Summary
The rearrangement and expression of immunoglobulin genes are regulated by enhancers and their binding transcriptional factors that activate or suppress the activities of the enhancers. The immunoglobulin κ (Igκ) gene locus has three important enhancers: the intrinsic enhancer (Ei), 3′ enhancer (E3′), and distal enhancer (Ed). Ei and E3′ are both required for Igκ gene rearrangement during early stages of B‐cell development, whereas optimal expression of the rearranged Igκ gene relies on both E3′ and Ed. The transcription factor YY1 affects the expression of many genes involved in B‐cell development, probably by mediating interactions between their enhancers and promoters. Herein, we found that YY1 binds to the E3ʹ enhancer and suppresses Igκ expression in B lymphoma cells by epigenetically modifying the enhancer. Knocking down YY1 enhanced Igκ expression, which was associated with increased levels of E2A (encoded by the TCF3 gene) and its binding to the E3ʹ enhancer. Moreover, in germinal centre B cells and plasma cells, YY1 expression was reversely associated with Igκ levels, implying that YY1 might facilitate antibody affinity maturation in germinal centre B cells through the transient attenuation of Igκ expression.
Keywords: E2A, E3′ enhancer, epigenetic modification, Igκ gene, YY1
Abbreviations
- ChIP
chromatin immunoprecipitation assays
- E3′
3′ enhancer
- Ed
distal enhancer
- Ei
intrinsic enhancer
- FACS
fluorescence‐activated cell sorting
- GC
germinal centre
- IgH
immunoglobulin heavy chain
- IgL
immunoglobulin light chain
- PBS
phosphate‐buffered saline
- PCR
polymerase chain reaction
- RT
reverse transcription
- SHM
somatic hypermutation
- siRNA
small interfering RNA
Introduction
The expression of immunoglobulin genes, including the immunoglobulin heavy chain gene (IgH) and the immunoglobulin light chain gene (IgL), is critical for successful B‐cell development. During early B‐cell development, IgH gene rearrangement takes place at the pro‐B cell stage before IgL rearrangement, which generally occurs in the pre‐B compartment.1 In the two IgL genes, the immunoglobulin κ (Igκ) locus rearranges before the immunoglobulin λ (Igλ) locus, and most B cells (~ 95%) express Igκ as the light chain; only ~ 5% of B cells express Igλ as an attempt to rescue B cells that would otherwise undergo apoptosis due to an unproductive Igκ rearrangement. Upon completion of the IgL rearrangement, two identical heavy chains and two identical light chains form the B‐cell antigen receptor, and pre‐B cells develop into immature B cells, which then exit the bone marrow to become mature peripheral B cells.2
The rearrangement and expression of both the IgH and IgL genes are strictly controlled and coordinated through their unique gene structures and a sophisticated transcriptional factors network.3 Using models, the mechanisms by which IgH and Igκ are regulated have been extensively investigated. Specifically, three enhancers have been identified in the Igκ gene, the intronic enhancer (Ei),4 3′ enhancer (E3′)5 and distal enhancer (Ed).6 Ei and E3′ are both required for Igκ gene rearrangement during the early stages of B‐cell development,7 whereas E3ʹ and Ed each play quantitative roles in the rearranged gene expression.8 Although we have greatly enhanced our understanding of the roles of Igκ enhancers in gene regulation using individual or double‐enhancer knockout mouse models, the key regulators and mechanisms that orchestrate the activities of these enhancers, especially in human B cells, are not fully understood.
YY1 is a multifunctional transcription factor that exhibits positive and negative control on a large number of genes through its ability to initiate, activate, or repress transcription depending upon the context in which it binds.9, 10 The ablation of YY1 in the B lineage leads to a blocked transition from pro‐B to pre‐B cells, partially by impairing chromatin contraction at the IgH locus and gene rerrangement.11 In germinal centre (GC) B cells, YY1 DNA binding sites are enriched within the promoters of a group of genes that were significantly up‐regulated or down‐regulated in GC B cells compared with other B‐cell compartments.12 The deletion of YY1 in GC B cells results in increased apoptosis in GC B cells, leading to an impaired GC reaction.13, 14, 15 Using mouse models in which YY1 was deleted at various B‐cell development stages, Kleiman et al.16 demonstrated that the YY1 knockout not only completely prevented the differentiation of GC B cells but also inhibited other stages of B‐cell development, with transitional 1 (T1) cells being the most YY1‐dependent subset. These data suggested that YY1 might regulate B‐cell function at all developmental stages.
Pan et al.17 investigated the function of YY1 in Igκ gene rearrangement and found that the YY1 REPO domain was not required for IgH rearrangement but was crucial for the normal Igκ repertoire, suggesting a direct role of YY1 in Igκ locus structure and rearrangement. In line with that, a recent study revealed that YY1 contributes to enhancer–promoter structural interactions in a manner that is analogous to the DNA interactions mediated by the transcriptional repressor CTCF.18 In mouse pre‐B cells, YY1 binds to E3ʹ and negatively regulates the enhancer's activity in Igκ rearrangement.19 However, whether YY1 has any impact on Igκ expression has not been investigated. Here, we found that YY1 binds to the human E3′ enhancer and inhibits Igκ expression by inducing the suppressive epigenetic modifications of the enhancer. In contrast, knocking down YY1 enhanced Igκ expression, which was associated with increased levels of E2A expression and its recruitment to E3′. These results shed light on a novel mechanism by which YY1 regulates Igκ expression and B‐cell development.
Materials and methods
Cell culture
The HEK‐293T cell line was purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China) and cultured in Dulbecco's modified Eagle's medium (Gibco, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS). The human diffuse large B‐cell lymphoma cell line HBL‐1 was kindly provided by Dr Xiaodong Yang from the Shanghai Institute of Immunology, and the cells were cultured in RPMI‐1640 (Gibco) supplemented with 10% FBS. The Daudi cells (B lymphoblast, CCL‐213) were purchased from the American Type Culture Collection (Manassas, VA) and cultured in RPMI‐1640 supplemented with 10% FBS. All cell lines were cultivated at 37° in 5% CO2 and humidity around 95%.
RT‐PCR and real‐time PCR
Total RNA was prepared using the TRIzol reagent (Invitrogen, Carlsbad, CA), and cDNA was synthesized using the SuperScript VILO cDNA Synthesis Kit (Invitrogen). Polymerase chain reaction (PCR) was performed using Ex Taq™ HS (TaKaRa Bio, Otsu, Japan). Real‐time quantitative PCR was performed by using a LightCycler Systems (Roche Molecular Systems, Indianapolis, IN) and SYBR green dye. For data analysis, the method was used to calculate the fold changes. β‐Actin expression was set to be unaffected under our treatment conditions and used as a reference gene. Each experiment was run in triplicate, and the error bars represent the range of fold changes calculated from the three or four independent experiments. The sequences for primers used for the reverse transcription (RT‐) PCR or real‐time PCR are listed in the Supplementary material (Table S1).
Western blotting
Western blotting was performed using whole‐cell lysates. Aliquots of total protein (20–50 μg per lane) were electrophoresed on 10% sodium dodecyl sulphate–polyacrylamide gradient gels and transferred to nitrocellulose membranes (Millipore, Bedford, MA). The membranes were incubated at 4° overnight with anti‐GAPDH, ‐YY1, ‐E2A or ‐Igκ monoclonal antibodies (all purchased from Abcam, Cambridge, MA). After rinsing in buffer wash, the membranes were incubated with a horseradish peroxidase‐conjugated secondary antibody (Santa Cruz Biotechnology, Dallas, TX) diluted 1 : 10 000 to 1 : 30 000 followed by development with enhanced chemiluminescence reagents (Amersham, Little Chalfont, UK).
Transfection and plasmids
YY1‐small interfering RNA (siRNA), E2A‐siRNA and control non‐specific siRNA were designed by and purchased from Shanghai GenePharma Co., Ltd (Shanghai, China). The human YY1‐expressing vector was purchased from Shanghai Genechem Co., Ltd (Shanghai, China). For transfection, 2 × 106 cells were resuspended in 100 μl of buffer (Engreen, Beijing, China), and electro‐transfection was performed with 2·5 μg of plasmid using a Celetrix Electroporator (Celetrix, LLC, Manassas, VA). After transfection, the cells were cultured in six‐well plates and harvested after 48–72 hr for analysis.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) analysis was performed following a protocol provided by Upstate Biotechnology (Lake Placid, NY). Briefly, HBL‐1 cells were cross‐linked by adding 1·0% formaldehyde buffer and then lysed with 1% sodium dodecyl sulphate lysis buffer. The cell lysates were sonicated to shear the DNA to lengths between 200 and 1000 bp. The resulting chromatin solution was incubated overnight at 4° with ChIP‐grade antibodies specific for YY1, E2A or a rabbit immunoglobulin control (Abcam). The chromatin–antibody complex was incubated with Protein‐A agarose for 2 hr at 4° and then washed extensively. The input or immunoprecipitated chromatin–antibody complex was incubated at 65° overnight to reverse the cross‐linking. After proteinase K digestion for 1 hr, the DNA was collected using Qiagen spin columns (Qiagen, Hilden, Germany) and then analysed by PCR for 30 cycles or real‐time PCR using human E3′‐specific primers (see Supplementary material, Table S1).
Fluorescence‐activated cell sorting with flow cytometry
C57BL/6 mice were purchased from the Laboratory for Animal Science of Shanghai Medical College, Fudan University (Shanghai, China). All experiments were performed according to the Institutional Guidelines for Animal Care and Use and were approved by the Animal Experimentation Ethics Committee of Nantong University. Peyer's patch lymph nodes were collected from the wild‐type C57BL/6 mice, and the single‐cell suspension was prepared in the following staining buffer: phosphate buffered saline (PBS) supplemented with 2% FBS. Fluorescein isothiocyanate‐labelled B220, phycoerythrin‐labelled anti‐Igκ, and Alexa Fluor 647‐labelled anti‐GL7 antibodies were added to 106 cells resuspended in 100 μl of staining buffer and then incubated at 4° for 30 min. After staining, the cells were washed with staining buffer three times and analysed with a BD FACSCalibur instrument (Becton Dickinson, Franklin Lakes, NJ). Germinal centre B cells (B220+ GL7+) and non‐GC B cells (B220+ GL7−) from the Peyer's patch lymph nodes were sorted on a MoFlo machine (Dako Cytomation, Carpinteria, CA). To isolate splenic GC B cells and plasma cells, 10‐ to 16‐week‐old C57BL/6 mice were immunized intraperitoneally with 2 × 108 sheep red blood cells (Sigma‐Aldrich, St Louis, MO) emulsified in CFA (Complete Freund's Adjuvant) (Sigma‐Aldrich). The spleens were harvested 5 days post immunization, and the GC B cells (B220+ GL7+) and plasma cells (B220low and negative CD138high) were sorted on the MoFlo machine. All antibodies were purchased from BD Bioscience (San Jose, CA).
Statistical analysis
All statistical analyses were carried out using graphpad prism for Windows (GraphPad, San Diego, CA). The quantitative variables were analysed by Student's t‐test, Fisher's exact test or the chi‐squared test. All statistical analyses were two‐sided, and P < 0·05 was considered statistically significant.
Results
YY1 suppresses Igκ expression in human B lymphoma cells
To examine the impact of YY1 on Igκ expression, exogenous YY1 was introduced into B lymphoma HBL‐1 cells, which express moderate levels of YY1 and Igκ. Three days post transfection, the levels of YY1 and Igκ were determined using real‐time PCR and Western blot. Igκ expression was significantly reduced in the YY1‐overexpressing HBL‐1 cells (YY1OE) compared with that in the control cells transfected with empty vectors (Fig. 1a,b). The reduced Igκ mRNA levels in the YY1OE cells suggested that YY1 may suppress Igκ expression at the transcriptional level. We next knocked down YY1 using siRNA and then measured Igκ expression in the HBL‐1 cells. As shown in Fig. 1(c,d), knocking down YY1 significantly increased Igκ expression compared with that in the control cells. Similar results were obtained with human B lymphoblast Daudi cells (see Supplementary material, Fig. S1a,b). These data suggested that YY1 functions as a negative regulator of Igκ expression in HBL‐1 cells.
Figure 1.
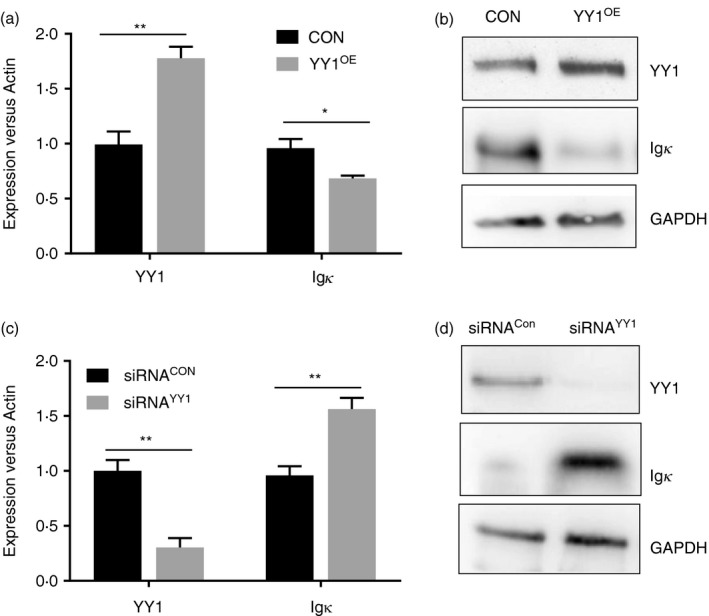
YY1 suppresses Igκ expression in HBL‐1 cells. (a, b) Compared with that in the control HBL‐1 cells (CON), the cells over‐expressing YY1 (YY1OE) expressed lower levels of Igκ, as indicated by real‐time PCR or Western blot. (c, d) Knocking down YY1 using small interfering RNA (siRNA) in HBL‐1 cells increased Igκ expression, as indicated by real‐time PCR and Western blot (*P < 0·05, **P < 0·01 compared with the control group).
Binding of YY1 and E2A to human E3′
Previously, a YY1 binding site was identified downstream of the mouse E3ʹ core and was thought to negatively regulate the enhancer's activity.19 In agreement with this, the YY1 binding site in the mouse E3ʹ was identified using the JASPAR program (http://jaspar.genereg.net/), which makes predictions based on published collections of experimentally defined transcription factor binding sites.20 For human E3ʹ, the JASPAR program identified six potential YY1 sites (see Supplementary material, Fig. S2). E2A is a known positive regulator of Igκ rearrangement21, 22 and is an essential regulator of progenitor B‐cell, GC B‐cell and plasma cell development.23 A previous study showed that E2A binds to multiple regulatory elements across the human Igκ gene, including E3ʹ.22, 24 The JASPAR program identified four potential E2A binding sites in the human E3ʹ (see Supplementary material, Fig. S2). Interestingly, one predicted E2A binding site overlapped with a predicted YY1 binding site (see Supplementary material, Fig. S2), implying that the two factors might compete with each other for binding to that sequence. To examine whether YY1 and E2A are recruited to human E3′, we performed ChIP assays with HBL‐1 cells. As shown in Fig. 2(a), E3′ was clearly enriched in the genomic DNA precipitated by the YY1 or E2A antibody but not in that precipitated by the control IgG, indicating that YY1 and E2A both bind to E3ʹ in vivo.
Figure 2.
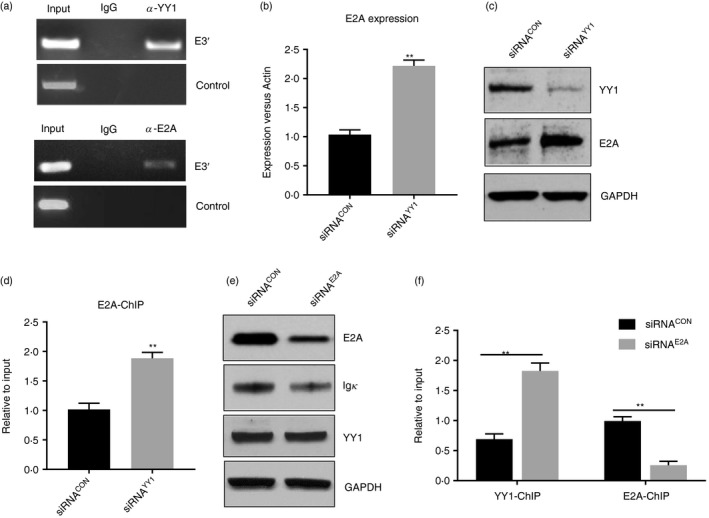
YY1 and E2A bind to E3′ and regulate the enhancer's activity. (a) To perform the ChIP assay, genomic DNA (input) was precipitated with antibodies specific for YY1, E2A or control IgG, and the E3′ sequence was detected using a PCR assay. Both the YY1 and E2A antibodies enriched E3′ from the genomic DNA. In contrast, we did not detect binding of the two factors in a control region 4·5 kb downstream of human E3′ (Chr2: 88840810–88841276), which is not conserved between humans and mice. The primers used in the ChIP assay for E3′ and the negative control are listed in the Supplementary material (Table S1). (b, c) Knocking down YY1 increased the levels of E2A in the HBL‐1 cells, as indicated by the real‐time PCR assay and Western blot analysis. (d) The ChIP assay with the HBL‐1 cells indicated that knocking down YY1 enhanced the recruitment of E2A to E3ʹ. (e) Knocking down E2A using a small interfering RNA (siRNA) in HBL‐1 cells suppressed Igκ expression compared with that in the control cells. (f) The ChIP assay showed that YY1 was recruited more in the E2A siRNA‐treated cells than in the control cells. The results also demonstrated that knocking down E2A significantly reduced its binding to E3ʹ (*P < 0·05, **P < 0·01 compared with the control group).
YY1 suppresses E2A expression and its recruitment to E3′
To examine whether a relationship exists between YY1 and E2A in regulating E3ʹ activity and Igκ, we measured the levels of E2A in YY1 siRNA‐treated cells (siRNAYY1) compared with those in control siRNA‐treated cells (siRNACON). As shown in Fig. 2(b,c), knocking down YY1 enhanced the expression of E2A, as indicated by real‐time PCR and Western blot. We then performed a ChIP assay and found that the binding of E2A to E3ʹ was increased in the YY1 siRNA‐treated cells compared with that in the control cells (Fig. 2d). To test whether E2A is required for Igκ expression, we treated HBL‐1 cells with siRNA to knock down E2A. Igκ expression was reduced in the E2A siRNA‐treated cells compared with that in the control cells (Fig. 2e). Although knocking down E2A did not change the levels of YY1 (Fig. 2e), it increased the recruitment of YY1 to E3ʹ (Fig. 2f), suggesting that competition might exist between these two transcriptional factors for binding to E3′. Taken together, these results suggested that YY1 might regulate E3′ activity by suppressing E2A expression and interfering with its recruitment to E3′.
YY1 and E2A epigenetically modify E3′
Activated or silenced enhancers can be epigenetically marked with H3K27ac or H3K27me3.25 In addition, previous studies suggest that YY1 interacts with the histone acetyltransferase p300 or H3K27me3 methyltransferase Ezh2, depending on the cellular context.10 To evaluate the epigenetic impact of YY1 binding, ChIP assays were performed using HBL‐1 cells to detect H3K27ac and H3K27me3 in E3′. As shown in Fig. 3(a), the levels of H3K27ac increased, while H3K27me3 decreased, in the YY1 siRNA‐treated cells compared with levels in the control cells. Interestingly, knocking down E2A reduced H3K27ac in E3ʹ, but the levels of H3K27me3 were not significantly changed compared with those in the control cells (Fig. 3b). These results suggested that YY1 and E2A might regulate Igκ expression through epigenetically modifying E3′.
Figure 3.
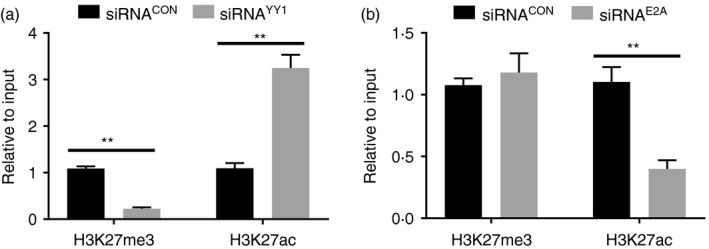
YY1 and E2A regulate E3′ activity via epigenetic modifications. (a) The ChIP assay was performed with HBL‐1 cells to detect H3K27me3 and H3K27ac, and the results indicated that knocking down YY1 suppressed H3K27me3 but enhanced H3K27ac in E3′. (b) The ChIP assay with HBL‐1 cells indicated that knocking down E2A suppressed H3K27ac levels but not H3K27me3 levels in E3ʹ (*P < 0·05, **P < 0·01 compared with the control group).
Igκ expression is down‐regulated in GC B cells
YY1 is reported to be highly expressed in GC B cells.26 To examine the relationship between YY1 and Igκ in GC B cells or non‐GC B cells, mouse Peyer′s patch lymph nodes were isolated from the small intestine according to a previously established procedure.27 The cell suspension was harvested, and the surface expression of the B‐cell marker B220, the GC B‐cell marker GL7, and Igκ were determined by fluorescence‐activated cell sorting (FACS). As shown in Fig. 4(a,b), the GC B cells (B220+ GL7+) expressed low levels of surface Igκ compared with those in the non‐GC B cells (B220+ GL7−). Total RNA was isolated from the FACS‐sorted GC B cells and non‐GC B cells, and the levels of Igκ and YY1 were measured by real‐time PCR. Igκ was markedly down‐regulated, whereas YY1 expression was increased in the GC B cells compared with in the non‐GC B cells (Fig. 4c,d). We found no difference in E2A expression between the GC B cells and non‐GC B cells (Fig. 4d).
Figure 4.
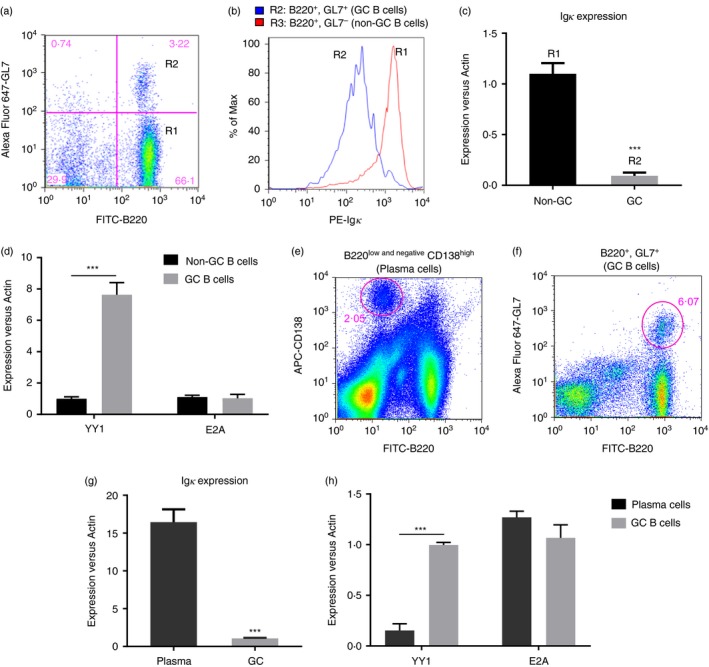
Igκ expression is down‐regulated in germinal centre (GC) B cells. (a) A single‐cell suspension was prepared from Peyer's patch lymph nodes and analysed with FACS using fluorescein isothiocyanate (FITC) ‐labelled B220, phycoerythrin (PE) ‐labelled Igκ and Alex‐Fluor‐647‐labelled GL7. The FACS assay readily distinguished the non‐GC B cells (B220+ GL7−, R1) from the GC B cells (B220+ GL7+, R2). (b) The fluorescence intensity of the surface Igκ on the non‐GC B cells from the GC B cells was measured, and the results indicated that the GC B cells expressed less Igκ than the non‐GC B cells. (c) The GC and non‐GC cells were sorted by FACS, and real‐time PCR was performed to measure the mRNA levels of Igκ, YY1 and E2A. The expression of Igκ was down‐regulated in the GC B cells compared with that in the non‐GC B cells (***P < 0·001, GC B cells versus non‐GC B cells). (d) In contrast, the expression of YY1 was significantly higher in GC B cells compared wth that in non‐GC B cells. No difference in E2A expression was observed between the two populations (***P < 0·001, GC B cells versus non‐GC B cells). (e, f) To prepare the splenic plasma and GC B cells, a single‐cell suspension was prepared from the spleen collected 5 days post‐immunization with sheep red blood cells (SRBCs), and the cells were stained with FITC‐B220 plus APC‐CD138 or FITC‐B220 plus Alex‐Fluor‐647‐labelled GL7. The plasma cells (B220low and negative CD138high) and GC B cells (B220+ GL7+) were sorted by FACS according to their surface markers. (g) Real‐time PCR indicated that Igκ expression was markedly increased in plasma cells compared with that in GC B cells (***P < 0·001, plasma cells versus GC B cells). (h) The plasma cells expressed less YY1 than the GC B cells, whereas there was no difference in the levels of E2A between the two populations (***P < 0·001, plasma cells versus GC B cells).
As YY1 exhibited inhibitory effects on Igκ expression, we wondered whether the level of YY1 was suppressed in plasma cells in which the immunoglobulin genes were highly transcribed. To this end, mice were immunized with sheep red blood cells to boost the population of plasma cells as previously described.28 Five days after immunization, the spleens were collected, and the plasma cells (B220low and negative CD138high) and GC B cells (B220+ GL7+) were sorted by FACS (Fig. 4e,f). Real‐time PCR was performed to measure Igκ, YY1 and E2A expression. As expected, Igκ expression was dramatically up‐regulated, whereas YY1 was reduced in the plasma cells compared with in the GC B cells (Fig. 4g). No significant difference in E2A levels was identified between the GC B cells and plasma cells (Fig. 4h), which was consistent with previous observations that E2A and E2‐2 (encoded by the TCF4 gene) are essential for both GC B‐cell and plasma cell development and that the levels of E2A remain high at both stages of B‐cell development.23
Discussion
YY1 has long been believed to play an important role in immunoglobulin gene regulation and B‐cell development. In the present study, we found that YY1 binds to human E3′ and inhibits E3′ activity by inducing suppressive epigenetic modifications. YY1 is the only known mammalian Polycomb Group (PcG) PcG protein with DNA binding site specificity.29 PcG proteins comprise a family of proteins involved in many biological processes, such as haematopoietic development, epigenetic chromosomal condensation, and stable transcriptional repression.30 YY1 is reported to interact with another PcG protein, Ezh2, which is part of PRC2 (polycomb repressive complex 2) and catalyses the trimethylation of H3K27 (H3K27me3).31, 32 Reduced H3K27me3 and enhanced H3K27ac of E3′ were observed upon knocking down YY1, suggesting that Ezh2 might be involved in the inhibitory effects of YY1 in Igκ expression. E2A reportedly promotes the chromatin accessibility of Igκ and increases histone acetylation.24, 33 Knocking down E2A inhibited the H3K27ac of E3ʹ as well as the expression of Igκ. In addition, knocking down YY1 up‐regulated E2A expression and increased its binding to E3ʹ. Conversely, knocking down E2A increased YY1 binding to E3ʹ; however, it did not affect YY1 expression. These ChIP assays clearly suggested the reciprocal binding of YY1 and E2A in E3ʹ. Several potential binding sites for both factors exist in E3ʹ, including the potential YY1/E2A‐overlapping binding site shown in the Supplementary material (Fig. S2). Although this site is within a protein‐binding region identified by Judde and Max using in vivo footprinting,34 mutating this site does not result in increased enhancer activity in reporter assays.34, 35 Therefore, more analyses are required in the future to elucidate the underlying mechanism and pinpoint the location of the binding sites. Nevertheless, our results suggest that Igκ gene expression is regulated by a balance between YY1 and E2A, possibly through the relative availability of the factors and their competition for E3′ binding, which determines the epigenetic status of the enhancer and, hence, the expression levels of Igκ.
Interestingly, Igκ expression was transiently down‐regulated in the mouse GC B cells compared with that in the non‐GC B cells. We previously reported that terminally differentiated plasma cells expressed a higher level of Igκ than naive B cells,28 suggesting that the expression levels of Igκ at different developmental stages were controlled in accordance with their function. During the humoral immune response, activated B cells proliferate rapidly upon antigen recognition, forming GCs. In GCs, B cells undergo somatic hypermutation (SHM), affinity maturation and class‐switch recombination to generate long‐lived memory B cells and plasma cells. Antibody affinity maturation is achieved via SHM and clonal selection; new immunoglobulin molecules result from SHM replacing the original molecules on the cell surface, and the cells that obtain immunoglobulin molecules with high antigen affinity are selected for further proliferation, whereas the cells with low‐affinity immunoglobulin molecules undergo apoptosis.36 We speculate that in GC B cells, diminished Igκ expression may be required for the replacement by new immunoglobulin molecules that have undergone SHM. A quick turnover of immunoglobulin may facilitate the antibody affinity maturation process, which relies on the interaction between the new immunoglobulin and the corresponding antigen provided on the follicular dendritic cells in the GC.37 The GC B cells that express high‐affinity immunoglobulin develop into plasma cells, wherein YY1 expression is diminished, and high levels of E2A are maintained,12 presumably for the task of producing the maximal amount of immunoglobulin. In summary, we found that both YY1 and E2A bind to E3′ of the human Igκ locus and regulate Igκ expression through epigenetic modifications. Our results also imply a novel mechanism by which YY1 contributes to antibody maturation by transiently inhibiting Igκ expression in GC B cells.
Disclosures
The authors have no conflicts of interest to declare.
Supporting information
Figure S1. YY1 suppresses Igκ expression in Daudi cells.
Figure S2. Schematic diagram of the human Igκ locus and the sequence of E3ʹ are shown.
Table S1. The list of sequence of primers (5′→3′)
Acknowledgements
XZ and XW designed the experiments and wrote the manuscript. XZ and WX performed the experiments and analysed the data. JZ, YZ, XS, YH, YQ and XD helped perform the experiments. This work was supported by the National Nature Science Foundation of China (grant numbers 81471539, 81571527, 81771681, 21607082), the Jiangsu Province Science & Technology Department Foundation (grant numbers BK20141236, BK20160414), the Natural Science Foundation of the Higher Education Institutions of Jiangsu Province (grant number 15KJB310012), and the Nantong City Science & Technology Projects (grant number MS12015062).
Contributor Information
Xiaorong Zhou, Email: Zhou-xiaorong@163.com.
Xiaoying Wang, Email: wxy_ntu@163.com.
References
- 1. Nemazee D. Mechanisms of central tolerance for B cells. Nat Rev Immunol 2017; 17:281–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pelanda R, Torres RM. Receptor editing for better or for worse. Curr Opin Immunol 2006; 18:184–90. [DOI] [PubMed] [Google Scholar]
- 3. Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol 2011; 11:251–63. [DOI] [PubMed] [Google Scholar]
- 4. Xu Y, Davidson L, Alt FW, Baltimore D. Deletion of the Igκ light chain intronic enhancer/matrix attachment region impairs but does not abolish VκJκ rearrangement. Immunity 1996; 4:377–85. [DOI] [PubMed] [Google Scholar]
- 5. Meyer KB, Neuberger MS. The immunoglobulin κ locus contains a second, stronger B‐cell‐specific enhancer which is located downstream of the constant region. EMBO J 1989; 8:1959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xiang Y, Garrard WT. The Downstream Transcriptional Enhancer, Ed, positively regulates mouse Ig κ gene expression and somatic hypermutation. J Immunol 2008; 180:6725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inlay M, Alt FW, Baltimore D, Xu Y. Essential roles of the κ light chain intronic enhancer and 3ʹ enhancer in κ rearrangement and demethylation. Nat Immunol 2002; 3:463–8. [DOI] [PubMed] [Google Scholar]
- 8. Zhou X, Xiang Y, Garrard WT. The Igκ gene enhancers, E3ʹ and Ed, are essential for triggering transcription. J Immunol 2010; 185:7544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Deng Z, Cao P, Wan MM, Sui G. Yin Yang 1: a multifaceted protein beyond a transcription factor. Transcription 2010; 1:81–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Q, Stovall DB, Inoue K, Sui G. The oncogenic role of Yin Yang 1. Crit Rev Oncog 2011; 16:163–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu H, Schmidt‐Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H et al Yin Yang 1 is a critical regulator of B‐cell development. Genes Dev 2007; 21:1179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Green MR, Monti S, Dalla‐Favera R, Pasqualucci L, Walsh NC, Schmidt‐Supprian M et al Signatures of murine B‐cell development implicate Yy1 as a regulator of the germinal centre‐specific program. Proc Natl Acad Sci USA 2011; 108:2873–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zaprazna K, Atchison ML. YY1 controls immunoglobulin class switch recombination and nuclear activation‐induced deaminase levels. Mol Cell Biol 2012; 32:1542–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pan X, Jones M, Jiang J, Zaprazna K, Yu D, Pear W et al Increased expression of PcG protein YY1 negatively regulates B cell development while allowing accumulation of myeloid cells and LT‐HSC cells. PLoS One 2012; 7:e30656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Trabucco SE, Gerstein RM, Zhang H. YY1 regulates the germinal centre reaction by inhibiting apoptosis. J Immunol 2016; 197:1699–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kleiman E, Jia H, Loguercio S, Su AI, Feeney AJ. YY1 plays an essential role at all stages of B‐cell differentiation. Proc Natl Acad Sci USA 2016; 113:E3911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pan X, Papasani M, Hao Y, Calamito M, Wei F, Quinn Iii WJ et al YY1 controls Igκ repertoire and B‐cell development, and localizes with condensin on the Igκ locus. EMBO J 2013; 32:1168–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS et al YY1 is a structural regulator of enhancer‐promoter loops. Cell 2017; 171:1573–88 e1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park K, Atchison ML. Isolation of a candidate repressor/activator, NF‐E1 (YY‐1, Δ), that binds to the immunoglobulin κ 3ʹ enhancer and the immunoglobulin heavy‐chain mu E1 site. Proc Natl Acad Sci USA 1991; 88:9804–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bryne JC, Valen E, Tang MH, Marstrand T, Winther O, da Piedade I et al JASPAR, the open access database of transcription factor‐binding profiles: new content and tools in the 2008 update. Nucleic Acids Res 2008; 36:D102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pongubala JM, Atchison ML. Functional characterization of the developmentally controlled immunoglobulin κ 3ʹ enhancer: regulation by Id, a repressor of helix‐loop‐helix transcription factors. Mol Cell Biol 1991; 11:1040–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Greenbaum S, Zhuang Y. Identification of E2A target genes in B lymphocyte development by using a gene tagging‐based chromatin immunoprecipitation system. Proc Natl Acad Sci USA 2002; 99:15030–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wohner M, Tagoh H, Bilic I, Jaritz M, Poliakova DK, Fischer M et al Molecular functions of the transcription factors E2A and E2‐2 in controlling germinal centre B cell and plasma cell development. J Exp Med 2016; 213:1201–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakamoto S, Wakae K, Anzai Y, Murai K, Tamaki N, Miyazaki M et al E2A and CBP/p300 act in synergy to promote chromatin accessibility of the immunoglobulin κ locus. J Immunol 2012; 188:5547–60. [DOI] [PubMed] [Google Scholar]
- 25. Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell 2013; 49:825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Banerjee A, Sindhava V, Vuyyuru R, Jha V, Hodewadekar S, Manser T et al YY1 is required for germinal centre B cell development. PLoS One 2016; 11:e0155311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lefrancois L, Lycke N. Isolation of mouse small intestinal intraepithelial lymphocytes, Peyer's patch, and lamina propria cells. Curr Protoc Immunol 2001; Chapter 3: Unit 3 19. [DOI] [PubMed] [Google Scholar]
- 28. Zhou X, Xiang Y, Ding X, Garrard WT. A new hypersensitive site, HS10, and the enhancers, E3ʹ and Ed, differentially regulate Igκ gene expression. J Immunol 2012; 188:2722–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Atchison L, Ghias A, Wilkinson F, Bonini N, Atchison ML. Transcription factor YY1 functions as a PcG protein in vivo . EMBO J 2003; 22:1347–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Di Croce L, Helin K. Transcriptional regulation by Polycomb group proteins. Nat Struct Mol Biol 2013; 20:1147–55. [DOI] [PubMed] [Google Scholar]
- 31. Van Kruijsbergen I, Hontelez S, Veenstra GJ. Recruiting polycomb to chromatin. Int J Biochem Cell Biol 2015; 67:177–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B et al Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 2002; 111:197–208. [DOI] [PubMed] [Google Scholar]
- 33. Lazorchak AS, Wojciechowski J, Dai M, Zhuang Y. E2A promotes the survival of precursor and mature B lymphocytes. J Immunol 2006; 177:2495–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Judde JG, Max EE. Characterization of the human immunoglobulin κ gene 3ʹ enhancer: functional importance of three motifs that demonstrate B‐cell‐specific in vivo footprints. Mol Cell Biol 1992; 12:5206–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pongubala JM, Atchison ML. Activating transcription factor 1 and cyclic AMP response element modulator can modulate the activity of the immunoglobulin κ 3ʹ enhancer. J Biol Chem 1995; 270:10304–13. [DOI] [PubMed] [Google Scholar]
- 36. Suan D, Sundling C, Brink R. Plasma cell and memory B cell differentiation from the germinal centre. Curr Opin Immunol 2017; 45:97–102. [DOI] [PubMed] [Google Scholar]
- 37. Zhang Y, Garcia‐Ibanez L, Toellner KM. Regulation of germinal centre B‐cell differentiation. Immunol Rev 2016; 270:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. YY1 suppresses Igκ expression in Daudi cells.
Figure S2. Schematic diagram of the human Igκ locus and the sequence of E3ʹ are shown.
Table S1. The list of sequence of primers (5′→3′)


