Abstract
Introduction
Hypervirulent capsular type K1 Klebsiella pneumoniae strains of clonal complex 23 (CC23) are associated with severe community-acquired pyogenic liver abscesses, often complicated by metastatic infections and significant mortality. The majority of hypervirulent strains reported are susceptible to most antibiotics except ampicillin. To the best of our knowledge, this is the first case of New Delhi metallo-β-lactamase (blaNDM)-producing hypervirulent K. pneumoniae from the UK.
Case presentation
We present a case of pyogenic liver abscess in a 63-year-old female of Bangladeshi origin, with a recent diagnosis of pancreatic cancer. The patient was treated with piperacillin/tazobactam and blood cultures grew a fully susceptible Escherichia coli. Despite antimicrobial therapy and drainage of the abscess, the patient continued to deteriorate and died on day seven of admission. The fluid drained from the liver abscess grew a fully susceptible E. coli and a multi-drug-resistant K. pneumoniae. Two weeks prior to admission, a rectal screening swab grew a metallo-β-lactamase-producing K. pneumoniae. Molecular characterization revealed that both the K. pneumoniae isolates belonged to the hypervirulent K1 cluster of CC23, sequence type 23. The isolate from the rectal screen was positive for the blaNDM metallo-β-lactamase gene.
Conclusion
The emergence of carbapenemase-producing hypervirulent K. pneumoniae strains presents a new and significant threat to global public health. Management of these infections will be extremely challenging due to the limited treatment options available and they are likely to be associated with an even greater mortality.
Keywords: pyogenic liver abscess, hypervirulent Klebsiella pneumoniae, K1 capsular type, clonal complex 23, carbapenemase, metallo-β-lactamase
Introduction
Hypervirulent K1 Klebsiella pneumoniae strains are associated with severe community-acquired pyogenic liver abscesses, often complicated by metastatic infections including bacteraemia, necrotizing fasciitis, pneumonia, endopthalmitis and meningitis [1]. These infections frequently occur in younger, previously healthy adults and are associated with significant mortality [2]. This invasive syndrome emerged in the 1980s and was primarily reported in South-East Asia, but there are increasing numbers of cases being reported worldwide [1–3]. Virulence factors include capsule overproduction facilitating evasion of the immune system, and production of siderophores, which are particularly effective in sequestering iron. Some of the virulence genes are carried on a plasmid, which is also found in diverse hypervirulent strains of other capsular types, particularly K2 [2, 3].
The majority of hypervirulent K. pneumoniae isolates reported have been susceptible to most antibiotics except ampicillin [3, 4]. Whole-genome sequencing has been used to distinguish clonal groups of multi-drug-resistant and hypervirulent K. pneumoniae and although these populations are largely non-overlapping, isolates with combined virulence and resistance determinants have been detected [5]. Carbapenemase-producing K. pneumoniae have emerged worldwide over the last ten years [6]. These strains and their resistance plasmids have the capacity to disseminate rapidly and cause outbreaks in healthcare settings. They are frequently resistant to multiple antibiotic classes and infections can be associated with a higher mortality compared to infections with susceptible strains [7]. Hypervirulent strains of carbapenemase-producing K. pneumoniae have recently been reported in Argentina and China [8, 9]. To the best of our knowledge, this is the first reported case of New Delhi metallo-β-lactamase (blaNDM)-producing hypervirulent K. pneumoniae from the UK.
Case report
A 63-year-old female of Bangladeshi origin presented to an Emergency Department in London, United Kingdom, in 2015 with a four day history of fever, rigors and right upper quadrant pain. She had been recently diagnosed with metastatic pancreatic cancer, and had an internal biliary stent inserted six weeks prior to admission. Two weeks prior to admission, the patient had attended hospital as a day case for a liver biopsy, at which time a rectal screening swab for carbapenemase-producing organisms was taken, which grew K. pneumoniae. Automated antimicrobial-sensitivity testing (AST) was performed using the Becton Dickinson Phoenix platform (BD Diagnostics) according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines. The isolate was resistant to co-amoxiclav, piperacillin/tazobactam, ceftriaxone, ceftazidime, temocillin, ertapenem, meropenem, gentamicin, amikacin, tobramycin, ciprofloxacin, levofloxacin, trimethoprim/sulfamethoxazole and fosfomycin, and susceptible to colistin, tigecycline and nitrofurantoin. Minimum inhibitory concentrations (MICs) as determined by Etest (bioMérieux) were 128 µg/ml for meropenem and 8 µg/ml for ertapenem. A positive result for the combination disc synergy test (Rosco Diagnostica) between meropenem and dipicolinic acid indicated the possible presence of a metallo-β-lactamase.
Investigations
On admission, the patient was septic, with a temperature of 38 °C, a heart rate of 125 beats per min, blood pressure of 106/44 mmHg and a respiratory rate of 25 breaths per min. On examination, the patient had mild right upper quadrant tenderness and initial investigations revealed a white cell count of 2.8×1010 cells/L (2.5×1010 neutrophils/L), a C-reactive protein level of 318 mg/L and a normal chest X-ray. Blood cultures were taken and, after discussion with the microbiology team, the patient was started on piperacillin/tazobactam, 4.5 g intravenously, every 8 h, for presumed biliary sepsis. On day one of admission, the blood cultures grew Escherichia coli, which was found to be fully susceptible to all antibiotics tested including β-lactams, aminoglycosides and fluoroquinolones on day two.
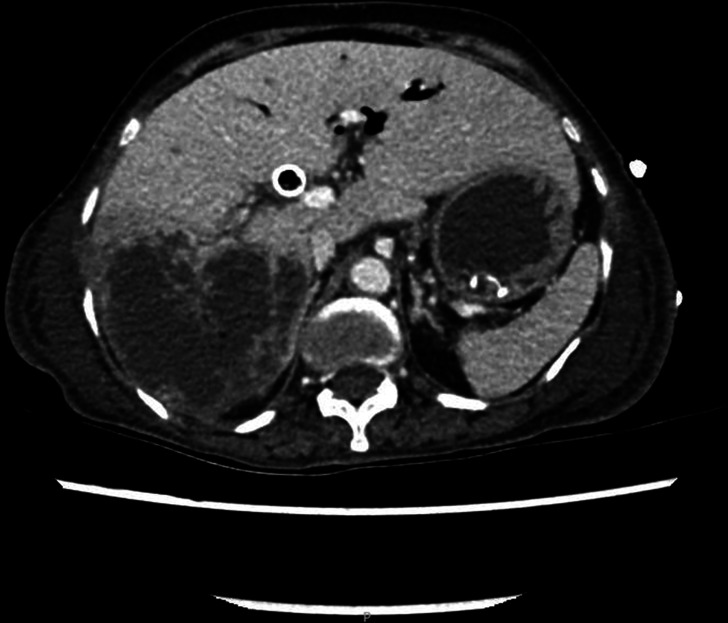
On day three of admission, a computed tomography scan of the chest, abdomen and pelvis revealed a large multi-septate liver abscess within the right lobe measuring 8.6×8.5×14 cm, with a patent biliary stent and stable appearances of the large pancreatic head mass encasing the superior mesenteric vessels (Fig. 1). A pig-tail drain was inserted into the abscess under ultrasound-guidance, and heavily blood-stained fluid was drained and sent to the microbiology laboratory.
Fig. 1.
Computed tomography scan of the chest, abdomen and pelvis revealing a large (8.6×8.5×14 cm) multi-septate liver abscess within the right lobe.
Diagnosis
The fluid drained from the liver abscess grew E. coli and K. pneumoniae. The E. coli demonstrated the same susceptibility profile as the E. coli previously isolated from blood cultures. The K. pneumoniae was resistant to the same antibiotics as the K. pneumoniae previously isolated from the rectal screen, apart from ertapenem and meropenem, to which it was sensitive. MICs as determined by Etest (bioMérieux) were 0.047 µg/ml for meropenem and 0.047 µg/ml for ertapenem. Combination disc synergy test between cefpodoxime and cefpodoxime/clavulanic acid indicated the presence of an extended-spectrum β-lactamase.
Treatment
On admission, the patient was treated empirically with piperacillin/tazobactam, 4.5 g intravenously, every 8 h, for presumed biliary sepsis. This was changed to co-amoxiclav, 1.2 g intravenously, every 8 h based on the AST profile of the E. coli isolated from blood cultures.
Outcome and follow-up
Despite drainage and antimicrobial therapy active against the E. coli grown from blood cultures, the patient continued to deteriorate. The hepato-pancreato-biliary surgical team did not consider the patient to be an appropriate candidate for surgery and she died on day seven of admission. AST results for the K. pneumoniae isolated from the liver abscess fluid were not available until the day the patient died.
The K. pneumoniae isolates from the rectal screening swab and liver abscess were referred to the national reference laboratory for further molecular investigation. Molecular characterization was undertaken using nine-locus variable number tandem repeat (VNTR) analysis (at loci A, E, H, J, K, D, N1, N2 and N4) and multiplex-PCR for capsular type-specific and virulence gene targets, as described previously [10, 11]. Multiplex-PCR was used to screen for KPC, OXA-48-like, NDM and VIM acquired carbapenemase genes, as described previously [12]. Next-generation sequencing (NGS) was also performed by the Genomic Services and Development Unit (National Infection Service, Public Health England, London, UK) on an Illumina Hi-Seq platform following extraction on a QIASymphony instrument.
Both isolates were confirmed as capsular type K1, and VNTR and NGS identified both strains as belonging to the K1 cluster of clonal complex 23 (CC23) (VNTR profile 3,5,2,7,0,1,4,4,1), both of classical sequence type 23 (ST23). Both isolates were positive for wcaG, which encodes capsular fucose synthesis and is always associated with K1 isolates, and negative for rmpA, a positive regulator of extracapsular polysaccharide synthesis. The isolate from the rectal screening swab was positive for the blaNDM metallo-β-lactamase gene (confirmed as blaNDM-1 by NGS), but the isolate from the liver abscess was negative for all acquired carbapenemase genes tested, in keeping with the phenotypic antibiotic-susceptibility results. NGS revealed the presence of virulence determinants typically associated with hypervirulent K1 K. pneumoniae strains of CC23 in both isolates. These included: allS, which is associated with allantoin metabolism, various iron acquisition systems, aerobactin (iucABCD-iutA), the ferric uptake operon kfuABC, yersiniabactin (ybtAEUPQRSTX), colibactin (clbABCDEFGHIJKLMNOPQR), salmochelin (iroBCDN), a tellurite resistance operon (terABCDEWXZ) and rmpA2 (a variant of rmpA).
Discussion
As far as we are aware, this is the first reported case of carbapenemase-producing ST23 K. pneumoniae in the UK. Although reports of carbapenemase-producing hypervirulent K. pneumoniae strains have so far been scarce, the emergence of such strains is particularly concerning due to the combination of hypervirulence and multi-drug resistance. Management of carbapenemase-producing hypervirulent K. pneumoniae infections will be extremely challenging due to the limited treatment options available and these strains are likely to be associated with an even greater mortality. Rapid diagnosis and appropriate management has the potential to improve outcomes and minimize the risk of metastatic complications [1]. The prospect of potentially untreatable invasive infection in young, previously healthy adults presents a new and significant threat to global public health.
Funding information
The authors received no specific grant from any funding agency.
Acknowledgements
We acknowledge the Royal Free Hospital Microbiology and Hepato-pancreato-biliary Surgery teams, Public Health England AMRHAI and Genomic Services and Development Unit.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
We were unable to obtain patient consent as the patient died. We have removed all patient identifiable data from this manuscript.
Footnotes
Abbreviations: AMRHAI, Antimicrobial Resistance and Healthcare Associated Infections; AST, antimicrobial-sensitivity testing; CC23, clonal complex 23; NGS, next-generation sequencing; VNTR, variable number tandem repeat; MIC, minimum inhibitory concentration; EUCAST, European Committee on Antimicrobial Susceptibility Testing; ST23, sequence type 23.
References
- 1.Siu LK, Yeh KM, Lin JC, Fung CP, Chang FY. Klebsiella pneumoniae liver abscess: a new invasive syndrome. Lancet Infect Dis. 2012;12:881–887. doi: 10.1016/S1473-3099(12)70205-0. [DOI] [PubMed] [Google Scholar]
- 2.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, et al. Mapping the evolution of hypervirulent Klebsiella pneumoniae. MBio. 2015;6:e00630. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luo Y, Wang Y, Ye L, Yang J. Molecular epidemiology and virulence factors of pyogenic liver abscess causing Klebsiella pneumoniae in China. Clin Microbiol Infect. 2014;20:O818–O824. doi: 10.1111/1469-0691.12664. [DOI] [PubMed] [Google Scholar]
- 5.Bialek-Davenet S, Criscuolo A, Ailloud F, Passet V, Jones L, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20:1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect. 2014;20:821–830. doi: 10.1111/1469-0691.12719. [DOI] [PubMed] [Google Scholar]
- 7.Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, et al. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol. 2009;30:972–976. doi: 10.1086/605922. [DOI] [PubMed] [Google Scholar]
- 8.Cejas D, Fernández Canigia L, Rincón Cruz G, Elena AX, Maldonado I, et al. First isolate of KPC-2-producing Klebsiella pneumonaie sequence type 23 from the Americas. J Clin Microbiol. 2014;52:3483–3485. doi: 10.1128/JCM.00726-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang R, Lin D, Chan EW, Gu D, Chen GX, et al. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2016;60:709–711. doi: 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turton JF, Perry C, Elgohari S, Hampton CV. PCR characterization and typing of Klebsiella pneumoniae using capsular type-specific, variable number tandem repeat and virulence gene targets. J Med Microbiol. 2010;59:541–547. doi: 10.1099/jmm.0.015198-0. [DOI] [PubMed] [Google Scholar]
- 11.Al-Agamy MH, Shibl AM, Elkhizzi NA, Meunier D, Turton JF, et al. Persistence of Klebsiella pneumoniae clones with OXA-48 or NDM carbapenemases causing bacteraemias in a Riyadh hospital. Diagn Microbiol Infect Dis. 2013;76:214–216. doi: 10.1016/j.diagmicrobio.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Ellington MJ, Findlay J, Hopkins KL, Meunier D, Alvarez-Buylla A, et al. Multicentre evaluation of a real-time PCR assay to detect genes encoding clinically relevant carbapenemases in cultured bacteria. Int J Antimicrob Agents. 2016;47:151–154. doi: 10.1016/j.ijantimicag.2015.11.013. [DOI] [PubMed] [Google Scholar]