Abstract
Multimodal, imaging-genomics techniques offer a platform for understanding genetic influences on brain abnormalities in psychiatric disorders. Such approaches utilize the information available from both imaging and genomics data and identify their association. Particularly for complex disorders such as schizophrenia, the relationship between imaging and genomic features may be better understood by incorporating additional information provided by advanced multimodal modeling. In this study, we propose a novel framework to combine features corresponding to functional magnetic resonance imaging (functional) and single nucleotide polymorphism (SNP) data from 61 schizophrenia (SZ) patients and 87 healthy controls (HC). In particular, the features for the functional and genetic modalities include dynamic (i.e., time-varying) functional network connectivity (dFNC) features and the SNP data, respectively. The dFNC features are estimated from component time-courses, obtained using group independent component analysis (ICA), by computing sliding-window functional network connectivity, and then estimating subject specific states from this dFNC data using a k-means clustering approach. For each subject, both the functional (dFNC states) and SNP data are selected as features for a parallel ICA (pICA) based imaging-genomic framework. This analysis identified a significant association between a SNP component (defined by large clusters of functionally related SNPs statistically correlated with phenotype components) and time-varying or dFNC component (defined by clusters of related connectivity links among distant brain regions distributed across discrete dynamic states, and statistically correlated with genomic components) in schizophrenia. Importantly, the polygenetic risk score (PRS) for SZ (computed as a linearly weighted sum of the genotype profiles with weights derived from the odds ratios of the psychiatric genomics consortium (PGC)) was negatively correlated with the significant dFNC component, which were mostly present within a state that exhibited a lower occupancy rate in individuals with SZ compared with HC, hence identifying a potential dFNC imaging biomarker for schizophrenia. Taken together, the current findings provide preliminary evidence for a link between dFNC measures and genetic risk, suggesting the application of dFNC patterns as biomarkers in imaging genetic association study.
Keywords: Multimodal analysis, resting-state fMRI, schizophrenia, single nucleotide polymorphism, dynamic functional connectivity, parallel ICA
1. INTRODUCTION
Understanding how genetic risk factors are translated into clinical symptoms, disease progression and response to treatment of complex mental disorders, such as schizophrenia (SZ), is challenging. SZ is a serious, highly heritable (i.e. proportion of variance explained by genetic factors; SZ heritability rate is about 80% (Sullivan et al., 2003)) genetically complex disorder with disruptions in brain structure and function, and cognition (Ellison-Wright and Bullmore, 2010; Keshavan et al., 2011; Lichtenstein et al., 2009; Meda et al., 2012). Family and twin studies have shown evidence of moderate to high heritable component related to most psychiatric disorders (Kendler and Eaves,2005). The concordance rate is defined by the probability that a second twin will progress to a disorder given the first examined twin already has the disorder, and for monozygotic twins it has been found to be about 40%−50% for schizophrenia (Cardno and Gottesman, 2000). These results indicate that the unaffected twins might carry a heritable genetic risk for schizophrenia without expressing the disease.
An efficient strategy to unravel the genetic risk factors of SZ is through investigating the effects of genetic variations on intermediate phenotypes such as aberrant brain structure and function, as they are more related to biological mechanisms compared to behavioral measures. An intermediate phenotype or so-called “endophenotype” represents more biologically defined levels, such as cellular, neuronal level or neurocognitive measures, and is thought to reflect the genetic effects more prominently than disease entities such as SZ (Gottesman and Gould, 2003; Gottesman and Shields, 1972). Brain function measured by fMRI data has been proposed as an intermediate phenotype for genetic studies of mental illness, and patients with psychiatric disorders indeed present brain structural and functional idiosyncrasies, which may underlie the clinical manifestations. Such brain related idiosyncrasies in psychiatric disorders include significantly less gray matter and abnormal regional activations during cognitive task performance (Harris et al., 2006; Ivleva et al., 2012; Manoach et al., 2000; Monks et al., 2004). Moreover, disrupted patterns in resting state inter-network functional connectivity have been reported in a wide range of psychiatric disorders, including schizophrenia and bipolar disorder (Bassett et al., 2012; Calhoun et al., 2012; Jafri et al., 2008; Manoliu et al., 2013; Meda et al., 2012; Pearlson and Calhoun, 2009; Rashid et al., 2014), Alzheimer’s disease and mild cognitive impairment (Petrella et al., 2011) and dyslexia (Koyama et al., 2010). Interestingly, studies have suggested that resting state networks are heritable (Fu et al., 2015; Glahn et al., 2010). Further, a recent study on the UK Biobank participants suggested lower rate of heritability for connectivity edges, but higher heritability for independent component analysis (ICA) extracted connectivity components (Elliott et al.,2017). In particular, the default-mode network connectivity was reported to have a heritability of 0.42 (Glahn et al., 2010).
Recent resting state and task-based fMRI studies examining dynamic (i.e., time-varying) functional network connectivity (dFNC) offer additional information beyond conventional, static or average functional network connectivity (sFNC) (Calhoun et al., 2014; Hutchison et al., 2013; Preti et al., 2017; Rashid et al., 2016). The time-varying or dFNC approach focuses on identifying whole brain transient and recurring patterns in temporal coupling in the human brain (Allen et al., 2014; Calhoun et al., 2014; Rashid et al., 2014). The temporal dynamics of the identified components’ timecourses are commonly characterized by the sliding window approach to estimate the connectivity as moving between stable dFNC patterns or states (Allen et al., 2014; Sakoglu et al., 2010). These dFNC states have been shown to be stably present in the data, reoccurring over time, and disrupted by null models, providing further evidence of replicability of dFNC states (Abrol et al., 2016; Abrol et al., 2017; Miller et al., 2017).
Before the availability of low-cost genomic technologies, most genetic association studies focused on exploring candidate genes involvement in a target disease. The advance in technologies has made it possible to study the whole genome, a large portion of which have not been previously studied or understudied for a particular disease (Hirschhorn and Daly, 2005) and therefore, offers substantial potential for identifying cluster of genetic variants associated with complex genetic disorders (Hindorff et al., 2009). The conventional SNP or candidate gene analysis using predefined reference set of SNPs or genes only investigates the SNPs (or genes) of interest. Therefore, it cannot provide a comprehensive genetic risk profile for a complex trait like SZ. Genome-wide association studies (GWAS) can solve this problem, however, it will not shed light on the interplay among genetic variables unless additional analysis (e.g., pathway analysis) is employed for interpretation.
There has been a tremendous growth of interest in the application of multivariate, multimodal techniques, in areas such as imaging-genetics, to understand interactions between neuronal and genetic mechanisms of schizophrenia. The complexity of the genetics in schizophrenia strongly supports a genomic approach, i.e. an approach that can capture not only a single genetic effect but also the relationship between multiple genetic factors and phenotypic information. Imaging genomics method integrates genetic with structural and functional neuroimaging data to explore individuals with multiple genetic risk variants that relate to a psychiatric disorder. Multivariate multimodal techniques provide additional statistical power by extracting genetic and imaging data to capture the aggregated effect of multiple genetic factors and brain regions and assess the co-varying pattern of these data modalities (Calhoun and Sui, 2016; Chen et al., 2013; Liu and Calhoun, 2014). Multivariate data fusion techniques enable us to assess multiple variables simultaneously, and offer some additional benefits over the use of univariate techniques. The interpretation of results from multivariate data-fusion approaches is relatively simple due to co-varying nature of the variables (i.e., regions of interest or ROIs) as measures from patterns are explored rather than a huge number of univariate paired relationships (Calhoun and Sui, 2016). Further, using multivariate strategy replicability of the results can be improved. A number of multivariate data-fusion approaches have been proposed including parallel independent component analysis (pICA) (Liu et al., 2009; Pearlson et al., 2015), joint independent component analysis (jICA) (Calhoun et al., 2006), multiset canonical correlation analysis (mCCA) (Correa et al., 2007), partial least squares (PLS) (Chen et al., 2009), linked ICA (Groves et al., 2011), and coefficient constrained ICA (CC-ICA) (Sui et al., 2009).
The pICA approach, a data-driven technique, is constructed upon the conventional ICA algorithm to jointly conduct separate multivariate analysis in two different data modalities while also optimizing the inter-modality association (Liu and Calhoun, 2014).pICA has been applied to incorporate genetic and neuroimaging data including gray matter density, gray matter volume, and task-related activation (Gupta et al., 2014; Meda et al., 2014; Pearlson et al., 2015). In the context of imaging-genetics study, pICA uses higher order statistics to identify the independent brain networks, associated genetic variants, and their interrelationships (Jagannathan et al., 2010; Liu et al., 2009). Similar to the conventional ICA approach, pICA can identify hidden linear mixture of factors within the genetic and neuroimaging data, while also eliminating unwanted artifacts from the data. The results from the pICA provide independent components (ICs) from both data sets (neuroimaging and genetics) that are inter-linked while maximizing the linkage function in a joint estimation approach (Liu et al., 2009; Meda et al., 2010). There are three criteria that pICA optimizes simultaneously, including identifying a set of maximally independent functional neuroimaging factors, identifying a set of maximally independent genetic variants, and identifying the inter-modality linking relationship between the identified genetic and neuroimaging components. Furthermore, pICA also provides loading parameters for each component that reflect the component’s influence in individual subjects (Liu et al., 2009), and interrelationships between neuroimaging and genetic components can be statistically tested and compared between groups.
In the current work, we studied 61 SZ patients and 87 healthy controls with genome-wide single nucleotide polymorphism (SNP) and fMRI data following denoising and quality control (QC) assessment (where SNP data went through several QC steps, imputation and post-imputation QC steps as described in section 2.2 and Figure S1, and fMRI data went through pre- and post-processing steps as described in section 2.3 and Figure S2 to ensure data quality). We focused on feature based fusion analysis of an array of genetic variants (SNP) and functional (fMRI) images hypothesizing a significant correspondence between genomic factors and brain function. Our expectation is that the relationship between SNP patterns and time-varying functional connectivity is a more natural way to analyze the data which will improve our understanding of both genomic factors and functional connectivity measures. We estimate the functional features for neuroimaging data as subject-specific states that are revealed from the dynamic FNC data using a sliding window plus clustering approach (Allen et al., 2014; Rashid et al., 2014; Sakoğlu et al., 2010). The SNP data were first pre-filtered using results from an independent GWAS study to locate risk variants based on a large cohort. These risk loci were then simultaneously analyzed for multivariate associations with the derived functional features from fMRI data using the pICA data fusion algorithm. Further, we explored the association between genetic risk and SNP and functional connectivity features. Polygenic risk score (PRS) estimates the aggregate risk of a set of SNPs, by computing a linearly weighted sum of the genetic data where weights are derived from the effect sizes of the individual SNPs obtained from univariate analysis. This approach enables us to study genomic patterns grouped that co-vary across subjects with maximally independent dynamic FNC patterns. To guard against overfitting, the identified dFNC-SNP associations were evaluated with a permutation test.
2. MATERIALS AND METHODS
2.1. Participants
We used resting-state functional magnetic resonance imaging (fMRI) data and single nucleotide polymorphism (SNP) data obtained from 87 healthy controls (61 males, 26 females; mean age 36.83) and 61 age- and gender matched patients with SZ (52 males, 9 females; mean age 38.36) during eyes closed condition as part of the multi-site fBIRN project (Potkin and Ford, 2008). Informed consent was obtained from each participant prior to scanning in accordance with the Internal Review Boards of corresponding institutions.
2.2. SNP Data Preprocessing and Feature Selection
DNA was extracted from blood and saliva samples collected from the participants. No significant difference was noted in genotyping call rates between blood and saliva samples. DNA extraction was performed at the University of California at Irvine and genotyping was performed by Illumina (SD) using a custom made assay including the Infinium MEGAex chip as well all SNPs on the Psych chip. The SNP data went through quality control (QC), imputation and post-imputation QC as described in (Chen et al.,2018), which is briefly summarized here. PLINK (Purcell et al., 2007) was employed for pre- and post-imputation QC (Chen et al., 2012), including: (a) gender consistency check, (b) sample relatedness (not closer than second degree relatives), (c) genotyping call rate (>90% at both the individual and SNP level), (d) Hardy-Weinberg equilibrium in the control population (p <1 × 10 −6), (e) minor allele frequency (MAF > 0.05). Then imputation was conducted with SHAPEIT used for pre-phasing (Delaneau et al., 2011), IMPUTE2 for imputation (Marchini and Howie, 2010), and the 1000 Genomes data as the reference panel (Altshuler et al., 2012). Only markers with high imputation qualities (INFO score > 0.95) were retained. Missing calls were replaced using high linkage disequilibrium (LD) loci if available or otherwise removed. A total of 977,242 SNP loci were obtained after post-imputation QC and LD pruning (r2 < 0.9), and discrete numbers were then assigned to the categorical genotypes: 0 (no minor allele), 1 (one minor allele), and 2 (two minor alleles). Population structure was corrected with PCA (Price et al.,2006). A pre-filtering (i.e., pre-selection) step was conducted leveraging the Psychiatric Genomic Consortium SZ GWAS (Ripke et al., 2014) based on SZ relevance (i.e., the p- values reported by the PGC SZ GWAS). This resulted in a total of pre-filtered 1546 SNPs, discriminating patients from controls via a univariate SNP-wise test with p-values less than 5× 10−7 in the GWAS.
Imaging data processing and feature selection
2.3.1. fMRI Data Collection and Preprocessing
Imaging data was collected on 3T Siemens Tim Trio Systems (all but one site) or on a 3T General Electric Discovery MR750 scanner. Resting state fMRI scans were acquired using a standard gradient-echo echo planar imaging paradigm: FOV of 220 × 220 mm (64 × 64 matrix), TR = 2 s, TE = 30 ms, FA = 77°, 162 volumes, 32 sequential ascending axial slices of 4 mm thickness and 1 mm skip. Subjects had their eyes closed during the resting state scan.
Data processing was performed using a combination of toolboxes (AFNI, SPM, GIFT) and custom code written in Matlab. We performed rigid body motion correction using the INRIAlign (Freire and Mangin, 2001) toolbox in SPM to correct for subject head motion followed by slice-timing correction to account for timing differences in slice acquisition.
Then the fMRI data were despiked using AFNI’s 3dDespike algorithm to mitigate the impact of outliers. The fMRI data were subsequently warped to a Montreal Neurological Institute (MNI) template (Calhoun et al., 2017), and resampled to 3 mm3 isotropic voxels. Instead of Gaussian smoothing, we smoothed the data to 6 mm full width at half maximum (FWHM) using AFNI3s BlurToFWHM algorithm, which performs smoothing by a conservative finite difference approximation to the diffusion equation. This approach has been shown to reduce scanner specific variability in smoothness providing a “smoothness equivalence” to data across sites (Friedman et al., 2006). Each voxel time course was variance normalized by removing the mean and dividing by the standard deviation at each voxel prior to performing group independent component analysis as this has shown to better decompose subcortical sources in addition to cortical networks. Note that, no significant group difference in signal-to-noise ratio (SNR) was found (p=0.5, Figure S4).
2.3.2. Group ICA, Intrinsic Connectivity Network (ICN) Selection and Post processing
After preprocessing the data, functional data from both control and patient groups were analyzed using spatial group independent component analysis (GICA) framework as implemented in the GIFT software (Calhoun and Adali, 2012; Calhoun et al., 2001; Erhardt et al., 2011). Spatial ICA decomposes the subject data into linear mixtures of spatially independent components that exhibit a unique time course profile. A subject- specific data reduction step was first used to reduce 162 time point data into 120 direction of maximal variability using principal component analysis. Then subject-reduced data were concatenated across time and a group data PCA step reduced this matrix further into 100 components along directions of maximal group variability. Note that, group PCA on the entire dataset was performed to avoid biasing the solution to group differences (Erhardt et al., 2011). One hundred independent components were obtained from the group PCA reduced matrix using the infomax algorithm (Bell and Sejnowski, 1995). To ensure stability of estimation, we repeated the ICA algorithm 20 times in ICASSO, and aggregate spatial maps were estimated as the modes of component clusters (Ma et al., 2011). Subject specific spatial maps (SMs) and time courses (TCs) were obtained using the spatiotemporal regression back reconstruction approach (Calhoun et al., 2001; Erhardt et al., 2011) implemented in the GIFT software.
Subject specific SMs and TCs underwent post-processing as described in our earlier work (Allen et al., 2012). Briefly, we obtained one sample t-test maps for each SM across all subjects and thresholded these maps to obtain regions of peak activation clusters for that component; we also computed mean power spectra of the corresponding TCs. We visually inspected and identified a set of components as intrinsic connectivity networks (ICNs) if their peak activation clusters fell on GM (gray matter) and showed less overlap with known vascular, susceptibility, ventricular, and edge regions corresponding to head motion. We also ensured that the mean power spectra of the selected ICN time courses showed higher low frequency spectral power. This selection procedure resulted in 47 ICNs out of the 100 independent components obtained. The ICNs were assessed and distributed into sub-cortical (SC), auditory (AUD), visual (VIS), sensorimotor (SM), attention/cognitive control (CC), default-mode (DMN), and cerebellar (CB) network domains.
Subject-specific TCs corresponding to the retained ICNs underwent additional post processing steps. The TCs were detrended to remove any existing linear, quadratic or cubic low frequency trends originating from scanner drift, orthogonalized with respect to estimated subject motion and realignment parameters, and despiked using AFNI’s 3dDespike function to replace outlier points with values estimated from third order spline fit to neighboring portions of the TCs.
2.3.3. FC Estimation and Clustering
Similar to our previous work (Damaraju et al., 2014) time-varying FNC was estimated by sliding a window of length 22 TRs (44 s) in steps of 1 TR (2 s). This sliding window analysis used a tapered window generated by convolving a rectangular window of length 22 TRs (44 s) with a Gaussian window of standard deviation equal to 3 TRs. To characterize the full covariance matrix, we estimated covariance from the regularized precision matrix or the inverse covariance matrix (Smith et al., 2011). Following the graphical LASSO method (Friedman et al., 2008), we placed a penalty on the L1 norm of the precision matrix to promote sparsity. The regularization parameter lambda was optimized separately for each subject by evaluating the log-likelihood of unseen data (windowed covariance matrices from the same subject) in a cross-validation framework. Final dynamic FC estimates for each window, were concatenated to form a C x C x W array representing the changes in covariance (correlation) between components as a function of time.
Next, we selected windows of higher variability as subject exemplars and used K-means clustering to obtain group centroids. We determined the number of clusters to be five using the elbow criterion of the cluster validity index, which is computed as the ratio between within-cluster distances to between-cluster distance. These centroids are then used as starting points to cluster all of the dynamic FNC data. Group- and subject- specific centroids were computed for further analyses (Allen et al., 2014).
2.4. Proposed pICA framework
Figure 1 and Figure S5 provides an illustration of the proposed framework. pICA was performed through the Fusion ICA Toolbox (FIT, http://mialab.mrn.org/software/fit) using the imaging (dynamic states) and genomic (SNP array) features. The algorithm was configured with a threshold of 0.25 for constrained correlations to avoid false positive associations and to only constrain one pair of components (Liu et al., 2009). A SNP component is defined by large clusters of functionally related SNPs statistically correlated with phenotype components, whereas a dFNC component is defined by clusters of related connectivity links among distant brain regions distributed across 5 discrete dynamic states, and statistically correlated with genomic components (Table S2). An endurance parameter was set to −5 × 10−4 to control the decreasing slope of the entropy term and to mitigate against over fitting. For the fMRI modality (dFNC features), the component number was predefined as 15, and for genomic modality (SNP data) the component number was predefined as 25 based on pICA model stability, which was validated using the permutation test. The parallel ICA estimates components by maximizing component independence within each data modality, and maximizing the inter-modality association assessed based on component loadings from the two modalities. In the current work, parallel ICA extracted 15 dFNC components and 25 SNP components, which led to a total of 375 SNP-dFNC pairs for which the associations were assessed. Significant SNP-dFNC associations were then identified based on Bonferroni correction (p < 0.05/375).
Figure 1: An overview of proposed imaging genetics approach.
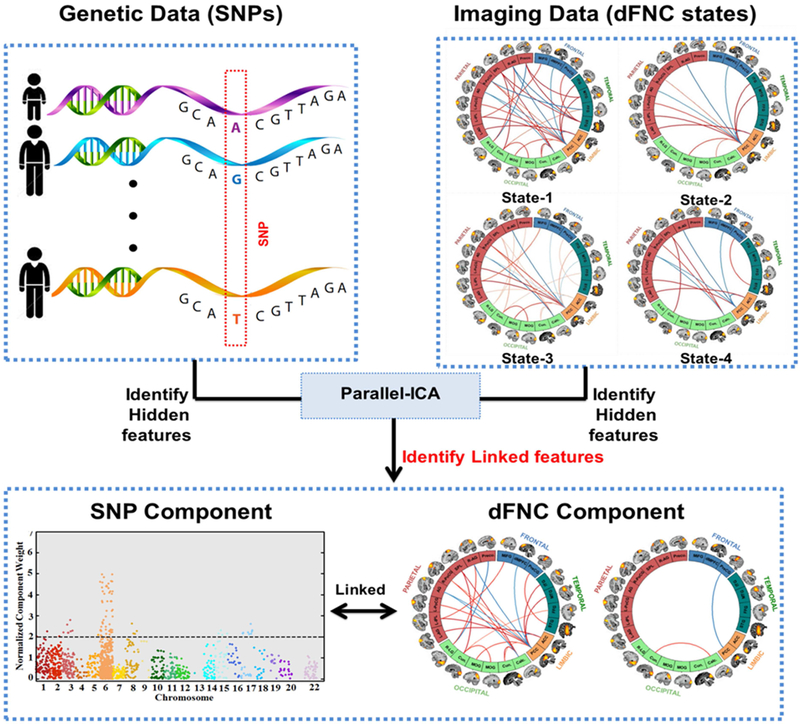
Genomic features (preselected SNPs) and imaging features (dynamic connectivity states estimated using sliding-window k-means clustering approach) are used in a parallel independent component analysis (pICA) framework, and the component showing significant association between genomic and imaging features is identified.
We performed a 1000-run permutation test to assess the validity and stability of identified dFNC-SNP association by investigating the occurrences of inter-modality correlations by chance in permuted dFNC and SNP datasets (Chen et al., 2012). The null distribution was constructed with the top correlation obtained from each test run. We then counted the instances with correlations greater than that observed from the original data and calculated the two-tail probability as the significance level.
2.5. Polygenic Risk Score Analysis
Additionally, we computed the polygenic risk score (PRS) for SZ of the set of thresholded SNPs (i.e., top SNPs after thresholding the SNP component), which was a linearly weighted sum of the genotype profiles with weights derived from the odds ratios of the PGC SZ GWAS (Purcell, 2009; Ripke et al., 2014). The loading parameters from significant pICA paired components were then assessed for associations with PRS for SZ using Pearson’s and Spearmann’s rank order correlations.
2.6. Partial Replication Study
While an independent dataset with exact imaging and genomic features was unavailable for replication purpose, we implemented our proposed model on a separate, independent dataset for partial replication. The resting-state fMRI data were collected while the participants had their eyes open (as opposed to our main analyses where resting-state fMRI data were collected in eyes closed condition). We used resting-state functional magnetic resonance imaging (fMRI) data and single nucleotide polymorphism (SNP) data obtained from a local COBRE (center of biomedical research excellence) study. After preprocessing, we obtained data from a total of 135 participants (72 healthy controls and 63 SZ patients matched for age) for which both fMRI and SNP data were collected.
Using the same steps mentioned for our main analyses, we preprocessed both fMRI and SNP data. As the genomic features, we identified the same 1546 pre-selected SNPs as our main analyses. In order to identify the similar imaging features (i.e., dFNC states), we implemented the following two-step replication: (i) using a spatially constrained ICA approach, we computed subject specific maps and timecourses for the 49 ICNs and computed the sliding-window based dynamic connectivity matrices for each subject, and (ii) using k-means clustering approach while using the original dFNC states (i.e., from our main study) as stating points for clusters, we identified 5 cluster centroids/dFNC states for each subject. These dFNC states were then used as the imaging features for our proposed pICA framework.
The proposed algorithm proceeds in two stages, consisting of independent component computation at group and subject levels, respectively. First, a group-level ICA is performed to extract information in the form of group ICs from preprocessed fMRI data of multiple subjects. Then the group information is used as guidance for the subject-level ICA, computed separately on the fMRI data of each subject (Du and Fan, 2013; Du et al., 2015; Lin et al., 2010).
A list of key acronyms and definitions can be found in Tables S1 and S2.
3. RESULTS
Figure 2 demonstrates the dFNC states for HC and SZ participants. The pICA framework identified one dFNC-SNP component pair with (i) significant correlation between the functional and genetic components, (ii) significant group difference in the functional component loading scores, and (iii) significant correlations between polygenic risk scores (PRS) and functional and genetic components. Figure 3 and Figure S3 show the significant component pair, with the connectivity strengths for the parallel co-varying functional component’s inter-regional links or connections and Manhattan plot showing the SNP component. In order to display the top 5% of the connectivity links and SNPs, only links with connectivity strengths and SNPs with genomic component weights higher or lower than a cutoff value are shown. In particular, after converting to z-scores the genomic (SNP) component is thresholded at |z|>2 to retain only the top 5% SNPs, whereas the inter-regional connectivity strengths from the functional component, are thresholded at |z|>3 to retain only the top 5% connectivity links. For both functional and genetic components, we will hereafter refer to the retained inter-regional connections and top SNPs as “top links” and “top SNPs”, respectively.
Figure 2: Dynamic connectivity states from clustering approach for k=5.
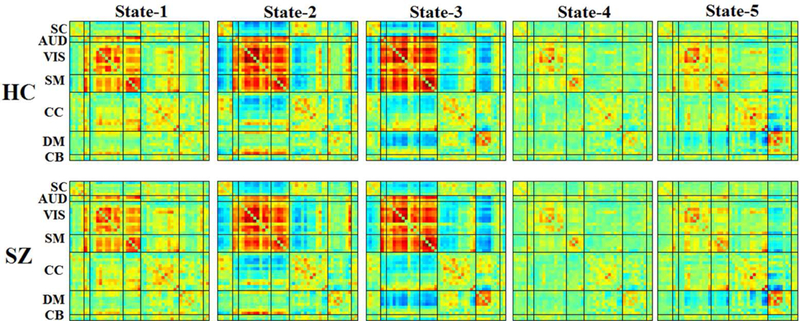
Group- specific centroids of the dynamic states for healthy (HC) and schizophrenia (SZ) are obtained from k-means clustering.
Figure 3: Parallel ICA component.
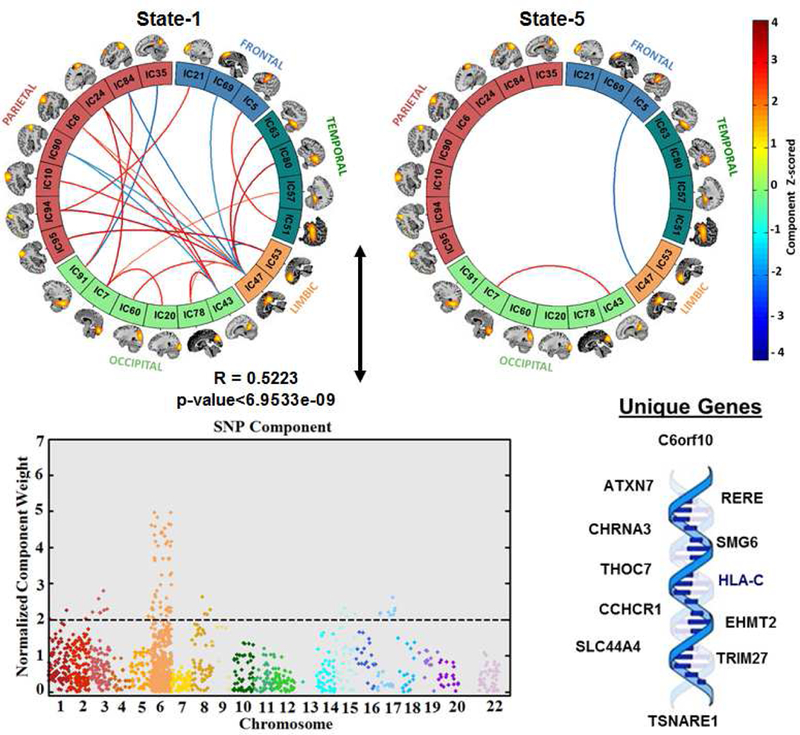
Results from pICA showing significantly correlated dFNC component (top), and SNP component (bottom). For the dFNC component the significant links (i.e., functional connections with highest connectivity strengths thresholded using z-scores of connectivity strengths: |z| > 3) and their connectivity strengths for the functional component are shown. For the SNP component, a Manhattan plot highlighting the thresholded SNPs at |z| > 2, and the unique genes that these thresholded SNPs are mapped into are shown.
3.1. Parallel Significant Component
As illustrated in Figure 3 and Figure S3, the number of significant connections in the linked functional component were high for the temporal, parietal, limbic and occipital regions in state 1, whereas state 5 only had two significant connections, and state 2, 3 and 4 had none. The peak activation coordinates of the significant links identified in state 1 and state 5 are presented in Table 4. In terms of occupancy rate, state 1 was dominated by the healthy subjects (HC=22%; SZ=14%), whereas state 5 was dominated by schizophrenia patients (HC=16%; SZ=27%). Note that, a positive connectivity is defined as the positive correlation strength between two brain networks (ICNs), and depicted with red color within the connectogram plots (i.e., the greater the positive connectivity strength is, the darker red color is used as per the color bar), whereas a negative connectivity is defined as the negative or anti-correlation strength between two brain networks (ICNs), and depicted with blue color within the connectogram plots (i.e., the greater the negative connectivity strength is, the darker blue color is used as per the color bar)
Table 4:
Peak activations of ICN spatial maps related to significant links identified from pICA framework. For more information on ICNs, see (Damaraju et cl., 2014).
| BA | Nv | Tmax. | Coordinate | |
|---|---|---|---|---|
| Auditory | ||||
| IC51 | ||||
| R Middle T emporal Gyrus | 21 | 440 | 32.6 | 63, −15, −9 |
| L Middle T emporal Gyrus | 21 | 300 | 30.4 | −60, −18, −6 |
| Visual | ||||
| IC57 | ||||
| L Parahippocampal Gyrus | 37 | 234 | 37.5 | −24, −45, −12 |
| R Parahippocampal Gyrus | 37 | 206 | 39.3 | 30, −45, −12 |
| IC7 | ||||
| R Cuneus | 17 | 855 | 54.4 | 3, −84, 6 |
| IC20 | ||||
| R Middle Frontal Gyrus | 10 | 332 | 31.2 | −30, −93, −6 |
| L Superior Frontal Gyrus | 10 | 278 | 29.4 | 30, −90, 5 |
| IC78 | ||||
| R Cuneus | 18 | 949 | 29 | 3, −87, 21 |
| IC43 | ||||
| R Calcarine Gyrus | 30 | 952 | 23.2 | 15, −63, 9 |
| IC24 | ||||
| R Superior Parietal Lobule | 7 | 768 | 25.3 | −32, −88, −1 |
| IC91 | ||||
| R Lingual Gyrus | 19 | 277 | 29.9 | 27, −66, −6 |
| IC60 | ||||
| R Precuneus | 19 | 357 | 39.8 | 30, −78, 33 |
| L Cuneus | 19 | 278 | 33.6 | −27, −78, 27 |
| IC80 | ||||
| R Middle T emporal Gyrus | 22 | 414 | 38.4 | 54, −51, 12 |
| L Middle T emporal Gyrus | 22 | 185 | 28.1 | −54, −51, 9 |
| Sensorimotor | ||||
| IC6 | ||||
| Right Postcentral Gyrus | 3 | 622 | 39.8 | 42, −21, 54 |
| IC10 | ||||
| L Precentral Gyrus | 4 | 598 | 39.6 | −36, −24, 51 |
| IC5 | ||||
| R Precentral Gyrus | 6 | 328 | 47.4 | 54, −6, 27 |
| L Precentral Gyrus | 6 | 288 | 49.6 | −54, −9, 30 |
| Cognitive Control | ||||
| IC63 | ||||
| L Fusiform Gyrus | 37 | 236 | 35.6 | −42, −57, −12 |
| R Fusiform Gyrus | 37 | 89 | 28.1 | 45, −54, −12 |
| IC35 | ||||
| R Middle Frontal Gyrus | 10 | 332 | 31.2 | 33, 54, 12 |
| L Superior Frontal Gyrus | 10 | 278 | 29.4 | −33, 45, 21 |
| IC47 | ||||
| Cingulate Gyrus | 23 | 621 | 47.4 | 0, −36, 27 |
| IC94 | ||||
| R Inferior Parietal Lobule | 40 | 374 | 34.9 | −42, −42, 45 |
| IC21 | ||||
| R Middle Frontal Gyrus | 10 | 332 | 31.2 | 33, 54, 12 |
| L Superior Frontal Gyrus | 10 | 278 | 29.4 | −33, 45, 21 |
| Default-mode | ||||
| IC84 | ||||
| R Angular Gyrus | 40 | 443 | 45.8 | 51, −60, 39 |
| IC90 | ||||
| R Angular Gyrus | 39 | 213 | 36.1 | 45, −75, 30 |
| L Superior Occipital Gyrus | 19 | 89 | 26.6 | −36, −81, 30 |
| IC53 | ||||
| L Anterior Cingulate Gyrus | 32 | 742 | 43.5 | −3, 48, 12 |
| IC69 | ||||
| R Medial Frontal Gyrus | 8 | 443 | 45.8 | 3,42, 45 |
| IC95 | ||||
| L Angular Gyrus | 40 | 555 | 43 | −48, −63, 42 |
BA=Brodmann area; Nv=number of voxels in each cluster; Tmax.=maximum t-statistic in each cluster; Coordinate = max coordinate (mm) in MNI space, following LPI convention.
The significant SNP component was first z-scored and then absolute z-scored values were used to represent the normalized component weight. The SNP component consists of peak weights of SNPs mostly located in chromosome 6 (and few at chromosomes 1, 2, 3,5, 15 and 17). The significant SNP component was predominantly contributed to by 80 SNPs (Table 5; top 5% based on absolute values of the component weights). 24 SNPs were mapped to 13 unique genes using UCSC hg19 assembly (http://hgdownload.cse.ucsc.edu/), while the rest were from inter-genic regions.
Table 5:
Top 80 SNPs ID, chromosome number (Chr. #) and base position
| SNP ID | Chr. # | Base Position |
|---|---|---|
| rs10779702 | chrl | 8423510 |
| rs3765971 | chrl | 8445360 |
| rs6709720 | chr2 | 57967563 |
| rs832190 | chr3 | 63842629 |
| rs704373 | chr3 | 63867355 |
| rs56293138 | chr3 | 63888935 |
| rs59971314 | chr3 | 63914618 |
| rs7615475 | chr3 | 63926661 |
| rs3774720 | chr3 | 63951765 |
| rs13272 | chr3 | 63986170 |
| rs13160798 | chr5 | 152276252 |
| rs150082944 | chr6 | 28728722 |
| rs115003944 | chr6 | 28734806 |
| rs115544230 | chr6 | 28742340 |
| rs116558610 | chr6 | 28773983 |
| rs148659564 | chr6 | 28817300 |
| rs138562260 | chr6 | 28854772 |
| rs115018585 | chr6 | 28862617 |
| rs115673158 | chr6 | 28877773 |
| rs2233956 | chr6 | 31081205 |
| rs3130566 | chr6 | 31102618 |
| rs114523252 | chr6 | 31116636 |
| rs115997058 | chr6 | 31142265 |
| rs139358712 | chr6 | 31147476 |
| rs150576357 | chr6 | 31149520 |
| rs150602567 | chr6 | 31154434 |
| rs114702079 | chr6 | 31154493 |
| rs116340174 | chr6 | 31161577 |
| rs114974300 | chr6 | 31163638 |
| rs149973721 | chr6 | 31166352 |
| rs115250945 | chr6 | 31178279 |
| rs116311494 | chr6 | 31178328 |
| rs115883612 | chr6 | 31183509 |
| rs141197791 | chr6 | 31184877 |
| rs114272705 | chr6 | 31191070 |
| rs139837897 | chr6 | 31195996 |
| rs115761897 | chr6 | 31197074 |
| rs114504365 | chr6 | 31200764 |
| rs145495788 | chr6 | 31201357 |
| rs116254022 | chr6 | 31202680 |
| rs114151693 | chr6 | 31220273 |
| rs112684598 | chr6 | 31228634 |
| rs2524108 | chr6 | 31232451 |
| rs114031082 | chr6 | 31237061 |
| rs114700001 | chr6 | 31251477 |
| rs116824795 | chr6 | 31258255 |
| rs115612789 | chr6 | 31271700 |
| rs149248545 | chr6 | 31326324 |
| rs2395475 | chr6 | 31326920 |
| rs2523587 | chr6 | 31327400 |
| rs116778584 | chr6 | 31335901 |
| rs2763981 | chr6 | 31840021 |
| rs652888 | chr6 | 31851234 |
| rs142520578 | chr6 | 31861815 |
| rs114425641 | chr6 | 32200670 |
| rs116153975 | chr6 | 32226126 |
| rs143312186 | chr6 | 32229238 |
| rs150597224 | chr6 | 32237221 |
| rs145499705 | chr6 | 32237260 |
| rs116522184 | chr6 | 32237463 |
| rs115430867 | chr6 | 32247710 |
| rs116105456 | chr6 | 32252507 |
| rs114513253 | chr6 | 32270283 |
| rs116434681 | chr6 | 32298484 |
| rs10957102 | chr8 | 60544006 |
| rs7465167 | chr8 | 143315080 |
| rs8180995 | chr8 | 143326237 |
| rs10875482 | chr8 | 143330385 |
| rs564585 | chr15 | 78886227 |
| rs7359276 | chr15 | 78892661 |
| rs11637630 | chr15 | 78899719 |
| rs8042059 | chr15 | 78907859 |
| rs1532292 | chr17 | 2097483 |
| rs11078865 | chr17 | 2109109 |
| rs9906500 | chr17 | 2129210 |
| rs9893527 | chr17 | 2164210 |
| rs170044 | chr17 | 2197502 |
| rs216200 | chr17 | 2199846 |
| rs2224770 | chr17 | 2205923 |
| rs391300 | chr17 | 2216258 |
| rs114151693 | chr6 | 31220273 |
The correlation between the significant (i.e., after the correlations identified by pICA which were controlled for diagnosis were corrected for multiple comparisons), parallel co-varying dFNC and SNP components was: Pearson’s r= 0.5223 (p< 6.95 × 10-9) and Spearmann’s: r = 0.4952 (p < 6.95 × 10-9). In a 1000-run permutation test, the absolute values of the dFNC-SNP top correlations ranged from 0.15 to 0.67 with a median of 0.26, yielding a p-value of 1.1 × 10-2.
3.2. Group Differences in Component Loadings
Figure 4a presents the scatterplot of the significant parallel components’ loading parameters, with a pattern of positively increasing relationship between these two loading parameters. Figures 4b and 4c show the group-wise loading parameters in healthy control and schizophrenia populations estimated from dFNC and SNP components, respectively. The ANOVA showed a significant group difference in the dFNC loading parameter (p=0.008), with a lower dFNC mean loading in the SZ group (mean=0.0066, standard deviation=0.0159) compared to the control group (mean=0.0152, standard deviation=0.0212). There was no significant group difference in the SNP loading parameter (p=0.243), although the group mean SNP loading was lower in the SZ group (mean=−0.0265, standard deviation=0.0977) compared to the healthy group (mean=- 0.0069, standard deviation=0.1012). This validates our pICA approach as typically between group effect sizes for imaging data are much larger than those for genomic data.
Figure 4: Loading parameter from the significant parallel ICA component.
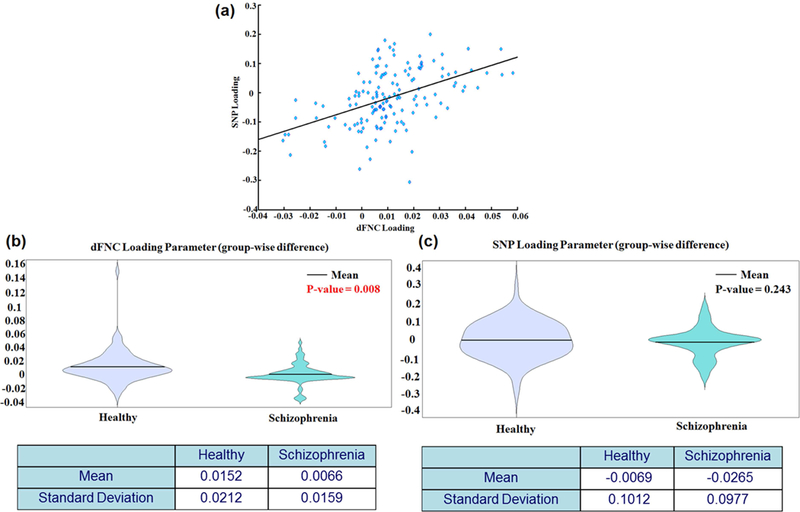
Scatterplot showing dFNC loading parameters versus SNP loading parameters from the significant pICA component (top), group-wise violin plots of dFNC loading parameters (middle, left) and SNP loading parameters (middle, right).
3.3. Risk Score and Component Loadings
We also assessed the correlations between the polygenic risk scores (PRS) and both dFNC and SNP components’ loading parameters. The PRS was computed from the top SNPs (i.e., 80 SNPs after thresholding the SNP component at |z|>2)) of the linked SNP component. The PRS computed using the top SNPs ranged between 0 and −8. The scatterplots of PRS versus component loadings are shown in Figure 5. The correlation between the risk scores and dFNC loadings was: Pearson’s r= −0.26 (p=0.0017); Spearmann’s r=−0.2743 (p=7.7230e-4), whereas the correlation between the risk scores and SNP loadings was: Pearson’s r= −0.5401 (p=1.4×10-12); Spearmann’s r=−0.5406 (p =< 6.95 × 10-9). Results show that, as the risk of SZ increases, the loading coefficients associated with the dFNC component decrease, suggesting that the SZ patients tend to under-utilize the vast amount of connections available through the brain regions, particularly SZ patients have the poor utilization of the top links in state 1 (mostly among temporal, parietal, limbic and occipital regions, see Figure S3 and Table 4 for more details). Interestingly, the SZ group showed low occupancy rate in state 1 compared to the HC group (HC=22%; SZ=14%) suggesting lower occupancy of that state may be a marker of schizophrenia risk.
Figure 5: Component loading parameters and polygenic risk scores.
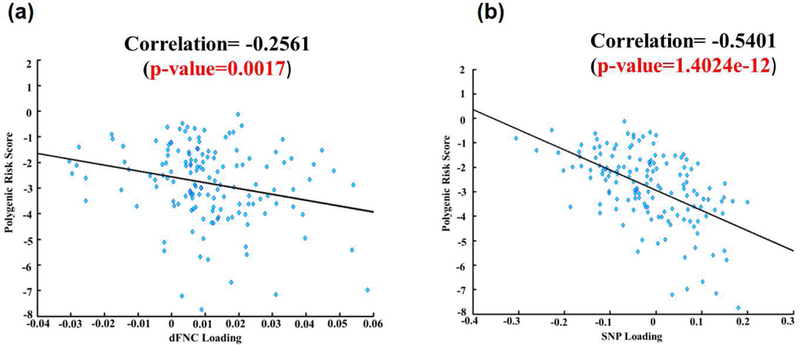
Scatterplots showing polygenic risk scores versus (a) dFNC loading parameters and (b) SNP loading parameter for the significant pICA component.
3.4. Pathway Analysis Using Top Genes
We examined the genetic architecture of our findings using Reactome Pathway Analysis (RPA: https://www.reactome.org), where the 13 genes were compared with the preselected SNPs as background. The results obtained from RPA are provided in Table 2. Further, GeneMANIA (Mostafavi et al., 2008) was used to identified the interactions among the query genes. We also analyzed our query genes to find the biological functions of these top genes, which are provided in Table 3.
Table 2:
Reactome Pathway Analysis using the top genes
| Canonical Pathway | P-value | Gene Name |
|---|---|---|
| Neuronal System | 0.604202 | CHRNA3 |
| Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell | 0.313669 | CHRNA3 |
| Transmission across Chemical Synapses | 0.431053 | CHRNA3 |
| Immune System | 9.28E-05 | HLA-C |
| Metabolism | 0.064322 | SLC44A4, ATXN7 |
| Gene Expression | 0.866556 | EHMT2, THOC7, SMG6 |
Table 3:
Biological functions of the top genes
| Functions | P-value | Gene Name |
|---|---|---|
| Neurological Disease, Psychological Disorders | 4.64E-02 | CHRNA3 |
| Hereditary Disorder, Neurological Disease | 6.78E-04 | ATXN7 |
| Molecular Transport, RNA Trafficking | 3.10E-04 | SMG6, THOC7 |
| Visual System Development and Function | 4.45E-02 | ATXN7 |
| Cell Morphology, Embryonic Development | 6.76E-03 | CCHCR1 |
| Nervous System Development and Function, Tissue Morphology | 1.55E-02 | ATXN7 |
| Developmental Disorder, Neurological Disease | 3.34E-02 | SMG6 |
| Immunological Disease | 1.08E-02 | RERE |
| Immunological Disease, Inflammatory Disease, Inflammatory Response | 6.78E-04 | HLA-C |
3.5. Parallel Significant Component From Partial Replication Study
The results from pICA framework captured a significant imaging (dFNC) and genomic (SNPs) component pair with a correlation of r= 0.22 (p=0.0099). The correlation between the main (fBIRN) and replication dataset () SNPs components was 0.28 (p<0.0005). However, the correlation between dFNC components was not significant (r= 0.01,p=0.46). We speculate that, due to different imaging data acquisition conditions (eyes closed versus eyes open) (Wu et al., 2010), we were unable to replicate the imaging part of the pICA component. Future work should implement replication using an independent dataset with similar features in order to systematically verify our findings from this pilot study.
In summary, here we have investigated the genomic underpinnings of dysfunctional dynamic FNC in SZ. A multivariate approach, pICA, was used to extract SNP and dFNC components, and retrieve intermodality associations. We investigated the relationship between genomic data and time-varying FNC measures and explored characteristic brain abnormalities with regard to schizophrenia. By employing a novel approach, we first estimated relatively lower-dimensional feature spaces from the high-dimensional fMRI and SNP array data. Next, we performed a multivariate data fusion approach on these estimated lower-dimensional feature spaces by implementing a pICA approach to extract corresponding co-varying genetic and functional brain components and evaluated associations between the components. Due to the limited sample size compared to genome-wide SNPs, we preselected 1546 SZ risk loci based upon large-scale GWAS in the published PGC study (Ripke et al., 2014) to focus the dFNC-SNP association analysis on polymorphisms likely relevant to SZ. One significant dFNC-SNP pair was identified and the permutation test indicated a low possibility that the observed correlation was due to overfitting, though the current result still awaits further independent validation.
4. DISCUSSION
In this study, we have investigated the relationship between neuroimaging and genomic features in patients with SZ. In particular, our results captured one pICA component pair that exhibited significant correlation between their underlying modality-specific components, highlighting significant group differences in time-varying FNC component loadings, and significant negative correlations between polygenic risk scores (PRSs) and both dFNC and SNP component. The pICA dFNC and SNP components showed a significant positive correlation of r=0.5223 (p<6.95e−09), as depicted in Figure 4a. This implies that, individuals that load more heavily on the top SNPs (and therefore the unique genes that these SNPs are map into) show more increased (or more decreased) functional connectivity among brain networks that underlie the functional component capturing top positive (and negative) connectivity strengths (i.e., top links in state 1 and 5).
Substantial correspondence with previously reported studies in literature could also be drawn for few evaluated top inter-regional (i.e., inter-ICN) links in the estimated functional components. First, the functional component for the retained parallel source showed top connectivity links in time-varying connectivity states 1 and 5. Most of the top inter-ICN links are captured in state 1, a state more often occupied by healthy individuals, where both positive and negative connectivity strengths across various network domains can be observed (Figure 3). In particular, a CC domain component, IC47 (cingulate gyrus), showed inter-ICN link with components from several domains including VIS (IC60: right precuneus/left cuneus; IC78: right cuneus and IC 80: middle temporal gyrus), SM (IC63: fusiform gyrus; IC6: right postcentral gyrus and IC10: left precentral gyrus) and DMN (IC53: left anterior cingulate gyrus; IC69: right medial frontal gyrus; IC84: right angular gyrus and IC90: right angular gyrus). Additionally, in this state, ICNs in the DMN domain showed connectivity within themselves and with ICNs from CC and VIS domains as well. For example, one of the DMN ICNs, IC84, highlighted by the brain regions in right angular gyrus (parietal component), showed positive connectivity weight with an ICN from the VIS domain characterized by calcarine gyrus (IC91, occipital component) and negative connectivity weight with a CC ICN, cingulate gyrus (IC47, limbic component). Another DMN ICN, IC90, encompassing the right angular gyrus also showed negative connectivity weights with two ICNs from the VIS and CC domains, right calcarine gyrus (IC43, occipital component) and cingulate gyrus (IC47, limbic component). Moreover, two DMN ICNs, IC53 (left anterior cingulate gyrus, limbic component) and IC95 (left angular gyrus, parietal component) have shown positive connectivity weight between themselves. These results are in line with a previous reports that have shown aberrant connectivity patterns within regions including angular gyrus, calcarine gyrus and cingulate sub-regions in schizophrenia patients (Rashid et al., 2014; Wang et al., 2015). Indeed, the roles of angular gyrus in language processing, memory and social cognition have been extensively reported (Binder et al., 2009; Clos et al., 2014; Hall et al., 2005). Disrupted DMN connectivity in schizophrenia patients has been extensively reported by several recent studies (Garrity et al., 2007; Ongur et al., 2010; Rashid et al., 2014). Thus, further follow-up examination of these top links involving the DMN ICNs is warranted. Interestingly, four temporal lobe components, IC63 (fusiform gyrus), IC80 (middle temporal gyrus), IC51 (middle temporal gyrus) and IC57 (parahippocampal gyrus) showed connectivity with limbic (IC47: cingulate gyrus), frontal (IC5: precentral gyrus) and occipital (IC7: right cuneus) components. Previous studies have reported disrupted temporal lobe connectivity in schizophrenia (Alonso-Solis et al., 2015; Barta et al., 1990; £etin et al., 2014; Ford et al., 2002; Peters et al., 2016; Wolf et al., 2007), suggesting that components within the temporal lobe play an important role in schizophrenia disease etiology. However, further investigation is required to validate the role of temporal lobe dysfunction in schizophrenia with regard to the current imaging-genomics framework.
Dynamic connectivity state 1 also captured a CC ICN, left precuneus (IC35, parietal component) showing negative connectivity weight with a VIS component, right lingual gyrus (IC91, occipital component). Patterns of disrupted connectivity in schizophrenia within the precuneus and lingual gyrus have been reported (Kühn and Gallinat, 2011; Rashid et al., 2014; Zhu et al., 2017). Precuneus is involved in episodic memory (Rugg and Henson, 2002), mental imagery recall (Fletcher et al., 1996), and self-processing operations (Cavanna and Trimble, 2006), whereas lingual gyrus is involved in visual processing (Bogousslavsky et al., 1987). In state 2, positive connectivity weight between two VIS domain ICNs, right cuneus (IC7, occipital component) and right calcarine gyrus (IC43, occipital component), and negative connectivity weight between a CC domain ICN, cingulate gyrus (IC47, limbic component) and a SM domain component, precentral gyrus (IC5, frontal component) have been observed. While we evaluate only a few interesting links within the scope of the current work, there is much more that could be done to explore these results to further expand our understanding of the genomic- connectivity relationships and further contribute to characterization of schizophrenia.
While inter-modality association is assessed based on loading vectors, interpretation of source of variance is based on component scores. Figure S3(a) demonstrates the top connectivity features obtained with thresholding the dFNC component scores at |z| > 3. In the ICA framework, the corresponding loading vector largely captures the covariation of these top features. Figure S3(b) shows that these top features majorly comprise regional connectivities of State-1 and State-5, involving mostly brain regions of temporal, parietal, limbic and occipital areas (see Table 4 for details on the top brain regions). In parallel, the SNP loading vector primarily reflects the covariation among the 80 top SNPs obtained with thresholding the SNP component scores at |z| > 2, as shown in Figure S3(c). Collectively, the identified SNP-dFNC association indicates that the across-subject covariation pattern of the top 80 SNPs explains 11.55% of the variance in the across- subject covariation pattern of the top connectivity feature (i.e., top links). Our results imply that the identified 13 genes mapped from the thresholded 80 SNPs derived from the significant pICA component (for details on the derivation steps of these thresholded SNPs and the mapped genes, see Figure S5) might mediate the dysfunctional connectivity links (observed among the thresholded dFNC component’s connectivity links found in State-1 and State-5, Figure S3) among these brain regions in patients with SZ.
The identified pICA component revealed significant group differences in dFNC loading parameters, where patients with SZ exhibited a significantly lower group mean compared to the HC group. This could imply that, for the significant links within dFNC component, SZ individuals have diminished capability to establish links between these brain regions and share information. We also assessed the correlations between the polygenic risk scores (PRS) and both dFNC and SNP components’ loading parameters. Also, the correlations between the polygenic risk scores (PRS) and dFNC loadings were significantly negative (Figure 5). For dFNC component, thresholded top links were observed mostly in state 1, a state that is dominated by the healthy controls in terms of occupancy rate (Table 1; HC=22% and SZ=14%). This could imply that, as the polygenic risk scores increase for individuals (as measured using PRS), chances to load onto the links within state 1 decrease.
Table 1:
Significant dynamic FNC cells and group-wise occupancy rate across each state
| State# | #dFNC Cells | Occupancy (HC/SZ) |
|---|---|---|
| 1 | 23 | 22%/14% |
| 2 | 0 | 32%/11% |
| 3 | 0 | 20%/18% |
| 4 | 0 | 9%/32% |
| 5 | 2 | 16%/27% |
Using GeneMANIA (Mostafavi et al., 2008) we estimated the percentages of co expression and genetic interaction among the 13 query genes (where co-expression measures whether the genes have similar expression levels across conditions and therefore are linked, and genetic interaction assesses whether the genes are functionally associated by identifying if perturbing one gene would result in perturbations to a second gene). The result obtained from GeneMANIA shows that 40.49 % of the 13 listed genes show co-expression. There is 59.51% genetic interaction among the query genes. A Reactome Pathway Analysis (RPA) (Croft et al., 2013) further shows that these selected genes are involved in different cellular processes including the immune system, metabolism and neuronal systems (Table 2). Among the top 13 genes, CHRNA3, ATXN7 and SMG6 are found to be involved in neurological, psychological and developmental disorders, while RERE and HLA-C are involved in immunological disease (Table 3). Indeed, previous evidence indicated that sensorimotor gating is influenced by variations of the CHRNA3 gene, and might affect the course and severity of schizophrenia (Petrovsky et al., 2010). Moreover, a recent pICA based work explored the genetic control over the DMN in SZ and psychotic bipolar disorder patients, and identified the underlying biological pathways and neurodevelopment/transmission processes including PKA, immune response signaling, NMDA-related long-term potentiation, axon guidance, and synaptogenesis that significantly influenced DMN disconnectivity (Meda et al., 2014). Taken together, the proposed pICA framework presents a powerful way to understand how disrupted connectivity in SZ can be mediated by underlying genes, which could lead us to potential disease-specific biomarkers.
6. LIMITATION AND FUTURE DIRECTION
This is the first study to examine the link between genetic data and resting-state dynamic functional connectivity in schizophrenia and healthy controls using a simultaneous ICA (pICA) approach. The samples in this study, although albeit small, were well characterized, and the pICA approach is a powerful method to examine dFNC-SNP relationships. While the current findings unravel few important aspects of the proposed model, there were some limitations. First, sample sizes in this study were small, so future studies should endeavor to replicate these findings in larger samples. Second, for our replication study, one dataset acquired fMRI data while participants were resting with their eyes closed (fBIRN), and the other dataset acquired fMRI data while participants were resting with their eyes open (COBRE). Recent studies investigating dynamic FNC found difference in patterns of connectivity measure between eyes open and eyes condition (Allen et al., 2018). While we have partially replicated our pICA findings, future studies should investigate independent dataset with same acquisition paradigms (i.e., eyes closed) for full replication. Finally, based on the known dynamic nature of the brain and the unconstrained nature of resting fMRI data, we utilized dFNC as our imaging modality-related features. As a future investigation, an extension of this work should compare performance of the proposed pICA framework using sFNC versus dFNC features. The dFNC-SNP association presented here is the first step showing the strength of the proposed framework, and future work will involve investigation of other potential connectivity references, for association with genomic data.
7. CONCLUSION
In summary, using a multivariate pICA based fusion approach, we propose a framework to identify and investigate parallel co-varying SNP and dFNC components that might uncover genetic contributions to disrupted dynamic connectivity in schizophrenia. We demonstrate that studying such interactions can provide a powerful way of evaluating gene-brain relationships to characterizing schizophrenia and other mental illnesses.
Supplementary Material
8.
FUNDING
This work was supported by the National Institutes of Health (NIH) grants (P20GM103 472/5P20RR021938, R01EB005846, 1R01EB006841, R01REB020407, R01EB000840) and the National Science Foundation (NSF) grant #1539067.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrol A, Chaze C, Damaraju E, Calhoun VD, 2016. The chronnectome: Evaluating replicability of dynamic connectivity patterns in 7500 resting fMRI datasets. Engineering in Medicine and Biology Society (EMBC), 2016. IEEE; 38th Annual International Conference of the. IEEE, pp. 5571–5574. [DOI] [PubMed] [Google Scholar]
- Abrol A, Damaraju E, Miller RL, Stephen JM, Claus ED, Mayer AR, Calhoun VD, 2017. Replicability of time-varying connectivity patterns in large resting state fMRI samples. Neuroimage 163, 160–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Damaraju E, Eichele T, Wu L, Calhoun V, 2018. EEG signatures of dynamic functional network connectivity states. Brain topography 31, 101–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD, 2014. Tracking whole-brain connectivity dynamics in the resting state. Cerebral cortex 24, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Solís A, Vives-Gilabert Y, Grasa E, Portella MJ, Rabella M, Sauras RB, … Pérez V, 2015. Resting-state functional connectivity alterations in the default network of schizophrenia patients with persistent auditory verbal hallucinations. Schizophrenia research 161, 261–268. [DOI] [PubMed] [Google Scholar]
- Altshuler DM, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A, Clark AG,… Consortium GP, 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barta PE, Pearlson GD, Powers RE, Richards SS, Tune LE, 1990. Auditory hallucinations and smaller superior temporal gyral volume in schizophrenia. The American journal of psychiatry 147, 1457. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Nelson BG, Mueller BA, Camchong J, Lim KO, 2012. Altered resting state complexity in schizophrenia. Neuroimage 59, 2196–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AJ, Sejnowski TJ, 1995. An information-maximization approach to blind separation and blind deconvolution. Neural computation 7, 1129–1159. [DOI] [PubMed] [Google Scholar]
- Binder JR, Desai RH, Graves WW, Conant LL, 2009. Where is the semantic system? A critical review and meta-analysis of 120 functional neuroimaging studies. Cerebral cortex 19, 2767–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogousslavsky J, Miklossy J, Deruaz J-P, Assal G, Regli F, 1987. Lingual and fusiform gyri in visual processing: a clinico-pathologic study of superior altitudinal hemianopia. Journal of Neurology, Neurosurgery & Psychiatry 50, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun V, Adali T, Giuliani N, Pekar J, Kiehl K, Pearlson G, 2006. Method for multimodal analysis of independent source differences in schizophrenia: combining gray matter structural and auditory oddball functional data. Human brain mapping 27, 47–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, 2012. Multisubject independent component analysis of fMRI: a decade of intrinsic networks, default mode, and neurodiagnostic discovery. IEEE reviews in biomedical engineering 5, 60–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar J, 2001. A method for making group inferences from functional MRI data using independent component analysis. Human brain mapping 14, 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Miller R, Pearlson G, Adali T, 2014. The chronnectome: time- varying connectivity networks as the next frontier in fMRI data discovery. Neuron 84, 262–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Sui J, 2016. Multimodal fusion of brain imaging data: A key to finding the missing link (s) in complex mental illness. Biological psychiatry: cognitive neuroscience and neuroimaging 1, 230–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Sui J, Kiehl K, Turner JA, Allen EA, Pearlson G, 2012. Exploring the psychosis functional connectome: aberrant intrinsic networks in schizophrenia and bipolar disorder. Frontiers in psychiatry 2, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Wager TD, Krishnan A, Rosch KS, Seymour KE, Nebel MB, …Kiehl K, 2017. The impact of T1 versus EPI spatial normalization templates for fMRI data analyses. Human brain mapping 38, 5331–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardno AG, Gottesman II, 2000. Twin studies of schizophrenia: from bow - and - arrow concordances to star wars Mx and functional genomics. American Journal of Medical Genetics Part A 97, 12–17. [PubMed] [Google Scholar]
- Cavanna AE, Trimble MR, 2006. The precuneus: a review of its functional anatomy and behavioural correlates. Brain 129, 564–583. [DOI] [PubMed] [Google Scholar]
- Çetin MS, Christensen F, Abbott CC, Stephen JM, Mayer AR, Canive JM, …Calhoun VD, 2014. Thalamus and posterior temporal lobe show greater internetwork connectivity at rest and across sensory paradigms in schizophrenia. Neuroimage 97, 117–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Calhoun VD, Lin D, Perrone-Bizzozero NI, Bustillo JR, Pearlson GD, … Ehrlich S, 2018. Shared Genetic Risk of Schizophrenia and Gray Matter Reduction in 6p22. 1. Schizophrenia bulletin Epub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2012. Multifaceted genomic risk for brain function in schizophrenia. Neuroimage 61, 866–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Calhoun VD, Pearlson GD, Perrone-Bizzozero N, Sui J, Turner JA, …Cañive JM, 2013. Guided exploration of genomic risk for gray matter abnormalities in schizophrenia using parallel independent component analysis with reference. Neuroimage 83, 384–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Reiman EM, Huan Z, Caselli RJ, Bandy D, Ayutyanont N, Alexander GE, 2009. Linking functional and structural brain images with multivariate network analyses: a novel application of the partial least square method. Neuroimage 47, 602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clos M, Langner R, Meyer M, Oechslin MS, Zilles K, Eickhoff SB, 2014. Effects of prior information on decoding degraded speech: an fMRI study. Human brain mapping 35, 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa N, Adali T, Calhoun VD, 2007. Performance of blind source separation algorithms for fMRI analysis using a group ICA method. Magnetic resonance imaging 25, 684–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, …Kamdar MR, 2013. The Reactome pathway knowledgebase. Nucleic acids research 42, D472–D477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damaraju E, Allen EA, Belger A, Ford J, McEwen S, Mathalon D, …Preda, A., 2014. Dynamic functional connectivity analysis reveals transient states of dysconnectivity in schizophrenia. Neuroimage: Clinical 5, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF, 2011. A linear complexity phasing method for thousands of genomes. Nat Methods 9, 179–181. [DOI] [PubMed] [Google Scholar]
- Du Y, Fan Y, 2013. Group information guided ICA for fMRI data analysis. Neuroimage 69, 157–197. [DOI] [PubMed] [Google Scholar]
- Du Y, Pearlson GD, Liu J, Sui J, Yu Q, He H, …Calhoun VD, 2015. A group ICA based framework for evaluating resting fMRI markers when disease categories are unclear: application to schizophrenia, bipolar, and schizoaffective disorders. Neuroimage 122, 272–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott L, Sharp K, Alfaro-Almagro F, Douaud G, Miller K, Marchini J, Smith S, 2017. The genetic basis of human brain structure and function: 1,262 genome-wide associations found from 3,144 GWAS of multimodal brain imaging phenotypes from 9,707 UK Biobank participants. BioRxiv, 178806. [Google Scholar]
- Ellison-Wright I, Bullmore E, 2010. Anatomy of bipolar disorder and schizophrenia: a meta-analysis. Schizophrenia research 117, 1–12. [DOI] [PubMed] [Google Scholar]
- Erhardt EB, Rachakonda S, Bedrick EJ, Allen EA, Adali T, Calhoun VD, 2011. Comparison of multi - subject ICA methods for analysis of fMRI data. Human brain mapping 32, 2075–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher P, Shallice T, Frith C, Frackowiak R, Dolan R, 1996. Brain activity during memory retrieval: The influence of imagery and semantic cueing. Brain 119, 1587–1596. [DOI] [PubMed] [Google Scholar]
- Ford JM, Mathalon DH, Whitfield S, Faustman WO, Roth WT, 2002. Reduced communication between frontal and temporal lobes during talking in schizophrenia. Biological psychiatry 51, 485–492. [DOI] [PubMed] [Google Scholar]
- Freire L, Mangin J-F, 2001. Motion correction algorithms may create spurious brain activations in the absence of subject motion. Neuroimage 14, 709–722. [DOI] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R, 2008. Sparse inverse covariance estimation with the graphical lasso. Biostatistics 9, 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L, Glover GH, Krenz D, Magnotta V, 2006. Reducing inter-scanner variability of activation in a multicenter fMRI study: role of smoothness equalization. Neuroimage 32, 1656–1668. [DOI] [PubMed] [Google Scholar]
- Fu Y, Ma Z, Hamilton C, Liang Z, Hou X, Ma X, …Wang Y, 2015. Genetic influences on resting - state functional networks: A twin study. Human brain mapping 36, 3959–3972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity AG, Pearlson GD, McKiernan K, Lloyd D, Kiehl KA, Calhoun VD, 2007. Aberrant “default mode” functional connectivity in schizophrenia. American Journal of Psychiatry 164, 450–457. [DOI] [PubMed] [Google Scholar]
- Glahn DC, Winkler A, Kochunov P, Almasy L, Duggirala R, Carless M, …Smith S, 2010. Genetic control over the resting brain. Proceedings of the National Academy of Sciences 107, 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Gould TD, 2003. The endophenotype concept in psychiatry: etymology and strategic intentions. American Journal of Psychiatry 160, 636–645. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Shields J, 1972. A polygenic theory of schizophrenia. International Journal of Mental Health 1, 107–115. [Google Scholar]
- Groves AR, Beckmann CF, Smith SM, Woolrich MW, 2011. Linked independent component analysis for multimodal data fusion. Neuroimage 54, 2198–2217. [DOI] [PubMed] [Google Scholar]
- Gupta CN, Calhoun VD, Rachakonda S, Chen J, Patel V, Liu J, …Arias-Vasquez A, 2014. Patterns of gray matter abnormalities in schizophrenia based on an international mega-analysis. Schizophrenia bulletin 41, 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DA, Fussell C, Summerfield AQ, 2005. Reading fluent speech from talking faces: typical brain networks and individual differences. Journal of Cognitive Neuroscience 17, 939–953. [DOI] [PubMed] [Google Scholar]
- Harris GJ, Chabris CF, Clark J, Urban T, Aharon I, Steele S, …Tager-Flusberg G, 2006. Brain activation during semantic processing in autism spectrum disorders via functional magnetic resonance imaging. Brain and cognition 61, 54–68. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA, 2009. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proceedings of the National Academy of Sciences 106, 9362–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ, 2005. Genome-wide association studies for common diseases and complex traits. Nature Reviews Genetics 6, 95. [DOI] [PubMed] [Google Scholar]
- Hutchison RM, Womelsdorf T, Allen EA, Bandettini PA, Calhoun VD, Corbetta M, … Gonzalez-Castillo J, 2013. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage 80, 360–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivleva EI, Bidesi AS, Thomas BP, Meda SA, Francis A, Moates AF, … Tamminga CA, 2012. Brain gray matter phenotypes across the psychosis dimension. Psychiatry Research: Neuroimaging 204, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafri MJ, Pearlson GD, Stevens M, Calhoun VD, 2008. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. Neuroimage 39, 1666–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagannathan K, Calhoun VD, Gelernter J, Stevens MC, Liu J, Bolognani F, … Pearlson GD, 2010. Genetic associations of brain structural networks in schizophrenia: a preliminary study. Biological psychiatry 68, 657–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler K, Eaves L, 2005. Psychiatric genetics (review of psychiatry). Arlington, VA: American Psychiatric Publishing, Inc. [Google Scholar]
- Keshavan MS, Morris DW, Sweeney JA, Pearlson G, Thaker G, Seidman LJ, … Tamminga C, 2011. A dimensional approach to the psychosis spectrum between bipolar disorder and schizophrenia: the Schizo-Bipolar Scale. Schizophrenia research 133, 250–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama MS, Kelly C, Shehzad Z, Penesetti D, Castellanos FX, Milham MP, 2010. Reading networks at rest. Cerebral cortex 20, 2549–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühn S, Gallinat J, 2011. Resting-state brain activity in schizophrenia and major depression: a quantitative meta-analysis. Schizophrenia bulletin 39, 358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenstein P, Yip BH, Björk C, Pawitan Y, Cannon TD, Sullivan PF, Hultman CM, 2009. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. The Lancet 373, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin QH, Liu J, Zheng YR, Liang H, Calhoun VD, 2010. Semiblind spatial ICA of fMRI using spatial constraints. Human brain mapping 31, 1076–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Calhoun VD, 2014. A review of multivariate analyses in imaging genetics. Frontiers in neuroinformatics 8, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pearlson G, Windemuth A, Ruano G, Perrone - Bizzozero NI, Calhoun V, 2009. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Human brain mapping 30, 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Correa NM, Li X-L, Eichele T, Calhoun VD, Adali T, 2011. Automatic identification of functional clusters in FMRI data using spatial dependence. IEEE Transactions on Biomedical Engineering 58, 3406–3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoach DS, Gollub RL, Benson ES, Searl MM, Goff DC, Halpern E, … Rauch SL, 2000. Schizophrenic subjects show aberrant fMRI activation of dorsolateral prefrontal cortex and basal ganglia during working memory performance. Biological psychiatry 48, 99–109. [DOI] [PubMed] [Google Scholar]
- Manoliu A, Riedl V, Doll A, Bauml JG, Muhlau M, Schwerthoffer D, … Bauml J,2012. Insular dysfunction reflects altered between-network connectivity and severity of negative symptoms in schizophrenia during psychotic remission. Frontiers in human neuroscience 7, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchini J, Howie B, 2010. Genotype imputation for genome-wide association studies. Nat Rev Genet 11, 499–511. [DOI] [PubMed] [Google Scholar]
- Meda SA, Gill A, Stevens MC, Lorenzoni RP, Glahn DC, Calhoun VD, … Thaker G, 2012. Differences in resting-state functional magnetic resonance imaging functional network connectivity between schizophrenia and psychotic bipolar probands and their unaffected first-degree relatives. Biological psychiatry 71, 881–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda SA, Jagannathan K, Gelernter J, Calhoun VD, Liu J, Stevens MC, Pearlson GD, 2010. A pilot multivariate parallel ICA study to investigate differential linkage between neural networks and genetic profiles in schizophrenia. Neuroimage 53, 1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meda SA, Ruano G, Windemuth A, O’Neil K, Berwise C, Dunn SM, … Sprooten E, 2014. Multivariate analysis reveals genetic associations of the resting default mode network in psychotic bipolar disorder and schizophrenia. Proceedings of the National Academy of Sciences 111, E2066–E2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RL, Adali T, Levin-Schwartz Y, Calhoun VD, 2017. Resting-state fMRI dynamics and null models: perspectives, sampling variability, and simulations. BioRxiv, 153411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks PJ, Thompson JM, Bullmore ET, Suckling J, Brammer MJ, Williams SC, … Louise Harrison C, 2004. A functional MRI study of working memory task in euthymic bipolar disorder: evidence for task - specific dysfunction. Bipolar Disorders 6, 550–564. [DOI] [PubMed] [Google Scholar]
- Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q, 2008. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome biology 9, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongur D, Lundy M, Greenhouse I, Shinn AK, Menon V, Cohen BM, Renshaw PF, 2010. Default mode network abnormalities in bipolar disorder and schizophrenia. Psychiatry Research: Neuroimaging 183, 59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlson G, Calhoun VD, 2009. Convergent approaches for defining functional imaging endophenotypes in schizophrenia. Frontiers in human neuroscience 3, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearlson GD, Calhoun VD, Liu J, 2015. An introductory review of parallel independent component analysis (p-ICA) and a guide to applying p-ICA to genetic data and imaging phenotypes to identify disease-associated biological pathways and systems in common complex disorders. Frontiers in genetics 6, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Shao J, Scherr M, Schwerthoffer D, Zimmer C, Forstl H, … Koch, K., 2016. More consistently altered connectivity patterns for cerebellum and medial temporal lobes than for amygdala and striatum in schizophrenia. Frontiers in human neuroscience 10, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella J, Sheldon F, Prince S, Calhoun V, Doraiswamy P, 2011. Default mode network connectivity in stable vs progressive mild cognitive impairment. Neurology 76, 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovsky N, Quednow BB, Ettinger U, Schmechtig A, Mossner R, Collier DA, … Kumari, V., 2010. Sensorimotor gating is associated with CHRNA3 polymorphisms in schizophrenia and healthy volunteers. Neuropsychopharmacology 35, 1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Ford JM, 2008. Widespread cortical dysfunction in schizophrenia: the FBIRN imaging consortium. Schizophrenia bulletin 35, 15–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preti MG, Bolton TA, Van De Ville D, 2017. The dynamic functional connectome: State-of-the-art and perspectives. Neuroimage 160, 41–54. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D, 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38, 904–909. [DOI] [PubMed] [Google Scholar]
- Purcell S, 2009. International Schizophrenia ConsortiumWray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, … Daly MJ, 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid B, Arbabshirani MR, Damaraju E, Cetin MS, Miller R, Pearlson GD, Calhoun VD, 2016. Classification of schizophrenia and bipolar patients using static and dynamic resting-state fMRI brain connectivity. Neuroimage 134, 645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid B, Damaraju E, Pearlson GD, Calhoun VD, 2014. Dynamic connectivity states estimated from resting fMRI Identify differences among Schizophrenia, bipolar disorder, and healthy control subjects. Frontiers in human neuroscience 8, 897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JT, Farh K-H, Holmans PA, … Huang H,Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugg MD, Henson RN, 2002. Episodic memory retrieval: an (event-related) functional neuroimaging perspective. The cognitive neuroscience of memory encoding and retrieval, 3–37. [Google Scholar]
- Sakoglu U, Pearlson GD, Kiehl KA, Wang YM, Michael AM, Calhoun VD, 2010. A method for evaluating dynamic functional network connectivity and taskmodulation: application to schizophrenia. Magnetic Resonance Materials in Physics, Biology and Medicine 23, 351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, … Woolrich MW, 2011. Network modelling methods for FMRI. Neuroimage 54, 875–891. [DOI] [PubMed] [Google Scholar]
- Sui J, Adali T, Pearlson GD, Calhoun VD, 2009. An ICA-based method for the identification of optimal FMRI features and components using combined group- discriminative techniques. Neuroimage 46, 73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Kendler KS, Neale MC, 2003. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Archives of general psychiatry 60, 1187–1192. [DOI] [PubMed] [Google Scholar]
- Wang D, Zhou Y, Zhuo C, Qin W, Zhu J, Liu H, …Yu C, 2015. Altered functional connectivity of the cingulate subregions in schizophrenia. Translational psychiatry 5, e575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf DH, Gur RC, Valdez JN, Loughead J, Elliott MA, Gur RE, Ragland JD, 2007. Alterations of fronto-temporal connectivity during word encoding in schizophrenia. Psychiatry Research: Neuroimaging 154, 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Eichele T, Calhoun VD, 2010. Reactivity of hemodynamic responses and functional connectivity to different states of alpha synchrony: a concurrent EEG- fMRI study. Neuroimage 52, 1252–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Tang Y, Zhang T, Li H, Tang Y, Li C, …Wang J, 2017. Reduced functional connectivity between bilateral precuneus and contralateral parahippocampus in schizotypal personality disorder. BMC psychiatry 17, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


