Abstract
Gut dysbiosis induced by high fat diet (HF) or obesity is a predisposing factor to develop diverse inflammatory diseases. Polyphenols and fibers, often eaten together, have been reported to have prebiotic actions, but their health promoting benefits still need to be further characterized and defined. This study attempted to understand how polyphenol rutin and polysaccharide inulin influence intestinal health in mouse model fed a HF (60 kcal%) diet. A total of 48 C57BL/6J mice were divided into four groups fed with a low fat (10% kcal%) control diet (LC), a high fat control diet (HC), a high-fat diet supplemented with rutin (HR), or a high-fat diet supplemented rutin and inulin (HRI) for 20 weeks. Rutin supplementation reduced the HF diet-induced increase of Firmicutes/Bacteroidetes (F/B) ratio, Deferribacteraceae population and plasma lipopolysaccharide (LPS) (p < 0.05); ameliorated inflammation as indicated by the decreased circulating inflammatory cytokines (p < 0.05) and the reduced expressions of intestinal inflammatory mediators (p < 0.05); and attenuated the endoplasmic reticulum (ER) stress in Paneth cells as indicated by the decreased expressions of the ER markers (p < 0.05). Compared to the rutin supplementation alone, the co-administration of rutin with inulin improved the utilization of rutin as indicated by its decreased excretion, suppressed a number of harmful bacteria including Deferribacteraceae and Desulfovibrionaceae (p < 0.05), and further reduced the expression of the key inflammatory cytokine TNF-α and increased the production of butyrate, despite the supplementation of inulin reversed the decrease of body weight induced by rutin supplementation due to an increased food intake. Taken together, our data demonstrated that rutin supplementation ameliorated the inflammatory status and ER stress in Paneth cells under a HF-induced obese state, and its co-administration with inulin further mitigated the inflammatory status, indicating the potential to combine polyphenol rutin and the polysaccharide inulin as a dietary strategy to ameliorate gut dysbiosis, to improve inflammatory status and thereby to reduce medical disorders associated with HF-induced obesity.
Keywords: obesity, rutin, inulin, gut dysbiosis, inflammation, Paneth cells
Introduction
The prevalence of obesity has dramatically increased and has more than doubled since 1980 (Gregg and Shaw, 2017). Currently, one third of the world’s population is overweight or considered obese, and a further increase is even predicted, ∼50% by 2030 (Kelly et al., 2008; Finkelstein et al., 2012). Obesity has now been considered as a major public health issue since it is associated with an array of medical complications, including increased risk of type 2 diabetes, hypertension, cardiovascular disease, inflammatory bowel disease, and cancers (Hotamisligil, 2006). One of the responsible mechanisms for this relationship is the systemic inflammatory status associated with high fat diet or obesity. Recently, growing evidence has implicated the intestinal immune system as an important contributor to those diseases (Garidou et al., 2015; Luck et al., 2015; Monteiro-Sepulveda et al., 2015; Winer et al., 2016).
A close relationship between the gut microbiota and obesity has been demonstrated in both human and animals, with enriched Firmicutes and reduced Bacteroidetes phyla in obesity (Ley et al., 2005, 2006). Our recent research revealed that high fat (HF) diet stimulated intestinal inflammation via altering gut microbiota, and it occurred prior to the potential influence by circulating inflammatory cytokines (Guo et al., 2017), indicating, in addition to adipose tissue, HF per se also drives intestinal inflammation via altering gut microbiota. Data from Lee et al. (2017) reiterated the role of HF on intestinal inflammation and health by showing that HF-fed mice were more susceptible to experimental colitis, and exhibited severe colonic inflammation, accompanied by the expansion of selected pathobionts such as Atopobium sp. and Proteobacteria.
A number of studies have demonstrated that a variety of polyphenols and dietary fibers exert the properties to mitigate the microbial dysbiosis produced by high-fat diets (Delzenne et al., 2011; Karlsson et al., 2013; Etxeberria et al., 2015). For instances, the beneficial Bacteroidetes community is favored by wine vinegar, polyphenol-rich fruits and green tea (Lee et al., 2006; Cerezo et al., 2008; Selma et al., 2009). Quercetin or grape polyphenols attenuated Firmicutes/Bacteroidetes ratio and suppressed the growth of bacterial species associated to diet-induced obesity, resulting in lower intestinal and systemic inflammation (Etxeberria et al., 2015; Roopchand et al., 2015). Non-digestible dietary fiber exerts a significant role in promoting beneficial bacteria and enhance microbial diversity in the gut (Cani et al., 2007), which might contribute to ameliorate obesity and obesity associated disorders (Dewulf et al., 2011).
Polyphenols and dietary fiber may interact to mediate gut microbiota. Jaganath et al. (2006) have shown that combining fermentable carbohydrates accelerate the breakdown of the polyphenol rutin in an in vitro model of colonic fermentation (Hou et al., 2015). Moreover, a range of fermentable fibers inhibited phenolic acid production from rutin (Mansoorian et al., 2015). Also, polyphenols could influence the fermentation of the dietary fiber as polyphenols have been shown to have both anti-bacterial (Taguri et al., 2004) and prebiotic actions (Tuohy et al., 2012). Thus, there would be a reciprocal interaction between dietary polyphenols and fibers in the mediation of gut mcirobiota. The present study was to assess the potential of rutin and its co-administration with inulin to ameliorate HF-diet-induced gut microbiota dysbiosis and inflammation.
Materials and Methods
Animal Study
The animal protocol was approved by the Institutional Animal Care and Use Committee of Chengdu University. Forty-eight male C57BL/6J mice (4 weeks old) were randomly distributed into four groups: the low-fat control group (LC; n = 12), mice were fed a low-fat diet (LF) with 10% kcal from fat (D12450B; Research Diets Inc.); the high-fat control group (HC; n = 12), mice were fed a high-fat (HF) diet with 60% kcal from fat (D12492; Research Diets Inc.); a rutin group (HR; n = 12), mice fed a HF diet with rutin 6.4 mg/g diet; and a rutin + inulin group (HRI; n = 12), mice fed a HF diet with rutin 6.4 mg/g diet and inulin (30 mg/ml) via drinking water. The doses of rutin (about 0.01 mol/kg diet) and inulin were chosen based on previous studies (Jurgoński et al., 2012; Hoek-van den Hil et al., 2013, 2015; Hamilton et al., 2017). Rutin (≥97% purity) was supplied by Chengdu Okay Pharmaceutical Co. Ltd (Sichuan, China); and inulin (≥95% purity) was supplied by Rui Hu Biological Co. Ltd (Qinghai, China).
The mice were fed ad libitum and housed under a 12 h light/12 h dark cycle. Body weights and food intake were recorded every other week. During the 19th week, food and feces samples were collected, and feces excretion was recorded. After 20 weeks of feeding, all mice were euthanized with CO2. The blood sample was collected via heart puncture after euthanasia, and plasma was separated and stored at −80°C for later analysis of inflammatory cytokines and biochemical parameters. Following with the heart puncture, the abdomen was opened and visceral fat pads were harvested. Samples of the intestinal and colonic contents were collected, frozen immediately with liquid nitrogen, and stored at −80°C for further microbial abundance and SCFA analysis. A segment (∼2 cm) of ileum was excised and formalin-fixed for immunohistochemistry, and the intestinal mucosa was collected as described in a previous publication (Guo et al., 2017), and then immediately frozen with liquid nitrogen and stored at −80°C for later real-time PCR analysis.
16S rRNA Gene Sequencing and Analysis of Microbial Profile
16S rRNA gene sequencing was performed on the small intestinal contents from 32 C57BL/6J mice (n = 8 mice per diet group). Bacterial DNA was extracted from the intestinal contents using the QIAamp DNA Stool Mini Kit (Qiagen, Germany) according to the manufacturer’s instructions. The concentration and purity of extracted DNA were determined using the NanoDrop-2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States).
The V4 region of the 16S rRNA gene was amplified used universal bacterial primers 515F (5′-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) with Phusion High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA, United States) followed by purification with Qiagen Gel Extraction Kit (Qiagen, Germany). The sequencing libraries were prepared using TruSeq DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, United States), sequenced on an Illumina HiSeq 2500 platform (San Diego, CA, United States), and 250 bp paired-end reads were generated. Paired-end reads were merged using FLASH (Baltimore, MD, United States), and subsequently raw tags were filtered with a specific standard to obtain the high-quality tags using QIIME version 1.7.0 (Caporaso et al., 2010). Sequences with ≥97% similarity were assigned to the same operational taxonomic units (OTUs) using Uparse software version 7.0.1001 (Edgar, 2013), and were annotated with taxonomic information based on the RDP classifier version 2.2 (Wang et al., 2007) algorithm using the Greengene database. OTU abundance data was normalized using a standard sequence number corresponding to the sample with least sequences, and subsequent analysis of alpha diversity and beta diversity were performed with QIIME. To visualize the sample differences, principal coordinate analysis (PCoA) was performed with weighted Unifrac (Lozupone and Knight, 2005). The clustering of samples was explained with the principal coordinate (PC) values. Differences in OTU abundance between groups were identified using LDA (Linear Discriminant Analysis) Effect Size (LEfSe) (Segata et al., 2011)1.
Gene Expression in Small Intestine Mucosa
The mRNA expressions of inflammatory factors, 4 anti-microbial peptides, α-defensin 5, lysozyme, angiogenin 4 (ANG 4) and regenerating islet derived 3-gamma (Reg IIIγ), and ER stress markers in the intestine were measured by real-time PCR. Briefly, total RNA was extracted from the small intestine mucosa with Trizol (Invitrogen, Carlsbad, CA, United States); the concentrations of total RNA were determined spectrophotometrically (NanoDrop-2000, Thermo Fisher Scientific, Waltham, MA, United States), and cDNA was synthesized with the ExScriptTM RT-PCR kit (TaKaRa, Dalian, China). Real-time PCR was performed on the ViiATM 7 System (Applied Biosystems, Foster City, CA, United States) utilizing the following thermal cycling conditions: 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Primer sequences were chosen according to our previous studies (Liu et al., 2016; Guo et al., 2017) and listed in Supplementary Data (Table S1).
Immunohistochemistry of Lysozyme
Immunohistochemical analysis was performed on the formalin-fixed sections of ileum. The paraffin-embedding slides were deparaffinized in xylene, followed by rehydration in ethanol. After blocking non-specific antibody binding with 5% bovine serum albumin, sections were incubated with the specific first antibody of rabbit monoclonal antibody lysozyme (ab108508; Abcam, Cambridge, United Kingdom) followed with the second antibody goat anti-rabbit IgG HRP. Immunoreactivity was detected with horseradish peroxidase-conjugated anti-goat EnVision kit (DAKO). All slides were counterstained with hematoxylin. Paneth cells were identified microscopically by their location just at the base of small intestinal crypts of Lieberkuhn (Elphick and Mahida, 2005).
Blood Plasma Analysis
For detecting inflammatory cytokines in the blood, a ProcartaPlex Mouse High Sensitivity Panel (5 plex) from Thermo Fisher Scientific for 5 cytokines, IFN-γ, IL-2, IL-4, IL-6, and TNF-α, was selected for mice, and the assays were performed on the MAGPIXTM platforms (Luminex, Austin, TX, United States) following the manufacturer’s instruction. Plasma leptin level was determined using a leptin ELISA kit (Multisciences, Hangzhou, China) according to the manufacturer’s instructions. Triglycerides and total cholesterol contents were quantified by Beckman Coulter DXC 600 Pro (Beckman Coulter, Inc., Brea, CA, United States) using standard spectrophotometric assays. Plasma LPS level was analyzed using a mouse LPS ELISA kit (Cusabio, Wuhan, China). All standards and samples were measured in duplicate.
Short-Chain Fatty Acid (SCFA) Analysis
Short-chain fatty acids (acetate, propionate, and butyrate) in colonic contents were determined. Colonic content samples (150 mg) were vortexed with 350 μl deionized water and 2-ethylbutyric acid (internal standard, Sigma Chemical), after which 500 μl 2.5 M sulphuric acid was added to the sample. SCFA were then extracted with diethyl ether, silylated with n-(tert-butyldimethylsilyl)-n-methyltrifluoroacetamide (Sigma Chemical), and then centrifuged (5,000 × g, 10 min). The supernatant was filtered through a 0.45 μm filter, and 1 μl of clear filter solution was directly injected into gas chromatograph system (Agilent Technologies) for analysis, which was performed as described by Sheveleva and Ramenskaya (2010).
Rutin Content and Rutin Degradation Analysis
Rutin content was measured according to Araujo et al. (2013). The food samples or feces samples were mixed with the 60% methanol solution, and the suspension was extracted under ultrasonication for 50 min and subsequently centrifuged for 10 min at 5,000 × g. The supernatant filtered through a 0.45 μm filter was used in high performance liquid chromatography (Shimadzu Co., Kanagawa, Japan) analysis. After detecting the rutin content in feed and feces, the degradation of rutin in the gut was determined as follows:
Statistical Analysis
Data are expressed as means ± SEM. Data analysis was performed using SPSS 15.0 software (Chicago, IL, United States). Comparisons between groups were made using ANOVA followed by post hoc test and associations were assessed by the linear regression. Correlation and regression analysis of the microbiome and relative parameters were conducted using Spearman’s rank correlation coefficient. For the gene expression data analysis, the expression of each gene was normalized to the housekeeping gene β-actin (CtTarget gene-Ctβ–actin). Statistical analyses were performed based on ΔCt. The relative abundance of relative gene expression was reported as 2−ΔΔCt, where ΔΔCt = ΔCtExperiment-ΔCtControl.
Results
Food Consumption and Animal Growth
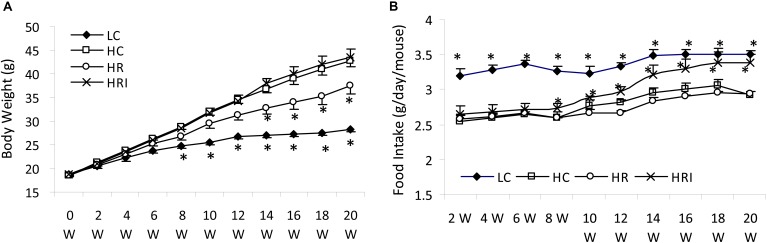
The body weights between the HC and HRI groups were similar (p > 0.05), but higher than that of LC group after 8 weeks and that of HR group after 14 weeks (p < 0.05, Figure 1A). As expected, the LC group had the highest food intake during the experiment and the HC group has the lowest (Figure 1B). The HR group had the similar food intake to the HC group (p > 0.05) whereas the further supplementation with inulin (the HRI group) increased food intake gradually, reaching to a significant level at 8 weeks when compared to the HC and HR group (p < 0.01), and close to LC after 16 weeks.
FIGURE 1.
Effect of high fat diet, rutin and inulin supplementation on body weight (A) and food intake (B). Rutin supplement ameliorated the elevation of body weight induced by high-fat diet, but its co-administration with inulin elevated gradually food intake toward no significant effect on body weight compared with HC. Black star indicated there was a significant difference compared with HC (p < 0.05).
Plasma Metabolic Parameters
High fat diet (the HC group) increased plasma cholesterol, triglycerides, leptin and LPS contents when compared to the LC group (Table 1). The rutin supplementation numerically reduced the plasma triglyceride (HR vs. HC), and its co-administration with inulin magnified this effect into a statistically significant level (p < 0.05, HRI vs. HC). Surprisingly, the cholesterol contents were not reduced by rutin (HR vs. HC) and even increased by inulin (HRI vs. HC). The leptin content was reduced by rutin supplementation and the LPS contents were significantly reduced in HR and HRI groups when compared to the HC group (p < 0.05).
Table 1.
Effect of high fat diet, rutin, and inulin supplementation on plasma biochemical parameters.
| LC | HC | HR | HRI | |
|---|---|---|---|---|
| Plasma Cholesterol (mmol/L) | 4.45 ± 0.11a | 6.48 ± 0.15b | 6.33 ± 0.52b | 7.63 ± 0.27c |
| Plasma TG (mmol/L) | 0.73 ± 0.03a | 1.19 ± 0.04b | 1.06 ± 0.03bc | 1.00 ± 0.02c |
| Plasma Leptin (ng/ml) | 1.89 ± 0.24a | 22.22 ± 2.10b | 14.56 ± 2.83c | 21.87 ± 2.04b |
| Plasma LPS (EU/L) | 772.6 ± 32.8a | 969.6 ± 36.3b | 772.4 ± 54.8a | 817.2 ± 52.3a |
LC, low-fat control group; HC, high-fat control group; HR, the group fed high-fat diet with rutin; HRI, the group fed high-fat diet with rutin and inulin. In each line, the values with different letters indicated significant difference among groups (p < 0.05).
The Inflammatory Factors in the Blood and Small Intestine
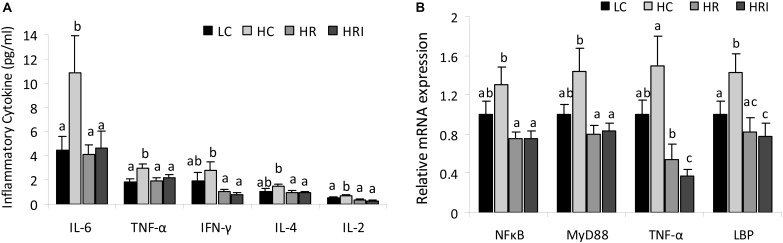
Five plasma inflammatory cytokines, IFN-γ, IL-4, IL-6, IL-2, and TNF-α, were measured using the ProcartaPlex Multiplex Immunoassay (Figure 2A). Compared to the LC group, the high fat diet (the HC group) increased plasma levels of those cytokines, with statistically significant differences for IL-6, TNF-α and IL-2 (p < 0.05), and the supplementation with rutin or rutin + inulin significantly reduced the plasma concentrations of those cytokines (HR and HRI vs. HC, p < 0.01). For the inflammatory mediators (Figure 2B), NFκB, MyD 88, TNF-α and LBP, the mRNA levels of them were higher in the HC group when compared to the LC group with a statistical difference for LBP, the gene for LPS binding protein (p < 0.05). The rutin supplement or its combination with inulin suppressed the production of these mediators (HR and HRI vs. HC, p < 0.05), and notably the combinatorial supplementation of rutin and inulin significantly reduced the intestinal expression of TNF-α (HRI vs. HR, p < 0.05).
FIGURE 2.
Effect of high fat diet, rutin and inulin supplementation on inflammatory cytokine profile. (A) Plasma inflammatory cytokine levels across the groups; (B) mRNA expression levels of inflammatory mediators in the small intestine tissue. The values with different letters indicated significant difference among groups (p < 0.05).
Expression of Paneth Cell AMPs and ER Stress Markers
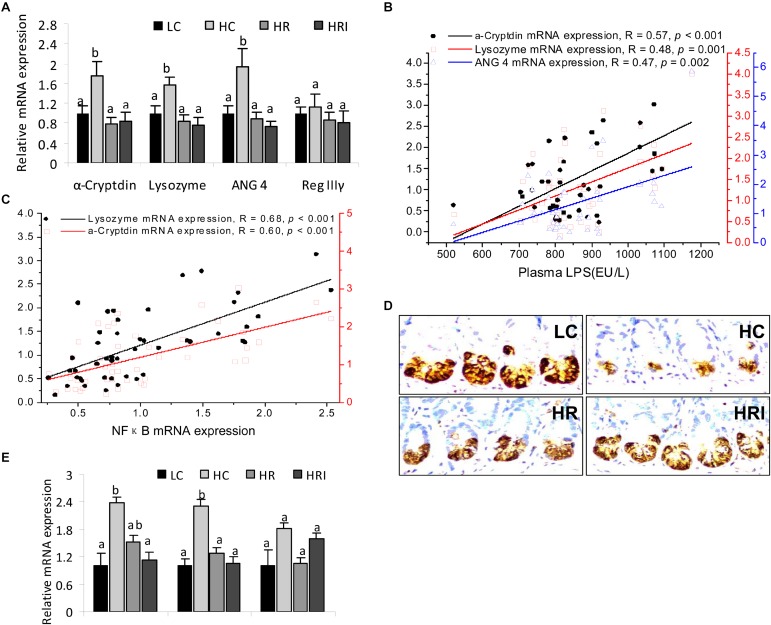
The relative mRNA expression levels of Paneth cell antimicrobial peptides (AMPs) were detected in the small intestinal tissue (Figure 3A). Except for Reg IIIγ, high fat diet (the HC group) significantly increased mRNA expressions of Paneth AMPs: α-cryptdin (1.8-fold), lysozyme (1.6-fold), and ANG 4 (1.9-fold) (p < 0.05). The mRNA expression levels of these Paneth cell AMPs were positively correlated with the plasma LPS level (Figure 3B, p < 0.01) and the gene expressions of inflammatory mediators NFκB (Figure 3C, p < 0.01).
FIGURE 3.
Effect of high fat diet, rutin, and inulin supplementation on the expression of Paneth cell antimicrobial peptides (AMPs) and ER stress biomarkers in intestine tissue. (A) mRNA expression of Paneth cell AMPs; (B) the correlations of Paneth cell AMPs with plasma LPS; (C) the correlations of Paneth cell AMPs with mRNA expressions of NFκB; (D) Immunohistochemtry for lysozyme; (E) mRNA expression of ER stress biomarkers. The values with different letters indicated significant difference among groups (p<0.05).
However, when we measured the protein level of lysozyme by immunohistochemistry (Figure 3D) the results indicated HC had lower protein expression in Paneth cells when compared to the LC group, indicating discordance between protein expression and mRNA expression of lysozyme in the Paneth cells. Since endoplasmic reticulum directly participates in the translational expression of those AMPs and may be responsible for the inconsistency at the protein and mRNA level, we therefore detected mRNA expression of ER stress markers in the intestine tissue, such as chaperone protein binding protein (BiP), activating transcription factor 4 (ATF4) and c/EBP-homologous protein (CHOP). The results showed higher mRNA expressions of BiP and ATF4 in HC when compared to the LC group (Figure 3E).
With the addition of rutin or its combination with inulin ameliorated the elevation of the mRNA expression of Paneth AMPs and ER stress biomarkers, and reduction of the lysozyme protein expression induced high-fat diet (Figures 3A,D,E).
The Abundances of Gut Microflora
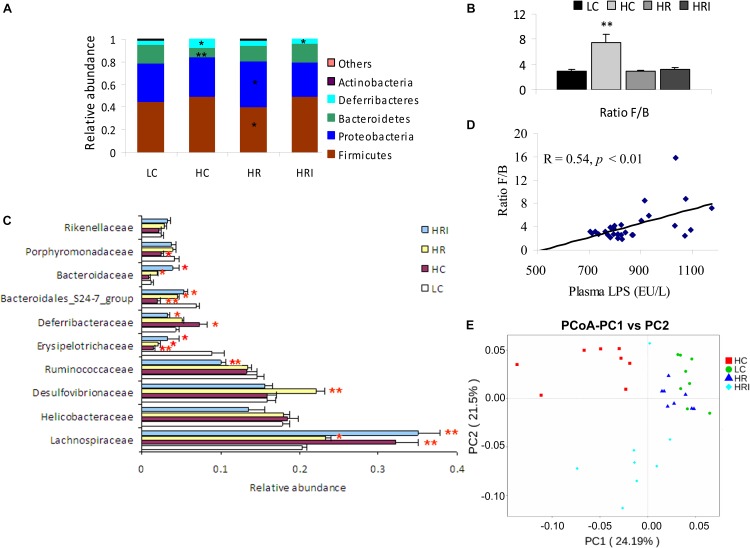
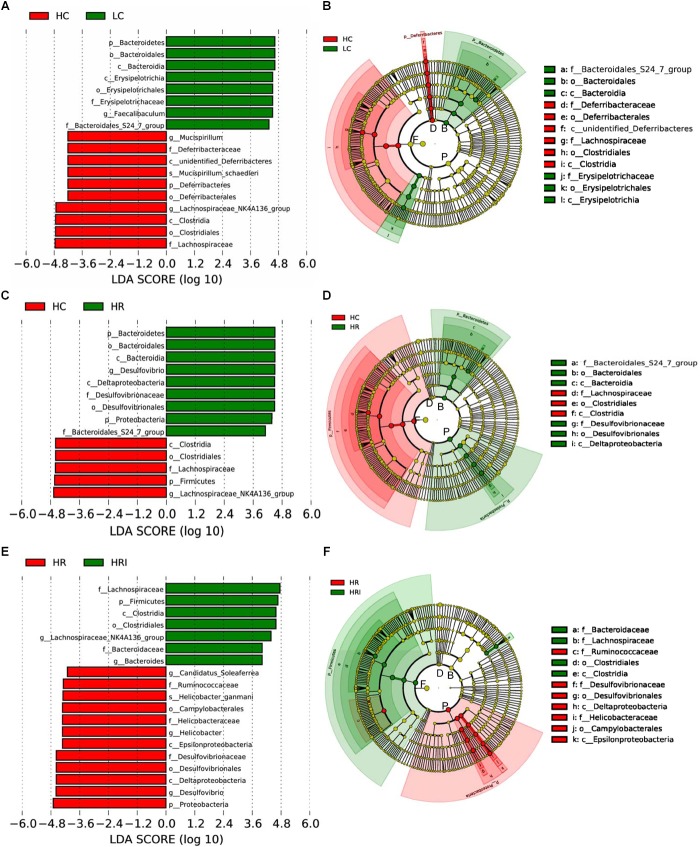
High fat diet (the HC group) significantly reduced the Bacteroidetes (p < 0.01)and its families (especially Bacteroidales_ S24-7 group, p < 0.001, Porphyromonadaceae, p < 0.05), in favor of the presence of Lachnospiraceae family in Firmicutes (p < 0.01) and Deferribacteres (largely due to increase of Deferribacteraceae family, p < 0.05), and resulted in an increased Firmicutes/Bacteroidetes (F/B) ratio (p < 0.01) (Figures 4A–C, 5A,B). However, high fat diet reduced Erysipelotrichaceae in Firmicutes phylum (p < 0.01) in the intestine when comparing to the LC group (Figure 4C). Correlation analysis indicated F/B ratio was positively associated with plasma LPS level (Figure 4D, p < 0.01). Rutin supplementation or its combinational supplement with inulin promoted the growth of the most families of Bacteroidetes (Figures 4A,C), such as Bacteroidales_S24-7 group (HR and HRI vs. HC, p < 0.001), Bacteroidaceae (HR and HRI vs. HC, p < 0.01), Porphyromonadaceae (HR vs. HC, p < 0.01; HRI vs. HC, p = 0.09), and Rikenellaceae (HR vs. HC, p < 0.05; HRI vs. HC, p = 0.05), and decreased the F/B ratio (Figure 4B, HR and HRI vs. HC, p < 0.01) and Deferribacteraceae population (Figures 4A,C, HR vs. HC, p = 0.05; HRI vs. HC, p < 0.01). Rutin supplementation alone (the HR group) reduced the number of Firmicutes (p < 0.01), especially Lachnospiraceae family (p < 0.05), and promoted the growth of Proteobacteria phylum (p < 0.05) (largely due to Desulfovibrionaceae family) in comparison with HC group (Figures 4A,C, 5C,D). The further supplementation with inulin (the HRI group) altered the composition of gut microflora (Figures 4C, 5E,F): increased the proportions of Lachnospiraceae (p < 0.01) and Bacteroidaceae (p < 0.05), and suppressed Desulfovibrionaceae (p < 0.001), Ruminococcaceae (p < 0.01), and Deferribacteraceae (p < 0.001) at family level; at genus level, elevated (p < 0.05) the abundances of Lachnospiraceae_NK4A136_group, Lachnoclostridium, Roseburia, Blautia, Bacteroides and Lactobacillus, and reduced (p < 0.01) Desulfovibrio Ruminiclostridium_9 and Mucispirillum (Supplementary Figure S1) when compared with HR group.
FIGURE 4.
Effect of high fat diet, rutin and inulin supplementation on the composition of gut microflora. (A) Taxonomic composition at the phylum level, the abundance ratio of each phylum was based on total microorganisms; (B) Firmicutes/Bacteroidetes (F/B) ratio; (C) The relative abundance of top 10 families; (D) Ratio F/B was positively related with plasma LPS; (E) Principal coordinate analysis (PCoA) of 16S sequences from 32 intestine content samples of four treatments based on Weighted Unifrac. The microbiome of HR was similar to LC, HF feeding reduced significantly the abundance of Bacteroidetes and its families, favoring the presence of Lachnospiraceae family in Firmicutes and Deferribacteres, and resulted in higher Firmicutes/Bacteroidetes (F/B) ratio in intestine, while HR and HRI group increased the abundance of Bacteroidetes and its families, and attenuated the rise of F/B ratio and Deferribacteres, and HR group increased the growth of Proteobacteria and its family. ∗ indicated significant difference at p < 0.05, and ∗∗ indicated significant difference at p < 0.01 compared with LC.
FIGURE 5.
Linear discriminant analysis (LDA) effect size (LEfSe) analyses compared the alterations in microbiome according to the diet. Output showing effect size of significantly enriched taxa when comparing groups: HC vs. LC (A), HC vs. HR (B) and HR vs. HRI (C). Significant taxa plotted onto a cladogram when comparing groups: HC vs. LC (D), HC vs. HR (E), and HR vs. HRI (F). F, Firmicutes; P, Proteobacteria; B, Bacteroidetes; D, Deferribacteres; p_, phylum; c_, class; o_, order; f_, family; g_genus, s_, species.
The characteristics of the gut microbiota in LC group and rutin-supplemented mice were similar according to PCoA plot (Figure 4E), besides rutin supplementation increased significantly Proteobacteria phylum (Figure 4A), and they were far away from HC and HRI groups.
The Content of SCFA and Rutin in Feces
High fat diet feeding (the HC group) reduced acetate, propionate, butyrate and their total contents in colonic contents when compared to the LC control group (Table 2). The supplementation of rutin or its combination with inulin significantly attenuated the reduction of SCFA. When compared to the HC group, the rutin supplementation (HR group) increased acetate and propionate contents (p < 0.05), and its co-administration with inulin (the HRI group) increased the production of all 3 SCFAs (p < 0.05). It is highly noteworthy that the co-administration with inulin (the HRI group) further significantly increased the production of butyrate (p < 0.05), which is considered as the key SCFA, and numerically increased propionate (p = 0.10) when compared to the HR group.
Table 2.
Effect of high fat diet, rutin and inulin supplementation on colonic contents of SCFA in mice (μmol/g).
| LC | HC | HR | HRI | |
|---|---|---|---|---|
| Acetate | 9.52 ± 0.88a | 7.04 ± 0.48b | 9.23 ± 0.99a | 9.20 ± 0.78a |
| Propionate | 1.43 ± 0.23a | 0.95 ± 0.08b | 1.53 ± 0.15a | 1.92 ± 0.19a |
| Butyrate | 2.75 ± 0.79a | 1.12 ± 0.05b | 1.28 ± 0.17b | 1.79 ± 0.20c |
| Total | 13.69 ± 1.23a | 9.10 ± 0.45b | 12.04 ± 1.12a | 12.91 ± 1.06a |
In each line, the values with different letters indicated significant difference among groups (p<0.05).
The HRI group indicated lower rutin left in feces (2.16 ± 0.47 vs. 2.90 ± 0.09 mg/g, p < 0.05), and increased the catabolism (the % of degradation) of rutin (95.37 ± 0.79 vs. 92.42 ± 0.64%, p < 0.05) in the feces compared with HR group.
Discussion
The dysbiosis of gut microbiota is an important mediator in the high fat-induced inflammation, and thus manipulation of the gut microbiota, using prebiotics, may provide novel preventive strategies for obesity-associated inflammation and medical disorders. Both polyphenols and fibers, often eaten together, have been reported to have prebiotic actions. The present study demonstrated that the supplementation of polyphenol rutin suppressed the rise of F/B ratio, Deferribacteracea population and plasma LPS induced by HF diet, and thereby mitigated systematic and intestinal inflammation, and ER stress in Paneth cells. The co-administration with polysaccharide inulin improved the utilization of rutin and further increased the production of butyrate and reduced the expression of the key inflammatory cytokine TNF-α, indicating the polyphenol rutin and polysaccharide inulin can be utilized combinatorially as a dietary strategy to ameliorate gut dysbiosis and inflammation associated with HF-induced obesity.
In the present study, after feeding a 60 kcal% HF diet for 20 weeks, the animals exhibited a significant increase of body weight with increased plasma cholesterol, triglycerides, leptin and LPS contents, and displayed a systematic and intestinal inflammation as indicated by increased circulating inflammatory cytokines and elevated expressions of inflammatory mediators in the intestinal epithelial cells. Meanwhile, HF diet significantly increased mRNA expressions of Paneth AMPs, which were positively associated with plasma LPS and inflammatory mediators, suggesting that a critical role of Paneth cell AMPs in promoting obesity-associated inflammation. HF diet also induced a significantly increase of ER stress as indicated by the higher mRNA expressions of ER stress biomarkers, BiP and ATF4, which resulted in a discordance between protein expression and mRNA expression of lysozyme. Similarly, Hodin et al. (2011) reported ER stress was apparent in Paneth cells in obese subjects, indicated by diminished AMPs protein expression and increased mRNA levels of their corresponding genes.
Corresponding to those metabolic and inflammatory changes, a shift of the gut microbiota was observed in the animals fed with HF diet comparing to the LF diet group, with elevated F/B ratio, similar to the alteration observed in obese individuals induced by high-fat (Zhang et al., 2012; Guo et al., 2017) and high fat/high-sucrose diet (Parks et al., 2013). The increase of Firmicutes in HC was largely due to increase of Lachnospiraceae in this study, which was similar to results of Han et al. (2018) indicating Lachnospiraceae is the main intestinal bacterial family, and accounts over thirty percent of the total in the microbiota. Correlation analysis indicated F/B ratio and abundance of Lachnospiraceae (R = 0.41, p < 0.05) were correlated with plasma LPS in the present study. These findings were consistent with previous studies, which demonstrated that HF diet-induced gut microbiota dysbiosis (shifting to Firmicutes) led to an increase in gut permeability and plasma LPS concentration, and thereby promoted a low-grade inflammation (Cani et al., 2007, 2008; Creely et al., 2007).
Rutin supplementation (HR group) or its co-administration with inulin (HRI group) mitigated the increase of plasma triglycerides or leptin, attenuated inflammatory status, and improved the ER stress in Paneth cells induced by high-fat diet (the HC group) in this study. These data suggested that the inflammatory status and ER stress in Paneth cells in an obese state could be compromised by rutin supplementation or its co-administration with inulin in HF diet.
The characteristics of the gut microbiota in rutin-supplemented mice were similar to LC group according to PCoA plot, suggesting the addition of rutin could attenuate gut dysbiosis induced by HF diet. Rutin supplementation in HF diet suppressed the reduction of Bacteroidetes and its families, and ameliorated the elevation of Lachnospiraceae, Firmicutes/Bacteroidetes (F/B) ratio, Deferribacteraceae and plasma LPS induced by high-fat diet. Our finding was supported by a number of previous studies that demonstrated that the influences of polyphenols on intestinal bacteria: purple lettuces administration, which contains high flavonoid content, decreased Lachnospiraceae and Deferribacteraceae (Han et al., 2018); regular wine vinegar ingestion or polyphenol-rich fruits and green tea favored the growth of Bacteroidetes community (Lee et al., 2006; Cerezo et al., 2008; Selma et al., 2009); and supplementation of quercetin or grape polyphenols stimulated the proliferation of bifidobacteria and attenuated the rise of F/B ratio and thereby ameliorated obesity-associated inflammation and metabolic disorders (Parkar et al., 2013; Etxeberria et al., 2015; Roopchand et al., 2015).
Polyphenols and dietary fiber are often eaten together, and interact to mediate gut microbiota. In the present study, the co-administration of polysaccharide inulin with rutin increased the breakdown of the rutin, indicated by the lower rutin left in feces when comparing the HRI with the HR group. This result was consistent with a prior in vitro study, which demonstrated that fermentable fibers could speed up the breakdown of the rutin (Jaganath et al., 2006). As expected, the combinatorial supplementation of inulin with rutin magnified the effects of rutin supplementation alone: elevated the production of butyrate and reduced the intestinal expression of TNF-α (p < 0.05).
Unfavorably, in this study, rutin supplement ameliorated the increase of body weight triggered by high-fat diet, while the co-administration with inulin reversed the decrease of body weight induced by rutin supplementation. This might largely be due to food intake elevated by sweet taste of inulin. Nevertheless, the combination of rutin and inulin further elevated the growth of Bacteroidaceae and Lactobacillus, and recovered the numbers of SCFA -producing bacterium Lachnospiraceae reduced by rutin addition (Figure 4C and Supplementary Figure S1). The bloom of Lachnospiraceae was largely accounted by increases of Lachnospiraceae_NK4A136_group, Lachnoclostridium, Roseburia and Blautia, most of which are butyrate producers (Million et al., 2018). Previous studies also demonstrated that inulin supplementation favored Lactobacillus and SCFA-producing bacteria Lachnospiraceae or Roseburia, and Bacteroides (Aguirre et al., 2016; Zhang et al., 2018). The HRI group also reduced a number of harmful bacteria including Deferribacteraceae and Desulfovibrionaceae. The further decrease in Deferribacteraceae in HRI was largely accounted for reduction of Mucispirillum genus in this study, which is known as mucin degrader and associated with early disruption of the colonic surface mucus layer, prior to the onset of symptomatic colitis (Belzer et al., 2014). Most members of Desulfovibrionaceae are reported as lipopolysaccharide (LPS) producers and lead to a low grade and chronic inflammation in obese objects (Xiao et al., 2014; Zhang-Sun et al., 2015). These results were in agreement with previous studies that reported Mucispirillum (belong to Deferribacteracea) and Desulfovibrionaceae abundances were significantly suppressed by inulin treatment (Yu et al., 2018; Zhang et al., 2018).
Conclusion
In summary, the present study demonstrated that rutin supplementation suppressed the rise of F/B ratio, Deferribacteracea and plasma LPS induced by HF diet, and thereby mitigated systematic and intestinal inflammation, and ER stress in Paneth cells. The co-administration of rutin with inulin improved the utilization of rutin as indicated by its decreased excretion, further suppressed a number of harmful bacteria, reduced the expression of the key inflammatory cytokine TNF-α and increased the production of butyrate. Taken together, our data demonstrated the potential to combine polyphenol rutin and the polysaccharide inulin as a dietary strategy to ameliorate gut dysbiosis, to improve inflammatory status and thereby to reduce medical disorders associated with HF-induced obesity.
Ethics Statement
This study was carried out in accordance with the laboratory animal-guildline for ethical review for animal welfare in China. The animal protocol was approved by the Institutional Animal Care and Use Committee of Chengdu University (Permission No. 2018-23-01).
Author Contributions
XG and ZL designed the research. YL and JL performed the research. RT and SY analyzed the data. XG wrote the paper. ZL supervised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The handling Editor declared a shared affiliation, though no other collaboration, with one of the authors SY at the time of review.
Acknowledgments
We acknowledge the Novogene Genome Sequencing Company (Beijing, China) for 16S rRNA gene sequencing and analysis of microbial profile.
Funding. This project was supported in part by National Natural Science Foundation of China (31401488, XG) and USDA grant (2014-67017-21762, ZL).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02651/full#supplementary-material
References
- Aguirre M., Bussolo de Souza C., Venema K. (2016). The gut microbiota from lean and obese subjects contribute differently to the fermentation of arabinogalactan and inulin. PLoS One 11:e0159236. 10.1371/journal.pone.0159236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo K. C., de M. B., Costa E. M., Pazini F., Valadares M. C., de Oliveira V. (2013). Bioconversion of quercetin and rutin and the cytotoxicity activities of the transformed products. Food Chem. Toxicol. 51 93–96. 10.1016/j.fct.2012.09.015 [DOI] [PubMed] [Google Scholar]
- Belzer C., Gerber G. K., Roeselers G., Delaney M., DuBois A., Liu Q., et al. (2014). Dynamics of the microbiota in response to host infection. PLoS One 9:e95534. 10.1371/journal.pone.0095534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani P. D., Bibiloni R., Knauf C., Waget A., Neyrinck A. M., Delzenne N. M., et al. (2008). Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes Metab. Res. Rev. 57 1470–1481. 10.2337/db07-1403 [DOI] [PubMed] [Google Scholar]
- Cani P. D., Neyrinck A. M., Fava F., Knauf C., Burcelin R. G., Tuohy K. M., et al. (2007). Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 50 2374–2383. 10.1007/s00125-007-0791-0 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo A. B., Tesfaye W., Torija M. J., Mateo E., García-Parrilla M. C., Troncoso A. M. (2008). The phenolic composition of red wine vinegar produced in barrels made from different woods. Food Chem. 109 606–615. 10.1016/j.foodchem.2008.01.013 [DOI] [Google Scholar]
- Creely S. J., McTernan P. G., Kusminski C. M., Fisher F. M., Da Silva N. F., Khanolkar M., et al. (2007). Lipopolysaccharide activates an innate immune system response in human adipose tissue in obesity and type 2 diabetes. Am. J. Physiol. Endocrinol. Metab. 292 E740–E747. 10.1152/ajpendo.00302.2006 [DOI] [PubMed] [Google Scholar]
- Delzenne N. M., Neyrinck A. M., Backhed F., Cani P. D. (2011). Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat. Rev. Endocrinol. 7 639–646. 10.1038/nrendo.2011.126 [DOI] [PubMed] [Google Scholar]
- Dewulf E. M., Cani P. D., Neyrinck A. M., Possemiers S., Van Holle A., Muccioli G. G., et al. (2011). Inulin-type fructans with prebiotic properties counteract GPR43 overexpression and PPARgamma-related adipogenesis in the white adipose tissue of high-fat diet-fed mice. J. Nutr. Biochem. 22 712–722. 10.1016/j.jnutbio.2010.05.009 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10 996–998. 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Elphick D. A., Mahida Y. R. (2005). Paneth cells: their role in innate immunity and inflammatory disease. Gut 54 1802–1809. 10.1136/gut.2005.068601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxeberria U., Arias N., Boque N., Macarulla M. T., Portillo M. P., Martinez J. A., et al. (2015). Reshaping faecal gut microbiota composition by the intake of trans-resveratrol and quercetin in high-fat sucrose diet-fed rats. J. Nutr. Biochem. 26 651–660. 10.1016/j.jnutbio.2015.01.002 [DOI] [PubMed] [Google Scholar]
- Finkelstein E. A., Khavjou O. A., Thompson H., Trogdon J. G., Pan L., Sherry B., et al. (2012). Obesity and severe obesity forecasts through 2030. Am. J. Prev. Med. 42 563–570. 10.1016/j.amepre.2011.10.026 [DOI] [PubMed] [Google Scholar]
- Garidou L., Pomié C., Klopp P., Waget A., Charpentier J., Aloulou M., et al. (2015). The gut microbiota regulates intestinal CD4 T cells expressing RORγt and controls metabolic disease. Cell Metab. 22 100–112. 10.1016/j.cmet.2015.06.001 [DOI] [PubMed] [Google Scholar]
- Gregg E. W., Shaw J. E. (2017). Global health effects of overweight and obesity. N. Engl. J. Med. 377 80–81. 10.1056/NEJMe1706095 [DOI] [PubMed] [Google Scholar]
- Guo X., Li J., Tang R., Zhang G., Zeng H., Wood R. J., et al. (2017). High fat diet alters gut microbiota and the expression of Paneth cell-antimicrobial peptides preceding changes of circulating inflammatory cytokines. Mediators Inflamm. 2017:9474896. 10.1155/2017/9474896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton M. K., Ronveaux C. C., Rust B. M., Newman J. W., Hawley M., Barile D., et al. (2017). Prebiotic milk oligosaccharides prevent development of obese phenotype, impairment of gut permeability, and microbial dysbiosis in high fat-fed mice. Am. J. Physiol. Gastrointest. Liver Physiol. 312 474–487. 10.1152/ajpgi.00427.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Zhao C., He X., Sheng Y., Ma T., Sun Z., et al. (2018). Purple lettuce (Lactuca sativa L.) attenuates metabolic disorders in diet induced obesity. J. Funct. Foods 45 462–470. 10.1016/j.jff.2018.04.027 [DOI] [Google Scholar]
- Hodin C. M., Verdam F. J., Grootjans J., Rensen S. S., Verheyen F. K., Dejong C. H., et al. (2011). Reduced Paneth cell antimicrobial protein levels correlate with activation of the unfolded protein response in the gut of obese individuals. J. Pathol. 225 276–284. 10.1002/path.2917 [DOI] [PubMed] [Google Scholar]
- Hoek-van den Hil E. F., Keijer J., Bunschoten A., Vervoort J. J., Stankova B., Bekkenkamp M., et al. (2013). Quercetin induces hepatic lipid omega-oxidation and lowers serum lipid levels in mice. PLoS One 8:e51588. 10.1371/journal.pone.0051588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek-van den Hil E. F., van Schothorst E. M., van der Stelt I., Swarts H. J., van Vliet M., Amolo T., et al. (2015). Direct comparison of metabolic health effects of the flavonoids quercetin, hesperetin, epicatechin, apigenin and anthocyanins in high-fat-diet-fed mice. Genes Nutr. 10:469. 10.1007/s12263-015-0469-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotamisligil G. S. (2006). Inflammation and metabolic disorders. Curr. Opin. Clin. Nutr. Metab. Care 444 459–464. 10.1038/nature05485 [DOI] [PubMed] [Google Scholar]
- Hou M., Combet E., Edwards C. A. (2015). Glucose fermentation does not impact on in vitro bacterial metabolism of hesperidin. Proc. Nutr. Soc. 74:E106 10.1017/S0029665115001214 [DOI] [Google Scholar]
- Jaganath I. B., Mullen W., Edwards C. A., Crozier A. (2006). The relative contribution of the small and large intestine to the absorption and metabolism of rutin in man. Free Radic. Res. 40 1035–1046. 10.1080/10715760600771400 [DOI] [PubMed] [Google Scholar]
- Jurgoński A., Juśkiewicz J., Kowalska K., Zduńczyk Z. (2012). Does dietary inulin affect biological activity of a grapefruit flavonoid-rich extract? Nutr. Metab. 9:31. 10.1186/1743-7075-9-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson F., Tremaroli V., Nielsen J., Backhed F. (2013). Assessing the human gut microbiota in metabolic diseases. Diabetes Metab. Res. Rev. 62 3341–3349. 10.2337/db13-0844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T., Yang W., Chen C. S., Reynolds K., He J. (2008). Global burden of obesity in 2005 and projections to 2030. Int. J. Obesity 32 1431–1437. 10.1038/ijo.2008.102 [DOI] [PubMed] [Google Scholar]
- Lee H. C., Jenner A. M., Low C. S., Lee Y. K. (2006). Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota. Res. Microbiol. 157 876–884. 10.1016/j.resmic.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Lee J. C., Lee H. Y., Kim T. K., Kim M. S., Park Y. M., Kim J., et al. (2017). Obesogenic diet-induced gut barrier dysfunction and pathobiont expansion aggravate experimental colitis. PLoS One 12:e0187515. 10.1371/journal.pone.0187515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Bäckhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102 11070–11075. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444 1022–1023. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- Liu W., Crott J. W., Lyu L., Pfalzer A. C., Li J., Choi S.-W., et al. (2016). Diet- and genetically-induced obesity produces alterations in the microbiome, inflammation and Wnt pathway in the intestine of Apc+/1638N mice: comparisons and contrasts. J. Cancer 7 1780–1790. 10.7150/jca.15792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71 8228–8235. 10.1128/aem.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck H., Tsai S., Chung J., Clemente-Casares X., Ghazarian M., Revelo Xavier S., et al. (2015). Regulation of obesity-related insulin resistance with gut anti-inflammatory agents. Cell Metab. 21 527–542. 10.1016/j.cmet.2015.03.001 [DOI] [PubMed] [Google Scholar]
- Mansoorian B., Garcia A. L., Combet E., Edwards C. A. (2015). Dietary fiber reduced phenolic acid production from rutin in an ex vivo fermentation model. Proc. Nutr. Soc. 74:E47 10.1017/S0029665115000622 [DOI] [Google Scholar]
- Million M., Tomas J., Wagner C., Lelouard H., Raoult D., Gorvel J.-P. (2018). New insights in gut microbiota and mucosal immunity of the small intestine. Hum. Microbiome J. 7-8 23–32. 10.1016/j.humic.2018.01.004 [DOI] [Google Scholar]
- Monteiro-Sepulveda M., Touch S., Mendes-Sá C., André S., Poitou C., Allatif O., et al. (2015). Jejunal T cell inflammation in human obesity correlates with decreased enterocyte insulin signaling. Cell Metab. 22 113–124. 10.1016/j.cmet.2015.05.020 [DOI] [PubMed] [Google Scholar]
- Parkar S. G., Trower T. M., Stevenson D. E. (2013). Fecal microbial metabolism of polyphenols and its effects on human gut microbiota. Anaerobe 23 12–19. 10.1016/j.anaerobe.2013.07.009 [DOI] [PubMed] [Google Scholar]
- Parks B. W., Nam E., Org E., Kostem E., Norheim F., Hui S. T., et al. (2013). Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 17 141–152. 10.1016/j.cmet.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopchand D. E., Carmody R. N., Kuhn P., Moskal K., Rojas-Silva P., Turnbaugh P. J., et al. (2015). Dietary polyphenols promote growth of the gut bacterium akkermansia muciniphila and attenuate high-fat diet-induced metabolic syndrome. Diabetes Metab. Res. Rev. 64 2847–2858. 10.2337/db14-1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. 10.1186/gb-2011-12-6-r60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selma M. V., Espin J. C., Tomas-Barberan F. A. (2009). Interaction between phenolics and gut microbiota: role in human health. J. Agric. Food Chem. 57 6485–6501. 10.1021/jf902107d [DOI] [PubMed] [Google Scholar]
- Sheveleva M. A., Ramenskaya G. V. (2010). Gas chromatographic analysis of short-chain fatty acids in the standardization of medicinal formulations based on bacterial substrates. Pharm. Chem. J. 44 334–336. 10.1007/s11094-010-0462-1 [DOI] [Google Scholar]
- Taguri T., Tanaka T., Kouno I. (2004). Antimicrobial activity of 10 different plant polyphenols against bacteria causing food-borne disease. Biol. Pharm. Bull. 27 1965–1969. [DOI] [PubMed] [Google Scholar]
- Tuohy K. M., Conterno L., Gasperotti M., Viola R. (2012). Up-regulating the human intestinal microbiome using whole plant foods, polyphenols, and/or fiber. J. Agric. Food Chem. 60 8776–8782. 10.1021/jf2053959 [DOI] [PubMed] [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73 5261–5267. 10.1128/aem.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winer D. A., Luck H., Tsai S., Winer S. (2016). The intestinal immune system in obesity and insulin resistance. Cell Metab. 23 413–426. 10.1016/j.cmet.2016.01.003 [DOI] [PubMed] [Google Scholar]
- Xiao S., Fei N., Pang X., Shen J., Wang L., Zhang B., et al. (2014). A gut microbiota-targeted dietary intervention for amelioration of chronic inflammation underlying metabolic syndrome. FEMS Microbiol. Ecol. 87357–367. 10.1111/1574-6941.12228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Zhao J., Xu Z., Chen Y., Shao T., Long X., et al. (2018). Inulin from Jerusalem artichoke tubers alleviates hyperlipidemia and increases abundance of bifidobacteria in the intestines of hyperlipidemic mice. J. Funct. Foods 40 187–196. 10.1016/j.jff.2017.11.010 [DOI] [Google Scholar]
- Zhang C., Zhang M., Pang X., Zhao Y., Wang L., Zhao L. (2012). Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 6 1848–1857. 10.1038/ismej.2012.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Yu H., Xiao X., Hu L., Xin F., Yu X. (2018). Inulin-type fructan improves diabetic phenotype and gut microbiota profiles in rats. PeerJ 6:e4446. 10.7717/peerj.4446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang-Sun W., Augusto L. A., Zhao L., Caroff M. (2015). Desulfovibrio desulfuricans isolates from the gut of a single individual: structural and biological lipid A characterization. FEBS Lett. 589 165–171. 10.1016/j.febslet.2014.11.042 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.