Abstract
The finding that the adult mammalian brain is still capable of producing neurons has ignited a new field of research aiming to identify the molecular mechanisms regulating adult neurogenesis. An improved understanding of these mechanisms could lead to the development of novel approaches to delay cognitive decline and facilitate neuroregeneration in the adult human brain. Accumulating evidence suggest microRNAs (miRNAs), which represent a class of post-transcriptional gene expression regulators, as crucial part of the gene regulatory networks governing adult neurogenesis. This review attempts to illustrate how miRNAs modulate key processes in the adult neurogenic niche by interacting with each other and with transcriptional regulators. We discuss the function of miRNAs in adult neurogenesis following the life-journey of an adult-born neuron from the adult neural stem cell (NSCs) compartment to its final target site. We first survey how miRNAs control the initial step of adult neurogenesis, that is the transition of quiescent to activated proliferative adult NSCs, and then go on to discuss the role of miRNAs to regulate neuronal differentiation, survival, and functional integration of the newborn neurons. In this context, we highlight miRNAs that converge on functionally related targets or act within cross talking gene regulatory networks. The cooperative manner of miRNA action and the broad target repertoire of each individual miRNA could make the miRNA system a promising tool to gain control on adult NSCs in the context of therapeutic approaches.
Keywords: microRNAs, adult neurogenesis, hippocampus, neural stem cells, neurons, feedback loops, miRNA convergence
Introduction
Neural stem cells (NSCs) are self-renewing, multipotent progenitors that generate all neurons and glial cells of the mammalian central nervous system (CNS). In the adult mammalian brain, a limited number of adult neural stem cells (aNSCs) persists in the subventricular zone (SVZ) of the lateral ventricles and the subgranular zone (SGZ) in the hippocampal dentate gyrus as the two main neurogenic niches (Figures 1A–C). The composition and functionality of these germinal zones has been most extensively studied in rodents (reviewed by Kempermann et al., 2015; Pino et al., 2017). Similar regions containing neurogenic progenitor cells have been described in the SVZ and dentate gyrus of the adult human brain (Eriksson et al., 1998; Roy et al., 2000; Nunes et al., 2003). Neurogenesis within the human SVZ declines during infancy (Sanai et al., 2011), whereas several studies have pointed to a quite substantial generation of dentate granule neurons in humansthroughout life (Spalding et al., 2013; Boldrini et al., 2018). This view has been challenged by a recent report suggesting that hippocampal neurogenesis decreases dramatically after the first years of life in both humans and macaques [Sorrells et al. (2018); see also Kempermann et al. (2018) for a statement regarding the recent discussion on the relevance of adult hippocampal neurogenesis in humans]. Nevertheless, there is a strong interest in understanding the mechanisms regulating adult neurogenesis (reviewed by Kempermann et al., 2015; Peng and Bonaguidi, 2018). Research in this regard is motivated by the fact that the hippocampus is involved in memory and learning, which led to the hypothesis that adult neurogenesis could play an important role in cognition (reviewed by Gonçalves et al., 2016). Furthermore, a series of studies revealed that adult hippocampal neurogenesis in rodents can be modulated by experiential and environmental conditions as well as by aging (reviewed by Toda and Gage, 2018). Altered hippocampal neurogenesis has been linked to a number of pathological conditions, such as ischemia- or epilepsy-induced insults, mood disorders, and neurodegenerative diseases (reviewed by Pino et al., 2017).
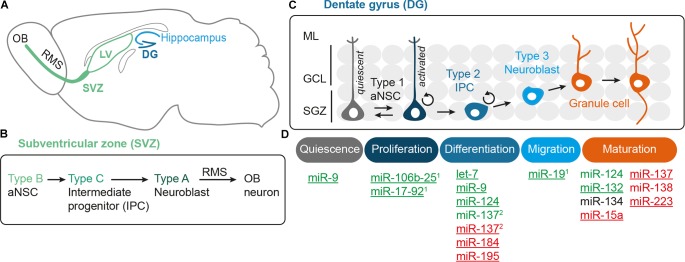
FIGURE 1.
MicroRNAs as key regulators of the multistep process of adult neurogenesis. (A) The adult mouse brain contains two main neurogenic niches, i.e., the subventricular zone (SVZ) of the lateral ventricle (LV), which produces neurons for the olfactory bulb (OB), and the subgranular zone (SGZ) of the hippocampal dentate gyrus (DG). (B) The adult neural stem cells (aNSCs) of the subventricular zone are called type B cells. They give rise to intermediate progenitors (IPCs, type C cells), which after a few cell cycles develop into type A neuroblasts that migrate along the rostral migratory stream (RMS) to the OB before terminal differentiation (Doetsch et al., 1997). (C) The aNSCs of the SGZ of the dentate gyrus are called type 1 cells and the IPCs to which they develop are designated as type 2, the neuroblasts as type 3 cells (Seri et al., 2004). The neuroblasts generated from the SGZ migrate up into the granule cell layer (GCL), where they maturate into granule cells, extend dendrites into the molecular layer (ML), and grow axons. (D) Adult neurogenesis is a multistep process that begins with activation of quiescent aNSCs to re-enter cell cycling, followed by a proliferation phase of aNSCs, which eventually develop into IPCs and further differentiate into neuroblasts. The last steps of adult neurogenesis involve neuroblast migration and functional maturation into terminally differentiated neurons. For all these steps, regulatory miRNAs have been identified that either have a positive (green), negative (red), or modulatory (black) effect. Underlined miRNAs have been studied in the context of adult neurogenesis, whereas the function of the other miRNAs has been inferred from studies addressing developmental neurogenesis. The miRNAs miR-106b-25(1), miR-17-92(1) and miR-19(1) belong to the family of polycistronic miR-17 clusters. There are divergent results for miRNA-137(2), which was found to have a pro-differentiation effect in embryonic NSCs (Sun et al., 2011), but an anti-differentiation effect in adult NSCs (Szulwach et al., 2010).
Adult neurogenesis involves multiple steps that have to be tightly regulated, i.e., aNSC activation, proliferation, differentiation into neural progeny as well as survival, migration, and functional maturation of the adult-born neurons (Figure 1D). Recent evidence indicate that microRNAs (miRNAs) can be placed in midst of the regulatory mechanisms operating in adult neurogenesis (reviewed by Lopez-Ramirez and Nicoli, 2014; Murao et al., 2016; Encinas and Fitzsimons, 2017). miRNAs are short (22 nucleotide long) single-stranded RNA molecules that post-transcriptionally repress gene expression by complementary binding to mRNA targets (reviewed by Bartel, 2009). miRNA genes are transcribed as hairpin-shaped transcripts, called pri-miRNAs, which are sequentially processed to liberate the mature miRNAs (Figure 2). Some pri-miRNAs are polycistronic and encode for several mature miRNAs (reviewed by Olive et al., 2015). The final and essential cleavage step of miRNA biogenesis is carried out by the ribonuclease Dicer, and genetic ablation studies for Dicer have been used to assess the overall importance of the miRNA system. Mature miRNAs are then loaded onto Argonaut (Ago) proteins to form the RNA-induced silencing complex (RISC) through which they target mRNAs for translational inhibition or mRNA degradation. miRNAs often come as families whose members share a common seed sequence and are thought to have similar functions (Figure 2) (reviewed by Ha and Kim, 2014). During brain development, miRNAs regulate neural progenitor proliferation, neurogenic and gliogenic differentiation as well as maturation and functional integration of neurons (reviewed by Bian et al., 2013; Barca-Mayo and De Pietri Tonelli, 2014; Stappert et al., 2014). Likewise, miRNAs play important roles in regulating cell fate decisions in the adult SVZ and SGZ (reviewed by Lopez-Ramirez and Nicoli, 2014; Murao et al., 2016) and have been linked to diseases associated with these compartments, e.g., epilepsy (Bielefeld et al., 2017), stroke (Khoshnam et al., 2017), and neurodegenerative disorders (Qiu et al., 2014).
FIGURE 2.
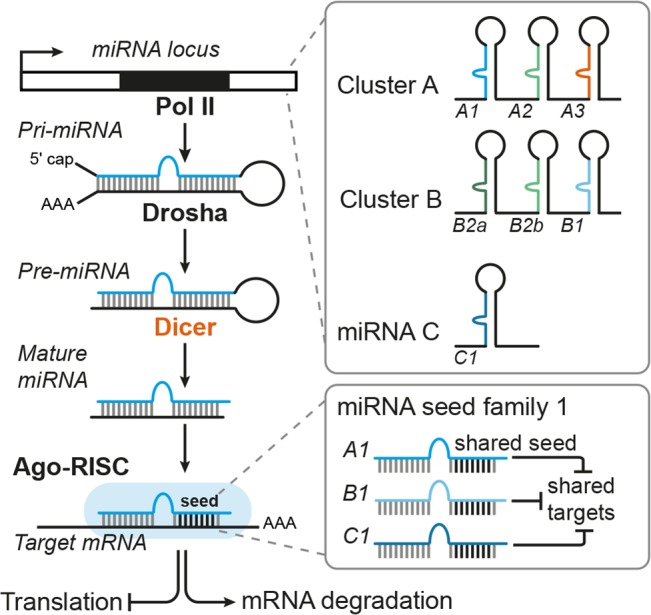
Canonical miRNA biogenesis and function. miRNA genes are transcribed by RNA Polymerase II (Pol II) and then processed by the sequential action of Drosha and Dicer to liberate the mature miRNA duplex, which is then loaded onto Argonaut proteins (Ago) to form the RNA-induced silencing complex (RISC). As part of the RISC, miRNAs can bind to target mRNAs whereby binding of the miRNA seed sequence is the key requirement for mRNA target recognition. Thus, miRNAs with the same seed sequence are thought to target overlapping sets of mRNAs and are assigned to the same miRNA seed family. miRNA genes can be either monocistronic (miRNA C) or polycistronic (clusters A and B) containing several miRNAs that are usually expressed as one pri-miRNA transcript. Some polycistronic clusters comprise miRNA homologs that have emerged via duplication events (cluster B with miR-B2a, B2b as homologous miRNAs) whereas other clusters consist of miRNAs belonging to different seed families (here exemplarily designated as seed family 1 (in blue) with miRNAs A1, B1, C1, and seed family 2 (in green) with miRNAs A2, B2a, and B2b). For more details on miRNA biogenesis and polycistronic miRNA clusters, see the reviews from Ha and Kim (2014) and Olive et al. (2015), respectively.
In many cases, miRNAs act in concert with transcription factors and chromatin modifiers to control gene expression in NSCs (of embryonic or adult origin), thereby affecting NSC number and their ability to generate differentiated progeny.
In this review, we discuss the physiological role of miRNAs during adult neurogenesis following the route from aNSC maintenance to neuronal differentiation and maturation of newborn neurons. Given its discussed relevance for human neurogenesis and cognition, we will mainly focus on hippocampal neurogenesis. However, in some cases we will also refer to data generated in other compartments, e.g., the adult SVZ or the developing brain, to highlight important principles of miRNA function during neurogenesis (see Figure 1D and Tables 1, 2 for an overview of the functions of the miRNAs presented here). In the first part, we depict how miRNAs control the balance between quiescent and activated aNSCs contributing to homeostasis and plasticity in response to neurogenic stimuli. In the second part, we describe how miRNAs influence the neurogenic output of the aNSC niche by modulating neuronal differentiation, survival, and functional integration. In this context, we delineate the interactions of miRNAs with gene regulatory networks controlling adult neurogenesis and focus on miRNAs that converge by targeting functionally associated genes. We end by summarizing the diverse roles of miRNAs during adult neurogenesis and discuss the importance of target multiplicity and miRNA cooperativity as key features of the miRNA system. Finally, we speculate how miRNAs may contribute to aNSC heterogeneity and give an outlook on how knowledge on miRNA-based regulation could be further increased and eventually exploited to facilitate neuroregeneration in the adult human brain.
Table 1.
MicroRNAs modulating proliferation and differentiation during adult neurogenesis.
| MicroRNA | Main model system | Observed main effect | miRNA regulator | Target mRNA | Reference |
|---|---|---|---|---|---|
| let-7b | Mouse primary neonatal forebrain NSCs | let-7 decreases NSC proliferation | Hmga2 | Nishino et al., 2008 | |
| Mouse embryonic VZ, primary adult forebrain NSCs | let-7 promotes neuronal differentiation | Tlx, CyclinD1 (Ccnd1) | Zhao et al., 2010 | ||
| miR-17-92 cluster | Nestin-CreER miR-17-92 KO/OE mice, adult hippocampus | miR-17-92 promotes aNSC proliferation and rescues stress-induced impairment of neurogenesis | Sgk1 | Jin et al., 2016 | |
| miR-25 (miR-106b-25) cluster | Mouse primary adult forebrain NSCs | miR-25 promotes aNSC proliferation | FOXO3 | Foxo3 (predicted) | Brett et al., 2011 |
| miR-9 | Mouse embryonic VZ, primary adult forebrain NSCs | miR-9 promotes neuronal differentiation | TLX | Tlx | Zhao et al., 2009 |
| Human embryonal carcinoma cell line | miR-9/9∗ and the REST silencing complex form a double negative feedback loop | REST | Rest, CoRest (Rcor2) | Packer et al., 2008 | |
| Human neuroblastoma cell line | miR-9 expression is inhibited by Rest in undifferentiated cells and promoted by CREB in differentiated cells, miR-9 targets Rest forming a feedback loop | REST, CREB | Rest | Laneve et al., 2010 | |
| Mouse primary neonatal NSCs, adult SVZ | miR-9 OE promotes neuronal differentiation; miR-9 decreases Notch signaling dependent on FOXO1 | FOXO1 (predicted) | Foxo1 | Kim et al., 2015 | |
| Adult zebrafish brain | Nuclear localized non-canonical miR-9 maintains aNSC quiescence | TNRC6 (for nuclear shuttling) | Notch signaling (indirect positive effect) | Katz et al., 2016 | |
| miR-124 | Mouse embryonal carcinoma cell line | REST prevents miR-124 expression in neural progenitors | REST | Conaco et al., 2006 | |
| Chick neural tube, mouse embryonal carcinoma cell line | miR-124 promotes neuronal differentiation | REST | Scp1 | Visvanathan et al., 2007 | |
| Mouse adult SVZ: injection of miR-124 OE retrovirus, infusion of miR-124 inhibitor | miR-124 promotes neuronal differentiation and is necessary for SVZ regeneration | Sox9 | Cheng et al., 2009 | ||
| Mouse adult SVZ: injection of miR-124 OE/sponge lentivirus | miR-124 OE leads to precocious neuronal differentiation and aNSC exhaustion; miR-124 inhibition represses neuronal differentiation and promotes glial differentiation | Akerblom et al., 2012 | |||
| miR-137 | Mouse primary adult forebrain NSCs, DG retroviral injection | miR-137 OE promotes aNSC proliferation and represses neuronal differentiation | MECP2 | Ezh2 | Szulwach et al., 2010 |
| Mouse embryonic NSCs, in utero electroporation into the lateral ventricle | miR-137 OE inhibits NSC proliferation and accelerates neuronal differentiation | TLX (via LSD1) | Lsd1 | Sun et al., 2011 | |
| miR-184 | Mouse primary adult DG NSCs, DG retroviral injection | miR-184 promotes aNSC proliferation | MBD1 | Numbl | Liu et al., 2010 |
| miR-195 | Mouse primary adult DG NSCs, DG retroviral injection | miR-195 promotes aNSC proliferation | MBD1 | Mbd1 | Liu et al., 2013 |
Overview of miRNAs (in ascending numerical order) discussed in this review that modulate proliferation and differentiation of (adult) neural progenitor cells, their upstream regulators, target mRNAs, and the model systems they have been studied in. Findings relating to adult neurogenesis are highlighted in bold. (a) NSC, (adult) neural stem cells; DG, dentate gyrus; KO, knockout; OE, overexpression; SVZ, subventricular zone; VZ, ventricular zone.
Table 2.
Putative impact of miRNAs on neuronal migration and neuronal morphogenesis in the context of adult neurogenesis.
| MicroRNA | Main model system | Observed main effect | miRNA regulator | Target mRNA | Reference |
|---|---|---|---|---|---|
| miR-15a | Floxed Mecp2 mice, DG retroviral grafting of Cre-GFP/miR-15a sponge, primary neonatal cortical/hippocampal neurons | miR-15a impairs dendrite maturation, miR-15a inhibition rescues Mecp2-deficiency-induced neuronal maturation deficits | MECP2 | Bdnf | Gao et al., 2015 |
| miR-19 | Mouse primary adult DG NSCs, DG and SVZ retroviral injection | miR-19 promotes migration of neurons | Rapgef2 | Han et al., 2016 | |
| miR-124 | Rat primary embryonic hippocampal neurons | miR-124 increases axonal and dendrite complexity | Rhog | Franke et al., 2012 | |
| miR-132 | Rat primary neonatal cortical neurons | CREB induces miR-132 expression downstream of BDNF, miR-132 promotes neurite outgrowth | CREB | p250Gap (Arhgap32) | Vo et al., 2005 |
| Rat primary neonatal cortical neurons | miR-132 and MECP2 form a feedback mechanism via BDNF | BDNF via CREB | Mecp2 | Klein et al., 2007 | |
| Rat primary neonatal hippocampal neurons | miR-132 promotes dendrite growth and spine maturation | p250Gap (Arhgap32) | Wayman et al., 2008 | ||
| Floxed miR-213/132 mice and GFP-Cre DG retroviral injection | Deletion of miR-132 decreases dendrite length and arborization | Magill et al., 2010 | |||
| miR-134 | Rat primary embryonic cortical/hippocampal neurons | miR-134 decreases size of dendritic spines | BDNF (indirect effect) | Limk1 | Schratt et al., 2006 |
| Sirt1 KO/Nestin-Cre mice, adult hippocampal lentiviral injection | miR-134 knockdown rescues Sirt1-deficiency-induced LTP and memory defects | SIRT1 | Creb | Gao et al., 2010 | |
| Mecp2 KO/OE mice, mouse primary embryonic cortical neurons | Mecp2 OE inhibits dendrite growth and pri-miR-134 processing; miR-134 OE rescues dendrite growth defect | MECP2 | Cheng T.-L. et al., 2014 | ||
| miR-134 part of miR-379-410 cluster | Rat primary embryonic cortical/hippocampal neurons | miR-379-410 expression is regulated by neuronal activity via MEF2; miR-134 promotes dendrite outgrowth | MEF2 | Pum2 | Fiore et al., 2009 |
| miR-137 | Mouse DG retroviral injection, primary embryonic hippocampal neurons | miR-137 OE inhibits dendrite morphogenesis | Mib1 | Smrt et al., 2010 | |
| miR-138 | Rat primary embryonic cortical/hippocampal neurons | miR-138 decreases size of dendritic spines | Apt1 | Siegel et al., 2009 | |
| miR-223 | Retroviral injection of miR-223 sponge into mouse DG, miR-223 OE in human fetal NPCs | miR-223 inhibits dendrite outgrowth | Harraz et al., 2014 | ||
List of the miRNAs (in ascending numerical order) and their respective up-stream regulators and target mRNAs associated with regulating neuron migration and morphogenesis during neurogenesis. Findings directly related to adult neurogenesis are highlighted in bold. DG, dentate gyrus; KO, knockout; LTP, long-term potentiation; NSC, neural stem cells; OE, overexpression; SVZ, subventricular zone.
MicroRNAs Regulating Activation and Proliferation of Adult Neural Stem Cells
Adult NSCs reside in a specialized microenvironment, the stem cell niche, which is composed of different cell types and extracellular matrix molecules and provides extracellular signals to regulate NSC homeostasis and differentiation (reviewed by Llorens-Bobadilla and Martin-Villalba, 2017). In the currently prevalent view, aNSCs are slowly dividing radial glia-like precursor cells that express Nestin, Gfap, as well as Sox2 and are in close contact with the vasculature. The radial glia-like precursor cells of the dentate gyrus SGZ are further characterized by basal processes that span the granule cell layer. The radial glia-like NSCs (SVZ: type B cells, SGZ: type 1 cells) generate fast-dividing committed intermediate progenitors (IPCs, SVZ: type C cells, SGZ: type 2 cells), which then give rise to neuroblasts (SVZ: type A cells, SGZ: type 3 cells) (Doetsch et al., 1997; Seri et al., 2004; and reviewed by Pino et al., 2017) (see Figures 1B,C for an overview of the lineage relationships and nomenclature in the SVZ and SGZ). The SVZ neuroblasts migrate along the rostral migratory stream to the olfactory bulb where they differentiate into olfactory interneurons. The dentate gyrus neuroblasts generated from the SGZ move up into the granule cell layer before they differentiate into mature granule neurons (Figures 1A–C) (reviewed by Pino et al., 2017).
The majority of aNSCs is quiescent, which seems to be important to ensure long-term homeostasis of the niche throughout the life span of the organism. However, a small fraction of aNSCs has been shown to proliferate quite rapidly, and it has been suggested that aNSCs transition between activated proliferative and quiescent states (Figure 1C; Lugert et al., 2010). The number of aNSCs shuttling between these two states is modulated by neurogenic stimuli and during aging (van Praag et al., 1999; Lugert et al., 2010; Encinas et al., 2011). In fact, neurogenic activity seems to decline with age, whereby it is still unresolved whether this is due to an exhaustion of aNSCs, which once activated eventually lose their self-renewal capacity and terminally differentiate (Encinas et al., 2011; Ziebell et al., 2018), or an increased quiescence of the aNSCs pool (Lugert et al., 2010; and reviewed by Giachino and Taylor, 2015).
MicroRNAs Controlling the Balance Between Quiescent and Activated Proliferative aNSCs
There are many factors known to control the proliferative capacity of aNSCs (reviewed by Beckervordersandforth et al., 2015) including transcription factors and signaling pathways, such as Notch, BMP, Wnt, and insulin/IGF signaling. While for many of the players involved regulatory miRNAs have been identified, we will focus on such miRNA–target interactions that have been explicitly studied in the context of adult neurogenesis.
One pathway that is regulated by many miRNAs and plays a key role in controlling aNSCs proliferation is Notch signaling (reviewed by Giachino and Taylor, 2015). High Notch levels protect aNSC from activation as shown by studies in mouse SVZ (Kawaguchi et al., 2013) and SGZ (Semerci et al., 2017) as well as in zebrafish aNSCs (Chapouton et al., 2010). While studying adult neurogenesis in the zebrafish brain, which exhibits a widespread neurogenic capacity and contains regions similar to the rodent SVZ and SGZ, Katz et al. (2016) found that miR-9 potentiates Notch signaling to maintain aNSC quiescence. This function of miR-9 stands in apparent contrast to its previously described pro-differentiation effect in embryonic (reviewed by Roese-Koerner et al., 2017) and adult NSCs (Zhao et al., 2009; Kim et al., 2015) (see also Table 1). The proposed role of miR-9 during aNSC quiescence in zebrafish may, however, reflect a non-canonical function of miR-9 since Katz et al. (2016) found that miR-9 is actively transported to the nucleus by binding of miR-9-Ago complexes to the shuttle protein TNRC6 and that this nuclear concentration is critical for aNSC quiescence. Interestingly, nuclear localization of miR-9 was also detected in a subset of cells in the SVZ and SGZ neurogenic niche in the mouse brain, suggesting that the role of miR-9 in regulating aNSC quiescence might be conserved (Katz et al., 2016). Furthermore, nuclear concentration of miR-9 was also found to be increased in older versus younger mice, pointing to an age-dependent shift of the subcellular localization of miR-9 (Katz et al., 2016).
Another example of a miRNA-target pair implicated in aNSC maintenance is let-7 and Hmga2. miRNA let-7 and its target gene Hmga2 show an inverse expression pattern in the murine SVZ during aging (Nishino et al., 2008). HMGA2 is a member of the high mobility group protein family that modulates gene expression as part of the enhanceosome (reviewed by Pfannkuche et al., 2009). Nishino et al. (2008) showed that HMGA2 promotes the proliferative capacity of fetal and adult NSCs by repressing expression of Ink4a/Arf locus, which encodes for the cell cycle inhibitors p16 (Cdkn2a) and p19 (Cdkn2d). However, HMGA2 seems not to bind to the Ink4a/Arf locus and it is not yet clear how HMGA2 represses expression of Ink4/Arf. Expression of Hmga2 declines during aging, and premature loss of Hmga2 impairs self-renewal in in vitro cultured forebrain aNSCs, which may contain both SVZ and SGZ aNSCs. Nishino et al. (2008) further found that Hmga2 expression is regulated by let-7, whose expression increases with age. Furthermore, overexpression of let-7 decreases the proliferative capacity of in vitro cultured forebrain aNSCs mimicking the effect of Hmga2 loss. Thus, the interaction of HMGA2 and let-7 may couple aNSC self-renewal and aging.
Quiescence of aNSCs is also regulated by FOXO3, which was initially identified as an age- and longevity-associated factor downstream of insulin/IGF signaling (reviewed by Martins et al., 2015). FOXO3 is necessary to maintain quiescent aNSCs in both the SVZ and SGZ (Paik et al., 2009; Renault et al., 2009) and was suggested to interact with the polycistronic miR-106b-25 cluster. This cluster is encoded in an intronic region of the protein-coding Mcm7 gene, which is one of the FOXO3 target genes (Brett et al., 2011). In turn, miR-25, generated by the miR-106b-25 cluster, is critical for the proliferative capacity of in vitro cultured forebrain aNSC and is predicted to target Foxo3 mRNA. Thus, miR-25 and FOXO3 might form a feedback loop regulating the self-renewal capacity of aNSCs (Brett et al., 2011). Unfortunately, the impact of miR-25 on Foxo3 has not yet been experimentally validated. Nevertheless, an interaction between IGF signaling, FOXO3, and miR-25 to regulate NSC proliferation seems an attractive scenario, which might even be evolutionary conserved as the C. elegans ortholog of miR-25, cel-miR-253, has been shown to couple proliferation of blast cells to the nutritional state downstream of IGF signaling (Kasuga et al., 2013). Interestingly, ectopic expression of miR-25 can even re-instate cell cycling of post-mitotic zebrafish neurons (Rodriguez-Aznar et al., 2013). The relevant target gene in this context is p57 (Cdkn1c), which has been shown to pace adult hippocampal neurogenesis in mice by controlling aNSCs quiescence (Furutachi et al., 2013).
Taken together, these findings indicate the importance of miRNAs in regulating activation and proliferation of aNSCs, which is key for long-term homeostasis of the adult neurogenic niche.
MicroRNAs Regulating aNSC Activation Downstream of Neurogenic Modulators
Hippocampal neurogenesis is modulated by several physiological stimuli, such as physical exercise, environmental conditions, learning, and aging. Physical activation, for instance, has been shown to promote neurogenesis by inducing cell cycle entry of quiescent aNSCs (Lugert et al., 2010), while exposing mice to enriched environment enhances the survival of new neurons (van Praag et al., 1999). Interestingly, it has been demonstrated that exposure of mice to an enriched environment also leads to altered miRNA expression profiles in the hippocampus (Barak et al., 2013). In total, 29% of the miRNAs, including miR-124, were found to be down-regulated in response to enriched environment, while 8% (including miR-132 as the most increased miRNA) were up-regulated (Barak et al., 2013). Many of the differentially expressed miRNAs also showed an inverse differential expression in a mouse model for Alzheimer’s disease, which has been associated with impaired adult hippocampal neurogenesis. These data point to an important role of miRNAs in conferring plasticity of hippocampal neurogenesis in response to modulatory signals.
In further support of this, the polycistronic miR-17-92 cluster, which shares miRNA seed family members with the miR-106b-25 cluster, was recently shown to regulate hippocampal neurogenesis in a mouse model of chronic stress (Jin et al., 2016). Stress and mood-related disorders have adverse effects on the neurogenic activity of the hippocampus, while antidepressant treatment may reverse this impairment. In fact, mood and psychiatric disorders have been associated with dysfunctional hippocampal neurogenesis (reviewed by Mahar et al., 2014). Mouse transgenic gain- and loss-of-function models revealed that miR-17-92 is required for aNSC proliferation in the dentate gyrus and that alterations of miR-17-92 expression have a strong impact on hippocampal neurogenesis and evoke changes in stress and anxiety-related behavioral tests. Mice depleted for miR-17-92 showed a reduced hippocampal neurogenic activity and exhibited anxiety-like behavior. It was further shown that miR-17-92 targets Sgk1, a downstream effector of glucocorticoid receptor signaling involved in cellular stress response, and that miR-17-92 can rescue the impairment of neural progenitor proliferation induced by corticosterone treatment (Jin et al., 2016). Expression of miR-17-92 itself is down-regulated upon chronic stress suggesting miR-17-92 as a physiological effector of stress-impaired hippocampal neurogenesis. Of note, miR-17-92 expression in aNSCs might change during aging as indicated by a recent study reporting a reduced miR-17-92 expression in neurogenic niches of old versus young N. furzeri fish (Terzibasi Tozzini et al., 2014).
MicroRNAs Modulate Adult Neuronal Differentiation by Acting in Concert With Gene Expression Regulators
Activated aNSCs give rise to IPCs, which then differentiate into neuroblasts that further mature into the respective neurons, i.e., olfactory bulb neurons (generated from the SVZ) and dentate granule cells (generated from the SGZ) (Figure 1). miRNAs have been shown to regulate the transition from proliferation to neuronal differentiation both during embryonic and adult neurogenesis. In this context, miRNAs are often found to interact with gene expression regulators forming feedback loops as we delineate in the following paragraphs. In fact, a recurrent feature of the miRNA’s mode of action is that several miRNAs act on the same transcriptional regulator, which in turn modulates the expression of its regulatory miRNAs, thereby forming feedback loops to fine-tune gene expression (reviewed by Arora et al., 2013; Lopez-Ramirez and Nicoli, 2014; Osella et al., 2014; Murao et al., 2016).
MicroRNA-124 and miR-9 Are Part of a Gene Regulatory Network Controlling Developmental and Adult Neurogenesis
One of the most studied miRNAs expressed in the brain are miR-124 and miR-9, which are not only able to induce neuronal differentiation of embryonic NSCs (reviewed by, e.g., Akerblom and Jakobsson, 2014; Roese-Koerner et al., 2013) but can even instruct a neurogenic gene expression program in non-neuronal cells (Yoo et al., 2011). Both miR-124 and miR-9 are widely expressed in the mouse adult brain including the hippocampus (Bak et al., 2008) and play important roles during adult neurogenesis. miRNA-124 has been shown to promote neuronal differentiation in the mouse SVZ by targeting the transcription factor SOX9 (Cheng et al., 2009). Stable overexpression of miR-124 in the mouse SVZ initially boosts neuronal differentiation but ultimately leads to premature exhaustion of the aNSC pool and loss of neurogenic activity (Akerblom et al., 2012). miRNA-9 is also expressed in the adult mouse SVZ, where it promotes neuronal differentiation (Zhao et al., 2009; Kim et al., 2015). One of the bona fide miR-9 targets identified in this context is Foxo1, which is related to Foxo3, and was shown to maintain aNSC self-renewal by acting in concert with Notch signaling (Kim et al., 2015). These findings are in contrast to the suggested role of miR-9 in maintaining quiescence in the zebrafish neurogenic niche (Katz et al., 2016), and it is not clear whether miR-9 impacts on both aNSC quiescence and differentiation in the adult mouse brain.
miRNA-124 and miR-9 have been shown to interact with an overlapping set of gene expression regulators that play important roles during NSC differentiation (reviewed by Stappert et al., 2014). Several of the transcription factors interacting with miR-124 and miR-9, e.g., REST and TLX, are also relevant in the context of adult neurogenesis. We will center our discussion on the miRNA-based circuitry formed around REST and TLX (Figures 3A,B).
FIGURE 3.
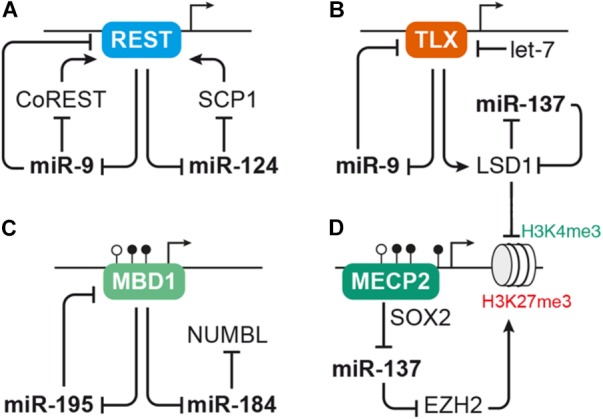
MicroRNAs interact with transcription factors and epigenetic regulators to control aNSC proliferation and differentiation. (A) The transcriptional repressor REST serves as regulatory hub for several miRNAs, including pro-neural miR-9 and miR-124. REST represses expression of miR-9 and miR-124, which in turn interfere with the activity of the Rest complex by targeting Rest or its cofactors CoRest and Scp1. (B) Another transcription factor interacting with miRNAs in the context of NSC self-renewal and differentiation is TLX. TLX has a direct repressive effect on miR-9 expression and also inhibits expression of miR-137 via recruiting the histone demethylase LSD1. The activity of TLX itself is regulated by miR-9 as well as let-7, which both target Tlx, and miR-137, which targets Lsd1. (C–D) miRNAs often interact with epigenetic regulators, such as mediators of DNA methylation and histone modifiers. (C) Expression of miR-184 and miR-195 is suppressed by binding of the methyl-CpG-binding protein MBD1 to their genomic loci. miRNA-195, in turn, binds to Mbd1 mRNA forming a negative feedback loop, while miR-184 targets Numbl. (D) MECP2 cooperates with the neural progenitor transcription factor SOX2 to repress miR-137 expression. miRNA-137 may regulate global gene transcription through repressing the expression of histone modifiers, i.e., EZH2, which induces repressive H3K27 trimethylation (me3) marks, and LSD1, which erases activating H3K4me3 marks. LSD1 and MECP2 are also recruited by the REST/CoREST complex, thus forming another node through which the miRNAs discussed here may cross talk with each other. For sake of simplicity, these interactions were not depicted here. For a detailed review on the cross talk between miRNAs and epigenetic regulators, see Jobe et al. (2012).
Both miR-124 and miR-9 are connected to the anti-neural transcription factor REST (Conaco et al., 2006; Visvanathan et al., 2007; Packer et al., 2008; Laneve et al., 2010). While in undifferentiated cells high levels of REST prevent the expression of miR-9 and miR-124, these miRNAs are up-regulated during differentiation and enforce their own expression by targeting Rest and its cofactors CoRest (Rcor2) and Scp1 (Figure 3A; Conaco et al., 2006; Visvanathan et al., 2007; Packer et al., 2008; Laneve et al., 2010). In the murine dentate gyrus, Rest is down-regulated during the transition of aNSCs to neuroblasts before it is up-regulated again in mature hippocampal granule cells (Gao et al., 2011). Adult NSC-specific depletion of Rest in the dentate gyrus results in premature differentiation and exhaustion of the aNSC pool (Gao et al., 2011). Of note, Rest-depleted aNSCs also show tremendous differences in their miRNA expression profile suggesting that dysregulation of miRNAs may at least in part underlie the imbalance of aNSC maintenance and differentiation induced by Rest-deficiency (Gao et al., 2012).
Another important transcription factor interacting with miRNAs to control NSC proliferation and differentiation is the nuclear receptor TLX (Nr2e1). TLX is expressed in neurogenic regions during development and adulthood, and its presumed function is to prevent premature differentiation and maintain the undifferentiated self-renewing state of NSCs (reviewed by Islam and Zhang, 2015). Conditional ablation of Tlx in dentate gyrus aNSCs does not induce differentiation, but results in precocious cell cycle exit of aNSCs that enter a quiescent inactive state (Shi et al., 2004; Zhang et al., 2008; Niu et al., 2011). Studies performed in mouse primary forebrain aNSCs revealed that miR-9 and let-7 promote neuronal and glial differentiation by targeting Tlx (Zhao et al., 2009, 2010). In utero electroporation into the lateral ventricles to overexpress miR-9 or let-7 during embryonic development resulted in a similar effect with a decrease of proliferative cells and an increase of differentiating, migrating cells (Zhao et al., 2009, 2010), indicating that miR-9 and let-7 induce differentiation both during developmental and adult neurogenesis. Interestingly, TLX has a direct repressive effect on miR-9 expression forming a negative feedback loop (Figure 3; Zhao et al., 2009). Furthermore, TLX also represses miR-137 by recruiting the histone demethylase LSD1 (Kdm1a) to the miR-137 locus. Lsd1 is highly expressed in mouse aNSCs and declines during neuronal differentiation. Inhibition of LSD1 results in a decrease of aNSC proliferation in vitro as well as in the adult dentate gyrus (Sun et al., 2010). Experiments in embryonic NSCs revealed that miR-137 promotes neuronal differentiation by targeting Lsd1 (Sun et al., 2011), thus adding another loop to the feedback circuity formed around TLX (Figure 3B).
MicroRNAs Interacting With Epigenetic Regulators to Balance aNSC Proliferation and Differentiation
Another recurring theme in the context of miRNA-based regulation is the interplay of miRNAs and epigenetic regulators, such as chromatin modifiers (reviewed by Jobe et al., 2012; Lopez-Ramirez and Nicoli, 2014; Murao et al., 2016). miRNA-137, for instance, was reported to target the histone methyltransferase EZH2. EZH2 is part of the polycomb group protein complex involved in epigenetic remodeling by histone methylation. In line with that, overexpression of miR-137 in aNSCs resulted in an overall reduction of H3K27 tri-methylation (Szulwach et al., 2010). The same study also reported an increase of aNSC proliferation at the expense of neuronal differentiation upon miR-137 overexpression (Szulwach et al., 2010), which is opposed to the pro-differentiation function of miR-137 reported in embryonic NSCs (Sun et al., 2011). It was further shown that expression of miR-137 in aNSCs depends on the action of another epigenetic regulator, the DNA methyl-CpG-binding protein MECP2, which in cooperation with SOX2 retains miR-137 expression (Szulwach et al., 2010). Thus, miR-137 may act in concert with several epigenetic regulators (LSD1, MECP2, and EZH2) to control global gene expression (Figures 3B,D). The question is, though, which of these targets are responsible for the distinct effects of miR-137 during proliferation and differentiation of NSCs.
Another example for the interaction of miRNAs with epigenetic regulators is the interaction of the methyl-CpG-binding protein MBD1 with miR-184 and miR-195 in dentate gyrus aNSCs (Figure 3C; Liu et al., 2010, 2013). Mbd1 is abundantly expressed in the adult brain with highest concentrations in the SGZ of the dentate gyrus. In line with that, Mbd1-deficient mice display reduced adult neurogenic activity and impaired hippocampal function (Zhao et al., 2003). Adult NSCs lacking Mbd1 accumulate at the level of IPCs and fail to transition to the neuronal fate (Jobe et al., 2017). Besides regulating the expression of lineage differentiation-associated protein-coding genes (Jobe et al., 2017), MBD1 was also found to repress the expression of miR-184 and miR-195 by direct binding to their proximal genomic regions (Liu et al., 2010, 2013). Both miR-184 and miR-195 have been shown to promote aNSC proliferation at the expense of neuronal differentiation in vitro as well as in vivo. Bona fide targets identified in this context were Numbl for miR-184 (Liu et al., 2010) and Mbd1 for miR-195, representing yet another example of a negative feedback loop to reinforce miRNA expression (Liu et al., 2013). Additionally, there seems to be some functional redundancy between the two MBD1-regulated miRNAs, miR-195 and miR-184, as the effect induced by miR-195 overexpression could be rescued by miR-184 inhibition (Liu et al., 2013).
MicroRNAs Regulating Survival in the Adult Neurogenic Niche
An important feature determining the neurogenic output of aNSC is the survival of their neuronal progeny. In fact, under normal conditions, the majority of newborn dentate gyrus neurons undergoes apoptosis within the first days of their life with apoptotic neurons being cleared out by microglia (Sierra et al., 2010; reviewed by Kim and Sun, 2011). Several miRNAs have been shown to regulate survival-associated genes, making them an interesting target for neuroprotective or cell replacement strategies in neurodegenerative diseases (Zhang et al., 2018), stroke (Sun et al., 2017), and epilepsy (Schouten et al., 2015; Yuan et al., 2016; Bielefeld et al., 2017).
A series of studies has been focusing on the impact of global miRNA loss using conditional knockout mouse (cKO) lines for Dicer, the key enzyme of miRNA biogenesis (e.g., Davis et al., 2008; De Pietri Tonelli et al., 2008; Hébert et al., 2010; Konopka et al., 2010; Li et al., 2011; Cheng S. et al., 2014). While these studies came to sometimes contradictory results [see Barca-Mayo and De Pietri Tonelli (2014) for a detailed comparison], many of them report that Dicer depletion in the embryonic mouse forebrain reduces forebrain growth, increases apoptosis, and leads to premature death of the animals (Davis et al., 2008; De Pietri Tonelli et al., 2008; Hébert et al., 2010; Li et al., 2011). Likewise, specific deletion of Dicer in forebrain (cortical and hippocampal) neurons at postnatal stages also resulted in enhanced apoptosis and neuronal loss (Konopka et al., 2010; Cheng S. et al., 2014). Taken together, these findings indicate that a functional miRNA system is critical for neuron survival, but they do not provide direct information on the impact of the miRNA system on the generation and survival of adult-born neurons.
This question was recently assessed by two studies, in which Dicer was specifically deleted in aNSCs (Cernilogar et al., 2015; Pons-Espinal et al., 2017). In the first study, Dicer was inactivated by delivery of Cre recombinase into primary neural progenitors isolated from the adult SVZ or by retroviral injection into the adult hippocampus of Dicerflox/flox mice. Both in vitro as well as in vivo an increase in doublecortin Dcx expression was noted, hinting to an enhanced neuronal differentiation upon Dicer deletion (Cernilogar et al., 2015). However, the authors also observed a reduced viability of Dicer-depleted cells in their in vivo experiments. Pons-Espinal et al. (2017) used the Split-Cre approach developed by Beckervordersandforth et al. (2014) to selectively inactivate Dicer in aNSCs in the dentate gyrus and performed experiments on primary aNSCs isolated from Dicerflox/flox mice that were nucleofected with Cre recombinase. In contrast to the earlier study by Cernilogar et al. (2015), they observed an impaired neurogenic activity and a bias toward astrocytic differentiation. Furthermore, they noted a reduced survival of Dicer-depleted aNSCs both in vivo as well as in vitro, while the proliferative capacity of in vitro cultured aNSCs was not affected. This is in line with previous studies on embryonic NSCs reporting that the ability for NSC self-renewal in culture does not depend on Dicer activity (De Pietri Tonelli et al., 2008; Andersson et al., 2010). These Dicer cKO embryonic NSCs were, however, compromised with regard to their differentiation capacity (Andersson et al., 2010). Furthermore an increased cell loss was observed during differentiation of Dicer-depleted NSCs (embryonic or adult) indicating that differentiated cells are more sensitive toward global miRNA loss than undifferentiated cells (De Pietri Tonelli et al., 2008; Pons-Espinal et al., 2017).
These two findings prompted the hypothesis that a functional miRNA system is particularly important for cell fate transitions (De Pietri Tonelli et al., 2008; Andersson et al., 2010). Following this notion, Pons-Espinal et al. (2017) focused on a pool of 11 miRNAs (including miR-124 and miR-134) that was found to be highly up-regulated during early neuronal differentiation and asked whether these miRNAs could rescue the bias toward astrocytic differentiation and the impaired neuronal differentiation of Dicer cKO aNSCs (Pons-Espinal et al., 2017). Interestingly, they found that only combined delivery of all 11 miRNAs, but not subsets of them, was able to rescue the Dicer deletion phenotypes (Pons-Espinal et al., 2017). By combining proteomics and in silico miRNA target gene prediction they further found that quite a number of targets are shared by at least 2 of the 11 miRNAs, which led them to speculate that these 11 miRNAs may have a cooperative function targeting an overlapping set of genes to induce neuronal differentiation of aNSC. In fact, cooperative binding of several miRNAs to the same target mRNA is an important feature of the miRNAs’ mode of action and may create functional redundancy (reviewed by Barca-Mayo and De Pietri Tonelli, 2014). However, the data generated by Pons-Espinal et al. (2017) suggest that the level of redundancy among the 11 miRNA is rather low since only the combination of all miRNAs was able to compensate for Dicer cKO and not the individual miRNAs.
MicroRNAs Regulating Migration and Neurite Morphogenesis of Adult-Born Neurons
MicroRNAs also influence functional integration of the neuronal progeny, i.e., migration to their final homing site, neuronal morphogenesis, and synaptogenesis. Immature neuroblasts that arise from the SGZ first have to migrate up into the granule cell layer before they start extending dendrites toward the molecular layer of the dentate gyrus and grow axons that target the CA3 region of the hippocampus, approximately 1 week after their birth (Figure 1C) (Zhao et al., 2006). The maturation process in adult neurogenesis differs from that of embryonic neurogenesis in that adult-born neurons have to integrate into already existing coordinated neuronal networks and that they receive synaptic inputs early on. As shown for mouse hippocampal neurogenesis, immature adult granule cells initially receive excitatory GABAergic inputs, which become inhibitory by 2 weeks after their birth (reviewed by Gonçalves et al., 2016). Around the same time, immature adult-born granule cells also receive glutamatergic input, start developing dendritic spines, and establish efferent and afferent synapses with the local neuron network (reviewed by Deng et al., 2010). Neurons that fail to develop strong synaptic connections will undergo selective apoptosis (Tashiro et al., 2006; and reviewed by Kim and Sun, 2011). Finally, by 8 weeks after their birth, adult-born granule neurons are considered to be fully mature and are indistinguishable from their earlier-born neighbors (Laplagne et al., 2006; and reviewed by Deng et al., 2010).
The molecular mechanisms guiding adult-generated granule cells to their final homing site in the granule cell layer and their mode of migration have not yet been completely resolved. Interestingly, it has been reported that adult-born dentate gyrus neuroblasts first undergo tangential migration along the blood vasculature in the SGZ followed by radial migration to reach the granule cell layer (Sun et al., 2015). While miRNAs have been shown to regulate various genes involved in neuron migration during development (reviewed by Rajman and Schratt, 2017), there is only one study that specifically assessed the impact of miRNAs in the context of adult-born neuron migration (Han et al., 2016). In this study, Han et al. (2016) discovered that elevated levels of miR-19 increased cell migration of in vitro cultured hippocampal neural progenitors. They further showed that ectopic expression of miR-19 in the dentate gyrus triggered newborn granule cells to migrate deeper into the granule cell layer. Likewise, neuroblasts generated from the SVZ were found to cover longer distances within the RMS upon miR-19 overexpression. These data indicate that miR-19 promotes migration of adult-born neurons (Han et al., 2016). This function seems to be at least in part mediated by miR-19 targeting Rapgef2, which regulates cell migration by modulating the activity of RAP proteins (Han et al., 2016). Knock-down of Rapgef2 in neural progenitors mimicked the effect of miR-19 overexpression as it promoted migration of in vitro cultured neural progenitors and newborn granule cells in the dentate gyrus granule cell layer. Since abnormal migration of adult-born hippocampal neurons was also described in schizophrenia (Duan et al., 2007; Kim et al., 2009), and rare inherited copy number variants of RAPGEF2 have been associated with familiar schizophrenia (Xu et al., 2009), Han et al. (2016) went on to study the role of miR-19 and RAPGEF2 in schizophrenia patient-derived hippocampal neural progenitor cells (SZ-NPCs). Indeed, they found that miR-19 is up-regulated in SZ-NPCs and that the regulation of RAPGEF2 by miR-19 is conserved in humans (Han et al., 2016). miRNA-19 belongs to the family of polycistronic miR-17 clusters, including also miR-17-92 and miR-106b-25b, which have been shown to be important for aNSC proliferation (Brett et al., 2011; Jin et al., 2016). Hence, it would be interesting to assess whether the other members of the miR-17 clusters are also altered in schizophrenia and whether they contribute to the disease.
After the immature neuroblasts have reached the granule cell layer, they develop into mature neurons, extend dendrites and axons, and establish synaptic contacts. Numerous miRNA-target pairs have been identified to regulate neurite outgrowth and morphogenesis as well as synaptic plasticity (e.g., reviewed by Bicker et al., 2014; Hu and Li, 2017; Rajman and Schratt, 2017). Some miRNAs even show a specific synaptic localization, and several miRNAs are regulated in response to neuronal activity (reviewed by Hu and Li, 2017). Here, we focus on those miRNAs that have been shown to regulate neuronal morphogenesis of hippocampal neurons. Due to the limited number of studies addressing morphogenesis of adult-generated neurons, we also include studies on the role of miRNAs regulating neurite formation and maturation in the developing hippocampus (see Table 2 for an overview of the miRNAs discussed). In the following paragraphs, we highlight examples of miRNAs that converge on functionally related targets or are regulated by the same set of transcription factors (Figure 4).
FIGURE 4.
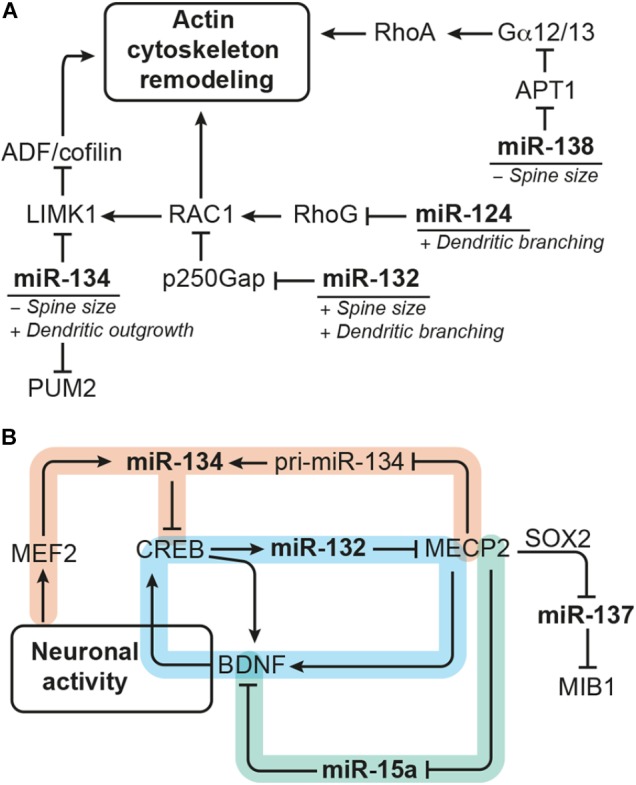
miRNAs modulating dendrite morphogenesis converge by targeting components of the actin remodeling complex and cross talk with each other via neuronal activity-dependent gene regulatory networks. (A) miRNA-124, miR-132, miR-134, and miR-138 have divergent effects on dendrite morphogenesis by targeting different components of the actin remodeling pathway. miRNA-124 and miR-134 reduce the activity of RAC1/LIMK1 signaling, whereby miR-124 targets RhoG and miR-134 targets Limk1. miRNA-132, in contrast, increases RAC1/LIMK1 activity by inhibiting p250Gap-mediated repression of RAC1. miRNA-138 increases Gα12/13 palmitoylation and downstream RhoA signaling by targeting the mRNA coding for the depalmitoylation enzyme APT1. An additional target of miR-134 is the translational repressor PUM2 (Pumilio2) through which miR-134 promotes overall dendritogenesis. (B) Many of the miRNAs modulating dendrite morphogenesis engage in regulatory circuits with transcription regulators that respond to neuronal activity. One such regulatory circuit is formed around miR-132 (highlighted in blue), whose expression is induced by CREB downstream of neuronal activity. miRNA-132 itself may influence CREB activation via interfering with MECP2-induced expression of Bdnf, which forms a positive feedback loop with CREB. The regulation of miR-134 expression (in orange) seems to be more complex: miR-134 expression was found to be induced by MEF2 downstream of neuronal activity and inhibited by MECP2, which interferes with pri-miR-134 processing. miRNA-134, in turn, targets Creb and may thereby provide a negative feedback signal for activity-induced gene expression and also influence miR-132 transcription. Another regulatory hub involves miR-15a (in green), the expression of which is inhibited by MECP2. miRNA-15a targets Bdnf mRNA, thereby providing an alternative route through which MECP2 promotes Bdnf expression. MECP2 further represses expression of miR-137 by cooperating with SOX2. miRNA-137 inhibits neuronal maturation and dendrite morphogenesis by targeting the ubiquitin ligase MIB1.
One common pathway through which miRNAs modulate neuronal morphogenesis is Rho GTPase signaling (Figure 4A). Rho GTPase-mediated remodeling of the actin cytoskeleton is important for regulating several aspects of neuronal morphogenesis (reviewed by Stankiewicz and Linseman, 2014) including dendritic spine maturation of hippocampal neurons (Vadodaria et al., 2013). miRNA-124 promotes axonal and dendrite complexity of rat embryonic hippocampal neurons by inhibiting expression of Rhog GTPase (Franke et al., 2012). miRNA-134 negatively affects the size of dendritic spines of in vitro cultured embryonic rat hippocampal neurons by targeting Limk1, which regulates actin filament dynamics via inhibition of ADF/cofilin (Schratt et al., 2006). In addition, miR-134 was shown to have a growth-promoting effect on dendrites, which is mediated by Pumilio2 (Pum2), a translational repressor involved in dendritogenesis (Fiore et al., 2009). miRNA-132 enhances dendrite growth and spine maturation of in vitro cultured neonatal rat hippocampal neurons by inhibiting expression of the Rho GTPase activating protein p250Gap (Arhgap32), which results in an increased RAC actin remodeling signal and LIMK1 activation (Wayman et al., 2008). Furthermore, miR-138 has been described to negatively regulate the size of dendritic spines by targeting the mRNA coding for the depalmitoylation enzyme APT1 (Siegel et al., 2009). Inhibition of Apt1 expression by miR-138 leads to an increased palmitolyation and membrane-tethering of G protein Gα12/13, which activates RhoA signaling – another component of the actin remodeling pathway.
MicroRNA expression is modulated by transcriptional regulators, and many of the miRNAs involved in dendrite morphogenesis have been found to be targets of neuronal activity-associated gene regulatory networks. The transcription factors operating in those gene regulatory networks are by themselves often subject to miRNA regulation. Experiments in rat hippocampal neurons showed that expression of the polycistronic miR-379-410 cluster, which also contains miR-134, is induced by the transcription factor MEF2 in response to membrane depolarization or BDNF treatment (Fiore et al., 2009). However, BDNF treatment was also shown to relieve miR-134-mediated repression of Limk1 in dendrites of embryonic rat hippocampal neurons via a yet unknown mechanism (Schratt et al., 2006). Another miRNA up-regulated in response to neuronal activity is miR-132, the expression of which is induced by the transcription factor CREB. CREB contributes to activity-induced refinement of dendrite morphology and is regulated by miR-134 (Gao et al., 2010). In addition, miR-132 fine-tunes BDNF-mediated CREB activation by targeting Mecp2, which induces Bdnf (Klein et al., 2007). Recently, it has also been shown that MECP2 can directly interact with the miRNA processing machinery to inhibit the expression of mature miR-134 (Cheng T.-L. et al., 2014). Thus, miR-132 and miR-134 may cross talk in the context of dendrite morphogenesis through CREB and MECP2 (Figure 4B). MECP2 also interacts with SOX2 to retain the expression of miR-137 (Szulwach et al., 2010), which negatively regulates neuronal maturation and dendrite morphogenesis by targeting the mRNA coding for ubiquitin ligase MIB1 (Smrt et al., 2010). MECP2 was further shown to act as a repressor of miR-15a expression, and miR-15a inhibits dendrite maturation of developmental- and adult-born hippocampal neurons by targeting Bdnf (Gao et al., 2015). Thus, MECP2-mediated repression of miR-15a represents an indirect route to promote Bdnf expression downstream of MECP2 (Figure 4B).
It is important to note that most of the experiments discussed above were performed in primary cultures of rodent embryonic or neonatal hippocampal neurons (Table 2), thus addressing the effect of miRNAs on neurons born in an early developmental phase. However, the same miRNA-target pairs might be also involved in regulating neurite morphogenesis during adult neurogenesis. Depletion of the miR-132 genomic loci in dentate gyrus aNSCs, for instance, was shown to reduce dendrite complexity of neuronal progenies demonstrating that miR-132 has a positive effect on dendrite maturation both during embryonic and adult hippocampal neurogenesis (Magill et al., 2010). In contrast, miR-137 and miR-15a were demonstrated to have a negative effect on dendrite maturation of adult-born dentate granule cells (Smrt et al., 2010; Gao et al., 2015). Another miRNA specifically studied in the context of adult neurogenesis and neuronal maturation is miR-223. Inhibition of miR-223 in dentate gyrus aNSCs by retroviral miRNA sponge injection resulted in an increased dendrite length, while overexpression of miR-223 in human embryonic stem cell-derived neurons had the opposite effect, suggesting that miR-223 also regulates neuronal morphogenesis both during developmental and adult neurogenesis (Harraz et al., 2014).
Taken together, these examples of miRNAs-target pairs that cross talk at different hierarchy levels illustrate once more the complexity and the importance of the miRNA system for adult neurogenesis. Furthermore, many of the miRNAs involved in neuronal morphogenesis discussed above have also been linked to neurological disorders, like Alzheimer’s disease (miR-132, miR-138), Huntington’s disease (miR-132, miR-137, miR-138), psychiatric disorders (miR-132, miR-134, miR-137, miR-138), and epilepsy (miR-134, miR-138) (reviewed by, e.g., Bicker et al., 2014). It is therefore tempting to speculate that dysregulation of miRNAs and altered neuronal morphogenesis may contribute to the disease pathology.
A Synopsis of the Diverse Roles of MicroRNAs in Adult Neurogenesis
Adult neurogenesis is a multistep process comprising activation of quiescent aNSCs, their differentiation into committed progenitor cells, neuronal survival, migration and functional integration of newborn neurons (Figure 1D). Transitions along these steps are accompanied by dynamic gene expression changes (Llorens-Bobadilla et al., 2015; Shin et al., 2015; Dulken et al., 2017; and reviewed by Beckervordersandforth et al., 2015). As discussed in this review and also in other reports (e.g., reviewed by Lopez-Ramirez and Nicoli, 2014; Murao et al., 2016), miRNAs are an integral part of the gene regulatory networks driving these changes. Although not all interactions discussed here have been experimentally validated in the context of adult neurogenesis, many of the individual factors involved, e.g., miR-124, miR-9, let-7, and miR-137 on the one side and REST, TLX, and LSD1 on the other side, have been shown to play important roles during adult neurogenesis (see Figure 3 and Table 1 for an overview). Thus, the emerging picture is that miRNAs are frequently engaged in feedback loops with transcription factors and epigenetic regulators importantly involved in regulating adult neurogenesis.
By doing so, miRNAs provide an additional layer to control gene expression programs and may help to ensure the robustness of such programs by dampening perturbations and reducing noise (reviewed by Arora et al., 2013; Lopez-Ramirez and Nicoli, 2014; Osella et al., 2014; Murao et al., 2016). However, miRNAs are also involved in remodeling gene expression programs during neural lineage progression (reviewed by Herranz and Cohen, 2010; Peláez and Carthew, 2012). miRNAs may even exert an instructive effect on cell fate as impressively demonstrated by the finding that miR-9/9∗ and miR-124 can induce neuronal conversion of fibroblasts (Yoo et al., 2011). Furthermore, global miRNA loss by Dicer depletion seems to evoke stronger effects in differentiating cells than in self-renewing NSCs (derived from either embryonic and adult origin), suggesting that cell fate transitions show a particular dependency on miRNA-based regulation (De Pietri Tonelli et al., 2008; Andersson et al., 2010; Pons-Espinal et al., 2017).
However, some of the miRNA functions might be context-dependent. For instance, miR-9 was found to maintain quiescence of aNSCs in the zebrafish brain (Katz et al., 2016), while in mouse aNSCs it promotes neuronal differentiation of aNSCs (Zhao et al., 2009; Kim et al., 2015). These divergent observations might be due to the different experimental model systems employed but may also reflect two different modes of action of miR-9, i.e., cytoplasmic miR-9 promotes neuronal differentiation via canonical targeting of Tlx and Foxo1 (Zhao et al., 2009; Kim et al., 2015), while nuclear miR-9 maintains aNSC quiescence (Katz et al., 2016). Another example of a miRNA eliciting context-dependent effects is miR-137, which was found to promote differentiation of embryonic NSCs (Sun et al., 2010) but inhibits differentiation of adult NSCs (Szulwach et al., 2010).
MicroRNAs Converge on Shared Targets to Control Adult Neurogenesis
Many miRNAs have been reported to have rather mild effects on their target genes (Baek et al., 2008; Selbach et al., 2008). Yet, each miRNA might target several hundreds of mRNAs (Lewis et al., 2005; Lim et al., 2005). This “multiplicity” of miRNA targets means that, although the effect of an individual miRNA on a given target might be rather weak, this miRNA might still exert a meaningful biological effect by acting on different genes with overlapping functions (Barca-Mayo and De Pietri Tonelli, 2014; Fischer et al., 2015). Another important feature of miRNA function is “cooperativity,” which describes that most mRNAs possess binding sites for multiple miRNAs (Barca-Mayo and De Pietri Tonelli, 2014; Schmitz et al., 2014; Fischer et al., 2015). Cooperative binding of several miRNAs to the same target mRNA creates functional redundancy and might compensate for the rather mild repression mediated by individual miRNAs on that given target. It has been suggested that by the interplay of multiplicity and cooperativity, miRNAs may have “converging functions” defined as synergic action of a single miRNA or several miRNAs on multiple targets that belong to the same pathway or are exerting redundant functions (Barca-Mayo and De Pietri Tonelli, 2014). For instance, miR-9 and miR-124 have been shown to drive neuronal differentiation of NSCs by converging on Rest, Baf53a, and components of the Notch signaling cascade as common target genes (reviewed by Stappert et al., 2014). Other examples of miRNAs with convergent functions and mentioned in this review in the context of adult neurogenesis are let-7 and miR-9, which both target Tlx (Figure 3B and Table 1; Zhao et al., 2009, 2013), and miR-184 and miR-195, which are both regulated by MBD1 and promote aNSC proliferation (Figure 3C and Table 1; Liu et al., 2010, 2013). Furthermore, several miRNAs (i.e., miR-124, miR-132, miR-134, and miR-138) have been found to be involved in dendrite morphogenesis by targeting components of the actin remodeling pathway as common denominator (Figure 4A and Table 2; Schratt et al., 2006; Wayman et al., 2008; Siegel et al., 2009; Franke et al., 2012). Another mechanism to coordinate miRNA function is to couple the expression of several miRNAs to a common transcription regulator as shown for MECP2, which regulates the expression of various miRNAs involved in dendrite morphogenesis (Figure 4B; Szulwach et al., 2010; Cheng T.-L. et al., 2014; Gao et al., 2015). miRNA cooperativity may be also reflected by the genomic localization of miRNA genes (reviewed by Olive et al., 2015). Many miRNAs are located in polycistronic clusters that encode for members of different miRNA seed families (Figure 2), as is the case for the paralog clusters miR-17-92, miR-160b-25, and miR-106a-363. Interestingly, these clusters encode for miRNAs with distinct functions in adult neurogenesis. For example, miR-25 promotes aNSC proliferation (Brett et al., 2011), whereas miR-19 enhances neuroblast migration (Han et al., 2016; Figure 1D). It would be interesting to assess the extent of co-expression of these miRNAs and their seed family members in the aNSC compartments.
Taken together, these examples illustrate how miRNAs act in concert with gene regulatory networks and also cooperate with each other by targeting functionally related genes. However, most of the reports mentioned above have focused on the action of a single miRNA-target pair. Future studies should also investigate the cooperative function of miRNAs in the context of adult neurogenesis, as it was addressed by Pons-Espinal et al. (2017).
MicroRNAs May Contribute to Heterogeneity in the aNSC Niche
Neural progenitors within the adult neurogenic niche are a heterogeneous population that can be distinguished by their cell cycle status (quiescence versus activated cells), by their differentiation potential (neurogenic or gliogenic), and their differentiation stage (aNSCs, IPCs, and neuroblasts) (reviewed by Giachino and Taylor, 2015; Bonaguidi et al., 2016). miRNAs might be importantly involved in conferring aNSC heterogeneity. miRNA-9, for instance, is specifically found in the nucleus of a subset of quiescent aNSCs in the adult brain of zebrafish and mouse (Katz et al., 2016). Disruption of the nuclear localization of miR-9 leads to an increased activation of aNSCs suggesting that this particular expression pattern of miR-9 is crucial for aNSC quiescence (Katz et al., 2016). In addition, miRNAs are involved in biasing embryonic NSCs to either neurogenic or gliogenic differentiation (reviewed by Rajman and Schratt, 2017; Shimazaki and Okano, 2018). It is well perceivable that these miRNAs might have a similar role in aNSCs. In fact, it is not yet clear whether aNSCs are truly multipotent or whether several neural precursor populations with variable differentiation potencies (neurogenic or gliogenic) exist within the NSC niche (reviewed by Bonaguidi et al., 2016), and it might well be that miRNAs could contribute to this heterogenic differentiation potency. Indeed, in their aNSC-specific Dicer-knockout model, Pons-Espinal et al. (2017) discovered a shift toward astrogial differentiation at the expense of neuronal differentiation, which could be rescued by combined delivery of 11 miRNAs.
Recent advances regarding single cell tracing and single cell transcriptomics have led to the assignment of specific gene expression profiles to different cell states and further demonstrated the presence of diverse cell states along the process of adult neurogenesis (Llorens-Bobadilla et al., 2015; Shin et al., 2015; Dulken et al., 2017). These analyses revealed that aNSCs do not exist in only two stages (quiescent and activated aNSCs) but instead move through a continuum of different stages during activation (Llorens-Bobadilla et al., 2015; Dulken et al., 2017). It was further shown that aNSC activation is associated with increased protein synthesis (Llorens-Bobadilla et al., 2015; Shin et al., 2015) as well as with vast expression changes in genes associated with energy metabolism, transcriptional regulation, and signaling pathway integration (Shin et al., 2015). Since miRNAs provide an important mechanism to control mRNA–protein output, and many signaling pathway components as well as transcription factors are regulated by miRNAs, it would be interesting to also analyze miRNA profiles at a single cell level and to assess to what extent miRNA expression in the adult neurogenic niche reflects aNSC heterogeneity.
MicroRNAs May Contribute to Homeostasis in the aNSC Niche in Healthy and Disease Conditions
Although there is currently a controversial discussion about the extent and role of adult neurogenesis in humans (Boldrini et al., 2018; Sorrells et al., 2018; and reviewed by Kempermann et al., 2018), there remains a strong interest to decipher the molecular mechanisms governing adult neurogenesis and to identify novel tools to gain control over this process. This direction of research is driven by the idea to recruit aNSCs as endogenous cell source replacing the cells lost due to aging, acute lesions, or neurodegenerative diseases (reviewed by Lindvall and Kokaia, 2010). Since miRNAs have been shown to regulate aNSC activation, proliferation, and differentiation and may thereby contribute to the homeostatic regulation in the adult neurogenic niche, they could be envisioned as targets to harness aNSCs for therapeutics approaches. Promising candidates might be miR-9, let-7, miR-106b-25, and miR-17-92, which regulate the balance between quiescence and activation of aNSCs (Nishino et al., 2008; Brett et al., 2011; Jin et al., 2016; Katz et al., 2016). miRNAs might be even involved in conferring plasticity of adult hippocampal neurogenesis in response to environmental signals as shown for miR-17-92 and stress (Jin et al., 2016). Furthermore, miRNAs might also impact on age-dependent decline of adult neurogenesis, and a number of miRNAs have been found to interact with important aNSC regulators that are also associated with aging, including HMGA2, FOXO1, FOXO3, and TLX (Nishino et al., 2008; Zhao et al., 2009, 2010; Brett et al., 2011; Kim et al., 2015). In addition, a functional miRNA system seems to be important to sustain adult neurogenesis and survival within the neurogenic niche (Cernilogar et al., 2015; Pons-Espinal et al., 2017). Finally, numerous miRNAs are dysregulated in pathophysiological conditions associated with dysfunctional hippocampal neurogenesis, such as epilepsy (reviewed by Bielefeld et al., 2017), stroke (reviewed by Khoshnam et al., 2017), and neurodegenerative diseases (reviewed by Qiu et al., 2014). These miRNAs might represent promising targets to not only tackle the primary cause of the disease but to also counteract the disease-induced impairment of adult neurogenesis.
From an evolutionary perspective, it is noteworthy that adult neurogenesis appears to be less pronounced in human compared to rodent brain (e.g., Sanai et al., 2011; Sorrells et al., 2018). The miRNA-target pairs discussed in this review were all identified in rodents, but considering their sequence similarity they should be largely conserved in humans. Nevertheless, there is quite some evolutionary pressure on the miRNA regulome as indicated by the presence of primate-specific miRNAs (Awan et al., 2017), the evolution of miRNA binding sites on mRNA targets (Gardner and Vinther, 2008), and the acquisition of novel factors regulating miRNA-associated processes across evolution (Pratt and Price, 2016). Thus, elucidating the differences in miRNA-based regulation of murine versus human adult neurogenesis might eventually enable the promotion of adult neurogenesis in humans and their exploitation for regenerative purposes.
Author Contributions
LS, FK, and OB wrote the manuscript and designed the figures.
Conflict of Interest Statement
OB is a co-founder of and has stock in Life & Brain GmbH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. Work in the laboratory of OB was supported by the EU [Seventh Framework Program HEALTH-F4-2013-602278-NeuroStemCellRepair; Horizon2020 grant 667301-COSYN and European Cooperation in Science and Technology (COST) grant CA16210-MINDDS]; the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (grant 01ZX1314A-IntegraMent); the German Federal Ministry of Education and Research (BMBF) grant 01EK1603A-Neuro2D3; the North Rhine Westphalian Program “LifeSciences.NRW”; European Regional Development Fund (EFRE) (grant EFRE-0800978-SCF III); the National Institutes of Health (award 1R01NS100514) and the Stem Cell Network North Rhine Westphalia (start-up financing for interdisciplinary and multi-site projects-323-40000513). LS was supported by the BONFOR program.
References
- Akerblom M., Jakobsson J. (2014). MicroRNAs as neuronal fate determinants. Neuroscientist 20 235–242. 10.1177/1073858413497265 [DOI] [PubMed] [Google Scholar]
- Akerblom M., Sachdeva R., Barde I., Verp S., Gentner B., Trono D., et al. (2012). MicroRNA-124 is a subventricular zone neuronal fate determinant. J. Neurosci. 32 8879–8889. 10.1523/JNEUROSCI.0558-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson T., Rahman S., Sansom S. N., Alsiö J. M., Kaneda M., Smith J., et al. (2010). Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS One 5:e13453. 10.1371/journal.pone.0013453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora S., Rana R., Chhabra A., Jaiswal A., Rani V. (2013). miRNA-transcription factor interactions: a combinatorial regulation of gene expression. Mol. Genet. Genomics 288 77–87. 10.1007/s00438-013-0734-z [DOI] [PubMed] [Google Scholar]
- Awan H. M., Shah A., Rashid F., Shan G. (2017). Primate-specific long non-coding RNAs and microRNAs. Genom. Proteom. Bioinform. 15 187–195. 10.1016/j.gpb.2017.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Villén J., Shin C., Camargo F. D., Gygi S. P., Bartel D. P. (2008). The impact of microRNAs on protein output. Nature 455 64–71. 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak M., Silahtaroglu A., Møller M., Christensen M., Rath M. F., Skryabin B., et al. (2008). MicroRNA expression in the adult mouse central nervous system. RNA 14 432–444. 10.1261/rna.783108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak B., Shvarts-Serebro I., Modai S., Gilam A., Okun E., Michaelson D. M., et al. (2013). Opposing actions of environmental enrichment and Alzheimer’s disease on the expression of hippocampal microRNAs in mouse models. Transl. Psychiatry 3:e304. 10.1038/tp.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barca-Mayo O., De Pietri Tonelli D. (2014). Convergent microRNA actions coordinate neocortical development. Cell. Mol. Life Sci. 71 2975–2995. 10.1007/s00018-014-1576-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D. P. (2009). MicroRNAs: target recognition and regulatory functions. Cell 136 215–233. 10.1016/j.cell.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R., Deshpande A., Schäffner I., Huttner H. B., Lepier A., Lie D. C., et al. (2014). In vivo targeting of adult neural stem cells in the dentate gyrus by a split-cre approach. Stem Cell Rep. 2 153–162. 10.1016/j.stemcr.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R., Zhang C.-L., Lie D. C. (2015). Transcription-factor-dependent control of adult hippocampal neurogenesis. Cold Spring Harb. Perspect. Biol. 7:a018879. 10.1101/cshperspect.a018879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S., Xu T.-L., Sun T. (2013). Tuning the cell fate of neurons and glia by microRNAs. Curr. Opin. Neurobiol. 23 928–934. 10.1016/j.conb.2013.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicker S., Lackinger M., Weiß K., Schratt G. (2014). MicroRNA-132, -134, and -138: a microRNA troika rules in neuronal dendrites. Cell. Mol. Life Sci. 71 3987–4005. 10.1007/s00018-014-1671-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielefeld P., Mooney C., Henshall D. C., Fitzsimons C. P. (2017). miRNA-mediated regulation of adult hippocampal neurogenesis; implications for epilepsy. BPL 3 43–59. 10.3233/BPL-160036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrini M., Fulmore C. A., Tartt A. N., Simeon L. R., Pavlova I., Poposka V., et al. (2018). Human hippocampal neurogenesis persists throughout aging. Stem Cell 22 589.e5–599.e5. 10.1016/j.stem.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonaguidi M. A., Stadel R. P., Berg D. A., Sun J., Ming G.-L., Song H. (2016). Diversity of neural precursors in the adult mammalian brain. Cold Spring Harb. Perspect. Biol. 8:a18838-20. 10.1101/cshperspect.a018838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett J. O., Renault V. M., Rafalski V. A., Webb A. E., Brunet A. (2011). The microRNA cluster miR-106b∼25 regulates adult neural stem/progenitor cell proliferation and neuronal differentiation. Aging 3 108–124. 10.18632/aging.100285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernilogar F. M., Di Giaimo R., Rehfeld F., Cappello S., Lie D. C. (2015). RNA interference machinery-mediated gene regulation in mouse adult neural stem cells. BMC Neurosci. 16:60. 10.1186/s12868-015-0198-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapouton P., Skupien P., Hesl B., Coolen M., Moore J. C., Madelaine R., et al. (2010). Notch activity levels control the balance between quiescence and recruitment of adult neural stem cells. J. Neurosci. 30 7961–7974. 10.1523/JNEUROSCI.6170-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L.-C., Pastrana E., Tavazoie M., Doetsch F. (2009). miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12 399–408. 10.1038/nn.2294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., Zhang C., Xu C., Wang L., Zou X., Chen G. (2014). Age-dependent neuron loss is associated with impaired adult neurogenesis in forebrain neuron-specific Dicer conditional knockout mice. Int. J. Biochem. Cell Biol. 57 186–196. 10.1016/j.biocel.2014.10.029 [DOI] [PubMed] [Google Scholar]
- Cheng T.-L., Wang Z., Liao Q., Zhu Y., Zhou W.-H., Xu W., et al. (2014). MeCP2 suppresses nuclear microRNA processing and dendritic growth by regulating the DGCR8/Drosha complex. Dev. Cell 28 547–560. 10.1016/j.devcel.2014.01.032 [DOI] [PubMed] [Google Scholar]
- Conaco C., Otto S., Han J.-J., Mandel G. (2006). Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. U.S.A. 103 2422–2427. 10.2307/30048143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis T. H., Cuellar T. L., Koch S. M., Barker A. J., Harfe B. D., McManus M. T., et al. (2008). Conditional loss of dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 28 4322–4330. 10.1523/JNEUROSCI.4815-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D., Pulvers J. N., Haffner C., Murchison E. P., Hannon G. J., Huttner W. B. (2008). miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development 135 3911–3921. 10.1242/dev.025080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W., Aimone J. B., Gage F. H. (2010). New neurons and new memories: how does adult hippocampal neurogenesis affect learning and memory? Nat. Rev. Neurosci. 11 339–350. 10.1038/nrn2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doetsch F., García-Verdugo J. M., Alvarez-Buylla A. (1997). Cellular composition and three-dimensional organization of the subventricular germinal zone in the adult mammalian brain. J. Neurosci. 17 5046–5061. 10.1523/JNEUROSCI.17-13-05046.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X., Chang J. H., Ge S., Faulkner R. L., Kim J. Y., Kitabatake Y., et al. (2007). Disrupted-in-schizophrenia 1 regulates integration of newly generated neurons in the adult brain. Cell 130 1146–1158. 10.1016/j.cell.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulken B. W., Leeman D. S., Boutet S. C., Hebestreit K., Brunet A. (2017). Single-cell transcriptomic analysis defines heterogeneity and transcriptional dynamics in the adult neural stem cell lineage. Cell Rep. 18 777–790. 10.1016/j.celrep.2016.12.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Encinas J. M., Fitzsimons C. P. (2017). Gene regulation in adult neural stem cells, current challenges and possible applications. Adv. Drug Deliv. Rev. 120 118–132. 10.1016/j.addr.2017.07.016 [DOI] [PubMed] [Google Scholar]
- Encinas J. M., Michurina T. V., Peunova N., Park J.-H., Tordo J., Peterson D. A., et al. (2011). Division-coupled astrocytic differentiation and age-related depletion of neural stem cells in the adult hippocampus. Cell Stem Cell 8 566–579. 10.1016/j.stem.2011.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson P. S., Perfilieva E., Björk-Eriksson T., Alborn A. M., Nordborg C., Peterson D. A., et al. (1998). Neurogenesis in the adult human hippocampus. Nat. Med. 4 1313–1317. 10.1038/3305 [DOI] [PubMed] [Google Scholar]
- Fiore R., Khudayberdiev S., Christensen M., Siegel G., Flavell S. W., Kim T.-K.et al. (2009). Mef2-mediated transcription of the miR379–410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 28 697–710. 10.1038/emboj.2009.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer S., Handrick R., Aschrafi A., Otte K. (2015). Unveiling the principle of microRNA-mediated redundancy in cellular pathway regulation. RNA Biol. 12 238–247. 10.1080/15476286.2015.1017238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke K., Otto W., Johannes S., Baumgart J., Nitsch R., Schumacher S. (2012). miR-124-regulated RhoG reduces neuronal process complexity via ELMO/Dock180/Rac1 and Cdc42 signalling. EMBO J. 31 2908–2921. 10.1038/emboj.2012.130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S., Matsumoto A., Nakayama K. I., Gotoh Y. (2013). p57 controls adult neural stem cell quiescence and modulates the pace of lifelong neurogenesis. EMBO J. 32 970–981. 10.1038/emboj.2013.50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Wang W.-Y., Mao Y.-W., Gräff J., Guan J.-S., Pan L., et al. (2010). A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature 466 1105–1109. 10.1038/nature09271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Su J., Guo W., Polich E. D., Magyar D. P., Xing Y., et al. (2015). Inhibition of miR-15a promotes BDNF expression and rescues dendritic maturation deficits in MeCP2-deficient neurons. Stem Cells 33 1618–1629. 10.1002/stem.1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Ding P., Hsieh J. (2012). Profiling of REST-dependent microRNAs reveals dynamic modes of expression. Front. Neurosci. 6:67. 10.3389/fnins.2012.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Ure K., Ding P., Nashaat M., Yuan L., Ma J., et al. (2011). The master negative regulator REST/NRSF controls adult neurogenesis by restraining the neurogenic program in quiescent stem cells. J. Neurosci. 31 9772–9786. 10.1523/JNEUROSCI.1604-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner P. P., Vinther J. (2008). Mutation of miRNA target sequences during human evolution. Trends Genet. 24 262–265. 10.1016/j.tig.2008.03.009 [DOI] [PubMed] [Google Scholar]
- Giachino C., Taylor V. (2015). Notching up neural stem cell homogeneity in homeostasis and disease. Front. Neurosci. 8:32. 10.3389/fnins.2014.00032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves J. T., Schafer S. T., Gage F. H. (2016). Adult neurogenesis in the hippocampus: from stem cells to behavior. Cell 167 897–914. 10.1016/j.cell.2016.10.021 [DOI] [PubMed] [Google Scholar]
- Ha M., Kim V. N. (2014). Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 15 509–524. 10.1038/nrm3838 [DOI] [PubMed] [Google Scholar]
- Han J., Kim H. J., Schafer S. T., Paquola A., Clemenson G. D., Toda T., et al. (2016). Functional implications of miR-19 in the migration of newborn neurons in the adult brain. Neuron 91 79–89. 10.1016/j.neuron.2016.05.034 [DOI] [PubMed] [Google Scholar]
- Harraz M. M., Xu J.-C., Guiberson N., Dawson T. M., Dawson V. L. (2014). MiR-223 regulates the differentiation of immature neurons. Mol. Cell Ther. 2 18–19. 10.1186/2052-8426-2-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hébert S. S., Papadopoulou A. S., Smith P., Galas M.-C., Planel E., Silahtaroglu A. N., et al. (2010). Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum. Mol. Genet. 19 3959–3969. 10.1093/hmg/ddq311 [DOI] [PubMed] [Google Scholar]
- Herranz H., Cohen S. M. (2010). MicroRNAs and gene regulatory networks: managing the impact of noise in biological systems. Genes Dev. 24 1339–1344. 10.1101/gad.1937010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Li Z. (2017). miRNAs in synapse development and synaptic plasticity. Curr. Opin. Neurobiol. 45 24–31. 10.1016/j.conb.2017.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M. M., Zhang C.-L. (2015). TLX: a master regulator for neural stem cell maintenance and neurogenesis. Biochim. Biophys. Acta 1849 210–216. 10.1016/j.bbagrm.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Kim S.-N., Liu X., Zhang H., Zhang C., Seo J.-S., et al. (2016). miR-17-92 cluster regulates adult hippocampal neurogenesis, anxiety, and depression. Cell Rep. 16 1653–1663. 10.1016/j.celrep.2016.06.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe E. M., Gao Y., Eisinger B. E., Mladucky J. K., Giuliani C. C., Kelnhofer L. E., et al. (2017). Methyl-CpG-binding protein MBD1 regulates neuronal lineage commitment through maintaining adult neural stem cell identity. J. Neurosci. 37 523–536. 10.1523/JNEUROSCI.1075-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobe E. M., McQuate A. L., Zhao X. (2012). Crosstalk among epigenetic pathways regulates neurogenesis. Front. Neurosci. 6:59. 10.3389/fnins.2012.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga H., Fukuyama M., Kitazawa A., Kontani K., Katada T. (2013). The microRNA miR-235 couples blast-cell quiescence to the nutritional state. Nature 497 503–506. 10.1038/nature12117 [DOI] [PubMed] [Google Scholar]
- Katz S., Cussigh D., Urbán N., Blomfield I., Guillemot F., Bally-Cuif L., et al. (2016). A nuclear role for miR-9 and argonaute proteins in balancing quiescent and activated neural stem cell states. Cell Rep. 17 1383–1398. 10.1016/j.celrep.2016.09.088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi D., Furutachi S., Kawai H., Hozumi K., Gotoh Y. (2013). Dll1 maintains quiescence of adult neural stem cells and segregates asymmetrically during mitosis. Nat. Commun. 4:1880. 10.1038/ncomms2895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G., Gage F. H., Aigner L., Song H., Curtis M. A., Thuret S., et al. (2018). Human adult neurogenesis: evidence and remaining questions. Cell Stem Cell 23 25–30. 10.1016/j.stem.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G., Song H., Gage F. H. (2015). Neurogenesis in the adult hippocampus. Cold Spring Harb. Perspect. Biol. 7:a18812-14. 10.1101/cshperspect.a018812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnam S. E., Winlow W., Farbood Y., Moghaddam H. F., Farzaneh M. (2017). Emerging roles of microRNAs in ischemic stroke: as possible therapeutic agents. J. Stroke 19 166–187. 10.5853/jos.2016.01368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-Y., Hwang I., Muller F. L., Paik J.-H. (2015). Functional regulation of FoxO1 in neural stem cell differentiation. Cell Death. Differ. 22 2034–2045. 10.1038/cdd.2015.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Duan X., Liu C. Y., Jang M.-H., Guo J. U., Pow-anpongkul N., et al. (2009). DISC1 regulates new neuron development in the adult brain via modulation of AKT-mTOR signaling through KIAA1212. Neuron 63 761–773. 10.1016/j.neuron.2009.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. R., Sun W. (2011). Programmed cell death during postnatal development of the rodent nervous system. Dev. Growth Differ. 53 225–235. 10.1111/j.1440-169X.2010.01226.x [DOI] [PubMed] [Google Scholar]
- Klein M. E., Lioy D. T., Ma L., Impey S., Mandel G., Goodman R. H. (2007). Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 10 1513–1514. 10.1038/nn2010 [DOI] [PubMed] [Google Scholar]
- Konopka W., Kiryk A., Novak M., Herwerth M., Parkitna J. R., Wawrzyniak M., et al. (2010). MicroRNA loss enhances learning and memory in mice. J. Neurosci. 30 14835–14842. 10.1523/JNEUROSCI.3030-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laneve P., Gioia U., Andriotto A., Moretti F., Bozzoni I., Caffarelli E. (2010). A minicircuitry involving REST and CREB controls miR-9-2 expression during human neuronal differentiation. Nucleic Acids Res. 38 6895–6905. 10.1093/nar/gkq604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplagne D. A., Espósito M. S., Piatti V. C., Morgenstern N. A., Zhao C., van Praag H., et al. (2006). Functional convergence of neurons generated in the developing and adult hippocampus. PLoS Biol. 4:e409. 10.1371/journal.pbio.0040409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Burge C. B., Bartel D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120 15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Li Q., Bian S., Hong J., Kawase-Koga Y., Zhu E., Zheng Y., et al. (2011). Timing specific requirement of microRNA function is essential for embryonic and postnatal hippocampal development. PLoS One 6:e26000. 10.1371/journal.pone.0026000.g009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., et al. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433 769–773. 10.1038/nature03315 [DOI] [PubMed] [Google Scholar]
- Lindvall O., Kokaia Z. (2010). Stem cells in human neurodegenerative disorders — Time for clinical translation? J. Clin. Invest. 120 29–40. 10.1172/JCI40543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Teng Z.-Q., McQuate A. L., Jobe E. M., Christ C. C., von Hoyningen-Huene S. J., et al. (2013). An epigenetic feedback regulatory loop involving microRNA-195 and MBD1 governs neural stem cell differentiation. PLoS One 8:e51436. 10.1371/journal.pone.0051436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Teng Z.-Q., Santistevan N. J., Szulwach K. E., Guo W., Jin P., et al. (2010). Epigenetic regulation of miR-184 by MBD1 governs neural stem cell proliferation and differentiation. Cell Stem Cell 6 433–444. 10.1016/j.stem.2010.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Martin-Villalba A. (2017). Adult NSC diversity and plasticity: the role of the niche. Curr. Opin. Neurobiol. 42 68–74. 10.1016/j.conb.2016.11.008 [DOI] [PubMed] [Google Scholar]
- Llorens-Bobadilla E., Zhao S., Baser A., Saiz-Castro G., Zwadlo K., Martin-Villalba A. (2015). Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Stem Cell 17 329–340. 10.1016/j.stem.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Lopez-Ramirez M. A., Nicoli S. (2014). Role of miRNAs and epigenetics in neural stem cell fate determination. Epigenetics 9 90–100. 10.4161/epi.27536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugert S., Basak O., Knuckles P., Haussler U., Fabel K., Götz M., et al. (2010). Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell 6 445–456. 10.1016/j.stem.2010.03.017 [DOI] [PubMed] [Google Scholar]
- Magill S. T., Cambronne X. A., Luikart B. W., Lioy D. T., Leighton B. H., Westbrook G. L., et al. (2010). microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl. Acad. Sci. U.S.A. 107 20382–20387. 10.1073/pnas.1015691107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahar I., Bambico F. R., Mechawar N., Nobrega J. N. (2014). Stress, serotonin, and hippocampal neurogenesis in relation to depression and antidepressant effects. Neurosci. Biobehav. Rev. 38 173–192. 10.1016/j.neubiorev.2013.11.009 [DOI] [PubMed] [Google Scholar]
- Martins R., Lithgow G. J., Link W. (2015). Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell 15 196–207. 10.1111/acel.12427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murao N., Noguchi H., Nakashima K. (2016). Epigenetic regulation of neural stem cell property from embryo to adult. NEPIG 5 1–10. 10.1016/j.nepig.2016.01.001 [DOI] [Google Scholar]
- Nishino J., Kim I., Chada K., Morrison S. J. (2008). Hmga2 promotes neural stem cell self-renewal in young but not old mice by reducing p16Ink4a and p19Arf expression. Cell 135 227–239. 10.1016/j.cell.2008.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W., Zou Y., Shen C., Zhang C. L. (2011). Activation of postnatal neural stem cells requires nuclear receptor TLX. J. Neurosci. 31 13816–13828. 10.1523/JNEUROSCI.1038-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes M. C., Roy N. S., Keyoung H. M., Goodman R. R., McKhann G., Jiang L., et al. (2003). Identification and isolation of multipotential neural progenitor cells from the subcortical white matter of the adult human brain. Nat. Med. 9 439–447. 10.1038/nm837 [DOI] [PubMed] [Google Scholar]
- Olive V., Minella A. C., He L. (2015). Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Sci. Signal. 8:re2. 10.1126/scisignal.2005813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osella M., Riba A., Testori A., Corà D., Caselle M. (2014). Interplay of microRNA and epigenetic regulation in the human regulatory network. Front. Genet. 5:345 10.3389/fgene.2014.00345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer A. N., Xing Y., Harper S. Q., Jones L., Davidson B. L. (2008). The bifunctional microRNA miR-9/miR-9∗ regulates REST and CoREST and is downregulated in Huntington’s disease. J. Neurosci. 28 14341–14346. 10.1523/JNEUROSCI.2390-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik J.-H., Ding Z., Narurkar R., Ramkissoon S., Muller F., Kamoun W. S., et al. (2009). FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell 5 540–553. 10.1016/j.stem.2009.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peláez N., Carthew R. W. (2012). Biological robustness and the role of microRNAs: a network perspective. Curr. Top. Dev. Biol. 99 237–255. 10.1016/B978-0-12-387038-4.00009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Bonaguidi M. A. (2018). Function and dysfunction of adult hippocampal neurogenesis in regeneration and disease. Am. J. Pathol. 188 23–28. 10.1016/j.ajpath.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfannkuche K., Summer H., Li O., Hescheler J., Dröge P. (2009). The high mobility group protein HMGA2: a co-regulator of chromatin structure and pluripotency in stem cells? Stem Cell Rev. Rep. 5 224–230. 10.1007/s12015-009-9078-9 [DOI] [PubMed] [Google Scholar]
- Pino A., Fumagalli G., Bifari F., Decimo I. (2017). New neurons in adult brain: distribution, molecular mechanisms and therapies. Biochem. Pharmacol. 141 4–22. 10.1016/j.bcp.2017.07.003 [DOI] [PubMed] [Google Scholar]
- Pons-Espinal M., de Luca E., Marzi M. J., Beckervordersandforth R., Armirotti A., Nicassio F., et al. (2017). Synergic functions of miRNAs determine neuronal fate of adult neural stem cells. Stem Cell Rep. 8 1046–1061. 10.1016/j.stemcr.2017.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt T., Price D. J. (2016). Junk DNA used in cerebral cortical evolution. Neuron 90 1141–1143. 10.1016/j.neuron.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Qiu L., Zhang W., Tan E.-K., Zeng L. (2014). Deciphering the function and regulation of microRNAs in Alzheimer’s disease and Parkinson’s disease. ACS Chem. Neurosci. 5 884–894. 10.1021/cn500149w [DOI] [PubMed] [Google Scholar]
- Rajman M., Schratt G. (2017). MicroRNAs in neural development: from master regulators to fine-tuners. Development 144 2310–2322. 10.1242/dev.144337 [DOI] [PubMed] [Google Scholar]
- Renault V. M., Rafalski V. A., Morgan A. A., Salih D. A. M., Brett J. O., Webb A. E., et al. (2009). FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell 5 527–539. 10.1016/j.stem.2009.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Aznar E., Barrallo-Gimeno A., Nieto M. A. (2013). Scratch2 prevents cell cycle re-entry by repressing miR-25 in postmitotic primary neurons. J. Neurosci. 33 5095–5105. 10.1523/JNEUROSCI.4459-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roese-Koerner B., Stappert L., Brüstle O. (2017). Notch/Hes signaling and miR-9 engage in complex feedback interactions controlling neural progenitor cell proliferation and differentiation. Neurogenesis 4:e1313647. 10.1080/23262133.2017.1313647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roese-Koerner B., Stappert L., Koch P., Brüstle O., Borghese L. (2013). Pluripotent stem cell-derived somatic stem cells as tool to study the role of microRNAs in early human neural development. Curr. Mol. Med. 13 707–722. 10.2174/1566524011313050003 [DOI] [PubMed] [Google Scholar]
- Roy N. S., Wang S., Jiang L., Kang J., Benraiss A., Harrison-Restelli C., et al. (2000). In vitro neurogenesis by progenitor cells isolated from the adult human hippocampus. Nat. Med. 6 271–277. 10.1038/73119 [DOI] [PubMed] [Google Scholar]
- Sanai N., Nguyen T., Ihrie R. A., Mirzadeh Z., Tsai H.-H., Wong M., et al. (2011). Corridors of migrating neurons in the human brain and their decline during infancy. Nature 478 382–386. 10.1038/nature10487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz U., Lai X., Winter F., Wolkenhauer O., Vera J., Gupta S. K. (2014). Cooperative gene regulation by microRNA pairs and their identification using a computational workflow. Nucleic Acids Res. 42 7539–7552. 10.1093/nar/gku465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schouten M., Fratantoni S. A., Hubens C. J., Piersma S. R., Pham T. V., Bielefeld P., et al. (2015). MicroRNA-124 and -137 cooperativity controls caspase-3 activity through BCL2L13 in hippocampal neural stem cells. Sci. Rep. 5:12448. 10.1038/srep12448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt G. M., Tuebing F., Nigh E. A., Kane C. G., Sabatini M. E., Kiebler M., et al. (2006). A brain-specific microRNA regulates dendritic spine development. Nature 439 283–289. 10.1038/nature04367 [DOI] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature 455 58–63. 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- Semerci F., Choi W. T., Bajic A., Thakkar A., Encinas J. M., Depreux F., et al. (2017). Lunatic fringe-mediated Notch signaling regulates adult hippocampal neural stem cell maintenance. eLife 6:e1002466. 10.7554/eLife.24660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seri B., Garcia-Verdugo J. M., Collado-Morente L., McEwen B. S., Alvarez-Buylla A. (2004). Cell types, lineage, and architecture of the germinal zone in the adult dentate gyrus. J. Comp. Neurol. 478 359–378. 10.1002/cne.20288 [DOI] [PubMed] [Google Scholar]
- Shi Y., Chichung Lie D., Taupin P., Nakashima K., Ray J., Yu R. T., et al. (2004). Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature 427 78–83. 10.1038/nature02211 [DOI] [PubMed] [Google Scholar]
- Shimazaki T., Okano H. (2018). Heterochronic microRNAs in temporal specification of neural stem cells: application toward rejuvenation. NPJ Aging Mech Dis. 2:15014. 10.1038/npjamd.2015.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J., Berg D. A., Zhu Y., Shin J. Y., Song J., Bonaguidi M. A., et al. (2015). Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Stem Cell 17 360–372. 10.1016/j.stem.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel G., Obernosterer G., Fiore R., Oehmen M., Bicker S., Christensen M., et al. (2009). A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat. Cell Biol. 11 705–716. 10.1038/ncb1876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra A., Encinas J. M., Deudero J. J. P., Chancey J. H., Enikolopov G., Overstreet-Wadiche L. S., et al. (2010). Microglia shape adult hippocampal neurogenesis through apoptosis-coupled phagocytosis. Cell Stem Cell 7483–495. 10.1016/j.stem.2010.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt R. D., Szulwach K. E., Pfeiffer R. L., Li X., Guo W., Pathania M., et al. (2010). MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells 28 1060–1070. 10.1002/stem.431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrells S. F., Paredes M. F., Cebrian-Silla A., Sandoval K., Qi D., Kelley K. W., et al. (2018). Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature 555 377–381. 10.1038/nature25975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding K. L., Bergmann O., Alkass K., Bernard S., Salehpour M., Huttner H. B., et al. (2013). Dynamics of hippocampal neurogenesis in adult humans. Cell 153 1219–1227. 10.1016/j.cell.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz T. R., Linseman D. A. (2014). Rho family GTPases: key players in neuronal development, neuronal survival, and neurodegeneration. Front. Cell Neurosci. 8:314. 10.3389/fncel.2014.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stappert L., Roese-Koerner B., Brüstle O. (2014). The role of microRNAs in human neural stem cells, neuronal differentiation and subtype specification. Cell Tissue Res. 359 47–64. 10.1007/s00441-014-1981-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Alzayady K., Stewart R., Ye P., Yang S., Li W., et al. (2010). Histone demethylase LSD1 regulates neural stem cell proliferation. Mol. Cell. Biol. 30 1997–2005. 10.1128/MCB.01116-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G., Ye P., Murai K., Lang M.-F., Li S., Zhang H., et al. (2011). miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat. Commun. 2:529. 10.1038/ncomms1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G. J., Zhou Y., Stadel R. P., Moss J., Yong J. H. A., Ito S., et al. (2015). Tangential migration of neuronal precursors of glutamatergic neurons in the adult mammalian brain. Proc. Natl. Acad. Sci. U.S.A. 1129484–9489. 10.1073/pnas.1508545112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L., Zhao M., Wang Y., Liu A., Lv M., Li Y., et al. (2017). Neuroprotective effects of miR-27a against traumatic brain injury via suppressing FoxO3a-mediated neuronal autophagy. Biochem. Biophys. Res. Commun. 4821141–1147. 10.1016/j.bbrc.2016.12.001 [DOI] [PubMed] [Google Scholar]
- Szulwach K. E., Li X., Smrt R. D., Li Y., Luo Y., Lin L., et al. (2010). Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J. Cell Biol. 189 127–141. 10.1083/jcb.200908151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro A., Sandler V. M., Toni N., Zhao C., Gage F. H. (2006). NMDA-receptor-mediated, cell-specific integration of new neurons in adult dentate gyrus. Nature 442 929–933. 10.1038/nature05028 [DOI] [PubMed] [Google Scholar]
- Terzibasi Tozzini E., Savino A., Ripa R., Battistoni G., Baumgart M., Cellerino A. (2014). Regulation of microRNA expression in the neuronal stem cell niches during aging of the short-lived annual fish Nothobranchius furzeri. Front. Cell Neurosci. 8:51. 10.3389/fncel.2014.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda T., Gage F. H. (2018). Review: adult neurogenesis contributes to hippocampal plasticity. Cell Tissue Res. 373 693–709. 10.1007/s00441-017-2735-4 [DOI] [PubMed] [Google Scholar]
- Vadodaria K. C., Brakebusch C., Suter U., Jessberger S. (2013). Stage-specific functions of the small Rho GTPases Cdc42 and Rac1 for adult hippocampal neurogenesis. J. Neurosci. 33 1179–1189. 10.1523/JNEUROSCI.2103-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Praag H., Kempermann G., Gage F. H. (1999). Running increases cell proliferation and neurogenesis in the adult mouse dentate gyrus. Nat. Neurosci. 2 266–270. 10.1038/6368 [DOI] [PubMed] [Google Scholar]
- Visvanathan J., Lee S., Lee B., Lee J. W., Lee S. K. (2007). The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21 744–749. 10.1101/gad.1519107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N., Klein M. E., Varlamova O., Keller D. M., Yamamoto T., Goodman R. H., et al. (2005). A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 102 16426–16431. 10.1073/pnas.0508448102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman G. A., Davare M., Ando H., Fortin D., Varlamova O., Cheng H. Y., et al. (2008). An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. U.S.A. 105 9093–9098. 10.1073/pnas.0803072105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B., Woodroffe A., Rodriguez-Murillo L., Roos J. L., van Rensburg E. J., Abecasis G. R., et al. (2009). Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc. Natl. Acad. Sci. U.S.A. 106 16746–16751. 10.1073/pnas.0908584106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., et al. (2011). MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476 228–231. 10.1038/nature10323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J., Huang H., Zhou X., Liu X., Ou S., Xu T., et al. (2016). MicroRNA-132 interact with p250GAP/Cdc42 Pathway in the hippocampal neuronal culture model of acquired epilepsy and associated with epileptogenesis process. Neural Plast. 2016:5108489. 10.1155/2016/5108489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C.-L., Zou Y., He W., Gage F. H., Evans R. M. (2008). A role for adult TLX-positive neural stem cells in learning and behaviour. Nature 451 1004–1007. 10.1038/nature06562 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu C., Wang J., Li Q., Ping H., Gao S., et al. (2018). MiR-299-5p regulates apoptosis through autophagy in neurons and ameliorates cognitive capacity in APPswe/PS1dE9 mice. Sci. Rep. 6:24566. 10.1038/srep24566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S., Lang M.-F., Yang S., Li W., et al. (2010). MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. U.S.A. 107 1876–1881. 10.1073/pnas.0908750107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G., Li S., Shi Y. (2009). A feedback regulatory loop involving microRNA-9 and nuclear receptor TLX in neural stem cell fate determination. Nat. Struct. Mol. Biol. 16 365–371. 10.1038/nsmb.1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Sun G., Ye P., Li S., Shi Y. (2013). MicroRNA let-7d regulates the TLX/microRNA-9 cascade to control neural cell fate and neurogenesis. Sci. Rep. 3:1329. 10.1038/srep01329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Teng E. M., Summers R. G., Ming G.-L., Gage F. H. (2006). Distinct morphological stages of dentate granule neuron maturation in the adult mouse hippocampus. J. Neurosci. 26 3–11. 10.1523/JNEUROSCI.3648-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X., Ueba T., Christie B. R., Barkho B., McConnell M. J., Nakashima K., et al. (2003). Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function. Proc. Natl. Acad. Sci. U.S.A. 100 6777–6782. 10.1073/pnas.1131928100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell F., Dehler S., Martin-Villalba A., Marciniak-Czochra A. (2018). Revealing age-related changes of adult hippocampal neurogenesis using mathematical models. Development 145:dev153544. 10.1242/dev.153544 [DOI] [PMC free article] [PubMed] [Google Scholar]