Abstract
Detection of RAS and BRAF mutations is essential to determine the optimal treatment strategy for metastatic colorectal cancer (CRC). We prospectively evaluated the MEBGEN RASKET-B KIT (RASKET-B), a novel multiplex kit, simultaneously detecting 48 types of RAS mutations and the BRAF V600E mutation using Luminex xMAP technology. The aim was to obtain market approval for RASKET-B as an in vitro diagnostic (IVD) option in Japan. Genomic DNA was extracted from 302 formalin-fixed paraffin-embedded tissues obtained from CRC patients. The primary endpoints were the concordance rate (CR) between the results from RASKET-B and the previously approved IVD kit (RASKET) for RAS mutations, and CR between the results from RASKET-B and direct sequencing (DS) for BRAF mutations. The secondary endpoints included the CR between RASKET-B and DS for RAS mutations and between RASKET-B and the pyrosequencing (PYRO) for the BRAF V600E mutation. Among the 302 samples, 142 RAS mutations (47%) and 18 BRAF V600E mutations (6.0%) were detected by RASKET-B. All mutations detected in the recruited patients were mutually exclusive. Both RAS and BRAF mutation rates were statistically higher in right-sided than left-sided CRC. The CR between RASKET-B and RASKET for RAS gene and RASKET-B and DS for BRAF V600E mutation was 100% for both (95% CI: 99%-100%). The results from RASKET-B were also highly concordant with DS for RAS (97.4%) and with PYRO for the BRAF (V600E) gene (99.7%). RASKET-B thus provides rapid, precise, and simultaneous detection of RAS and BRAF mutations in CRC.
Introduction
RAS (KRAS and NRAS) mutations are present in approximately 50% to 55% of colorectal cancer (CRC) cases. The clinical significance of the detection of RAS mutations has been previously established as a required test prior to the initiation of anti–epidermal growth factor receptor (EGFR) antibody therapy to predict the efficacy in metastatic CRC [1], [2], [3], [4], [5]. Prospective-retrospective biomarker analyses in randomized clinical trials have consistently demonstrated that anti-EGFR antibodies, cetuximab and panitumumab, are unlikely to benefit patients with KRAS exon 3 and 4 and NRAS exons 2, 3, and 4 mutations, in addition to those with a KRAS exon 2 mutation [6], [7], [8]. Moreover, recent results from clinical trials revealed that overall survival is possibly better when patients are treated with anti-EGFR therapy as a first-line treatment than when treated with bevacizumab in the RAS wild-type population [9], [10]. This suggests that RAS mutation status has a large impact on the treatment decision in patients with metastatic CRC.
Many studies have reported that the BRAF V600E mutation is detected in approximately 5%-12% of metastatic CRC patients. RAS and BRAF V600E mutations are almost mutually exclusive [11]. Unlike RAS mutations, the predictive value of BRAF mutations for anti-EGFR mAb efficacy is less certain. On the other hand, the BRAF V600E mutation leads to a poor prognosis or rapid progression, regardless of treatment in metastatic CRC [12], [13]. Recently, the possibility was reported that triplet chemotherapy combining 5-fluorouracil, oxaliplatin, and irinotecan (FOLFOXIRI) with bevacizumab is more effective than other chemotherapies for patients with the BRAF V600E mutation [14], [15], and both European Society for Medical Oncology (ESMO) consensus guidelines and pan-Asian adapted ESMO consensus guidelines recommend FOLFOXIRI plus bevacizumab as the preferred choice for these patients [16], [17]. Therefore, the BRAF mutation status should be assessed before starting the first-line chemotherapy. The latest edition of the Japanese Society of Medical Oncology Clinical Guidelines: Molecular Testing for Colorectal Cancer Treatment states that proper testing for BRAF V600E mutation and mismatch repair deficiency is necessary in addition to testing for RAS mutation [18].
We previously reported that the MEBGEN RASKET KIT (RASKET) is useful for rapid detection of 48 types of mutations in codons 12, 13, 59, 61, 117, and 146 of KRAS and NRAS using PCR-reverse sequence specific oligonucleotide (PCR-rSSO) and xMAP technology [19]. The RASKET clinical validation study confirmed the precise detection of RAS mutations, with a concordance rate (CR) of 98.4% between the RASKET KIT and direct sequencing in RAS mutations (UMIN000011781). The RASKET KIT was approved in Japan as an in vitro diagnostic (IVD) and has become widely used in daily practice and is recognized as an RAS testing platform in Japan.
As mentioned above, the detection of RAS and BRAF mutations is an essential step for decision-making regarding therapeutic approaches and predicting resistance to EGFR-targeted therapy. The PCR-rSSO and xMAP technologies allow multiplex molecular testing in a single well. It would be clinically beneficial to develop a new kit for the simultaneous detection of BRAF V600E mutations and RAS gene mutations. In this study, we evaluated the newly developed MEBGEN RASKET-B KIT (RASKET-B) to detect 48 different RAS amino acid mutations and the BRAF V600E mutation in CRC patients. This study was performed as a registration trial for regulatory approval of the kit in Japan.
Material and Methods
Patients and Tumor Samples
The RASKET-B study used the identical cohort and the DNA sample sets that were used in the RASKET study (Study ID: UMIN 000011784) [19]. Briefly, the eligibility criteria for patients were 1) histologically confirmed adenocarcinoma of colorectal origin, 2) age ≥20 years at the time of informed consent, and 3) patients' written consent for participation in the study. Patients with insufficient amounts of formalin-fixed paraffin-embedded (FFPE) tissues, those with an undetermined RAS status by the RASKET kit in the previous RASKET study, and those who withdrew consent were excluded from the RASKET-B study. One central pathologist assigned for the study microscopically confirmed cancer in each patient, classified the tumor into the appropriate histologic type, calculated the tumor area ratio and tumor cell ratio, and then marked the tumor area on the prepared hematoxylin and eosin–stained slides for manual microdissection (MMD).
Study Design
All specimens were anonymized. Only the participating affiliations were able to access patients' information using a correspondence table, which was only available at each study site to eliminate any disclosure to outsiders. Sample anonymity was the task of one employee of G&G Science Co., Ltd., who was not involved in the study.
The set of extracted DNA from the FFPE specimens was sent to three different reference laboratories (G&G Science Co., Ltd.; Health Sciences Research Institute, Inc.; and SRL Inc.), where independent assays were performed with RASKET-B, direct sequencing (DS), and pyrosequencing (PYRO), respectively. All samples were deidentified and blinded to the tissue genotype and clinical characteristics of each patient.
The primary endpoints in this study were the CR between results from RASKET-B and RASKET for RAS mutations, and the CR between RASKET-B and DS for BRAF mutations. The results of RAS mutations by the RASKET study [19] were used for comparison with those obtained by RASKET-B. As the secondary endpoints, we determined the CR between results from RASKET-B and DS for the RAS gene, and results from RASKET-B and PYRO for the BRAF gene. In addition, the accuracy of genotyping was evaluated by comparing data between RASKET-B and DS. The original and revised protocols were approved by the ethical committees in each of the participating affiliations. The study was conducted in accordance with the Declaration of Helsinki and ethical guidelines for clinical research.
Direct Sequencing
After the pathological confirmation of cancer in each patient, 10-μm–thick sections were processed by MMD. DNA extraction was performed with QIAamp DNA FFPE Tissue Kit (Qiagen, Venlo, Netherlands) according to the manufacturer's protocol and as previously reported [19], [20]. Briefly, each extracted DNA was amplified using six sets of primers to amplify exon 2, exon 3, and exon 4 in KRAS and NRAS, and exon 15 in BRAF. The mutations in these regions were detected using the BigDye Terminator Cycle Sequencing Kit (Thermo Fischer Scientific, Waltham, MA).
Pyrosequencing
DNA samples were analyzed for codon 600 in exon 15 of BRAF with the Therascreen BRAF Pyro Kit (Qiagen) as described by the manufacturer's protocol. The PYRO method was performed without MMD. For one patient with a discrepancy between RASKET-B and PYRO, MMD was additionally performed for PYRO to carefully confirm the existence of BRAF-mutated tumor cells.
Assay with MEBGEN RASKET-B KIT
Extracted DNA samples were diluted to a concentration of 10-20 ng/μl with sterile TE buffer (1 mmol/l Tris–HCL [pH = 8.0], 0.1 mmol/L EDTA). Assays with the RASKET-B kit (MBL, Nagoya, Japan) were performed according to the manufacturer's protocol. Briefly, a 5-μl template of each sample was mixed with 20 μl master mix, including primers, Taq DNA polymerase, and uracil-DNA-glycosylase. Reactions were heated for 5 minutes at 40°C and 2 minutes at 95°C; 10 repeating cycles of 94°C for 20 seconds and 62°C for 30 seconds; 45 repeating cycles of 90°C for 20 seconds, 60°C for 30 seconds, and 72°C for 30 seconds; and finally 72°C for 1 minute and 94°C for 10 minutes. Each amplification product was then hybridized to mutation detection probes and immobilized with color-coded beads. Five microliters of PCR products and 45 μl of hybridization solution containing probe-coupled beads were hybridized at 95°C for 2 minutes followed by 55°C for 30 minutes. After washing, the PCR amplification-bead complexes were reacted with streptavidin-phycoerythrin at 52°C for 15 minutes. Using the Luminex 100/200 system (Luminex, Austin, TX), the median fluorescence intensity was determined for the color-coded beads and PE, which represented the types of RAS mutations and their signal intensities, respectively. UniMAG (MBL, Nagoya Japan) data analysis software was used to analyze the raw data from Luminex 100/200. Thus, using the RASKET-B KIT, we simultaneously examined 12 types of RAS exon 2 (G12S, G12C, G12R, G12D, G12 V, G12A, G13S, G13C, G13R, G13D, G13V, and G13A), 8 types of RAS exon 3 (A59T, A59G, Q61K, Q61E, Q61L, Q61P, Q61R, and Q61H), 4 types of RAS exon 4 (K117N, A146T, A146P, and A146V) mutations, and BRAF exon 15 (V600E). The evaluation criterion for the performance of the RASKET-B was CR ≥90% with the DS, PYRO, and RASKET reference assays in the primary and secondary endpoint analyses.
TaqMan Assays
In the case of any controversial data between the RASKET-B and the reference assays, we confirmed the results with TaqMan Mutation Detection Assays (Thermo Fisher Scientific) [21].
Statistical Analysis
The number of specimens required for the RASKET-B study was estimated as the acceptable number of specimens to satisfy that the CR between the RASKET and the reference assays would be >90%, at the lower limit of the 95% confidence interval. We then determined the number of specimens for the RASKET-B study needed to exceed 278. Patients' demographic and disease characteristics were reported as standard statistics. Statistical analysis was carried out using StatFlex (Artech Co. Ltd., Osaka, Japan). The RASKET-B study is registered with UMIN22742.
Results
Patients
Tissues were obtained from 309 consenting patients with histologically confirmed CRC. Insufficient amounts of FFPE were available in four patients, and RAS gene status in three patients was not reportable using RASKET in the previous RASKET study. Therefore, 302 patients were eligible for the primary endpoint analysis (Figure 1) in the RASKET-B study. Patient data are provided in Table 1.
Figure 1.
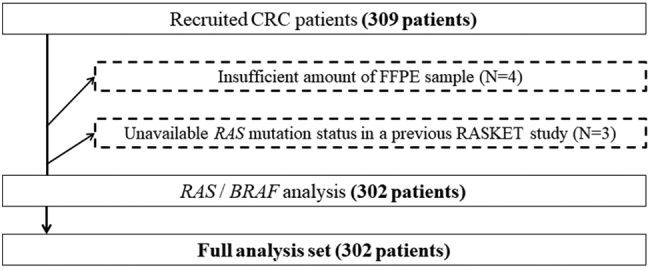
Study design and patient eligibility for primary and secondary endpoint analyses. Among 309 assessable samples, 4 were excluded because of an insufficient amounts of FFPE samples, and 3 were excluded because of unavailability of RAS mutation status in the previous study.
Table 1.
Patient Characterization in the Blinded Clinical Evaluation Study for the MEBGEN RASKET-B KIT
| Total (N = 302) |
||
|---|---|---|
| Age, years | Median (minimum-maximum) | 64 (26-89) |
| Sex | Male | 178 |
| Female | 124 | |
| Primary tumor location | Right-sided colon* | 62 |
| Left-sided colon† | 98 | |
| Rectum‡ | 142 | |
| Stage | Stage 0-I | 33 |
| Stage II | 48 | |
| Stage III | 125 | |
| Stage IV | 96 | |
| Histologic type | Well to moderately differentiated adenocarcinoma | 273 |
| Poorly differentiated or mucinous | 29 | |
| Tumor area ratio (%) | Median (minimum-maximum) | 50 (5-100) |
| Tumor cell ratio (%) | Median (minimum-maximum) | 50 (5-90) |
Cecum, ascending colon, and transverse colon.
Descending colon and sigmoid colon.
Rectum (Ra, and Rb). Ra: upper rectum (above peritoneal reflection), Rb: lower rectum (below peritoneal reflection).
Frequency of RAS and BRAF Mutations in MEBGEN RASKET-B KIT
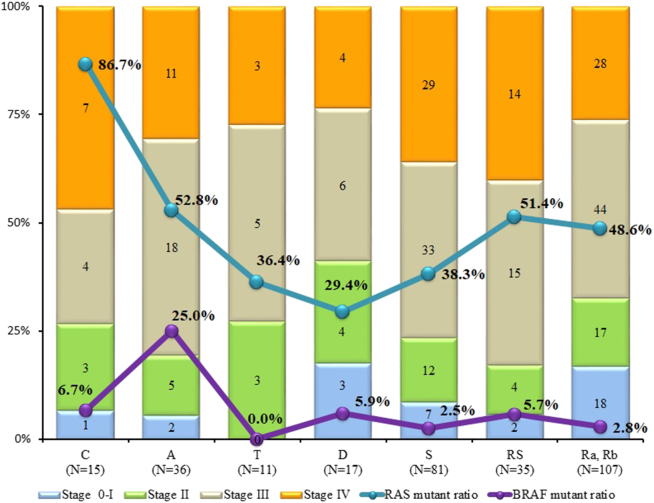
Among the 302 samples, RASKET-B detected 142 RAS mutations [113 (37.4%) KRAS exon 2, 5 (1.7%) KRAS exon 3, 10 (3.3%) KRAS exon 4, 6 (2.0%) NRAS exon 2, 8 (2.6%) NRAS exon 3] and 18 (6.0%) BRAF exon 15 mutations (Table 2). All mutations detected from the recruited patients were mutually exclusive. Both RAS and BRAF mutation rates were statistically higher in colon cancer on the right side than on the left side. Especially, the frequency of BRAF mutations among patients with RAS wild type was six-fold higher on the right side than on the left side (Table 3). Among patients with ascending colon cancer, 25% (9/36) had BRAF V600E mutations, a higher frequency than the other tumor locations (Figure 2). In terms of histologic types, the frequency of BRAF V600E mutations was significantly higher in patients with poorly differentiated or mucinous colon cancer. Also, the frequency of BRAF V600E mutations in females was not significant but tended to be higher than in males (P = .0745, Table 3).
Table 2.
Frequency of Breakdown of RAS and BRAF Mutations Detected in Colorectal Cancer Patients
| Mutation Status | No. of Cases | Proportion Among 302 Cases |
|---|---|---|
| WT RAS or BRAF | 142 | 47.0% |
| KRAS exon 2 mutant | 113 | 37.4% |
| p.G12S | 5 | 1.7% |
| p.G12C | 8 | 2.6% |
| p.G12R | 4 | 1.3% |
| p.G12D | 44 | 14.6% |
| p.G12 V | 23 | 7.6% |
| p.G12A | 6 | 2.0% |
| p.G12A, p.G12R | 1 | 0.3% |
| p.G13D | 20 | 6.6% |
| p.G12D, p.G13D | 2 | 0.7% |
| Other KRAS exon 2 mutant* | 0 | 0.0% |
| KRAS exon 3 mutant | 6 | 2.0% |
| p.A59E | 1 | 0.3% |
| p.Q61H | 5 | 1.7% |
| Other KRAS exon 3 mutant* | 0 | 0.0% |
| KRAS exon 4 mutant | 10 | 3.3% |
| p.K117N | 2 | 0.7% |
| p.A146T | 6 | 2.0% |
| p.A146P | 1 | 0.3% |
| p.A146V | 1 | 0.3% |
| NRAS exon 2 mutant | 6 | 2.0% |
| p.G12D | 4 | 1.3% |
| p.G12V | 2 | 0.7% |
| Other NRAS exon 2 mutants | 0 | 0.0% |
| NRAS exon 3 mutant | 8 | 2.6% |
| p.Q61K | 2 | 0.7% |
| p.Q61L | 5 | 1.7% |
| p.Q61R | 1 | 0.3% |
| Other NRAS exon 3 mutant† | 0 | 0.0% |
| NRAS exon 4 mutant‡ | 0 | 0.0% |
| BRAF V600E mutant | 18 | 6.0% |
*KRAS p.G13S, p.G13R, p.G13V, and p.G13A.
**KRAS p.A59T, p.A59G, p.Q61K, p.Q61E, p.Q61L, p.Q61P, and p.Q61R.
§RAS p.G12S, pG12C, p.G12R, and p.G12A.
†NRAS p.A59T, p.A59G, pQ61E, p.Q61P, and Q61H.
‡NRAS p.K117N, p.A146T, p.A146P, and p.A146V.
Table 3.
Correlation Between Tumor Location Side and Mutation Status, Sex, or Tissue Type
| n |
RAS |
BRAF |
Braf mutant/ras Wild Type |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild Type |
Mutant |
P Value | Wild Type |
Mutant |
P Value | ||||
| 160 | 142 (47.0%) | 284 | 18 (6.0%) | 18/160 (11.3%) | |||||
| Age, median (minimum-maximum) | 64 (33-89) | 63 (26-84) | P = .275 | 63 (26-85) | 70 (46-89) | P = .0077 | |||
| Sex (%) | Male | 178 | 101 | 77 (43.3%) | P = .117 | 171 | 7 (3.9%) | P = .0745 | 7/77 (9.1%) |
| Female | 124 | 59 | 65 (52.4%) |
113 | 11 (8.9%) | 11/59 (18.6%) | |||
| Histologic types (%) | Well to moderately differentiated adenocarcinoma | 273 | 144 | 129 (47.3%) | P = .804 | 263 | 10 (3.7%) | P < .0001 | 10/144 (6.9%) |
| Poorly differentiated or Mucinous | 29 | 16 | 13 (44.8%) | 21 | 8 (38.1%) | 8/16 (50%) | |||
| Primary tumor location (%) | Right-sided colon | 62 | 26 | 36 (58.1%) | P = .0506 | 52 | 10 (16.1%) | P < .0001 | 10/26 (38.5%) |
| Left-sided colon and Rectum | 240 | 134 | 106 (44.2%) | 232 | 8 (3.3%) | 8/134 (6.0%) | |||
Each parameter was analyzed via the Fisher's test, except age, which was analyzed via the Student's t test.
Figure 2.
Frequency of RAS and BRAF mutations for each tissue location and stage.
Concordance of RAS and BRAF Status
In the primary endpoint analysis, the CR between results concerning RAS mutation status obtained from RASKET-B and RASKET was 100% (302/302) (95% CI: 98.8%-100%) (Table 4A). Concerning the BRAF mutation status, the CR between results obtained from RASKET-B and DS was also 100% (302/302) (95% CI: 98.8%-100%) (Table 4B).
Table 4A.
Consistency of RAS Mutation Status (Primary Endpoint Analysis)
| RASKET |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| RASKET-B | Positive | 142 | 0 | 142 |
| Negative | 0 | 160 | 160 | |
| Total | 142 | 160 | 302 | |
| Overall agreement percentage | 100% (95% CI, 98.8%-100%) | |||
| Positive agreement percentage | 100% (95% CI, 98.8%-100%) | |||
| Negative agreement percentage | 100% (95% CI, 98.8%-100%) | |||
Table 4B.
Consistency of BRAF V600E Mutation Status (Primary Endpoint Analysis)
| DS |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| RASKET-B | Positive | 18 | 0 | 18 |
| Negative | 0 | 284 | 284 | |
| Total | 18 | 284 | 302 | |
| Overall agreement percentage | 100% (95% CI, 98.8%-100%) | |||
| Positive agreement percentage | 100% (95% CI, 98.8%-100%) | |||
| Negative agreement percentage | 100% (95% CI, 98.8%-100%) | |||
In the secondary endpoint analyses of all samples, the CR between RASKET-B and DS was 97.4% (294/302) (95% CI: 94.9%-98.9%) in RAS (Table 5A). Among the eight samples with conflicting results between RASKET-B and DS, six samples were positive with RAS mutations (two cases with KRAS G12D, two cases with KRAS G13D, one case with KRAS G12R, and one case with KRAS Q61H) in RASKET-B but negative in DS. Genotypic results of these samples in RASKET-B were consistent with RASKET and with results from the TaqMan Mutation Detection Assay, which is a more sensitive method (cutoff levels 0.1%-1%). The other two samples were negative with RAS mutations in RASKET-B and positive with DS. One of the samples had two mutations in KRAS codon 11 and codon 12 (G12C). The other sample had a KRAS mutation in A59E, which was not covered by RASKET-B (Table 6).
Table 5A.
Consistency Between RASKET-B and DS in RAS Gene Mutations (Secondary Endpoint-1 Analysis)
| DS |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| RASKET-B | Positive | 136 | 6 | 142 |
| Negative | 2 | 158 | 160 | |
| Total | 138 | 164 | 302 | |
| Overall agreement percentage | 97.4% (95% CI, 94.9%-98.9%) | |||
| Positive agreement percentage | 95.8% (95% CI, 92.1%-99.5%) | |||
| Negative agreement percentage | 98.8% (95% CI, 96.4%-100%) | |||
Table 6.
Discrepancy Samples in RAS Gene Mutation Detection Between the RASKET-B KIT and DS
| RASKET-B | DS | Percentage of Mutant DNA in TaqMan Detection Assays |
|---|---|---|
| KRAS p.G12D | WT | 2.81% (KRAS p.G12D) |
| KRAS p.G12D | WT | 1.78% (KRAS p.G12D) |
| KRAS p.G12R | WT | 0.31% (KRAS p.G12R) |
| KRAS p.G13D | WT | 0.41% (KRAS p.G13D) |
| KRAS p.G13D | WT | 2.03% (KRAS p.G13D) |
| KRAS p.Q61H | WT | 38.9% (KRAS p.Q61H) |
| WT |
KRAS p.A11A KRAS p.G12C |
Not tested |
| WT | KRAS p.A59E | Not tested |
For secondary endpoint analysis of BRAF V600E mutation detection, the result of RASKET-B was compared with PYRO. The CR was 99.7% (301/302) (95% CI: 98.2%-100%) (Table 5B). One sample was positive with BRAF V600E by RASKET-B and negative by PYRO without MMD. We also performed PYRO assays with MMD and then confirmed the detection of a mutation in BRAF. The percentage of BRAF mutant alleles in the sample transcript was 11.2%.
Table 5B.
Consistency Between RASKET-B and PYRO in BRAF Gene Mutations (Secondary Endpoint-1 Analysis)
| PYRO |
||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| RASKET-B | Positive | 17 | 1 | 18 |
| Negative | 0 | 284 | 284 | |
| Total | 18 | 284 | 302 | |
| Overall agreement percentage | 99.7% (95% CI, 98.2%-100%) | |||
| Positive agreement percentage | 94.4% (95% CI, 80.2%-100%) | |||
| Negative agreement percentage | 100% (95% CI, 98.8%-100%) | |||
Genotyping Performance in RASKET-B
One hundred and fifty-seven specimens with positive RAS or BRAF mutation results by both the RASKET-B and DS were included. The concordance of each genotype for the overall population assessed by RASKET-B and DS was 100% (157/157) (95% CI: 98.3%-100%) (data not shown).
Discussion
This study is the first to demonstrate the clinical usefulness of the RASKET-B, which can simultaneously detect RAS and BRAF mutations using the PCR-rSSO and xMAP technologies. For RAS genes, we compared the clinical significance of the RASKET-B to RASKET (previously confirmed and approved in Japan [19]) and DS with MMD. The overall CRs of RAS gene detections were 100% and 97.4%, respectively. For the BRAF gene, the results from the RASKET-B were compared to DS with MMD and PYRO; CRs were 100% and 99.7%, respectively. The CRs satisfied the predefined criteria. Based on these results, the RASKET-B was approved by the Ministry of Health, Labour, and Welfare of Japan as an IVD kit for simultaneous determination of both RAS and BRAF mutation status in FFPE and fresh frozen tissues of CRC patients on 05 December 2017.
The presently detected frequency of RAS mutations obtained using the RASKET-B agrees with those reported in several previous studies [2], [3], [6], [7], [22], while the frequency of BRAF mutations was slightly lower than that in Western countries [23], [24], [25], [26]. The detection limit of RASKET-B was approximately 1%-5% (Supplementary Table 1, A, B, and C), which is identical toRASKET [19]. This detection sensitivity is similar to that of other allele-specific PCR-based technologies, which suggests that the discrepancy of the BRAF mutation rate was not due to sensitivity differences. The frequency of BRAF mutations in Asian countries is approximately 5% [12], [27], [28], [29], [30], [31], which is consistent with the present study. The BRAF frequency was higher in the right-sided CRC and in females, which is consistent with a previous report [32].
In this study, we observed several inconsistencies between the results of the RASKET-B and the reference assays. Six specimens with positive results in the RASKET-B and negative results in DS were confirmed to be RAS mutation positive via the TaqMan method. The discrepancy may be mainly caused by the sensitivity difference: the detection limit of RASKET-B is higher than the DS (> 10%). In fact, five of six cases included a smaller amount of mutant DNA. For the BRAF gene, one sample showed discrepant results between RASKET-B and PYRO, possibly due to its small tumor ratio (tumor cell ratio 70%, tumor area ratio 15%). MMD additionally performed with PYRO showed a positive BRAF mutation result.
Conversely, among the two specimens with a negative result in RASKET-B and a positive result in DS, one sample had a KRAS A59E mutation that was not reported in the PRIME study [3]. The other false-negative sample had a double mutation in KRAS codon 11 and codon 12 (G12C). Based on the assay principle, PCR amplifications including codon 11 mutations cannot hybridize to the detection beads for codons 12 and 13 because the codons are adjacent. However, this would have little impact in clinical practice due to the very rare frequency [1], [3], [7], [8]. Thus, this kit can provide clinically appropriate detection of RAS and BRAF mutations.
The Luminex xMAP technology is widely applied for multiplex molecular testing, such as tissue and virus genotyping, which requires differential detections from a number of similar sequences [33], [34]. Additionally, for the amplification of multiple genes or their regions, any possible cross-reactions should be minimized to provide appropriate assays. The RASKET-B allows the simultaneous PCR in the same well of nine regions (four regions in each of KRAS and NRAS genes and one region in the BRAF gene) with few cross-reactions. The turnaround time for detection of both RAS and BRAF gene mutations is approximately 4.5 hours, regardless of the number of samples (<96). Thus, the RASKET-B can potentially solve unmet medical needs of clinicians and reference laboratories, as it can be designed for the rapid, high-throughput, and multiplex detection of all RAS and BRAF mutations.
There were some limitations in this study. First, all samples were obtained through surgery and not biopsies. Also, there may be a bias in that only samples with available RAS mutation data were recruited in this study. Poor quality DNA extracted from FFPE tissues could possibly lead to an inaccurate result. However, a sensitivity of 1%-5% in the RASKET-B would be enough to provide RAS/BRAF mutation status in biopsy samples and in clinical practice. Another limitation was that the RASKET-B was designed to detect only V600E mutations in the BRAF gene. This is because the clinical significance of other BRAF mutations still remains unclear in CRC. Even so, the RASKET-B could provide results of RAS and BRAF (V600E) mutations simultaneously with lower cost and a shorter turnaround time compared to other methods, such as next-generation sequencing.
In conclusion, clinical evaluation of the MEBGEN RASKET-B KIT met the predefined primary and secondary endpoints and displayed a high CR with existing RAS and BRAF assays. The RASKET-B provides rapid and precise detection of RAS and BRAF mutations from FFPE tissue from CRC patients.
Conflict of Interest
The study was designed under the responsibility of MBL and was funded by MBL, Japan. The RASKET-B and RASKET were provided by MBL. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Acknowledgements
We thank the patients and their families for their support and participation in this trial.
Footnotes
[Clinical trial identification] Study ID: UMIN000022742.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2018.10.004.
Appendix A. Supplementary data
Supplementary material
References
- 1.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(10):1626–1634. doi: 10.1200/JCO.2007.14.7116. [Available from] [DOI] [PubMed] [Google Scholar]
- 2.Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27(5):663–671. doi: 10.1200/JCO.2008.20.8397. [Available from] [DOI] [PubMed] [Google Scholar]
- 3.Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010;28(31):4697–4705. doi: 10.1200/JCO.2009.27.4860. [Available from] [DOI] [PubMed] [Google Scholar]
- 4.Lièvre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Côté JF, Tomasic G, Penna C, Ducreux M. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66(8):3992–4005. doi: 10.1158/0008-5472.CAN-06-0191. [Available from] [DOI] [PubMed] [Google Scholar]
- 5.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–762. doi: 10.1016/S1470-2045(10)70130-3. [Available from] [DOI] [PubMed] [Google Scholar]
- 6.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [Available from] [DOI] [PubMed] [Google Scholar]
- 7.Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmüller C, Kahl C, Seipelt G. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomized, open-label, phase 3 trial. Lancet Oncol. 2014;15:1065–1075. doi: 10.1016/S1470-2045(14)70330-4. [Available from] [DOI] [PubMed] [Google Scholar]
- 8.Schwartzberg LS, Rivera F, Karthaus M, Fasola G, Canon JL, Hecht JR, Yu H, Oliner KS, Go WY. PEAK: a randomised, multicenter phase II study of panitumumab plus modified fluorouracil, leucovorin, and oxaliplatin (mFOLFOX6) or bevacizumab plus mFOLFOX6 in patients with previously untreated, unresectable, wild-type KRAS exon 2 metastatic colorectal cancer. J Clin Oncol. 2014;32:2240–2247. doi: 10.1200/JCO.2013.53.2473. [Available from] [DOI] [PubMed] [Google Scholar]
- 9.Stintzing S, Modest DP, Rossius L, Lerch MM, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T. FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab for metastatic colorectal cancer (FIRE-3): a post-hoc analysis of tumour dynamics in the final RAS wild-type subgroup of this randomised open-label phase 3 trial. Lancet Oncol. 2016;17:1426–1434. doi: 10.1016/S1470-2045(16)30269-8. [Available from] [DOI] [PubMed] [Google Scholar]
- 10.Rivera F, Karthaus M, Hecht JR, Sevilla I, Forget F, Fasola G, Canon JL, Guan X, Demonty G, Schwartzberg LS. Final analysis of the randomised PEAK trial: overall survival and tumour responses during first-line treatment with mFOLFOX6 plus either panitumumab or bevacizumab in patients with metastatic colorectal carcinoma. Int J Color Dis. 2017;32:1179–1190. doi: 10.1007/s00384-017-2800-1. [Available from] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [Available from] [DOI] [PubMed] [Google Scholar]
- 12.Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M, Kondo C, Mizota A, Utsunomiya S, Muro K. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 2011;104:856–862. doi: 10.1038/bjc.2011.19. [Available from] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kadowaki S, Kakuta M, Takahashi S, Takahashi A, Arai Y, Nishimura Y, Yatsuoka T, Ooki A, Yamaguchi K, Matsuo K. Prognostic value of KRAS and BRAF mutations in curatively resected colorectal cancer. World J Gastroenterol. 2015;21:1275–1283. doi: 10.3748/wjg.v21.i4.1275. [Available from] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loupakis F, Cremolini C, Masi G, Lonardi S, Zagonel V, Salvatore L, Cortesi E, Tomasello G, Ronzoni M, Spadi R. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med. 2014;371:1609–1618. doi: 10.1056/NEJMoa1403108. [Available from] [DOI] [PubMed] [Google Scholar]
- 15.Cremolini C, Loupakis F, Antoniotti C, Lupi C, Sensi E, Lonardi S, Mezi S, Tomasello G, Ronzoni M, Zaniboni A. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015;16:1306–1315. doi: 10.1016/S1470-2045(15)00122-9. [Available from] [DOI] [PubMed] [Google Scholar]
- 16.Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson A, Bodoky G. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27(8):1386–1422. doi: 10.1093/annonc/mdw235. [Available from] [DOI] [PubMed] [Google Scholar]
- 17.Yoshino T, Arnold D, Taniguchi H, Pentheroudakis G, Yamazaki K, Xu RH, Kim TW, Ismail F, Tan IB, Yeh KH. Pan-Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: a JSMO-ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann Oncol. 2018;29:44–70. doi: 10.1093/annonc/mdx738. [Available from] [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki K, Taniguchi H, Yoshino T, Akagi K, Ishida H, Ebi H, Nakatani K, Muro K, Yatabe Y, Yamaguchi K. Japanese Society of Medical Oncology clinical guidelines: molecular testing for colorectal cancer treatment, third edition. Cancer Sci. 2018;109(6):2074–2079. doi: 10.1111/cas.13617. [Available from] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshino T, Muro K, Yamaguchi K, Nishina T, Denda T, Kudo T, Okamoto W, Taniguchi H, Akagi K, Kajiwara T. Clinical validation of a multiplex kit for RAS mutations in colorectal cancer: results of the RASKET (RAS KEy Testing) prospective, multicenter study. EBioMedicine. 2015;2:317–323. doi: 10.1016/j.ebiom.2015.02.007. [Available from] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [Available from] [DOI] [PubMed] [Google Scholar]
- 21.Didelot A, Le Corre D, Luscan A, Cazes A, Pallier K, Emile JF, Laurent-Puig P, Blons H. Competitive allele specific TaqMan PCR for KRAS, BRAF and EGFR mutation detection in clinical formalin fixed paraffin embedded samples. Exp Mol Pathol. 2012;92:275–280. doi: 10.1016/j.yexmp.2012.03.001. [Available from] [DOI] [PubMed] [Google Scholar]
- 22.Watanabe T, Yoshino T, Uetake H, Yamazaki K, Ishiguro M, Kurokawa T, Saijo N, Ohashi Y, Sugihara K. KRAS mutational status in Japanese patients with colorectal cancer: results from a nationwide, multicenter, cross-sectional study. Jpn J Clin Oncol. 2013;43:706–712. doi: 10.1093/jjco/hyt062. [Available from] [DOI] [PubMed] [Google Scholar]
- 23.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, Slattery ML. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65(14):6063–6069. doi: 10.1158/0008-5472.CAN-05-0404. [Available from] [DOI] [PubMed] [Google Scholar]
- 24.Barault L, Charon-Barra C, Jooste V, de la Vega MF, Martin L, Roignot P, Rat P, Bouvier AM, Laurent-Puig P, Faivre J. Hypermethylator phenotype in sporadic colon cancer: study on a population-based series of 582 cases. Cancer Res. 2008;68(20):8541–8546. doi: 10.1158/0008-5472.CAN-08-1171. [Available from] [DOI] [PubMed] [Google Scholar]
- 25.Jover R, Nguyen TP, Pérez-Carbonell L, Zapater P, Payá A, Alenda C, Rojas E, Cubiella J, Balaguer F, Morillas JD. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 2011;140(4):1174–1181. doi: 10.1053/j.gastro.2010.12.035. [Available from] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lochhead P, Kuchiba A, Imamura Y, Liao X, Yamauchi M, Nishihara R, Qian ZR, Morikawa T, Shen J, Meyerhardt JA. Microsatellite instability and BRAF mutation testing in colorectal cancer prognostication. J Natl Cancer Inst. 2013;105(15):1151–1156. doi: 10.1093/jnci/djt173. [Available from] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Cho NY, Choi M, Yoo EJ, Kim JH, Kang GH. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathol Int. 2008;58(2):104–113. doi: 10.1111/j.1440-1827.2007.02197.x. [Available from] [DOI] [PubMed] [Google Scholar]
- 28.Min BH, Bae JM, Lee EJ, Yu HS, Kim YH, Chang DK, Kim HC, Park CK, Lee SH, Kim KM. The CpG island methylator phenotype may confer a survival benefit in patients with stage II or III colorectal carcinomas receiving fluoropyrimidine-based adjuvant chemotherapy. BMC Cancer. 2011;11:344. doi: 10.1186/1471-2407-11-344. [Available from] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakanishi R, Harada J, Tuul M, Zhao Y, Ando K, Saeki H, Oki E, Ohga T, Kitao H, Kakeji Y. Prognostic relevance of KRAS and BRAF mutations in Japanese patients with colorectal cancer. Int J Clin Oncol. 2013;18(6):1042–1048. doi: 10.1007/s10147-012-0501-x. [Available from] [DOI] [PubMed] [Google Scholar]
- 30.Kawazoe A, Shitara K, Fukuoka S, Kuboki Y, Bando H, Okamoto W, Kojima T, Fuse N, Yamanaka T, Doi T. A retrospective observational study of clinicopathological features of KRAS, NRAS, BRAF and PIK3CA mutations in Japanese patients with metastatic colorectal cancer. BMC Cancer. 2015;15:258. doi: 10.1186/s12885-015-1276-z. [Available from] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujiyoshi K, Yamamoto G, Takenoya T, Takahashi A, Arai Y, Yamada M, Kakuta M, Yamaguchi K, Akagi Y, Nishimura Y. Metastatic pattern of stage IV colorectal cancer with high-frequency microsatellite instability as a prognostic factor. Anticancer Res. 2017;37(1):239–247. doi: 10.21873/anticanres.11313. [DOI] [PubMed] [Google Scholar]
- 32.Gonsalves WI, Mahoney MR, Sargent DJ, Nelson GD, Alberts SR, Sinicrope FA, Goldberg RM, Limburg PJ, Thibodeau SN, Grothey A. Patient and tumor characteristics and BRAF and KRAS mutations in colon cancer, NCCTG/Alliance N0147. J Natl Cancer Inst. 2014;106(7) doi: 10.1093/jnci/dju106. [Available from] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itoh Y, Mizuki N, Shimada T, Azuma F, Itakura M, Kashiwase K, Kikkawa E, Kulski JK, Satake M, Inoko H. High-throughput DNA typing of HLA-A, -B, -C, and -DRB1 loci by a PCR-SSOP-Luminex method in the Japanese population. Immunogenetics. 2005;57:717–729. doi: 10.1007/s00251-005-0048-3. [Available from] [DOI] [PubMed] [Google Scholar]
- 34.Ozaki S, Kato K, Abe Y, Hara H, Kubota H, Kubushiro K, Kawahara E, Inoue M. Analytical performance of newly developed multiplex human papillomavirus genotyping assay using Luminex xMAP technology (Mebgen HPV Kit) J Virol Methods. 2014;204:73–80. doi: 10.1016/j.jviromet.2014.04.010. [Available from] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material