Abstract
Yarrowia lipolytica, which is model oleaginous yeast with high industrial interest, was cultivated on fatty substrates. Data concerning fatty acid composition of both substrate and yeast lipids and comparisons of the experimental data with model predictions presented in “Biomodification of fats and oils and scenarios of adding value on renewable fatty materials through microbial fermentations: Modelling and trials with Yarrowia lipolytica” (Vasiliadou et al., 2018) were provided. Furthermore, the total yeast lipids were fractionated into their main fractions, that is, phospholipids, glucolipids plus sphingolipids and neutral lipids, and the fatty acid composition of each lipid fraction was reported.
Abbreviations: t (h), fermentation time; L, cellular lipid; x, cell mass; NL, neutral lipids; G+S, glycolipids plus sphingolipids; P, phospholipids
Specifications table
| Subject area | Biotechnology, Chemistry |
| More specific subject area | Lipid Biotechnology |
| Type of data | Tables, figures |
| How data was acquired | The yeast Yarrowia lipolytica was cultivated on fatty substrates and the fatty acid composition of both the extracellular and intracellular lipids, as well as of their fractions was determined using an Agilent 7890 A device Gas Chromatography (Agilent Technologies, Shanghai, China). |
| Data format | Raw samples were collected during growth of Y. lipolytica and processed. Substrate and cellular lipids were purified and analysed. |
| Experimental factors | Different fatty materials were used as substrates for Y. lipolytica. |
| Experimental features | Various fats of plant (i.e., olive, sunflower, palm and linseed) and animal (i.e., cod liver and beef tallow) origin were used as carbon substrates for Y. lipolytica. Cultures, carried out in 250-mL Erlenmeyer flasks, were incubated in a rotary shaker (ZHWY211C, Zhicheng, Shanghai, China) at 180 rpm and T=28±1 °C. |
| Data source location | University of Patras, Greece |
| Data accessibility | The data are available in this article |
| Related research article | [1] Vasiliadou et al., 2018 “Biomodification of fats and oils and scenarios of adding value on renewable fatty materials through microbial fermentations: Modelling and trials with Yarrowia lipolytica.” Journal of Cleaner Production, 200, 1111–1129. |
Value of the data
-
•
The data can be used in order to identify the fatty acid specificity of Yarrowia lipolytica.
-
•
The composition of lipids (i.e., mainly neutral) accumulated in Y. lipolytica can be pre-determined.
-
•
New biomodification processes of common fats can be designed.
1. Data
The data article includes Table 1 reporting fatty acid composition of lipid fractions of Yarrowia lipolytica growing on olive oil, linseed oil, palm oil, sunflower oil, cod liver oil, and beef tallow, and two Figures showing: (1) Experimental data and theoretical predictions of the fatty acid composition of extracellular and intracellular lipids of Y. lipolytica and (2) theoretical fatty acid profiles of the free fatty acid fraction released in the growth medium during growth of Y. lipolytica on the above mentioned fatty substrates.
Table 1.
Fatty acid composition of lipids accumulated in Yarrowia lipolytica growing on various fats of plant or animal origin.
| Culture on olive oil | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| t (h) | L/x %, w/w | Lipid fractions | % in total lipids | C16:0 | C16:1 | C18:0 | C18:1 | C18:2 | Others |
| 109 | 28.0 | NL | 94.0 | 7.3 | 5.6 | 1.2 | 70.0 | 15.1 | 0.7 |
| G + S | 4.2 | 9.2 | 5.4 | 2.7 | 65.9 | 15.2 | 0.6 | ||
| P | 1.9 | 11.7 | 7.7 | 0.7 | 44.0 | 35.7 | 0.4 | ||
| 335 | 13.0 | NL | 96.0 | 7.2 | 10.0 | 1.8 | 60.9 | 19.2 | 1.0 |
| G + S | 2.7 | 14.6 | 10.9 | 1.2 | 52.5 | 18.9 | 1.9 | ||
| P | 1.4 | 10.6 | 13.3 | 2.1 | 49.7 | 22.8 | 1.6 | ||
| Culture on linseed oil | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| t (h) | L/x %, w/w | Lipid fractions | % in total lipids | C16:0 | C16:1 | C18:0 | C18:1 | C18:2 | C18:3α | Others |
| 72 | 21.9 | NL | 86.5 | 4.3 | 1.6 | 1.3 | 15.1 | 18.9 | 58.6 | 0.2 |
| G + S | 10.3 | 5.5 | 1.6 | 2.0 | 21.2 | 17.9 | 36.3 | 15.6 | ||
| P | 3.3 | 16.1 | 2.8 | 2.4 | 25.5 | 20.9 | 24.7 | 7.6 | ||
| 263 | 12.2 | NL | 92.0 | 4.6 | 4.5 | 2.3 | 21.2 | 19.4 | 47.9 | 0.2 |
| G + S | 4.6 | 10.2 | 5.4 | 7.5 | 24.8 | 16.8 | 35.0 | 0.4 | ||
| P | 3.7 | 15.8 | 7.9 | 1.6 | 29.5 | 17.0 | 28.2 | – | ||
| Culture on palm oil | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| t (h) | L/x %, w/w | Lipid fractions | % in total lipids | C16:0 | C16:1 | C18:0 | C18:1 | C18:2 | Others |
| 67 | 28.5 | NL | 90.2 | 23.6 | 3.9 | 2.1 | 51.1 | 19.2 | 0.2 |
| G + S | 5.5 | 23.8 | 1.9 | 3.8 | 39.1 | 15.8 | 15.6 | ||
| P | 4.3 | 15.8 | 7.2 | 1.3 | 31.5 | 39.2 | 5.0 | ||
| 238 | 6.9 | NL | 94.7 | 21.2 | 6.0 | 5.5 | 44.3 | 22.7 | 0.4 |
| G + S | 2.6 | 24.0 | 4.4 | 3.8 | 40.7 | 24.3 | 2.8 | ||
| P | 2.8 | 15.8 | 7.2 | 1.3 | 31.5 | 39.2 | 5.0 | ||
| Culture on sunflower oil | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| t (h) | L/x %, w/w | Lipid fractions | % in total lipids | C16:0 | C16:1 | C18:0 | C18:1 | C18:2 | Others |
| 72 | 25.5 | NL | 87.5 | 5.2 | 1.9 | 2.8 | 30.9 | 55.7 | 3.4 |
| G + S | 10.0 | 5.1 | 2.0 | 2.3 | 24.8 | 32.6 | 33.1 | ||
| P | 2.5 | 13.2 | 6.1 | 1.7 | 28.0 | 41.6 | 9.3 | ||
| 357 | 4.6 | NL | 84.4 | 4.2 | 4.3 | 2.2 | 30.0 | 55.1 | 4.2 |
| G + S | 11.8 | 9.2 | 3.4 | 4.5 | 24.4 | 25.1 | 33.3 | ||
| P | 3.8 | 10.8 | 7.3 | 0.9 | 31.0 | 38.0 | 12.0 | ||
| Culture on cod liver oil | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| t (h) | L/x %,w/w | Lipid fractions | % in total lipids | C16:0 | C16:1 | C18:0 | C18:1 | C18:2 | C18:3 | C20:1 | C20:5 | C22:6 | Others |
| 72 | 25.5 | NL | 89.6 | 16.0 | 14.5 | 3.0 | 31.7 | 6.9 | 9.0 | 7.4 | 2.1 | 3.0 | 6.5 |
| G + S | 7.8 | 7.6 | 5.5 | 2.2 | 18.3 | 7.6 | 4.2 | 16.6 | – | 3.9 | 34.1 | ||
| P | 2.6 | 10.8 | 11.7 | 2.7 | 41.0 | 22.4 | 3.1 | 3.6 | – | 0.2 | 4.4 | ||
| 357 | 4.6 | NL | 86.0 | 11.1 | 17.2 | 3.4 | 37.7 | 10.1 | 2.5 | 5.4 | 0.5 | 0.7 | 11.5 |
| G+S | 10.4 | 11.0 | 9.9 | 3.5 | 31.6 | 6.8 | 7.1 | 9.9 | 2.1 | 1.0 | 17.1 | ||
| P | 3.6 | 8.1 | 13.8 | 1.5 | 44.7 | 21.7 | 3.0 | – | – | 0.5 | 6.7 | ||
| Culture on beef tallow | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| t (h) | L/x %,w/w | Lipid fractions | % in total lipids | C16:0 | C16:1 | C18:0 | C18:1 | C18:2 | Others |
| 96 | 2.1 | NL | 88.8 | 13.9 | 7.1 | 40.3 | 30.6 | 4.9 | 3.2 |
| G + S | 5.6 | 15.7 | 4.5 | 32.3 | 22.8 | 4.9 | 19.8 | ||
| P | 5.6 | 13.6 | 14.1 | 10.5 | 28.0 | 21.0 | 12.8 | ||
| 235 | 9.5 | NL | 95.5 | 15.3 | 9.9 | 26.4 | 36.6 | 7.3 | 4.5 |
| G + S | 2.5 | 11.7 | 5.6 | 22.5 | 24.0 | 7.3 | 28.9 | ||
| P | 2.0 | 12.9 | 12.7 | 3.4 | 37.3 | 24.8 | 9.0 | ||
Culture conditions: pH 6.0 ± 0.5; T = 28 °C; agitation rate 280 rpm. Data represent means of two replicates.
2. Experimental design, material, and methods
The yeast Y. lipolytica ACA-DC 50109 was used in the current investigation. The strain was maintained on potato dextrose agar (PDA, Conda, Madrid, Spain) at 7 ± 1 °C and re-cultured twice a month.
The growth media contained (in g/L): MgSO4.7H2O (Fluka, Steinheim, Germany), 1.5; KH2PO4 (Fluka), 7.0; Na2PO4 (Fluka), 2.0; CaCl2.2H2O (Carlo Erba, Rodano, Italy), 0.1; ΖnSO4.7H2O (Merck, Darmstadt, Germany), 0.001; CuSO4.5H2O (BDH, Poole, England), 0.0001; Co(NO3)3.3H2O (Merck), 0.0001; MnSO4.5H2O (Fluka), 0.0001; (NH4)2SO4 (Fluka), 0.5; yeast extract (Sigma, Steinheim, Germany), 2.0. Various commercial fats of plant (i.e., olive, sunflower, palm, and linseed) and animal (i.e., cod liver and beef tallow) origin were used as carbon and energy sources at a concentration of 25 g/L.
Experiments were performed in 250-mL Erlenmeyer flasks. The flasks containing 50±1 mL of growth media were sterilized at 121 °C for 20 min and thereafter inoculated with 1 mL of a mid-exponential phase pre-culture containing 4×106 cells/mL. The cultures were incubated in a rotary shaker (ZHWY211C, Zhicheng, Shanghai, China) at 180 rpm and T = 28 ± 1 °C.
Determination of extracellular and intracellular lipids was performed as described in [2]. Intracellular lipids were fractionated as described in [3]. Fatty acid moieties of both extracellular and intracellular lipids and their fractions were converted into fatty acid methyl-esters (FAMEs) and analysed by using a Gas Chromatography (GC; Agilent 7890 A device, Agilent Technologies, Shanghai, China) as described in [4].
The predictions have been obtained using the mathematical model which is presented in [1].
Experiments were performed in duplicate. Data represent means of two replicates. (Fig. 1, Fig. 2)
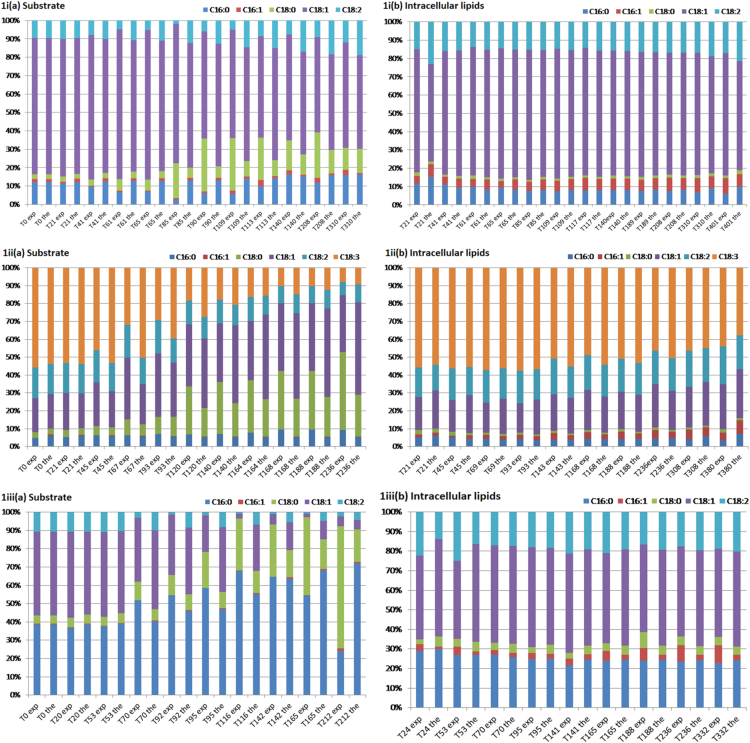
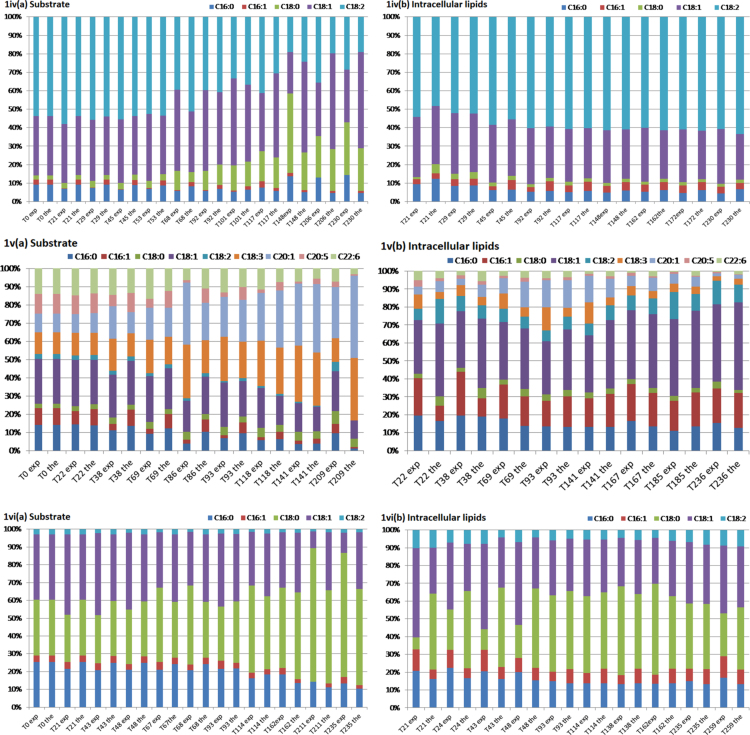
Fig. 1.
Experimental data and theoretical predictions of the fatty acid composition (%) of extracellular (a) and intracellular (b) lipids of Yarrowia lipolytica cultivated on: (i) Olive oil, (ii) linseed oil, (iii) palm oil, (iv) sunflower oil, (v) cod liver oil, and (vi) beef tallow. Culture conditions: As in Table 1.
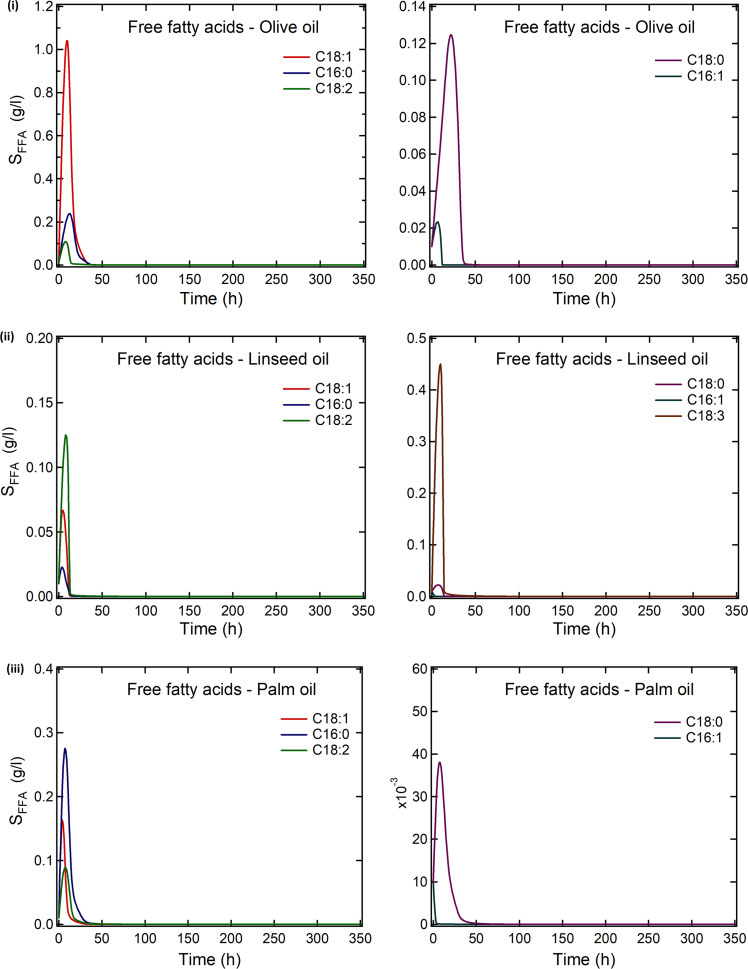
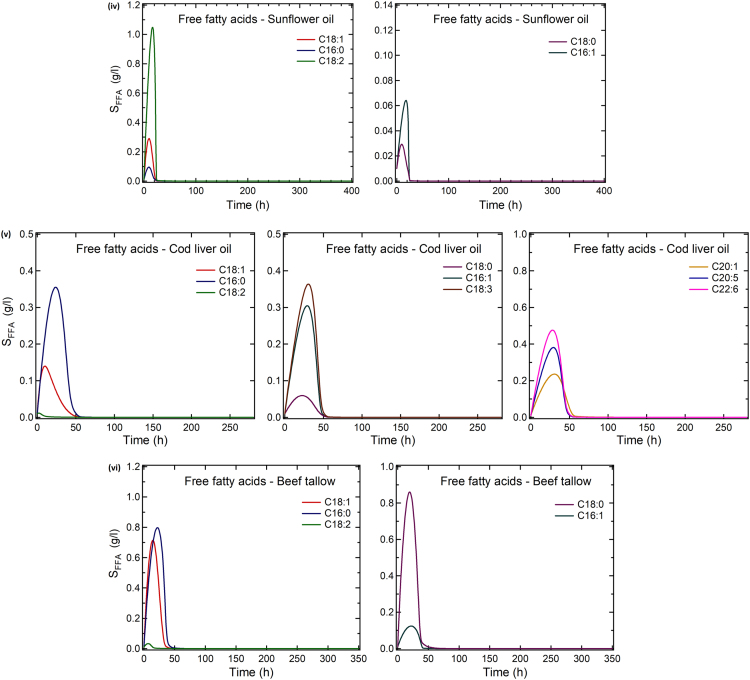
Fig. 2.
Theoretical fatty acid profiles of the free fatty acid fraction released in the growth medium (g/l) vs. time when Yarrowia lipolytica was cultivated on: (i) Olive oil, (ii) linseed oil, (iii) palm oil, (iv) sunflower oil, (v) cod liver oil, and (vi) beef tallow. Culture conditions: As in Table 1.
Acknowledgments
We acknowledge support of this work by the project “INVALOR: Research Infrastructure for Waste Valorization and Sustainable Management” (MIS 5002495) which is implemented under the Action “Reinforcement of the Research and Innovation Infrastructure”, funded by the Operational Programme "Competitiveness, Entrepreneurship and Innovation" (NSRF 2014–2020) and co-financed by Greece and the European Union (European Regional Development Fund).
Footnotes
Transparency data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.10.116.
Transparency document. Supplementary material
Supplementary material
References
- 1.Vasiliadou I., Bellou S., Daskalaki A., Tomaszewska- Hetman L., Chatzikotoula C., Kompoti B., Papanikolaou S., Vayenas D., Pavlou S., Aggelis G. Biomodification of fats and oils and scenarios of adding value on renewable fatty materials through microbial fermentations: modelling and trials with Yarrowia lipolytica. J. Clean. Prod. 2018;200:1111–1129. [Google Scholar]
- 2.Bellou S., Makri A., Sarris D., Michos K., Rentoumi P., Celik A., Papanikolaou S., Aggelis G. The olive mill wastewater as substrate for single cell oil production by Zygomycetes. J. Biotechnol. 2014;170:50–59. doi: 10.1016/j.jbiotec.2013.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Bellou S., Triantaphyllidou I.-E., Mizerakis P., Aggelis G. High lipid accumulation in Yarrowia lipolytica cultivated under double limitation of nitrogen and magnesium. J. Biotechnol. 2016;234:116–126. doi: 10.1016/j.jbiotec.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Dourou M., Aggeli D., Papanikolaou S., Aggelis G. Critical steps in carbon metabolism affecting lipid accumulation and their regulation in oleaginous microorganisms. Appl. Microbiol. Biotechnol. 2018;102:2509–2523. doi: 10.1007/s00253-018-8813-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material