Figure 2.
Fitting the Input of a Biophysical Model to In Vivo Dendritic Recordings
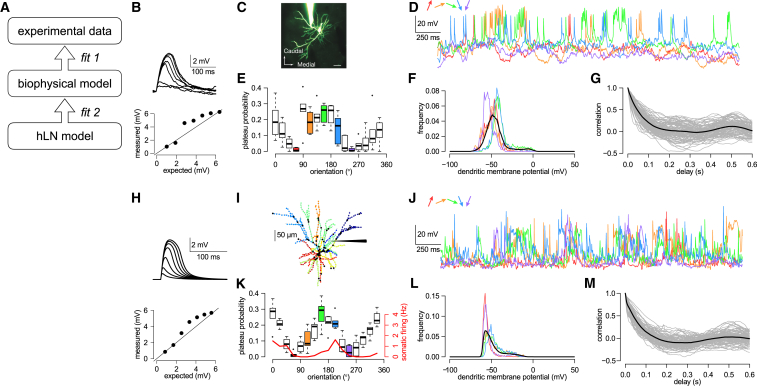
(A) Logic of the approach. We first matched the dendritic and the somatic response of a detailed biophysical model to in vivo data (fit 1). This step was required because there are no experimental measurements of the spatiotemporal activation profile for all synapses of a neuron in vivo and its corresponding output. Next, we tuned the parameters of the phenomenological hLN model to match the somatic membrane potential time course of the biophysical model in response to known synaptic inputs (fit 2).
(B) Experimental data showing nonlinear dendritic integration in a layer 2/3 pyramidal neuron in vitro (reanalyzed from Branco et al., 2010). Top: somatic responses to 1–7 glutamate uncaging events at 1 ms intervals on a single dendritic branch. Bottom: measured response amplitudes as a function of the response amplitude expected from linear integration.
(C) Two-photon microscopy image (maximum intensity projection) of an Alexa Fluor 594-filled layer 2/3 pyramidal neuron in the mouse visual cortex during a dendritic patch-clamp recording in vivo (scale bar, 20 μm). Reproduced from Smith et al. (2013).
(D) Examples of membrane potential recordings from a single dendrite in response to differently oriented drifting gratings (colors). Experimental data are from Smith et al. (2013). The same dendrite is analyzed in (E)–(G).
(E) Orientation tuning of plateau potentials in the dendritic branch. Boxplots show median, quartiles, and range of data; open circles indicate outliers.
(F) Histogram of the dendritic membrane potential for different sample input orientations (colors as in D and E) and the average across all different orientations (black).
(G) Auto-correlation of the dendritic membrane potential (gray, individual traces for each orientation and repetition; black, average).
(H) Nonlinear dendritic integration in a biophysical model layer 2/3 pyramidal neuron (analyzed as in B).
(I) Morphology of a reconstructed L2/3 pyramidal neuron and the distribution of inhibitory (black dots) and excitatory synapses (synapses with the same color received correlated inputs). Schematic electrode points to dendrite analyzed in (J)–(M).
(J) Membrane potential traces recorded in a model dendritic branch in response to sustained, in vivo-like inputs corresponding to different orientations (colors as in D).
(K–M) Orientation tuning (K), membrane potential histogram (L), and auto-correlation (M) of the model dendrite. Colors and symbols are as in (E)–(G); red line in (K) shows somatic orientation tuning. Boxplots in (K) show median, quartiles, and range of data; open circles indicate outliers.