained maximum levels after 8–10 h under similar conditions40,41. We confirmed that wild-type plants subjected to short-term and long-term stress accumulated greater than 10 times the level of ABA compared to plants from control treatments (Fig. 1b). Note that ABA levels in wild type plants were not equivalent between the two control conditions, possibly because different treatments were applied (liquid media for short-term stress and solid media for long term stress). Although we observed a slight increase in ABA content in stressed aba3 and aba2 mutant plants, the levels were much lower than those in wild-type plants in all conditions tested.
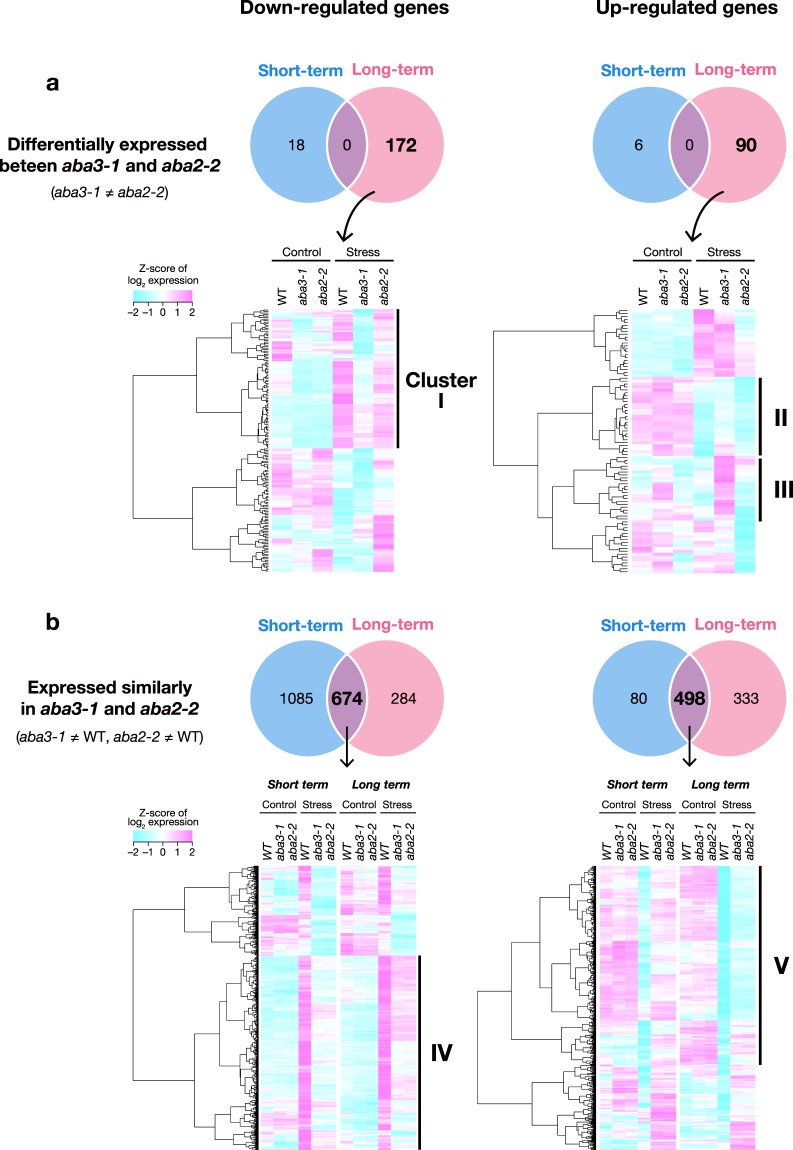
We first selected genes that were differentially expressed (more than 1.5-fold) between aba3 and aba2 mutant plants under osmotic stress (Fig. 2). Only a small number of genes (18 and six down-regulated and up-regulated genes, respectively) were differentially expressed between aba3 and aba2 after the short-term stress treatment. In contrast, 262 genes (172 down- and 90 up-regulated genes) were identified as differentially expressed genes (DEGs) during the long-term stress treatment (Fig. 2a).
Figure 2.
Genome-wide gene expression under short-term and long-term stress. (a) Genes that are differentially expressed between aba3-1 and aba2-2 plants in response to osmotic stress (aba3-1 ≠ aba2-2). Numbers of genes whose expression levels were significantly different (±1.5 fold) between aba3-1 and aba2-2 plants under short-term or long-term stress are shown in the Venn diagrams (upper). The identified DEGs were classified into several clusters based on their expression in wild-type (WT), aba3-1 and aba2-2 plants by hierarchical clustering analysis (lower). (b) Genes that are expressed similarly in aba3-1 and aba2-2 plants but differentially between aba3-1 and wild-type plants (aba3-1 ≠ WT), and between aba2-2 and wild-type plants (aba2-2 ≠ WT). Numbers of genes whose expression levels were significantly different (±1.5 fold) in both aba3-1 and aba2-2 plants, compared to wild type, under short-term or long-term stress are shown in the Venn diagrams (upper). The identified DEGs were classified into several clusters based on their expression in wild-type (WT), aba3-1 and aba2-2 plants by hierarchical clustering analysis (lower).
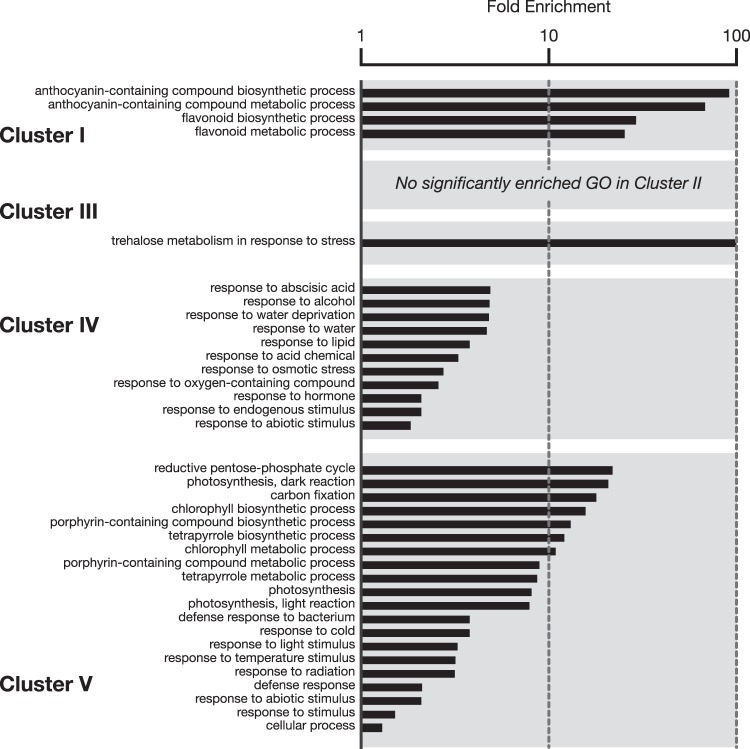
We then used hierarchical clustering analysis to classify the DEGs (independently for down- and up-regulated genes) from the long-term stress response into several groups according to their expression patterns in wild-type, aba3 and aba2 plants under both non-stress and stress conditions. Using this approach, we also identified genes that were differentially expressed between wild-type and aba3 plants (Cluster I, II and III in Fig. 2a): these genes may require functional ABA3 but not ABA for their correct expression. Interestingly, GO enrichment analysis showed that in Cluster I, genes related to flavonoid and anthocyanin biosynthesis were enriched significantly (Fig. 3 and Supplementary Table S2). Cluster I is composed of 91 genes down-regulated specifically in aba3 plant under stress. Expression of the genes in Cluster II were down-regulated by osmotic stress, however, the levels were relatively high in aba3 compared with those in wild-type and aba2 plants. Cluster III was composed of genes whose expression was not changed in wild-type plants under osmotic stress, but of genes whose expression was up-regulated in the aba3 mutant in the presence and/or absence of osmotic stress. Although genes involved in trehalose metabolism were over-represented significantly in Cluster III, no significant enrichment in GO terms was observed for Cluster II.
Figure 3.
Functional classification of DEGs. DEGs in each cluster presented in Fig. 2 were classified according to the functional categories of the GO biological processes. Values are fold enrichment of each GO term (FDR, P < 0.05).
We also identified genes that were differentially expressed in both aba3 and aba2 mutants when compared with wild-type plants (Cluster IV and V): these genes are potentially regulated in an ABA-dependent manner (Fig. 2b). As expected, GO enrichment analysis showed that genes involved in abiotic stress responses, including ABA-responsive genes, were over-represented in these clusters (Fig. 3). In total, more than 2900 genes were identified as DEGs under either short-term or long-term stress.
ABA-dependent and -independent metabolomic responses to osmotic stress mediated by the ABA3 enzyme
To further explore the unknown functions of the ABA3 enzyme during stress responses, we performed metabolome analysis in addition to transcriptome analysis. The impact of the aba3 mutation on the endogenous levels of more than 500 primary and secondary metabolites was investigated using a widely targeted metabolomics platform, as described in42. As a result, 19 compounds were found to be differentially accumulated (more than 1.5-fold) in aba3 plants, compared to both wild-type and aba2 plants, under short-term and/or long-term stress. These compounds were named differentially accumulated metabolites (DAMs) (Supplementary Fig. S4 and Supplementary Table S3). As expected from previous reports22,29,43, aba3 plants under non-stress and stress (both short-term and long-term) conditions specifically accumulated high levels of xanthine, which is the substrate of XDH, whereas endogenous levels of allantoic acid, a downstream metabolite of xanthine, were greatly reduced. In addition, a purine metabolite, 3’,5’-cyclic guanosine monophosphate (cGMP), was accumulated at lower levels in aba3 plants irrespective of the presence or absence of stress treatments. Interestingly, we also identified several compounds related to flavonoid and anthocyanin biosynthesis (shikimic acid, quercitrin, and quercetin-Rha-Glc-Rha) as DAMs. These results support the observation that genes related to flavonoid and anthocyanin biosynthesis were differentially expressed in aba3 plants compared to wild-type and aba2 plants.
ABA3 has an important role in activating the later steps of the flavonoid biosynthesis pathway
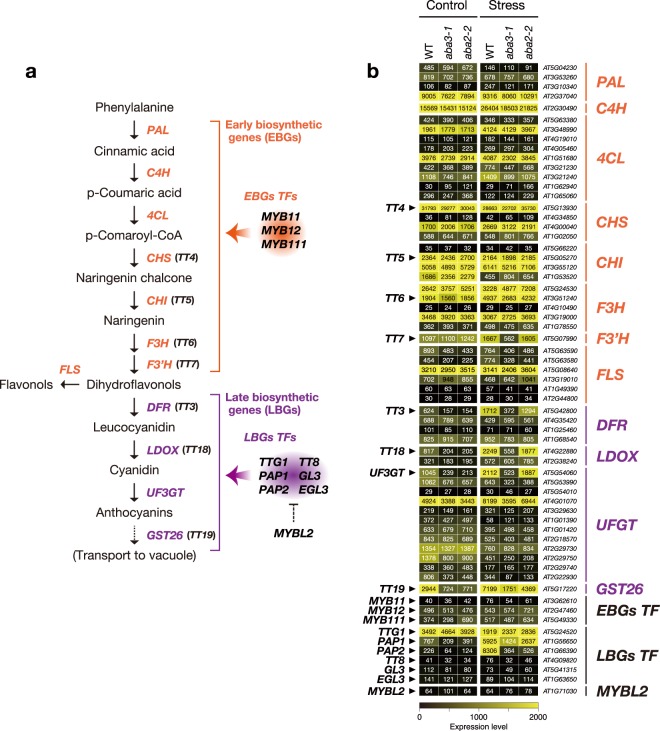
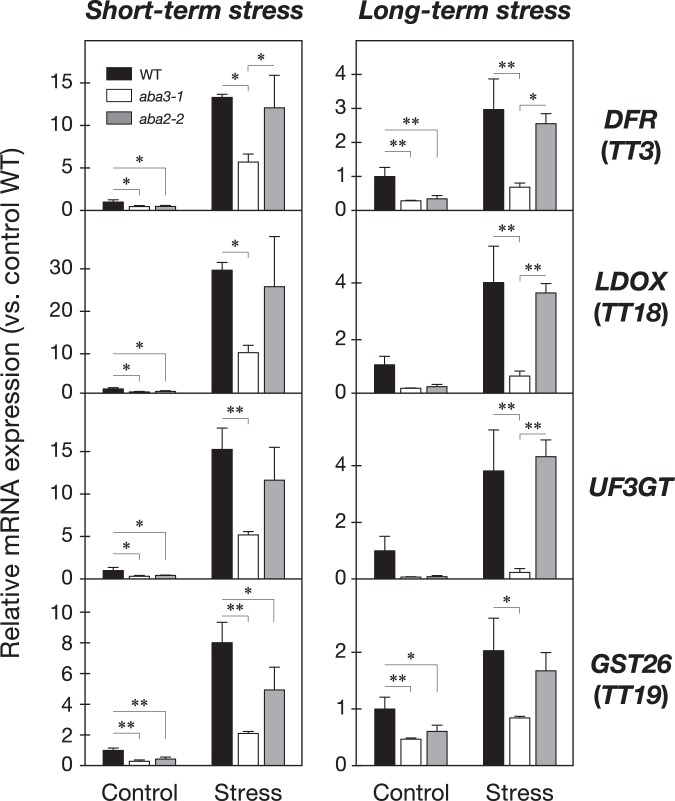
Based on the results obtained from the transcriptome and metabolome analyses, we focused on the role of the ABA3 enzyme in the regulation of flavonoid biosynthesis during stress responses. The expression levels of 57 genes encoding 47 enzymes and 10 transcription factors, involved in the 10 reactions of the main flavonoid biosynthesis pathway determined by microarray analysis, were compared among wild-type, aba3 and aba2 plants before and after long-term stress treatment (Fig. 4a,b). Interestingly, we found that transcript levels of four TRANSPARENT TESTA genes TT7, TT3, TT18 and TT19, which encode flavonoid 3’-hydroxylase (F3’H), dihydroflavonol 4-reductase (DFR), leucoanthocyanidin dioxygenase (LDOX), and glutathione S-transferase 26 (GST26), respectively, were reduced specifically in aba3 plants. In addition, a gene encoding UDP-glucosyltransferase (UDP glucose-flavonoid 3-O-glucosyltransferase; UFGT) (AT5G54060), involved in glycosylation of flavonoids, was expressed at lower levels in aba3 plants than in wild-type and aba2 plants under stress (see the list of down-regulated DEGs in Supplementary Table S2). Further qRT-PCR analysis confirmed that the induction of four genes (TT3, TT18, TT19 and AT5G54060) under short-term and long-term stress conditions, observed in wild-type and aba2 plants, was reduced in aba3 plants (Fig. 5). Interestingly, the four genes are located in the later steps of the flavonoid biosynthesis pathway and are known to play central roles in the production and accumulation of anthocyanins in vegetative tissues in response to environmental cues such as poor nutrition, high light, cold, salt, drought, and oxidative stress44–47.
Figure 4.
Effects of ABA3 disruption on flavonoid biosynthesis. (a) Flavonoid biosynthetic pathway. Orange and purple letters indicate the early biosynthetic genes (EBGs) and the late biosynthetic genes (LBGs), respectively. Genes encoding transcription factors (TFs) that regulate the flavonoid biosynthetic pathway are also shown on the right. (b) Expression of genes involved in flavonoid biosynthesis under long-term stress. Values are average signal intensities obtained from three independent microarray data experiments. PAL, Phenylalanine ammonia-lyase; C4H, Cinnamate 4-hydroxylase; 4CL, 4-Coumarate:CoA ligase; CHS, Chalcone synthase; CHI, chalcone isomerase; F3H, flavonol 3-hydroxylase; F3′H, flavonol 3′-hydroxylase; FLS, flavonol synthase; DFR, dihydroflavonol-4-reductase; LDOX, leucoanthocyanidin dioxygenase; UF3GT, UDP-glucose flavonoid-3-O glucosyltransferase; GST26, Glutathione S-transferase 26.
Figure 5.
Expression levels of LBGs under short-term and long-term stress. Relative expression levels of four LBGs in wild-type (WT), aba3-1 and aba2-2 plants were analyzed by qRT-PCR. Values are means (±SD) of three biological replicates (*P < 0.05, **P < 0.01 by Tukey’s multiple comparison test). DFR, dihydroflavonol-4-reductase; LDOX, leucoanthocyanidin dioxygenase; UF3GT, UDP-glucose flavonoid-3-O glucosyltransferase; GST26, Glutathione S-transferase 26.
ABA3 contributes to anthocyanin accumulation under oxidative stress
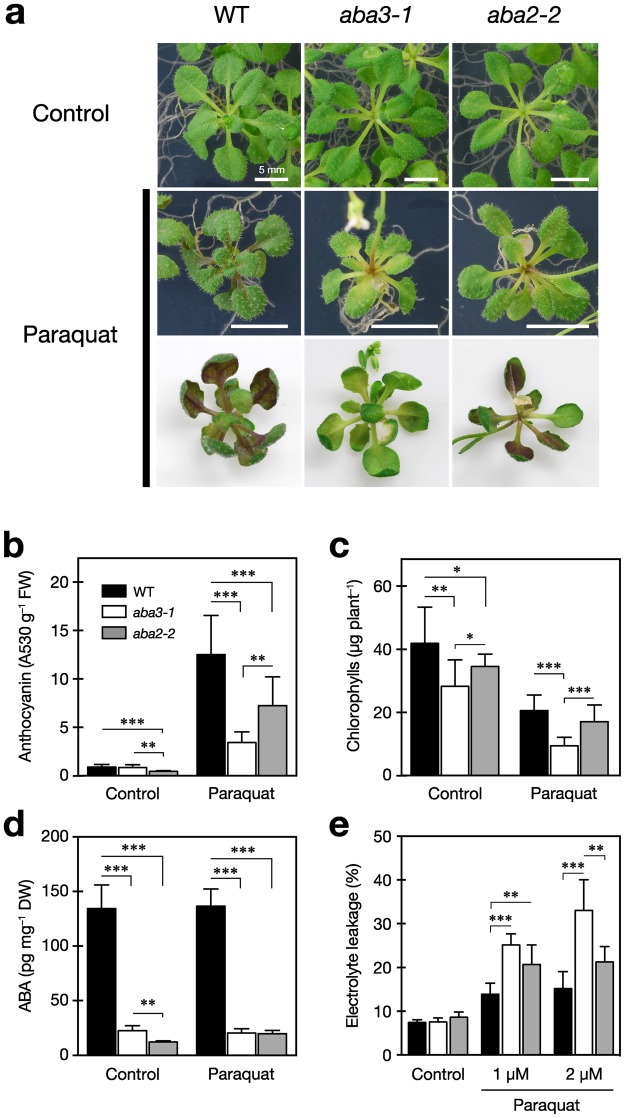
Previous in vitro and in vivo studies have shown that flavonoid compounds, including their glycosides such as anthocyanins, have various bioactivities48–52. Of particular relevance to this study, it has been reported that anthocyanins exert antioxidant activities against ROS generated by stresses53,54. In Arabidopsis, it has been reported that oxidative stress markedly induced anthocyanin accumulation with the up-regulation of genes involved in the later steps of flavonoid biosynthesis, namely late biosynthesis genes (LBGs) such as TT3 and TT18 (Fig. 4a)55. Thus, we speculated that expression of LBGs under osmotic stress might be related to protection against oxidative stress. Since mRNA levels of LBGs were lower in aba3 plants compared to wild-type and aba2 plants under stress, we quantified endogenous levels of anthocyanins in wild-type, aba3 and aba2 plants that had been subjected to oxidative stress. After two weeks of oxidative stress treatment induced by paraquat, which is a well-known ROS generator, wild-type plants accumulated considerable amounts of anthocyanin pigments in their leaves (Fig. 6a,b). Anthocyanin levels in aba2 plants were slightly lower than those in wild-type plants before and after paraquat treatment, suggesting that ABA promotes anthocyanin accumulation to some extent. However, interestingly, anthocyanins accumulated at much lower levels in aba3 plants, compared with wild-type and aba2 plants, after oxidative stress treatment. In addition to the reduced anthocyanin pigmentation, aba3 plants exhibited a typical symptom of oxidative injuries, reduced chlorophyll content. This was significantly lower in aba3 plants than in wild-type and aba2 plants, especially under oxidative stress (Fig. 6c). Consequently, we determined cell membrane stability to estimate cellular damage caused by the stress and observed a higher rate of electrolyte leakage in aba3 plants compared to that in both wild-type and aba2 plants (Fig. 6e). Contrary to these aba3-specific phenotypes related to oxidative stress, the endogenous ABA content in aba3 plants was reduced to a similar level to that of aba2 plants, when compared with wild-type plants, in the presence or absence of oxidative stress (Fig. 6d). Finally, we confirmed that the phenotypes observed in aba3 (aba3-1) plants were not allele-specific phenomena (Supplementary Fig. S5). These combined results suggest that the ABA3 enzyme regulates anthocyanin accumulation and hence oxidative stress tolerance independently from its role in ABA biosynthesis. Similarly, exogenous ABA application did not complement the aba3-specific phenotypes although the treatment enhanced paraquat-induced accumulation of anthocyanins (Supplementary Fig. S6a). It is also known that ABA3 is essential to produce allantoin, another bioactive small molecule involved in plant responses to abiotic stresses30,35. However, exogenous application of allantoin did not affect the phenotype observed in aba3 plants, suggesting that this compound, like ABA, is also not involved in ABA3-specific stress responses (Supplementary Fig. S6b).
Figure 6.
Reduced anthocyanin accumulation in aba3-1 plants under oxidative stress. (a) Representative photos of wild-type (WT), aba3-1, and aba2-2 seedlings after paraquat treatment. Twelve-d-old seedlings were transferred onto 1/2MS media containing 1 µM paraquat and photos were taken two weeks later (bars; 5 mm). (b) Anthocyanin contents in the aerial parts of wild-type (WT), aba3-1, and aba2-2 seedlings after paraquat treatment. (c) Chlorophyll contents in the aerial parts of WT, aba3-1, and aba2-2 seedlings after paraquat treatment. (d) ABA contents in the aerial parts of WT, aba3-1, and aba2-2 seedlings after paraquat treatment. (e) Electrolyte leakage from the shoots of WT, aba3-1, and aba2-2 seedlings after paraquat treatment. Values are the mean (±SD) of at least three biological replicates (*P < 0.05, **P < 0.01, ***P < 0.001 by Tukey’s multiple comparison test).
AO and XDH are not involved in the ABA3-dependent accumulation of anthocyanins under oxidative stress
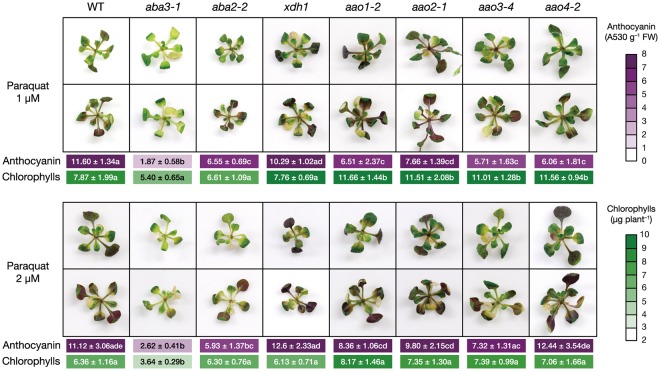
AO and XDH are enzymes that require sulfurated MoCo, which is produced by the reaction catalyzed by MOCOS (encoded by ABA3 in Arabidopsis), as a prosthetic group22. There are four and two genes encoding AO and XDH, respectively, in Arabidopsis. In addition to its role in ABA biosynthesis, AO catalyzes the oxidation of a variety of aldehydes present in plants. The loss of XDH function not only results in the reduction of allantoin levels but also has a more global effect on purine metabolism. Therefore, we hypothesized that additional functions of AO or XDH might be involved in the ABA-independent oxidative stress responses mediated by the ABA3 enzyme. Consequently, we investigated the effects of oxidative stress on mutants defective in each of four AOs (aao1, aao2, aao3 and aao4) and in the major XDH isoform, XDH1 (xdh1) (Leon-Kloosterziel, et al.20 Watanabe, et al.35 Nambara, et al.56 Seo, et al.57 and Supplementary Fig. S1). We found that all these mutants contained significantly higher levels of anthocyanins and chlorophylls than aba3 plants after paraquat treatment (Fig. 7). These results suggest that ABA3 plays a role in anthocyanin accumulation in response to oxidative stress or in protection of chlorophyll from oxidative stress even in the absence of AO and XDH.
Figure 7.
Anthocyanin and chlorophyll contents in mutants defective in Mo enzymes. Twelve-d-old seedlings of wild-type (WT), aba3-1, aba2-2, xdh1, aao1-2, aao2-1, aao3-4, and aao4-2 were transferred onto 1/2MS medium containing 1 µM or 2 µM paraquat and anthocyanin and chlorophyll contents in the aerial parts were determined two weeks later. Representative photos of the mutant seedlings were also taken two weeks after paraquat treatment. Values are the mean (±SD) of three biological replicates. Different letters indicate statistically significant differences (P < 0.05) by Tukey’s multiple comparison test.
Discussion
It is well known that the Arabidopsis ABA3 enzyme plays an essential role in ABA biosynthesis because the enzyme (AO) that catalyzes the last step of ABA biosynthesis requires sulfurated MoCo, synthesized by ABA3, for its enzymatic activity25. It was also reported recently that XDH, whose activity also depends on sulfurated MoCo, affects ABA accumulation indirectly35. In the present study, we first conducted transcriptome analyses using wild type, aba3 and a typical ABA-deficient mutant aba2 to elucidate currently unidentified ABA-independent physiological functions of ABA3. ABA2 is a member of the SDR family, which is composed of 56 gene products in Arabidopsis, however, the enzyme specifically involved in ABA biosynthesis appears to be encoded by a single gene (ABA2)58. Although ABA3 is also encoded by a single gene, endogenous ABA levels in aba3-1 plants were slightly higher than those in aba2-2 plants (Fig. 1b). It is possible that a shunt pathway via abscisic alcohol contributed to the accumulation of ABA in aba3-1 plants to some extent59. Comparison of the transcriptome between wild-type, aba3 and aba2 plants revealed that many genes were commonly up- and down-regulated by osmotic stress in aba3 and aba2 plants compared to wild type (Fig. 2b). This suggests that reduced endogenous ABA levels in aba3 and aba2 plants had a significant impact on global gene expression under the stress conditions studied here. Correspondingly, genes that are related to ABA and/or stress responses were significantly enriched in the gene clusters (Fig. 3). Nevertheless, we also identified genes whose expression was differentially regulated between aba3 and aba2 plants under osmotic stress (Fig. 2a). These genes may be regulated in an ABA3- or ABA2-dependent manner. Further hierarchical clustering analysis identified at least three gene clusters (Cluster I, II and III) whose expression was deregulated in aba3 plants alone. Interestingly, Cluster I, which contained genes whose expression was induced by osmotic stress in wild-type and aba2, but not in aba3 plants, was enriched in GO terms related to the biosynthesis and metabolism of flavonoids and anthocyanins (Fig. 3). The main flavonoid biosynthesis pathway is divided into two parts, early and late (Fig. 4a). Among 34 genes encoding the early biosynthesis pathway enzymes, only one gene (TT7) was clearly expressed at a lower level in aba3 plants than in wild-type or aba2 plants under stress (Fig. 4b). In contrast, under the same stress, four out of 19 LBGs were down-regulated in aba3 plants but not in aba2, compared to wild-type plants (Fig. 4b). Lower expression of the gene encoding a MYB-type transcription factor (TF), PRODUCTION OF ANTHOCYANIN PIGMENT (PAP) 1, in aba3 plants may account for this observation, since PAP1 regulates the expression of LBGs by forming a protein complex composed of a bHLH-type TF [GLABRA3 (GL3), ENHANCER OF GLABRA3 (EGL3) or TT8] and a WD40-type TF (TTG1) (Fig. 4a)60. Consistent with these observations, several compounds related to flavonoid and anthocyanin biosynthesis were differentially accumulated in aba3 plants compared to wild-type and aba2 plants (Supplementary Fig. S4 and Supplementary Table S3). In contrast, expression of a PAP1 homologue, PAP2, was similarly reduced both in aba3 and aba2 plants compared to wild type (Fig. 4b), suggesting that anthocyanin biosynthesis is also regulated by ABA. In fact, it has been reported that endogenous anthocyanin levels increase in response to various stresses and that several mutants defective in ABA biosynthesis or signaling contained reduced anthocyanin levels compared to wild type55,61,62.
It is well known that osmotic stress induces ROS production and hence oxidative stress63. It is also known that anthocyanins have antioxidant activities in vitro and act as a ROS scavenger in plants exposed to osmotic or oxidative stress53–55. Thus, although it is possible that the oxidative damage given to the plants was not equivalent between the PEG treatment and paraquat treatment, we speculated that ABA3 might induce the anthocyanin production in response to ROS accumulation, which leads to protection of the cells from oxidative damage. Our present study showed that the increase in anthocyanin levels in response to oxidative stress, which is normally observed in wild-type plants, was diminished in aba3 plants (Fig. 6a,b). Interestingly, however, endogenous ABA levels in wild type did not increase in response to oxidative stress (Fig. 6d), suggesting that basal levels of ABA present in non-stressed plants affected the stress-induced accumulation of anthocyanins. In contrast, although aba2 plants accumulated significantly higher levels of anthocyanins compared to aba3 plants under oxidative stress, ABA levels in aba3 plants were slightly higher or comparable to those in aba2 plants (Fig. 6d). These results support the hypothesis that anthocyanin accumulation is regulated partly by the ABA3 enzyme, but independently from its role in ABA biosynthesis. In addition to reduced anthocyanin accumulation, aba3 plants exhibited a bleaching phenotype in the presence of paraquat, indicating that the mutant is indeed sensitive to oxidative stress (Fig. 6c).
One reason for the reduced anthocyanin levels in aba3 plants could be the down-regulation of PAP1. However, the mechanisms by which ABA3 regulates its expression are totally unknown. As mentioned above, it is unlikely that ABA could explain all the defects observed in aba3 plants. This idea is supported by the observation that the reduced anthocyanin accumulation in aba3 plants was not restored to the wild-type level by exogenous ABA application (Supplementary Fig. S6). One possible explanation is that the ABA3 enzyme is involved in the production of a bioactive compound that somehow induces anthocyanin accumulation, since MoCo synthesized by ABA3 is required for the activity of several enzymes. It has been reported that allantoin, which is an intermediate in the purine catabolism pathway, promotes the production of free bioactive ABA from its inactive glucose-conjugated form35. Mutants defective in XDH contained reduced allantoin levels and showed stress-sensitive phenotypes that can be partly explained by altered ABA levels. It is possible that allantoin has additional biological activities that enhance anthocyanin biosynthesis. Indeed, allantoin activates the JA response, and JAs play important roles in stress-induced accumulation of anthocyanins32,64,65. Endogenous levels of the bioactive hormone JA-Ile and its precursor JA were altered in aba3 plants compared to wild-type plants (Supplementary Fig. S8). However, their levels were comparable between aba3 and aba2 plants, suggesting that JAs are also not involved in the ABA3-dependent accumulation of anthocyanins. In addition, exogenous application of allantoin did not rescue the aba3-specific reduction in anthocyanin levels (Supplementary Fig. S6). It is also known that xanthine-derived compounds other than allantoin, such as uric acid, have antioxidant activities66. However, the xdh1 mutant accumulated anthocyanins at a similar level to wild-type plants under oxidative stress (Fig. 7). Some aldehyde compounds, which are potential substrates of AO, are also bioactive. Of these, α,β-unsaturated carbonyls, such as acrolein, are regarded as highly reactive aldehydes, named reactive carbonyl species (RCS). RCS function as signaling molecules, but excess amounts of aldehydes are cytotoxic due to their high electrophilicity, leading to protein modification and gene expression associated with stress responses67–70. It has been reported that AAO4 plays a key role in silique senescence by regulating cellular RCS levels during seed development27. However, anthocyanin levels in the aao4 mutant plants, as well as in the aao1, aao2 and aao3 single mutants, were much higher than those in aba3 plants (Fig. 7). Together, these data suggest that AO and XDH are not absolutely required for the ABA3-dependent regulation of anthocyanin biosynthesis. We speculate that there is an unidentified Mo enzyme, although we cannot exclude the possibilities that multiple AO and XDH isoforms regulate this process redundantly. Since ABA3 is induced by osmotic stress and ABA (Xiong, et al.21 and Supplementary Fig. S2), it is possible that the loss of these enzymatic activities altered the ABA3-dependent (but AO and XDH-independent) stress responses. It is worth noting that the cyclic purine nucleotide cGMP, which is known to act as a second messenger like Ca2+, accumulated to a lesser extent in aba3 plants compared to wild-type and aba2 plants under stress and non-stress conditions (Supplementary Fig. S4b and Supplementary Fig. S7c). In several plant species, the involvement of cGMP and its nitrated derivative, 8-nitro-cGMP, has been reported in the regulation of several important physiological processes such as hormone signaling, ion transport, expression of stress-related genes, stomatal movement, and light response47,71–79. Interestingly, it was observed in cultured cells of soybean that the expression of anthocyanin biosynthesis genes was induced in response to exogenous cGMP application, resulting in an increased anthocyanin level80. Thus, it is possible that the lower anthocyanin levels in aba3 plants are due to reduced levels of cGMP, although the mechanisms involved are unknown.
Our metabolome analyses showed that, in addition to anthocyanins, some compounds related to oxidative stress, such as GSH and pyrimidine precursors, differentially accumulated in aba3 plants compared to wild-type and aba2 plants under oxidative stress (Supplementary Fig. S7b,c). Cellular redox imbalance damages DNA, lipids and proteins and triggers ROS signaling and scavenging systems such as the ascorbate-GSH/GSSG cycle81. The central metabolites in pyrimidine biosynthesis, N-Carbamoyl-l-aspartate and orotate, are involved in electron transfer and redox balance in mitochondria82,83. These results suggest that the ABA3 enzyme plays important roles in oxidative stress responses thorough multiple processes. This idea is supported by the results from the multidimensional scaling analysis based on metabolite contents, in which aba3 plants responded to oxidative stress in a very different manner to wild-type and aba2 plants (Supplementary Fig. S7a). Currently, however, it is difficult to discriminate between the primary or direct effects caused by the loss of ABA3 functions and the accompanying secondary or indirect effects.
In conclusion, our study demonstrated the existence of an ABA-independent stress response pathway that is mediated by the ABA3 enzyme and suggested the presence of an unidentified Mo enzyme(s) that produces functional metabolite(s) involved in plant stress responses. We speculate that plants have evolved a system to regulate multiple metabolic pathways with a single enzyme (ABA3) to allow them to respond globally to, and to cope with, their ever-changing environment.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana (L.) Heynh. [accession Columbia-0 (Col-0)] was used as the wild type for all experiments in this study. Mutant lines used in the experiments were as follows: aba3-120, aba3-7 (SALK_054454)84, aba3–8 (SAIL_576_D01), aba2-256, xdh1 (SALK_148366)35, aao1-2 (SALK_069221)26, aao2-1 (SALK_104895; Supplementary Fig. S1), aao3-4 (SALK_072361)57, and aao4-2 (SALK_057531)27. Homozygous aba3-8 and aao2-1 mutants were selected by PCR using primer combinations designed by the T-DNA Primer Design Tool (http://signal.salk.edu/tdnaprimers.2.html).
After surface sterilization with 70% (v/v) ethanol and then with 5% (v/v) NaClO (0.25% active chlorine) containing 1% (w/v) SDS, seeds were sown on half-strength Murashige-Skoog (1/2MS) media with 1% (w/v) sucrose and 0.8% (w/v) agar. To break dormancy, the seeds were incubated at 4 °C for 3 d in the dark, and the plates were placed in growth chambers at 22 °C under long-day conditions (16 h : 8 h, light : dark, 100 µmol photons m−2 s−1).
For osmotic stress treatments, 10-d-old seedlings were floated on MES buffer (pH 5.7) containing 25% (w/v) PEG for 3 h (short-term stress) or placed on PEG-infused 1/2MS plates40 for 8 h (long-term stress). MES buffer and 1/2MS plates were used as controls for the short-term and long-term stress treatments, respectively. For oxidative stress treatments, 12-d-old seedlings were transferred to 1/2MS plates supplemented with 1 µM or 2 µM paraquat and incubated for two weeks under continuous light.
Microarray and qRT-PCR analyses
Total RNA was prepared from whole seedlings using the PureLink Plant RNA reagent (Thermo Fisher Scientific, Waltham, USA) according to the manufacturer’s instructions. For microarray analysis, the quality of RNA was checked with an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, USA). Cy3-labeled complementary RNA (cRNA) probes were synthesized from 400 ng total RNA using the Quick Amp labeling kit (Agilent Technologies). The probes were hybridized to a custom Agilent 8 × 60 k array using the Gene Expression Hybridization kit (Agilent Technologies). Microarray chips were scanned using a Microarray scanner G2505B (Agilent Technologies). Expression levels were normalized by the Robust Multiarray Average (RMA) algorithm provided by the LIMMA package of the Bioconductor project and further analyzed using R statistical software ver. 3.4.0 (R Core Team, Vienna, Austria) and RStudio ver. 1.0.143 (RStudio Team, Boston, USA). Experiments were performed in triplicate, and differentially expressed genes (DEGs) between aba3-1 and aba2-2 mutants were identified by one-way ANOVA with the Benjamini-Hochberg False Discovery Rate (BH-FDR) controlled at 1% followed by the Student’s t-test with the BH-FDR controlled at 5%. Hierarchical clustering analysis with Euclidean distance and Ward linkage, and heat map visualization was performed using the gplots package85 in the R software. Gene ontology (GO) analysis was performed on the Gene Ontology Consortium web site (http://www.geneontology.org)86. All microarray data have been deposited in the Gene Expression Omnibus (GEO) database under the accession number GSE110079. For qRT-PCR analysis, total RNA was reverse-transcribed using the ReverTra Ace qPCR RT Master Mix with the gDNA Remover kit (Toyobo, Osaka, Japan) to generate cDNA. Quantification of mRNA levels was performed using the Mx3000P QPCR System (Agilent Technologies). All PCR amplifications contained 1 x SYBR Green Master Mix (THUNDERBIRD SYBR qPCR Mix; Toyobo), 0.2 µM reverse and forward primers and cDNA in a total volume of 20 µl. The sequences of primers used in this study are presented in Supplementary Table S1. Expression levels were normalized with the levels of 18S rRNA.
Metabolome analysis
Widely targeted metabolome analysis using ultra-performance liquid chromatography coupled with tandem mass spectrometry (UPLC-MS/MS) was performed as described previously42. Samples were extracted with 80% (v/v) methanol containing 0.1% (v/v) acetic acid with internal standards, and targeted metabolites were separated and detected on a Xevo TQ-S LC-MS system (Waters, Mississauga, Canada). Metabolites whose endogenous levels were significantly altered in aba3-1 plants compared with aba2-2 plants were identified by one-way ANOVA with the BH-FDR controlled at 1% followed by the Student’s t-test with the BH-FDR controlled at 5%. Multidimensional scaling analysis based on metabolite contents was conducted using the R software.
Measurement of anthocyanin and chlorophyll content
Endogenous levels of anthocyanins and chlorophylls were measured as described previously87,88 with some modifications. Briefly, the aerial parts of wild-type and mutant plants exposed to oxidative stress for two weeks were harvested and homogenized in 90% methanol using a TissueLyser II (Qiagen, Hilden, Germany). The extracts were mixed with an equal volume of 10% (v/v) acetic acid (for anthocyanins) or 10 x volume of 90% methanol (for chlorophylls). After centrifugation at 15,000 rpm for 10 min at 4 °C, the absorbance of supernatants was measured using the 96-well microplate reader SAFIRE (TECAN, Männedorf, Switzerland). Anthocyanin and chlorophyll contents were calculated according to formulae as previously described87,88.
Hormone measurements
Endogenous ABA, JA, and JA-Ile were extracted with 80% (v/v) acetonitrile containing 1% (v/v) acetic acid from whole wild-type and mutant seedlings after freeze-drying. Hormone contents were determined using a UPLC-MS/MS system consisting of a quadrupole/ time-of-flight tandem mass spectrometer (Triple TOF 5600, SCIEX, Concord, Canada), and a Nexera UPLC system (Shimadzu Corp., Kyoto, Japan) as described previously89.
Measurement of electrolyte leakage
Electrolyte leakage was measured as described previously90. Aerial parts from three wild-type and mutant seedlings were incubated in 5 ml sterilized deionized water for 4 h at room temperature and the conductivities of the solutions were measured with the electrical conductivity meter Twin Cond (HORIBA, Kyoto, Japan). The water containing the plant tissues was then autoclaved at 121 °C for 20 min and the conductivities of the solutions were measured. Electrolyte leakage of the samples (%) was calculated based on the conductivities after autoclaving.
Statistical analysis
One-way ANOVA with the BH-FDR controlled at 1% followed by the Student’s t-test with the BH-FDR controlled at 5% was used to detect DEGs and DAMs. Significant differences among three or more groups were evaluated by Tukey’s multiple comparison test using the statistical software Prism 6 ver. 6 (GraphPad Software Inc., California, USA).
Native PAGE and activity staining
AO activities were determined by in-gel activity staining after native PAGE as described previously24. Total protein was extracted from 10-d-old whole wild-type and mutant seedlings grown on 1/2MS media. Forty µg of protein was separated on 7.5% polyacrylamide gels without SDS, and then the gels were incubated at 30 °C for 60 min in buffer [100 µM potassium phosphate buffer (pH 7.5), 0.1 mM phenazine methosulfate, 0.4 mM 3(4,5-dimethylthiazolyl-2)2,5-diphenyltetrazolium bromide] containing 1 mM 2-naphthaldehyde as a substrate.
Electronic supplementary material
Acknowledgements
This work was supported by a Grant-in-Aid for Japan Society for the Promotion of Science Fellows (JSPS KAKENHI Grant Number 15J09097 to S.W.), a grant from The Japanese Society for Chemical Regulation of Plants (to S.W.) and the Japan Science and Technology Agency (JST), Core Research for Evolutionary Science and Technology (CREST; JPMJCR13B4 to M.Seki).
Author Contributions
S.W. and M.Seo designed the study. S.W. performed most of the experiments. M.T., A.M. and M.Seki contributed to the microarray analysis. M.Sato., Y.S. and M.Y.H. conducted the widely targeted metabolome analysis. Y.K. conducted the phytohormone analysis. S.W., A.S. and M.Seo. analyzed the data. S.W. and M.Seo wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-34862-1.
References
- 1.Schwarz G, Mendel RR, Ribbe MW. Molybdenum cofactors, enzymes and pathways. Nature. 2009;460:839–847. doi: 10.1038/nature08302. [DOI] [PubMed] [Google Scholar]
- 2.Reiss J, Hahnewald R. Molybdenum cofactor deficiency: Mutations in GPHN, MOCS1, and MOCS2. Hum Mutat. 2011;32:10–18. doi: 10.1002/humu.21390. [DOI] [PubMed] [Google Scholar]
- 3.Kugler S, Hahnewald R, Garrido M, Reiss J. Long-term rescue of a lethal inherited disease by adeno-associated virus-mediated gene transfer in a mouse model of molybdenum-cofactor deficiency. Am J Hum Genet. 2007;80:291–297. doi: 10.1086/511281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gabard J, et al. Genetic-Analysis of Nitrate Reductase Deficient Mutants of Nicotiana-Plumbaginifolia - Evidence for 6 Complementation Groups among 70 Classified Molybdenum Cofactor Deficient Mutants. Molecular & General Genetics. 1988;213:206–213. doi: 10.1007/BF00339583. [DOI] [Google Scholar]
- 5.Gupta, U. C. & Gupta, U. C. In Molybdenum in Agriculture (ed. Umesh C. Gupta) Ch. 10, 160-170 (Cambridge University Press 1997).
- 6.Barker, A. V. & Pilbeam, D. J. Handbook of Plant Nutrition. (CRC Press 2007).
- 7.Kaufholdt D, et al. Identification of a protein-protein interaction network downstream of molybdenum cofactor biosynthesis in Arabidopsis thaliana. J Plant Physiol. 2016;207:42–50. doi: 10.1016/j.jplph.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 8.Schwarz G, et al. The molybdenum cofactor biosynthetic protein Cnx1 complements molybdate-repairable mutants, transfers molybdenum to the metal binding pterin, and is associated with the cytoskeleton. Plant Cell. 2000;12:2455–2472. doi: 10.1105/tpc.12.12.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campbell WH. Nitrate Reductase Structure, Function And Regulation: Bridging the Gap between Biochemistry and Physiology. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:277–303. doi: 10.1146/annurev.arplant.50.1.277. [DOI] [PubMed] [Google Scholar]
- 10.Eilers T, et al. Identification and biochemical characterization of Arabidopsis thaliana sulfite oxidase. A new player in plant sulfur metabolism. J Biol Chem. 2001;276:46989–46994. doi: 10.1074/jbc.M108078200. [DOI] [PubMed] [Google Scholar]
- 11.Mendel R-R, Müller AJ. A Common Genetic Determinant of Xanthine Dehydrogenase and Nitrate Reductase in Nicotiana tabacum. Biochemie und Physiologie der Pflanzen. 1976;170:538–541. doi: 10.1016/S0015-3796(17)30254-8. [DOI] [Google Scholar]
- 12.Koshiba T, Saito E, Ono N, Yamamoto N, Sato M. Purification and Properties of Flavin- and Molybdenum-Containing Aldehyde Oxidase from Coleoptiles of Maize. Plant Physiol. 1996;110:781–789. doi: 10.1104/pp.110.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruenewald S, et al. The fourth molybdenum containing enzyme mARC: cloning and involvement in the activation of N-hydroxylated prodrugs. J Med Chem. 2008;51:8173–8177. doi: 10.1021/jm8010417. [DOI] [PubMed] [Google Scholar]
- 14.Bittner F. Molybdenum metabolism in plants and crosstalk to iron. Front Plant Sci. 2014;5:28. doi: 10.3389/fpls.2014.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swenson TL, Casida JE. Aldehyde oxidase importance in vivo in xenobiotic metabolism: imidacloprid nitroreduction in mice. Toxicol Sci. 2013;133:22–28. doi: 10.1093/toxsci/kft066. [DOI] [PubMed] [Google Scholar]
- 16.Coelho C, et al. Structural insights into xenobiotic and inhibitor binding to human aldehyde oxidase. Nat Chem Biol. 2015;11:779–783. doi: 10.1038/nchembio.1895. [DOI] [PubMed] [Google Scholar]
- 17.Pryde DC, et al. Aldehyde oxidase: an enzyme of emerging importance in drug discovery. J Med Chem. 2010;53:8441–8460. doi: 10.1021/jm100888d. [DOI] [PubMed] [Google Scholar]
- 18.Sagi M, Fluhr R, Lips SH. Aldehyde oxidase and xanthine dehydrogenase in a flacca tomato mutant with deficient abscisic acid and wilty phenotype. Plant Physiol. 1999;120:571–578. doi: 10.1104/pp.120.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leydecker MT, Moureaux T, Kraepiel Y, Schnorr K, Caboche M. Molybdenum Cofactor Mutants, Specifically Impaired in Xanthine Dehydrogenase Activity and Abscisic Acid Biosynthesis, Simultaneously Overexpress Nitrate Reductase. Plant Physiol. 1995;107:1427–1431. doi: 10.1104/pp.107.4.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leon-Kloosterziel KM, et al. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 1996;10:655–661. doi: 10.1046/j.1365-313X.1996.10040655.x. [DOI] [PubMed] [Google Scholar]
- 21.Xiong L, Ishitani M, Lee H, Zhu JK. The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell. 2001;13:2063–2083. doi: 10.1105/tpc.13.9.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bittner F, Oreb M, Mendel RR. ABA3 is a molybdenum cofactor sulfurase required for activation of aldehyde oxidase and xanthine dehydrogenase in Arabidopsis thaliana. J Biol Chem. 2001;276:40381–40384. doi: 10.1074/jbc.C100472200. [DOI] [PubMed] [Google Scholar]
- 23.Sekimoto H, et al. Molecular cloning and characterization of aldehyde oxidases in Arabidopsis thaliana. Plant Cell Physiol. 1998;39:433–442. doi: 10.1093/oxfordjournals.pcp.a029387. [DOI] [PubMed] [Google Scholar]
- 24.Seo M, et al. Abscisic aldehyde oxidase in leaves of Arabidopsis thaliana. Plant J. 2000;23:481–488. doi: 10.1046/j.1365-313x.2000.00812.x. [DOI] [PubMed] [Google Scholar]
- 25.Seo M, et al. The Arabidopsis aldehyde oxidase 3 (AAO3) gene product catalyzes the final step in abscisic acid biosynthesis in leaves. Proc Natl Acad Sci USA. 2000;97:12908–12913. doi: 10.1073/pnas.220426197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bottcher C, et al. The Biosynthetic Pathway of Indole-3-Carbaldehyde and Indole-3-Carboxylic Acid Derivatives in Arabidopsis. Plant Physiol. 2014;165:841–853. doi: 10.1104/pp.114.235630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava S, et al. Aldehyde Oxidase 4 Plays a Critical Role in Delaying Silique Senescence by Catalyzing Aldehyde Detoxification. Plant Physiol. 2017;173:1977–1997. doi: 10.1104/pp.16.01939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith PM, Atkins CA. Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiol. 2002;128:793–802. doi: 10.1104/pp.010912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakagawa A, Sakamoto S, Takahashi M, Morikawa H, Sakamoto A. The RNAi-mediated silencing of xanthine dehydrogenase impairs growth and fertility and accelerates leaf senescence in transgenic Arabidopsis plants. Plant Cell Physiol. 2007;48:1484–1495. doi: 10.1093/pcp/pcm119. [DOI] [PubMed] [Google Scholar]
- 30.Brychkova G, Alikulov Z, Fluhr R, Sagi M. A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. Plant J. 2008;54:496–509. doi: 10.1111/j.1365-313X.2008.03440.x. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe S, Nakagawa A, Izumi S, Shimada H, Sakamoto A. RNA interference-mediated suppression of xanthine dehydrogenase reveals the role of purine metabolism in drought tolerance in Arabidopsis. FEBS Lett. 2010;584:1181–1186. doi: 10.1016/j.febslet.2010.02.023. [DOI] [PubMed] [Google Scholar]
- 32.Takagi H, et al. Allantoin, a stress-related purine metabolite, can activate jasmonate signaling in a MYC2-regulated and abscisic acid-dependent manner. J Exp Bot. 2016;67:2519–2532. doi: 10.1093/jxb/erw071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gus’kov EP, et al. Allantoin as a vitamin. Dokl Biochem Biophys. 2004;398:320–324. doi: 10.1023/B:DOBI.0000046649.11374.8d. [DOI] [PubMed] [Google Scholar]
- 34.Watanabe S, Kounosu Y, Shimada H, Sakamoto A. Arabidopsis xanthine dehydrogenase mutants defective in purine degradation show a compromised protective response to drought and oxidative stress. Plant. Biotechnology. 2014;31:173–178. [Google Scholar]
- 35.Watanabe S, et al. The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant Cell Environ. 2014;37:1022–1036. doi: 10.1111/pce.12218. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz SH, Leon-Kloosterziel KM, Koornneef M, Zeevaart JA. Biochemical characterization of theaba2 and aba3 mutants in Arabidopsis thaliana. Plant Physiol. 1997;114:161–166. doi: 10.1104/pp.114.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhong R, Thompson J, Ottesen E, Lamppa GK. A forward genetic screen to explore chloroplast protein import in vivo identifies Moco sulfurase, pivotal for ABA and IAA biosynthesis and purine turnover. Plant J. 2010;63:44–59. doi: 10.1111/j.1365-313X.2010.04220.x. [DOI] [PubMed] [Google Scholar]
- 38.Iuchi S, et al. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001;27:325–333. doi: 10.1046/j.1365-313x.2001.01096.x. [DOI] [PubMed] [Google Scholar]
- 39.Winter D, et al. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One. 2007;2:e718. doi: 10.1371/journal.pone.0000718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verslues PE, Agarwal M, Katiyar-Agarwal S, Zhu J, Zhu JK. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006;45:523–539. doi: 10.1111/j.1365-313X.2005.02593.x. [DOI] [PubMed] [Google Scholar]
- 41.Verslues PE. ABA and cytokinins: challenge and opportunity for plant stress research. Plant Mol Biol. 2016;91:629–640. doi: 10.1007/s11103-016-0458-7. [DOI] [PubMed] [Google Scholar]
- 42.Sawada Y, et al. Widely targeted metabolomics based on large-scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant Cell Physiol. 2009;50:37–47. doi: 10.1093/pcp/pcn183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hesberg C, Hansch R, Mendel RR, Bittner F. Tandem orientation of duplicated xanthine dehydrogenase genes from Arabidopsis thaliana: differential gene expression and enzyme activities. J Biol Chem. 2004;279:13547–13554. doi: 10.1074/jbc.M312929200. [DOI] [PubMed] [Google Scholar]
- 44.Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol. 2002;5:218–223. doi: 10.1016/S1369-5266(02)00256-X. [DOI] [PubMed] [Google Scholar]
- 45.Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013;18:477–483. doi: 10.1016/j.tplants.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 46.Xu W, Dubos C, Lepiniec L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015;20:176–185. doi: 10.1016/j.tplants.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 47.Teng Y, Xu W, Ma M. cGMP is required for seed germination in Arabidopsis thaliana. J Plant Physiol. 2010;167:885–889. doi: 10.1016/j.jplph.2010.01.015. [DOI] [PubMed] [Google Scholar]
- 48.Merzlyak MN, Chivkunova OB, Solovchenko AE, Naqvi KR. Light absorption by anthocyanins in juvenile, stressed, and senescing leaves. J Exp Bot. 2008;59:3903–3911. doi: 10.1093/jxb/ern230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chalker-Scott L. Do anthocyanins function as osmoregulators in leaf tissues? Advances in Botanical Research. 2002;37:103–127. doi: 10.1016/S0065-2296(02)37046-0. [DOI] [Google Scholar]
- 50.Brown DE, et al. Flavonoids act as negative regulators of auxin transport in vivo in arabidopsis. Plant Physiol. 2001;126:524–535. doi: 10.1104/pp.126.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hondo T, et al. Structural basis of blue-colour development in flower petals from Commelina communis. Nature. 1992;358:515–518. doi: 10.1038/358515a0. [DOI] [Google Scholar]
- 52.Takeda K. Blue metal complex pigments involved in blue flower color. Proc Jpn Acad Ser B Phys Biol Sci. 2006;82:142–154. doi: 10.2183/pjab.82.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agati G, Azzarello E, Pollastri S, Tattini M. Flavonoids as antioxidants in plants: location and functional significance. Plant Sci. 2012;196:67–76. doi: 10.1016/j.plantsci.2012.07.014. [DOI] [PubMed] [Google Scholar]
- 54.Nakabayashi R, et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014;77:367–379. doi: 10.1111/tpj.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu Z, Mahmood K, Rothstein SJ. ROS Induces Anthocyanin Production Via Late Biosynthetic Genes and Anthocyanin Deficiency Confers the Hypersensitivity to ROS-Generating Stresses in Arabidopsis. Plant Cell Physiol. 2017;58:1364–1377. doi: 10.1093/pcp/pcx073. [DOI] [PubMed] [Google Scholar]
- 56.Nambara E, Kawaide H, Kamiya Y, Naito S. Characterization of an Arabidopsis thaliana mutant that has a defect in ABA accumulation: ABA-dependent and ABA-independent accumulation of free amino acids during dehydration. Plant Cell Physiol. 1998;39:853–858. doi: 10.1093/oxfordjournals.pcp.a029444. [DOI] [PubMed] [Google Scholar]
- 57.Seo M, et al. Comparative studies on the Arabidopsis aldehyde oxidase (AAO) gene family revealed a major role of AAO3 in ABA biosynthesis in seeds. Plant Cell Physiol. 2004;45:1694–1703. doi: 10.1093/pcp/pch198. [DOI] [PubMed] [Google Scholar]
- 58.Gonzalez-Guzman M, et al. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell. 2002;14:1833–1846. doi: 10.1105/tpc.002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rock CD, Heath TG, Gage DA, Zeevaart JA. Abscisic alcohol is an intermediate in abscisic Acid biosynthesis in a shunt pathway from abscisic aldehyde. Plant Physiol. 1991;97:670–676. doi: 10.1104/pp.97.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu W, et al. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014;202:132–144. doi: 10.1111/nph.12620. [DOI] [PubMed] [Google Scholar]
- 61.Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl):S15–45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huijser C, et al. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. Plant Journal. 2000;23:577–585. doi: 10.1046/j.1365-313x.2000.00822.x. [DOI] [PubMed] [Google Scholar]
- 63.Smirnoff N. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytologist. 1993;125:27–58. doi: 10.1111/j.1469-8137.1993.tb03863.x. [DOI] [PubMed] [Google Scholar]
- 64.Loreti E, et al. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008;179:1004–1016. doi: 10.1111/j.1469-8137.2008.02511.x. [DOI] [PubMed] [Google Scholar]
- 65.Laurie-Berry N, Joardar V, Street IH, Kunkel BN. The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae. Mol Plant Microbe Interact. 2006;19:789–800. doi: 10.1094/MPMI-19-0789. [DOI] [PubMed] [Google Scholar]
- 66.Ames BN, Cathcart R, Schwiers E, Hochstein P. Uric acid provides an antioxidant defense in humans against oxidant- and radical-caused aging and cancer: a hypothesis. Proc Natl Acad Sci USA. 1981;78:6858–6862. doi: 10.1073/pnas.78.11.6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Farmer EE, Davoine C. Reactive electrophile species. Curr Opin Plant Biol. 2007;10:380–386. doi: 10.1016/j.pbi.2007.04.019. [DOI] [PubMed] [Google Scholar]
- 68.Mano J, Nagata M, Okamura S, Shiraya T, Mitsui T. Identification of oxidatively modified proteins in salt-stressed Arabidopsis: a carbonyl-targeted proteomics approach. Plant Cell Physiol. 2014;55:1233–1244. doi: 10.1093/pcp/pcu072. [DOI] [PubMed] [Google Scholar]
- 69.Biswas MS, Mano J. Reactive Carbonyl Species Activate Caspase-3-Like Protease to Initiate Programmed Cell Death in Plants. Plant Cell Physiol. 2016;57:1432–1442. doi: 10.1093/pcp/pcw053. [DOI] [PubMed] [Google Scholar]
- 70.Mueller MJ, Berger S. Reactive electrophilic oxylipins: pattern recognition and signalling. Phytochemistry. 2009;70:1511–1521. doi: 10.1016/j.phytochem.2009.05.018. [DOI] [PubMed] [Google Scholar]
- 71.Barnes SA, McGrath RB, Chua NH. Light signal transduction in plants. Trends Cell Biol. 1997;7:21–26. doi: 10.1016/S0962-8924(97)10043-5. [DOI] [PubMed] [Google Scholar]
- 72.Donaldson L, Ludidi N, Knight MR, Gehring C, Denby K. Salt and osmotic stress cause rapid increases in Arabidopsis thaliana cGMP levels. FEBS Lett. 2004;569:317–320. doi: 10.1016/j.febslet.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 73.Isner JC, Nuhse T, Maathuis FJ. The cyclic nucleotide cGMP is involved in plant hormone signalling and alters phosphorylation of Arabidopsis thaliana root proteins. J Exp Bot. 2012;63:3199–3205. doi: 10.1093/jxb/ers045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Joudoi T, et al. Nitrated cyclic GMP modulates guard cell signaling in Arabidopsis. Plant Cell. 2013;25:558–571. doi: 10.1105/tpc.112.105049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maathuis FJ. cGMP modulates gene transcription and cation transport in Arabidopsis roots. Plant J. 2006;45:700–711. doi: 10.1111/j.1365-313X.2005.02616.x. [DOI] [PubMed] [Google Scholar]
- 76.Pagnussat GC, Lanteri ML, Lamattina L. Nitric oxide and cyclic GMP are messengers in the indole acetic acid-induced adventitious rooting process. Plant Physiol. 2003;132:1241–1248. doi: 10.1104/pp.103.022228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pasqualini S, et al. Ozone and nitric oxide induce cGMP-dependent and -independent transcription of defence genes in tobacco. New Phytol. 2009;181:860–870. doi: 10.1111/j.1469-8137.2008.02711.x. [DOI] [PubMed] [Google Scholar]
- 78.Szmidt-Jaworska A, Jaworski K, Tretyn A, Kopcewicz J. The involvement of cyclic GMP in the photoperiodic flower induction of Pharbitis nil. J Plant Physiol. 2004;161:277–284. doi: 10.1078/0176-1617-01122. [DOI] [PubMed] [Google Scholar]
- 79.Wu Y, Hiratsuka K, Neuhaus G, Chua NH. Calcium and cGMP target distinct phytochrome-responsive elements. Plant J. 1996;10:1149–1154. doi: 10.1046/j.1365-313X.1996.10061149.x. [DOI] [PubMed] [Google Scholar]
- 80.Suita K, et al. Cyclic GMP acts as a common regulator for the transcriptional activation of the flavonoid biosynthetic pathway in soybean. Planta. 2009;229:403–413. doi: 10.1007/s00425-008-0839-5. [DOI] [PubMed] [Google Scholar]
- 81.Mittler Ron, Vanderauwera Sandy, Gollery Martin, Van Breusegem Frank. Reactive oxygen gene network of plants. Trends in Plant Science. 2004;9(10):490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 82.Zrenner R, Stitt M, Sonnewald U, Boldt R. Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol. 2006;57:805–836. doi: 10.1146/annurev.arplant.57.032905.105421. [DOI] [PubMed] [Google Scholar]
- 83.Lin Michael T., Beal M. Flint. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443(7113):787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 84.Jia LG, et al. Modulation of anti-oxidation ability by proanthocyanidins during germination of Arabidopsis thaliana seeds. Mol Plant. 2012;5:472–481. doi: 10.1093/mp/ssr089. [DOI] [PubMed] [Google Scholar]
- 85.Warnes, G. R., Bolker, B. & Bonebakker, L. R. R. G. T. C. 2016. gplots: various R programming tools for plotting data. R package version 3.0.1 (2016).
- 86.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lichtenthaler HK. Chlorophylls and Carotenoids - Pigments of Photosynthetic Biomembranes. Methods in Enzymology. 1987;148:350–382. doi: 10.1016/0076-6879(87)48036-1. [DOI] [Google Scholar]
- 88.Nakata, M. & Ohme-Takagi, M. Quantification of Anthocyanin Content. Bio-Protocol4(7), e1098 (2014).
- 89.Kanno Y, et al. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun. 2016;7:13245. doi: 10.1038/ncomms13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jambunathan N. Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. Methods Mol Biol. 2010;639:292–298. doi: 10.1007/978-1-60761-702-0_18. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.