Abstract
The complexity of eukaryotic cells is underscored by the compartmentalization of chemical signals by phospholipid membranes. A grand challenge of synthetic biology is building life from the ‘bottom-up’, for the purpose of generating systems simple enough to precisely interrogate biological pathways or for adapting biology to perform entirely novel functions. Achieving compartmentalization of chemistries in an addressable manner is a task exquisitely refined by nature and embodied in a unique membrane remodelling machinery that pushes membranes away from the cytosol, the ESCRT-III (endosomal sorting complex required for transport-III) complex. Here, we show efforts to engineer a single ESCRT-III protein merging functional features from its different components. The activity of such a designed ESCRT-III is shown by its ability to drive the formation of compartments encapsulating fluorescent cargo. It appears that the modular nature of ESCRT-III allows its functional repurposing into a minimal machinery that performs sophisticated membrane remodelling, therefore enabling its use to create eukaryotic-like multi-compartment architectures.
Keywords: membranes, ESCRT, compartmentalization, artificial cells
1. Introduction
A major challenge in bottom-up synthetic biology is the engineering of artificial cells [1–3]. Inspired by biological cells, these are compartmentalized architectures containing functional chemistries in communication with their external environment [4]. The development of artificial cells promises novel, highly adaptive and multi-functional chemical systems with wide-ranging potential applications in the chemical [5], biotechnological [6] and medical industries [7], as well as contributing to our developing rudimentary understanding of these fundamental units of life [8]. Most commonly, compartmentalization strategies for artificial cells closely mimic the natural membrane architectures of their living counterparts with much of the early work in the endeavour focused on single membrane-bound compartments, comparable to a prokaryotic cell [9]. More recently, however, the spotlight has started to shift towards multi-compartment systems, analogous with eukaryotic cells, with the prospect of increased sophistication and complexity of function [10–13].
In nature, membrane compartmentalization is controlled by the action of ordered protein assemblies that can bend, push or pull the phospholipid bilayer and ultimately bud away vesicles [14]. This provides inspiration for de novo fabrication of multi-compartment membrane-based systems by repurposing these natural protein complexes in vitro. We focus our attention on the ESCRT complex (endosomal sorting complex required for transport) involved in the formation of multivesicular bodies (MVBs), which have architectural similarity to the systems we wish to engineer. In particular, ESCRT-III is strongly implicated in the membrane remodelling capabilities of ESCRTs [15].
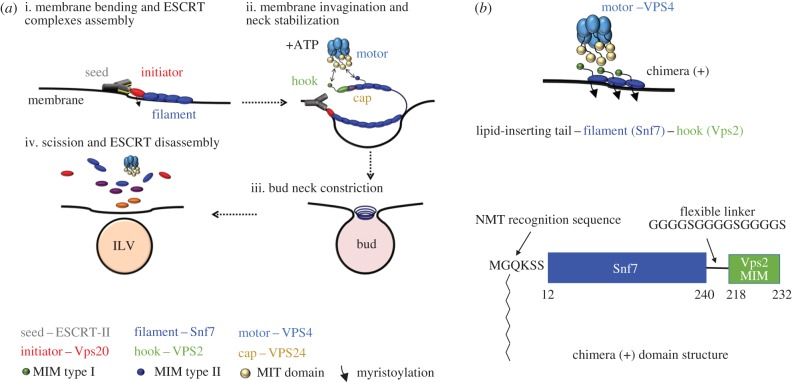
ESCRT-III proteins assemble on the cytosolic face of MVB to perform membrane remodelling and scission of newly formed intraluminal vesicles (ILVs) (figure 1) [16–18]. ESCRT-III is evolutionary conserved and its complexity decreases going from eukaryota to archea, where there are only three components to the complex [19]. In Saccharomyces cervisiae, the complex consists of core subunits, namely Vps20, Snf7, Vps24 and Vps2 [20]. The main component Snf7 is able to assemble into spiral filaments when in contact with phospholipid membranes (figure 1a; ‘filament’), but in the cytoplasm, all ESCRT-III maintains an autoinhibited conformation that keeps them soluble [21]. The Snf7 filament is held into place by short N-terminal regions that contain hydrophobic amino acid sequences capable to insert into the membrane [22]. However, it is upstream complexes such as ESCRT-II (figure 1a; ‘seed’) that initiate membrane invagination and bind to ‘initiator’ Vps20 nucleating ESCRT-III assembly at regions of negative membrane curvature (figure 1a) [23–26]. Vps20 is the only ESCRT-III component that appears to be myristoylated in vivo, possibly to increase its membrane-binding affinity and nucleate complex assembly. Upon binding to Vps20, Snf7 is capable of self-oligomerization into circular filaments growing radially, a process that is ‘capped’ by the subunits Vps24 and Vps2 [27] (figure 1a; ‘cap’). Vps2 molecules work as strong binders [28] (figure 1a; ‘adaptor’) for the AAA+ ATPase Vps4 (figure 1a; ‘motor’). In vivo, Vps4 hexamers, formed in the presence of ATP, use N-terminal microtubule interacting and trafficking (MIT) domains to anchor the type I MIT-interacting motifs (MIM) of Vps2. MIM motifs are also present in Snf7 and Vps20, but have lower affinity for Vps4 (type II MIM) [29,30]. ATP hydrolysis induces inter-subunit conformational changes within Vps4, which mechanically extracts ESCRT-III components from the assembled filament. To date, two possible mechanisms have been proposed for membrane remodelling. In the ‘purse-string’ model, flat ESCRT-III spirals accumulate elastic energy, which is released upon ESCRT-III disassembly by Vps4 and used to deform membranes [31]. In the ‘dome’ model, ESCRT-III forms cylindrical spirals ending in dome-shaped structures, with the spirals' external surfaces interacting with the membrane. The domes are lined with Vps2 and Vps24 molecules, which would drive disassembly of the dome by recruiting Vps4 [32,33]. Ultimately, these processes dynamically remodel nanoscale ESCRT-III spirals into structures with supposedly incrementally smaller size, which restrict the neck of the vesicle performing scission of the membrane at this point. Membrane invagination and bud formation by ESCRT-II coupled to bud neck restriction by ESCRT-III and Vps4 action result in ILV formation within endosomal organelles in vivo [23,34]. Recycling of ESCRT-III filaments by Vps4 makes the complex competent for a second round of membrane remodelling and ILV formation [29]. It is possible to reconstitute membrane remodelling [35] and ILV formation from giant unilamellar vesicles (GUVs) using purified ESCRT-III and ESCRT-II components, in vitro [18,36]. Furthermore, Vps4 recycling action on ESCRT-III has the potential to afford multiple rounds of ILVs, opening the possibility to exploit these proteins to generate multi-compartment architectures within a larger membrane structure, segregating different chemical cargo.
Figure 1.
(a) The core membrane scission machinery ESCRT-III generates ILVs. (i) Membrane bending by ESCRT-II and seeding of ESCRT-III assembly. (ii) The subunit Vps20 initiates Snf7 polymerization into a spiral filament (rectangle shaded blue) and induces membrane budding. Vps24 stops filament elongation and the spiral filament drives membrane buckling into an ILV. (iii) Neck constriction of the budded vesicle occurs via Vps4-mediated ESCRT-III polymer shortening. (iv) Full ESCRT-III disassembly in preparation for a second round of ILV formation. (b) The myristoylated Snf7–Vps2 chimera is labelled as ‘chimera (+)’ in all figures for clarity (‘chimera (−)’ indicates the absence of myristoylation). Chimera (+) should anchor the membrane via an N-terminal myristoyl group, assemble on the membranes via the full-length Snf7 domain and have the ability to bind the Vps4 enzyme. (c) The chimera protein is designed by fusion of the Vps4 binding region of Vps2 (MIM type I) to the C-terminus of the filament subunit Snf7, via a flexible linker. An N-myristoyl transferase recognition sequence replaces the first 11 residues of the Snf7 N-terminus.
The complexity of the ESCRT system does not make it readily amenable to large-scale preparation for in vitro remodelling of GUVs. Therefore, we set out to create an all-in-one ESCRT component capable to perform membrane budding and scission. Here, we show encouraging data, suggesting that a very basic design incorporating the key elements for membrane insertion and oligomerization can indeed remodel phospholipid membranes in vitro.
2. Material and methods
2.1. Plasmids, recombinant proteins expression and purification
The synthetic gene for the Snf7–Vps2 chimera (electronic supplementary material), with the Saccharomyces cerevisiae N-myristyl transferase (NMT) recognition sequence MGQKSS replacing the first 11 residues of Snf7, was synthesized and subcloned by DC Biosciences (Dundee, UK) into a modified pET32a with a C-terminal hexa-histidine tag. Plasmids containing genes encoding the Vps4 (pGST-Vps4; Addgene plasmid # 21495), Snf7(pMBP-HIS2-Snf7; Addgene plasmid # 21492), Snf7–Vps2 chimera and NMT (pNMT; Addgene plasmid # 42578) [37] were transformed into competent JM109 cells and grown for 16 h at 25°C in 2xYT autoinduction media, containing trace metals (Formedium). For the chimera to be myristoylated, the media were supplemented with myristic acid (10 mg l−1) and ZnSO4 (0.1 mM).
Cells from 1 l of culture were resuspended in 15 ml of ice-cold PBS (50 mM NaPO4 pH 7.4, 150 mM NaCl) with EDTA-free protease inhibitor cocktail (Roche). Cells were sonicated on ice for 30 s with 30 s rest on ice and clarified by centrifugation at 30 000g for 30 min at 4°C. For the Snf7–Vps2 chimera, the supernatant was applied to Profinity IMAC Ni-charged resin (Biorad) pre-equilibrated in PBS and incubated at 4°C for 1 h with gentle rotation. The resin was washed with at least five column volumes of PBS and a final wash of one column volume of PBS with 50 mM imidazole. The bound protein was eluted with PBS containing 300 mM imidazole and then applied to a pre-equilibrated (PBS) Superdex 75 30/100 GL (GE Healthcare) size exclusion column. The peak corresponding to the monomeric protein was separated into aliquots, flash frozen in liquid nitrogen and stored at −80°C.
The supernatant from bacterial cells overexpressing the Snf7 protein fused to maltose-binding protein was filtered through a 0.45 µM membrane and applied to 5 × 1 ml MBPTrap HP (GE Healthcare) at a flow rate of 1 ml min−1. After seven column volumes of PBS, MBP–Snf7 was eluted with three column volumes of PBS containing 10 mM maltose. The MBP tag was removed by incubation overnight with excess TEV protease and 1 mM DTT. After cleavage, the tag and TEV were removed by passing through an Ni2+ column (both TEV and MBP contain a histidine tag) monomeric protein was resolved from aggregates using SEC.
2.2. Liquid chromatography–mass spectrometry of proteins
Prior to mass spectrometry, proteins were separated on an Agilent 260 Infinity liquid chromatography instrument. About 1 µl of protein sample (in PBS) was injected onto a Phenomenex Aeris Widepore column (3.6 u, XB-C18, 50 mm × 2.2 mm) with a flow rate of 0.4 ml min−1. Proteins were eluted by performing a linear gradient from 95% solvent A (0.1% formic acid) to 95% solvent B (acetonitrile/0.1% formic acid) over 15 min.
Mass spectrometry was performed on an in line Agilent 6530 Q-ToF mass spectrometer in electrospray (+ESI) ionization mode (with source settings as follows: drying gas temperature 350°C, 11 l min−1; nebulizer 45 psi; capillary voltage 4000 V). Data were analysed using Agilent MassHunter Qualitative Analysis B.06.00 software with a maximum deconvolution algorithm.
2.3. Co-sedimentation assays with Folch liposomes
Folch homogenates from bovine brain extracts were purchased from Sigma-Aldrich. Liposomes were formed by dehydration and rehydration in PBS, sonicated for 5 min and passed through five freeze–thaw cycles. About 3 µM protein and 5 µl of 1 mg ml−1 liposome solution were incubated for 15 min and directly centrifuged in a TLA-100 (Beckman Coulter) for 15 min at 100 000 r.p.m. at 4°C. The supernatant (S) and pellet (P) were immediately separated and analysed using SDS–PAGE and proteins identified with Sypro Ruby protein gel stain (Sigma).
2.4. Electroformation of giant unilamellar vesicles
About 15 µl of a 0.7 mM solution of the desired lipid mixture (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC, 61.9 mol%), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-l-serine (POPS, 10 mol%), cholesterol (25 mol%), 1,2-dioleoyl-sn-glycero-3-phospho-(1′-myo-inositol-3′-phosphate) (PI(3)P, 3 mol%) and lissamine-rhodamine-PE (0.1 mol%), from Avanti Polar Lipids) in chloroform was applied to the conductive surface of indium–tin oxide (ITO)-coated glass slides (surface resistivity 8–12 Ω sq−1, Sigma-Aldrich product no. 703192), using a syringe in a meandering pattern so as to achieve an even coating of lipids. The resulting lipid deposits were briefly dried using a stream of dry N2 gas. Two such slides were applied to a silicon rubber gasket with their conductive, lipid-coated, sides facing the interior of the resulting chamber, approximate volume 500 µl and held in place with a clip. A length of copper tape applied to the gasket provided electrical contact between the conductive sides of each slide, but isolated from the interior of the chamber. The chamber was then filled with an approximately 600 mM sucrose solution and the aperture in the gasket was sealed with a silicon rubber plug. The copper contacts were attached to a function generator and an AC voltage of 3 V (peak-to-peak) was applied to the chamber, at a frequency of 10 Hz sinusodial and maintained for 2 h. The frequency was then reduced to zero incrementally over approximately 10 min, before the solution in the chamber was harvested using a syringe needle.
2.5. Confocal microscopy intraluminal vesicle counting assay
Eight-well glass bottom imaging chambers (ibidi GmbH) were prepared by passivation of the interior glass surface by incubation overnight in 10% bovine serum albumin solution, followed by copious rinsing with MilliQ water.
Tris buffer solutions containing the desired mixtures of proteins and a membrane-impermeable fluorescent dye (Cascade blue-labelled dextran, Mr approx. 10 000) were prepared in Eppendorf tubes to give a final volume of 160 µl, to which was added 40 µl of GUV suspension. The resulting 200 µl solution of GUVs and proteins was gently mixed by repeated inversion of the tube before being transferred to the imaging chambers. After an incubation period of 20 min, imaging was conducted using a Zeiss LSM 880 laser scanning confocal microscope. A tile scanning technique was employed to capture a cross-section of a large number of GUVs and avoid double counting of fast-moving ILVs as can be the case when imaging an entire GUV using a z-stack experiment. The pinhole was adjusted to give a section depth of 3.1 µm. A manual count was performed of ILVs with cascade blue fluorescence in their lumen, indicating that they formed after the addition of the proteins and contain extravesicular bulk medium.
The total volume of GUV lumens observed was determined using the Fiji image analysis software to determine the total GUV lumen volume (total combined lumen area, multiplied by the section depth of 3.1 µm) and was then divided by the volume of a typical 20 µm diameter GUV (volume of a 20 µm sphere) to give the number of ‘GUV volume equivalents’ observed, one volume equivalent being the volume of an idealized 20 µm diameter spherical vesicle. The number of ILVs is thus expressed as ILVs per GUV volume equivalent.
Additionally, the number of mature ILVs versus nascent ILV buds was assessed by counting ILVs that appear to be free-floating in the GUV lumen and those that are clearly in contact with, or in close proximity to the ‘parent’ GUV membrane. ‘Free-floating’ ILVs were counted as those more than 1 µm from the GUV membrane and those closer as ‘buds’. Total numbers of free-floating ILVs versus buds were then used to calculate a percentage for each sample.
3. Results
3.1. Snf7–Vps2 chimera design
The modular nature of ESCRT-III components is such that it is possible to swap between the proteins' functional features such as membrane binding, and recognition motifs for Vps4 and still end up with active ESCRT-III chimeras [38]. We hypothesized that a unique ESCRT-III component could be designed by combining the element of membrane recognition by Vps20 (figure 1b), into the oligomerization potential offered by the Snf7 sequence. The ESCRT-III component that seeds the complex, Vps20, is myristoylated at its N-terminus region providing an additional feature that confers high affinity for membrane binding. We therefore swapped the N-terminal hydrophobic helical segment of Snf7 [22] with the Vps20 myristoylation sequence. The residues removed provide the natural insertion motif that is being replaced with the lipidation to increase stability at the membrane. This sequence is modified by the enzyme N-myristoyl transferase to transfer a myristoyl moiety onto the glycine present in the sequence. Additionally, we sought to introduce a higher affinity for Vps4 binding. The MIM motif of Vps2 was fused at the C-terminus of the Snf7 molecule in order for this chimera to bind to the MIT domain of Vps4 (figure 1c). This sequence was fused to the C-terminal of Snf7 via a flexible linker to provide accessibility to the Vps4 ATPase. The myristoylation of this designed ESCRT-III should confer more stability on the membrane than the wild-type Snf7 has on its own via the N-terminal helix and make ESCRT-II function redundant.
3.2. Snf7–Vps2 chimera myristoylation
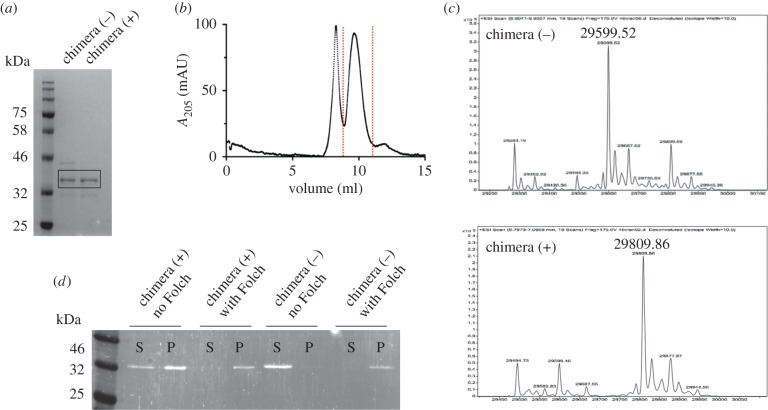
The covalent attachment of myristic acid to an N-terminal glycine residue of a protein is called N-myristoylation. The ESCRT-III subunit Vps20 is myristoylated in yeast, a modification that appears to be necessary for its localization and membrane association [39]. An hexapeptide grafted in the N-terminal sequence of Vps20 acts as recognition sequence for the N-myristoyltransferase NMt1. This enzyme attaches a myristate moiety to the glycine contained within the recognition motif MGQKSS [40]. N-myristoylation of proteins in Escherichia coli cells is achieved by coexpression of heterologous NMt1 with the target protein [41]. Plasmids capable to co-express proteins NMt1 and the Snf7–Vps2 protein were used to produce the myristoylated chimera. Lipidation of the overexpressed protein occurs upon addition of myristic acid and ZnSO4 to the bacterial culture medium as previously demonstrated [37]. The non-myristoylated and myristoylated proteins can be isolated at high purity and are indistinguishable by molecular weight (figure 2a), with the lipidated form purifying as a monomer and higher oligomeric forms (figure 2b). Mass spectrometry of purified protein samples identifies the myristoyl chain successfully covalently bonded to the Snf7–Vps2 with a molecular weight of 29 809.6 Da (figure 2c).
Figure 2.
(a) SDS–PAGE showing the purity of chimera (−) and chimera (+) proteins. (b) Size exclusion chromatography of chimera (+). The fractions corresponding to the indicated peak have been used for GUV assays. (c) Mass spectrometry shows the addition of a myristoyl chain to chimera (−) to give chimera (+). (d) Co-sedimentation assays of chimera proteins with Folch liposomes. Chimera (−) is soluble in the absence of liposomes but sediments with liposomes when present. Chimera (+) solubility is reduced by the myristoyl tail but completely sediments with liposomes, indicating binding to membranes.
3.3. The myristoylated Snf7–Vps2 chimera binds to Folch liposomes
We sought to confirm the ability of binding to membranes by the myristoylated chimera using a co-sedimentation assay with Folch liposomes. Folch homogenates from bovine brain extract contain a mixture of lipids including phosphatidylinositol and phosphatidylserine, which are both believed to be required for ESCRT-III binding to membranes. In the assay, the non-myristoylated chimera is retained in the soluble fraction of the centrifugate when not in the presence of Folch liposomes but co-sediments with liposomes when these are present (figure 2d) showing a partial interaction with lipids. This result is surprising, given that the short N-terminal helical region responsible for Snf7 anchoring to membranes was replaced with a myristoylation sequence. However, the myristoylation sequence itself is partially hydrophobic, and ESCRT-III proteins interaction with membranes is in large part due to electrostatics, which may account for the membrane affinity of the unmodified protein. By contrast, the myristoylated chimera is present in the sedimented fraction also in the absence of liposomes, thus suggesting a propensity to form supramolecular assemblies capable to sediment in the conditions of the assay. In the presence of liposomes, the myristoylated protein completely partitions in the sedimented fraction, thus confirming the ability of the lipid tail to increase the affinity for phospholipid membranes. It is clear, however, that the co-sedimentation assay cannot resolve the difference in affinity between the unmodified and myristoylated protein as both show significant binding to the membrane fraction.
3.4. The myristoylated Snf7–Vps2 chimera performs more efficient membrane remodelling than the parent ESCRT-III component Snf7
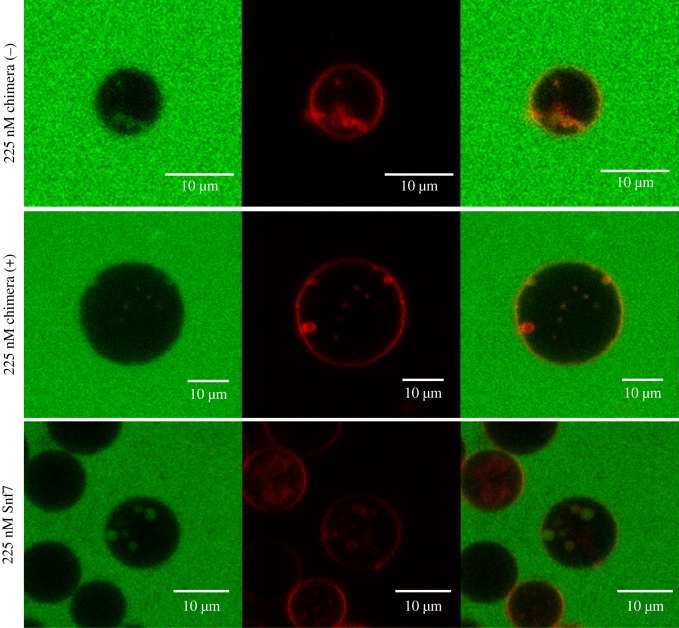
The membrane-binding ability showed by both lipidated and non-lipidated Snf7–Vps2 chimeras prompted us to investigate if these proteins were capable of performing membrane remodelling and the extent of the remodelling process. We therefore compared the ability of myristoylated and non-myristoylated chimeras to generate ILVs filled with bulk-phase within micrometre-size GUVs. The bulk-phase uptake GUV-based assay has been previously used to quantify ESCRT complexes activity [18]. In our assay, 0.1 mol% rhodamine labelled PE lipids (red) were incorporated within electroformed GUVs with a composition of 61.9 : 10 : 3 : 25 POPC : POPS : PI(3)P : cholesterol. Snf7 or Snf7–Vps2 chimeras were added, in separate experiments, to a solution of GUVs of matched osmolarity, containing 10 kDa dextran molecules labelled with cascade blue (false coloured green in images in figure 3) and the formation of ILVs filled with cascade blue dextran was observed.
Figure 3.
Representative confocal microscopy images of ILVs observed after the addition of chimera (+), chimera (−) or Snf7 at a concentration of 225 nM. Green, cascade blue-labelled dextran (approx. 10 000 Da); red, lissamine-rhodamine-PE, section depth = 3.1 µm, 40× objective.
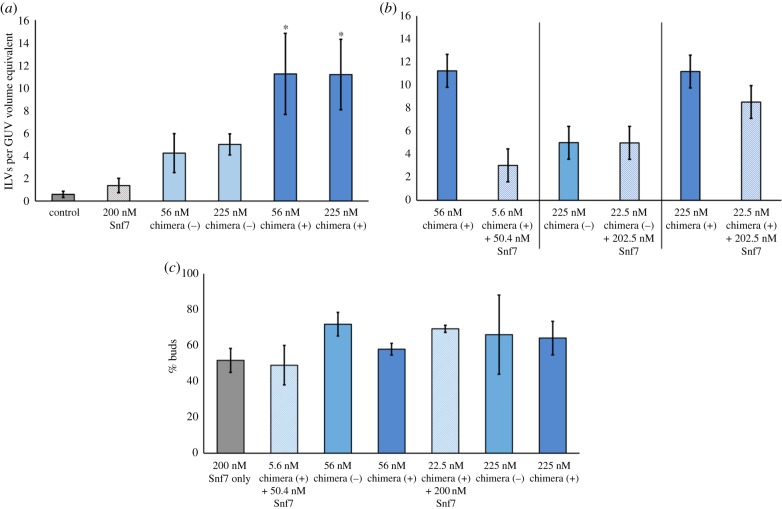
Incorporation of bulk-phase solution within internal compartments within a giant vesicle implies that (i) the proteins can invaginate the GUV membrane to generate inward budding and (ii) the membrane buds, which contain the extravesicular medium, are closed and in some cases severed at the neck. The efficiency of membrane remodelling and encapsulation of dextrans was quantified by scoring the number of green ILVs in a fixed size volume of solution containing GUVs (see Material and methods). The quantification of ILVs filled with the fluorescent dextran is therefore reported as ILVs per GUV volume equivalent. GUVs in a solution of dextrans prior to addition of protein (Snf7 or chimeras) did not contain any detectable green ILVs (figure 4a). However, upon the addition of 200 nM Snf7, an average of 1.4 ILVs (per GUV volume equivalent) containing fluorescent dextrans were generated. In comparison, an average of five ILVs per GUV volume equivalent were formed by a similar concentration of non-myristoylated Snf7–Vps2 chimera. Strikingly, myristoylation does make a notable difference to ILV generation, doubling the number of ILVs, in assays run with two different concentrations of lipidated protein versus the non-lipidated chimera (figure 4a). Both chimeras have significantly greater functional activity than Snf7 at approximately one-quarter the concentration (56 nM vs. 200 nM). By contrast, Vps20 does not induce any ILV formation at 160 nM, a concentration similar to the highest tested for the chimeras [18].
Figure 4.
Quantitative analysis of ILV formation—ILVs observed per typical GUV volume (20 µm diameter sphere). (a) Comparison of the extent of ILV formation by Snf7, non-myristoylated chimera (−) or myristoylated chimera (+), Control, no protein addition. The chimera (+) counts were analysed using a one-way ANOVA with Tukey multiple comparisons test (p < 0.05 for the 56.25 and 225 nM chimera (+) categories when compared with either control or 200 nM Snf7). (b) Pairwise comparison of the activity of pure chimera protein with a mixture of 90 mol% Snf7 + 10% Chimera protein (p < 0.05 for the 22.5 + 202 nM Snf7 chimera (+) category when compared with 200 nM Snf7). (c) Analysis of the proportion of ILV ‘buds’; those observed to be visibly incident on, or within 1 µm of the parent GUV membrane, as a percentage of the total ILVs observed.
ILVs formed by both Snf7 and the chimeras were typically in the range 1–2 µm in diameter, although smaller and larger individual ILVs can also be observed. We aimed to form mixed complexes by combining the chimeras with Snf7 in a 1 : 9 ratio based on the concept that the native filament forming Snf7 might form more active complexes when doped with lower chimera compositions. This turned out not to be the case (figure 4b), where Snf7/chimera complexes tended to present less ILV-forming activity than the equivalent concentration of pure chimera. However, these activities were still much enhanced when compared with Snf7 alone, demonstrating that Snf7 and chimeras do form mixed complexes with enhanced activity compared with Snf7 only. Despite this, there is no advantage gained in using this more complex mixture compared to the simpler, single-component chimera-only system.
We also observed a certain proportion of green ILVs still attached to the membrane both in the Snf7 only and in the chimera GUV population. This motivated us to perform an analysis of the proportion of vesicles still attached versus those free-floating within GUVs, which revealed no apparent difference between the ability of the chimeras and Snf7 to perform vesicle scission (figure 4c). This may indicate a thermally driven stochastic fission probability for nascent ILV buds (approx. 30–40% in this case) that is dominated by the properties of the membrane rather than the different properties of protein assemblies that drive the initial budding process. Note that for in vivo ESCRT systems, the ATPase Vps4 is required for efficient neck scission of ILV buds.
4. Conclusion
The membrane remodelling action of the ESCRT-III complex is unique in that this is the only known protein assembly capable of generating inward budding of the membrane, away from the side of the membrane from which the complex binds. In vivo, this topological process generates new membrane compartments encapsulating transmembrane protein cargo within MVBs. This process can be reconstituted in vitro using purified protein components, generating intraluminal compartments within GUVs that encapsulate constituents from the external media. ESCRT-III function requires the concerted action of four core subunits in addition to an AAA+ ATPase that is crucial to maintain the complex homeostasis. However, here we have shown preliminary data, suggesting that cargo encapsulation can be efficiently performed in vitro on model membranes using a single protein, engineered by merging functional motifs from some of the core ESCRT-III subunits.
This initial design does not appear to be capable of binding to the ATPase Vps4, as tested by standard protein pulldown assays (electronic supplementary material, figure S1). Using our current chimera protein, approximately 60–80% of membrane invaginations do not sever at the neck to form full ILVs. This indicates that neck scission can occur with acceptable efficiency in a lipid bilayer GUV, likely due to the energy barrier for the scission step being thermally accessible with a moderate probability. However, based on the inferred in vivo role of Vps4 in the final energy-dependent scission at the neck of ESCRT-generated membrane invaginations, a chimera capable of binding Vps4 might improve the efficiency of formation of fully mature ILVs in vitro.
Our current work is an encouraging starting point towards our goal of engineering a simple and efficient molecular machinery for on-demand generation of new membrane compartments within artificial cells, inspired by the function of native ESCRT-III proteins. We have shown that an ESCRT-III chimera protein is more efficient at forming ILVs than the core Snf7 subunit of ESCRT-III alone, which forms supramolecular spiral assemblies on the membrane. This is suggestive that an efficient in vitro machinery should be attainable using a very minimal number of engineered components, thus making it a viable approach for the wider bottom-up synthetic biology community. While our current chimera can generate ILVs, the efficiency of neck scission of bud-like invaginations can be improved. This encourages further engineering of an ESCRT-III chimera competent to bind the Vps4 ATPase as an efficient two-component membrane remodelling tool for artificial cell systems.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
The plasmids containing the Vps4 (pGST-Vps4; Addgene plasmid # 21495), N-methyl transferase (pNMT; Addgene plasmid # 42578) and Snf7 (pMBP-HIS2-Snf7; Addgene plasmid # 21492) were a gift from James Hurley. The plasmid containing the TEV protease (pET28-MBP-TEV Addgene plasmid #69929) was a gift from Zita Balklava and Thomas Wassmer [42].
Data accessibility
This article has no additional data.
Authors' contributions
C.J.M. and A.B. designed and carried out the experiments, performed data analysis and drafted the manuscript. P.A.B. and B.C. conceived of the study, designed the study, coordinated the study and wrote the manuscript. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
B.C. acknowledges the EPSRC grant EP/M027821/1 for funding. P.A.B. acknowledges the EPSRC grant EP/M027929/1 for funding.
References
- 1.Buddingh BC, van Hest JCM. 2017. Artificial cells: synthetic compartments with life-like functionality and adaptivity. Acc. Chem. Res. 50, 769–777. ( 10.1021/acs.accounts.6b00512) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blain JC, Szostak JW. 2014. Progress toward synthetic cells. Annu. Rev. Biochem. 83, 615–640. ( 10.1146/annurev-biochem-080411-124036) [DOI] [PubMed] [Google Scholar]
- 3.Hammer DA, Kamat NP. 2012. Towards an artificial cell. FEBS Lett. 586, 2882–2890. ( 10.1016/j.febslet.2012.07.044) [DOI] [PubMed] [Google Scholar]
- 4.Dzieciol AJ, Mann S. 2012. Designs for life: protocell models in the laboratory. Chem. Soc. Rev. 41, 79–85. ( 10.1039/c1cs15211d) [DOI] [PubMed] [Google Scholar]
- 5.Einfalt T, Goers R, Dinu IA, Najer A, Spulber M, Onaca-Fischer O, Palivan CG. 2015. Stimuli-triggered activity of nanoreactors by biomimetic engineering polymer membranes. Nano Lett. 15, 7596–7603. ( 10.1021/acs.nanolett.5b03386) [DOI] [PubMed] [Google Scholar]
- 6.Pohorille A, Deamer D. 2002. Artificial cells: prospects for biotechnology. Trends Biotechnol. 20, 123–128. ( 10.1016/S0167-7799(02)01909-1) [DOI] [PubMed] [Google Scholar]
- 7.Krinsky N, Kaduri M, Zinger A, Shainsky-Roitman J, Goldfeder M, Benhar I, Hershkovitz D, Schroeder A. 2018. Synthetic cells synthesize therapeutic proteins inside tumors. Adv. Healthc. Mater. 7, e1701163 ( 10.1002/adhm.201701163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salehi-Reyhani A, Ces O, Elani Y. 2017. Artificial cell mimics as simplified models for the study of cell biology. Exp. Biol. Med. 242, 1309–1317. ( 10.1177/1535370217711441) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roodbeen R, van Hest JCM. 2009. Synthetic cells and organelles: compartmentalization strategies. Bioessays 31, 1299–1308. ( 10.1002/bies.200900106) [DOI] [PubMed] [Google Scholar]
- 10.Beales PA, Vanderlick TK. 2014. Application of nucleic acid-lipid conjugates for the programmable organisation of liposomal modules. Adv. Colloid Interface Sci. 207, 290–305. ( 10.1016/j.cis.2013.12.009) [DOI] [PubMed] [Google Scholar]
- 11.Elani Y, Gee A, Law RV, Ces O. 2013. Engineering multi-compartment vesicle networks. Chem. Sci. 4, 3332–3338. ( 10.1039/C3SC51164B) [DOI] [Google Scholar]
- 12.Elani Y, Law RV, Ces O. 2014. Vesicle-based artificial cells as chemical microreactors with spatially segregated reaction pathways. Nat. Commun. 5, 5305 ( 10.1038/ncomms6305) [DOI] [PubMed] [Google Scholar]
- 13.Peters RJRW, Marguet M, Marais S, Fraaije MW, van Hest JCM, Lecommandoux S. 2014. Cascade reactions in multicompartmentalized polymersomes. Angew. Chem. Int. Ed. Engl. 53, 146–150. ( 10.1002/anie.201308141) [DOI] [PubMed] [Google Scholar]
- 14.Beales PA, Ciani B, Cleasby AJ. 2015. Nature's lessons in design: nanomachines to scaffold, remodel and shape membrane compartments. Phys. Chem. Chem. Phys. 17, 15 489–15 507. ( 10.1039/C5CP00480B) [DOI] [PubMed] [Google Scholar]
- 15.Henne WM, Stenmark H, Emr SD. 2013. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb. Perspect. Biol. 5, pii: a016766 ( 10.1101/cshperspect.a016766) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falguières T, Luyet P-P, Bissig C, Scott CC, Velluz M-C, Gruenberg J. 2008. In vitro budding of intralumenal vesicles into late endosomes is regulated by Alix and Tsg101. Mol. Biol. Cell 19, 4942–4955. ( 10.1091/mbc.E08-03-0239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saksena S, Wahlman J, Teis D, Johnson AE, Emr SD. 2009. Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136, 97–109. ( 10.1016/j.cell.2008.11.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. 2009. Membrane scission by the ESCRT-III complex. Nature 457, 172–177. ( 10.1038/nature07836) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samson RY, Obita T, Hodgson B, Shaw MK, Chong PL-G, Williams RL, Bell SD. 2011. Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol. Cell 41, 186–196. ( 10.1016/j.molcel.2010.12.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muzioł T, Pineda-Molina E, Ravelli RB, Zamborlini A, Usami Y, Göttlinger H, Weissenhorn W. 2006. Structural basis for budding by the ESCRT-III factor CHMP3. Dev. Cell 10, 821–830. ( 10.1016/j.devcel.2006.03.013) [DOI] [PubMed] [Google Scholar]
- 21.Zamborlini A, Usami Y, Radoshitzky SR, Popova E, Palu G, Göttlinger H. 2006. Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc. Natl Acad. Sci. USA 103, 19 140–19 145. ( 10.1073/pnas.0603788103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchkovich NJ, Henne WM, Tang S, Emr SD. 2013. Essential N-terminal insertion motif anchors the ESCRT-III filament during MVB vesicle formation. Dev. Cell 27, 201–214. ( 10.1016/j.devcel.2013.09.009) [DOI] [PubMed] [Google Scholar]
- 23.Teis D, Saksena S, Judson BL, Emr SD. 2010. ESCRT-II coordinates the assembly of ESCRT-III filaments for cargo sorting and multivesicular body vesicle formation. EMBO J. 29, 871–883. ( 10.1038/emboj.2009.408) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. 2002. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev. Cell 3, 283–289. ( 10.1016/S1534-5807(02)00219-8) [DOI] [PubMed] [Google Scholar]
- 25.Henne WM, Buchkovich NJ, Zhao Y, Emr SD. 2012. The endosomal sorting complex ESCRT-II mediates the assembly and architecture of ESCRT-III helices. Cell 151, 356–371. ( 10.1016/j.cell.2012.08.039) [DOI] [PubMed] [Google Scholar]
- 26.Im YJ, Wollert T, Boura E, Hurley JH. 2009. Structure and function of the ESCRT-II-III interface in multivesicular body biogenesis. Dev. Cell 17, 234–243. ( 10.1016/j.devcel.2009.07.008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mierzwa BE, et al. 2017. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol. 19, 787–798. ( 10.1038/ncb3559) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Obita T, Saksena S, Ghazi-Tabatabai S, Gill DJ, Perisic O, Emr SD, Williams RL. 2007. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449, 735–739. ( 10.1038/nature06171) [DOI] [PubMed] [Google Scholar]
- 29.Shestakova A, Hanono A, Drosner S, Curtiss M, Davies BA, Katzmann DJ, Babst M. 2010. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell 21, 1059–1071. ( 10.1091/mbc.E09-07-0572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hurley JH, Hanson PI. 2010. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 11, 556–566. ( 10.1038/nrm2937) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiaruttini N, Redondo-Morata L, Colom A, Humbert F, Lenz M, Scheuring S, Roux A. 2015. Relaxation of loaded ESCRT-III spiral springs drives membrane deformation. Cell 163, 866–879. ( 10.1016/j.cell.2015.10.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lata S, Schoehn G, Solomons J, Pires R, Göttlinger HG, Weissenhorn W. 2009. Structure and function of ESCRT-III. Biochem. Soc. Trans. 37, 156–160. ( 10.1042/BST0370156) [DOI] [PubMed] [Google Scholar]
- 33.Fabrikant G, Lata S, Riches JD, Briggs JA. G., Weissenhorn W, Kozlov MM. 2009. Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput. Biol. 5, e1000575 ( 10.1371/journal.pcbi.1000575) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adell MAY, Vogel GF, Pakdel M, Müller M, Lindner H, Hess MW, Teis D. 2014. Coordinated binding of Vps4 to ESCRT-III drives membrane neck constriction during MVB vesicle formation. J. Cell Biol. 205, 33–49. ( 10.1083/jcb.201310114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Härtel T, Schwille P. 2014. ESCRT-III mediated cell division in Sulfolobus acidocaldarius—a reconstitution perspective. Front. Microbiol. 5, 257 ( 10.3389/fmicb.2014.00257) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wollert T, Hurley JH. 2010. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–869. ( 10.1038/nature08849) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlson LA, Hurley JH. 2012. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proc. Natl Acad. Sci. USA 109, 16 928–16 933. ( 10.1073/pnas.1211759109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Votteler J, Ogohara C, Yi S, Hsia Y, Nattermann U, Belnap DM, King NP, Sundquist WI. 2016. Designed proteins induce the formation of nanocage-containing extracellular vesicles. Nature 540, 292–295. ( 10.1038/nature20607) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Babst M, Katzmann DJ, Estepa-Sabal EJ, Meerloo T, Emr SD. 2002. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell 3, 271–282. [DOI] [PubMed] [Google Scholar]
- 40.Ashrafi K, Farazi TA, Gordon JI. 1998. A role for Saccharomyces cerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J. Biol. Chem. 273, 25 864–25 874. ( 10.1074/jbc.273.40.25864) [DOI] [PubMed] [Google Scholar]
- 41.Glück JM, Hoffmann S, Koenig BW, Willbold D. 2010. Single vector system for efficient N-myristoylation of recombinant proteins in E. coli. PLoS ONE 5, e10081 ( 10.1371/journal.pone.0010081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Currinn H, Guscott B, Balklava Z, Rothnie A, Wassmer T. 2015. APP controls the formation of PI(3,5)P vesicles through its binding of the PIKfyve complex. Cell. Mol. Life Sci. 73, 393–408. ( 10.1007/s00018-015-1993-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This article has no additional data.