Highlights
-
•
Here we highlight our recent advances in de novo enzyme design.
-
•
Heme C-binding maquettes with minimal complexity serve as promiscuous and efficient enzymes.
-
•
Well-defined substrate binding sites are possibly not required for efficient oxidoreductase catalysis.
Abstract
Though established 40 years ago, the field of de novo protein design has recently come of age, with new designs exhibiting an unprecedented level of sophistication in structure and function. With respect to catalysis, de novo enzymes promise to revolutionise the industrial production of useful chemicals and materials, while providing new biomolecules as plug-and-play components in the metabolic pathways of living cells. To this end, there are now de novo metalloenzymes that are assembled in vivo, including the recently reported C45 maquette, which can catalyse a variety of substrate oxidations with efficiencies rivalling those of closely related natural enzymes. Here we explore the successful design of this de novo enzyme, which was designed to minimise the undesirable complexity of natural proteins using a minimalistic bottom-up approach.
Current Opinion in Structural Biology 2018, 51:149–155
This review comes from a themed issue on Engineering & design
Edited by Giovanna Ghirlanda and Ivan Korendovych
For a complete overview see the Issue and the Editorial
Available online 10th May 2018
https://doi.org/10.1016/j.sbi.2018.04.008
0959-440X/© 2018 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/4.0/).
The design of novel proteins
De novo protein design has its roots in the 1970s with the design of functional peptides that aimed to understand the rules governing the relationship of amino acid sequence with higher order structure and function [1, 2, 3]. Since then, the field has flourished with advances in technology, protein structure prediction and recombinant protein expression [4••], with designs that mimic [5, 6], supersede [7] or perform chemistry not seen in nature [8]. Through this we may further our understanding of how natural proteins operate [9], or augment the functional possibilities currently available to us from nature's repertoire of proteins. The simplicity of most de novo proteins is an advantage over the often complex and intricate characteristics and interactions of natural proteins; millennia of natural selection have imprinted and consolidated this complexity on natural proteins, with many individual amino acids becoming irreversibly dependent on each other [10, 11]. Therefore, it is not always straightforward to replicate function in a de novo designed protein by importing natural sequences. In this review we discuss a bottom-up approach to de novo protein design, based on bundles of four alpha-helices, in which the complexity of natural proteins is avoided.
The first de novo four-helix bundle proteins were designed in the late 1980s by DeGrado in which repeated amino acid heptads form a structure with each individual amino acid having a well-defined role [12]. Heptads of amino acid residues with high helical forming propensities form two turns of an alpha helix, helix length can be tailored by building up a series of heptad repeats, and linked with loops containing residues with low helix-forming propensities. Protein folding is driven by the exclusion of water from the protein core through the patterning of polar and nonpolar residues [12]. These simple principles have formed the basis of many de novo protein designs, although there are designs that are made up of beta-strand elements [13, 14, 15].
Novel protein scaffolds may be designed rationally to achieve a particular function, and/or use directed evolution to evolve towards or refine the desired activity. Developments in high throughput techniques have facilitated the construction and screening of large protein libraries [16, 17], while the use of computational design has increasingly allowed us to design scaffolds whose experimentally determined structure remains faithful to that of the intended design [15]. While many de novo proteins were designed with a specific function in mind [6], other functionalities were more serendipitous [18]. Many designs take inspiration from natural structures [19], or incorporate natural sequences [20].
In this review we discuss a strategy for the successful design of de novo enzymes, and in particular we discuss our use of the maquette approach [21•], in which we design an evolutionary naïve, robust structure built from the minimum number of amino acids possible, and use a cofactor to imprint function [22••]. In this review we challenge some design principles surmised from natural proteins: complexity, specificity, and a defined structure, and examine whether they are strictly necessary for an effective de novo-designed biological catalyst.
The maquette approach to protein design
Maquettes are de novo-designed self-assembling peptide scaffolds pioneered by Dutton and colleagues. They are designed bottom-up without mimicking natural sequences with the intention of minimising the complexity present in naturally evolved proteins, and are subject to iterative rounds of design with significant engineering freedom [21•, 23, 24]. Maquette functions to date are diverse, recent examples include light harvesting [25] and subsequent energy transfer [26], oxygen binding [27], oxidation and oxidative dehalogenation catalysis [22••], amphiphilic maquettes for transmembrane electron transfer [28], and magnetic field sensing [29].
Function is conferred onto a maquette scaffold through the incorporation of cofactor molecules, and many maquette designs contain heme. A large amount of natural proteins contain heme, exhibiting an exceptionally diverse range of functions including electron transfer, catalysis, sensing, and transport [30], and in fact heme may have been utilised by early enzymes as a way to incorporate activity [18, 31]. Many heme-containing proteins are alpha helical structures, including simple 4-helix bundles [18, 30], the scaffold used in most maquette designs. Heme has therefore proven a useful cofactor to incorporate into artificial proteins, particularly due to the ease by which it can be incorporated: heme B can be ligated through two histidine residues on the interior faces of neighbouring helices, and multiple hemes can be bound within a monomeric scaffold [32, 33]. The first maquette design was based on the bis-histidine heme binding sites in the respiratory bc1 complex [19], with the heme-ligating residues located along the hydrophobic helix interfaces.
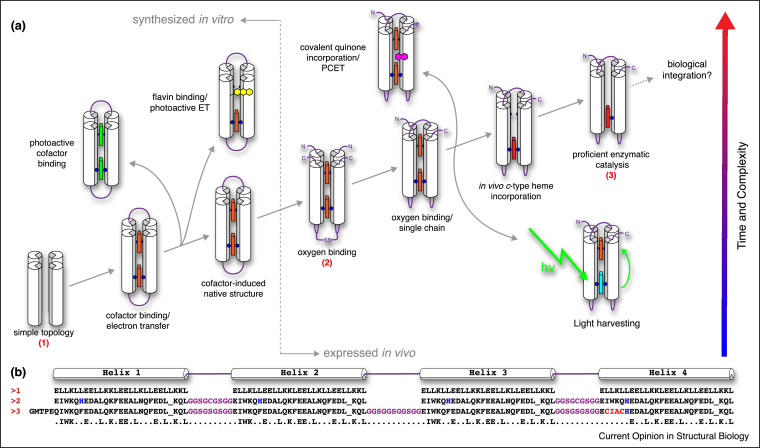
Early maquettes were dimeric, comprising synthesized peptides each with 2 helices connected by linking loops. A subsequent design, HP7 (Figure 1(2)) has an O2-binding heme and features helix–loop–helix monomers linked by a covalent disulphide bond ‘candelabra’ geometry [34, 35]. More recent maquettes have utilised single polypeptide chains (Figure 1(3)) thereby avoiding the symmetry-induced constraints of earlier designs; these scaffolds have been used to reproduce oxidoreductase functions with activities comparable to their natural counterparts [22••, 36, 37]. Single-chain scaffolds have advantages including the ability to incorporate single site mutations or covalent modifications, and they can be expressed in vivo [36].
Figure 1.
The evolution of C45 from humble beginnings. (a) Evolutionary diagram displaying the broad design strokes and functions of the maquettes from a simple featureless 4-helix bundle (1) [35], to the oxygen binding HP7 maquette (2) [35], to the functional de novo enzyme C45 (3) [22••]. (b) Sequence alignment of three maquettes from the evolutionary diagram of C45. Heme-ligating histidines are highlighted in blue, CXXC from the c-type cytochrome consensus motif (CXXCH) is highlighted in red, and the interhelical loops are displayed in purple.
The maquette approach has proven effective in conferring function onto a simple scaffold, thus challenging the necessity of the complexity observed in natural proteins.
How important is protein complexity in catalysis?
While natural proteins can be complex, de novo protein design has proven that we can build catalytic function onto a relatively simple scaffold. Many successful de novo protein designs that can perform catalysis are made up of bundles of alpha helices, with activities including the hydration of CO2 [6], catalase activity [38], enantiospecific hydrolysis [39•], oxidation and hydroxylation [40, 41], and there are some that can rescue auxotroph E. coli strains [39•]. De novo catalytic proteins have been assembled from more complex structures, such as the hydrolytic barrel designed by Burton and colleagues [42]. However, an advantage of the bottom-up minimalist approach taken in maquette design is that more complex structures and catalytic function can be built up iteratively. In this way it is easy to determine the function of any one amino acid, facilitating the addition of mutations. We propose that a simple helix bundle is the best starting point for many forms of catalysis, particularly as it has been demonstrated that alpha helical bundle proteins are amenable to expression in vivo [22••], whereas more complex designs may require peptide synthesis or full assembly in vitro.
While there are many types of catalytic activity that can be performed by de novo designed proteins, the remainder of this review will focus on oxidoreductase activity. Oxidoreductases are an exceptionally large and important enzyme family, with many functions involving the transfer of electrons from a donor molecule to an acceptor. Heme-containing peroxidases that catalyse substrate oxidation coupled to H2O2 reduction coordinate the catalytic heme by a single histidine side chain, leaving the 6th coordination site free to bind a substrate molecule [43]; the simplicity of this design lends itself to incorporation into a de novo scaffold. It is well known that the requirements for peroxidase activity are minimal. For example, the mimochrome family of artificial proteins which have been designed to maintain the properties of heme within a minimal protein scaffold [44]. They consist of two polypeptide chains <14 residues long, and heme. When the heme is 5-coordinate, mimochromes have peroxidase ability, oxidising ABTS in the presence of H2O2 [45]. Not only is such a small scaffold capable of facilitating catalytic ability, mutations to the sequence can fine tune the reactivity.
There has been much interest in incorporating these characteristics into maquettes [27, 33, 37]. De novo proteins containing heme C are advantageous in that the irreversible covalent binding of the heme to the protein backbone facilitates purification of the functional holoprotein after expression, and, importantly, provide an opportunity for supporting a 5-coordinate heme, with one site free for substrate binding and catalysis.
B-type heme-binding maquettes can be converted to a covalently-bound c-type by using the conserved c-type binding motif, CX1X2CH [36]. Despite their unnatural protein sequences, c-type maquettes are fully assembled in E. coli through the addition of a periplasmic export tag and the co-expression of the type I c-type cytochrome maturation (Ccm) machinery [22••, 36]. This results in the covalent binding of heme through the heme vinyl groups and protein cysteine residues [36]. The maquette C45 arose from the mutation of previous c-type maquette designs to produce a single-chain maquette with a mono-histidine ligated heme [22••]. C45 has the basic requirements of a peroxidase in that the heme cofactor is solvent exposed and can bind peroxides on its distal coordination site. Furthermore, the reaction between C45 and H2O2/ABTS (2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)) follows the kinetics of natural peroxidases, even matching the catalytic efficiency of horse radish peroxidase (HRP) operating at its optimum.
The ability to perform efficient catalysis even in an extremely simplified scaffold compared to the complexity of natural proteins leads us to pose the question: how important is a defined structure in catalysis?
How important is a defined structure in catalysis?
Protein design has utilised the basic requirements for protein folding in which thermodynamic requirements for the assembly of secondary structure are satisfied by using simple heptad patterning of the residues comprising the alpha helices. The folding of alpha helical units into a bundle may then follow the principles of natural proteins in which the hydrophobic interior residues are hidden from the aqueous solvent [12]. Such structure is necessary over a disordered unfolded peptide sequence to ensure stability, to facilitate structure prediction and to prevent aggregation, for example for in vivo expression.
In nature, structure is often very important for catalysis, in which the precise arrangement of active site amino acid side chains is imperative [46, 47]. It is thought that the active site of an enzyme stabilises the transition state over the substrate, thus lowering the activation energy required for the reaction [48, 49]. There is debate as to the importance or extent of binding-induced conformational changes to align active site residue side chains for effective catalysis [50, 51]. Regardless of the precise malleability of the active site, in many cases amino acid sidechains must form precise interactions with the substrate to facilitate catalysis, and may involve acting as donors or acceptors of protons, electrons or other groups [52, 53]; electrostatic interactions are important for transition state stabilisation [54]. There are forms of catalysis in which the precise spatial arrangement of amino acid side chains at the substrate binding site is not so important, and the precise location of substrate binding is ill-defined [55]. Where catalytic cofactors are utilised, their properties must be modulated, which is often achieved through the precise alignment and the effects of nearby amino acids [56]. This has presented a challenge when it comes to the design of de novo enzymes, although advances in computing power for design and structure prediction are facilitating this [4••]. In the case of C45, despite the dynamic nature of the protein, certain characteristics imply that it is a stable, water impenetrable structure [22••]. Similar structural characteristics have been observed in other de novo [36] and natural proteins [57]. In many of these cases this may be due to the substrate conferring structural homogeneity on the active site, it remains to be experimentally determined whether this is the case for C45.
It has been proposed that conformational flexibility may have been an important mechanism in the evolution of new reactivities in early enzymes [58]. It may therefore be prudent to follow the example of nature by replicating the characteristics of early enzymes in de novo designs before iteration and directed evolution to refine and expand function. Additionally, de novo proteins could serve as models for early enzymes to gain insights into the evolution of modern enzymes.
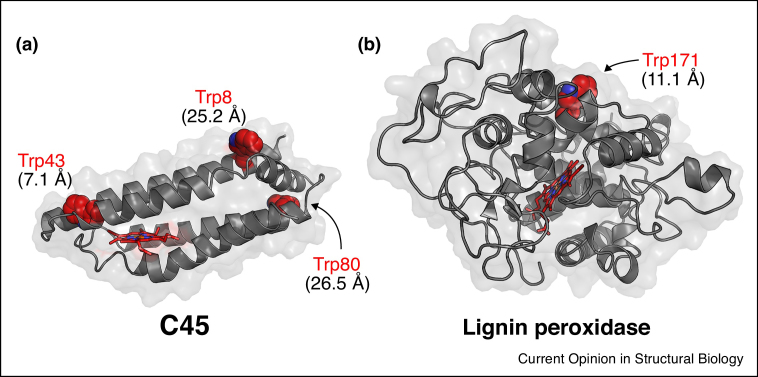
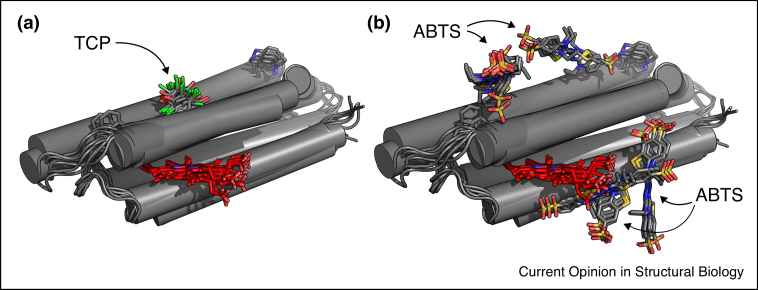
Some natural peroxidases, such as ascorbate peroxidase [59], possess defined substrate-binding sites whereas others, such as lignin peroxidase, have buried hemes and do not have a well-defined cavity within the protein but instead bind the substrate on the surface [55]. In the case of surface-bound substrate, there is evidence to suggest that long-range electron transfer pathways exist to link the substrate to the buried heme via a catalytic surface tryptophan residue [60, 61, 62], and C45 does have surface Trp residues that could fulfil this role (Figure 2a), as does lignin peroxidase [63, 64] (Figure 2b). In the case of C45, it performs efficient catalysis without a specific binding site, and its substrate ABTS binds over the protein surface (Figure 3b). By contrast, 2,4,6-trichlorophenol (TCP), to which C45 has a lower activity, may bind in a specific place (Figure 3a). These findings indicate that it may be that binding of the heme and its immediate environment is more important for catalysis than a snug binding pocket for the substrate.
Figure 2.
Surface tryptophan residues in both C45 (a) and lignin peroxidase (PDB: 1B82) (b) which potentially participate in long-range electron transfer from a surface-bound substrate to the protein-bound heme. Numbers in parentheses represent the edge-to-edge distances between the tryptophan side chains and the conjugated porphyrin system of the bound hemes.
Figure 3.
Computational analysis of potential C45-substrate binding sites. While 2,4,6-trichlorophenol (TCP) (a) appears to preferentially bind in one position on C45, the larger ABTS (2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)) molecule (b) appears to bind indiscriminately across the surface. The data presented is derived from docking analysis performed by the Bristol University Docking Engine (BUDE) [22••] in conjunction with molecular dynamics simulations. Overlays are shown here for representative low energy binding poses for TCP and ABTS.
While it has been suggested that the design of recessed cavities in de novo enzymes is important [4••], the success of C45 shows that they are not always necessary for efficient catalysis. Instead it may be profitable to focus de novo design efforts on long-range efficient and rapid electron transfer from surface-bound substrates. Furthermore, recent research by the DeGrado group has highlighted the importance of designing the whole protein as a unit, with features far from the active site having an impact on activity [65•].
It is worth noting that defined structure may not be essential for de novo protein functions other than catalysis. The Hecht group have used a library approach of de novo sequences to identify proteins that can rescue auxotroph E. coli strains. Many of these proteins act on gene regulation [66] and, there is evidence for some of these structures that, despite being highly stable, they do not form well ordered structures in vitro [67].
The advantages of substrate promiscuity in de novo protein design
The lack of highly specific substrate binding sites in many natural peroxidases can lead to broad substrate promiscuity, which we have observed in the case of C45, which can catalyse a variety of peroxidase substrates including guaiacol, reactive blue 4, and halogenated phenols [22••]. As theorised by Roy Jensen, primordial enzymes were catalytically promiscuous, then evolved to perform more specific and/or active function [68]. Thus, we may take inspiration from nature and design de novo enzymes without specificity, then evolve to improve or narrow selectivity. This theory of the evolution of proteins from generalists to specialists [69] has been tested using de novo protein design by the Hecht group [18, 70]. This was done through the use of combinatorial libraries of protein sequences designed with binary patterning principles, in which the polar and non polar nature of residues is selected and patterned to build a particular secondary structure — in this case 4-helix bundles. The majority of the proteins that were expressed could bind b-type heme, and most of those exhibited peroxidase activity and exhibited catalytic promiscuity. A small amount of the de novo proteins were hydrolases with lipase and esterase activity (activities that do not depend on bound heme). These results demonstrate that achieving catalytic ability and/or substrate binding is not difficult in unevolved protein sequences [18].
In many natural enzymes specificity is often vitally important to stringently distinguish between substrates, such is the case with restriction endonucleases [71, 72]. But for the purposes of de novo enzymes, this fidelity may not be strictly necessary. For example, in an industrial reaction in which only one substrate is fed into the reaction, tight control over promiscuity is not necessary as long as the desired product is produced effectively and efficiently. In fact promiscuity may be an advantage as one enzyme may be able to perform various different reactions as desired, providing the reaction energetically favours the creation of the product over the back reaction. Many natural proteins exhibit substrate promiscuity, including HRP, making it useful for many biotechnological applications [73].
While the substrate promiscuity of C45 may be advantageous, it also lends the opportunity to employ both rational protein design and directed evolution methodologies to optimize the catalytic chassis towards a selected substrate or chemical mechanism. This will be facilitated by the fact that C45 is fully assembled (and functional) in vivo.
Conclusions and future directions for de novo enzyme design
C45 is an example of how a catalytically productive de novo enzyme was achieved with relative ease, without a defined substrate binding site or strict specificity. The simplicity of the C45 design provides a basic and flexible scaffold which lends itself to further modifications to achieve new functions. For an example, we anticipate that C45 may perform other natural and artificial reactions, such as carbene and nitrene transfer, and may provide a platform for accessing powerful hydroxylase chemistries.
Based on the success of the C45 design we propose that future catalytic protein design will be aided by taking a bottom-up approach, using simple scaffolds as a starting point. Rather than focussing on highly specific and efficient catalysis, we should take inspiration from the evolution of natural proteins, in which promiscuity provides the starting point for refined reactivities. While the design of active sites is a worthwhile goal, in certain cases they may not be necessary and instead treating the protein as a reactive surface with networks of interactions across the structure may be beneficial.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgments
Funding: this work was supported by the BBSRC [BB/M02315X/1].
References
- 1.Chakravarty P.K., Mathur K.B., Dhar M.M. Synthesis of a decapeptide with glycosidase activity. Experientia. 1973;29:786–788. [Google Scholar]
- 2.Gutte B., Daumigen M., Wittschieber E. Design, synthesis and characterization of a 34-residue polypeptide that interacts with nucleic-acids. Nature. 1979;281:650–655. doi: 10.1038/281650a0. [DOI] [PubMed] [Google Scholar]
- 3.DeGrado W.F., Raleigh D.P., Handel T. De novo protein design: what are we learning? Curr Opin Struct Biol. 1991;1:984–993. [Google Scholar]
- 4••.Huang P.S., Boyken S.E., Baker D. The coming of age of de novo protein design. Nature. 2016;537:320–327. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]; A review discussing how advances in understanding and technologies have contributed to recent advances in the design of de novo protein structures and functions.
- 5.Zastrow M.L., Peacock A.F.A., Stuckey J.A., Pecoraro V.L. Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nat Chem. 2012;4:118–123. doi: 10.1038/nchem.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cangelosi V.M., Deb A., Penner-Hahn J.E., Pecoraro V.L. A de novo designed metalloenzyme for the hydration of CO2. Angew Chem Int Ed. 2014;53:7900–7903. doi: 10.1002/anie.201404925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher M.A., McKinley K.L., Bradley L.H., Viola S.R., Hecht M.H. De novo designed proteins from a library of artificial sequences function in Escherichia coli and enable cell growth. PLoS One. 2011;6 doi: 10.1371/journal.pone.0015364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siegel J.B., Zanghellini A., Lovick H.M., Kiss G., Lambert A.R., Clair J.L.S., Gallaher J.L., Hilvert D., Gelb M.H., Stoddard B.L. Computational design of an enzyme catalyst for a stereoselective bimolecular Diels–Alder reaction. Science. 2010;329:309–313. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richardson J.S., Richardson D.C. The de novo design of protein structures. Trends Biochem Sci. 1989;14:304–309. doi: 10.1016/0968-0004(89)90070-4. [DOI] [PubMed] [Google Scholar]
- 10.Muller H.J. The relation of recombination to mutational advance. Mutat Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 11.Dutton P.L., Moser C.C. Engineering enzymes. Faraday Discuss. 2011;148 doi: 10.1039/c005523a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regan L., Degrado W.F. Characterization of a helical protein designed from 1st principles. Science. 1988;241:976–978. doi: 10.1126/science.3043666. [DOI] [PubMed] [Google Scholar]
- 13.MacDonald J.T., Kabasakal B.V., Godding D., Kraatz S., Henderson L., Barber J., Freemont P.S., Murray J.W. Synthetic beta-solenoid proteins with the fragment-free computational design of a beta-hairpin extension. Proc Natl Acad Sci U S A. 2016;113:10346–10351. doi: 10.1073/pnas.1525308113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcos E., Basanta B., Chidyausiku T.M., Tang Y.F., Oberdorfer G., Liu G.H., Swapna G.V.T., Guan R.J., Silva D.A., Dou J.Y. Principles for designing proteins with cavities formed by curved beta sheets. Science. 2017;355:201–206. doi: 10.1126/science.aah7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang P.S., Feldmeier K., Parmeggiani F., Velasco D.A.F., Hocker B., Baker D De novo design of a four-fold symmetric TIM-barrel protein with atomic-level accuracy. Nat Chem Biol. 2016;12 doi: 10.1038/nchembio.1966. 29-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoegler K.J., Hecht MH: A de novo protein confers copper resistance in Escherichia coli. Protein Sci. 2016;25:1249–1259. doi: 10.1002/pro.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chevalier A., Silva D.A., Rocklin G.J., Hicks D.R., Vergara R., Murapa P., Bernard S.M., Zhang L., Lam K.H., Yao G.R. Massively parallel de novo protein design for targeted therapeutics. Nature. 2017;550 doi: 10.1038/nature23912. 74-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel S.C., Bradley L.H., Jinadasa S.P., Hecht M.H. Cofactor binding and enzymatic activity in an unevolved superfamily of de novo designed 4-helix bundle proteins. Protein Sci. 2009;18:1388–1400. doi: 10.1002/pro.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robertson D.E., Farid R.S., Moser C.C., Urbauer J.L., Mulholland S.E., Pidikiti R., Lear J.D., Wand A.J., Degrado W.F., Dutton P.L. Design and synthesis of multi-heme proteins. Nature. 1994;368:425–431. doi: 10.1038/368425a0. [DOI] [PubMed] [Google Scholar]
- 20.Mahendran K.R., Niitsu A., Kong L.B., Thomson A.R., Sessions R.B., Woolfson D.N., Bayley H. A monodisperse transmembrane alpha-helical peptide barrel. Nat Chem. 2017;9:411–419. doi: 10.1038/nchem.2647. [DOI] [PubMed] [Google Scholar]
- 21•.Ennist N., Mancini J., Auman B.D., Bialas C.J., Iwanicki M., Esipova T., Discher B., Moser C.C., Leslie Dutton P. 2017. Maquette strategy for creation of light- and redox-active proteins. 10.1142/9789813230309_0001. [Google Scholar]; A summary of the maquette approach to date, detailing recent and future advances.
- 22••.Watkins D.W., Jenkins J.M.X., Grayson K.J., Wood N., Steventon J.W., Le Vay K.K., Goodwin M.I., Mullen A.S., Bailey H.J., Crump M.P. Construction and in vivo assembly of a catalytically proficient and hyperthermostable de novo enzyme. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00541-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; Design and activity of the C45 maquette.
- 23.Lichtenstein B.R., Farid T.A., Kodali G., Solomon L.A., Anderson J.L.R., Sheehan M.M., Ennist N.M., Fry B.A., Chobot S.E., Bialas C. Engineering oxidoreductases: maquette proteins designed from scratch. Biochem Soc Trans. 2012;40:561–566. doi: 10.1042/BST20120067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibney B.R., Rabanal F., Skalicky J.J., Wand A.J., Dutton P.L. Design of a unique protein scaffold for maquettes. J Am Chem Soc. 1997;119:2323–2324. [Google Scholar]
- 25.Kodali G., Mancini J.A., Solomon L.A., Episova T.V., Roach N., Hobbs C.J., Wagner P., Mass O.A., Aravindu K., Barnsley J.E. Design and engineering of water-soluble light-harvesting protein maquettes. Chem Sci. 2017;8:316–324. doi: 10.1039/c6sc02417c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mancini J.A., Kodali G., Jiang J.B., Reddy K.R., Lindsey J.S., Bryant D.A., Dutton P.L., Moser C.C. Multi-step excitation energy transfer engineered in genetic fusions of natural and synthetic light-harvesting proteins. J R Soc Interf. 2017;14 doi: 10.1098/rsif.2016.0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watkins D.W., Armstrong C.T., Beesley J.L., Marsh J.E., Jenkins J.M.X., Sessions R.B., Mann S., Anderson J.L.R. A suite of de novo c-type cytochromes for functional oxidoreductase engineering. Biochim Biophys Acta-Bioenerg. 2016;1857:493–502. doi: 10.1016/j.bbabio.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Goparaju G., Fry B.A., Chobot S.E., Wiedman G., Moser C.C., Dutton P.L., Discher B.M. First principles design of a core bioenergetic transmembrane electron-transfer protein. Biochim Biophys Acta-Bioenerg. 2016;1857:503–512. doi: 10.1016/j.bbabio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bialas C., Jarocha L.E., Henbest K.B., Zollitsch T.M., Kodali G., Timmel C.R., Mackenzie S.R., Dutton P.L., Moser C.C., Hore P.J. Engineering an artificial flavoprotein magnetosensor. J Am Chem Soc. 2016;138:16584–16587. doi: 10.1021/jacs.6b09682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reedy C.J., Gibney B.R. Heme protein assemblies. Chem Rev. 2004;104:617–649. doi: 10.1021/cr0206115. [DOI] [PubMed] [Google Scholar]
- 31.Moffet D.A., Certain L.K., Smith A.J., Kessel A.J., Beckwith K.A., Hecht M.H. Peroxidase activity in heme proteins derived from a designed combinatorial library. J Am Chem Soc. 2000;122:7612–7613. [Google Scholar]
- 32.Shifman J.M., Gibney B.R., Sharp R.E., Dutton P.L. Heme redox potential control in de novo designed four-alpha-helix bundle proteins. Biochemistry. 2000;39:14813–14821. doi: 10.1021/bi000927b. [DOI] [PubMed] [Google Scholar]
- 33.Farid T.A., Kodali G., Solomon L.A., Lichtenstein B.R., Sheehan M.M., Fry B.A., Bialas C., Ennist N.M., Siedlecki J.A., Zhao Z. Elementary tetrahelical protein design for diverse oxidoreductase functions. Nat Chem Biol. 2013;9 doi: 10.1038/nchembio.1362. 826-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koder R.L., Valentine K.G., Cerda J., Noy D., Smith K.M., Wand A.J., Dutton P.L. Nativelike structure in designed four alpha-helix bundles driven by buried polar interactions. J Am Chem Soc. 2006;128:14450–14451. doi: 10.1021/ja064883r. [DOI] [PubMed] [Google Scholar]
- 35.Koder R.L., Anderson J.L.R., Solomon L.A., Reddy K.S., Moser C.C., Dutton P.L. Design and engineering of an O-2 transport protein. Nature. 2009;458 doi: 10.1038/nature07841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anderson J.L.R., Armstrong C.T., Kodali G., Lichtenstein B.R., Watkins D.W., Mancini J.A., Boyle A.L., Farid T.A., Crump M.P., Moser C.C. Constructing a man-made c-type cytochrome maquette in vivo: electron transfer, oxygen transport and conversion to a photoactive light harvesting maquette. Chem Sci. 2014;5:507–514. doi: 10.1039/C3SC52019F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watkins D.W., Armstrong C.T., Anderson J.L.R. De novo protein components for oxidoreductase assembly and biological integration. Curr Opin Chem Biol. 2014;19:90–98. doi: 10.1016/j.cbpa.2014.01.016. [DOI] [PubMed] [Google Scholar]
- 38.Olson T.L., Espiritu E., Edwardraja S., Canarie E., Flores M., Williams J.C., Ghirlanda G., Allen J.P. Biochemical and spectroscopic characterization of dinuclear Mn-sites in artificial four-helix bundle proteins. Biochim Biophys Acta-Bioenerg. 2017;1858:945–954. doi: 10.1016/j.bbabio.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 39•.Donnelly A.E., Murphy G.S., Digianantonio K.M., Hecht M.H. A de novo enzyme catalyzes a life-sustaining reaction in Escherichia coli. Nat Chem Biol. 2018 doi: 10.1038/nchembio.2550. [DOI] [PubMed] [Google Scholar]; Mutation and selection of a alpha helical bundle protein from a combinational library that had a non-catalytic life-sustaining function resulted in a design that could rescue an auxotroph E. coli strain by performing in vivo catalysis.
- 40.Snyder R.A., Betzu J., Butch S.E., Reig A.J., DeGrado W.F., Solomon E.I. Systematic perturbations of binuclear non-heme iron sites: structure and dioxygen reactivity of de novo Due Ferri proteins. Biochemistry. 2015;54:4637–4651. doi: 10.1021/acs.biochem.5b00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snyder R.A., Butch S.E., Reig A.J., DeGrado W.F., Solomon E.I. Molecular-level insight into the differential oxidase and oxygenase reactivities of de novo Due Ferri proteins. J Am Chem Soc. 2015;137:9302–9314. doi: 10.1021/jacs.5b03524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Burton A.J., Thomson A.R., Dawson W.M., Brady R.L., Woolfson D.N. Installing hydrolytic activity into a completely de novo protein framework. Nat Chem. 2016;8:837–844. doi: 10.1038/nchem.2555. [DOI] [PubMed] [Google Scholar]
- 43.Poulos T.L. Heme enzyme structure and function. Chem Rev. 2014;114:3919–3962. doi: 10.1021/cr400415k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lombardi A., Nastri F., Pavone V. Peptide-based heme-protein models. Chem Rev. 2001;101:3165–3189. doi: 10.1021/cr000055j. [DOI] [PubMed] [Google Scholar]
- 45.Nastri F., Lista L., Ringhieri P., Vitale R., Faiella M., Andreozzi C., Travascio P., Maglio O., Lombardi A., Pavone V. A heme-peptide metalloenzyme mimetic with natural peroxidase-like activity. Chem — Eur J. 2011;17:4444–4453. doi: 10.1002/chem.201003485. [DOI] [PubMed] [Google Scholar]
- 46.Gutteridge A., Thornton J. Conformational change in substrate binding, catalysis and product release: an open and shut case? FEBS Lett. 2004;567:67–73. doi: 10.1016/j.febslet.2004.03.067. [DOI] [PubMed] [Google Scholar]
- 47.Osuna S., Jimenez-Oses G., Noey E.L., Houk K.N. Molecular dynamics explorations of active site structure in designed and evolved enzymes. Acc Chem Res. 2015;48:1080–1089. doi: 10.1021/ar500452q. [DOI] [PubMed] [Google Scholar]
- 48.Pauling L. Nature of forces between large molecules of biological interest. Nature. 1948;161:707–709. doi: 10.1038/161707a0. [DOI] [PubMed] [Google Scholar]
- 49.Bugg T.D.H. The development of mechanistic enzymology in the 20th century. Nat Prod Rep. 2001;18:465–493. doi: 10.1039/b009205n. [DOI] [PubMed] [Google Scholar]
- 50.Koshland D.E. The key–lock theory and the induced fit theory. Angew Chem-Int Ed. 1994;33:2375–2378. [Google Scholar]
- 51.Michel D. Conformational selection or induced fit? New insights from old principles. Biochimie. 2016;128:48–54. doi: 10.1016/j.biochi.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 52.Hempel J., Kuo I., Perozich J., Wang B.C., Lindahl R., Nicholas H. Aldehyde dehydrogenase — maintaining critical active site geometry at motif 8 in the class 3 enzyme. Eur J Biochem. 2001;268:722–726. doi: 10.1046/j.1432-1327.2001.01926.x. [DOI] [PubMed] [Google Scholar]
- 53.Kim T.W., Brieba L.G., Ellenberger T., Kool E.T. Functional evidence for a small and rigid active site in a high fidelity DNA polymerase — probing T7 DNA polymerase with variably sized base pairs. J Biol Chem. 2006;281:2289–2295. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 54.Burschowsky D., van Eerde A., Okvist M., Kienhofer A., Kast P., Hilvert D., Krengel U. Electrostatic transition state stabilization rather than reactant destabilization provides the chemical basis for efficient chorismate mutase catalysis. Proc Natl Acad Sci U S A. 2014;111:17516–17521. doi: 10.1073/pnas.1408512111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miki Y., Calvino F.R., Pogni R., Giansanti S., Ruiz-Duenas F.J., Martinez M.J., Basosi R., Romero A., Martinez A.T. Crystallographic, kinetic, and spectroscopic study of the first ligninolytic peroxidase presenting a catalytic tyrosine. J Biol Chem. 2011;286:15525–15534. doi: 10.1074/jbc.M111.220996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paoli M., Marles-Wright J., Smith A. Structure–function relationships in heme-proteins. DNA Cell Biol. 2002;21:271–280. doi: 10.1089/104454902753759690. [DOI] [PubMed] [Google Scholar]
- 57.Schulenburg C., Stark Y., Kunzle M., Hilvert D. Comparative laboratory evolution of ordered and disordered enzymes. J Biol Chem. 2015;290:9310–9320. doi: 10.1074/jbc.M115.638080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Risso V.A., Martinez-Rodriguez S., Candel A.M., Kruger D.M., Pantoja-Uceda D., Ortega-Munoz M., Santoyo-Gonzalez F., Gaucher E.A., Kamerlin S.C.L., Bruix M. De novo active sites for resurrected Precambrian enzymes. Nat Commun. 2017;8 doi: 10.1038/ncomms16113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharp K.H., Mewies M., Moody P.C.E., Raven E.L. Crystal structure of the ascorbate peroxidase–ascorbate complex. Nat Struct Biol. 2003;10:303–307. doi: 10.1038/nsb913. [DOI] [PubMed] [Google Scholar]
- 60.Mayo S.L., Ellis W.R., Crutchley R.J., Gray H.B. Long-range electron-transfer in heme-proteins. Science. 1986;233:948–952. doi: 10.1126/science.3016897. [DOI] [PubMed] [Google Scholar]
- 61.Perez-Boada M., Ruiz-Duenas F.J., Pogni R., Basosi R., Choinowski T., Martinez M.J., Piontek K., Martinez A.T. Versatile peroxidase oxidation of high redox potential aromatic compounds: site-directed mutagenesis, spectroscopic and crystallographic investigation of three long-range electron transfer pathways. J Mol Biol. 2005;354:385–402. doi: 10.1016/j.jmb.2005.09.047. [DOI] [PubMed] [Google Scholar]
- 62.Gray H.B., Winkler J.R. Long-range electron transfer. Proc Natl Acad Sci U S A. 2005;102:3534–3539. doi: 10.1073/pnas.0408029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doyle W.A., Blodig W., Veitch N.C., Piontek K., Smith A.T. Two substrate interaction sites in lignin peroxidase revealed by site-directed mutagenesis. Biochemistry. 1998;37:15097–15105. doi: 10.1021/bi981633h. [DOI] [PubMed] [Google Scholar]
- 64.Blodig W., Smith A.T., Doyle W.A., Piontek K. Crystal structures of pristine and oxidatively processed lignin peroxidase expressed in Escherichia coli and of the W171F variant that eliminates the redox active tryptophan 171. Implications for the reaction mechanism. J Mol Biol. 2001;305:851–861. doi: 10.1006/jmbi.2000.4346. [DOI] [PubMed] [Google Scholar]
- 65•.Polizzi N.F., Wu Y.B., Lemmin T., Maxwell A.M., Zhang S.Q., Rawson J., Beratan D.N., Therien M.J., DeGrado W.F. De novo design of a hyperstable non-natural protein–ligand complex with sub-angstrom accuracy. Nat Chem. 2017;9:1157–1164. doi: 10.1038/nchem.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]; Design of a de novo protein with an apolar folded cores which supports the cofactor binding region, mimicking natural proteins, thus illustrating the importance of designing the whole protein as a unit.
- 66.Digianantonio K.M., Korolev M., Hecht M.H. A non-natural protein rescues cells deleted for a key enzyme in central metabolism. ACS Synth Biol. 2017;6:694–700. doi: 10.1021/acssynbio.6b00336. [DOI] [PubMed] [Google Scholar]
- 67.Murphy G.S., Greisman J.B., Hecht M.H. De novo proteins with life-sustaining functions are structurally dynamic. J Mol Biol. 2016;428:399–411. doi: 10.1016/j.jmb.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jensen R.A. Enzyme recruitment in evolution of new function. Ann Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- 69.O’Loughlin T.L., Patrick W.M., Matsumura I. Natural history as a predictor of protein evolvability. Protein Eng Des Sel. 2006;19:439–442. doi: 10.1093/protein/gzl029. [DOI] [PubMed] [Google Scholar]
- 70.Smith B.A., Mularz A.E., Hecht M.H. Divergent evolution of a bifunctional de novo protein. Protein Sci. 2015;24:246–252. doi: 10.1002/pro.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pingoud A., Jeltsch A. Recognition and cleavage of DNA by type-II restriction endonucleases. Eur J Biochem. 1997;246:1–22. doi: 10.1111/j.1432-1033.1997.t01-6-00001.x. [DOI] [PubMed] [Google Scholar]
- 72.Saravanan M., Vasu K., Nagaraja V. Evolution of sequence specificity in a restriction endonuclease by a point mutation. Proc Natl Acad Sci U S A. 2008;105:10344–10347. doi: 10.1073/pnas.0804974105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Z.F., Lv Y., Zhu A.Q., An Z. One-enzyme triple catalysis: employing the promiscuity of horseradish peroxidase for synthesis and functionalization of well-defined polymers. ACS Macro Lett. 2018;7:1–6. doi: 10.1021/acsmacrolett.7b00950. [DOI] [PubMed] [Google Scholar]