Abstract
Many biophysical techniques that are available to study the structure, function and dynamics of cellular constituents require modification of the target molecules. Site-specific labelling of a protein is of particular interest for fluorescence-based single-molecule measurements including single-molecule FRET or super-resolution microscopy. The labelling procedure should be highly specific but minimally invasive to preserve sensitive biomolecules. The modern molecular engineering toolkit provides elegant solutions to achieve the site-specific modification of a protein of interest often necessitating the incorporation of an unnatural amino acid to introduce a unique reactive moiety. The Amber suppression strategy allows the site-specific incorporation of unnatural amino acids into a protein of interest. Recently, this approach has been transferred to the mammalian expression system. Here, we demonstrate how the combination of unnatural amino acid incorporation paired with current bioorthogonal labelling strategies allow the site-specific engineering of fluorescent dyes into proteins produced in the cellular environment of a human cell. We describe in detail which parameters are important to ensure efficient incorporation of unnatural amino acids into a target protein in human expression systems. We furthermore outline purification and bioorthogonal labelling strategies that allow fast protein preparation and labelling of the modified protein. This way, the complete eukaryotic proteome becomes available for single-molecule fluorescence assays.
Keywords: Bioorthogonal chemistry, Unnatural amino acids, Amber suppression, eGFP, Genetic code expansion
Highlights
-
•
Optimal conditions for incorporation of unnatural amino acids in mammalian cells.
-
•
Fast small scale purification/labelling for unnatural amino acid-carrying proteins.
-
•
Evaluation of biorthogonal labelling reactions for dye engineering into proteins.
1. Introduction
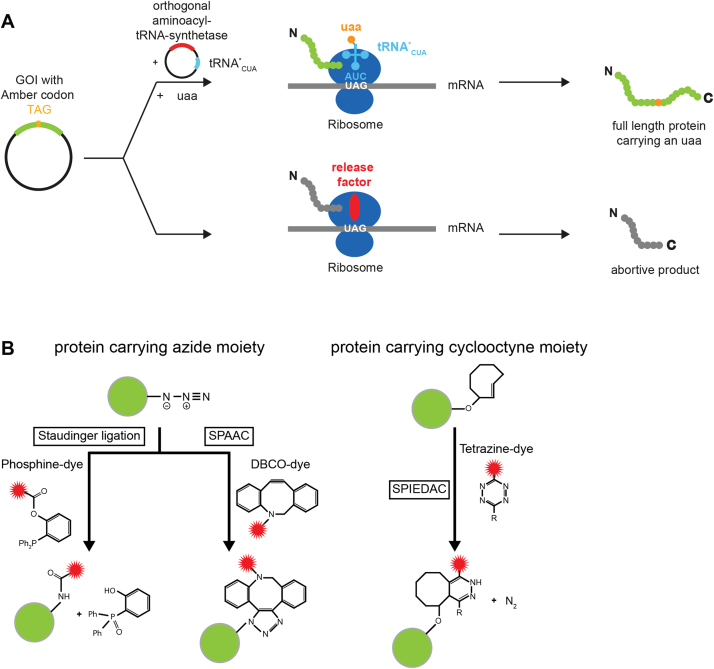
The possibility to engineer a non-canonical amino acid carrying a unique reactive side chain into a human protein of interest is highly advantageous for biochemical and biophysical studies as suitable probes like fluorescent dyes, biotins, lipids, sugars, EPR labels, crosslinkers etc can be attached to this protein via bioorthogonal chemistries [1], [2]. One example is super-resolution microscopy that allows to image (biological) structures with unprecedented spatiotemporal resolutions in the low nanometre range providing new opportunities to study and quantitatively monitor biomolecules (for a recent review see [3]). In this context, highly specific and dense labelling of the biological structure with fluorophores is required to achieve an optimal resolution. Site-specific engineering of molecular handles and fluorophores is also desired to equip proteins to be used for other biophysical techniques (e.g. single-molecule FRET) and nanobiotechnology. In the context of nanobiotechnology, it is the aim to produce functional nanostructures built of a programmable nanomaterial and a functional biomolecule to create nanometre-sized machineries with high precision [4], [5]. This approach often requires the physical and ideally site-specific connection of a biomolecule with a nanostructure. Hence, the site-directed engineering of biophysical probes, molecular handles and modifications has increasing importance to expand the structural and chemical diversity of proteins, which in turn increases the flexibility and chemical space for the design of biological nanostructures and biophysical assays [2], [6], [7], [8], [9], [10]. However, the site-specific modification of (especially human) proteins is still a challenging task that, among others, can be achieved via bioorthogonal chemistries based on an unnatural amino acid (uaa) that has been incorporated into the protein of interest. The uaa carries a unique reactive moiety that can be exploited for highly selective bioorthogonal reactions [11], [12], [2]. An uaa can be genetically encoded by reprogramming a rarely used codon or a non-sense codon in the gene of interest [10], [13]. In bacterial and mammalian expression systems, the Amber stop codon (TAG) is frequently exploited for this purpose. E. coli rarely makes use of this codon and human cells utilise this codon with lowest frequency to terminate translation (TAG: 23%, TAA: 30%, TGA: 47% in Homo sapiens) [14], [15]. In order to repurpose the Amber codon, a bioorthogonal tRNA / aminoacyl-tRNA synthetase (aaRS) pair is required that allows the coupling of the uaa with high efficiency and extraordinary selectivity to a tRNA that recognises the Amber codon thereby avoiding the abortion of translation by the release factor at this site (Fig. 1A).
Fig. 1.
(A) Incorporation of an unnatural amino acid (uaa) into a protein using the Amber suppressor strategy. Site-specific introduction of the Amber stop codon (TAG) into the gene of interest (GOI) usually leads to the abortion of translation at this site and consequently results in a shortened protein of interest (POI). Co-transformation of additional plasmids harbouring an engineered orthogonal aminoacyl tRNA synthetase (aaRS) / tyrosyl- tRNA (tRNA*CUA) pair charges the additional tRNA with the unnatural amino acid. The tRNA is optimised to work efficiently in the cellular translation system leading to the incorporation of the uaa into the protein strand and allows expression of the full length protein. (B) Protein labelling schemes based on bioorthogonal reaction schemes: the Staudinger-Bertozzi ligation between a phosphine and an organic azide (left), the strain-promoted azide-alkin cycloaddition (SPAAC) reaction between a cyclooctyne and an azide (middle) and the strain-promoted inverse Diels Alder cycloaddition (SPIEDAC) between a tetrazine and a cycloalkene. The green sphere symbolises the protein to which the fluorophore is coupled. The red sphere indicates the fluorescent dye linked to the protein via the bioorthogonal reaction. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.).
The uaa introduces a distinct reactive group into the protein that can be utilised for coupling reactions, among them biocompatible variations of the highly specific and catalyst-free click reactions (e.g. strain-promoted azide-alkyne cycloaddition [SPAAC], and the strain-promoted inverse electron demand Diels-Alder click reaction [SPIEDAC]) as well as the Staudinger ligation (reviewed for example in Lang et al. [16]) (Fig. 1B). Efficient uaa incorporation was only recently accomplished in human cells [14], [17], [18], [19], [20], [21], [22] opening up the possibility to directly label a protein of interest with organic dyes in human cells to perform for example direct super-resolved imaging of cellular structures [23], [24], [18], [25]. Moreover, uaa incorporation also allowed the site-specific fluorescent labelling of eukaryotic proteins for (single-molecule) FRET measurements, glycol-engineering, and the site-specific crosslinking of biomolecular complexes [1], [26]. Notably, uaa incorporation into human proteins expressed in their native cellular environment ensures that the protein adopts its native fold and carries all functionally important posttranslational modifications as we have recently demonstrated for the human Argonaute 2 protein [26]. This is a major step forward as it enables single-molecule FRET measurements with human proteins that hitherto evaded single-molecule interrogation as they often cannot be produced in recombinant form.
Here, we explain step-by-step how unnatural amino acids can be efficiently incorporated into proteins that are expressed in human cells. We describe in detail the major obstacles and pitfalls and evaluate the incorporation efficiencies for different unnatural amino acids and cell lines using different transfection reagents. Moreover, we describe a fast and efficient workflow that allows the fluorescent labelling and purification of these proteins and evaluate coupling efficiencies of different bioorthogonal reactions. Taken together, we wish to provide a practical guide for the efficient incorporation of unnatural amino acids in human expression systems to allow researchers to exploit bioorthogonal chemistries in their field of research.
2. Material and methods
2.1. Generation of expression plasmids
The eGFP expression plasmid that contains the eGFP Wildtype (WT) sequence was purchased from addgene (#13031). The Amber codon and the FLAG-tag was introduced at the given site using the QuikChange II site-directed mutagenesis kit (Agilent). The C-terminal His-tag was introduced making use of the Fusion® HD cloning kit (Takara Bio USA, Inc.).
2.2. Cell culture
HEK 293 T and HeLa cells were grown on Dulbecco's Modified Eagle Medium (DMEM) high glucose (4.5 g/l) buffered with HEPES without phenol red (Thermo Fisher). Additionally, 1% (v/v) Penicillin/Streptomycin and 10% (v/v) Fetal Bovine Serum (Sigma-Aldrich) was added to the growth media. Cells were cultivated under standard conditions (37 °C and 5% CO2). Upon cultivation, cells were passaged every 2–3 days using 1 ml Trypsin-EDTA (Sigma-Aldrich) to detach PBS-washed cells from a 10 cm dish and an appropriate amount of cells was transferred to a new 10 cm dish containing 10 ml of growth medium for passaging.
2.3. Transfection of cells
For optimal DNA transfection conditions, a suitable number of HEK 293 T or HeLa cells were seeded into a culture dish to reach a confluency of ~70–80% on the day of transfection. For a 24-well culture dish ~ 70 000 cells, for a 6-well culture dish ~ 200,000 cells were seeded per well and ~ 1.5 × 106 cells were used for a 10 cm culture dish. Prior transfection, the culture medium was exchanged with fresh untreated growth medium or growth medium containing an unnatural amino acid (see the following chapter: incorporation of unnatural amino acids). In this study, either Lipofectamine® 2000 or JetPRIME® was used to transfect human cells.
2.4. Transfection with Lipofectamine® 2000
If not noted otherwise, cells were transfected according to supplier's information (Thermo Fisher). The transfection mix was prepared from two separate solutions. For the transfection of cells grown in a 24-well format, 50 µl and for cells grown in a 6-well format, 150 µl of serum-free Opti-MEM™ medium was used to dilute 500 or 2500 ng of plasmid DNA, respectively. In a second solution, containing equal volumes of Opti-MEM™, 1–2.5 µl (24 well) or 5–12.5 µl of Lipofectamine® 2000 were added and mixed with the DNA-solution. After 5 min of incubation, the transfection mix was directly transferred to the cells and expression of proteins was performed for at least 16 h.
2.5. Transfection with JetPRIME®
If not noted otherwise, cells were transfected according to supplier's information (Polyplus-transfection, VWR). Briefly, 0.5 (24 well format), 2.0 (6 well format) or 10.0 µg (10 cm dish) of plasmid-DNA was diluted in 50 µl, 200 or 500 µl of JetPRIME-buffer, respectively. After dilution, 1, 4 or 20 µl of JetPRIME (1:2 DNA to jetPRIME ratio (w/v)) transfection reagent was directly added to the DNA-solution and incubated for 10 min at room temperature. The transfection mix was directly transferred to the cells and expression of proteins was performed for at least 16 h.
2.6. Incorporation of unnatural amino acids into proteins
In this study, we used two different systems that expand the genetic code of a human cell and allow the incorporation of either trans-cyclooctene L-lysine (TCO*A) or p-Azido-L-phenylalanine (AzF) into eGFP [26]. TCO incorporation was mediated by an orthogonal tRNA/ aaRS pair from Methanosarcina mazei co-encoded on a single plasmid (plasmid was supplied by Edward Lemke, EMBL Heidelberg, Germany) [27]. Incorporation of AzF is based on the orthogonal tRNATyrCUA (tRNA fusion construct from H. sapiens and Bacillus stearothermophilus) and the E. coli aaRS encoded on two separate plasmids (plasmid was supplied by T. Huber, Rockefeller University New York, USA) [28]. For transfection of human cells, conditions that were introduced above were used. In addition to the plasmids that encode the respective orthogonal tRNA and aaRS pair, the eGFPT39Amber expression vector was transfected. The total amount of plasmid DNA and transfection reagent was identical to the procedures described above.
TCO*A and AzF were obtained from SiChem (Bremen, Germany) and ChemImpex International (Wood Dale, USA), respectively. The uaas were dissolved in 200 mM NaOH and 15% DMSO to yield a 100 mM stock solution that can be stored at − 20 °C. At the day of transfection, a working solution of 25 mM uaa was generated by a 1:4 dilution with 1 M Hepes/KOH, pH 7.5. Prior transfection, the medium was exchanged to freshly prepared growth medium, containing the respective uaa in the desired concentration (50 µM were already sufficient for optimal incorporation of each uaa).
The efficiency of the uaa incorporation was monitored by fluorescence microscopy of living cells (Zeiss Axiovert 35 microscope connected to a Color view soft imaging system). For acquisition of fluorescence-images a filter-block (λex = 450–490 nm/ λem = 515–565 nm) was used and pictures of cells were taken with an exposure time of 5 s.
2.7. Purification and labelling of modified proteins
For protein extraction, harvested cells containing the modified protein were resuspended in 150 µl of IP-lysis buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 1% NP40, supplemented with cOmplete™ (Roche) protease inhibitor) per 1 × 106 cells and incubated on ice for 15 min. After incubation, cell debris was removed by centrifugation at 20,000 ×g for 15 min at 4 °C. The clarified supernatant was applied to either ANTI-FLAG® M2 beads (Sigma-Aldrich) for FLAG-tagged proteins or to Ni-NTA Magnetic Agarose Beads (Qiagen) for His6-tagged proteins. Beads were incubated with the cell lysate at 4 °C for 1–3 h in 1.5 ml reaction tubes while rotating overhead. Unbound proteins were removed by 3 wash steps with either HisA buffer (50 mM Tris/HCl pH 7.5, 200 mM NaCl, 20 mM imidazole and 0.5% Tween-20 (v/v)) for His-beads or FLAG-I buffer (50 mM Tris/HCl pH 7.5, 200 mM NaCl and 0.5% Tween-20 (v/v)) for FLAG-beads. Enriched, purified proteins were directly labelled while they were bound to the beads. Elution of eGFP was performed using either HisB (50 mM Tris-HCl pH 7.5, 200 mM NaCl, 250 mM imidazole and 0.5% Tween (v/v)) or FLAG-I buffer supplemented with 300 µg/µl 3xFLAG peptide (Sigma-Aldrich) for 60 min on ice, respectively. For labelling, a 10 mM stock solution of the respective dye was prepared by dissolving the dye in an appropriate volume of water-free DMSO (Phosphine-DL650 purchased from Thermo Fisher, DBCO-Cy5 and Tetrazine-Cy5 from Jena Biosciences).
For labelling, an equal volume to the volume of the beads was added, containing the respective dye concentration (in most cases 200 µM) and labelling was performed at 37 °C for 2 h while shaking the reaction tube (300 rpm). When uaa-modified eGFP was labelled on the beads, excessive dye was removed by 2–3 washing steps using HisA or FLAG-I that contain 2% (v/v) Tween-20. Another 4–5 washing steps using HisA or FLAG-I buffer and transfer to a new reaction tube were sufficient to remove most of the uncoupled dye molecules from the samples. Elution from the affinity beads was performed as described above.
2.8. Western blot analysis
Prior to Western blot analysis, samples were mixed with denaturing Laemmli Buffer (final concentration: 62.5 mM Tris/HCl pH 6.8, 10% glycerol, 2% SDS, 0.01% bromophenol blue supplemented with 2% β-mercaptoethanol) and samples were incubated at 95 °C for 5 min. Subsequently, samples were loaded onto a 12% SDS gel and separated in an electric field with a constant current of 30 mA per gel. After separation, proteins were transferred to an AmershamTM ProtranTM 0.45 µm NC nitrocellulose membrane (GE Healthcare) by semidry blotting in Towbin buffer (25 mM Tris-HCl pH 8:3, 192 mM glycine, 20% (v/v) methanol). For immunodetection of eGFP, a monoclonal mouse-anti-GFP antibody (#11814460001, Roche; 1:1000) was used. Immunodetection of α-Tubulin was performed with a monoclonal mouse-anti α-Tubulin antibody (#T90026, Sigma-Aldrich, 1:1000). An Alexa Fluor 647 labelled goat-anti-mouse secondary antibody (#A21236, Thermo, 1:10000) was used for detection of the antibody sandwich on a fluorescence scanner (FLA-5000, FUJIFILM, λex = 635 nm, filter module 557–270/01).
For the quantification of the Western blot signals, the open source code software ImageJ [29] was used. First, the background was subtracted with an ImageJ-integrated process and after processing, signal intensities were determined. In order to determine the expression efficiency of the eGFP-variants, the eGFP Western Blot signal was normalized to the respective α-Tubulin signal.
2.9. Native gel analysis
For native gel analysis, samples were mixed with 5x native loading dye (final concentrations: 60 mM Tris/HCl pH 6.8% and 12.5% glycerol) and loaded on a non-denaturing 15% PAA gel. Gel electrophoresis was performed using native Laemmli running buffer (24 mM Tris/HCl pH 8.3 and 192 mM Glycine) at 4 °C for 1–3 h. Fluorescent signals were visualised using a FLA7000 scanner (GE Healthcare) and finally gels were transferred to a staining solution (0.02% CBB-G250, 5% aluminium sulphate, 10% ethanol and 2% ortho-phosphoric acid) for colloidal Coomassie staining. After staining, gels were destained in destaining solution (10% Ethanol and 2% ortho-phosphoric acid) and visualised on a gel documentation device (BioDoc-It imaging system, UVP).
2.10. Determination of the FLAG-eGFP concentration after FLAG-IP
In order to determine eGFP yields, it was of interest to measure eGFP concentrations after enrichment of FLAG-eGFP using ANTI-FLAG® M2 agarose beads. Due to low protein yields, concentration determination via UV/Vis spectroscopy was difficult. Therefore, we used purified recombinant eGFP produced in E. coli and a linear range of known eGFP concentrations (0.2 – 1.4 µM) was loaded onto a native PAA gel to create a standard curve. The standard curve was employed to determine the concentration of a eGFP- elution sample derived from a FLAG-IP purification. eGFP signals were detected via a fluorescence scan (FLA7000, GE Healthcare, blue channel: λex = 473 nm, Y520 filter, red channel: λex = 635 nm, R670 filter), signal intensities were determined with ImageJ, and eGFP concentration from the FLAG-IPs were extrapolated from the standard curve.
2.11. Determination of labelling efficiency
For determination of the labelling efficiency, equal amounts of eGFPAzF bound to ANTI-FLAG® M2 agarose beads were incubated with increasing concentrations of fluorescent dyes (0–500 µM). After incubation at 37 °C for 2 h, excessive dye was removed by several wash steps using FLAG-I that contained 2% (v/v) Tween-20 and one final step using FLAG-I. Elution was performed with FLAG-I buffer containing 300 µg/µl 3xFLAG peptide and equal amounts from each sample were mixed with 5x native loading dye and loaded onto a native PAA gel. As labelled eGFP migrated faster in the native gel, labelling efficiency could be directly calculated as the ratio of the fluorescence signal of the dye-labelled eGFP and the fluorescing unlabelled eGFP.
2.12. Absorption spectroscopy analysis
All absorption spectroscopy measurements were performed on a Beckman DU® 640 spectrophotometer using a SUPRASIL® Quartz precision cell ultra-micro cuvette (Hellma, 10 × 4.5 mm, 120 µl). For the determination of eGFP concentrations from a single 6-well culture dish, cells were transfected either with wild-type (wt) eGFP or eGFPT39AzF in the presence of the AzF tRNA/aaRS expression system and 300 µM AzF. After 48 h of expression, cells were harvested and washed with PBS. The pellet was resuspended in 100 µl 50 mM Tris/HCl pH 7.5 and 150 mM NaCl and cells were lysed by sonication. Cell debris was removed by centrifugation (21,000 g, 30 min and 4 °C) and the supernatant was transferred to an ultra-micro cuvette and absorbance was measured at 488 nm. The total amount of eGFP per 6-well was determined using the Lambert-Beer law and an absorption coefficient of 56 000 M−1 cm−1 for eGFP.
3. Results and discussion
In order to exploit the experimental possibilities of an uaa-carrying human protein, the expression of the uaa-modified protein as well as purification and labelling procedures must be optimised and established. We guide the reader through such an expression, purification and labelling protocol that can be applied to any protein of choice pointing out important points to consider when planning the experiment as well as experimental tips and tricks.
3.1. Optimising the incorporation efficiency of unnatural amino acids in mammalian cells
The production of sufficient amounts of protein that carry the desired uaa is crucial for many downstream applications. Hence, it is important to optimise the uaa incorporation efficiency. Typically, the protein of interest will be cloned in a suitable expression vector that allows stable constitutive or inducible expression in mammalian cells. Please note, that the gene encoding the protein of interest (POI) must be free of an Amber stop codon. As 23% of human genes make use of the Amber stop codon [15], the WT sequence of the respective gene must be checked before cloning. If an Amber stop codon is present, it should be replaced by an alternative stop codon to allow the mutation of a natural codon to an Amber stop codon to facilitate the site-specific incorporation of the uaa. Several parameters, among them the choice of cell line, the transfection reagent, the uaa concentration, the uaa type, the expression plasmid to tRNA/aaRS ratio and the expression time influence the protein yield. Here, we screened transfection and expression conditions for two commonly used unnatural amino acids, namely p-Azido-L-phenylalanine (AzF) and trans-cyclooctene L-lysine (TCO*A). We chose the enhanced green fluorescent protein (eGFP) as model system as the successful incorporation of the uaa can be easily followed and quantified by the production of the fluorescent full-length eGFP (Fig. 2). We mutated the codon at position 39 to an Amber stop codon (TAG) to enable the incorporation of the uaa at this site instead of the naturally encoded threonine. eGFP expression is driven from the commercially available pcDNA3 expression plasmid, which ensures high constitutive expression levels in mammalian cells. For the incorporation of AzF two additional expression plasmids are required, one encoding the orthogonal tRNATyrCUA (a tRNA fusion construct from H. sapiens and Bacillus stearothermophilus) and a second plasmid for the expression of an additional aaRS from E. coli [28]. The orthogonal tRNA/aaRS pair from Methanosarcina mazei required for the incorporation of TCO*A is co-encoded on a single plasmid [14], [27], [17].
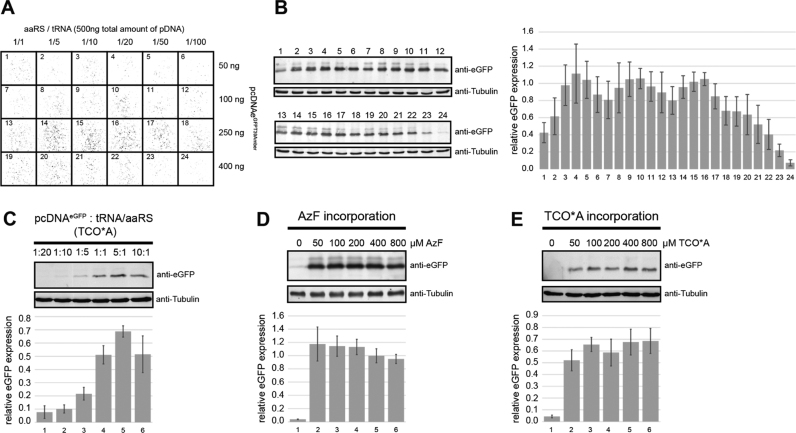
Fig. 2.
Optimisation of unnatural amino acid incorporation into eGFP using the Amber suppression strategy. (A) Fluorescence images of HEK 293 T cells expressing eGFPT39AzF at different plasmid ratios for the plasmids encoding the orthogonal tRNA, aminoacyl tRNA synthetase (aaRS) and the protein of interest (pcDNAeGFPT39Amber plasmid). In all cases, the p-azido-L-phenylalanine (AzF) concentration used was 300 µM. (B) eGFPAzF expression levels are detected by Western blot analysis for the conditions shown in (A). Quantification of eGFPAzF expression for the conditions chosen in (A). (C) eGFPT39TCO expression levels detected via Western blot at different plasmid ratios for the plasmid encoding the orthogonal tRNA/aaRS and the pcDNAeGFPT39Amber plasmid. In all cases, the TCO concentration was 300 µM. Quantification of eGFPAzF expression for the different plasmid ratios. (D and E) Addition of increasing amounts of AzF or TCO*A to the transfection mixture results in a maximum incorporation efficiency at uaa concentrations of > 50 µM, respectively. Western blots and quantification are analogous to (B-C). All eGFPuaa quantifications were normalized to α-tubulin expression.
First, we tested which plasmid ratios result in efficient uaa incorporation (Fig. 2A/B and C). In case of AzF incorporation, a pcDNAeGFP: tRNA plasmid: aaRS plasmid ratio of 10:9:1 or 10:9.5:0.5 yielded best results as detected by the number of fluorescent cells (imaged after 20 h) and Western blot analysis of cell lysates after 24 h (Fig. 2A/B). For the incorporation of TCO*A, a pcDNAeGFP: tRNA/aaRS plasmid ratio of 5:1 resulted in most efficient incorporation of the uaa (Fig. 2C). Incorporation of the uaa was only successful if all respective expression plasmids were transfected [26]. Expression yields are also dependent on the concentration of uaa added to the cells during transfection. Hence, we tested a wide range of AzF and TCO concentrations (0–800 µM). The uaa was added directly to the growth medium prior or during transfection (both works equally well). Expression of fluorescent eGFP was dependent on the presence of the unnatural amino acid and an AzF and TCO*A concentration of > 50–400 µM resulted in most efficient eGFP expression for both systems (Fig. 2D/E and Fig. S1). However, in our hands, TCO*A incorporation is less efficient than AzF incorporation (Fig. 2D/E). Recently, Nikic et al. found that the aaRS from Methanosarcina mazei used for the incorporation of TCO*A contains a nuclear localisation sequence and consequently, the aaRS is localised to the nucleus reducing uaa incorporation in the cytoplasm [25]. Engineering of a strong nuclear export sequence to the aaRS led to a predominantly cytoplasmatic localisation of the protein and a 14-fold increase in Amber suppression efficiencies. Usage of the optimised M. mazei aaRS should therefore lead to better expression of TCO-containing proteins.
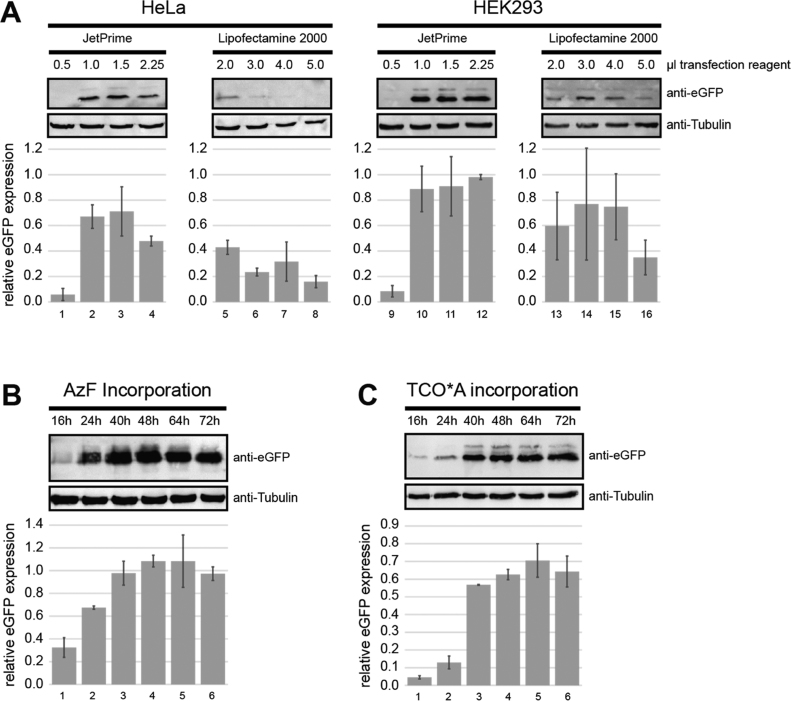
We furthermore tested whether the human cell line HeLa or HEK293 is more suited for uaa incorporation. We consistently found that the Amber suppression strategy works more efficiently in HEK293 cells (Fig. 3A). A wide range of transfection reagents can be used for mammalian cell transfection. In order to evaluate whether the choice of transfection reagent influences incorporation efficiencies, we tested Lipofectamine® and jetPRIME®, two frequently used transfection reagents. Interestingly, incorporation of TCO*A and AzF into eGFP was more effective when jetPRIME was used for transfection (Fig. 3A). This has been observed for expression in HeLa (lanes 1–8) as well as HEK293 cells (lanes 9–16). Finally, we quantified the eGFP expression level in a 16–72 h time-window after transfection of the cells. It is important to note that transfection combined with uaa treatment seemed to trigger cell death in HeLa cells as the cells detached from the culture dish approximately 24 h after transfection. In contrast, HEK293 cells grow normally for extended periods of time. Western blot analysis of cell lysates revealed that a maximum eGFPAzF and eGFPTCO expression level was reached after 48 and 64 h, respectively, when using HEK293 cells as expression system (Fig. 3B/C). Expression of wt eGFP yielded approximately 5 µg eGFP per 6-well (approximately three million cells) and expression of eGFPT39AzF yielded 1.2 µg eGFP per 6-well. eGFPT39TCO levels are significantly lower yielding only 11%, 21% and 51% of full-length eGFPT39TCO as compared to eGFPT39AzF after 16, 24 and 48 h of expression, respectively (Fig. S3).
Fig. 3.
Evaluation of transfection conditions and expression time. (A) eGFPAzF expression levels in HeLa (left) or HEK293 (right) cells. Cells were transfected with different amounts of Lipofectamine® or JetPrime® and eGFP expression was detected by Western blot analysis 24 h after transfection. P-azido-L-phenylalanine (AzF) concentration used for transfection was 300 µM. (B) Time-dependency of eGFPuaa expression as determined by Western blot analysis of cell lysates 16, 24, 40, 48, 64 and 72 h after transfection with JetPrime® (AzF and TCO*A concentration 300 µM each). All eGFPuaa quantifications were normalized to α-tubulin expression.
In conclusion, most efficient uaa incorporation and highest eGFPuaa yields were achieved when the unnatural amino acid AzF was incorporated. Optimal AzF incorporation efficiencies were observed if i) an equal amount of target protein expression plasmid and expression plasmid that encodes the bioorthogonal aaRS was used, ii) an uaa concentration of 50–400 µM was added to the cells, iii) JetPrime was used as transfection reagent, iv) HEK293 cells were used for protein expression and v) cells were harvested 48 h after transfection.
3.2. Purification of proteins carrying an unnatural amino acid
Prior to biophysical or biochemical characterisation of the POI, the protein should be purified to homogeneity. Here, a wealth of chromatographic purification schemes can be used. We established a small-scale purification scheme to separate uaa-modified proteins from the cellular content, which is especially important for subsequent (fluorescent) labelling of the protein via bioorthogonal chemistries. As the Amber stop codon is frequently used in human translation, there is a high number of endogenous proteins that also carry the uaa modification. Consequently, addition of a reactive fluorophore to the cell extract results in a broad range of unwanted labelled proteins (Fig. 4A). We furthermore found that cells derived from a single 6-well plate (approximately 2–3 × 106 cells) provide sufficient material for small-scale purification, which allows to follow the purification process on Coomassie-stained SDS polyacrylamide gels. Thereby, hour-long preparation and purification procedures that might negatively influence the activity or multimerisation state of a sensitive protein or protein complex are avoided. Here, we tested the histidine‐ and FLAG-tag that are commonly used for protein purification via affinity chromatography or immunoprecipitation, respectively. To this end, either the histidine‐ tag (eGFP6xHis) or a FLAG-tag (eGFPFLAG) was genetically engineered to the C-terminus of eGFP and purification was carried out using either Ni-NTA magnetic beads (eGFP6xHis) or anti-FLAG agarose beads (eGFPFLAG) in 1.5 ml reaction tubes. In general, a C-terminal tag is preferred as this allows the purification of the full-length protein. Please note that in some instances, the placement of a tag at either the N-terminus or C-terminus of a protein is detrimental to protein folding and/or function and if possible, the influence of the tag on a protein function should be evaluated via a functional assay. Ni-NTA purification of eGFP6xHis resulted in a number of contaminating proteins in the eluate fraction (Fig. 4B lanes 3 and 9). In contrast, the anti FLAG-bead eluate only showed two major bands (Fig. 4B lanes 6 and 12). One of the protein bands corresponds to eGFP as evident from immunodetection (not shown) and the fluorescence scan of an SDS gel on which the eluate fraction was analysed after on-bead labelling of eGFPT39*uaa (Fig. 4B). The protein with an apparent molecular weight of approximately 50 kDa is most likely a previously described contamination (the serine/threonine kinase STK38) of the FLAG-tag-based purification scheme [30]. In order to purify the POI to homogeneity, the protein can be equipped with two tags allowing a two-step purification [31]. We recently showed that the double-purification via first the FLAG-tag followed by a second purification step via the His-tag resulted in highly pure protein preparations suitable to perform bioorthogonal reactions [26]. Alternatively, the widely employed TAP (tandem affinity purification) tag could be used, which results in homogeneously pure protein samples [32]. The successful incorporation of the uaa and the identity of the protein should be checked employing mass spectrometry (MS). However, larger amounts of protein are required for the MS analysis. Moreover, if possible, the activity and folding of the protein should be checked to evaluate whether the introduction of an uaa changed the biochemical behaviour of the protein.
Fig. 4.
Purification and labelling of eGFPuaa. (A) Fluorescence scan of a SDS polyacrylamide gel loaded with HEK 293 T expressing eGFPT39Amber without (lane 1, 2) and with (lane 3, 4) p-azido-L-phenylalanine (AzF), and with (lane 3, 4) or without (lane 1,2) transfection of plasmids expressing tRNATyrCUA and aminoacyl tRNA synthetase (aaRS). Lane 2 and 4 shows cell lysates incubated with 400 µM DyLight650 (DL650), cell lysates in lane 1 and 3 were not incubated with the fluorophore that carries a phosphine group as reactive moiety. Lanes 5–8 show the respective Coomassie-stained gel. (B) Coomassie stain and fluorescence scan of an SDS gel loaded with site-specifically labelled eGFPT39AzF*DL650. Full-length eGFPT39AzF was purified and enriched via its C-terminal His- or FLAG-tag using anti FLAG agarose beads or Ni-NTA magnetic beads. On-bead labelling was performed using 200 µM of the phosphine-modified dye DyLight650 that reacts with the azide group of AzF. Thus, a fluorescent signal for eGFPT39AzF*DL650 could be detected after elution from the beads (Elu), while no signal was present in the input (Inp) or supernatant (Sup). (C) Titration of increasing concentrations (0–400 µM) of reactive dye (lane 2–7: phosphine-DL650, lane 9–14: DBCO-Cy5, lane 16–21: Tetrazine-Cy5) to C-terminally FLAG-tagged eGFPuaa variants (lane 1–14: eGFPAzF, lane 15–21: eGFPTCO*A). Reaction products are separated on a SDS polyacrylamide gel after incubation for 2 h at 37 °C. The arrow indicates the protein of interest eGFP. The star (*) indicates a labelled protein contamination that is most likely a previously described contamination (the serine/threonine kinase STK38) of the FLAG-tag-based purification scheme.
3.3. Suitable bioorthogonal chemistries for site-specific protein modification
Over the last years, new chemistries were established that allow the coupling of functional groups to a diverse range of sidechains programmed via the unnatural amino acids [1]. These developments aimed to render the coupling reaction to be highly specific, rapid and efficient under physiological conditions to ensure high labelling efficiencies and to preserve the native state of the protein. eGFPAzF was subjected to two reaction schemes: i) the relatively slow reaction between an azide and a phosphine group in a reaction called Staudinger Ligation [33] (low 10−3 M−1 s−1 range [34]) and ii) the slightly faster strain-promoted alkyne-azide cycloaddition (SPAAC) between the azide group and a strained cyclooctyne [35], [27] (approximately 10−2 to 1 M−1 s−1 [36], [27], [37]) (Fig. 1B). Both reactions follow second-order kinetics and hence, the reaction rate depends on the concentration of the reactants. Therefore, we tested different concentrations (5–400 µM) of the reactive dyes while keeping the eGFP concentrations constant (~ 0.6 µM) to determine optimal dye concentrations for the labelling reaction. We recommend to perform the labelling reaction directly on the beads used for purification of the protein. This way, the protein concentration is comparably high supporting the second-order reaction kinetics of both chemistries. But more importantly, the excess of dye used for the coupling reaction can be removed upon washing of the bead-coupled protein thereby avoiding time-consuming purification via additional chromatographic steps. Coupling of the dye via the Staudinger ligation required 200 µM of phosphine-dye to reach the maximum labelling degree (Fig. 4C, Fig. S2). Here, a labelling efficiency of about 70% was achieved (Fig. 4C). The reaction was completed after 2 h when the incubation was carried out at 37 °C. When the SPAAC reaction was employed, 200 µM of DBCO-modified dye was sufficient to reach the maximum labelling degree with a labelling efficiency of about 73% under otherwise identical conditions (Fig. 4C, Fig. S2). The fact that the azide group will be reduced in the chemical environment of the cell represents a drawback of azide-based coupling chemistries [12]. Hence, labelling efficiencies will never reach 100%. However, 100% labelling efficiency is often not necessary for many downstream applications. Additionally, we tested the strain-promoted inverse Diels Alder cycloaddition (SPIEDAC) between TCO and a tretrazine moiety. This reaction is known to be very fast with reaction rates of 1–104 M−1 s−1 [38] but it has to be noted that TCO can also undergo conversion to the less reactive cis-cyclooctene [39]. Here, maximum labelling was already achieved at very low concentrations of the tetrazine-coupled dye (5 µM) (Fig. 4C). Another factor that has to be considered is the specificity of the labelling reaction as off-target labelling and biophysical analysis of contaminating proteins alongside the target protein yields ambiguous results. In this respect, the SPAAC reaction yielded significantly more off-target labelling products than the Staudinger ligation and SPIEDAC reaction (Fig. 4C). The SPIEDAC reaction did not result in visible off-target labelling, rendering this reaction more suitable for downstream fluorescence-based analysis of the labelled protein. A second purification step can yield highly pure protein even when the SPAAC reaction or Staudinger Ligation was employed for labelling. This protein-specific purification can also be executed when using a target-specific antibody for single-molecule pulldown experiments to perform smFRET measurements on immobilised molecules thereby ensuring the selective immobilisation and analysis of the POI [26]. It is noteworthy, that – in contrast to the phosphine-modified dye - the DBCO- and tetrazine-modified dyes undergo extensive interactions with the agarose beads. Multiple washing steps did not lead to the removal of the dye and a high fraction of free dyes are found in the elution fraction alongside the POI. The presence of free dye can also cause problems in the downstream analysis of the protein.
For (single-molecule) FRET applications, the site-specific coupling of a donor and acceptor fluorophore is required. For the introduction of two distinct fluorophores into a single polypeptide chain, different strategies were developed [18]. One option is the use of a single cysteine and an uaa for labelling exploiting the different chemistries of the side chains for specific coupling [40]. Another possibility is the incorporation of different unnatural amino acids at the chosen sites. Here, a combination of different orthogonal pairs combined with respective non-sense codons (e.g. the amber and the opal stop codon) were shown to yield full-length proteins but with lower yields as compared to a single uaa incorporation event [22]. Alternatively, recoding of a conventional triplet stop codon in combination with the use of a quadruplet codon [41] (requiring an additional orthogonal ribosome) for the respective uaa and subsequent specific labelling was shown to lead to site-specific and quantitative labelling of a single protein with donor and acceptor fluorophore to allow FRET measurements [42], [43]. However, this approach was only realised in the E. coli expression system so far. Double-labelling can also be achieved when the same uaa is incorporated at two independent sites and donor and acceptor dye are added simultaneously to the protein [26]. This way, the protein will be stochastically labelled resulting in donor/donor, donor/acceptor and acceptor/acceptor pairing at the protein. Advanced single-molecule excitation schemes like ALEX (alternating laser excitation) [44], [45] or PIE (pulsed interleaved excitation) [46] allow the selection and subsequent analysis of the donor/acceptor population only. If there are no position-dependent effects on the dye, the distance between donor and acceptor is not changed if the coupling site of donor and acceptor is reversed. In our hands, stochastic labelling of the protein results in homogeneous FRET efficiency distributions as recently shown for Rpb4/7, a heterodimeric subcomplex of RNA polymerase, and human Argonaute 2 [26].
4. Conclusions
Recent developments in the field of genetic code expansion and bioorthogonal chemistries open up the possibility to engineer a non-canonical amino acid carrying a unique reactive side chain into a human protein of interest and to couple a wide variety of probes to this site including fluorescent dyes, biotins, lipids, sugars, EPR labels, crosslinkers via bioorthogonal chemistries [1], [2]. This methodological development opens up new analytical opportunities and is of special importance for biochemical and biophysical studies including single-molecule FRET measurements. We recently introduced the SLAM (site-specific labelling of endogenous mammalian proteins for smFRET measurements)-FRET workflow that overcomes many problems encountered so far when attempting to perform single-molecule FRET measurements with human proteins [26] (e.g. not all human proteins can be produced in insect cells that allow incorporation of unnatural amino acids; lack of human posttranslational modifications when producing the protein recombinantly in E. coli or insect cells; need for artificial crosslinking to stabilise a protein complex). The possibility to perform smFRET measurements with fully native human proteins or protein complexes with site-specifically engineered fluorophores allows integrated structure-function-dynamics investigations of human proteins and protein complexes [47], [48]. This approach can also provide the opportunity to screen small molecule libraries for compounds that prevent protein interactions for a given target in a high throughput format as a readout for the multimerisation status is now easily available even with minute amounts of protein (the readout would be either simple co-localisation of the donor and acceptor fluorophore or FRET). Due to minimal sample requirements smFRET measurements with labelled human proteins could be well adapted to 24-well or 96-well formats allowing high throughput screening and even automatized handling and preparation of samples.
Moreover, the possibility to introduce any desired modification in a site-specific manner paves the way for new mechanistic studies. For example, proteins can be site-specifically linked to lipids, the small protein SUMO (small ubiquitin-like modified), ubiquitin, sugars or biotins. These modifications influence the activity, conformation and cellular status of human proteins. This complexity of the eukaryotic proteomic space becomes accessible and can be studied in detail with site-specifically modified proteins. Also, spin labels for EPR measurements [49] or (photoactive) crosslinkers could be site-specifically introduced; proteins could be covalently linked for cryo-EM analysis or attached to surfaces or linkers for single-molecule force measurements via a biotin-streptavidin linkage for subsequent customised assays and as part of advanced nanostructures.
Acknowledgements
We gratefully acknowledge financial support by the Deutsche Forschungsgemeinschaft [DFG grant no. GR 3840/2-1 and SFB960-TP7 to D.G.]. Furthermore we would like to thank Thomas Sakmar and Edward Lemke for providing plasmids and Elisabeth Piechatschek and Elke Papst for technical assistance.
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.10.011.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2018.10.011.
Appendix A. Transparency document
Supplementary material
Appendix A. Supplementary material
Supplementary material
References
- 1.Koehler C., Sauter P.F., Wawryszyn M., Girona G.E., Gupta K., Landry J.J.M., Fritz M.H.-Y., Radic K., Hoffmann J.-E., Chen Z.A., Zou J., Tan P.S., Galik B., Junttila S., Stolt-Bergner P., Pruneri G., Gyenesei A., Schultz C., Biskup M.B., Besir H., Benes V., Rappsilber J., Jechlinger M., Korbel J.O., Berger I., Braese S., Lemke E.A. Genetic code expansion for multiprotein complex engineering. Nat. Methods. 2016;13:997–1000. doi: 10.1038/nmeth.4032. [DOI] [PubMed] [Google Scholar]
- 2.Neumann-Staubitz P., Neumann H. The use of unnatural amino acids to study and engineer protein function. Curr. Opin. Struct. Biol. 2016;38:119–128. doi: 10.1016/j.sbi.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Vangindertael J., Camacho R., Sempels W., Mizuno H., Dedecker P., Janssen K.P.F. An introduction to optical super-resolution microscopy for the adventurous biologist. Methods Appl. Fluoresc. 2018;6:22003. doi: 10.1088/2050-6120/aaae0c. [DOI] [PubMed] [Google Scholar]
- 4.Lowe C.R. Nanobiotechnology: the fabrication and applications of chemical and biological nanostructures. Curr. Opin. Struct. Biol. 2000;10:428–434. doi: 10.1016/s0959-440x(00)00110-x. [DOI] [PubMed] [Google Scholar]
- 5.Castillo-León J., Svendsen W.E., editors. Micro and Nanofabrication Using Self-Assembled Biological Nanostructures. William Andrew, Elsevier; Kidlington, England, Amsterdam, Boston, Heidelberg, London, New York, Oxford, Paris, San Diego, San Francisco, Singapore, Sydney, Tokyo: 2015. [Google Scholar]
- 6.Debelouchina G.T., Muir T.W. A molecular engineering toolbox for the structural biologist. Q. Rev. Biophys. 2017;50:e7. doi: 10.1017/S0033583517000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rahim M.K., Kota R., Lee S., Haun J.B. Bioorthogonal chemistries for nanomaterial conjugation and targeting. Nanotechnol. Rev. 2013:2. [Google Scholar]
- 8.Chen Y., Xianyu Y., Wu J., Yin B., Jiang X. Click chemistry-mediated nanosensors for biochemical assays. Theranostics. 2016;6:969–985. doi: 10.7150/thno.14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott T.S., Townsley F.M., Bianco A., Ernst R.J., Sachdeva A., Elsässer S.J., Davis L., Lang K., Pisa R., Greiss S., Lilley K.S., Chin J.W. Proteome labeling and protein identification in specific tissues and at specific developmental stages in an animal. Nat. Biotechnol. 2014;32:465–472. doi: 10.1038/nbt.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chin J.W. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 11.McKay C.S., Finn M.G. Click chemistry in complex mixtures: bioorthogonal bioconjugation. Chem. Biol. 2014;21:1075–1101. doi: 10.1016/j.chembiol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lang K., Chin J.W. Cellular incorporation of unnatural amino acids and bioorthogonal labeling of proteins. Chem. Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]
- 13.Davis L., Chin J.W. Designer proteins: applications of genetic code expansion in cell biology. Nat. Rev. Mol. Cell Biol. 2012;13:168–182. doi: 10.1038/nrm3286. [DOI] [PubMed] [Google Scholar]
- 14.Liu W., Brock A., Chen S., Chen S., Schultz P.G. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat. Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 15.Sun J., Chen M., Xu J., Luo J. Relationships among stop codon usage bias, its context, isochores, and gene expression level in various eukaryotes. J. Mol. Evol. 2005;61:437–444. doi: 10.1007/s00239-004-0277-3. [DOI] [PubMed] [Google Scholar]
- 16.Lang K., Chin J.W. Bioorthogonal reactions for labeling proteins. ACS Chem. Biol. 2014;9:16–20. doi: 10.1021/cb4009292. [DOI] [PubMed] [Google Scholar]
- 17.Sakamoto K., Hayashi A., Sakamoto A., Kiga D., Nakayama H., Soma A., Kobayashi T., Kitabatake M., Takio K., Saito K., Shirouzu M., Hirao I., Yokoyama S. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nikić I., Lemke E.A. Genetic code expansion enabled site-specific dual-color protein labeling: superresolution microscopy and beyond. Curr. Opin. Chem. Biol. 2015;28:164–173. doi: 10.1016/j.cbpa.2015.07.021. [DOI] [PubMed] [Google Scholar]
- 19.Monahan S.L., Lester H.A., Dougherty D.A. Site-specific incorporation of unnatural amino acids into receptors expressed in mammalian cells. Chem. Biol. 2003;10:573–580. doi: 10.1016/s1074-5521(03)00124-8. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Z., Alfonta L., Tian F., Bursulaya B., Uryu S., King D.S., Schultz P.G. Selective incorporation of 5-hydroxytryptophan into proteins in mammalian cells. Proc. Natl. Acad. Sci. USA. 2004;101:8882–8887. doi: 10.1073/pnas.0307029101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Serfling R., Coin I. Incorporation of Unnatural amino acids into proteins expressed in mammalian cells. Methods Enzymol. 2016;580:89–107. doi: 10.1016/bs.mie.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Serfling R., Lorenz C., Etzel M., Schicht G., Böttke T., Mörl M., Coin I. Designer tRNAs for efficient incorporation of non-canonical amino acids by the pyrrolysine system in mammalian cells. Nucleic Acids Res. 2018;46:1–10. doi: 10.1093/nar/gkx1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nikić-Spiegel I. Genetic code expansion- and click chemistry-based site-specific protein labeling for intracellular DNA-PAINT imaging. Methods Mol. Biol. (Clifton N. J.) 2018;1728:279–295. doi: 10.1007/978-1-4939-7574-7_18. [DOI] [PubMed] [Google Scholar]
- 24.Raulf A., Spahn C.K., Zessin P.J.M., Finan K., Bernhardt S., Heckel A., Heilemann M. Click chemistry facilitates direct labelling and super-resolution imaging of nucleic acids and proteins. RSC Adv. 2014;4:30462–30466. doi: 10.1039/c4ra01027b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikić I., Estrada Girona G., Kang J.H., Paci G., Mikhaleva S., Koehler C., Shymanska N.V., Ventura Santos C., Spitz D., Lemke E.A. Debugging eukaryotic genetic code expansion for site-specific click-PAINT super-resolution microscopy. Angew. Chem. (Int. Ed. Engl.) 2016;55:16172–16176. doi: 10.1002/anie.201608284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gust A., Jakob L., Zeitler D., Bruckmann A., Kramm K., Willkomm S., Tinnefeld P., Meister G., Grohmann D. Site-specific labelling of native mammalian proteins for single-molecule FRET measurements. Chembiochem: Eur. J. Chem. Biol. 2018 doi: 10.1002/cbic.201700696. [DOI] [PubMed] [Google Scholar]
- 27.Plass T., Milles S., Koehler C., Szymański J., Mueller R., Wiessler M., Schultz C., Lemke E.A. Amino acids for Diels-Alder reactions in living cells. Angew. Chem. (Int. Ed. Engl.) 2012;51:4166–4170. doi: 10.1002/anie.201108231. [DOI] [PubMed] [Google Scholar]
- 28.Tian H., Naganathan S., Kazmi M.A., Schwartz T.W., Sakmar T.P., Huber T. Bioorthogonal fluorescent labeling of functional G-protein-coupled receptors. Chembiochem: Eur. J. Chem. Biol. 2014;15:1820–1829. doi: 10.1002/cbic.201402193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellacheruvu D., Wright Z., Couzens A.L., Lambert J.-P., St-Denis N.A., Li T., Miteva Y.V., Hauri S., Sardiu M.E., Low T.Y., Halim V.A., Bagshaw R.D., Hubner N.C., Al-Hakim A., Bouchard A., Faubert D., Fermin D., Dunham W.H., Goudreault M., Lin Z.-Y., Badillo B.G., Pawson T., Durocher D., Coulombe B., Aebersold R., Superti-Furga G., Colinge J., Heck A.J.R., Choi H., Gstaiger M., Mohammed S., Cristea I.M., Bennett K.L., Washburn M.P., Raught B., Ewing R.M., Gingras A.-C., Nesvizhskii A.I. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods. 2013;10:730–736. doi: 10.1038/nmeth.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimple M.E., Brill A.L., Pasker R.L. Overview of affinity tags for protein purification. Curr. Protoc. Protein Sci. 2013;73 doi: 10.1002/0471140864.ps0909s73. (Unit 9.9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y. The tandem affinity purification technology: an overview. Biotechnol. Lett. 2011;33:1487–1499. doi: 10.1007/s10529-011-0592-x. [DOI] [PubMed] [Google Scholar]
- 33.Saxon E., Bertozzi C.R. Cell surface engineering by a modified Staudinger reaction. Science (New York, N.Y.) 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 34.Lin F.L., Hoyt H.M., van Halbeek H., Bergman R.G., Bertozzi C.R. Mechanistic investigation of the staudinger ligation. J. Am. Chem. Soc. 2005;127:2686–2695. doi: 10.1021/ja044461m. [DOI] [PubMed] [Google Scholar]
- 35.Agard N.J., Prescher J.A., Bertozzi C.R. A strain-promoted 3 + 2 azide-alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 2004;126:15046–15047. doi: 10.1021/ja044996f. [DOI] [PubMed] [Google Scholar]
- 36.Gordon C.G., Mackey J.L., Jewett J.C., Sletten E.M., Houk K.N., Bertozzi C.R. Reactivity of biarylazacyclooctynones in copper-free click chemistry. J. Am. Chem. Soc. 2012;134:9199–9208. doi: 10.1021/ja3000936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Agard N.J., Baskin J.M., Prescher J.A., Lo A., Bertozzi C.R. A comparative study of bioorthogonal reactions with azides. ACS Chem. Biol. 2006;1:644–648. doi: 10.1021/cb6003228. [DOI] [PubMed] [Google Scholar]
- 38.Blackman M.L., Royzen M., Fox J.M. Tetrazine ligation: fast bioconjugation based on inverse-electron-demand Diels-Alder reactivity. J. Am. Chem. Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rossin R., van den Bosch S.M., Hoeve W., ten, Carvelli M., Versteegen R.M., Lub J., Robillard M.S. Highly reactive trans-cyclooctene tags with improved stability for Diels-Alder chemistry in living systems. Bioconj. Chem. 2013;24:1210–1217. doi: 10.1021/bc400153y. [DOI] [PubMed] [Google Scholar]
- 40.Tyagi S., Lemke E.A. Genetically encoded click chemistry for single-molecule FRET of proteins. Methods Cell Biol. 2013;113:169–187. doi: 10.1016/B978-0-12-407239-8.00009-4. [DOI] [PubMed] [Google Scholar]
- 41.Neumann H., Wang K., Davis L., Garcia-Alai M., Chin J.W. Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature. 2010;464:441–444. doi: 10.1038/nature08817. [DOI] [PubMed] [Google Scholar]
- 42.Wang K., Sachdeva A., Cox D.J., Wilf N.M., Lang K., Wallace S., Mehl R.A., Chin J.W. Optimized orthogonal translation of unnatural amino acids enables spontaneous protein double-labelling and FRET. Nat. Chem. 2014;6:393–403. doi: 10.1038/nchem.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao H., Chatterjee A., Choi S.-h., Bajjuri K.M., Sinha S.C., Schultz P.G. Genetic incorporation of multiple unnatural amino acids into proteins in mammalian cells. Angew. Chem. (Int. Ed. Engl.) 2013;52:14080–14083. doi: 10.1002/anie.201308137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hohlbein J., Craggs T.D., Cordes T. Alternating-laser excitation: single-molecule FRET and beyond. Chem. Soc. Rev. 2014;43:1156–1171. doi: 10.1039/c3cs60233h. [DOI] [PubMed] [Google Scholar]
- 45.Kapanidis A.N., Laurence T.A., Lee N.K., Margeat E., Kong X., Weiss S. Alternating-laser excitation of single molecules. Acc. Chem. Res. 2005;38:523–533. doi: 10.1021/ar0401348. [DOI] [PubMed] [Google Scholar]
- 46.Müller B.K., Zaychikov E., Bräuchle C., Lamb D.C. Pulsed interleaved excitation. Biophys. J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schrimpf W., Barth A., Hendrix J., Lamb D.C. PAM: a Framework for integrated analysis of imaging, single-molecule, and ensemble fluorescence data. Biophys. J. 2018;114:1518–1528. doi: 10.1016/j.bpj.2018.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dimura M., Peulen T.O., Hanke C.A., Prakash A., Gohlke H., Am Seidel C. Quantitative FRET studies and integrative modeling unravel the structure and dynamics of biomolecular systems. Curr. Opin. Struct. Biol. 2016;40:163–185. doi: 10.1016/j.sbi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 49.Kucher S., Korneev S., Tyagi S., Apfelbaum R., Grohmann D., Lemke E.A., Klare J.P., Steinhoff H.-J., Klose D. Orthogonal spin labeling using click chemistry for in vitro and in vivo applications. J. Magn. Reson. (San. Diego Calif.: 1997) 2017;275:38–45. doi: 10.1016/j.jmr.2016.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material