Summary
One of the primary objectives of wheat breeding was to increase grain yield. Floral abortion during the stem elongation phase (SEP) leads to a loss of more than 50% of the grain number potential. In this study, we quantified 75 plant growth‐associated traits at seven stages during the SEP and mapped 15 696 single nucleotide polymorphism (SNP) markers in 210 accessions of wheat (Triticum aestivum). Our genomewide association study identified trait‐associated SNPs that are shared among various stages of the SEP, as well as SNPs that are shared between plant growth traits and grain yield in the field. The genomic selection analysis shows variation among the prediction abilities of various traits and stages. Furthermore, we found that the allelic variants of Ppd‐D1 (chromosome 2D) and Rht‐D1 (chromosome 4D) loci affect some plant growth traits (e.g. leaf area and spike length). These results have identified a narrow time window within the SEP in which plant growth traits can be manipulated to alter grain yield. This suggests that there may be multiple ways to regulate plant growth during the SEP, to ultimately influence grain number in wheat.
Keywords: genomic selection, GWAS, plant growth, stem elongation, wheat
Introduction
World food security is a serious and pressing contemporary issue (Godfray et al., 2010; Grassini et al., 2013; Schmidhuber and Tubiello, 2007; Tilman et al., 2011). The demand for crop production is predicted to increase as the global population increases (Beddington et al., 2012; Challinor et al., 2014; Lobell et al., 2011). Wheat (Triticum aestivum L.) is an important crop that provides 55% of the carbohydrates and 20% of the calories consumed by people worldwide (Breiman and Graur, 1995). Therefore, increasing wheat yield remains one of the main objectives of breeding efforts. An individual wheat spike generally consists of 20–30 spikelets, and an individual spikelet generally contains 2–4 grains at physiological maturity. The indeterminate nature of wheat spikelets enables the formation of 8–12 floret primordia within each spikelet, which is referred to as the grain number potential (Ferrante et al., 2013; Gonzalez et al., 2011; Guo and Schnurbusch, 2015). However, fewer than 50% of the floret primordia survive to develop into fertile florets at anthesis, so that most of the grain yield potential is lost during the stem elongation phase (SEP, Figure 1).
Figure 1.
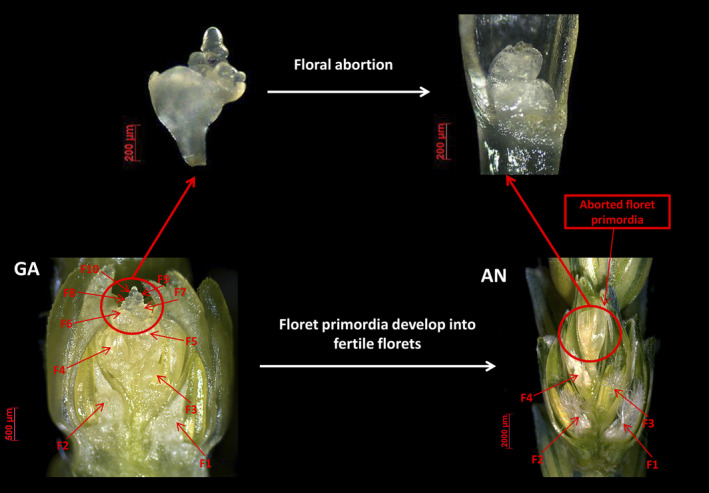
The loss of grain number yield potential (floral abortion) during stem elongation phase in wheat.
During the SEP, wheat floret primordia undergo a complicated process of development, including abortion of some primordia. Reducing the loss of floret primordia during the SEP and increasing the number of fertile florets at anthesis are important for improving grain number in wheat. Increasing the partitioning of assimilate into developing spikes during the SEP can prevent the loss of floret primordia and fertile florets in wheat (Bancal, 2009; Dreccer et al., 2014; Gonzalez et al., 2011; Guo et al., 2016). Therefore, it is necessary to identify the associated critical stages of plant growth (such as leaf growth, spike growth) to be able to decrease floral abortion and improve wheat grain number.
Following Kirby and Appleyard (1987), and Zadoks et al. (1974), we divided the floral development and abortion process into seven stages: terminal spikelet (TS) stage, white anther (WA) stage, green anther (GA) stage, yellow anther (YA) stage, tipping (TP) stage, heading (HD) stage and anthesis (AN) (Figure 2) (Guo and Schnurbusch, 2015). At the TS stage, the immature spike is almost completely formed, no spikelets are initiated, and the last few primordia become the glumes and floret primordia of a terminal spikelet, which terminates rachis development. At the WA stage, the meristem of each spikelet has initiated more than eight floret primordia, the glumes partially enclose the florets, and the stamens of florets 1 and 2 cannot be seen as they are completely enclosed by their lemmas at the bottom of the spikelet. At the GA stage, the spikelet meristem stops producing floret primordia (Guo and Schnurbusch, 2015), and the most obvious trait at this stage is that the glumes cover the entire spikelet except for the tips of the most distal floret primordia. At the YA stage, the glumes are fully formed, the lemmas of the first three florets are visible, the lemma of floret 1 (F1) has a short awn point, the awns of floret 2 (F2) and F3 at the base of the spikelet are longer compared with F1, and florets 1, 2 and 3 at the base of the spikelet are well developed. At the TP stage, the first awns are visible and florets with visible anthers and ovaries develop further, whereas the apical floret primordia remain arrested in their growth. At the HD stage, half of the individual spike is visible, the F1 anthers reach their maximum size, and the glumes become stiffer. At the AN stage, the yellow anthers extrude from florets 1 and 2 at the base of the spikelet, and the florets that are still alive become fertile florets. Between HD and AN, the F1 ovaries develop rapidly, the aborted apical floret parts disappear completely, anthers turn completely yellow, and stigmatic hair becomes well developed. A significant increase in spike length and width can be observed between the TS and YA stages and the YA stage and AN (Figure 2).
Figure 2.
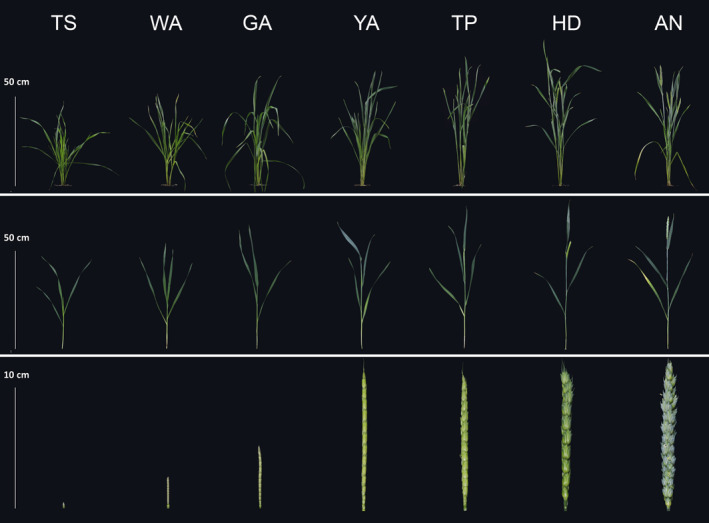
The seven floret development and abortion stages during stem elongation phase: TS, WA, GA, YA, TP, HD and AN.
Although features of floral development during the pre‐anthesis phase have been described in detail (Waddington et al., 1983), the temporal progression of accompanying traits (such as leaf area and spike length) during the seven stages has not been well documented. The interaction between floral development and the progression of accompanying traits may affect assimilate partitioning and the usage efficiency of developing florets, which ultimately influences wheat grain number.
Hence, we investigated the progression of the accompanying plant growth traits to obtain information on the dynamic changes in morphology (e.g. spike growth, leaf growth) within the substages of the SEP. We further showed an overlap in the single nucleotide polymorphism (SNP)‐associated traits between plant growth traits and grain yield, suggesting a genetic association between these traits. In addition, we identified SNPs that may be applied to genetically manipulate plant growth. The results from our genomic prediction analysis indicated the ability to predict plant growth traits.
Results
The results presented in this study will be useful for designing strategies to increase floret fertility by selecting plant growth traits with yield‐relevant effects during the SEP. Our results are divided into four parts: (i) trends in the plant growth traits among the seven SEP stages; (ii) identification of SNP–trait‐associated markers that are shared between different stages, shared between different traits at the same stage, and shared between plant growth and grain number traits; (iii) genomic selection analysis results; and (iv) effects of allelic variants of Ppd‐D1 (chromosome 2D) and Rht‐D1 (chromosome 4D) loci on plant growth traits.
Novel phenotyping to monitor plant growth during the stem elongation phase in wheat
According to the floral development and abortion process, we divided the SEP into seven stages: TS, WA, GA, YA, TP, HD and AN (Table 1). We monitored plant growth among to the seven specific stages by measuring 75 phenotypic traits: tiller number, leaf number, node number, spikelet number, plant height, spike length, leaf area, leaf dry weight (DW), main shoot DW and tiller DW during all seven stages, as well as spike DW at five stages (Table 1).
Table 1.
Plant growth traits and developmental stages in this study
| Plant growth trait | Plant growth stage |
|---|---|
| Tiller number | Terminal spikelet (TS) stage |
| Leaf number | White anther (WA) stage |
| Node number | Green anther (GA) stage |
| Plant height | Yellow anther (YA) stage |
| Spike length | Tipping (TP) stage |
| Spikelet number | Heading (HD) stage |
| Leaf area | Anthesis (AN) |
| Spike dry weight (DW) | |
| Leaf dry weight (DW) | |
| Main shoot dry weight (DW) | |
| Tiller dry weight (DW) |
All the traits were only measured on the main shoot, except tiller number and tiller DW. Tiller DW is the dry weight of a plant without the main shoot. Grain number indicates the grain number per spikelet/spike on the main shoot; grain area, length and width are measurements of individual grains on the main shoot. All of the plant growth traits were measured at each of the seven stages, except for spike DW, which was not measured at the TS or WA stages.
As shown in Figures 3 and S2, the maximum tiller number was observed during the WA stage and then declined until AN. The maximum leaf number, area and DW on the main shoot were observed at the GA stage and then declined until AN. The maximum node number was observed at the YA stage and remained constant thereafter. Plant height, main shoot DW and tiller DW generally increased over time at changing rates. These traits increased rapidly from the TS to WA stages and from the HD to AN stage, and increased slowly from WA to HD. Spike length increased rapidly from TS to YA, yet there was little change from YA to AN. Spike DW rapidly increased from TS to AN. Although the spikelet number appeared to increase within a narrow range, the differences between the stages were not significant (Figure S2).
Figure 3.
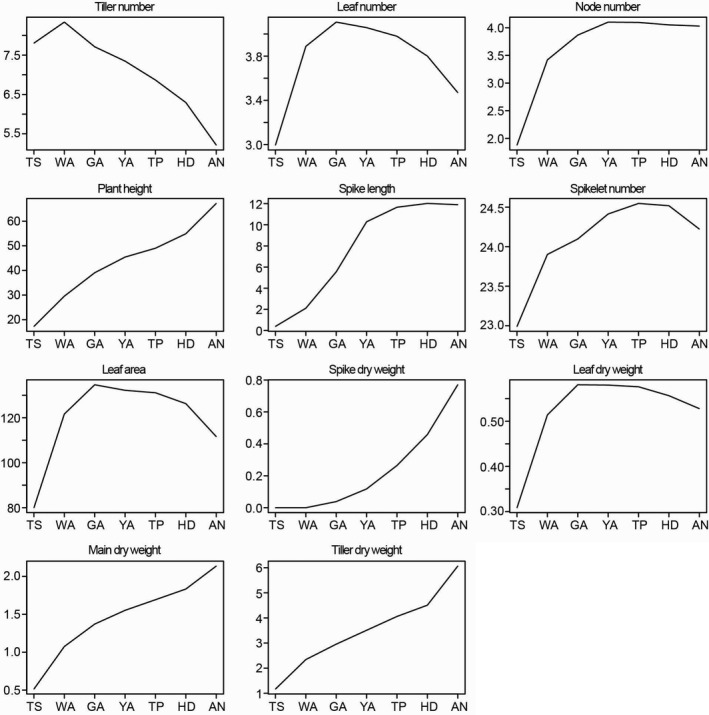
The trends of plant growth traits according to the seven stages during stem elongation phase.
The measured traits displayed variable trends among the seven developmental stages suggesting that the different traits peaked during different SEP stages (Figure 3). Moreover, most of the traits varied dramatically with variations in genotypes at the different stages in this study, and the variation in the traits changed among the stages (Figures 3 and S2). For example, tiller and node numbers displayed large variation during the early developmental stages including TS and WA, while the variation in these traits decreased during the late developmental stages including HD and AN (Figure S2). We surmised that the tiller abortion and achievement of stem elongation in the late stages led to the relatively narrow variation in tiller and node numbers. Spike dry weight, main shoot dry weight and tiller dry weight varied broadly in the late stages (e.g. HD, AN) compared with the early stages (e.g. TS, WA). The fast growth and large increase in these three traits during the booting stages (YA, TP, HD and AN) may explain the broad variation during the late stages. The ranges in variation observed among the tested traits (i.e. due to variations in genotype) provide sufficient space for the genetic analysis (e.g. GWAS).
Determining the shared SNP markers of the same plant growth traits between different stages
By dividing the SEP into seven stages according to the floral development and abortion process, we identified SNP–trait associations for plant growth traits at various stages for the first time. We also revealed differences in the SNP–trait associations among the seven stages.
Spike growth (leaf length of the main shoot) can be used as an example: we identified shared SNP markers (−log10(P)>4.0) among the seven stages (Table 2). Generally, we detected more SNP–spike length associations at the late development stages (e.g. HD, AN) than at the early stages, and the number of associations increased as plant growth progressed. The variation in SNP–spike length associations observed at different developmental stages can be exploited to manipulate spike length at specific stages, which is an important factor for determining grain yield. For example, we found that the largest increase in spike length occurs between the TS to YA stages, while the spike length changes little through the YA to AN stages (Figures 2 and 3). This suggests that the increase in spike length is associated with certain SNP markers during the early stages (e.g. 120.6 cM/608118822, 6A, WA), and the final spike length is associated with different SNP markers at late stages (e.g. 83.9 cM/92441199, 7D, HD, AN).
Table 2.
The genetic positions of SNPs that are associated with spike length of main shoot among the seven developmental stages (−log10(P)>4.0)
| SNP marker genetic position (cM) | TS | WA | GA | YA | TP | HD | AN |
|---|---|---|---|---|---|---|---|
| 1A, 72.1 | AN | ||||||
| 1B, 91.2 | HD | ||||||
| 2A, 64.3–69.8 | TP | HD | AN | ||||
| 2B, 39.5 | TS | ||||||
| 2B, 72.2 | YA | ||||||
| 2D, 74.0–74.8 | YA | TP | HD | AN | |||
| 3A, 108.7 | WA | ||||||
| 3A, 118.7 | WA | ||||||
| 4A, 160.3–161.8 | TP | ||||||
| 5A, 27.0 | HD | ||||||
| 5A, 114.5 | HD | ||||||
| 5B, 2.3 | AN | ||||||
| 5B, 93.0–99.3 | WA | TP | HD | ||||
| 5B, 105.6–112.5 | TP | HD | AN | ||||
| 5B, 122.6–128.1 | YA | TP | HD | AN | |||
| 5B, 152.6–154.9 | GA | YA | TP | HD | AN | ||
| 6A, 6.7–7.5 | HD | AN | |||||
| 6A, 33.4 | HD | AN | |||||
| 6A, 120.6 | WA | ||||||
| 6B, 44.4–49.0 | HD | AN | |||||
| 6B, 55.2 | TP | HD | |||||
| 7B, 76.4 | TP | ||||||
| 7B, 129.6 | HD | ||||||
| 7D, 83.9 | HD | AN |
The positions marked with SEP stages indicate that the SNP–spike length associations were detected at those stages. The names of the SNP markers are included in Data S1. Genetic positions are shown in centiMorgans (cM). The physical positions are displayed in Data S1. AN, anthesis; GA, green anther; HD, heading; TP, tipping; TS, Terminal spikelet; WA, white anther; YA, yellow anther.
However, there are two additional explanations for differences in trait‐associated SNP markers among the SEP stages. (i) The strong variation in spike length at the early stages leads to a decrease in the number of associated SNP markers; (ii) most of the detected SNP–trait associations at the late developmental stages (e.g. TP, HD, AN tage) are related to the determination of spike length, but the most important effects on spike elongation may occur at different stages.
Information on the SNP–trait associations for all the plant growth traits across the seven stages is provided in Data S1. The differences in SNP–trait associations among the different stages for the same traits that we observed will provide opportunities to manipulate plant growth traits during specific developmental stages.
Association of plant growth traits between glasshouse and field data
As this experiment was conducted in glasshouse, we selected 30 genotypes from the 210 wheat accessions to measure the traits in field and glasshouse simultaneously to display the connections of data between field and glasshouse (Table 3). Generally, we observed a close association of data between the field and glasshouse conditions. Leaf dry weight, leaf area and main dry weight displayed strong correlations between field and glasshouse conditions across all seven SEP stages. Plant height and spikelet number were also closely associated in the field and glasshouse conditions across all seven stages, except for the first stage (TS). Tiller dry weight showed relatively close connection between field and glasshouse conditions at the early stages (i.e. TS, WA, GA, YA and TP), yet weak at the late stages (i.e. HD and AN). As expected, tiller number, leaf number and node number between field and greenhouse conditions were weakly correlated at most of the stages. The weak associations may be attributed to the sensitivity of leaf, tiller and node initiation to environments (Friend, 1965; Huibert and Neuteboom, 1998; Whitechurch et al., 2007).
Table 3.
The phenotypic association between field and greenhouse conditions at the seven developmental stages
| Traits | TS | WA | GA | YA | TP | HD | AN |
|---|---|---|---|---|---|---|---|
| Tiller number | 0.3366 | 0.4000 | 0.0890 | −0.0340 | 0.0324 | 0.1384 | −0.2198 |
| Leaf number | 0.2853 | 0.2289 | 0.2930 | 0.2624 | 0.3360 | −0.0765 | −0.1146 |
| Node number | 0.3420 | 0.2699 | 0.1557 | 0.3955 | −0.3001 | 0.0974 | −0.1830 |
| Plant height | −0.4574 | 0.7586 | 0.6620 | 0.7212 | 0.6580 | 0.5953 | 0.7978 |
| Spike length | −0.1727 | 0.0237 | 0.0619 | 0.4515 | 0.6180 | 0.6554 | 0.7944 |
| Spikelet number | −0.4228 | 0.7318 | 0.7461 | 0.6804 | 0.6725 | 0.7283 | 0.8038 |
| Leaf area | 0.5988 | 0.5263 | 0.4362 | 0.7277 | 0.5300 | 0.4815 | 0.6609 |
| Spike dry weight | 0.0159 | −0.0260 | 0.0077 | 0.4228 | 0.4058 | 0.4975 | 0.5160 |
| Leaf dry weight | 0.5352 | 0.7044 | 0.5428 | 0.7432 | 0.5071 | 0.5216 | 0.5767 |
| Main dry weight | 0.5196 | 0.8107 | 0.7195 | 0.6902 | 0.6297 | 0.6261 | 0.6762 |
| Tiller dry weight | 0.4811 | 0.5642 | 0.3507 | 0.3454 | 0.2648 | 0.0117 | −0.0303 |
AN, anthesis; GA, green anther; HD, heading; TP, tipping; TS, Terminal spikelet; WA, white anther; YA, yellow anther.
Identification of the shared SNP markers between plant growth traits and grain yield in the field
In this study, we also measured the grain yield in eight different field environments. We conducted a GWAS for grain yield in the field and plant growth traits in the glasshouse to identify the associated SNP markers (Table 4). We observed that the SNP markers on chromosome 6D (1.6–4.0 cM/3137516–105892172) associated with plant height at the TS, WA, GA and TP stages were associated with grain yield in the field. This indicates that the increase in plant height during the early stages is important for determining grain yield in the field in this population.
Table 4.
SNP markers associated with plant growth traits and grain yield in the field
| Marker | Linkage group | Genetic position | −log10(P) | Trait |
|---|---|---|---|---|
| AX‐94398509 | 6D | 1.6 | 4.81 | WA plant height |
| AX‐94506752 | 6D | 1.6 | 4.81 | WA plant height |
| AX‐95176310 | 6D | 1.6 | 4.81 | WA plant height |
| wsnp_CAP12_c720_382116 | 6D | 1.6 | 4.93 | TS plant height |
| wsnp_CAP12_c720_382116 | 6D | 1.6 | 7.07 | WA plant height |
| AX‐94398509 | 6D | 1.6 | 4.00 | Grain yield |
| AX‐94506752 | 6D | 1.6 | 4.00 | Grain yield |
| AX‐95176310 | 6D | 1.6 | 4.00 | Grain yield |
| RAC875_c57371_238 | 6D | 2.3 | 4.21 | TS plant height |
| RAC875_c57371_238 | 6D | 2.3 | 4.48 | WA plant height |
| RAC875_rep_c118305_446 | 6D | 2.3 | 4.21 | TS plant height |
| RAC875_rep_c118305_446 | 6D | 2.3 | 4.48 | WA plant height |
| BobWhite_c11808_975 | 6D | 4.0 | 7.01 | TS plant height |
| BobWhite_c11808_975 | 6D | 4.0 | 4.81 | TS main DW |
| BobWhite_c11808_975 | 6D | 4.0 | 4.61 | WA plant height |
| BobWhite_c11808_975 | 6D | 4.0 | 5.62 | GA plant height |
| BobWhite_c11808_975 | 6D | 4.0 | 4.84 | YA plant height |
| BobWhite_c11808_975 | 6D | 4.0 | 6.06 | TP plant height |
Genetic positions are shown in centiMorgans (cM). The physical positions are displayed in Data S1. GA, green anther; TP, tipping; TS, Terminal spikelet; WA, white anther; YA, yellow anther.
In previous study, we conducted the GWAS for the grain number per spikelet using the same wheat population (Guo et al., 2017). Here, we assessed the shared SNP markers between plant growth traits and grain number per spikelet (Data S1). A QTL located on 6A (120.6–121.4 cM/604887172–613957462) was shared by plant growth traits (WA leaf area, WA main shoot DW, AN main shoot DW, TS plant height, WA spike length and WA plant height) and grain number per spikelet. It suggests associations between plant growth at early developmental stages and grain setting in wheat.
Genomewide prediction of plant growth traits
As shown in Figure 4, the results of genomic prediction revealed a relatively high prediction ability (with an average of approximately 0.6) for plant height, leaf dry weight and main shoot dry weight at all seven developmental stages, suggesting the ability to estimate breeding values for these plant growth traits. The ability to predict spike length was variable among the different stages, with high prediction abilities for spike length at the WA, YA and TP stages. The highest spike length prediction ability was detected at the TS stage and then significantly decreased during the other stages. During the HD and AN stages, the highest prediction ability was observed for tiller DW, while the lowest prediction ability was detected for leaf area. These variable prediction abilities provide an opportunity to further select plant growth traits in wheat breeding.
Figure 4.
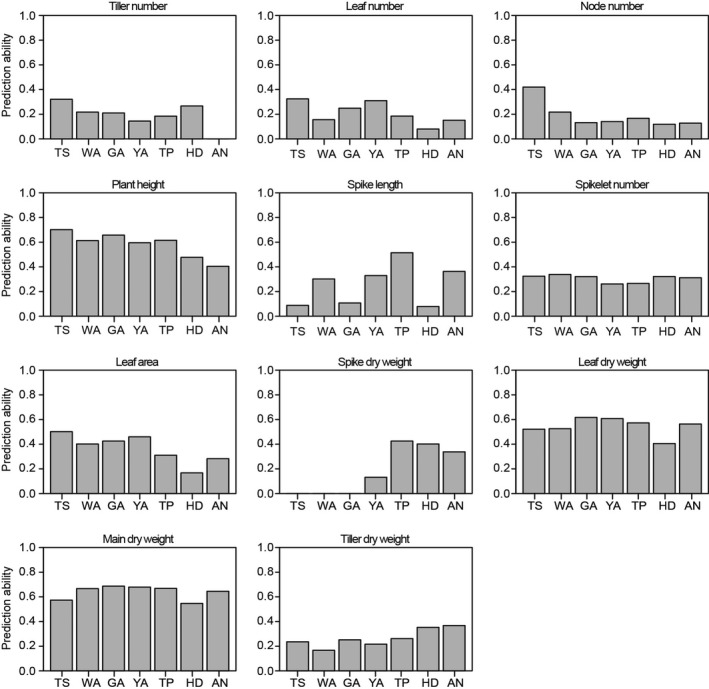
Prediction ability of plant growth traits revealed by genomic selection based on the SNP markers from 90k iSELECT chip.
The effects of the Ppd and Rht alleles on plant growth traits
Ppd and Rht are two historically important genes in wheat breeding. In this study, we assessed the effects of allelic variants of Ppd‐D1 (chromosome 2D) and Rht‐D1 (chromosome 4D) loci on plant growth traits. As an example of how these allelic variants of Ppd‐D1 and Rht‐D1 loci can influence plant growth traits, we assessed their influence on leaf area and spike length of the main shoot. As shown in Table 5, the day‐length‐sensitive allele of Ppd and the tall allele of Rht were significantly associated with higher leaf area. The effect of Rht alleles on leaf size may be attributed to their influence on stem and leaf growth.
Table 5.
Effects of Ppd and Rht alleles on leaf area
| Stage | Trait | Sensitivity | Insensitivity | T‐test‐P value | Dwarf allele | Tall allele | T‐test‐P value |
|---|---|---|---|---|---|---|---|
| AN | Leaf area | 115 ± 25 | 95 ± 16 | 5.04E‐05*** | 107 ± 23 | 123 ± 27 | 3.61E‐05*** |
| HD | Leaf area | 129 ± 26 | 106 ± 16 | 4.01E‐06*** | 122 ± 24 | 134 ± 30 | 2.25E‐03*** |
| TP | Leaf area | 134 ± 26 | 110 ± 21 | 2.52E‐06*** | 125 ± 24 | 143 ± 28 | 8.51E‐06*** |
| YA | Leaf area | 136 ± 28 | 104 ± 20 | 1.79E‐08*** | 125 ± 27 | 145 ± 31 | 2.31E‐05*** |
| GA | Leaf area | 138 ± 30 | 109 ± 20 | 1.20E‐06*** | 128 ± 28 | 149 ± 33 | 1.13E‐05*** |
| WA | Leaf area | 124 ± 28 | 93 ± 17 | 1.50E‐07*** | 114 ± 27 | 135 ± 28 | 3.04E‐06*** |
| TS | Leaf area | 83 ± 24 | 58 ± 10 | 7.99E‐08*** | 73 ± 20 | 94 ± 27 | 4.08E‐09*** |
AN, anthesis; GA, green anther; HD, heading; TP, tipping; TS, Terminal spikelet; WA, white anther; YA, yellow anther. *** suggests significance level of 0.001.
The Ppd sensitive allele was associated with significantly longer spikes at five of the seven stages (AN, HD, TP, YA and WA) (Table 6). This may be because the Ppd sensitive allele induced extension of the spike growth phase. The Rht tall allele is associated with an increase in spike length at the AN, HD, TP, GA and WA stages (Table 6). The effects of Ppd and Rht on all the plant growth traits were determined, and the information was displayed in Data S2. These data can be used in further investigations on the mechanism behind the effects of Ppd and Rht on grain yield and adaptation to variable environments.
Table 6.
Effects of Ppd and Rht alleles on spike length
| Stage | Trait | Sensitivity | Insensitivity | T‐test‐P value | Dwarf allele | Tall allele | T‐test‐P value |
|---|---|---|---|---|---|---|---|
| AN | Spike length | 11.99 ± 1.32 | 10.10 ± 1.21 | 8.85E‐12*** | 11.54 ± 1.47 | 12.06 ± 1.42 | 0.0229* |
| HD | Spike length | 12.09 ± 1.29 | 10.13 ± 1.12 | 5.21E‐13*** | 11.61 ± 1.39 | 12.15 ± 1.53 | 0.0186* |
| TP | Spike length | 11.94 ± 1.28 | 9.97 ± 1.05 | 1.73E‐13*** | 11.50 ± 1.39 | 12.00 ± 1.50 | 0.0278* |
| YA | Spike length | 10.49 ± 1.28 | 9.32 ± 1.18 | 6.01E‐06*** | 10.19 ± 1.26 | 10.60 ± 1.48 | 0.0513 |
| GA | Spike length | 5.52 ± 0.91 | 5.18 ± 0.78 | 0.0591 | 5.37 ± 0.81 | 5.74 ± 1.01 | 0.0074** |
| WA | Spike length | 2.08 ± 0.40 | 2.28 ± 0.41 | 0.0225* | 2.16 ± 0.40 | 2.00 ± 0.40 | 0.0128* |
| TS | Spike length | 0.37 ± 0.08 | 0.39 ± 0.08 | 0.1861 | 0.37 ± 0.08 | 0.39 ± 0.07 | 0.0771 |
AN, anthesis; GA, green anther; HD, heading; TP, tipping; TS, Terminal spikelet; WA, white anther; YA, yellow anther. *, **, and *** suggest significance levels of 0.05, 0.01, and 0.001, respectively.
Discussion
Plant growth during the SEP is important for the determination of grain number, and most of the grain number potential is lost during this phase. Therefore, this study was designed to uncover more detailed information on the seven SEP stages so that plant growth traits may be intricately manipulated to increase grain yield in wheat. Furthermore, highly plastic plant growth during the SEP provides the potential for the improvement of floret fertility in wheat. In this study, we identified SNP–trait associations that were shared among various SEP stages, and SNP–trait associations that were shared between plant growth traits and grain yield in field. The results of this study suggest that there may be several ways to increase floret fertility by manipulating specific stages during the SEP. However, it does not mean that the potential improvement of floret fertility will be easily achieved.
The general strategy for decreasing floral abortion and increasing floret fertility in wheat is to improve assimilate partitioning to the growing spikes during the SEP. Lengthening the duration of the SEP has been widely investigated as an alternative approach to increase the number of fertile florets at AN (Gonzalez et al., 2011; Miralles et al., 2000). Previous studies show that increasing the duration of the SEP under a short photoperiod results in an increase in the number of fertile florets due to an increase in assimilate partitioning to the spike at AN (Miralles et al., 2000; Slafer, 1996). In a previous study, we divided the SEP into seven floral development and abortion stages to investigate the time window of floral degradation in more detail (Guo and Schnurbusch, 2015). We found that delayed visible floral degradation may contribute to a higher numbers of fertile florets at AN (Guo and Schnurbusch, 2015). In the present study, we measured plant growth traits among the seven SEP stages to identify the most important stages for various important plant growth traits related to the determination of grain number. We found that the increase in spike length mainly occurs from the TS to GA stages, while the increase in spike dry weight mainly occurs from the YA to AN stages (Figures 2 and 3). These findings suggest that it may be beneficial to only increase the duration of the YA to AN stages, rather than the entire SEP, to improve spike dry weight at AN and also to validate our division of the SEP.
SNP–trait associations across developmental stages and traits have been previously reported in other species, which indicates that plant traits at the early developmental stages are related to grain yield traits at physiological maturity (Bac‐Molenaar et al., 2015; Fang et al., 2017; Sonah et al., 2015; Yang et al., 2014). Based on the division of the SEP and the assessment of genetic information (GWAS and genomic selection) in our study, we have uncovered the potential to genetically control plant growth by manipulating specific developmental stages to increase spike DW in wheat. In this way, genetically manipulating or mitigating the competition for assimilates between the main shoot and tillers, and between the spike and stem, may influence the determination of final grain yield traits.
During the SEP in wheat, manipulating the competition for assimilates between the tiller and main shoot growth affects assimilate partitioning to growing spikes. Excessive tiller growth (tiller number and tiller DW) in wheat leads to a reduction in grain yield because tillers compete for assimilates with the main shoot but are aborted before reaching physiological maturity and do not contribute to the final grain yield (Borras‐Gelonch et al., 2012; Evers et al., 2007; Thorne and Wood, 1987; Xie et al., 2016). In previous studies, tiller development in wheat was manipulated with a mutation (tin) that strongly reduced tillering (Kebrom and Richards, 2013; Kebrom et al., 2012; Kuraparthy et al., 2007). The tin mutants exhibit a greater rate of spikelet primordium initiation than the free‐tillering cultivars, resulting in a larger spike with more spikelets. Although the rate of leaf initiation is very similar, the later‐appearing leaves of the biculms are quite large compared with those from free‐tillering cultivars (Marshall and Boyd, 1985). The growth trait‐associated SNP markers detected by our GWAS (Data S1) may be used to control the main shoot:tiller DW ratio during early developmental stages to ultimately influence floret fertility. Furthermore, the tiller‐associated SNP markers (Data S1) may be used to manipulate tiller growth to further increase floret fertility.
Manipulating the Reduced height (Rht) genes has been the most successful method for adjusting the ratio between spike and stem DWs. The Green Revolution refers to the vast increases in grain yield achieved after the 1960s, which resulted from the introduction of dwarf mutations (Hedden, 2003; Peng et al., 1999; Saville et al., 2012). These dwarf alleles greatly enhanced assimilate partitioning towards the growing spikes in wheat (Abbate et al., 1998; Fischer, 2007; Foulkes et al., 2007). During this time, grain yield traits were substantially improved because more assimilates are allocated to the spikes (Fischer, 2011; Foulkes et al., 2011). We identified both shared and distinct SNP–trait associations for plant height, main shoot DW and the spike:stem DW ratio among the seven SEP developmental stages (Data S1). These data suggest that there may be benefits to genetically controlling these traits during specific stages in the SEP rather than during the entire SEP.
The concept of genomic prediction/selection was first proposed by Meuwissen et al. (2001). A prominent feature of genomic prediction/selection is the ability to estimate breeding values for quantitative traits based on whole genotypes through the simultaneous estimation of marker effects in a single step (Heslot et al., 2012). Previous studies have suggested that genomic prediction/selection has the potential to dramatically accelerate the breeding cycle, maintain genetic diversity within breeding programmes and increase genetic gains (Desta and Ortiz, 2014; Heslot et al., 2012; Zhao et al., 2012, 2015). In the present study, the results of genomic selection for plant growth according to the seven stages of the SEP revealed variable prediction abilities in wheat, suggesting that the predicted breeding values for plant growth traits vary among the developmental stages used in this study. For example, the ability to predict spike length was highly variable among the seven stages: it is low at the TS (0.08), GA (0.11) and HD (0.08) stages, but moderate (0.51) at the TP stage, indicating a relatively high ability to estimate breeding values for spike length at the TP stage specifically. Both high and low genomic prediction abilities of plant height were reported in previous work (Mirdita et al., 2015; Zhao et al., 2014). In this study, we found moderate genomic prediction abilities of plant height at different stages. He et al. (2016) reported that composition of training population, population structure and genomic prediction models can influence the results of prediction ability, which may be helpful for the explanation of variations for genomic prediction abilities of plant height. Tiller number declines from WA until AN stage, which will influence the resource allocation (Figure 2). Although the prediction ability of tiller number is low (Figure 4), we found that allelic variants of Rht‐D1 can significantly influence tiller number at the early developmental stages (e.g. TS, WA, GA) (Data S2). It implies that Rht‐D1 can be used to affect the variation of tiller number, which may further determine the assimilate distribution and grain yield‐related traits.
Taken together, the results of our genomic selection analysis may be used in the future to manipulate plant growth traits in wheat breeding. In summary, this study provides phenotypic and genotypic information related to wheat plant growth during the seven stages of the SEP. This information may aid in the implementation of wheat breeding strategies based on the manipulation of plant growth traits during the SEP.
Materials and methods
Plant materials and growth conditions
A total of 210 European hexaploid winter wheat cultivars were selected based on allelic variants of reduced height (Rht‐D1) (chromosome 4D) and photoperiod (Ppd‐D1) (chromosome 2D) loci. Seeds were sown into 96‐well trays and germinated in a glasshouse (photoperiod, 16‐h/8‐h, light/dark; temperature, 20 °C/16 °C, light/dark) for 14 days. At the two‐ to three‐leaf stage, seedlings were transferred to 4 °C to vernalize for 63 days. Vernalized seedlings were transferred to a hardening stage (photoperiod, 12‐h/12‐h, light/dark; temperature, 15 °C) for 7 days to gradually acclimatize. Finally, plants were transplanted into 0.5‐L pots (one plant per pot; 9 × 9 × 9 cm) in the glasshouse (photoperiod, 16‐h/8‐h, light/dark; temperature, 20 °C/6 °C, light/dark). Supplemental light (~250 μmol/m2/s photosynthetically active radiation) was supplied, and plants were irrigated when required.
Phenotyping
To determine the developmental stages, after 2 weeks of vernalization, plants were checked every 2 days until the starting stage (TS) was found. Then, we dissected the developing spikes/spikelets every 2 days from the terminal spikelet (TS, spikelet length around 0.5 mm) until anthesis (AN, spikelet length 1 cm) stage. When the appropriate stages were identified, three plants from each cultivar were randomly selected and their tiller number and tiller dry weight (DW) were measured. The leaf number, node number, plant height, spike length, spikelet number, leaf area, spike DW, leaf DW and main DW of plants in different developmental stages were measured from the main shoot. Tiller and leaf numbers indicate the numbers of ‘active’ green leaves and tillers.
The measurements of the maximum number of floret primordia, grain number, floret primordium loss, grain survival within individual spikelets and grain size traits (grain width, grain length, grain area and thousand kernel weight‐TKW) are described elsewhere (Guo et al., 2017). We also measured the grain yield in eight environments under field conditions in 2009 and 2010 (Schulthess et al., 2017).
Phenotypic data analyses
In this study, 76 traits, including 75 plant growth traits as well as grain yield in the field, were phenotyped.
After outlier tests (Anscombe and Tukey, 1963), we estimated the adjusted entry means for all of the 96 traits based on the following model:
where y im was the yield performance of nth observation for ith genotype, μ was the intercept, g i was the genetic effect of the ith genotype, and was the corresponding residual. Only the residual effect was treated as a random effect. The model was implemented using the software package ASRemL‐R 3.0 (Butler et al., 2009). After estimating the adjusted entry means, we estimated the Pearson correlation coefficients for plant growth traits across the seven different SEP stages during stem elongation and the overall mean of all evaluated genotypes to show growth of different parts within individual plants.
Genotyping
The 90k Infinium chip (90k iSELECT), a genotyping array including approximately 90 000 gene‐associated SNPs, was used to characterize genetic variation in allohexaploid and allotetraploid wheat populations. A total of 46 977 SNPs from the wheat 90k array were genetically mapped using a combination of eight mapping populations (Wang et al., 2014). Among these SNP markers, only 7934 mapped to the International Triticeae Initiative (ITMI) population based on recombinant inbred lines from a cross between the W7984 (synthetic wheat) and Opata M85 (Roder et al., 1998) parents, and the unmapped markers were not used in the association analysis. From a large collection of 819 571 previously characterized wheat SNP markers, 35 143 markers that are highly suited for the genotyping of elite hexaploid wheat accessions were identified with the 35k Affymetrix chip. Among these markers, only 7762 SNP markers were mapped in the ITMI population, and the unmapped markers were not used in the association analysis (Allen et al., 2017). We genotyped the photoperiodism gene Ppd‐D1 (chromosome 2D) and the dwarfing gene Rht‐D1 (chromosome 4D) in the wheat populations used in this study. Markers for Ppd‐D1 (chromosome 2D) and Rht‐D1 (chromosome 4D) were used for genotyping as described previously (Ellis et al., 2002; Turner, 2005). Detailed information about the genotyping process can be found in Zanke et al. (2014). We got the permit from International Wheat Genome Sequencing Consortium (IWGSC) to use the physical positions of the SNP markers. Here, we displayed genetic positions as well as physical positions of all the SNP markers that were uniquely aligned to IWGSC RefSeq V1.0 for all the associated SNP markers in this study. Due to the limitation of aligning quality, physical positions have not been shown for some markers.
Genomewide association mapping analyses
To investigate the influence of population admixture, we first used the block relaxation algorithm in the ADMIXTURE program (Alexander et al., 2009) to estimate individual ancestry proportions given an optimal number (k = 10) of hypothetical ancestral populations. Then, we performed a principal component analysis (PCA) on the wheat population of 210 cultivars. The first 10 principal components were used for subsequent analysis analyses according to the ADMIXTURE results. The PCA results did not reveal any apparent population structure among the varieties (Figure S1). Therefore, we carried out a genomewide association study (GWAS) with a mixed linear model using the software package GenStat 17th edition (VSN International Ltd, Hemel Hempstead, UK). The kinship matrix was applied for the relationship model:
Only SNPs with minor allele frequencies ≥0.05 were used to carry out the GWAS. GWA mapping was conducted based on 15 696 mapped SNP markers (7934 SNP markers from the 90k Infinium chip and 7762 from the 35k Affymetrix chip) from the wheat population. We used the false discovery rate (FDR) to assess the threshold for all the traits. The genomewide significance threshold of the GWAS was determined to be P < 0.0001 (i.e. −log10 (P) > 4) for all investigated traits, which is above or close to the threshold of most traits. A genomewide pairwise linkage disequilibrium (LD) analysis was performed for the SNP markers using GENSTAT 17. LD was estimated with squared allele frequency correlations (r 2) between loci pairs. To investigate the average LD decay in each chromosome, the pattern and distribution of intrachromosomal LD were visualized with LD plots generated by GENSTAT. To determine the critical r 2 with a genetic distance between loci, the smoothed second‐degree LOESS curve was fitted using GENSTAT.
Genomic prediction analysis
Based on the adjusted entry means, we applied a RR‐BLUP (ridge regression best linear unbiased prediction; VanRaden, 2008; Zhao et al., 2015) for genomewide prediction. The model is as follows:
where Y are the adjusted entry means, 1 n is a vector of ones, n is the number of genotypes, refers to the overall mean, and is the additive effect. In the model, is a fixed effect and the rest are random effects that have normal distributions, and , where is the relationship matrices, and and are the variance of additive effects and the residuals. Details on how to calculate these relationship matrices are described by Zhao et al. (2015). The prediction ability was defined as the Pearson correlation coefficient between predicted values and the adjusted means. We used a fivefold cross validation, by estimating the marker effects on 80% of the data, and using marker data for the remaining 20%, to predict the values. Then predictions were correlated with the observed data and standardized by dividing by the square root of heritability. All computations were conducted using the software R.
Author contributions
T.S. conceived the project. T.S. and Z.G. designed the experiments. Z.G. conducted phenotyping measurements in the glasshouse trials. G.L and Z.G. managed the main data analysis, including GWAS and genomic selection analysis, figure design and generation, and Y.Z. and J.C.R. supervised the data analysis in R. M.S.R. and M.W.G. developed and mapped the SNP markers. Z.G., T.S. and G.L. wrote the manuscript, and Y.Z., J.C.R, M.S.R. and M.W.G. were involved in reviewing the manuscript.
Supporting information
Figure S1 Genetic population structure determined by principal component analysis with SNP markers.
Figure S2 The ranges for all the developmental traits at the seven stages during the stem elongation phase.
Data S1 The associated SNP markers of plant growth traits and grain yield in field.
Data S2 The allelic variants of Rht‐D1 and Ppd‐D1 on plant growth traits.
Acknowledgements
We thank Martin Mascher for aligning the markers to IWGSC RefSeq v1.0, Karin Lipfert and Heike Ernst for their help with figure design and Mechthild Pürschel for excellent technical assistance. The authors wish to thank the International Wheat Genome Sequencing Consortium for prepublication access to IWGSC RefSeq v1.0. The authors received financial support from a scholarship from the Chinese Scholarship Council to ZG, the HEISENBERG Program of the German Research Foundation (DFG), Grant no. SCHN 768/8‐1, to TS, the EU‐FP7 KBBE‐2011‐5 ‘ADAPTAWHEAT’ project (number 289842, to TS and MWG) and the German Bundesministerium für Bildung und Forschung (0315947A to MWG and 0315947B, to MSR). The authors declare no conflict of interest.
Contributor Information
Zifeng Guo, Email: guo@ipk-gatersleben.de.
Thorsten Schnurbusch, Email: thor@ipk-gatersleben.de.
References
- Abbate, P.E. , Andrade, F.H. , Lazaro, L. , Bariffi, J.H. , Berardocco, H.G. , Inza, V.H. and Marturano, F. (1998) Grain yield increase in recent argentine wheat cultivars. Crop Sci. 38, 1203–1209. [Google Scholar]
- Alexander, D.H. , Novembre, J. and Lange, K. (2009) Fast model‐based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, A.M. , Winfield, M.O. , Burridge, A.J. , Downie, R.C. , Benbow, H.R. , Barker, G.L.A. , Wilkinson, P.A. et al. (2017) Characterization of a Wheat Breeders’ Array suitable for high‐throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum). Plant Biotechnol. J. 15, 390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anscombe, F.J. and Tukey, J.W. (1963) The examination and analysis of residuals. Technometrics, 5, 141–160. [Google Scholar]
- Bac‐Molenaar, J.A. , Vreugdenhil, D. , Granier, C. and Keurentjes, J.J.B. (2015) Genome‐wide association mapping of growth dynamics detects time‐specific and general quantitative trait loci. J. Exp. Bot. 66, 5567–5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bancal, P. (2009) Early development and enlargement of wheat floret primordia suggest a role of partitioning within spike to grain set. Field. Crop. Res. 110, 44–53. [Google Scholar]
- Beddington, J.R. , Asaduzzaman, M. , Clark, M.E. , Bremauntz, A.F. , Guillou, M.D. , Howlett, D.J.B. , Jahn, M.M. et al. (2012) What next for agriculture after Durban? Science, 335, 289–290. [DOI] [PubMed] [Google Scholar]
- Borras‐Gelonch, G. , Rebetzke, G.J. , Richards, R.A. and Romagosa, I. (2012) Genetic control of duration of pre‐anthesis phases in wheat (Triticum aestivum L.) and relationships to leaf appearance, tillering, and dry matter accumulation. J. Exp. Bot. 63, 69–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman, A. and Graur, D. (1995) Wheat evolution. Isr. J. Plant Sci. 43, 85–98. [Google Scholar]
- Butler, D. , Cullis, B.R. , Gilmour, A. and Gogel, B. (2009) ASReml‐R reference manual. The State of Queensland, Department of Primary Industries and Fisheries, Brisbane.
- Challinor, A.J. , Watson, J. , Lobell, D.B. , Howden, S.M. , Smith, D.R. and Chhetri, N. (2014) A meta‐analysis of crop yield under climate change and adaptation. Nat. Clim. Chang. 4, 287–291. [Google Scholar]
- Desta, Z.A. and Ortiz, R. (2014) Genomic selection: genome‐wide prediction in plant improvement. Trends Plant Sci. 19, 592–601. [DOI] [PubMed] [Google Scholar]
- Dreccer, M.F. , Wockner, K.B. , Palta, J.A. , McIntyre, C.L. , Borgognone, M.G. , Bourgault, M. , Reynolds, M. et al. (2014) More fertile florets and grains per spike can be achieved at higher temperature in wheat lines with high spike biomass and sugar content at booting. Funct. Plant Biol. 41, 482–495. [DOI] [PubMed] [Google Scholar]
- Ellis, M.H. , Spielmeyer, W. , Gale, K.R. , Rebetzke, G.J. and Richards, R.A. (2002) “Perfect” markers for the Rht‐B1b and Rht‐D1b dwarfing genes in wheat. Theor. Appl. Genet. 105, 1038–1042. [DOI] [PubMed] [Google Scholar]
- Evers, J.B. , Vos, J. , Chelle, M. , Andrieu, B. , Fournier, C. and Struik, P.C. (2007) Simulating the effects of localized red: far‐red ratio on tillering in spring wheat (Triticum aestivum) using a three‐dimensional virtual plant model. New Phytol. 176, 325–336. [DOI] [PubMed] [Google Scholar]
- Fang, C. , Ma, Y. , Wu, S. , Liu, Z. , Wang, Z. , Yang, R. , Hu, G. et al. (2017) Genome‐wide association studies dissect the genetic networks underlying agronomical traits in soybean. Genome Biol. 18, 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante, A. , Savin, R. and Slafer, G.A. (2013) Floret development and grain setting differences between modern durum wheats under contrasting nitrogen availability. J. Exp. Bot. 64, 169–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, R.A. (2007) Understanding the physiological basis of yield potential in wheat. J. Agric. Sci. 145, 99–113. [Google Scholar]
- Fischer, R.A. (2011) Wheat physiology: a review of recent developments. Crop Pasture Sci. 62, 95–114. [Google Scholar]
- Foulkes, M.J. , Sylvester‐Bradley, R. , Weightman, R. and Snape, J.W. (2007) Identifying physiological traits associated with improved drought resistance in winter wheat. Field. Crop. Res. 103, 11–24. [Google Scholar]
- Foulkes, M.J. , Slafer, G.A. , Davies, W.J. , Berry, P.M. , Sylvester‐Bradley, R. , Martre, P. , Calderini, D.F. et al. (2011) Raising yield potential of wheat. III. Optimizing partitioning to grain while maintaining lodging resistance. J. Exp. Bot. 62, 469–486. [DOI] [PubMed] [Google Scholar]
- Friend, D.J.C. (1965) Tillering and leaf production in wheat as affected by temperature and light intensity. Can. J. Bot. 43, 1063–1076. [Google Scholar]
- Godfray, H.C.J. , Beddington, J.R. , Crute, I.R. , Haddad, L. , Lawrence, D. , Muir, J.F. , Pretty, J. et al. (2010) Food security: the challenge of feeding 9 billion people. Science, 327, 812–818. [DOI] [PubMed] [Google Scholar]
- Gonzalez, F.G. , Miralles, D.J. and Slafer, G.A. (2011) Wheat floret survival as related to pre‐anthesis spike growth. J. Exp. Bot. 62, 4889–4901. [DOI] [PubMed] [Google Scholar]
- Grassini, P. , Eskridge, K.M. and Cassman, K.G. (2013) Distinguishing between yield advances and yield plateaus in historical crop production trends. Nat. Commun. 4, 2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z.F. and Schnurbusch, T. (2015) Variation of floret fertility in hexaploid wheat revealed by tiller removal. J. Exp. Bot. 66, 5945–5958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z.F. , Slafer, G.A. and Schnurbusch, T. (2016) Genotypic variation in spike fertility traits and ovary size as determinants of floret and grain survival rate in wheat. J. Exp. Bot. 67, 4221–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, Z.F. , Chen, D.J. , Alqudah, A.M. , Roder, M.S. , Ganal, M.W. and Schnurbusch, T. (2017) Genome‐wide association analyses of 54 traits identified multiple loci for the determination of floret fertility in wheat. New Phytol. 214, 257–270. [DOI] [PubMed] [Google Scholar]
- He, S. , Schulthess, A.W. , Mirdita, V. , Zhao, Y. , Korzun, V. , Bothe, R. , Ebmeyer, E. et al. (2016) Genomic selection in a commercial winter wheat population. Theor. Appl. Genet. 129, 641–651. [DOI] [PubMed] [Google Scholar]
- Hedden, P. (2003) The genes of the green revolution. Trends Genet. 19, 5–9. [DOI] [PubMed] [Google Scholar]
- Heslot, N. , Yang, H.P. , Sorrells, M.E. and Jannink, J.L. (2012) Genomic selection in plant breeding: a comparison of models. Crop Sci. 52, 146–160. [Google Scholar]
- Huibert, J.B. and Neuteboom, J.H. . (1998) Morphological analysis of leaf and tiller number dynamics of wheat (Triticum aestivum L.): Responses to temperature and light intensity. Ann. Bot. 81, 131–139. [Google Scholar]
- Kebrom, T.H. and Richards, R.A. (2013) Physiological perspectives of reduced tillering and stunting in the tiller inhibition (tin) mutant of wheat. Funct. Plant Biol. 40, 977–985. [DOI] [PubMed] [Google Scholar]
- Kebrom, T.H. , Chandler, P.M. , Swain, S.M. , King, R.W. , Richards, R.A. and Spielmeyer, W. (2012) Inhibition of tiller bud outgrowth in the tin mutant of wheat is associated with precocious internode development. Plant Physiol. 160, 308–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby, E.J.M. and Appleyard, M. (1987) Cereal Development Guide, 2nd ed. UK, Stoneleigh: Arable Unit. [Google Scholar]
- Kuraparthy, V. , Sood, S. , Dhaliwal, H.S. , Chhuneja, P. and Gill, B.S. (2007) Identification and mapping of a tiller inhibition gene (tin3) in wheat. Theor. Appl. Genet. 114, 285–294. [DOI] [PubMed] [Google Scholar]
- Lobell, D.B. , Schlenker, W. and Costa‐Roberts, J. (2011) Climate trends and global crop production since 1980. Science, 333, 616–620. [DOI] [PubMed] [Google Scholar]
- Marshall, C. and Boyd, W.J.R. (1985) A comparison of the growth and development of biculm wheat lines with freely tillering cultivars. J. Agric. Sci. 104, 163–171. [Google Scholar]
- Meuwissen, T.H.E. , Hayes, B.J. and Goddard, M.E. (2001) Prediction of total genetic value using genome‐wide dense marker maps. Genetics, 157, 1819–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miralles, D.J. , Richards, R.A. and Slafer, G.A. (2000) Duration of the stem elongation period influences the number of fertile florets in wheat and barley. Aust. J. Plant Physiol. 27, 931–940. [Google Scholar]
- Mirdita, V. , He, S. , Zhao, Y. , Korzun, V. , Bothe, R. , Ebmeyer, E. , Reif, J.C. et al. (2015) Potential and limits of whole genome prediction of resistance to Fusarium head blight and Septoria tritici blotch in a vast Central European elite winter wheat population. Theor. Appl. Genet. 128, 2471–2481. [DOI] [PubMed] [Google Scholar]
- Peng, J.R. , Richards, D.E. , Hartley, N.M. , Murphy, G.P. , Devos, K.M. , Flintham, J.E. , Beales, J. et al. (1999) ‘Green revolution’ genes encode mutant gibberellin response modulators. Nature, 400, 256–261. [DOI] [PubMed] [Google Scholar]
- Roder, M.S. , Korzun, V. , Wendehake, K. , Plaschke, J. , Tixier, M.H. , Leroy, P. and Ganal, M.W. (1998) A microsatellite map of wheat. Genetics, 149, 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saville, R.J. , Gosman, N. , Burt, C.J. , Makepeace, J. , Steed, A. , Corbitt, M. , Chandler, E. et al. (2012) The ‘Green Revolution’ dwarfing genes play a role in disease resistance in Triticum aestivum and Hordeum vulgare. J. Exp. Bot. 63, 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhuber, J. and Tubiello, F.N. (2007) Global food security under climate change. Proc. Natl Acad. Sci. USA 104, 19703–19708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulthess, A.W. , Reif, J.C. , Ling, J. , Plieske, J. , Kollers, S. , Ebmeyer, E. , Korzun, V. et al. (2017) The roles of pleiotropy and close linkage as revealed by association mapping of yield and correlated traits of wheat (Triticum aestivum L.). J. Exp. Bot. 68, 4089–4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slafer, G.A. (1996) Differences in phasic development rate amongst wheat cultivars independent of responses to photoperiod and vernalization. A viewpoint of the intrinsic earliness hypothesis. J. Agric. Sci. 126, 403–419. [Google Scholar]
- Sonah, H. , O'Donoughue, L. , Cober, E. , Rajcan, I. and Belzile, F. (2015) Identification of loci governing eight agronomic traits using a GBS‐GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol. J. 13, 211–221. [DOI] [PubMed] [Google Scholar]
- Thorne, G.N. and Wood, D.W. (1987) Effects of radiation and temperature on tiller survival, grain number and grain‐yield in winter‐wheat. Ann. Bot. 59, 413–426. [Google Scholar]
- Tilman, D. , Balzer, C. , Hill, J. and Befort, B.L. (2011) Global food demand and the sustainable intensification of agriculture. Proc. Natl Acad. Sci. USA 108, 20260–20264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, A. (2005) The pseudo‐response regulator Ppd‐H1 provides adaptation to photoperiod in barley. Science, 310, 1031–1034. [DOI] [PubMed] [Google Scholar]
- VanRaden, P.M. (2008) Efficient methods to compute genomic predictions. J. Dairy Sci. 91, 4414–4423. [DOI] [PubMed] [Google Scholar]
- Waddington, S.R. , Cartwright, P.M. and Wall, P.C. (1983) A quantitative scale of spike initial and pistil development in barley and wheat. Ann. Bot. 51, 119–130. [Google Scholar]
- Wang, S.C. , Wong, D.B. , Forrest, K. , Allen, A. , Chao, S.M. , Huang, B.E. , Maccaferri, M. et al. (2014) Characterization of polyploid wheat genomic diversity using a high‐density 90 000 single nucleotide polymorphism array. Plant Biotechnol. J. 12, 787–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitechurch, E.M. , Slafer, G.A. and Miralles, D.J. (2007) Variability in the duration of stem elongation in wheat genotypes and sensitivity to photoperiod and vernalization. J. Agron. Crop Sci. 193, 131–137. [Google Scholar]
- Xie, Q. , Mayes, S. and Sparkes, D.L. (2016) Optimizing tiller production and survival for grain yield improvement in a bread wheat x spelt mapping population. Ann. Bot. 117, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W. , Guo, Z. , Huang, C. , Duan, L. , Chen, G. , Jiang, N. , Fang, W. et al. (2014) Combining high‐throughput phenotyping and genome‐wide association studies to reveal natural genetic variation in rice. Nat. Commun., 5, 5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zadoks, J.C. , Chang, T.T. and Konzak, C.F. (1974) A decimal code for the growth stages of cereals. Weed Research 14, 415–421. [Google Scholar]
- Zanke, C. , Ling, J. , Plieske, J. , Kollers, S. , Ebmeyer, E. , Korzun, V. , Argillier, O. , et al. (2014) Genetic architecture of main effect QTL for heading date in European winter wheat Frontiers in plant science 5, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.S. , Gowda, M. , Liu, W.X. , Wurschum, T. , Maurer, H.P. , Longin, F.H. , Ranc, N. et al. (2012) Accuracy of genomic selection in European maize elite breeding populations. Theor. Appl. Genet. 124, 769–776. [DOI] [PubMed] [Google Scholar]
- Zhao, Y. , Mette, M.F. , Gowda, M. , Longin, C.F.H. and Reif, J.C. (2014) Bridging the gap between marker‐assisted and genomic selection of heading time and plant height in hybrid wheat. Heredity (Edinb), 112, 638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y.S. , Li, Z. , Liu, G.Z. , Jiang, Y. , Maurer, H.P. , Wurschum, T. , Mock, H.P. et al. (2015) Genome‐based establishment of a high‐yielding heterotic pattern for hybrid wheat breeding. Proc. Natl Acad. Sci. USA 112, 15624–15629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Genetic population structure determined by principal component analysis with SNP markers.
Figure S2 The ranges for all the developmental traits at the seven stages during the stem elongation phase.
Data S1 The associated SNP markers of plant growth traits and grain yield in field.
Data S2 The allelic variants of Rht‐D1 and Ppd‐D1 on plant growth traits.


