Abstract.
Grand challenges stimulate advances within the medical imaging research community; within a competitive yet friendly environment, they allow for a direct comparison of algorithms through a well-defined, centralized infrastructure. The tasks of the two-part PROSTATEx Challenges (the PROSTATEx Challenge and the PROSTATEx-2 Challenge) are (1) the computerized classification of clinically significant prostate lesions and (2) the computerized determination of Gleason Grade Group in prostate cancer, both based on multiparametric magnetic resonance images. The challenges incorporate well-vetted cases for training and testing, a centralized performance assessment process to evaluate results, and an established infrastructure for case dissemination, communication, and result submission. In the PROSTATEx Challenge, 32 groups apply their computerized methods (71 methods total) to 208 prostate lesions in the test set. The area under the receiver operating characteristic curve for these methods in the task of differentiating between lesions that are and are not clinically significant ranged from 0.45 to 0.87; statistically significant differences in performance among the top-performing methods, however, are not observed. In the PROSTATEx-2 Challenge, 21 groups apply their computerized methods (43 methods total) to 70 prostate lesions in the test set. When compared with the reference standard, the quadratic-weighted kappa values for these methods in the task of assigning a five-point Gleason Grade Group to each lesion range from to 0.27; superiority to random guessing can be established for only two methods. When approached with a sense of commitment and scientific rigor, challenges foster interest in the designated task and encourage innovation in the field.
Keywords: grand challenge, multiparametric magnetic resonance images, prostate cancer, Gleason Grade Group, imaging biomarker, lesion classification
1. Introduction
1.1. Challenges
The medical imaging research community has been highly active in the field of computer-aided diagnosis (CAD), which incorporates radiomics, machine learning, and deep learning. The successful development of clinically useful algorithms in this domain, however, requires extensive resources and many years of dedication from committed research groups. Working independently, these groups typically dedicate considerable effort to acquiring sufficient patient data and providing the “ground truth” required for proper algorithm training and testing. Most of the time, these data do not become publicly available. The reporting of CAD research, therefore, typically does not allow a comparison of the relative merits of different approaches used by different research groups, which is known to depend on database composition, lesion subtlety, “truth” definition, and performance evaluation metric.1–3
Medical imaging grand challenges facilitate the direct comparison of different task-specific algorithms since, in the challenge paradigm, all algorithms must operate on a common image dataset and have their performance evaluated with the same metric. Through challenges, the most promising methods for a specific task may be identified. Challenges provide the resources necessary for friendly competition among research groups with the overall goal of generating interest and collaborations among investigators. A relevant grand challenge in this context (and the one that actually motivated the PROSTATEx Challenges) is PROMISE12,4,5 which compares interactive and (semi)-automated segmentation algorithms for magnetic resonance images (MRI) of the prostate. Since its creation in 2012, PROMISE12 has been online and has accumulated 45 submissions to date. This ongoing challenge provides a ranking of the latest technology in prostate MRI segmentation and has been referenced in 141 publications according to a recent Google Scholar search.6,7
SPIE, the international society for optics and photonics, and the American Association of Physicists in Medicine (AAPM) along with the National Cancer Institute (NCI) sponsored the LUNGx Challenge (the “SPIE-AAPM-NCI Lung Nodule Classification Challenge”) for the computerized classification of lung nodules as benign or malignant on diagnostic computed tomography scans in conjunction with the 2015 SPIE Medical Imaging Symposium.8,9 The success of this challenge motivated these organizations to host another collaborative challenge with timely clinical relevance; MRI of the prostate was identified as presenting a number of possible tasks around which a meaningful challenge could be developed.
1.2. Magnetic Resonance Imaging of the Prostate
Prostate cancer accounts for one in five diagnoses of cancer.10 Conventionally, biopsy has been used to diagnose prostate cancer. Prostate MRI, however, can reduce unnecessary biopsies by 25%, reduce over-diagnosis of clinically insignificant prostate cancer, and improve detection of clinically significant cancer,7,11 where a “clinically significant” prostate cancer is one for which the highest biopsy Gleason score is ≥7 (i.e., Gleason Grade Group >1). Prostate MRI captures multiple anatomic and functional parameters of the prostate and is, therefore, referred to as “multiparametric MRI” (mpMRI). Interpreting mpMRI of the prostate is difficult, and a scoring system [prostate imaging-reporting and data system (PI-RADS)] has been developed to help improve interpretation.12 Unfortunately, even experienced radiologists achieve only moderate reproducibility with PI-RADS.13
Computerized image interpretation systems are being developed to help improve human evaluation of mpMRI of the prostate and the relationship between imaging and pathologic assessment.14–25 A first mpMRI CAD system for classification15 had a standalone area under the receiver operating characteristic curve (AUC) value of 0.89 in the differentiation of manually delineated lesions as insignificant or significant prostate cancers. This system was used to help improve the performance of less-experienced radiologists () to approach that of experts ().16 Another system extracted texture features from two mpMRI sequences to distinguish cancer from benign lesions, a task that achieved an AUC value of 0.83.17 A CAD system trained to identify prostate cancers with a Gleason score of at least seven attained a per-patient AUC of 0.95, which was statistically significantly greater than the AUC value from scores assigned manually on a five-point Likert scale at biopsy.18 An AUC value of 0.95 in the differentiation of prostate cancer from normal foci within mpMRI images was achieved by a CAD system that also identified image-based features demonstrating moderate correlation with Gleason score.19 A commercial system generated less satisfactory results with an MRI-based prostate cancer detection sensitivity and specificity of 47% and 75%, respectively; a devised feature-based index for prostate cancer classification demonstrated an AUC value of 0.65 and failed to achieve a statistically significant correlation with Gleason score.20 A fully automated CAD detection and classification system to identify clinically significant prostate lesions26 in a simulated combined reading paradigm was shown to provide moderate improvement in reading performance;6 however, evaluation of an automated CAD detection system27 led Greer et al.28 to conclude: “CAD-assisted mpMRI improved sensitivity and agreement, but decreased specificity” in the task of detecting clinically significant prostate cancers. This conclusion demonstrates that CAD for prostate mpMRI is a very challenging topic. Many more prostate mpMRI CAD systems have been published, yet they have been neither clinically validated nor compared with other mpMRI CAD systems, thus making it impossible to compare systems or define progress in the field. A major impediment to progress has been the absence of a public mpMRI dataset of the prostate with well-defined performance metrics to compare mpMRI CAD systems. The PROSTATEx Challenges sought to overcome this problem.
1.3. Two-Part PROSTATEx Challenge
The two PROSTATEx Challenges were a collaborative effort sponsored by the SPIE, the AAPM, and the NCI. The first challenge, the PROSTATEx Challenge (the “SPIE-AAPM-NCI Prostate MR Classification Challenge”), involved quantitative image analysis methods for the diagnostic classification of clinically significant prostate lesions, whereas the PROSTATEx-2 Challenge (the “SPIE-AAPM-NCI Prostate MR Gleason Grade Group Challenge”) involved quantitative MRI biomarkers for the determination of Gleason Grade Group in prostate cancer. Both challenges sought to promote the advancement of image-based computational approaches to reduce unnecessary biopsies. The PROSTATEx Challenge was conducted prior to the 2017 SPIE Medical Imaging Symposium (Orlando, February 2017) and was featured at a special session during the Symposium, during which the two best-performing groups presented their methods (these groups were recognized at an awards ceremony and were provided with complimentary meeting registration). The PROSTATEx-2 Challenge was conducted prior to the 2017 AAPM Annual Meeting (Denver, July 2017), again with the two best-performing groups presenting their methods at a special session during the meeting (these two groups also received recognition at an awards ceremony and received complimentary meeting registration). The purpose of this paper is to (1) report the clinical motivation behind the radiologic tasks that the PROSTATEx Challenges were designed to promote, (2) describe the complementary aspects (along with the integration difficulties) of the two-part PROSTATEx Challenges, (3) describe the processes involved with the conduct of these two challenges, (4) report statistics for the numbers of participating groups, (5) summarize the overall results of the performance assessment metrics used in the PROSTATEx and PROSTATEx-2 Challenges, and (6) discuss the potential clinical implications for best-performing methods in both parts of the challenge.
2. Methods
2.1. Dataset
A 2012 prostate mpMRI cohort acquired from a single center (Radboud University Medical Center) was reused from a previous CAD study.26,29 Each mpMRI scan was read or supervised by an expert radiologist (20 years experience), who indicated point-based suspicious findings and assigned a PI-RADS score. Findings with a PI-RADS were referred to biopsy. All biopsies were performed under MRI guidance. Biopsy specimens were graded subsequently by a pathologist with over 20 years of experience, and these results were used as ground truth for the PROSTATEx Challenges.
Each mpMRI scan included multiple orthogonal T2-weighted, dynamic contrast-enhanced (DCE), and diffusion-weighted imaging (DWI). MRI hardware included a 3T MAGNETOM Trio and Skyra (Siemens Medical Systems). T2-weighted images were acquired using a turbo spin echo sequence and had a 2-D resolution of and a slice thickness of 3.6 mm. The DCE time series was acquired using a 3-D turbo flash gradient echo sequence with a resolution of and a temporal resolution of 3.5 s. The DWI series was acquired with a single-shot echo-planar imaging sequence with resolution, diffusion-encoding gradients in three directions, 3 b-values (50, 400, and ), and a computed apparent diffusion coefficient map. Ktrans images computed from the DCE images30 were included in mhd format. Location and a reference thumbnail image were provided for each lesion, and each lesion had a known pathology-defined Gleason Grade Group:31
-
•
Grade Group 1 (Gleason ): only individual discrete well-formed glands.
-
•
Grade Group 2 (Gleason score ): predominantly well-formed glands with lesser component of poorly formed/fused/cribriform glands.
-
•
Grade Group 3 (Gleason score ): predominantly poorly formed/fused/cribriform glands with lesser component of well-formed glands.
-
•
Grade Group 4 (Gleason score , , ): (1) only poorly formed/fused/cribriform glands, (2) predominantly well-formed glands and lesser component lacking glands, or (3) predominantly lacking glands and lesser component of well-formed glands.
-
•
Grade Group 5 (Gleason scores 9 and 10): lacks gland formation (or with necrosis) with or without poorly formed/fused/cribriform glands.
The Gleason Grade Group for each lesion, determined by the pathologist, provided the ground truth for the PROSTATEx-2 Challenge (Fig. 1). Recent evidence suggests a relationship between PI-RADS score based on mpMRI findings and pathology-based Gleason Grade Group.32 The median age of patients in this cohort was 66 years (range: 48 to 83 years). The median PSA level was (range: 1 to ). The percentages of patients with Gleason Grade Group 1, 2, 3, 4, and 5 were 32%, 36%, 16%, 9%, and 7%, respectively.
Fig. 1.
Example mpMRI images demonstrating lesions assigned to each of the five Gleason Grade Groups based on subsequent biopsy and pathologic analysis. An arrow indicates the location of the lesion in each image.
2.2. Challenge Logistics
The challenge was conducted using the MedICI platform33 developed through funding from NIH and Leidos. This open-source platform is built using a number of open-source modules including CodaLab,34 caMicroscope, ePad, 3-D slicer, and R. The platform supports user and data management, communications through mass emails and forums, and evaluation and visualization of results. This platform has been used to host a number of challenges including challenges sponsored by MICCAI, RSNA, and AAPM. The platform was used as a front-end for case download (images and associated metadata along with lesion information), distribution of information regarding challenge logistics (e.g., due dates and results format) and rules, and ongoing communication among participants and organizers through a discussion forum. All de-identified images and lesion information were stored on NCI’s publicly accessible The Cancer Imaging Archive (TCIA).35,36 The data will remain on TCIA in perpetuity.
Participants were required to create an account on the MedICI platform. The challenges were specifically designed so that a participant (either an individual or a group) could participate in PROSTATEx, PROSTATEx-2, or both. Although participants were allowed to download the training cases and test cases (once they were made available) for either challenge to assess the compatibility of the images and clinical tasks with their methods and software interfaces, participants were not allowed to withdraw from either challenge once they submitted test set results. Participants were not explicitly limited in the manner in which they could use the provided training set cases or in the use of independent cases for the training of their methods. No human manipulation of, or intervention with, the test set cases was allowed with the exception of manual or human-supervised delineation of the prostate boundary or the gross lesion margin; participants were on their honor to abide by these rules.
2.3. Quality Assurance
Quality assurance (QA) is important for both the participants and the organizers of a challenge. Well-curated data reduces the burden of (1) the intensive communication between the participants and the organizers when issues and inaccuracies are discovered, (2) correcting the database once the tight timetable of a challenge has been initiated, and (3) ensuring that any corrections are reliably distributed to all participants. A QA process following a data inspection protocol was performed on the image data and the associated metadata to provide reliable and high-quality data to the participants.
The first part of the QA process (1) verified the availability of all image sets and associated metadata for download from TCIA and (2) verified the technical logistics of the download process. The database was corrected for missing images and missing metadata. The procedure also verified that the downloaded images were not corrupted and could be displayed with a standard graphic user interface tool.
The second part of the QA process verified the accuracy of the provided metadata in relation to the images. The mpMRI dataset used in the challenges had a complex composition, containing multiple types of MRI scans and derivative image volumes in different cross-sectional views. The QA process verified that the provided lesion centroids pointed to the correct locations on the corresponding MRI images for every lesion. The process also verified that the provided thumbnails corresponded to the correct lesion locations across all MRI scans. The lesion centroid locations and thumbnails were corrected if necessary. In addition, the QA process periodically assessed whether the description and corresponding procedure for download, organization, and utilization of the image sets and the corresponding metadata were up-to-date and correct; consequently, the workflow, the data, and the data descriptions were improved continuously.
2.4. PROSTATEx
The PROSTATEx Challenge released a training set of cases in November 2016 that contained mpMRI scans of 330 prostate lesions along with spatial location coordinates, anatomic zone location, and known clinical significance of each lesion. Three weeks later the test set of cases was made available; the test set contained mpMRI scans of 208 prostate lesions (Table 1) with spatial location and anatomic zone, but the clinical significance information for these lesions was not included. After a period of five weeks, the results from each participant were submitted to the organizers in the form of a single real number on the range [0, 1] for each lesion representing the computer-determined likelihood of the lesion being clinically significant.
Table 1.
The numbers of prostate lesions and patients in the training and test sets for PROSTATEx and PROSTATEx-2.
| Number of lesions (training set/test set) | Number of patients (training set/test set) | |
|---|---|---|
| PROSTATEx | 330/208 | 204/140 |
| PROSTATEx-2 | 112/70 | 99/63 |
To assess performance in the task of distinguishing clinically significant from nonsignificant disease, receiver operating characteristic (ROC) analysis based on the proper binormal ROC model37,38 was used with AUC as the figure of merit. The performance of each method was compared with that of random guessing (), and statistical significance of differences in performance between methods was assessed. These analyses were performed through bootstrapping (1000 iterations) to estimate -values and 95% confidence intervals for differences in AUC. In this investigational study, corrections of statistical significance for multiple comparisons were not performed. To assess the potential synergy among methods used by participants, submissions were incrementally “fused” in order of decreasing performance by averaging the estimated likelihoods provided by each submission for each lesion. In other words, submissions were “fused” by averaging, for each lesion, the estimated probabilities of being clinically significant disease for the two top-performing methods, then for the three top-performing methods, and so on through inclusion of all methods, and this average at each iteration was used as the decision variable in ROC analyses.
2.5. PROSTATEx-2
The PROSTATEx-2 Challenge released a training set of cases in May 2017 that contained mpMRI scans of 112 prostate lesions (Table 1) and, for each lesion, included spatial location coordinates, anatomic zone location, and known Gleason Grade Group. Three weeks later, the test cases were made available with mpMRI scans of 70 prostate lesions along with spatial location and anatomic zone, but the Gleason Grade Group information for these lesions was not included. After eight weeks, each participant submitted their results to the organizers as a single ordinal value on the range [1, 5] for each lesion representing the computer-determined Gleason Grade Group. Cases used for the PROSTATEx-2 Challenge were a subset of the cases use for the PROSTATEx Challenge and included only clinically significant lesions.
To assess the agreement between the Gleason Grade Group for each lesion as estimated by each participant’s method and the “truth” obtained from pathology, the quadratic-weighted kappa was used.39 Kappa, unlike “accuracy,” accounts for agreement by chance and uses quadratic weights to more strongly penalize larger differences between the estimated Gleason Grade Group and the true Gleason Grade Group. Should multiple methods achieve the same kappa coefficient, the positive predictive value of each method’s ability to identify Gleason Grade would be used to break the tie (after all results were submitted, no tie breaker was needed). Analogous to the analysis for the first PROSTATEx Challenge, submissions were incrementally “fused” in order of decreasing performance by averaging (and rounding) the submitted estimates for the Gleason Grade Group by lesion. Bootstrapping (1000 iterations) was used to assess statistical significance of differences in performance, both for individual methods with respect to random guessing () and for “fused” methods with respect to the method achieving the highest kappa.
3. Results
3.1. PROSTATEx
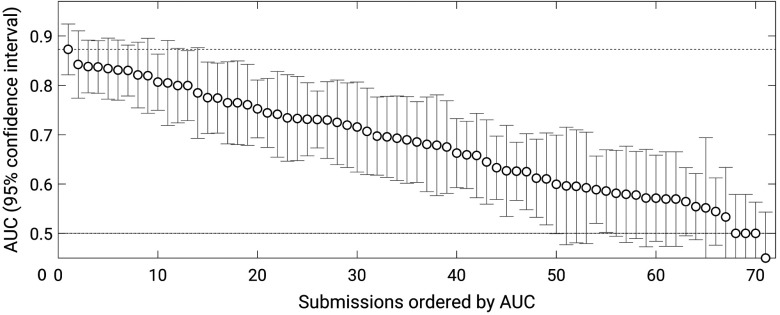
Thirty-two groups submitted results from a total of 71 methods (groups were allowed to submit results from up to three methods). Most, but not all, methods outperformed random guessing () (Fig. 2). The best-performing method obtained an AUC value of 0.87 (standard error 0.027), and the next three methods all achieved AUC values of 0.84 (with standard errors of 0.036, 0.032, and 0.032). Statistically significant differences in performance among these four best methods, however, were not observed. After ranking the methods by decreasing AUC, the first method that demonstrated a statistically significant difference in AUC with respect to the best-performing method had an AUC of 0.82 (standard error 0.034) (-value with respect to the winning submission 0.036; 95% confidence interval for the difference in AUC ).
Fig. 2.
AUC values achieved by the 71 methods that participated in the PROSTATEx Challenge with error bars indicating 95% confidence intervals.
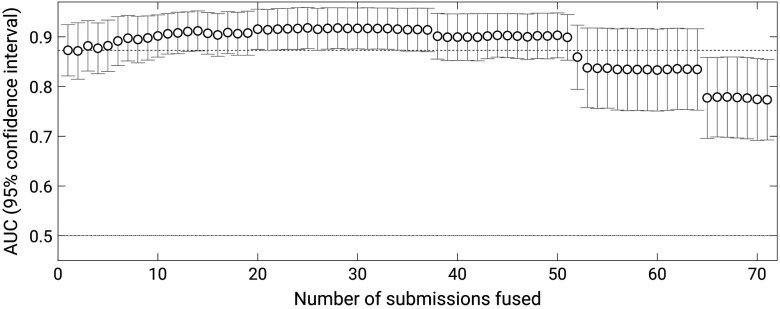
When methods were “fused,” the maximum performance was obtained for a fusion of the best 25 submissions, which obtained an AUC of 0.92 (standard error 0.077) (Fig. 3). The first instance in which the performance of fused methods exceeded that of the winning submission was when likelihoods of the best nine methods were averaged to achieve an AUC of 0.90 (standard error 0.076) (-value with respect to the winning submission 0.036; 95% confidence interval of the difference in AUC [0.002, 0.055]). As expected, the performance of “fused” methods leveled off with increasing number of submissions “fused” and subsequently declined when likelihoods of too many lower-performing methods were included in the average.
Fig. 3.
AUC values achieved by the incremental fusion of the 71 methods that participated in the PROSTATEx Challenge with error bars indicating 95% confidence intervals.
From among the participating groups that responded to a postchallenge questionnaire about their methods, most trained their system exclusively on the PROSTATEx training cases. Methods were equally divided between traditional feature extraction with an appropriate classifier and the use of a convolutional neural network. Different groups used various combinations of the provided mpMRI image sequences in their systems. The eight groups with a method that achieved one of the top six scores were invited to submit a conference proceeding paper; four groups accepted this invitation.40–43
3.2. PROSTATEx-2
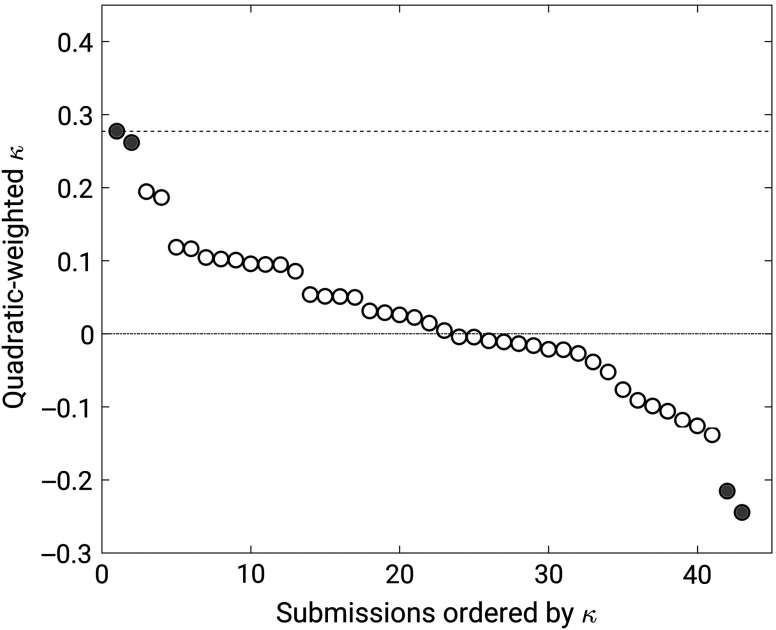
Twenty-one groups submitted results from a total of 43 methods. Superiority to random guessing could be established for only two methods, which achieved quadratic-weighted kappa values of 0.27 (95% confidence interval [0.06, 0.48]) and 0.26 (95% confidence interval [0.04, 0.47]) (Fig. 4). Two methods performed significantly worse than random guessing with quadratic-weighted kappa values of (95% confidence interval ) and (95% confidence interval ). The other methods failed to achieve statistically significant differences in performance with respect to random guessing (i.e., their 95% confidence intervals for kappa included zero).
Fig. 4.
Weighted kappa values achieved by the 43 methods that participated in the PROSTATEx-2 Challenge. Error bars were large and are not shown for clarity. Filled circles indicate methods that differed from random guessing ().
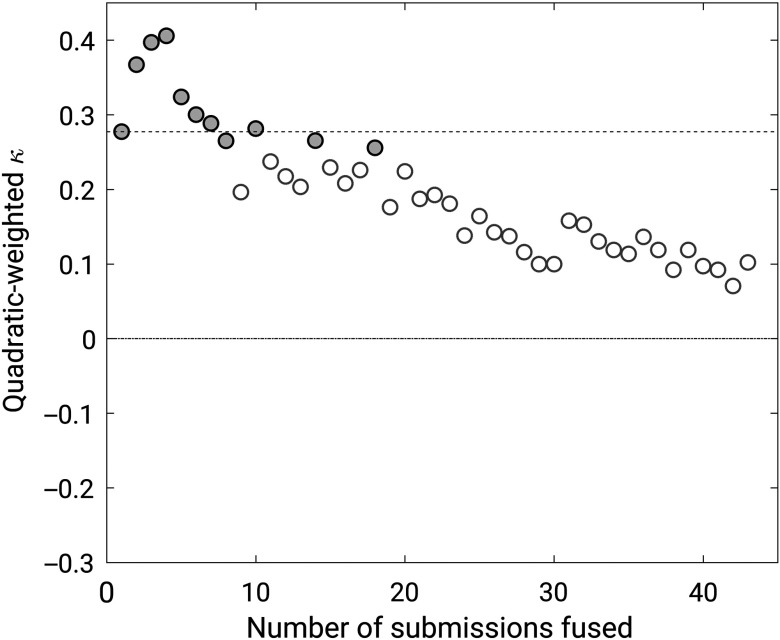
When methods were “fused,” the maximum performance was obtained for a fusion of the four best methods, which achieved a quadratic-weighted kappa of 0.40 (95% confidence interval [0.19, 0.55]) (Fig. 5). The difference with respect to the best-performing method, however, failed to reach statistical significance (95% confidence interval for the difference in kappa ).
Fig. 5.
Weighted kappa values achieved by the incremental fusion of the 43 methods that participated in the PROSTATEx-2 Challenge. Filled circles indicate fusions that differed from random guessing ().
From among the participating groups that responded to a postchallenge questionnaire about their methods, most trained their system exclusively on the PROSTATEx-2 training cases. Methods were equally divided between traditional feature extraction with an appropriate classifier and the use of a convolutional neural network. Different groups used various combinations of the provided mpMRI image sequences in their systems.
4. Discussion
The results achieved during the PROSTATEx Challenges are in agreement with clinical practice. The highest AUC value in the PROSTATEx Challenge (0.87) reflects promise for the potential of such computerized methods to reduce the number of unnecessary biopsies. The PROSTATEx-2 Challenge demonstrates that it is much more difficult for computerized methods to differentiate pathologic Gleason Grade Group than it is for methods to discriminate between clinically significant and clinically insignificant cancers in the same dataset of cases. The methods in this domain need further improvement before being used as an aid to assist with the reduction of unnecessary biopsies or as decision support in the management of prostate cancer. To further reduce biopsies, the community should focus on the discrimination of low- and high-grade cancers (i.e., clinical significance), which, similar to clinical practice, is a very difficult task.
The PROSTATEx-2 Challenge, in which participants attempted to assess a pathologic task based on radiologic data by developing computerized methods to assign a Gleason Grade Group to prostate lesions based on mpMRI, presented an especially challenging task, as evidenced by the resulting weak weighted kappa statistic values across all methods (the best-performing method achieved a weighted kappa value of 0.27). Rather than signifying a failed challenge, this result demonstrates that the development of imaging biomarkers for Gleason Grade Group (to potentially spare patients from biopsies) could benefit from more focused effort and resources. In effect, this “negative study” identified a line of investigation that is open for creative ideas. It should be noted that even among pathologists the unweighted kappa values for inter- and intraobserver Gleason score were found to be 0.54 and 0.66, respectively,44 which both serves as a target for how well a computerized method should be expected to perform and as an indication that the reference against which the methods in the challenge were compared is itself variable. For a more direct comparison of the present results with the findings of Melia et al.,44 the highest unweighted kappa value attained in the PROSTATEx-2 Challenge was 0.19; this value, it should be noted, is based on the five-point Gleason Grade Group, which has fewer gradations than the Gleason score used in Ref. 44.
A two-part challenge requires identification of two related clinical, pathologic, or radiologic tasks that may use the same dataset in such a way that the conduct of part 1 does not compromise part 2, either by biasing the results of part 2 or by giving groups that participated in part 1 an unfair advantage during part 2. The tasks could be related but independent, or the tasks could be sequential and complement each other. The PROSTATEx Challenges followed the latter approach: PROSTATEx was a classification task to differentiate between clinically significant and clinically insignificant prostate lesions, whereas PROSTATEx-2 sought an additional level of detail (Gleason Grade Group) for those lesions that were actually clinically significant. A two-part challenge provides an opportunity for participants to integrate their computational methods in a more broad clinical context while enhancing the utility of a valuable resource (the datasets).
Limitations of this study included the modest sizes of the datasets. The collection of a sufficient number of cases for training and testing remains a challenge for challenges, as does the resulting statistical conclusions that may be drawn. The 330/208 and 112/70 training/test prostate lesions for PROSTATEx and PROSTATEx-2, respectively, made achievement of statistically significant differences across methods difficult; each challenge had best-performing methods according to a rank ordering based on the respective performance metric, but statistically significant “winners” were not identifiable. It is worth noting that the sizes of the training and test sets in the PROSTATEx Challenge were nearly three times the sizes of the sets in the PROSTATEx-2 Challenge. The latter were a subset of the PROSTATEx datasets due to the nature of the different tasks in this two-part challenge. Ideally, to achieve reasonable statistical power and allow for participants to better train their methods, the PROSTATEx-2 datasets should have been larger than those for PROSTATEx since in PROSTATEx-2 participants were tasked with a five-category classification (Gleason Grade Group) rather than a two-class classification (clinically significant/insignificant) as in PROSTATEx.
Collecting, vetting, and organizing images along with associated metadata, clinical data (when appropriate), and “truth” is a tremendous task. QA processes to ensure the integrity of the provided data must be implemented for any well-run challenge. The single-radiologist and single-pathologist reference standard for the PROSTATEx Challenges was established in conjunction with an earlier published study,26 although such a standard has limitations. In a challenge setting, in which the overall goal is not necessarily determination of absolute clinical performance but is instead a comparison across methods, the impact of not having a definitive gold standard might be somewhat muted; however, incorporating multiple radiologists and potentially multiple pathologists in the reference process would have allowed incorporation of the variability inherent in these clinical decisions. Unfortunately, enhancing the reference standard through inclusion of multiple truthers was not possible within the timeframe of these challenges. The biopsy-based localization of prostate lesions could contribute to additional uncertainty in the reference standard; however, the biopsies obtained for this reference standard were performed using in-bore MRI, the results of which have shown an 88% correspondence with prostatectomy outcome,45 thus maximizing confidence that all biopsy scores accurately reflect true focal pathology.
Another limitation of this study was the lack of a requirement for participants to provide details regarding their methods. The results of challenges are most valuable to the medical imaging research community when the general algorithmic approach for methods that performed well (and those that did not perform well) is reported. Participants should support the intent of the challenge as a friendly competition among research groups with the overall goal of advancing the field. Public knowledge of promising methods would allow researchers (and funding agencies) to direct their effort and resources in a manner that achieves the greatest impact. To ensure documentation of each method’s general approach, the organizers of a challenge should require participants to submit a methodological summary at the time of results submission as a requirement for challenge participation. Perhaps future challenges could encourage (and eventually require) some forms of code sharing on the part of participating groups to further accelerate the advancement of the field.
5. Conclusion
The PROSTATEx Challenges provided annotated mpMRI datasets within a challenge framework that allowed for the comparison of state-of-the-art computational methods for prostate cancer diagnosis. The challenge results demonstrate that automated classification of clinically significant cancer seems feasible, but noninvasive prediction of aggressiveness is quite difficult. Various future research strategies should be considered. First, to allow future methods to join the challenge, the challenge has been made live at prostatex.grand-challenge.org so that researchers may continue to receive objective feedback, have the ability to reference their performance in scientific publications, and identify algorithms with generalizable performance. Second, two aspects of datasets are crucial: quality and quantity. Increasing dataset size is likely to increase performance, but there might be practical limitations on quality. MRI annotation was shown to be ambiguous in a few cases, and Gleason scoring has demonstrated variability as well; these issues likely present an upper limit to the performance achievable by the many top algorithms having similar performance. A consensus mechanism to update datasets (either in quality or quantity) in existing or new challenges is currently lacking; the scientific community should strive to develop such mechanisms.
Acknowledgments
The authors would like to express their appreciation to Angela Keyser and Shayna Knazik from the AAPM and Diane Cline and Robbine Waters from SPIE for their assistance in making these challenges a reality. The PROSTATEx Challenges were supported by SPIE, NCI, and AAPM. The authors are grateful to the groups that participated in the PROSTATEx Challenges, including groups led by Bejoy Abraham and Madhu S. Nair (University of Kerala), Jason Adleberg (Drexel University College of Medicine), Ruhallah Amandi and Mohammad Farhadi (Amirkabir University of Technology), Yoganand Balagurunathan (H.L. Moffitt Cancer Center), Adrian Barbu (Florida State University), Lei Bi (University of Sydney), Vy Bui (The Catholic University of America), Quan Chen (University of Virginia), King Chung Ho and Karthik Sarma (University of California, Los Angeles), Andy Kitchen, Panagiotis Korfiatis (Mayo Clinic), Peter S. LaViolette (Medical College of Wisconsin), Nathan Lay (National Institutes of Health), Chao Li (Huazhong University of Science and Technology), Qing Liang (Temple University), Saifeng Liu (The MRI Institute for Biomedical Research), Sean D. McGarry (Medical College of Wisconsin), Alireza Mehrtash (Brigham and Women’s Hospital), Mira Park (The University of Newcastle), N. Andres Parra (Moffitt Cancer Center), Yue Miao (University of Electronic Science and Technology of China), Hung Le Minh (Huazhong University of Science and Technology), Jin Qi and Miao Le (University of Electronic Science and Technology of China), Jarrel Chen Yi Seah (Alfred Health), Piotr Sobecki (Warsaw University of Technology), Radka Stoyanova (University of Miami), Yu Sun (Peter MacCallum Cancer Centre), Ning Wen (Henry Ford Health System), Xinran Zhong (University of California, Los Angeles), and Delong Zhu (Robotic Geometry Research Group CUHK). These (and the other) participating groups deserve recognition for their work to develop their prostate lesion classification and grading methods and for their contributions to the success of the challenges. This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. The challenge platform and JKC were funded in part by NIH/NCI U01CA154601, U24CA180927, and U24CA180918 and a contract (HHSN26120080001E) from Leidos Biomedical Research. This manuscript has been authored in part by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan. The mention of commercial products, their sources, or their use in connection with material reported herein is not to be construed as either an actual or implied endorsement of such products by the Department of Health and Human Services.
Biographies
Samuel G. Armato III is an associate professor of radiology and the Committee on Medical Physics at The University of Chicago. His research interests involve the development of computer-aided diagnostic methods for thoracic imaging, including automated lung nodule detection and analysis in CT scans, semiautomated mesothelioma tumor response assessment, image-based techniques for the assessment of radiotherapy-induced normal tissue complications, and the automated detection of pathologic change in temporal subtraction images.
Henkjan Huisman is an associate professor of radiology specializing in pelvic imaging biomarkers at the Radboud University Medical Center, The Netherlands. He has over 30 years of experience in research, development, and clinical implementation of computerized medical image analysis. The research team he leads is engaged in developing applications for MRI and ultrasound image analysis with a focus on prostate cancer imaging.
Karen Drukker is a research associate professor in the Department of Radiology at The University of Chicago. She has been actively involved in computer-aided diagnosis/radiomics research for over a decade. Her work has focused on multimodality detection/diagnosis/prognosis of breast cancer and on the performance evaluation of radiomics methods.
Lubomir Hadjiiski is a professor in the Department of Radiology at the University of Michigan. He has authored or coauthored more than 115 publications in peer-reviewed journals. His research interests include computer-aided diagnosis, neural networks, predictive models, image processing, medical imaging, and control systems. His current research involves design of decision support systems for detection and diagnosis of cancer in different organs and quantitative analysis of image biomarkers for treatment response monitoring.
Justin S. Kirby is a bioinformatics analyst at the Frederick National Laboratory for Cancer Research. His focus is on the developing informatics methods to improve reproducibility in imaging research through increased data and code sharing, as well as the adoption of structured reporting standards. His team manages The Cancer Imaging Archive, which provides free and open-access datasets of de-identified cancer images to researchers.
Nicholas Petrick received his BS degree from Rochester Institute of Technology in electrical engineering and his MS and PhD degrees from the University of Michigan in electrical engineering systems. He is a deputy director for the Division of Imaging, Diagnostics, and Software Reliability within the U.S. Food and Drug Administration, Center for Devices and Radiological Health, and is a member of the FDA Senior Biomedical Research Service. He is an SPIE fellow. His research interests include machine learning and artificial intelligence for medical data, computer-aided diagnosis, quantitative imaging and radiomic tools, and the development of statistical assessment approaches for validating device performance.
George Redmond is a program director for the Imaging Technology Development Branch of the NCI’s Cancer Imaging Program. He played a key role in several successful large enterprise systems development initiatives at NCI to improve the clinical trial process and advance the restructuring of the nation’s cancer clinical trials enterprise. He is the recipient of the prestigious NIH Director’s Award for the successful management and oversight of the Cancer Therapy Evaluation Program clinical trials management system.
Maryellen L. Giger is the A. N. Pritzker professor of radiology and the Committee on Medical Physics at The University of Chicago. Her research interests mainly involve the investigation of computer-aided diagnosis and radiomic methods for the assessment of risk, diagnosis, prognosis, and response to therapy of breast cancer on multimodality (mammography, ultrasound, and magnetic resonance) images. She is also involved in broad-based developments in computer vision and data mining of medical images.
Kenny Cha received his BSE, MSE, and PhD degrees from the University of Michigan in Biomedical Engineering. He is a staff fellow in the Division of Imaging, Diagnostics, and Software Reliability within the U.S. Food and Drug Administration, Center for Devices and Radiological Health. His research interests include artificial intelligence, machine learning, and deep learning for medical data, computer-aided diagnosis, and radiomics.
Artem Mamonov is a software engineer at the Center for Clinical Data Science (CCDS) at MGH. His work is focused on the development of scalable web-based applications for annotation of medical images and reports. He also works on the visualization of results from machine learning algorithms and the integration of these results into clinical workflows.
Jayashree Kalpathy-Cramer received his BTech degree in electrical engineering from IIT, Bombay, India, his MS and PhD degrees in electrical engineering from Rensselaer Polytechnic Institute, and his MS degree in biomedical informatics from Oregon Health and Science University. She is an associate professor of radiology at the Athinoula A. Center for Biomedical Imaging at MGH/Harvard Medical School. Her areas of research interest include medical image analysis, machine learning, and artificial intelligence for applications in radiology, oncology, and ophthalmology.
Keyvan Farahani is a program director for the Image-Guided Interventions Branch of the NCI’s Cancer Imaging Program. In this capacity, he is responsible for the development and management of NCI initiatives that address diagnosis and treatment of cancer and precancer through integration of advanced imaging and minimally invasive and noninvasive therapies. He has led the organization of brain tumor segmentation challenges at MICCAI 2013 to 2015. His graduate studies were in biomedical physics (UCLA, ‘93).
Disclosures
S. G. A., K. D., and M. L. G. receive royalties and licensing fees through The University of Chicago related to computer-aided diagnosis. M. L. G. is a stockholder in R2/Hologic, co-founder and equity holder in Quantitative Insights, shareholder in Qview, and receives royalties from Hologic, GE Medical Systems, MEDIAN Technologies, Riverain Medical, Mitsubishi, and Toshiba. It is the University of Chicago Conflict of Interest Policy that investigators disclose publicly actual or potential significant financial interest that would reasonably appear to be directly and significantly affected by the research activities.
References
- 1.Nishikawa R. M., et al. , “Effect of case selection on the performance of computer‐aided detection schemes,” Med. Phys. 21, 265–269 (1994). 10.1118/1.597287 [DOI] [PubMed] [Google Scholar]
- 2.Nishikawa R. M., et al. , “Variations in measured performance of CAD schemes due to database composition and scoring protocol,” Proc. SPIE 3338, 840–844 (1998). 10.1117/12.310894 [DOI] [Google Scholar]
- 3.Revesz G., Kundel H. L., Bonitatibus M., “The effect of verification on the assessment of imaging techniques,” Invest. Radiol. 18, 194–198 (1983). 10.1097/00004424-198303000-00018 [DOI] [PubMed] [Google Scholar]
- 4.Litjens G., et al. , “Evaluation of prostate segmentation algorithms for MRI: the PROMISE12 challenge,” Med. Image Anal. 18, 359–373 (2014). 10.1016/j.media.2013.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson E., et al. , “Designing image segmentation studies: statistical power, sample size and reference standard quality,” Med. Image Anal. 42, 44–59 (2017). 10.1016/j.media.2017.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Litjens G. J. S., et al. , “Clinical evaluation of a computer-aided diagnosis system for determining cancer aggressiveness in prostate MRI,” Eur. Radiol. 25, 3187–3199 (2015). 10.1007/s00330-015-3743-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasivisvanathan V., et al. , “MRI-targeted or standard biopsy for prostate-cancer diagnosis,” N. Engl. J. Med. 378, 1767–1777 (2018). 10.1056/NEJMoa1801993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Armato S. G., III, et al. , “The LUNGx challenge for computerized lung nodule classification: reflections and lessons learned,” J. Med. Imaging 2, 020103 (2015). 10.1117/1.JMI.2.2.020103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Armato S. G., III, et al. , “The LUNGx challenge for computerized lung nodule classification,” J. Med. Imaging 3, 044506 (2016). 10.1117/1.JMI.3.4.044506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siegel R. L., Miller K. D., Jemal A., “Cancer statistics, 2018,” CA Cancer J. Clin. 68, 7–30 (2018). 10.3322/caac.v68.1 [DOI] [PubMed] [Google Scholar]
- 11.Ahmed H. U., et al. , “Diagnostic accuracy of multi-parametric MRI and TRUS biopsy in prostate cancer (PROMIS): a paired validating confirmatory study,” Lancet 389, 815–822 (2017). 10.1016/S0140-6736(16)32401-1 [DOI] [PubMed] [Google Scholar]
- 12.Weinreb J. C., et al. , “PI-RADS Prostate Imaging—Reporting and Data System: 2015, Version 2,” Eur. Urol. 69, 16–40 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosenkrantz A. B., et al. , “Proposed adjustments to PI-RADS version 2 decision rules: impact on prostate cancer detection,” Radiology 283, 119–129 (2017). 10.1148/radiol.2016161124 [DOI] [PubMed] [Google Scholar]
- 14.Wang S., et al. , “Computer aided-diagnosis of prostate cancer on multiparametric MRI: a technical review of current research,” Biomed. Res. Int. 2014, 789561 (2014). 10.1155/2014/789561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vos P. C., et al. , “Computer-assisted analysis of peripheral zone prostate lesions using T2-weighted and dynamic contrast enhanced T1-weighted MRI,” Phys. Med. Biol. 55, 1719–1734 (2010). 10.1088/0031-9155/55/6/012 [DOI] [PubMed] [Google Scholar]
- 16.Hambrock T., et al. , “Prostate cancer: computer-aided diagnosis with multiparametric 3-T MR imaging—effect on observer performance,” Radiology 266, 521–530 (2013). 10.1148/radiol.12111634 [DOI] [PubMed] [Google Scholar]
- 17.Kwak J. T., et al. , “Automated prostate cancer detection using T2-weighted and high-b-value diffusion-weighted magnetic resonance imaging,” Med. Phys. 42, 2368–2378 (2015). 10.1118/1.4918318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dinh A. H., et al. , “Characterization of prostate cancer with Gleason score of at least 7 by using quantitative multiparametric MR imaging: validation of a computer-aided diagnosis system in patients referred for prostate biopsy,” Radiology 287, 525–533 (2018). 10.1148/radiol.2017171265 [DOI] [PubMed] [Google Scholar]
- 19.Peng Y., et al. , “Quantitative analysis of multiparametric prostate MR images: differentiation between prostate cancer and normal tissue and correlation with Gleason score—a computer-aided diagnosis development study,” Radiology 267, 787–796 (2013). 10.1148/radiol.13121454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thon A., et al. , “Computer aided detection in prostate cancer diagnostics: a promising alternative to biopsy? a retrospective study from 104 lesions with histological ground truth,” PLoS One 12, e0185995 (2017). 10.1371/journal.pone.0185995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Citak-Er F., et al. , “Final Gleason score prediction using discriminant analysis and support vector machine based on preoperative multiparametric MR imaging of prostate cancer at 3T,” Biomed. Res. Int. 2014, 690787 (2014) 10.1155/2014/690787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stoyanova R., et al. , “Association of multiparametric MRI quantitative imaging features with prostate cancer gene expression in MRI-targeted prostate biopsies,” Oncotarget 7, 53362–53376 (2016). 10.18632/oncotarget.v7i33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fehr D., et al. , “Automatic classification of prostate cancer Gleason scores from multiparametric magnetic resonance images,” Proc. Natl. Acad. Sci. U. S. A. 112, E6265–E6273 (2013). 10.1073/pnas.1505935112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadinski M., et al. , “Pilot study of the use of hybrid multidimensional T2-weighted imaging-DWI for the diagnosis of prostate cancer and evaluation of Gleason score,” AJR Am. J. Roentgenol. 207, 592–598 (2016). 10.2214/AJR.15.15626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parra N. A., et al. , “Automatic detection and quantitative DCE-MRI scoring of prostate cancer aggressiveness,” Front. Oncol. 7, 259 (2017). 10.3389/fonc.2017.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Litjens G., et al. , “Computer-aided detection of prostate cancer in MRI,” IEEE Trans. Med. Imaging 33, 1083–1092 (2014). 10.1109/TMI.2014.2303821 [DOI] [PubMed] [Google Scholar]
- 27.Lay N., et al. , “Detection of prostate cancer in multiparametric MRI using random forest with instance weighting,” J. Med. Imaging 4, 024506 (2017). 10.1117/1.JMI.4.2.024506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greer M. D., et al. , “Computer-aided diagnosis prior to conventional interpretation of prostate mpMRI: an international multi-reader study,” Eur. Radiol. 28, 4407–4417 (2018). 10.1007/s00330-018-5374-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Litjens G., et al. , “PROMISE12 Challenge Data,” The Grand Challenge Archive, https://grand-challenge.org (2012).
- 30.Futterer J. J., et al. , “Prostate cancer localization with dynamic contrast-enhanced MR imaging and proton MR spectroscopic imaging,” Radiology 241, 449–458 (2006). 10.1148/radiol.2412051866 [DOI] [PubMed] [Google Scholar]
- 31.Epstein J. I., et al. , “The 2014 International Society of Urologic Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: definition of grading patterns and proposal for a new grading system,” Am. J. Surg. Pathol. 40, 244–252 (2016). 10.1097/PAS.0000000000000530 [DOI] [PubMed] [Google Scholar]
- 32.Mehralivand S., et al. , “Prospective evaluation of PI-RADS Version 2 using the International Society of Urological Pathology Prostate Cancer Grade Group System,” J. Urol. 198, 583–590 (2017). 10.1016/j.juro.2017.03.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.https://github.com/MedICI-NCI/MedICI.
- 34.https://github.com/codalab/codalab-competitions.
- 35.Clark K., et al. , “The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository,” J. Digital Imaging 26, 1045–1057 (2013). 10.1007/s10278-013-9622-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Litjens G., et al. , “PROSTATEx Challenge Data,” The Cancer Imaging Archive (2017). 10.7937/K9TCIA [DOI] [Google Scholar]
- 37.Pesce L. L., et al. , “Reliable and computationally efficient maximum-likelihood estimation of ‘proper’ binormal ROC curves,” Acad. Radiol. 14, 814–829 (2007). 10.1016/j.acra.2007.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metz C. E., “ROC methodology in radiologic imaging,” Invest. Radiol. 21, 720–733 (1986). 10.1097/00004424-198609000-00009 [DOI] [PubMed] [Google Scholar]
- 39.Cohen J., “A coefficient of agreement for nominal scales,” Educ. Psychol. Meas. 20, 37–46 (1960). 10.1177/001316446002000104 [DOI] [Google Scholar]
- 40.Kitchen A., Seah J., “Support vector machines for prostate lesion classification,” Proc. SPIE 10134, 1013427 (2017). 10.1117/12.2277120 [DOI] [Google Scholar]
- 41.Liu S., et al. , “Prostate cancer diagnosis using deep learning with 3D multiparametric MRI,” Proc. SPIE 10134, 1013428 (2017). 10.1117/12.2277121 [DOI] [Google Scholar]
- 42.Seah J. C. Y., Tang J. S. N., Kitchen A., “Detection of prostate cancer on multiparametric MRI,” Proc. SPIE 10134, 1013429 (2017). 10.1117/12.2277122 [DOI] [Google Scholar]
- 43.Mehrtash A., et al. , “Classification of clinical significance of MRI prostate findings using 3D convolutional neural networks,” Proc. SPIE 10134, 101342A (2017). 10.1117/12.2277123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melia J., et al. , “A UK-based investigation of inter- and intra-observer reproducibility of Gleason grading of prostatic biopsies,” Histopathology 48, 644–654 (2006). 10.1111/his.2006.48.issue-6 [DOI] [PubMed] [Google Scholar]
- 45.Hambrock T., et al. , “Prospective assessment of prostate cancer aggressiveness using 3-T diffusion-weighted magnetic resonance imaging-guided biopsies versus a systematic 10-core transrectal ultrasound prostate biopsy cohort,” Eur. Urol. 61, 177–184 (2012). 10.1016/j.eururo.2011.08.042 [DOI] [PubMed] [Google Scholar]