Abstract
The interaction between extrinsic factors and intrinsic signal strength governs thymocyte development, but the mechanisms linking them remain elusive. We report that mTORC1 couples microenvironmental cues with metabolic programs to orchestrate the reciprocal development of two fundamentally distinct T cell lineages, the αβ and γδ T cells. Developing thymocytes dynamically engage metabolic programs including glycolysis and oxidative phosphorylation, as well as mTORC1 signaling. Loss of RAPTOR-mediated mTORC1 activity impairs the development of αβ T cells but surprisingly promotes γδ T cell generation, associated with disrupted metabolic remodeling of oxidative and glycolytic metabolism. Mechanistically, we identify mTORC1-dependent control of reactive oxygen species (ROS) production as a key metabolic signal in mediating αβ and γδ T cell development, and perturbation of redox homeostasis impinges upon thymocyte fate decisions and mTORC1-associated phenotypes. Furthermore, single-cell RNA sequencing and genetic dissection reveal that mTORC1 links developmental signals from T cell receptors and NOTCH to coordinate metabolic activity and signal strength. Our results establish mTORC1-driven metabolic signaling as a decisive factor for reciprocal αβ and γδ T cell development, and provide insight into metabolic control of cell signaling and fate decisions.
One Sentence Summary:
Development of αβ and γδ T cells requires coupling of environmental signals with metabolic and redox regulation by mTORC1.
INTRODUCTION
The thymus supports and guides the generation of a diverse repertoire of T cells from precursors migrating from the bone marrow (1). These thymocytes undergo a series of well-characterized developmental processes and differentiate into two fundamentally distinct T cell lineages, the αβ and the γδ T cells, which serve critical immune functions (fig. S1A) (2, 3). The divergence of αβ and γδ T cells occurs during thymocyte development around a stage known as double-negative (DN) 3, a crucial checkpoint in which the common progenitors undergo rearrangement of the T cell receptor (TCR) β, TCRγ, and TCRδ chains through integration of diverse thymic environmental signals (2, 3). For αβ T cell commitment, the expression of a functional pre-TCR after a productive rearrangement of the TCRβ chain, together with NOTCH and other signals, induces a proliferative response accompanied by differentiation into double-positive (DP) cells (4). In contrast, NOTCH signaling plays a less important role in γδ T cell development (5, 6). Discrete models, such as the instructive and stochastic models, have been proposed to describe the contribution of TCR-mediated signals to T cell lineage choices (3). More recent studies point to a key role of signal strength, in particular the ERK, EGR1, and ID3 signaling axis (ERK/EGR1/ID3), in determining lineage choices of thymic progenitors (7–10). However, the mechanisms linking extracellular stimuli to intrinsic signal strength remain elusive.
Emerging studies reveal metabolic reprogramming as a fundamental requirement for T cell function and adaptive immune responses (11, 12). For thymocyte development, evidence to date is mainly focused on the trophic effects of NOTCH and IL-7 on cell survival and growth (13, 14), but it remains unknown whether a specific metabolic signal is linked to lineage decisions and how metabolic programs interplay with immune signals. The mechanistic target of rapamycin (mTOR) is a central sensor that integrates immune signals and metabolic cues in orchestrating T cell function and fate (15). Emerging studies also highlight the involvement of mTOR signaling in thymocyte development and T-cell acute lymphoblastic leukemia (T-ALL) progression (16–18). However, whether mTOR signaling is implicated in αβ and γδ T cell lineage choices remains unknown.
Capitalizing on genetic deletion of RAPTOR, here we report that RAPTOR-dependent mTORC1 is a central regulator of T cell development and lineage choices by coordinating environmental cues and cellular metabolic programs. We find that developing thymocytes engage dynamic regulation of metabolic and mTORC1 activation. Loss of RAPTOR impairs αβ T cell development and reciprocally promotes γδ T cell development. mTORC1 signaling and the transcription factor c-MYC (MYC) constitute a feed-forward loop that orchestrates thymocyte lineage choices. RAPTOR deficiency disrupts the balance of oxidative and glycolytic metabolism, and results in excessive production of ROS that contributes to thymocyte lineage choices. Combining single-cell RNA sequencing (scRNA-Seq) and transcriptome analysis and experimental validation, we revealed the key roles of mTORC1 in coordinating anabolic metabolism and signal strength. Our results establish metabolic control of signal strength and lineage choices as a fundamental mechanism in thymocyte development.
RESULTS
Metabolic and mTORC1 activities are dynamically regulated in T cell development
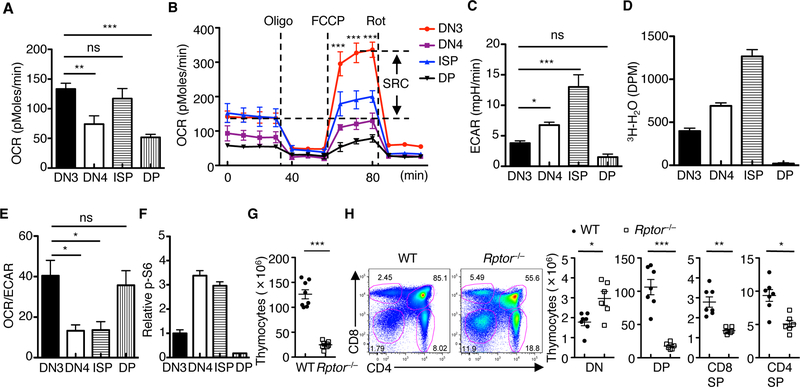
To understand metabolic regulation and requirements in developing thymocytes, we measured oxygen consumption rate (OCR) and extracellular acidification rate (ECAR), which respectively denote oxidative phosphorylation (OXPHOS) and glycolytic activities, in developing thymocytes. Specifically, we analyzed DN3 (the first stage with full T cell commitment (2)), DN4, and immature single-positive cells (ISP; the intermediate transitional stage between DN and DP stages), relative to quiescent DP thymocytes (fig. S1A). DN3 and ISP cells had the highest OCR at the basal level (Fig. 1A), whereas DN3 cells displayed an even greater spare respiratory capacity (SRC), a measure of how effectively the electron transport chain can respond to energy demand, than ISP cells (Fig. 1B). During the transition from DN3 to ISP cells, ECAR was progressively upregulated (Fig. 1C), indicating prominent upregulation of glycolysis in this process; this observation was confirmed by direct measurement of glycolytic activity (Fig. 1D). In contrast, DP cells markedly downregulated ECAR and glycolytic activity (Fig. 1, C and D). The dynamic regulation of OCR and ECAR in thymocyte subsets translated into a higher OCR/ECAR ratio in DN3 and DP cells (Fig. 1E), indicating the preferential use of oxidative metabolism over glycolysis by these cells.
Fig. 1. Dynamic regulation of cell metabolism and mTORC1 activity and the requirement of RAPTOR in thymocyte development.
(A and B) OCR in thymocyte subsets under basal condition (A) or in response to the indicated mitochondrial inhibitors (Oligo, Oligomycin; FCCP, carbonyl cyanide p-trifluoromethoxyphenylhydrazone; and Rot, Rotenone) (B). SRC, spare respiratory capacity (SRC of DN3 cells is shown). (C and D) Measurement of ECAR (C) and glycolytic activity (D) in freshly isolated thymocyte subsets. (E) Ratio of OCR to ECAR in thymocyte subsets. (F) Relative phosphorylation of S6 (mean fluorescence intensity (MFI) in DN3 cells is set to 1) in thymocyte subsets. (G) Total thymocyte cellularity of WT and Rptor–/– mice. Each symbol represents an individual mouse (n = 8 each group). (H) Flow cytometry of thymocyte subsets in WT and Rptor–/– mice (left). Right, cellularity of the indicated thymocyte subsets (DN, CD4–CD8–; DP, CD4+CD8+; CD8SP, CD4–CD8+TCRβ+; and CD4SP, CD4+CD8–TCRβ+). Each symbol represents an individual mouse (WT, n = 7; Rptor–/–, n = 6). Data are mean ± s.e.m. ns, not significant; *P < 0.05, **P < 0.001, and ***P < 0.0001. One-way analysis of variance (ANOVA) with Tukey test (A to E), or two-tailed unpaired t-test (G, H). Data are combination of three independent experiments (A to E), or representative of at least ten independent experiments (F to H). Numbers indicate percentage of cells in gates.
Metabolic activity is contingent upon nutrient uptake and mTOR activation, a crucial regulator of anabolic metabolism (15). Compared with DN3 and DP cells, DN4 and ISP cells showed higher expression of CD98 and CD71, the respective amino acid and iron transporters (fig. S1, B and C), and increased cell size (fig. S1D). Accordingly, DN4 and ISP cells had higher phosphorylation of S6, indicative of greater mTORC1 activity (Fig. 1F). Collectively, these results indicate the dynamic regulation of metabolic and mTORC1 activities in thymocyte development.
RAPTOR deficiency impairs DN to DP transition in αβ T cell development
To define the function of mTORC1 signaling in thymocyte development, we generated Rptorfl/flCd2-Cre (Rptor–/–) mice with the Cd2-Cre transgene (19) to initiate deletion of RAPTOR, an obligatory component of mTORC1, in early DN cells. Complete deletion of RAPTOR was observed in DN3 cells (fig. S2, A and B). RAPTOR deficiency resulted in greatly reduced thymocyte cellularity (Fig. 1G), including an approximately 10-fold reduction of DP cells as well as a considerable loss of CD4SP and CD8SP cells (Fig. 1H and fig. S2C). Consequently, T cell numbers in the spleen were significantly decreased (fig. S2D). RAPTOR deletion, however, caused a proportional increase of the DN compartment (Fig. 1H and fig. S2C), suggesting a possible blockade at the DN to DP transition.
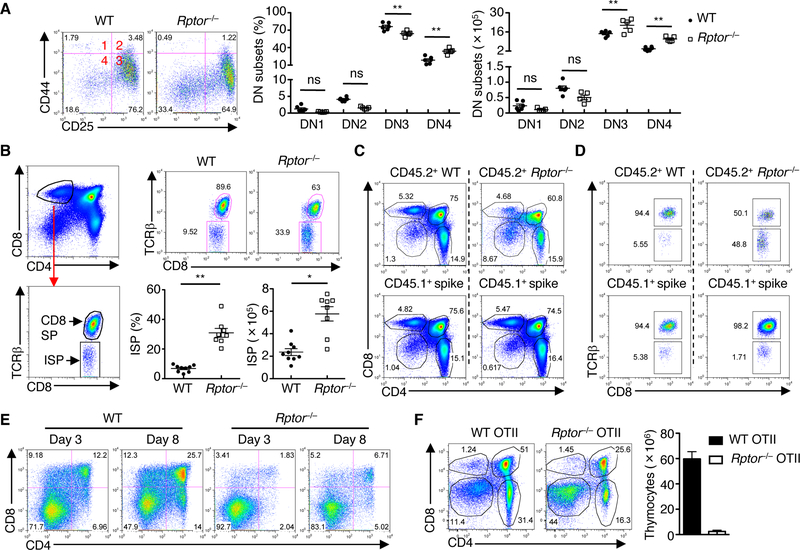
Hence, we performed detailed analysis of thymocyte subsets during the DN to DP transition. Although the frequency of Rptor–/– DN3 cells was reduced, their absolute number was elevated and this was in association with accumulation of total DN cells (Fig. 2A). In support of a role for RAPTOR in the DN to DP transition, Rptor–/– thymus contained significantly elevated frequency and cellularity of DN4 (Fig. 2A) and ISP cells (Fig. 2B). Next, we generated mixed bone marrow (BM) chimeras consisting of CD45.1+ (‘spike’) and CD45.2+ WT or Rptor–/– cells, and found that Rptor–/– but not WT or CD45.1+ cells showed increased DN and ISP cells, but reduced DP cells (Fig. 2, C and D). Finally, to directly assess the DN to DP transition, we co-cultured DN3 cells with OP9 stromal cells expressing the NOTCH ligand Delta-like 1 (OP9-DL1 cells), which represents a well-established in vitro system that models thymocyte development (5). In this system, Rptor–/– DN3 cells showed impaired capacity to progress into DP cells (Fig. 2E). Similarly, Rptor–/– ISP cells had defective development into DP cells (fig. S3A), consistent with the accumulation of ISP cells in vivo (Fig. 2B). Collectively, these results indicate that RAPTOR mediates the transition from DN to DP cells in a cell-autonomous manner.
Fig. 2. RAPTOR deficiency impairs the DN to DP transition in αβ T cell development.
(A) Flow cytometry of DN1–4 subsets (gated on lineage-negative DN thymocytes) in WT and Rptor–/– mice (left). Frequencies (middle) and cellularity (right) of DN subsets. Each symbol represents an individual mouse (WT, n = 6; Rptor–/–, n = 5). (B) Left, gating strategy for ISP cells (CD4– CD8+TCRβ–). Right, flow cytometry (upper panels) and frequency and cellularity (lower panels) of ISP cells in WT and Rptor–/– mice. Each symbol represents an individual mouse (WT, n = 9; Rptor–/–, n = 8). (C and D) Sublethally irradiated Rag1–/– mice were reconstituted with a 1:1 mixture of CD45.1+ BM and either CD45.2+ WT or Rptor–/– BM cells to generate mixed bone marrow chimeras. Flow cytometry of thymocyte subsets (C, upper panels) or ISP cells (D, upper panels) in CD45.2+ WT and Rptor–/– donor-derived cells, or in CD45.1+ spike-derived cells (C and D, lower panels). (E) Expression of CD4 and CD8 on WT and Rptor–/– DN3 cells co-cultured with OP9-DL1 cells for 3 or 8 days. (F) Flow cytometry of thymocyte subsets in WT OTII and Rptor–/– OTII mice (n = 4 each group) (left). Right, cellularity of total thymocytes. Data are mean ± s.e.m. ns, not significant; *P < 0.0005, and **P < 0.0001. One-way ANOVA with Tukey test (A), two-tailed Mann-Whitney test (B, frequency), or two-tailed unpaired t-test (B, cellularity). Data are representative of at least three independent experiments (A to F). Numbers indicate percentage of cells in quadrants or gates.
To explore the mechanistic basis, we examined the underlying cellular processes. Cell apoptosis, as indicated by staining of the active form of caspase-3, was not altered in any of the Rptor–/– subsets analyzed (fig. S3B). BrdU incorporation assay showed that RAPTOR deletion impaired proliferation of ISP cells, but not that of DN3 or DN4 cells (fig. S3C). The proliferative expansion in DN4 and ISP cells is known to be preceded by a period of quiescence at the DN3 stage (2). In fact, DN3 thymocytes are a mixture of two populations: small TCRβ-unrearranged DN3-E (DN3a) cells and large TCRβ-rearranged DN3-L (DN3b) cells that are beginning the progression toward DP cells (20, 21), which is associated with upregulated CD27 expression (fig. S3D). Compared with DN3a cells, DN3b cells had increased mTORC1 activity (fig. S3E). Further, RAPTOR deficiency impaired upregulation of cell size and expression of CD71 and CD98 as early as in DN3b cells, and these defects were exacerbated in DN4 and, more strikingly, in ISP cells (fig. S3F). However, induction of intracellular TCRβ (icTCRβ) proceeded largely normal in DN3b cells (fig. S3G). Despite the small reduction in icTCRβ expression in Rptor–/– ISP cells (fig. S3G), transgenic expression of pre-rearranged OTII (MHC Class II-restricted) TCRαβ chains failed to rescue thymocyte developmental defects (Fig. 2F). These results indicate that RAPTOR deficiency does not affect thymocyte survival or pre-TCR induction, but prevents subsequent upregulation of cell size and nutrient receptors, ultimately leading to defective proliferation and differentiation of DN into DP cells.
Given that ISP cells represent a critical stage in the transition from DN to DP cells and are dysregulated in the absence of RAPTOR, we compared the transcriptional profiles of ISP cells from WT and Rptor–/– mice. Gene set enrichment analysis (GSEA) (22) revealed that many downregulated gene sets in Rptor–/– cells were related to metabolic programs, including cholesterol biosynthesis and the respiratory electron transport chain (fig. S4A), and real-time PCR analysis of lipogenic genes verified these hits (fig. S4B). Ingenuity Pathway Analysis (IPA) predicted the inhibition of MYC and SREBP factors (fig. S4C), which serve as crucial activators of anabolic metabolism (11, 12), in the absence of RAPTOR. Altogether, consistent with defective cell proliferation, growth, and nutrient transporter expression (fig. S3), Rptor–/– ISP cells show impaired gene expression programs mediating lipogenic and other anabolic metabolism.
Loss of RAPTOR enhances γδ T cell development
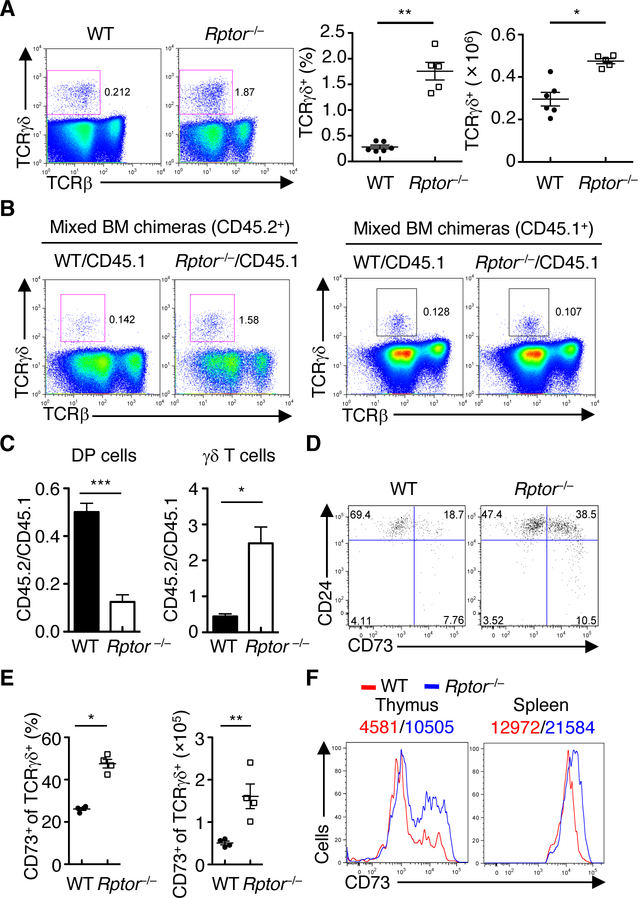
Aside from the αβ T cell lineage, the γδ T cells are the other fundamentally distinct T cell lineage derived from the common thymocyte progenitors such as DN3 cells (2, 3). Surprisingly, Rptor–/– mice had significantly increased frequency and cellularity of TCRγδ+ T cells (Fig. 3A), resulting in a profoundly enhanced ratio of TCRγδ+ to TCRβ+ T cells (fig. S5A). Rptor–/– γδ T cells had efficient deletion of Rptor (fig. S5B), and this was associated with reduced cell size and lower CD71 and CD98 expression (fig. S5C). Rptor–/– γδ T cells had slightly reduced Ki67 expression (fig. S5D), but largely normal caspase-3 activity (fig. S5E), indicating that the increase in the γδ T cell compartment was unlikely due to altered proliferation or apoptosis. The alteration in γδ T cell development was also observed in the previously mentioned mixed BM chimeras, in which CD45.2+ Rptor–/– donor cells had much higher frequency of γδ T cells than WT cells (Fig. 3, B and C), indicating that RAPTOR loss promotes γδ T cell development in a cell-autonomous manner (i.e. not secondary to the loss of DP thymocytes given that normal thymic cellularity is provided by the CD45.1+ spike cells). This conclusion was further verified in the OP9-DL1 co-culture system, in which Rptor–/– DN3a cells showed increased generation of γδ T cells than their WT counterparts (fig. S5, F to H), even in the presence of CD45.1+ spike cells (fig. S5I). Of note, CD73 has been recently identified as an important marker to define γδ T cell commitment (23). Rptor–/– mice had a greatly elevated percentage and cellularity of the CD73+ population in thymic γδ T cells (Fig. 3, D and E). Additionally, the expression level of CD73 was increased in RAPTOR-deficient γδ T cells from both the thymus and spleen (Fig. 3F) as well as in those generated in vitro in the OP9-DL1 system (fig. S5, F and I), further supporting the role of RAPTOR in constraining γδ T cell development. To assess the involvement of RAPTOR in the TCR repertoire of γδ T cells, we examined the usage of diverse TCRγ chains in γδ T cells from the thymus (24, 25), and found that the composition of various Vγ chains was comparable between WT and Rptor–/– γδ T cells (fig. S5J). In line with this finding, RAPTOR-deficient γδ T cells had largely normal proportions of Vγ1.1+ and Vγ2+ subsets in the thymus (fig. S5, K and L). Taken together, our results indicate that RAPTOR deficiency promotes γδ T cell commitment without affecting the usage of TCRγ chains.
Fig. 3. Loss of RAPTOR promotes γδ T cell development.
(A) Flow cytometry (left) of TCRγδ+ cells in WT and Rptor–/– thymocytes. Frequency (middle) and cellularity (right) of TCRγδ+ cells. Each symbol represents an individual mouse (WT, n = 6; Rptor–/–, n = 5). (B) Flow cytometry of TCRγδ+ cells derived from WT and Rptor–/– donor (CD45.2+, left) or spike (CD45.1+, right) cells in the reconstituted BM chimeras as described in Figure 2C. Flow cytometry of TCRγδ+ cells in WT and Rptor–/– donor-derived thymocytes in the reconstituted mixed BM chimeras. (C) Distribution of DP (left) and γδ T cells (right) between CD45.1+ and CD45.2+-derived cells in the mixed bone marrow chimeras generated as described in Figure 2C, after normalization against non-deleting DN1 thymocytes. (D) Expression of CD73 and CD24 on CD4–CD8–TCRγδ+ thymocytes from WT and Rptor–/– mice. (E) Frequency (left) and cellularity (right) of CD73+ TCRγδ+ cells in D. Each symbol represents an individual mouse (n = 4 each group). (F) Expression of CD73 on TCRγδ+ cells in the thymus and spleen of WT and Rptor–/– mice, with MFI plotted above graphs. Data are mean ± s.e.m. *P < 0.05, **P < 0.005, and ***P < 0.0005. Two-tailed Mann-Whitney test (A and E for frequency analyses), one-tailed unpaired t-test (C), or two-tailed unpaired t-test (A and E for cellularity). Data are representative of at least three independent experiments (A to F). Numbers indicate percentage of cells in quadrants or gates.
As an important regulator of mTORC1 signaling, the small G protein RHEB integrates different upstream signals from growth factor stimulation (15). However, Rhebfl/flCd2-Cre (Rheb–/–) mice had largely normal cellularity and frequencies of thymocyte subsets, including γδ T cells (fig. S6, A and B). On the other hand, even though RICTOR/mTORC2 is required for αβ T cell development (26, 27), loss of RICTOR (Rictorfl/flCd2-Cre or Rictor–/– mice) did not impact the frequency or cellularity of γδ T cells (fig. S6C). Deletion of RICTOR in the Rptor–/– background further reduced the cellularity of total thymocytes, including γδ T cells (fig. S6C), but it had no substantial rescue effects on the frequency of γδ T cells (fig. S6C) or CD73 expression (fig. S6D). Collectively, RAPTOR-mTORC1 exerts a unique and cell-autonomous role in constraining γδ T cell development.
The reciprocal interplay between mTORC1 and MYC shapes thymocyte development
Given the link of mTORC1 to cell growth and anabolic metabolism in thymocyte development (fig. S3 and S4), we next explored the functional importance of selective metabolic programs in T cell development by targeting key transcriptional regulators. RAPTOR deficiency is predicted to downregulate SREBP1 and SREBP2 activities (fig. S4C), both of which are dependent upon the scaffold protein SCAP that is crucial for lipogenic metabolism and proliferation of T cells in the periphery (28). Deletion of SCAP (Scapfl/flCd2-Cre or Scap–/– mice) had no substantial effects on thymocyte populations (fig. S7A), with normal frequencies of DN subsets (fig. S7B), ISP cells (fig. S7C), and γδ T cells observed (fig. S7D). Another metabolic regulator frequently associated with mTORC1 activity is the key glycolytic regulator HIF1α (29). HIF1α mediates metabolic reprogramming of TH17 cells (30) and effector CD8 T cells (31). While HIF1α expression is induced in developing thymocytes, deletion of HIF1α (Hif1afl/flCd2-Cre or Hif1a–/– mice) had no substantial impact on the frequencies of thymocyte subsets (fig. S7E) including γδ T cells (fig. S7F). These results indicate a dispensable role for SCAP-mediated lipogenesis and HIF1α function in thymocyte development.
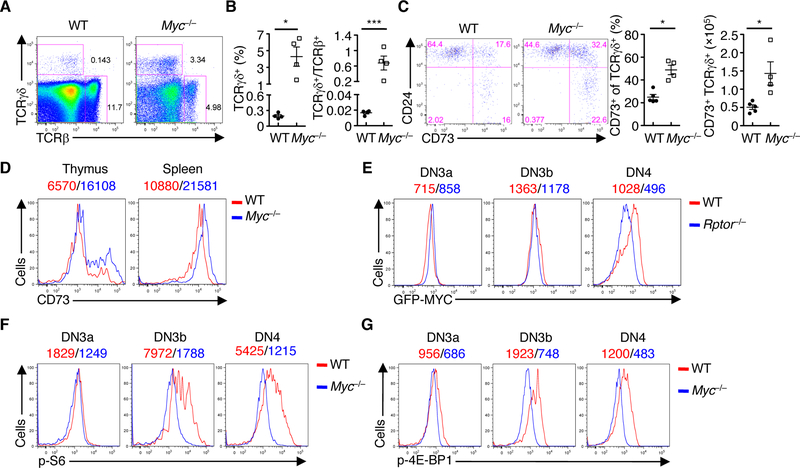
The metabolic regulator MYC was among the top hits of the potential upstream regulators in RAPTOR-deficient cells (fig. S4C). Although MYC is required for thymocyte development, its functions and mechanisms in αβ and γδ T cell fate decisions remain uncertain (4, 32–34). We found that deletion of MYC (Mycfl/flCd2-Cre or Myc–/– mice) greatly reduced cellularity of total thymocytes, and this was associated with reduced frequency of DP cells and increased frequency of DN cells (fig. S8A). Within the DN compartment, Myc–/– mice had increased frequency and number of DN3 cells (fig. S8, B and C). However, WT and Myc–/– DN3b cells had comparable induction of icTCRβ expression (fig. S8D), suggesting a largely normal β-selection. In contrast, Myc–/– mice had an elevated frequency of γδ T cells (Fig. 4, A and B) and ratio of TCRγδ+ to TCRβ+ cells (Fig. 4B). Despite undisturbed cellularity of γδ T cells in Myc–/– mice due to the drastic loss of total thymocytes (fig. S8E), Myc–/– γδ T cells had increased frequency and cellularity of the CD73+ population (Fig. 4C) as well as higher levels of CD73 expression (Fig. 4D), supporting a role for MYC in restricting γδ T cell development.
Fig. 4. Interplay between mTORC1 and MYC in developing thymocytes.
(A) Flow cytometry of TCRβ+ and TCRγδ+ cells in WT and Myc–/– thymocytes. (B) Frequency of TCRγδ+ cells (left) and the ratio of TCRγδ+ to TCRβ+ cells (right) in WT and Myc–/– thymocytes. Each symbol represents an individual mouse (n = 4 each group). (C) Expression of CD73 and CD24 on CD4– CD8–TCRγδ+ thymocytes from WT and Myc–/– mice (left). Frequency (middle) and cellularity (right) of CD73+ TCRγδ+ cells. Each symbol represents an individual mouse (WT, n = 5; Myc–/–, n = 4). (D) Expression of CD73 on TCRγδ+ cells in the thymus and spleen of WT and Myc–/– mice, with MFI plotted above graphs. (E) Expression of GFP-MYC in thymocyte subsets of WT and Rptor–/– mice, with MFI plotted above graphs. (F and G) Plots of p-S6 (F) and p-4E-BP1 (G) in thymocyte subsets of WT and Myc–/– mice, with MFI plotted above graphs. Data are mean ± s.e.m. *P < 0.05, **P < 0.005, and ***P < 0.0005. Two-tailed Mann-Whitney test (B and C for frequency analyses), or two-tailed unpaired t-test (B for ratio analyses, and C for cellularity). Data are representative of at least three independent experiments (A to E), or two (F, G) independent experiments. Numbers indicate percentage of cells in quadrants or gates.
We next investigated the interaction between RAPTOR and MYC in thymocyte development. First, we crossed Rptor–/– mice with GFP-MYC knockin mice (35) to track the dynamic regulation of MYC by means of GFP expression. WT DN3b cells upregulated expression of MYC, which was then downregulated in the DN4 stage (Fig. 4E). While Rptor–/– DN3a cells had normal GFPMYC expression, Rptor–/– DN3b cells had a modest reduction of GFP-MYC expression (Fig. 4E). This reduction was more pronounced in Rptor–/– DN4 cells (Fig. 4E), indicating that MYC acts downstream of RAPTOR. We next determined whether a reciprocal interaction exists; that is, whether MYC acts upstream of mTORC1 also. Indeed, Myc–/– DN3b and DN4 cells had markedly reduced phosphorylation of S6 and 4E-BP1 (Fig. 4, F and G). Therefore, mTORC1 and MYC constitute a reciprocal interaction loop in thymocyte development.
RAPTOR modulates ROS production in fate decisions of DN3 cells
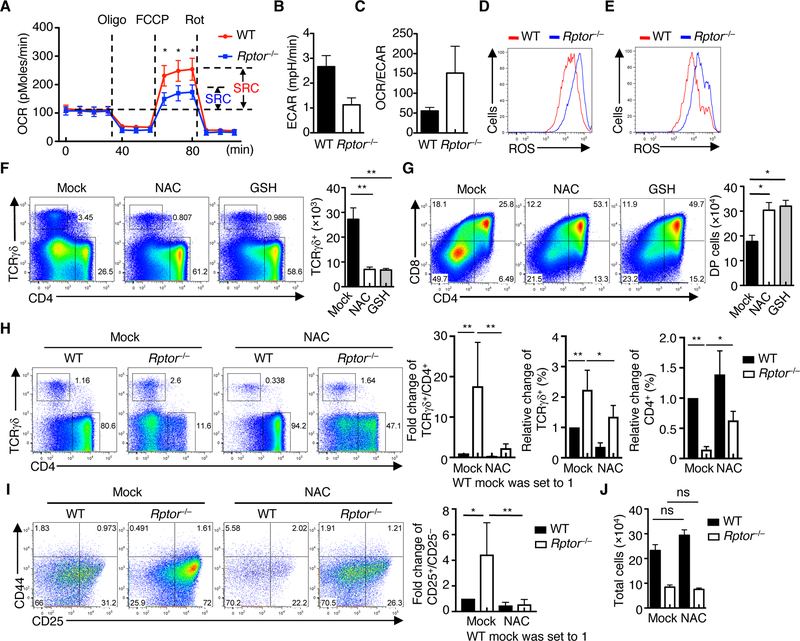
Given the dynamic metabolic regulation that occurs during T cell development (Fig. 1), we hypothesized that RAPTOR mediates metabolic reprogramming in DN3 cells, hence shaping their fate decisions. Compared with the WT counterparts, Rptor–/– cells had intact basal OCR (Fig. 5A) and expression of representative electron transport chain components (fig. S9A), albeit with a partial impairment in SRC (Fig. 5A). In contrast, Rptor–/– DN3 cells had reduced ECAR and glycolytic activity (Fig. 5B and fig. S9B). The increased ratio of OCR to ECAR in Rptor–/– DN3 cells (Fig. 5C) suggests disrupted control of bioenergetics and an increased reliance on oxidative metabolism to compensate for impaired glycolysis. Consistent with this idea, Rptor–/– DN3 cells, but not other subsets, had significantly increased ROS levels compared with WT counterparts (Fig. 5D and fig. S9C). The increased ROS induction was also observed in Rptor–/– γδ T cells (Fig. 5E). Other parameters associated with mitochondrial fitness, including mitochondrial mass and mitochondrial membrane potential, were comparable between WT and Rptor–/– cells (fig. S9, D and E). These results indicate that RAPTOR deficiency impairs metabolic remodeling and enhances ROS production in DN3 cells.
Fig. 5. RAPTOR controls metabolic balance between glycolysis and OXPHOS, and production of ROS in fate determination of DN3 cells.
(A) OCR in WT and Rptor–/– DN3 cells under basal conditions or in response to the indicated mitochondrial inhibitors (SRC of WT and Rptor–/– cells is shown). (B) Measurement of ECAR in WT and Rptor–/– DN3 cells. (C) Ratio of OCR to ECAR in WT and Rptor–/– DN3 cells. (D and E) Analysis of ROS production in DN3 cells (D) and TCRγδ+ cells (E) from WT and Rptor–/– mice. (F and G) WT DN3a cells were co-cultured with OP9-DL1 cells in the absence or presence of NAC or GSH for 3 days, followed by analysis of CD4 and TCRγδ expression (F) and CD4 and CD8 expression (G). Right, numbers of TCRγδ+ (F) and CD4+CD8+ DP cells (G) in the culture (n = 3 each group). (H) WT and Rptor–/– DN3a cells were co-cultured with OP9-DL1 cells in the absence or presence of NAC for 5 days, followed by analysis of CD4 and TCRγδ expression (left). Right, fold change of the ratio of TCRγδ+ to CD4+ cells (WT without NAC treatment is set to 1), and frequencies of TCRγδ+ and CD4+ cells (n = 3 each group). (I) WT and Rptor–/– DN3a cells were co-cultured with OP9-DL1 cells in the absence or presence of NAC for 5 days, followed by analysis of CD44 and CD25 expression (left). Right, fold change of the ratio of CD25+CD44– (DN3) to CD25–CD44– (DN4) populations of WT and Rptor–/– cells without or with NAC treatment (WT without NAC treatment is set to 1). (J) Cellularity of total cells in H. Data are mean ± s.e.m. ns, not significant; *P < 0.05, **P < 0.01, and ***P < 0.0001. One-way ANOVA with Turkey test (A, F to J). Data are the combination of three independent experiments (A to C), or representative of at least three (D to J) independent experiments. Numbers indicate percentage of cells in quadrants or gates.
We next assessed whether directly modulating ROS production and metabolic activity affects fate choices of DN3 cells. First, we co-cultured DN3a cells with OP9-DL1 cells in the presence of galactose (Gal) and galactose oxidase (GAO), known promoters of ROS production (36). Gal/GAO significantly elevated the generation of γδ T cells (fig. S10A), concomitant with impaired generation of αβ T cells (fig. S10A). Second, we used dichloroacetate (DCA), an inhibitor of the mitochondrial enzyme pyruvate dehydrogenase kinase (PDK), to activate pyruvate dehydrogenase (PDH) and hence shift glycolysis to fuel OXPHOS in the mitochondria (37). DCA treatment promoted the generation of γδ T cells at the expense of αβ T cells (fig. S10B), associated with increased ROS production (fig. S10C). To exclude the potential effects of the pharmacological agents on OP9-DL1 cells, we next used a stromal cell-free system for in vitro thymocyte differentiation (38). Gal/GAO and DCA markedly promoted generation of γδ T cells along with reduction of αβ T cells (fig. S10D), indicating that ROS signaling targets thymocytes but not stromal cells in altering fate choices. Moreover, addition of the ROS scavenger N-acetyl-L-cysteine (NAC) or glutathione (GSH) significantly diminished and promoted the generation of γδ (Fig. 5F) and αβ T cells (Fig. 5G), respectively, without substantially altering total cellularity of the culture (fig. S10E). Using the stromal cell-free system, we also observed the blocking effects of NAC and GSH on the generation of γδ T cells and their CD73 expression levels (fig. S10, F and G). We next hypothesized that neutralizing ROS production could partially rescue the altered fate choices of Rptor–/– DN3a cells. In support of this, NAC treatment significantly, though not completely, suppressed the generation of γδ T cells derived from Rptor–/– DN3a cells (Fig. 5H), while reciprocally restoring the defective αβ T cell development and the altered ratio of γδ to αβ T cells (Fig. 5H). Moreover, the impaired transition of Rptor–/– DN3 cells to DN4 cells (indicated by the ratio of DN3 (CD25+CD44–) to DN4 (CD25–CD44–) cells) was restored by NAC treatment (Fig. 5I). Of note, NAC treatment had no substantial effects on the reduced cellularity of Rptor–/– cells (Fig. 5J) or their survival (fig. S10H), indicating that ROS regulates lineage choices of DN3 cells independent of cell expansion. These results collectively indicate that mTORC1-dependent metabolic remodeling in DN3 cells regulates the production of ROS as a metabolic signal in dictating lineage choices.
RAPTOR coordinates intrathymic signals in DN cells
Multiple immune and developmental cues, including TCR, NOTCH, and IL-7 signaling, mediate lineage choices of DN3 cells (2, 3). We next sought to determine their individual contributions to mTORC1 activation and function. DN3 cells from Rag1-deficient mice, which lack TCR rearrangement and thus TCR signaling, had markedly reduced phosphorylation of S6 (fig. S11A). Also, Rag1-deficient DN3a cells cultured with OP9-DL1 cells were impaired to upregulate phosphorylation of S6 and 4E-BP1 (fig. S11B) compared to their WT counterparts, indicating that TCR signaling is required for mTORC1 activation. NOTCH is a pivotal factor in thymocyte development (3), however, the underlying mechanisms of NOTCH signaling in αβ and γδ T cell development remain to be fully understood (5, 6, 39, 40). Compared with OP9 cells (lacking NOTCH ligands), OP9-DL1 cells induced greater phosphorylation of S6 and 4E-BP1 (fig. S11C) in DN3a cells, in a process independent of cell survival (fig. S11D). Moreover, rapamycin-mediated inhibition of mTORC1 activity enhanced γδ T cell development and reciprocally inhibited αβ T cell development (fig. S11E). Although IL-7 signaling has been recently shown to induce trophic effects in developing thymocytes (14), addition of IL-7 did not affect mTORC1 activity in DN3 cells (fig. S11F). Our results indicate mTORC1 integrates TCR and NOTCH signals to mediate fate choices of DN3 cells.
To understand RAPTOR-dependent signaling pathways underlying T cell lineage choices, we compared transcriptional profiles of pre-selection DN3a cells from WT and Rptor–/– mice. GSEA revealed that NF-κB signaling, which has been implicated in thymocyte development by mediating pre-TCR, NOTCH, and cytokine signaling (41–43), was aberrantly upregulated in RAPTOR-deficient DN3a cells (fig. S12A). Moreover, Rptor–/– cells showed a significant enrichment of NOTCH signaling (fig. S12A), which can activate NF-κB (43) as well as the ERK-MAPK signaling cascade (44). Additionally, we compared transcriptional profiles of WT and Rptor–/– γδ T cells, and GSEA also revealed significant enrichment of NF-κB signaling in the absence of RAPTOR (fig. S12B). In support of this finding, Rptor–/– γδ T cells had elevated phosphorylation of p65 at steady state and upon anti-TCRγδ stimulation (fig. S12C). Altogether, these results indicate that RAPTOR deficiency alters signal transduction in thymocyte development.
scRNA-Seq reveals RAPTOR-dependent signaling circuits in thymocyte lineage choices
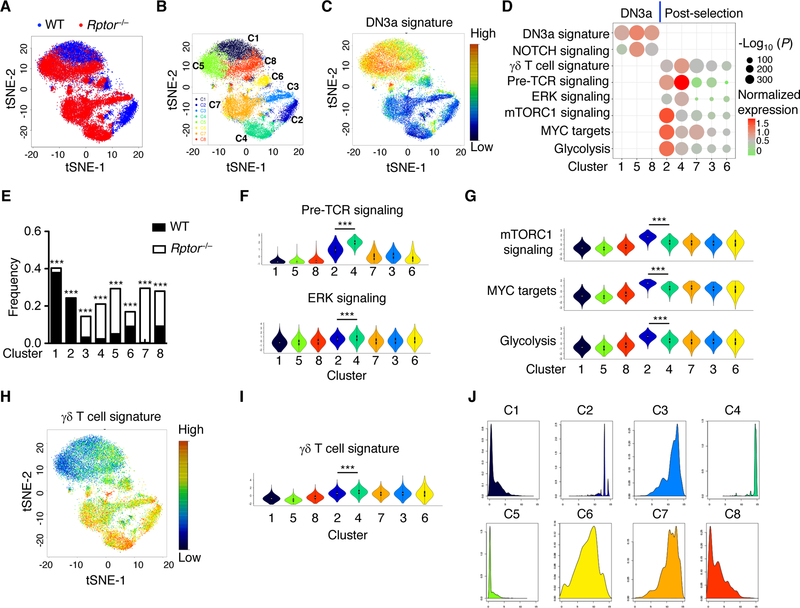
Recent advances in scRNA-Seq have enabled an unbiased characterization of immune cell subpopulations during developmental processes and immune responses (45). We performed scRNA-Seq (46) to further investigate RAPTOR-dependent signaling pathways and functional modules underlying thymocyte lineage decisions. We analyzed 21,332 late-stage DN thymocytes (DN3-DN4; Lin–CD4–CD8–CD44–), including 7,345 WT and 13,987 Rptor–/– cells, that passed quality control thresholds (Fig. 6A), and partitioned the cells into eight clusters in tSNE (‘t-distributed stochastic neighbor embedding’) plot through unsupervised clustering (Fig. 6B). To define the nature of these clusters, we calculated gene signature scores for DN3a cells based on the genes selectively expressed in DN3a cells in the public datasets (47), and then overlaid expression of the DN3a gene signature attributed to each cell onto the tSNE visualization. Clusters 1, 5, and 8 showed much greater expression of the DN3a signature than the remaining clusters (Fig. 6, C and D). High levels of Il2ra and Ptcra, which encode CD25 and pre-TCRα respectively and are known to be abundantly expressed in DN3a cells compared with more mature thymocytes (47), were also found in clusters 1, 5, and 8 (fig. S12D). These results support the classification of clusters 1, 5 and 8 as DN3a cells, whereas the remaining clusters likely represent post-selection thymocytes. Of note, Il2ra and Ptcra are target genes of NOTCH signaling, which is highly active in DN3a cells but downregulated after β-selection (47). Among these 3 DN3a clusters, cluster 5 showed increased expression of Il2ra, Ptcra, and additional NOTCH targets, including Notch1, Notch3, Dtx1, and Dtx3l (47) (fig. S12D) and, moreover, enhanced NOTCH signaling activity (fig. S12E). Interestingly, the majority of cells in cluster 5 were RAPTOR-deficient (Fig. 6E), while cluster 1 was dominated by WT cells, thereby verifying that deletion of RAPTOR rewires NOTCH signaling in DN3a cells. We next focused on transcriptional events in post-selection clusters. We found that the signature genes associated with pre-TCR signaling (i.e. genes induced in DN3b vs DN3a cells (47)) were highly enriched in cluster 4 (Fig. 6F and fig. S12F), which was predominantly comprised of RAPTOR-deficient cells, and, to a much lesser degree, in cluster 2, which was dominated by WT cells (Fig. 6E). In addition, the ERK signaling module was also higher in cluster 4 than in cluster 2 (Fig. 6F and fig. S12F), indicating the upregulation of TCR signaling in the absence of RAPTOR. In contrast, compared to cluster 2, cluster 4 showed lower signature scores of mTORC1 signaling, MYC targets, and glycolysis (Fig. 6G and fig. S12G), as well as expression of selective anabolic genes, including Ldha, Tpi1, Slc7a5, and Shmt2 (fig. S12H). Therefore, scRNA-Seq analysis suggests that RAPTOR coordinates TCR signaling and metabolic reprogramming in thymocyte development.
Fig. 6. scRNA-Seq reveals that RAPTOR coordinates immune signaling and anabolic metabolism in thymocyte fate decisions.
(A) Single-cell RNA sequencing of WT and RAPTOR-deficient DN cells, followed by a tSNE visualization of the 21,332 single cells analyzed. (B) tSNE visualization of eight clusters partitioned by unsupervised clustering. (C) tSNE visualization of DN3a gene signature expressed by individual cells. (D) Association of different gene signatures with the eight single-cell clusters, with the relative over-representation of gene signatures for a specific cluster determined using one-tailed Mann-Whitney test. Clusters 1, 5, and 8 contained DN3a cells, while the remaining clusters contained post-selection thymocytes. (E) Frequencies of WT and Rptor–/– cells in the different clusters. Clusters significantly enriched for presence of WT or Rptor–/– cells were marked. (F) Violin plots of signatures of pre-TCR and ERK signaling among the eight clusters. A violin plot combines the box plot and the local density estimation into a single display. The black bars and thin lines within the violin plots indicate the interquartile range (1st quantile – 3rd quantile) and the entire range of the data (up to 1.5 fold of interquartile range from 1st/3rd quantile), respectively, and the white dots in the center indicate the median values. (G) Violin plots of signatures of mTORC1 signaling, MYC targets, and glycolysis. (H and I) tSNE (H) and violin (I) plots of γδ T cell gene signature. (J) Pseudotime densities for the 8 clusters. Data are mean ± s.e.m. *P < 0.05, and ***P < 0.0001. One-tailed Mann-Whitney test (D), Logistic regression (E), two-tailed Mann-Whitney test (F, G, I). Data are one experiment (WT, n = 2; Rptor–/–, n = 3; A to J).
In further support of the enhanced γδ T cell development in RAPTOR-deficient mice, we found that the gene signature associated with γδ T cells (48) was upregulated in cluster 4, compared to cluster 2 (Fig. 6, H and I). To reconstruct the acquisition of γδ T cell fate over time, we analyzed our scRNA-Seq data using Monocle 2 (49), which provides pseudo-temporal ordering of cells relative to developmental progression (‘pseudotime’). This allowed us to derive a relative developmental trajectory that revealed pronounced differences in the pseudotime assignment of the 8 clusters (Fig. 6J and fig. S13, A). Specifically, we found that cells with early pseudotime assignment were mainly restricted to clusters 1, 5 and 8 (fig. S13A, blue arrow), consistent with the identification of these clusters as DN3a cells. This is further supported by the direct projection of the DN3a signature onto the pseudotime trajectory (fig. S13B), thereby corroborating our pseudotime analysis. In contrast, cells with late psedudotime were mostly enriched in cluster 4 and, to a lesser extent, in cluster 2 (fig. S13A, red arrows). Of note, the γδ T cell gene signature was highly enriched in cluster 4 (fig. S13C), which was primarily comprised of Rptor–/– cells (fig. S13D). Altogether, these results support the enhanced gene expression and developmental progression along the γδ T cell lineage of RAPTOR-deficient T cells.
RAPTOR tunes the strength of the ERK/EGR1/ID3 signaling pathway
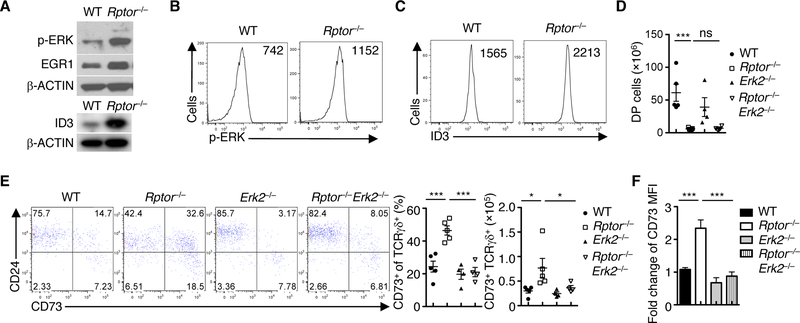
The lineage decision of developing thymocytes is governed by various intrinsic factors, with TCR signal strength being particularly important (2, 3). The γδ T cell lineage choice is characterized by strong signal strength, especially in regards to the ERK/EGR1/ID3 signaling axis, as opposed to αβ T cell development (7, 8). Our scRNA-Seq results suggest that RAPTOR coordinates metabolic reprogramming and signal strength in DN3 cells. Indeed, Rptor–/– DN3 cells had higher phosphorylation of ERK and expression of EGR1 and ID3 (Fig. 7, A to C) than did their WT counterparts. Ablation of ERK2 in a Rptor-deficient background did not substantially affect the reduction of DP thymocytes (Fig. 7D). However, it blocked the increased CD73+ γδ T cell population (Fig. 7E) and the elevated expression of CD73 (Fig. 7F), as compared to Rptor–/– mice. These results indicate that the increased signal strength due to RAPTOR deficiency contributes to γδ T cell development. In line with the intersection between RAPTOR/mTORC1 and MYC, Myc–/– DN3 cells displayed increased levels of ERK phosphorylation and ID3 expression (fig. S14, A and B). Interestingly, both MYC-deficient DN3a and DN3b cells had higher levels of ROS (fig. S14C), associated with increased expression of CD5 (fig. S14D). Furthermore, DCA, which altered the development of αβ and γδ T cells (fig. S10B), increased the expression of ID3 in DN3 cells co-cultured with OP9-DL1 cells (fig. S14E). The expression of CD73, which is induced by TCR-ERK signaling to promote γδ T cell commitment (9), was markedly enhanced by treatment with DCA (fig. S14F) or Gal/GAO (fig. S14G). Collectively, these results indicate that the interplay between mTORC1, MYC, and ROS metabolism shapes the activity of the ERK/EGR1/ID3 signaling axis and, therefore, the strength of TCR signals in dictating lineage choices of DN3 cells (fig. S15).
Fig. 7. RAPTOR tunes the strength of the ERK/EGR1/ID3 signaling axis in fate choices.
(A) Immunoblot analysis of p-ERK, EGR1, ID3 and β-ACTIN in WT and Rptor–/– DN3 cells. (B and C) Plots of p-ERK (B) and expression of ID3 (C) in WT and Rptor–/– DN3 cells, with MFI plotted in graphs. (D) Cellularity of CD4+CD8+ DP cells from the indicated mice. Each symbol represents an individual mouse (WT, n = 4; Rptor–/–, n = 3; Erk2–/–, n = 4; Rptor–/–Erk2–/–, n = 4). (E) Flow cytometry of CD73 and CD24 expression on TCRγδ+ thymocytes in the indicated mice (left). Right, frequency and cellularity of CD73+ TCRγδ+ cells. Each symbol represents an individual mouse (WT, n = 4; Rptor–/–, n = 3; Erk2–/–, n = 4; Rptor–/–Erk2–/–, n = 4). (F) Expression of CD73 on TCRγδ+ cells in E shown as the fold change (MFI on WT cells is set to 1). Data are mean ± s.e.m. *P < 0.05, and ***P < 0.0001. One-way ANOVA with Turkey test (D to F). Data are representative of at least three independent experiments (A to F).
DISCUSSION
Despite the emerging interest in immunometabolism, our understanding of whether and how metabolic intermediates or pathways modulate immunity through signaling mechanisms remains limited. Here we report that mTORC1-mediated integration of metabolic and signaling activities dictates the lineage choices of αβ and γδ T cells, and identify mTORC1-dependent control of ROS production as a key metabolic signal in this process. Such developmental function of mTORC1 signaling depends upon its interplay with MYC in a feed-forward loop, but not other metabolic regulators such as HIF1α and SCAP. Our findings indicate that regulation of metabolic rewiring and redox homeostasis by mTORC1 occurs in the lineage-specifying stage, DN3 cells, which likely reflects the cell context-specific nature of this control mechanism. These results point to metabolic processes constituting a fundamental mechanism that connects extrinsic signals with transcriptional events and fate decisions.
Recent extensive studies have advanced our understanding of metabolic reprogramming of T cell-mediated immune responses (11, 12). However, little is known about the metabolic programs in thymocyte development, although NOTCH and IL-7 have been shown to exert trophic effects in this process (13, 14). Here we found that DN3 cells predominately use OXPHOS over glycolysis, whereas ISP cells have high activities of both OXPHOS and glycolysis. In contrast, DP cells markedly downregulate these metabolic activities. The use of distinct metabolic programs by developing thymocytes likely fulfills the bioenergenetic and signaling requirements of the underlying developmental processes. Of note, these dynamic metabolic programs are associated with differential regulation of mTORC1 activity. Loss of RAPTOR and mTORC1 activity disrupts anabolic metabolism that supports population expansion, but also results in dysregulated ROS production. Furthermore, perturbation of redox homeostasis impinges upon thymocyte fate decisions and mTORC1-associated phenotypes. Specifically, enhancing or attenuating ROS production has a direct impact on αβ and γδ T cell development, and antioxidant treatment rescues, albeit incompletely, the differentiation defect of mTORC1-deficient thymocytes. These results reveal a selective role for redox regulation in mediating cell fate decisions downstream of the multifunctional kinase mTORC1, and establish metabolic signaling as a decisive factor for fate decisions of αβ and γδ T cells in thymocyte development.
Our scRNA-Seq analysis further highlights the interplay between TCR signaling and metabolic activity. Among DN3a cells, clusters dominated by Rptor–/– cells upregulate NOTCH signaling, while in post-selection cells, they upregulate TCR and ERK signaling but show dampened anabolic metabolism. Such unbiased analysis also shows the upregulation of γδ T cell gene signature in the Rptor–/–-dominant cluster, which is further supported by the late pseudotime assignment. We have also attempted to directly assess γδ T cell commitment using in vitro single-cell progenitor assay, but the proliferative defect in Rptor–/– cells prevents such analysis. Nonetheless, blocking excessive ERK signaling substantially rectifies the increased generation of committed γδ T cells and expression of CD73 in Rptor–/– cells, supporting our model that increased signal strength due to RAPTOR deficiency promotes γδ T cell development.
A feature of the adaptive immune system is the intertwining cycles of quiescence and activation that accompany developmental and functional progression of T lymphocytes through their lifespan. Thymic colonization of largely nondividing progenitor cells is followed by two waves of proliferation separated by a brief period of quiescence at the DN3 stage required for TCRβ chain rearrangement (2). T cells return to quiescence at the DP stage, which allows for their TCRα chain rearrangement. This quiescent status is maintained by mature thymocytes and peripheral naïve T cells until encounter with cognate antigen. Our data suggest that RAPTOR-dependent mTORC1 activation and metabolic reprogramming drive the exit of DN3 cells from quiescence, as Rptor–/– T cells cannot upregulate cell size or trophic receptors. Moreover, RAPTOR-deficient thymocytes are less efficient in population expansion, but this defect is observed mainly in ISP cells. Given the intricate link between cell proliferation and differentiation (4), such proliferative defect likely contributes to the developmental blockade and accumulation of ISP cells. For DN3 and DN4 cells in Rptor–/– mice, although their proliferation is largely normal, they already exhibit defective expression of trophic receptors and signaling events. These results indicate that the impaired cycling of Rptor–/– ISP cells is likely a consequence, not the cause, of loss of RAPTOR-dependent metabolic reprogramming and quiescence exit. The dispensable role of SCAP, an important regulator of cell proliferation (28), in thymocyte development further supports this notion.
In summary, our study reveals that dynamic regulation of metabolic programs and mTORC1 activity is required for thymocyte development. In the peripheral immune compartment, mTOR signaling exerts discrete effects on the fate decisions of effector and memory CD8+ T cells (50), and effector and regulatory CD4+ T cells (51). Interestingly, metabolic remodeling used by lineage-committed mature T cells to drive cell fates and immune functions following antigen stimulation is also adopted by developing T cells to determine lineage commitment itself in response to developmental signals and microenvironmental cues. We provide further evidence that mTORC1 signaling intersects with MYC function and ROS regulation, and coordinates TCR and NOTCH signaling and anabolic metabolism in dictating thymocyte fates. Collectively, our study establishes mTORC1 signaling as a developmental checkpoint that links intrathymic environmental signals and metabolic programs to drive thymocyte development. Future work is warranted to further dissect cell context-dependent regulation and physiologic effects of metabolic and redox controls in immune system development and activation.
Materials and Methods
Study design
Genetically engineered mouse strains were used to assess the role of RAPTOR and metabolic pathways in regulating the fate decisions of T cell lineages in the thymus. Mice 6–10 weeks old were used unless otherwise noted. The development of thymocytes was analyzed by various methods, including flow cytometry analysis, cell sorting, microarray, glucose uptake, Seahorse (for glycolysis and oxidative phosphorylation), glycolytic flux, immunoblot, real-time PCR, sequencing of TCRγ chains, and scRNA-Seq. Details are described in Supplementary Materials and Methods. The sample size and experimental replicates are indicated in the figure legends. Group sizes were determined based on published literatures with similar experiments.
Mice
C57BL/6, CD45.1+, Scapflox, Erk2flox, Cd2-Cre, and Rag1–/– mice were purchased from the Jackson Laboratory. Rptorflox, Rhebflox, Hif1aflox, Rictorflox, and Mycflox mice have been described previously (30, 52–55). Littermates (Cre– flox/flox and Cre+ non-floxed mice) were used as WT controls. Mice at 6–10 weeks old were used unless otherwise noted. All mice were kept in specific pathogen-free conditions in Animal Resource Center at St. Jude Children’s Research Hospital (SJCRH). Animal protocols were approved by Institutional Animal Care and Use Committee of SJCRH.
Supplementary Material
Acknowledgements:
The authors acknowledge M. Hendren, S. Rankin, B. Rhode and C. Cloer for animal colony management; R. Cross, G. Lennon and P. Ingle for cell sorting; J.C. Zuniga-Pflucker for kindly providing the OP9-DL1 cells; D. Kioussis for kindly providing the Cd2-Cre mice; and H. Zeng and Y. Wang for critical reading of the manuscript.
Funding:
This work was supported by US National Institutes of Health (AI101407, AI105887, CA176624, CA221290, and NS064599 to H.C.; AI107625 AI121832 to P.G.T.; and CA021765 to G.N.), and Arthritis National Research Foundation (to K.Y.).
Footnotes
Competing interests:
The authors declare no competing financial interests.
Data and materials availability:
Microarray data are available at GEO under accession number GSE86495.
References
- 1.Yui MA, Rothenberg EV, Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol 14, 529–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carpenter AC, Bosselut R, Decision checkpoints in the thymus. Nat Immunol 11, 666–673 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciofani M, Zuniga-Pflucker JC, Determining gammadelta versus alphass T cell development. Nat Rev Immunol 10, 657–663 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Kreslavsky T et al. , beta-Selection-induced proliferation is required for alphabeta T cell differentiation. Immunity 37, 840–853 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciofani M, Knowles GC, Wiest DL, von Boehmer H, Zuniga-Pflucker JC, Stage-specific and differential notch dependency at the alphabeta and gammadelta T lineage bifurcation. Immunity 25, 105–116 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Garbe AI, Krueger A, Gounari F, Zuniga-Pflucker JC, von Boehmer H, Differential synergy of Notch and T cell receptor signaling determines alphabeta versus gammadelta lineage fate. J Exp Med 203, 1579–1590 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haks MC et al. , Attenuation of gammadeltaTCR signaling efficiently diverts thymocytes to the alphabeta lineage. Immunity 22, 595–606 (2005). [DOI] [PubMed] [Google Scholar]
- 8.Hayes SM, Li L, Love PE, TCR signal strength influences alphabeta/gammadelta lineage fate. Immunity 22, 583–593 (2005). [DOI] [PubMed] [Google Scholar]
- 9.Lee SY et al. , Noncanonical Mode of ERK Action Controls Alternative alphabeta and gammadelta T Cell Lineage Fates. Immunity 41, 934–946 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zarin P, Wong GW, Mohtashami M, Wiest DL, Zuniga-Pflucker JC, Enforcement of gammadelta-lineage commitment by the pre-T-cell receptor in precursors with weak gammadelta-TCR signals. Proc Natl Acad Sci U S A 111, 5658–5663 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pearce EL, Poffenberger MC, Chang CH, Jones RG, Fueling immunity: insights into metabolism and lymphocyte function. Science 342, 1242454 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollizzi KN, Powell JD, Integrating canonical and metabolic signalling programmes in the regulation of T cell responses. Nat Rev Immunol 14, 435–446 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciofani M, Zuniga-Pflucker JC, Notch promotes survival of pre-T cells at the beta-selection checkpoint by regulating cellular metabolism. Nat Immunol 6, 881–888 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Boudil A et al. , IL-7 coordinates proliferation, differentiation and Tcra recombination during thymocyte beta-selection. Nat Immunol 16, 397–405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi H, Regulation and function of mTOR signalling in T cell fate decisions. Nat Rev Immunol 12, 325–338 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K et al. , Vital roles of mTOR complex 2 in Notch-driven thymocyte differentiation and leukemia. J Exp Med 209, 713–728 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang F et al. , A critical role for Rictor in T lymphopoiesis. J Immunol 189, 1850–1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshii T et al. , Loss of mTOR complex 1 induces developmental blockage in early T-lymphopoiesis and eradicates T-cell acute lymphoblastic leukemia cells. Proc Natl Acad Sci U S A 111, 3805–3810 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Boer J et al. , Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol 33, 314–325 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Hoffman ES et al. , Productive T-cell receptor beta-chain gene rearrangement: coincident regulation of cell cycle and clonality during development in vivo. Genes Dev 10, 948–962 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Taghon T, Yui MA, Pant R, Diamond RA, Rothenberg EV, Developmental and molecular characterization of emerging beta- and gammadelta-selected pre-T cells in the adult mouse thymus. Immunity 24, 53–64 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A et al. , Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coffey F et al. , The TCR ligand-inducible expression of CD73 marks gammadelta lineage commitment and a metastable intermediate in effector specification. J Exp Med 211, 329–343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dash P et al. , Paired analysis of TCRalpha and TCRbeta chains at the single-cell level in mice. The Journal of clinical investigation 121, 288–295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dash P, Wang GC, Thomas PG, Single-Cell Analysis of T-Cell Receptor alphabeta Repertoire. Methods Mol Biol 1343, 181–197 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Tang F et al. , A critical role for Rictor in T lymphopoiesis. J Immunol 189, 1850–1857 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee K et al. , Vital roles of mTOR complex 2 in Notch-driven thymocyte differentiation and leukemia. J Exp Med 209, 713–728 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kidani Y et al. , Sterol regulatory element-binding proteins are essential for the metabolic programming of effector T cells and adaptive immunity. Nat Immunol 14, 489–499 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duvel K et al. , Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Molecular cell 39, 171–183 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi LZ et al. , HIF1alpha-dependent glycolytic pathway orchestrates a metabolic checkpoint for the differentiation of TH17 and Treg cells. J Exp Med 208, 1367–1376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finlay DK et al. , PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J Exp Med 209, 2441–2453 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Douglas NC, Jacobs H, Bothwell AL, Hayday AC, Defining the specific physiological requirements for c-Myc in T cell development. Nat Immunol 2, 307–315 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Wong GW, Knowles GC, Mak TW, Ferrando AA, Zuniga-Pflucker JC, HES1 opposes a PTEN-dependent check on survival, differentiation, and proliferation of TCRbeta-selected mouse thymocytes. Blood 120, 1439–1448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dose M et al. , c-Myc mediates pre-TCR-induced proliferation but not developmental progression. Blood 108, 2669–2677 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang CY, Bredemeyer AL, Walker LM, Bassing CH, Sleckman BP, Dynamic regulation of c-Myc proto-oncogene expression during lymphocyte development revealed by a GFP-c-Myc knock-in mouse. Eur J Immunol 38, 342–349 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Sena LA et al. , Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gerriets VA et al. , Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J Clin Invest 125, 194–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tussiwand R et al. , The preTCR-dependent DN3 to DP transition requires Notch signaling, is improved by CXCL12 signaling and is inhibited by IL-7 signaling. Eur J Immunol 41, 3371–3380 (2011). [DOI] [PubMed] [Google Scholar]
- 39.Washburn T et al. , Notch activity influences the alphabeta versus gammadelta T cell lineage decision. Cell 88, 833–843 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Tanigaki K et al. , Regulation of alphabeta/gammadelta T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity 20, 611–622 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Aifantis I, Gounari F, Scorrano L, Borowski C, von Boehmer H, Constitutive pre-TCR signaling promotes differentiation through Ca2+ mobilization and activation of NF-kappaB and NFAT. Nat Immunol 2, 403–409 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Voll RE et al. , NF-kappa B activation by the pre-T cell receptor serves as a selective survival signal in T lymphocyte development. Immunity 13, 677–689 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Vacca A et al. , Notch3 and pre-TCR interaction unveils distinct NF-kappaB pathways in T-cell development and leukemia. The EMBO journal 25, 1000–1008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michie AM et al. , Constitutive Notch signalling promotes CD4 CD8 thymocyte differentiation in the absence of the pre-TCR complex, by mimicking pre-TCR signals. International immunology 19, 1421–1430 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Stubbington MJT, Rozenblatt-Rosen O, Regev A, Teichmann SA, Single-cell transcriptomics to explore the immune system in health and disease. Science 358, 58–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Macosko EZ et al. , Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mingueneau M et al. , The transcriptional landscape of alphabeta T cell differentiation. Nat Immunol 14, 619–632 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Narayan K et al. , Intrathymic programming of effector fates in three molecularly distinct gammadelta T cell subtypes. Nat Immunol 13, 511–518 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu X et al. , Reversed graph embedding resolves complex single-cell trajectories. Nat Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Araki K et al. , mTOR regulates memory CD8 T-cell differentiation. Nature 460, 108–112 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng H, Chi H, mTOR signaling in the differentiation and function of regulatory and effector T cells. Current opinion in immunology 46, 103–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang K, Neale G, Green DR, He W, Chi H, The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nat Immunol 12, 888–897 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang K et al. , T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity 39, 1043–1056 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng H et al. , mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature 499, 485–490 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y et al. , Tuberous sclerosis 1 (Tsc1)-dependent metabolic checkpoint controls development of dendritic cells. Proc Natl Acad Sci U S A 110, E4894–4903 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu G, Yang K, Burns S, Shrestha S, Chi H, The S1P(1)-mTOR axis directs the reciprocal differentiation of T(H)1 and T(reg) cells. Nat Immunol 11, 1047–1056 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.