Abstract
Merkel cell carcinoma (MCC) is a highly aggressive non-melanoma skin cancer of the elderly which is associated with the Merkel cell polyomavirus (MCPyV). MCC reveals a trilinear differentiation characterized by neuroendocrine, epithelial and pre/pro B-cell lymphocytic gene expression disguising the cellular origin of MCC. Here we investigated the expression of the neuroendocrine key regulators RE1 silencing transcription factor (REST), neurogenic differentiation 1 (NeuroD1) and the Achaete-scute homolog 1 (ASCL1) in MCC. All MCCs were devoid of REST and were positive for NeuroD1 expression. Only one MCC tissue revealed focal ASCL1 expression. This was confirmed in MCPyV-positive MCC cell lines. Of interest, MCPyV-negative cell lines did express REST. The introduction of REST expression in REST-negative, MCPyV-positive MCC cells downregulated the neuroendocrine gene expression. The lack of the neuroendocrine master regulator ASCL1 in almost all tested MCCs points to an important role of the absence of the negative regulator REST towards the MCC neuroendocrine phenotype. This is underlined by the expression of the REST-regulated microRNAs miR-9/9* in REST-negative MCC cell lines. These data might provide the basis for the understanding of neuroendocrine gene expression profile which is expected to help to elucidate the cellular origin of MCC.
Introduction
Merkel cell carcinoma (MCC) is a highly malignant non-melanoma skin cancer which predominantly arises in the sun-exposed skin of elderly patients [1], [2]. Next to UV exposure and age, MCC are associated with immune deficiencies and the presence of clonally integrated Merkel cell polyomavirus (MCPyV) [3], [4]. More than 80% of MCC are associated with MCPyV, and it has been shown that tumor cell proliferation of MCC is dependent on the expression of the oncogenic viral T antigens [5], [6], [7].
Although MCC accounts only for a minority of all cutaneous malignancies its incidence has increased worldwide and has tripled in the US and doubled in some European countries [8].
The 5-year survival of local MCC is 71% but only 20% in the presence of distant metastases [2]. Recent data of clinical trials on the use of immune checkpoint inhibitors in the treatment of patients with MCC stage IIIB/IV are promising [9], [10].
Despite the major progress that has been made during the past years concerning the understanding of the etiopathogenesis and treatment, the cellular origin of MCC remains enigmatic [11]. It has been postulated that MCC either originates from Merkel cells or epidermal/dermal stem cells [11], [12]. To date it is generally accepted that the post-mitotic Merkel cells do not constitute the cellular origin of MCC. Based on the frequently reported co-expression of PAX-5, TdT and immunoglobulins in MCCs, we have recently formulated the hypothesis that MCC originate from early B-cells, i.e. pre/pro B-cells [13], [14].
The repressor element 1 (RE1) silencing transcription factor (REST) is a master repressor of neuronal gene expression and neuronal programs in non-neuronal lineages [15], [16]. REST binds together with CoREST to the RE1-binding site of neuronal genes, which leads to the inhibition of the expression of these genes [17]. In the absence of REST, neuronal genes will be expressed. Among other genes, REST negatively controls the neuronal target genes encoding chromogranin A and synaptophysin [18], [19]. Although the absence of REST is insufficient to explain the full extent of chromogranin A expression, synaptophysin gene expression is predominantly regulated by REST [19]. REST has been proven to function as an oncogene in neural cells and as a tumor suppressor in non-neural cells [16]. In neoplastic neural cells, REST expression is switched on and is overexpressed, e.g. in medulloblastoma and in glioblastoma multiforme [20], [21]. In contrast, in non-neural tumors REST acts as a tumor suppressor, revealing deletions of the REST locus on chromosome 4 in a significant proportion of tumors [22].
REST expression is negatively reciprocal regulated by the neuronal development regulator microRNA-9 (miR-9) during neural differentiation [23], [24]. Further, its passenger strand miR-9* is downregulating CoREST: Recently it has been shown that miR-9 is upregulated in MCC [25]. Interestingly, it has been shown that miR-9 is activated by the human papillomavirus (HPV) E6 protein in cervical cancer [26]. In addition, REST has been shown to interact with other human DNA viruses, e.g. Herpes simplex virus and adenovirus [27], [28], [29].
The basic helix–loop–helix (BHLH) Achaete-scute homolog 1 (ASCL1) transcription factor is known as another master regulator of neuroendocrine differentiation and is detected in most neuroendocrine tumors as small cell lung cancer (SCLC) [30]. In addition, the transcription factor neurogenic differentiation 1 (NeuroD1) was considered as an alternative regulator of neuroendocrine differentiation.
In the present study, we assessed the expression of REST, NeuroD1 and ASCL1 in MCC and MCC cell lines. Moreover, the possible regulation of REST expression by promoter methylation was assessed with 5-aza-2′ deoxycytidine (5-aza-2′-dC) treatment and a methylation specific PCR of the REST promoter CpG islands. In addition, we tested the expression of miR-9/9* in REST-positive and REST-negative MCC cell lines in relation to MCPyV in order to understand in as much the regulation of neuroendocrine gene expression in MCC is affected by MCPyV.
Materials and Methods
Patient Samples
Twenty-eight formalin-fixed and paraffin-embedded (FFPE) primary and metastatic MCC tissues were obtained from the archives of the Department of Pathology, Maastricht University Medical Center + (Table 1). All use of tissue and patient data was in agreement with the Dutch Code of Conduct for Observational Research with Personal Data (2004) and Tissue (2001, "www.federa.org/sites/default/files/digital_version_first_part_code_in_uk_2011_12092012.pdf"). MCCs were previously diagnosed by histology and immunohistochemistry for CK20, CD56, synaptophysin and chromogranin A in routine diagnostic and have been reviewed by 2 experienced pathologists (VW, AZH).
Table 1.
Summary of the Clinicopathological Data of the MCC Patients and MCC Tissues Including the Results of the Immunohistochemical Assessment of REST, ASCL1 and NeuroD1
| ID | Gender | Age | location | Dx | Histo | MCPyV | REST | Ascl1 | NeuroD1 |
|---|---|---|---|---|---|---|---|---|---|
| Primary MCCs | |||||||||
| 1 | m | 63 | head | MCC | int. | pos. | − | − | ++ heter. |
| 2 | m | 92 | ear | MCC | s.c. | neg. | − | − | +++ |
| 3 | f | 85 | buttocks | MCC | s.c. | pos. | − | − | +++ |
| 4 | m | 69 | lip | MCC | int. | pos. | − | − | ++ |
| 5 | f | 93 | upper eye lid | MCC | int. | pos. | − | − | + |
| 6 | f | 60 | tongue | MCC | int./s.c | pos. | − | − | ++ heter. |
| 7 | m | 74 | upper leg | MCC | int. | pos. | − | − | +++ |
| 8 | m | 93 | head | MCC | int. | neg. | − | ++ foc. | +++ |
| 9 | f | 76 | buccal | MCC | int. | pos. | − | − | ++ |
| 10 | f | 91 | arm | MCC | int. | pos. | − | n.a. | n.a. |
| 11 | f | 83 | upper eye lid | MCC | int. | pos. | − | − | +++ |
| 12 | f | 76 | head | MCC | int. | pos. | − | n.a. | n.a. |
| 13 | m | 77 | neck | MCC | int. | neg. | − | − | +++ |
| 14 | m | 79 | head | MCC | int. | pos. | − | − | ++ |
| 15 | f | 71 | buccal | MCC | int. | pos. | − | − | ++ |
| 16 | m | 68 | buttocks | MCC | int. | pos. | − | − | +++ |
| 17 | f | 58 | buccal | MCC | int./s.c | pos. | − | − | +++ |
| 18 | m | 75 | upper leg | MCC | int | pos. | − | − | +++ |
| 19 | f | 65 | arm | MCC | int. | pos. | − | − | ++ |
| 20 | m | 63 | buccal | MCC | int. | neg. | − | − | ++ |
| Metastatic MCCs | |||||||||
| 21 | m | 71 | pancreas | MCC met. | int. | pos. | − | − | ++ |
| 22 | f | 75 | LN neck | MCC met. | int. | pos. | − | − | ++ |
| 23 | f | 66 | skin | MCC met. | int. | pos. | − | − | (+) |
| 24 | m | 74 | groin | MCC met. | int. | pos. | − | − | +++heter. |
| 25 | f | 67 | upper arm | MCC met. | int. | pos. | − | − | +++heter. |
| 26 | m | 65 | LN neck | MCC met. | int. | neg. | − | − | ++heter. |
| 27 | f | 43 | LN groin | MCC met. | int. | pos. | − | − | ++ |
| 28 | m | 69 | LN iliaca | MCC met. | int./s.c. | pos. | − | ++heter. | ++ |
ID = identity, M = male, F = female, Dx = diagnosis, MCC = Merkel cell carcinoma; Histo: histology; int = intermediate, s. c. = small cell, foc. = focal, n.a. = not applicable, pos. = positive, neg. = negative, − = no expression, + = weak expression, ++ = moderate expression, +++ = strong expression.
Cell Lines
The MCPyV-positive MCC cell lines MKL-1, MKL-2, WaGa, PeTa, BroLi, MS-1 the MCPyV-negative MCC cell lines MCC13, MCC26 and the B-ALL cell line REH were used. All MCC cell lines were kindly provided by Jürgen Becker (University Hospital Essen, Essen, Germany). REH was obtained from the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Germany.
The cell lines were cultured in Gibco® RPMI 1640 medium with 10% fetal calf serum (FCS) (Gibco®, ThermoFisher SCIENTIFIC, The Netherlands) in an incubator at 37°C and 5% CO2.
Immunohistochemistry
The expression of REST, ASCL1 and NeuroD1 was tested by immunohistochemistry (IHC) in 20 primary and 8 metastatic MCCs, in the MCPyV-positive MCC cell lines MKL-1, MKL-2, WaGa and in the MCPyV-negative MCC cell lines MCC13 and MCC26. In addition, the expression of these genes was assessed in the B-cell acute lymphoblastic leukemia (B-ALL) cell line REH. The following antibodies and dilutions were used in this study: Anti-MCPyV LT-antigen (clone: CM2B4) dilution 1:100, SANTA CRUZ BIOTECHNOLOGY, Germany; anti-REST (clone: CL0381) dilution 1:100, Sigma Aldrich, the Netherlands; anti-MASH1/Achaete-scute homologe1 (clone: 24B72D11.1) dilution 1:100, Abcam, UK; NeuroD1 (clone: 3H8) dilution 1:100, Abnova, Germany; anti-Cytokeratin 20 (clone: Ks 20.8) “ready to use antibody”, Dako, the Netherlands. All IHC stainings were conducted on a Dako Autostainer 48 Link using the EnVision FLEX Visualization Kit K8008 Dako as described previously and according to standard diagnostic routine protocols and manufacturer instructions [31]. The IHC double staining procedure of REST and Cytokeratin 20 (CK20) was performed manually by using the Dako Kit K8008 for anti-REST and K5005 for the staining of anti-CK20. The double staining method was adapted to the standard routine protocol.
REST-GFP Transfection
The cell line WaGa was transfected with human cDNA ORF GFP tagged clone NM_005612 from Origene. The expression vector led to a transient expression of REST-GFP. The transfection was performed by using Lipofectamine 3000 (ThermoFisher SCIENTIFIC, The Netherlands) according to the instructions of the manufacturer. The expression level changes of synaptophysin and chromogranin A were assessed by immunofluorescence microscopy by using a standard protocol. The cells were formalin fixed, permeabilized with 0,3% Triton X-100, blocked with 5% BSA, incubated with the first antibodies against chromogranin A 1:250 (SP11, ThermoFisher SCIENTIFIC, The Netherlands) or anti-synaptophysin 1:250 (SP12, ThermoFisher SCIENTIFIC, The Netherlands), stained with the second antibody anti Rabbit Texas Red conjugated (T-2767, ThermoFisher SCIENTIFIC, The Netherlands). The detection of the GFP tagged REST and fluorescence stained cells with the Leica microscope DM 5000 B (Leica, the Netherlands).
miRNA Isolation and miRNA Quantitative RT-PCR
The miRNA was extracted by using the miRNA isolation kit “NucleoSpin® miRNA” according to the manufacturers' instructions (Macherey Nagel, Germany). The miRNA concentration was measured with the Nanodrop 2000 (ThermoFisher SCIENTIFIC, The Netherlands). The cDNA from the isolated miRNAs was synthesized by using the Universal cDNA Synthesis Kit II from Exiqon, the Netherlands.
The expression of miR-9 and miR-9* was analyzed in means of the quantitative RT-PCR by using the specific LNA primers hsa-miR-5p (miR-9) and hsa-miR-3p (miR-9*) (Exiqon, the Netherlands). The miRNA expression level was normalized to the expression of miR-103 using the primer set hsa-miR-103a-3p. The qRT-PCR was performed by using the SYBR Green Supermix (Bio-Rad, Switzerland) on the CFX96 PCR Detection System (Bio-Rad, Switzerland) and recorded by the Bio-Rad CFX manager.
REST-, Synaptophysin-, and Chromogranin A- Gene Transcript Expression
RNA was extracted using the RNA isolation kit “NucleoSpin® RNA” according to the manufacturer's instructions (Macherey-Nagel, Germany). The RNA concentration was measured with the Nanodrop 2000 (ThermoFisher SCIENTIFIC, The Netherlands). RNA was converted into cDNA using the “iScript™ Select cDNA Synthesis Kit” (Bio-Rad, Switzerland). Exon 1 and exon 3 of REST were amplified using the R1 and R3 primers (Table S2). In addition, exon 1 and exon 3 spanning amplification of REST cDNA was performed using the REST forward (fw) primer and REST reverse (rv) primer (Table S2).
A quantitative RT-PCR was performed for the analysis of REST-, synaptophysin- and chromogranin A- expression in the cell lines MKL-1, MKL-2, WaGa, MCC13, MCC26 and REH. For this purpose, the qRT-PCR primer sets for REST, chromogranin A, synaptophysin were used. The resulting Ct values were normalized with the housekeeping genes β-actin or GAPDH (Table S2). The qRT-PCR was performed by using the SYBR Green Supermix (Bio-Rad, Switzerland) on the CFX96 PCR Detection System (Bio-Rad, Switzerland) and recorded by the Bio-Rad CFX manager. All used primers were obtained by Eurofins Genomics, Germany.
DNA-PCR
DNA isolation was performed using the Nucleospin Tissue DNA Kit (Macherey-Nagel, Germany) according to manufacturer's instructions. PCR was performed on a Veriti well thermal cycler (Applied Biosystems, the Netherlands) with an annealing temperature of 57°C for all combinations of primers and 35 cycles. The PCR products were sequenced using the Big Dye Sequencing Kit (ThermoFisher SCIENTIFIC, The Netherlands) according to manufacturers' instructions. All experiments were repeated at least 3 times.
Treatment of Cell Lines with 5-Aza-2′ Deoxycytidine
The cell lines MKL-1, MKL-2, MCC13 and MCC26 were treated with 0.5 μM, 1.0 μM and 2.0 μM of 5-aza-2′-dC (Sigma-Aldrich, the Netherlands) which was added every 24 hrs. In total, the cells were exposed to the demethylation agent for 96 h. After exposure the cells were formalin fixed and REST expression was tested by IHC.
Methylation Specific PCR (MSP)
The genomic DNA of all cell lines was extracted using the “NucleoSpin® Tissue” kit (Macherey-Nagel, Germany). The location of the CpG islands was previously described by Kreisler et al. [32] and was reproduced using the CpG island searcher [32], [33]. The gDNA was bisulfite converted using the EpiTect Bisulfite Kit (Qiagen, the Netherlands) according to the manufacturers' instruction. CpG islands were amplified using specific primers for both methylated and unmethylated specific primer (Table S2). As positive control, the unmethylated and the methylated Epitect Control DNAs (Qiagen, the Netherlands) were used.
Statistics
The significance of the qRT-PCR data was determined with Graphpad Prism 6 by using the One-Way ANOVA. Data were expressed as the mean ± standard deviation (SD). P-values of <0.05 were considered as statistically significant.
Results
Protein Expression of REST, ASCL1 and NeuroD1 in MCC
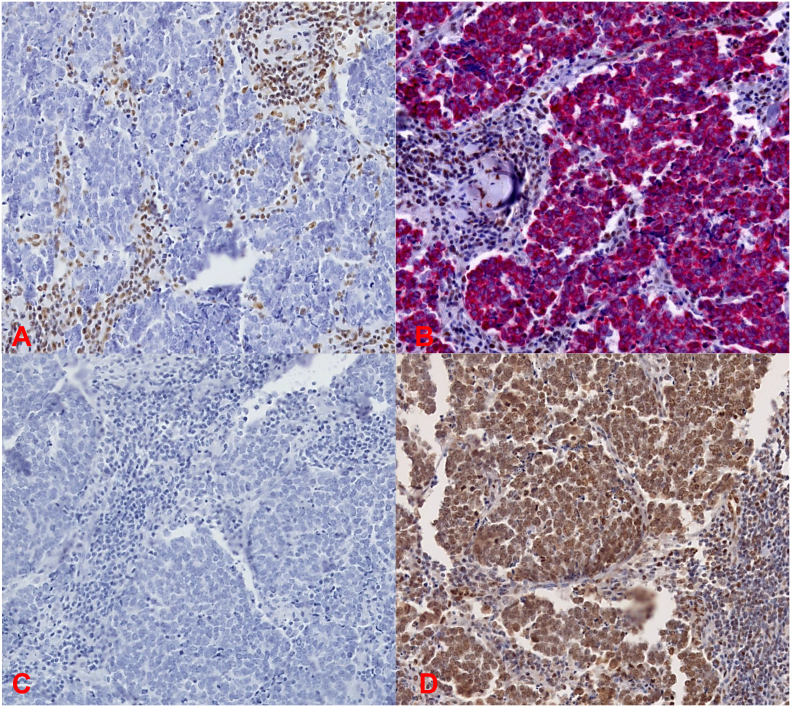
Of the 28 MCC tissues, 23 (82.1%) were MCPyV-positive and 5 (17.8%) MCPyV-negative as tested by IHC. All MCCs (Table 1) were completely devoid of REST expression as assessed by IHC (Figure 1, A and C). Double staining for CK20- and REST-expression of MCC tissues revealed a specific nuclear expression of REST within the tumor infiltrating lymphocytes but not in the MCC cells expressing CK20 (Figure 1B). All MCCs were negative for ASCL1, except one case which revealed focal expression for ASCL1 (ID8, Table 1). All MCCs revealed a specific moderate to strong nuclear expression of NeuroD1 (Table 1, Figure 1D).
Figure 1.
Immunohistochemistry staining of REST, ASCL1 and NeuroD1 in MCC ID 25.
A, IHC reveals no expression of REST expression in the MCC tissue. In B, double staining of CK20 (red) and REST (brown) confirms that the REST positive cells are not located in the MCC tissue. ID25 is negative for ASCL1 and positive for NeuroD1 as shown in C and D. The microphotographs were taken at 20x magnification.
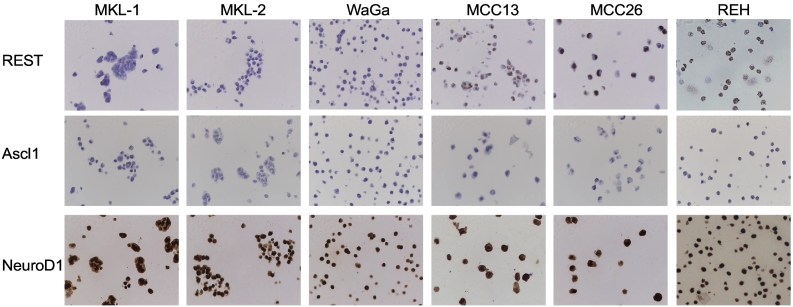
In addition, REST expression was assessed in MCC cell lines by IHC. MCPyV-positive cell lines (MKL-1, MKL-2, WaGa, PeTa, BroLi, MS-1) were negative for REST expression (Figure 2 and Supplemental Table S1). In contrast, the MCPyV-negative cell lines (MCC13, MCC26, REH) revealed a specific nuclear REST expression. In compliance with the MCC specimens, all cell lines were negative for ASCL1 expression, but revealed a strong and specific nuclear NeuroD1 expression in all cell lines. All MCPyV-positive cell lines showed a uniform expression pattern for the tested transcription factors. Therefore, for further experiments MKL-1, MKL-2 and WaGa cells were used.
Figure 2.
IHC staining of the MCC cell lines MKL-1, MKL-2, WaGa, MCC13, MCC26 and the B-ALL cell line REH for REST, ASCL1 and NeuroD1. Specific nuclear REST expression (brown) is found in the MCPyV-negative cell lines but not in MCPyV-positive cell lines MKL-1, WaGa and MKL-2. All cell lines tested were repeatedly negative for ASCL1 and positive for NeuroD1 by IHC. The microphotographs were taken at 40x magnification.
REST Transcript Gene Expression in MCC Cell Lines
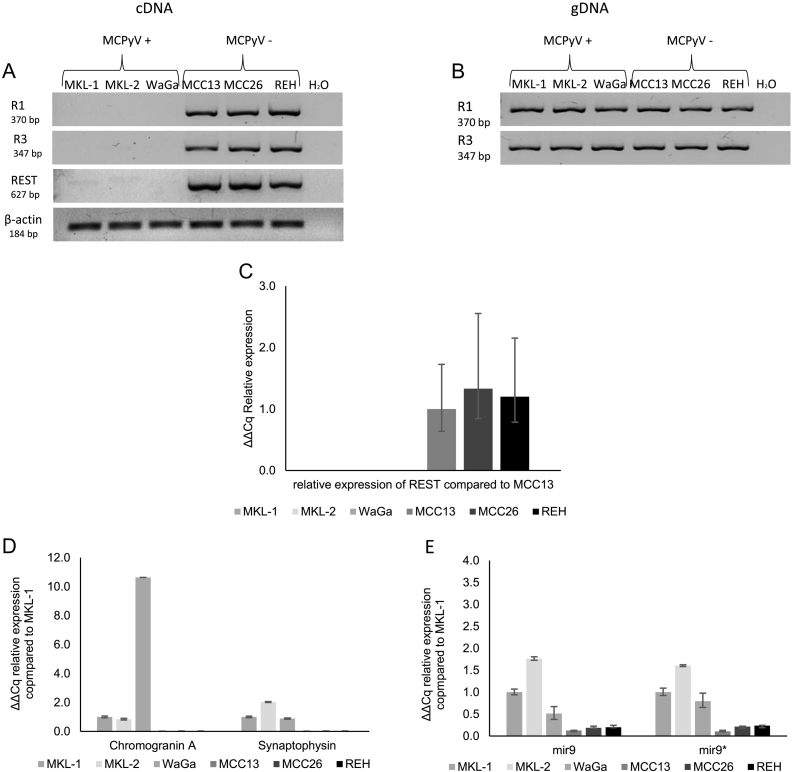
The presence of REST transcripts was assessed in MKL-1, MKL-2, WaGa, MCC13, MCC26 and REH cells (Figure 3). No transcript expression of the first (R1) and third (R3) exons and marginal, almost invisible presence of the RNA-specific REST transcript was observed in MKL-1, MKL-2 and WaGa. In contrast, the MCPyV-negative cell lines MCC13, MCC26 and REH revealed an evident and distinctive REST transcript expression. QRT-PCR for REST revealed that the expression of REST transcripts was under the signal threshold in MCPyV-positive MCC cells and thus negative. In contrast, MCPyV-negative cells revealed a high expression of REST transcripts (Figure 3, A and C).
Figure 3.
Analysis of REST, synaptophysin, chromogranin A and miR-9/9* expression.
No relevant REST transcripts were detected in MCPyV-positive MCC cell lines MKL-1, WaGa and MKL-2. Exon 1 (R1), exon 3 (R3) and intron-exon spanning amplification of exon 1 and exon3 (REST) MKL-1, MKL-2 and WaGa were not detectable using cDNA (A) but by using genomic DNA (B) level. The MCPyV-negative cell lines were positive for REST cDNA. The expression of REST, chromogranin A, synaptophysin and miR-9/9* were analyzed by means of a RT-qPCR. The cq values were normalized to MKL-1 and the sd of n = 2 is shown. Only for the REST expression the cq values were normalized to MCC13.
The REST RT-qPCR confirmed this result and shows a comparable REST transcript level in MCC13, MCC26 and REH (C). For chromogranin A and synaptophysin is the pattern vice versa. Whereas WaGa showed a 10-fold higher chromogranin A expression compared to the other MCPyV-positive cell lines (D). For The miR-9 and miR-9* expression the REST positive cell lines MCC13, MCC26 and REH showed an 84 to 76% respectively lower expression of miR-9 and miR-9* compared to the MCPyV-positive cell lines MKL-1, MKL-2 and WaGa. The p-value for the assessed genes was <0.5.
The sequencing of the cDNA amplification products for MCC13, MCC26 and REH showed the expression of the identical REST isoform (variant 1) in these three cell lines. The cDNA REST amplification product of MKL-2 (Figure 3A; Supplemental Figure S1) showed a faint PCR product, which was confirmed as REST by sequencing, carrying an insertion of 67-bp. This insertion had previously been reported in SCLC and is supposed to result in a truncated REST [34], [35].
Chromogranin A, Synaptophysin and miR-9/miR-9* Expression in MCC
The detected patterns of chromogranin A and synaptophysin expression levels in MCPyV-positive cell lines revealed an inverse correlation with REST expression (Figure 3D). Of interest, a 10-fold higher transcript expression level of chromogranin A was observed in WaGa cells compared to MKL-1 and MKL-2. Interestingly, the WaGa cells revealed an approximately 3-fold higher expression level of the MCPyV T antigens compared to MKL-1 and MKL-2 (Figure 3D and Supplemental Figure S2). MiR-9 expression in MCC cell lines was assessed in REST-negative, MCPyV-positive MCC cell lines MKL-1, MKL-2 and WaGa, and REST-positive, MCPyV-negative MCC cell lines MCC13 and MCC26 by means of qRT-PCR. The expression of miR-9 and miR-9* was normalized to MKL-1 (Figure 3E).
The REST-negative, MCPyV-positive, cell lines showed a high miR-9/miR-9* expression compared to the REST-positive, MCPyV-negative, MCC cell lines and the B-ALL cell line REH.
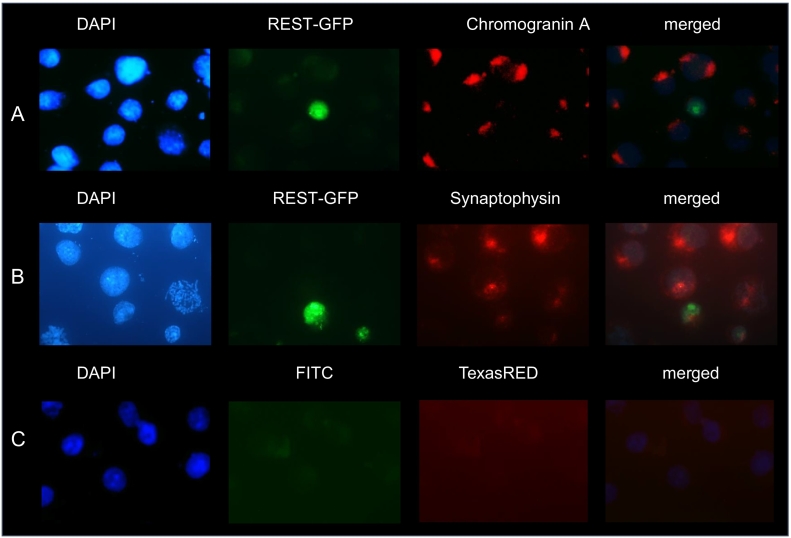
Effects of the Transient Expression of REST-GFP in WaGa Cells Assessed by IF
In order to determine the functional importance of REST expression in MCPyV-positive MCC cell lines, WaGa cells were transfected with a GFP-tagged REST and its transient expression was detected in the nuclei. The REST-GFP positive WaGa cells were stained for chromogranin A and synaptophysin. A significant decrease of chromogranin A and synaptophysin compared to the not transfected WaGa cells was observed (Figures 4 and S3) in the transfected cells compared to the non-transfected cells.
Figure 4.
REST-GFP transient expressing WaGa cells with downregulated chromogranin A and synaptophysin expression. WaGa nuclei were shown with DAPI. The transient expressed REST-GFP is detected by green fluorescence. Immunofluorescene staining of chromogranin A and synaptophysin reveal a cytoplasmic detection of both in WaGa cells und red flourescence. The merged picture shows no detection of chromogranin A and a weak detection of synaptophysin in WaGa cells with REST-GFP. The microphotographs were taken at 63x magnification.
Methylation of the REST Promoter in MCCs
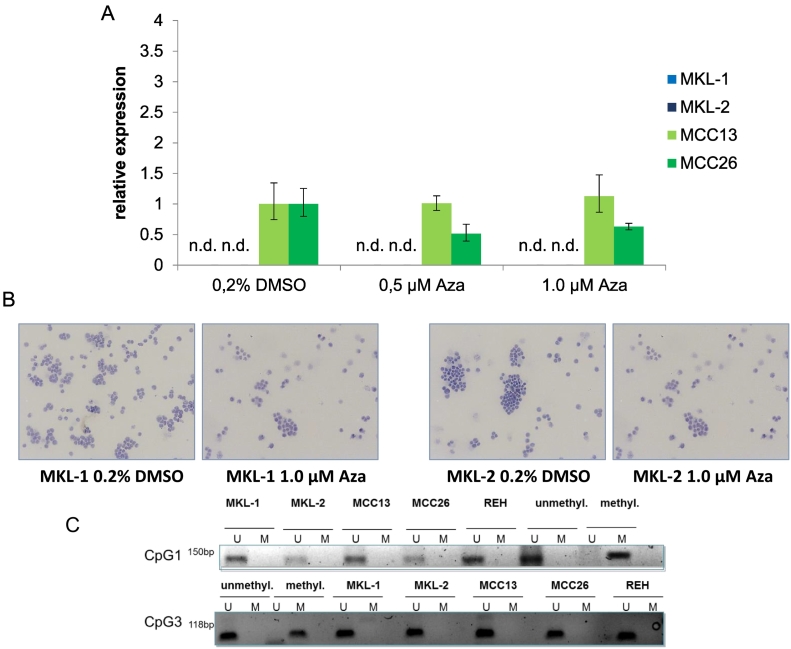
REST expression had been shown to be regulated by methylation of promoter CpG islands [32]. Since neither REST protein or transcript expression could be detected in the MCPyV-positive cell lines, MKL-1, MKL-2, MCC13, MCC26 and REH were treated with 5-aza-2′-dC (Figure 5A). Of interest, after 0.5 and 1.0 μM treatment with 5-aza-2′-dC MCC26 showed exclusively the response of approximate 50 to 60% respectively decrease of REST expression. However, due to the high standard deviation this failed to reach statistical significance. The expression of REST was not altered in the MCPyV-positive cell lines MKL-1 and MKL-2 by 5-aza2′ dC treatment as assessed by IHC and RT-PCR (Figure 5B). The REST expression in the other cell lines was not significantly altered by the 5-aza2 dC treatment.
Figure 5.
No methylation of REST CpGs in MCC cell lines. Q-RT-PCR of REST expression after 5 days treatment with 0.5 μM and 1 μM of 5-aza-2′-dC of MCC cell lines MKL-1, MKL-2, MCC13 and MCC26 and the BALL cell line REH revealed no increase in expression of REST in MCPyV-positive cell lines (A). The P value of the MKL-1/MKL-2 treatment was <0.01. For MCC26 the REST expression decreased up to 62% which failed to reach statistical significance. No REST protein is detectable in MKL-1 and MKL-2 after 2 μM treatment (B). The MSP for CpG 1 and 3 showed no methylation in all MCC cell lines and in the B-ALL cell line. The positive controls showed either a methylated or an unmethylated DNA fragment. M: methylated; U: unmethylated.
A methylation specific PCR was performed on the CpG islands 1 and 3 (Figure 5D) of the genomic DNA of the above-mentioned cell lines and no methylation was detected on both CpG islands in all tested cell lines.
Discussion
In the present study we investigated the expression of key regulator genes of neuroendocrine gene expression in MCC. For this purpose a large number of MCC tissues including MCC cell lines was tested for the expression of REST, ASCL1, and NeuroD1.
REST acts as an repressor of neuroendocrine gene expression, whereby loss of REST expression leads to the expression of synaptophysin and chromogranin A. In contrast, ASCL1 and NeuroD1, both proneural basic-helix–loop–helix proteins, have both been identified as activators of neuroendocrine lineage genes expression [36]. In contrast, ASCL1 induces synatophysin expression and NeuroD1 activates CD56 (NCAM1) expression [37], [38].
All MCC tissues and cell lines in this study were negative for ASCL1 expression except one MCC which revealed a focal expression of ASCL1 (Table 1). Ralston and colleagues (2008) have previously reported that MCCs were all tested negative for ASCL1 expression by IHC [39]. In contrast, Lewis et al. (2010) found that MCCs arising in the head and neck region were positive for ASCL1 expression [40]. Interestingly, the anti-ASCL antibody in this study and the two previous reports derived from the same antibody clone. Moreover, we show that ASCL1 expression is also absent in all tested MCC cell lines, irrespective of the MCPyV-status (Figure 2).
To the best of our knowledge we report for the first time that NeuroD1 is expressed to 100% in MCC tissues and cell lines irrespective of the MCPyV-status (Table 1, Figure 2). Interestingly, the acute B-cell lymphoblastic leukemia cell line REH also reveals a strong expression of NeuroD1, suggesting that NeuroD1 expression alone is not sufficient to induce neuroendocrine differentiation. This is supported by the fact that MCC13, MCC26 and REH were negative for the neuroendocrine genes chromogranin A and synaptophysin.
According to previous data, chromogranin A and synaptophysin gene expression is negatively regulated by REST [18], [41]. Therefore, we tested the expression of REST in MCC tissues and MCC cell lines (Table 1, Figure 2). None of the MCC in our study did reveal any REST expression in the MCC cells irrespective of the MCPyV-status.
The expression of REST-GFP in the MCPyV-positive MCC cell line WaGa and the subsequent REST induced decrease of synaptophysin and chromogranin A expression reveals that chromogranin A and synaptophysin are regulated by REST also in MCC cells (Figure 4) [18], [41]. Since MCC are devoid of neuroendocrine gene activator ASCL1 expression in combination with the lack of the neuroendocrine gene repressor REST expression strongly indicates that the neuroendocrine gene expression of synaptophysin and chromogranin A in MCC is mediated by the absence of REST. NeuroD1 expression in MCC alone apparently seems to be insufficient to induce the neuroendocrine gene expression of synaptophysin and chromogranin A in the MCPyV-negative MCC (Figure 3, C and D). According to previous reports one might speculate that the high NeuroD1 expression in MCC might be responsible for the known expression of NCAM1 (CD56) in MCCs [37], [42].
It is important to note, that the MCPyV-negative variant MCC cell lines MCC13 and MCC26 are currently controversially discussed in as much these cells lines indeed derive from MCC [43]. Further the absence of the neuroendocrine marker chromogranin A and synaptophysin can be explained by the expression of REST in these cells. In the context of the unknown cellular origin of MCC, it would be interesting to get more knowledge about the regulation of REST in MCC.
In this context we also assessed the methylation status of relevant REST promoter CpG islands. It has been shown previously in SCLC cells that REST expression is regulated by the CpG island methylation of the REST promoter. CpG1 and CpG3 islands were stronger methylated in SCLC REST-low expressing cell lines [32] (Figure 5). In our study, we did not observe methylation of CpG1 and CpG3 islands of the REST promoter in MCPyV-positive and -negative MCC cell lines. Moreover, the 5-aza-2′-dC treatment of the REST negative cell lines did not lead to REST expression which confirms the MSPs. Thus, the loss of REST expression in MCC is most likely not due to REST promoter methylation.
Notably, MCPyV-positive WaGa cells revealed an up to 10-fold higher expression of chromogranin A compared to the other MCPyV-positive MCC cell lines which was paralleled by also a 3-fold higher abundance of MCPyV T antigens. This might possibly hint to a direct or indirect activation of MCPyV T antigens on chromogranin A expression.
REST is not only negatively regulating the gene expression of chromogranin A and synaptophysin but also the expression of miR-9. Indeed, we could show that miR-9 and miR-9* are abundantly present in REST-negative MCPyV-positive MCC cell lines. Recently, miR-9 was found to be upregulated in 20 MCC tissues compared to cutaneous lesions of melanoma, squamous cell carcinoma, and basal cell carcinoma [25]. In cervical carcinoma and oropharyngeal squamous cell carcinoma it was shown that miR-9 is activated by HPV [26], [44]. It is tempting to speculate that a comparable mechanism might be applicable to MCPyV and miR-9 expression in MCC. In the context of the unknown cellular origin of MCC the expression of miR-9 and the absence of REST might be the first step towards understanding the regulation of neuroendocrine gene expression in MCC and might help to identify the cellular ancestry of MCC [13], [14].
Conclusion
MCCs reveal a unique expression pattern of the neuroendocrine key regulator genes REST, ASCL1 and NeuroD1, which is characterized by the lack of REST and ASCL1 expression and the presence of NeuroD1. The absence of REST expression in MCC and in MCPyV-positive MCC cell lines, in combination with REST expression in MCPyV-negative cell lines points to an important role of the MCPyV in the regulation of REST expression in MCC. Our data might provide the basis of MCPyV-related neuroendocrine gene expression in MCC.
The following are the supplementary data related to this article.
Blast of MKL-2 REST sequence. NCBI blast has revealed the identity of the truncated REST sequence which was detected in SCLC (A). The exons of the non-truncated and truncated splice variant are shown (B).
MCPyV expression level in all used cell lines. The expression of MCPyV sT and LT were analyzed by means of a qRT-PCR. The cq values were normalized to MKL-1 and the sd of n=2 is shown. The MCPyV-positive MCC cell lines reveal a different expression pattern of MCPyV T antigens whereas the distribution of LT and sT is comparable (A). In addition, the relative concentration of LT to sT were compared in B. ST is up to 60% less expressed compared to LT
Expression of chromogranin A and synaptophysin in WaGa cells as assessed by immunofluorescence. WaGa nuclei are shown with DAPI (blue). The specific cytoplasmic expression of chromogranin A and synaptophysin is reflected by red fluorescence. The merged picture reveals in all cells a strong expression of chromogranin A or synaptophysin. The microphotographs were taken at 63x magnification.
Summary of the IHC analysis for REST, ASCL1 and NeuroD1 in MCC cell lines and the B-ALL cell line REH, pos.= positive, neg.= negative, - = no expression, + = weak expression, ++ = moderate expression, +++ = strong expression
Used primer for all PCR applications
Acknowledgments
We would like to thank Dr. Christopher Buck (NIH Bethesda, Washington D.C.) for calling our attention to REST and for his inspiring discussions. We thank Dr. Judith Sluimer, (MUMC+, Department of Pathology) for the helpful discussions and support. We thank Mat Rousch (MUMC+, GROW-School for Oncology & Developmental Biology, Department of Pathology) for his assistance in the experimental set up.
Footnotes
Funding statement: This article has no funding source.
Disclosures: The authors have no conflict of interest to declare.
Prior presentation: This manuscript has not been presented anywhere.
Conflicts of interest: The authors declare no conflicts of interest.
This research was supported by RWTH Aachen University through Graduiertenförderung nach Richtlinien zur Förderung des wissenschaftlichen Nachwuchses (RFwN).
References
- 1.Calder KB, Smoller BR. New insights into Merkel cell carcinoma. Adv Anat Pathol. 2010;17(3):155–161. doi: 10.1097/PAP.0b013e3181d97836. [DOI] [PubMed] [Google Scholar]
- 2.Albores-Saavedra J, Batich K, Chable-Montero F, Sagy N, Schwartz AM, Henson DE. Merkel cell carcinoma demographics, morphology, and survival based on 3870 cases: a population based study. J Cutan Pathol. 2010;37:20–27. doi: 10.1111/j.1600-0560.2009.01370.x. [DOI] [PubMed] [Google Scholar]
- 3.Hughes MP, Hardee ME, Cornelius LA, Hutchins LF, Becker JC, Gao L. Merkel Cell Carcinoma: Epidemiology, Target, and Therapy. Curr Dermatol Rep. 2014;3:46–53. doi: 10.1007/s13671-014-0068-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng H, Shuda M, Chang Y, Moore PS. Clonal Integration of a Polyomavirus in Human Merkel Cell Carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kassem A, Schopflin A, Diaz C, Weyers W, Stickeler E, Werner M, Zur Hausen A. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res. 2008;68:5009–5013. doi: 10.1158/0008-5472.CAN-08-0949. [DOI] [PubMed] [Google Scholar]
- 6.Becker JC, Houben R, Ugurel S, Trefzer U, Pföhler C, Schrama D. MC polyomavirus is frequently present in Merkel cell carcinoma of European patients. J Invest Dermatol. 2009;129:248–250. doi: 10.1038/jid.2008.198. [DOI] [PubMed] [Google Scholar]
- 7.Houben R, Shuda M, Weinkam R, Schrama D, Feng H, Chang Y, Moore PS, Becker JC. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84:7064–7072. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schadendorf D, Lebbé C, zur Hausen A, Avril M-F, Hariharan S, Bharmal M, Becker JC. Merkel cell carcinoma: Epidemiology, prognosis, therapy and unmet medical needs. Eur J Cancer. 2017;71:53–69. doi: 10.1016/j.ejca.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 9.Nghiem PT, Bhatia S, Lipson EJ, Kudchadkar RR, Miller NJ, Annamalai L, Berry S, Chartash EK, Daud A, Fling SP. PD-1 blockade with pembrolizumab in advanced Merkel-cell carcinoma. N Engl J Med. 2016;374:2542–2552. doi: 10.1056/NEJMoa1603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaufman HL, Russell J, Hamid O, Bhatia S, Terheyden P, D'Angelo SP, Shih KC, Lebbé C, Linette GP, Milella M. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 2016;17:1374–1385. doi: 10.1016/S1470-2045(16)30364-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tilling T, Moll I. Which Are the Cells of Origin in Merkel Cell Carcinoma? J Skin Cancer. 2012;2012:6. doi: 10.1155/2012/680410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jankowski M, Kopinski P, Schwartz R, Czajkowski R. Merkel cell carcinoma: is this a true carcinoma? Exp Dermatol. 2014;23:792–794. doi: 10.1111/exd.12490. [DOI] [PubMed] [Google Scholar]
- 13.zur Hausen A, Rennspiess D, Winnepenninckx V, Speel E-J, Kurz AK. Early B-cell differentiation in merkel cell carcinomas: clues to cellular ancestry. Cancer Res. 2013;73:4982–4987. doi: 10.1158/0008-5472.CAN-13-0616. [DOI] [PubMed] [Google Scholar]
- 14.Sauer CM, Haugg AM, Chteinberg E, Rennspiess D, Winnepenninckx V, Speel EJ, Becker JC, Kurz AK, Zur Hausen A. Reviewing the current evidence supporting early B-cells as the cellular origin of Merkel cell carcinoma. Crit Rev Oncol Hematol. 2017;116:99–105. doi: 10.1016/j.critrevonc.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 15.Chong JA, Tapia-Ramírez J, Kim S, Toledo-Aral JJ, Zheng Y, Boutros MC, Altshuller YM, Frohman MA, Kraner SD, Mandel G. REST: a mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80(6):949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- 16.Westbrook TF, Hu G, Ang XL, Mulligan P, Pavlova NN, Liang A, Leng Y, Maehr R, Shi Y, Harper JW. SCF(βTRCP) controls oncogenic transformation and neural differentiation through REST degradation. Nature. 2008;452:370–374. doi: 10.1038/nature06780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- 18.Lietz M, Hohl M, Thiel G. RE-1 silencing transcription factor (REST) regulates human synaptophysin gene transcription through an intronic sequence-specific DNA-binding site. Eur J Biochem. 2003;270:2–9. doi: 10.1046/j.1432-1033.2003.03360.x. [DOI] [PubMed] [Google Scholar]
- 19.Kashiwagi K, Ishii J, Sakaeda M, Arimasu Y, Shimoyamada H, Sato H, Miyata C, Kamma H, Aoki I, Yazawa T. Differences of molecular expression mechanisms among neural cell adhesion molecule 1, synaptophysin, and chromogranin A in lung cancer cells. Pathol Int. 2012;62:232–245. doi: 10.1111/j.1440-1827.2011.02781.x. [DOI] [PubMed] [Google Scholar]
- 20.Conti L, Crisafulli L, Caldera V, Tortoreto M, Brilli E, Conforti P, Zunino F, Magrassi L, Schiffer D, Cattaneo E. REST controls self-renewal and tumorigenic competence of human glioblastoma cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0038486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuller GN, Su X, Price RE, Cohen ZR, Lang FF, Sawaya R, Majumder S. Many human medulloblastoma tumors overexpress repressor element-1 silencing transcription (REST)/neuron-restrictive silencer factor, which can be functionally countered by REST-VP16. Mol Cancer Ther. 2005;4:343–349. doi: 10.1158/1535-7163.MCT-04-0228. [DOI] [PubMed] [Google Scholar]
- 22.Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 23.Coolen M, Katz S, Bally-Cuif L. miR-9: a versatile regulator of neurogenesis. Front Cell Neurosci. 2013;7:220. doi: 10.3389/fncel.2013.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bi-functional microRNA miR-9/miR-9* regulates REST and CoREST and is down-regulated in Huntington's Disease. J Neurosci. 2008;28:14341–14346. doi: 10.1523/JNEUROSCI.2390-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ning MS, Kim AS, Prasad N, Levy SE, Zhang H, Andl T. Characterization of the Merkel cell carcinoma miRNome. J Skin Cancer. 2014;2014:9. doi: 10.1155/2014/289548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, Gao G, Hu X, Wang Y, Schwarz JK, Chen JJ, Grigsby PW, Wang X. Activation of miR-9 by human papillomavirus in cervical cancer. Oncotarget. 2014;5:11620–11630. doi: 10.18632/oncotarget.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu H, Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST–REST complex. Proc Natl Acad Sci U S A. 2007;104:17134–17139. doi: 10.1073/pnas.0707266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gu H, Liang Y, Mandel G, Roizman B. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci U S A. 2005;102:7571–7576. doi: 10.1073/pnas.0502658102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guan H, Ricciardi RP. Transformation by E1A oncoprotein involves ubiquitin-mediated proteolysis of the neuronal and tumor repressor REST in the nucleus. J Virol. 2012;86:5594–5602. doi: 10.1128/JVI.06811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Altree-Tacha D, Tyrrell J, Li F. mASH1 is highly specific for neuroendocrine carcinomas: an immunohistochemical evaluation on normal and various neoplastic tissues. Arch Pathol Lab Med. 2017;141:288–292. doi: 10.5858/arpa.2015-0489-OA. [DOI] [PubMed] [Google Scholar]
- 31.Kassem A, Schöpflin A, Diaz C, Weyers W, Stickeler E, Werner M, zur Hausen A. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the <em>VP1</em> gene. Cancer Res. 2008;68:5009–5013. doi: 10.1158/0008-5472.CAN-08-0949. [DOI] [PubMed] [Google Scholar]
- 32.Kreisler A, Strissel PL, Strick R, Neumann SB, Schumacher U, Becker CM. Regulation of the NRSF/REST gene by methylation and CREB affects the cellular phenotype of small-cell lung cancer. Oncogene. 2010;29:5828–5838. doi: 10.1038/onc.2010.321. [DOI] [PubMed] [Google Scholar]
- 33.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coulson JM, Edgson JL, Woll PJ, Quinn JP. A splice variant of the neuron-restrictive silencer factor repressor is expressed in small cell lung cancer: a potential role in derepression of neuroendocrine genes and a useful clinical marker. Cancer Res. 2000;60:1840–1844. [PubMed] [Google Scholar]
- 35.Chen G-L, Miller GM. Extensive alternative splicing of the repressor element silencing transcription factor linked to cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borromeo Mark D, Savage Trisha K, Kollipara Rahul K, He M, Augustyn A, Osborne Jihan K, Girard L, Minna John D, Gazdar Adi F, Cobb Melanie H. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep. 2016;16:1259–1272. doi: 10.1016/j.celrep.2016.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yazawa T. Recent advances in histogenesis research of lung neuroendocrine cancers: Evidence obtained from functional analyses of primitive neural/neuroendocrine cell-specific transcription factors. Pathol Int. 2015;65:277–285. doi: 10.1111/pin.12267. [DOI] [PubMed] [Google Scholar]
- 38.Osborne JK, Larsen JE, Shields MD, Gonzales JX, Shames DS, Sato M, Kulkarni A, Wistuba II, Girard L, Minna JD. NeuroD1 regulates survival and migration of neuroendocrine lung carcinomas via signaling molecules TrkB and NCAM. Proc Natl Acad Sci U S A. 2013;110:6524–6529. doi: 10.1073/pnas.1303932110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ralston J, Chiriboga L, Nonaka D. MASH1: a useful marker in differentiating pulmonary small cell carcinoma from Merkel cell carcinoma. Mod Pathol. 2008;21:1357–1362. doi: 10.1038/modpathol.2008.118. [DOI] [PubMed] [Google Scholar]
- 40.Lewis JS, Jr., Duncavage E, Klonowski PW. Oral cavity neuroendocrine carcinoma: a comparison study with cutaneous Merkel cell carcinoma and other mucosal head and neck neuroendocrine carcinomas. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2010;110:209–217. doi: 10.1016/j.tripleo.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 41.Wagoner MP, Gunsalus KTW, Schoenike B, Richardson AL, Friedl A, Roopra A. The transcription factor REST is lost in aggressive breast cancer. PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1000979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Osborne JK, Larsen JE, Gonzales JX, Shames DS, Sato M, Wistuba II, Girard L, Minna JD, Cobb MH. NeuroD1 regulation of migration accompanies the differential sensitivity of neuroendocrine carcinomas to TrkB inhibition. Oncogenesis. 2013;2 doi: 10.1038/oncsis.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daily K, Coxon A, Williams JS, Lee C-CR, Coit DG, Busam KJ, Brownell I. Assessment of cancer cell line representativeness using microarrays for Merkel cell carcinoma. J Invest Dermatol. 2015;135:1138–1146. doi: 10.1038/jid.2014.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gao G, Gay HA, Chernock RD, Zhang TR, Luo J, Thorstad WL, Lewis JS, Wang X. A microRNA expression signature for the prognosis of oropharyngeal squamous cell carcinoma. Cancer. 2013;119:72–80. doi: 10.1002/cncr.27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Blast of MKL-2 REST sequence. NCBI blast has revealed the identity of the truncated REST sequence which was detected in SCLC (A). The exons of the non-truncated and truncated splice variant are shown (B).
MCPyV expression level in all used cell lines. The expression of MCPyV sT and LT were analyzed by means of a qRT-PCR. The cq values were normalized to MKL-1 and the sd of n=2 is shown. The MCPyV-positive MCC cell lines reveal a different expression pattern of MCPyV T antigens whereas the distribution of LT and sT is comparable (A). In addition, the relative concentration of LT to sT were compared in B. ST is up to 60% less expressed compared to LT
Expression of chromogranin A and synaptophysin in WaGa cells as assessed by immunofluorescence. WaGa nuclei are shown with DAPI (blue). The specific cytoplasmic expression of chromogranin A and synaptophysin is reflected by red fluorescence. The merged picture reveals in all cells a strong expression of chromogranin A or synaptophysin. The microphotographs were taken at 63x magnification.
Summary of the IHC analysis for REST, ASCL1 and NeuroD1 in MCC cell lines and the B-ALL cell line REH, pos.= positive, neg.= negative, - = no expression, + = weak expression, ++ = moderate expression, +++ = strong expression
Used primer for all PCR applications