Abstract
Aspergillus nidulans has long-been used as a model organism to gain insights into the genetic basis of asexual and sexual developmental processes both in other members of the genus Aspergillus, and filamentous fungi in general. Paradigms have been established concerning the regulatory mechanisms of conidial development. However, recent studies have shown considerable genome divergence in the fungal kingdom, questioning the general applicability of findings from Aspergillus, and certain longstanding evolutionary theories have been questioned. The phylogenetic distribution of key regulatory elements of asexual reproduction in A. nidulans was investigated in a broad taxonomic range of fungi. This revealed that some proteins were well conserved in the Pezizomycotina (e.g. AbaA, FlbA, FluG, NsdD, MedA, and some velvet proteins), suggesting similar developmental roles. However, other elements (e.g. BrlA) had a more restricted distribution solely in the Eurotiomycetes, and it appears that the genetic control of sporulation seems to be more complex in the aspergilli than in some other taxonomic groups of the Pezizomycotina. The evolution of the velvet protein family is discussed based on the history of expansion and contraction events in the early divergent fungi. Heterologous expression of the A. nidulans abaA gene in Monascus ruber failed to induce development of complete conidiophores as seen in the aspergilli, but did result in increased conidial production. The absence of many components of the asexual developmental pathway from members of the Saccharomycotina supports the hypothesis that differences in the complexity of their spore formation is due in part to the increased diversity of the sporulation machinery evident in the Pezizomycotina. Investigations were also made into the evolution of sex and sexuality in the aspergilli. MAT loci were identified from the heterothallic Aspergillus (Emericella) heterothallicus and Aspergillus (Neosartorya) fennelliae and the homothallic Aspergillus pseudoglaucus (=Eurotium repens). A consistent architecture of the MAT locus was seen in these and other heterothallic aspergilli whereas much variation was seen in the arrangement of MAT loci in homothallic aspergilli. This suggested that it is most likely that the common ancestor of the aspergilli exhibited a heterothallic breeding system. Finally, the supposed prevalence of asexuality in the aspergilli was examined. Investigations were made using A. clavatus as a representative ‘asexual’ species. It was possible to induce a sexual cycle in A. clavatus given the correct MAT1-1 and MAT1-2 partners and environmental conditions, with recombination confirmed utilising molecular markers. This indicated that sexual reproduction might be possible in many supposedly asexual aspergilli and beyond, providing general insights into the nature of asexuality in fungi.
Key words: abaA, Asexuality, Aspergillus nidulans, brlA, Conidiation, Conidiophore, Development, Mating type, Sporulation, velvet
Introduction
Members of the genus Aspergillus have long-been used as model organisms to study developmental processes in filamentous fungi. This is due to their ease of cultivation and manipulation under laboratory conditions, the well-characterised morphology of asexual spore development, and the fact that they exhibit both homothallic (self-fertile) and heterothallic (obligate out-crossing) sexual breeding systems (Krijgsheld et al. 2013). The homothallic species A. nidulans in particular has been used extensively for investigations into the genetic basis of asexual and sexual sporulation, following its establishment as a model organism by Pontecorvo (1953). Studies have revealed a series of genetic pathways governing asexual and sexual reproduction, with ongoing research using a variety of –omic techniques to gain ever deeper insights into the precise molecular mechanisms of these pathways (Park and Yu, 2012, Dyer and O'Gorman, 2012).
Results from studies with A. nidulans have been considered to provide insights that are applicable to sporulation processes in the aspergilli as a whole, as well as being of relevance to the Pezizomycotina in general. However, it is possible that some aspects of the genetic regulation may be restricted to A. nidulans and its close relatives. Whole genome sequence has recently become available from both a wide taxonomic range of the aspergilli (de Vries et al. 2017) and the fungal kingdom in general, revealing considerable genome divergence within fungi. This now offers the opportunity to assess how widespread aspects of the regulatory pathways of reproduction in A. nidulans are in a broad biodiversity of fungi, as well as addressing certain specific questions concerning the control and evolution of asexual and sexual development in the aspergilli. These issues are investigated in detail in the present study under the common theme of reproduction, looking first at asexual and then later at sexual reproduction. Findings from both sets of analyses reveal how data obtained from Aspergillus species may, or may not, be of general relevance to understanding reproductive processes in other fungal taxa. The results presented follow on from initial work reported in the comparative genomics analysis of de Vries et al. (2017).
Asexual development in Aspergillus
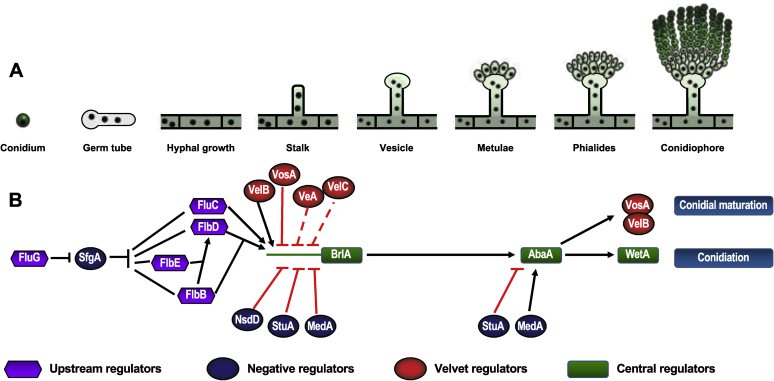
Aspergillus species are well known for the prolific production of asexual spores termed conidia. These are produced from conidiophores with a characteristic aspergillum-like morphology consisting of a foot cell, stalk and vesicle bearing metulae and phialides with radiating conidia (Fig. 1A), although rare exceptions do exist within the aspergilli with different conidial head morphologies (Yu, 2010, Samson et al., 2014). A. nidulans has been used as a model for the study of conidiation for many decades and consequently considerable knowledge has been accumulated about the regulatory pathways involved in this species (Adams et al., 1998, Etxebeste et al., 2010, Park and Yu, 2012). The initiation of conidiation involves the regulation of thousands of genes in A. nidulans (Garzia et al., 2013, Cánovas et al., 2014), of which there are a series of upstream activators and negative repressors, central regulators, as well as light-responsive and velvet regulators [Fig. 1B; also see Fig. 2A of de Vries et al. (2017)].
Fig. 1.
(A) A schematic presentation of conidiophore development of in A. nidulans. (B) A genetic model of the regulation of conidiophore development.
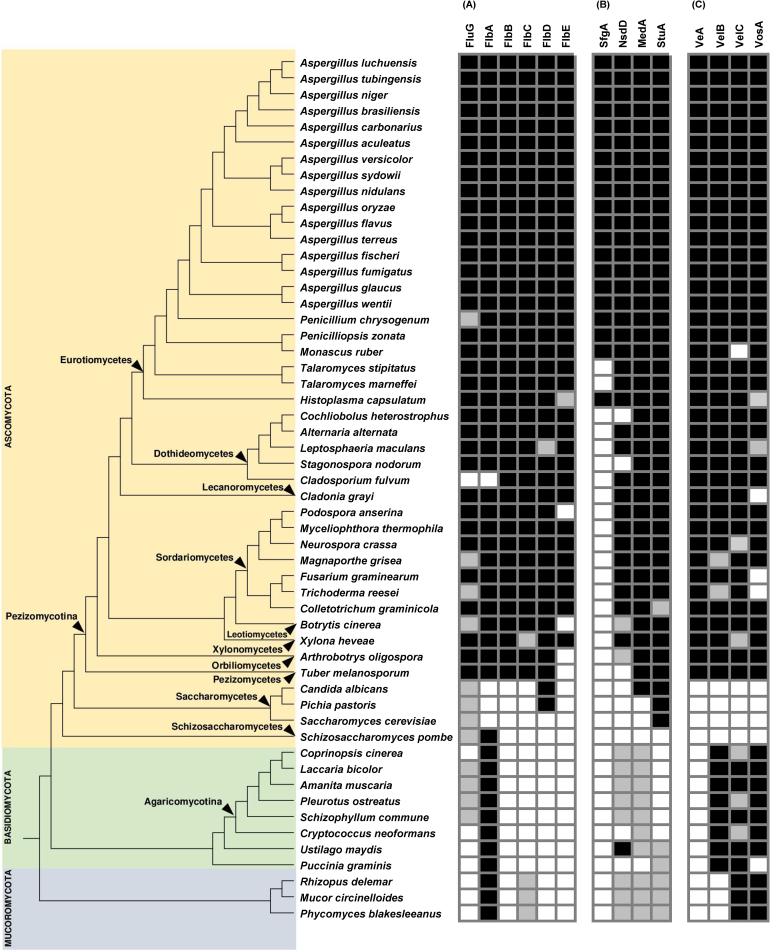
Fig. 2.
Distribution of proteins involved in the regulation of conidiation using A. nidulans proteins as bait. (A) Fluffy genes involved in the activation of the central developmental pathway of conidiation. (B) Repressors of conidiation and developmental modifiers. (C) Velvet proteins. The phylogenetic tree of species was estimated using Orthofinder. Black squares denote presence; white squares denote absence; and grey squares denote that the presence of proteins that Orthofinder predicts to belong to the orthogroup but they are either truncated with respect to the A. nidulans respective homologue or do not fulfill the domain architecture requirements. Absence in the Ascomycota clade was confirmed by tblastn searches in the corresponding genomes using the corresponding A. nidulans homologue.
The initiation of the conidial developmental pathway in A. nidulans is controlled by upstream developmental activators (UDAs), which consist of three genetic cascades containing the flbA (fluffy low BrlA expression), flbB/D/E and flbC genes. Upstream of the flbB/D/E and flbC modules lies fluG, which is an activator of the flb modules (Fig. 1B). FluG is responsible for the synthesis of an endogenous diffusible factor, with the meroterpenoid compound dehydroaustinol shown to induce conidiation in a ΔfluG mutant (Rodriguez-Urra et al. 2012). FluG is involved in the repression of the activity of SfgA (Seo et al. 2006). This step is crucial to initiate the conidiation machinery because SfgA itself is a repressor of the fluffy genes (Seo et al. 2006). Further repressors of conidiation active during vegetative growth are NsdD, VosA and two G-protein signalling pathways (Seo et al., 2006, Park and Yu, 2012, Lee et al., 2014, Lee et al., 2016).
The expression of the various fluffy genes ultimately activates the central regulatory pathway (CRP) composed sequentially of brlA, abaA and wetA (Adams et al., 1998, Etxebeste et al., 2010, Park and Yu, 2012) (Fig. 1B). Deletion of any of these genes leads to particular blocks in the proper development of conidiophores, resulting in abnormal morphologies termed bristle, abacus and wet-white, respectively (Yu 2010). The first genome sequencing projects of the aspergilli demonstrated that the CRP was also present in species such as A. fumigatus and A. niger, and it was suggested that the same pathway observed in A. nidulans was likely to control asexual sporulation in these species as well (Pel et al., 2007, Samson et al., 2009, Yu, 2010). Most recently, de Vries et al. (2017) investigated the presence of CRP in a broader range of Aspergillus and Pezizomycotina species. BrlA seemed to be limited to the Eurotiales, suggesting a specific role for conidiation in this group. By contrast, WetA was widely distributed in the Pezizomycotina, suggesting a general function for the synthesis of spore cell wall layers. Meanwhile, AbaA was widespread in the Ascomycota, Basidiomycota, and Mucoromycota, suggesting other general functions in fungal development. However, intriguingly the abaA gene was missing from Monascus ruber (a close relative of the aspergilli) and was not uniformly present in the fungal kingdom (de Vries et al. 2017).
Other proteins also influence conidial formation in A. nidulans. These include the transcription factors StuA and MedA, both of which have been termed developmental modifiers because they are required for the development of proper conidiophore morphology (Busby et al., 1996, Wu and Miller, 1997). A further major group is the velvet (Vel) proteins, which have been implicated in the regulation of developmental processes, and also secondary metabolism, and which are specific to fungi (Bayram & Braus 2012). The members of this family are characterized by the velvet domain comprising approximately 150 amino acids with a fold resembling the Rel homology domain (RHD) of the mammalian transcription factor NF-κB (Ahmed et al. 2013). The velvet proteins act downstream of the light receptor proteins LreA, LreB and FphA in A. nidulans to either promote or repress asexual or sexual reproduction, depending on the specific VeA, VelB or VelC protein (Bayram and Braus, 2012, Dyer and O'Gorman, 2012). The velvet regulators can interact both with each other and also with non-velvet proteins to control development and conidiation (Bayram & Braus, 2012).
Given this background, a main aim of the present study was to study the phylogenetic distribution of known regulators of conidiation in A. nidulans to determine how widespread the action of these proteins might be in the fungal kingdom. This included an analysis of the upstream regulators and repressors, the central regulatory pathway, and the possible expansion or contraction of the velvet family proteins. A second specific aim was to heterologously express abaA in M. ruber, to see to what extent this might impact on conidiophore morphology given the close taxonomic relatedness to the aspergilli and that abaA appears to be absent from the M. ruber genome whilst all other regulators are present (Vries et al. 2017).
Sexual development in Aspergillus
Whereas asexual reproduction is observed universally in the aspergilli (Raper and Fennell, 1965, Samson et al., 2014), sexual reproduction has only been observed in approximately 36 % of species (Dyer & O'Gorman 2011). Where sex occurs, it leads to the formation of ascospores within enclosed cleistothecia, which break down at maturity to passively release the sexual spores. The Aspergillus species with described sexual cycles are overwhelmingly homothallic in nature, with a ratio of approximately 13:1 homothallic: heterothallic taxa (Dyer & O'Gorman 2012). Despite the supposed monophyly of Aspergillus there is nevertheless a surprising diversity in the morphology of sexual structures within the genus compared to the more limited variation seen in conidial development (Samson et al. 2014). Up to 12 different sexual genera have been phylogenetically associated with Aspergillus asexual forms, being distinguished by morphological aspects of the cleistothecia such as the wall (peridium) composition and colour, and whether cleistothecia develop within larger surrounding sclerotia (Dyer, 2007, Peterson, 2008, Samson and Varga, 2010, Dyer and O'Gorman, 2012).
Numerous studies have been undertaken with A. nidulans to determine the genetic basis of sexual development, with over 70 genes now identified as having roles in various stages of the sexual process. These have been divided into genes encoding proteins involved with perception of environmental signals, mating and signal transduction, transcription factors and other regulatory proteins, endogenous physiological processes, and ascospore production and maturation (Dyer & O'Gorman 2012). Of particular note was the discovery that the breeding system of particular species is governed by the presence of mating-type (MAT) genes (Dyer et al. 2016). The genome of the homothallic model species A. nidulans was found to contain both MAT1 and MAT2 mating-type genes encoding alpha-domain and high-mobility group (HMG) domain transcription factors, respectively (Galagan et al., 2005, Paoletti et al., 2007). A similar discovery was later made for the homothallic Aspergillus (Neosartorya) fischeri and Aspergillus (Petromyces) alliaceus (Rydholm et al., 2007, Ramirez-Prado et al., 2008). Deletion of either MAT gene led to loss of self-fertility in A. nidulans, although deletion mutants were still able to outcross in a heterothallic fashion (Paoletti et al. 2007). Related work led to the identification of complementary MAT1-1 and MAT1-2 isolates in species such as A. fumigatus, A. parasiticus, A. flavus and A. lentulus (Paoletti et al., 2005, O'Gorman et al., 2009, Horn et al., 2009a, Horn et al., 2009b, Swilaiman et al., 2013). In these instances, isolates were found to have an idiomorphic MAT locus containing either a MAT1-1-1 alpha domain or a MAT1-2-1 HMG mating-type gene, respectively. Under suitable conditions a sexual cycle could be induced in all of these species, with successful crossing requiring isolates of compatible mating type to be present. This provided clear evidence of a heterothallic breeding system in all of these species, determined by the presence of either MAT1-1-1 or MAT1-2-1 genes in the genome of individual isolates. MAT genes were also shown to exhibit cross-species activity and influence gene expression in asexual species when heterologous genes were used in host MAT gene replacement experiments (Grobe and Krappmann, 2008, Pyrzak et al., 2008, Wada et al., 2012).
The observation that homothallism predominates in the genus has been used as evidence to suggest that this breeding system was ancestral in the aspergilli, with subsequent conversion to heterothallism through loss of self-fertility in the relatively few known heterothallic species (Geiser et al., 1996, Geiser et al., 1998, Varga et al., 2000). Furthermore, the fact that the majority of the aspergilli are only known to reproduce by asexual means has led to the theory that sexual reproduction (meiosis) has been lost multiple times in this group due to evolutionary selection for asexuality (Geiser et al. 1996). These hypotheses have been applied more generally to the evolution of sex and asexuality in fungi (Dyer & Kück 2017).
Given this background, an additional main aim of the present study was to determine whether homothallism is truly ancestral in the aspergilli, or whether the genus has heterothallic origins, building on recent findings by de Vries et al. (2017). To address this question, we examined how MAT locus architecture varied throughout the aspergilli, including the cloning of MAT loci from the first two ever identified heterothallic Aspergillus species, Aspergillus (Emericella) heterothallicus (Kwon & Raper 1967) and Aspergillus (Neosartorya) fennelliae (Kwon & Kim 1974), as well as the homothallic Aspergillus pseudoglaucus (=Eurotium repens) (Chen et al., 2017a, Chen et al., 2017b). A further specific aim was to determine whether asexuality truly dominates in the aspergilli, or whether supposed ‘asexual’ species might retain the potential for sexual reproduction. To address this, an analysis of MAT gene presence and recent breakthroughs in inducing sexual reproduction was used to investigate whether sex might be possible in A. clavatus, which lacks a known sexual morph, as a representative of the asexual aspergilli. This species was chosen due to its medical importance both as an opportunistic pathogen and producer of antibiotics (Bergel et al., 1944, Suzuki et al., 1971, Varga et al., 2007), as well as its importance as a spoilage organism (Varga et al. 2003). Indeed, these studies overall are of possible biotechnological and fundamental significance given that the sexual cycle provides a valuable tool for strain improvement and genetic analysis (Ashton & Dyer 2016), so it would be of great benefit if sex could be induced in supposed ‘asexual’ aspergilli.
Materials and methods
Bioinformatic analyses
Supplementary Table 1 lists species employed in bioinformatic analyses in this study. Filtered gene model derived proteomes were downloaded from the Mycocosm site (https://genome.jgi.doe.gov/programs/fungi/index.jsf) (Grigoriev et al. 2014). OrthoFinder version 2.2.0 (Emms & Kelly 2015) with default options was used to assess orthology among the 54 fungal proteomes. The resulting species tree was modified with Archaeopteryx (Han & Zmasek 2009) to fit the Mycocosm site evolutionary tree of the fungi, and a re-run of Orthofinder with the modified tree was used to estimate the reconciled trees for each orthogroup. Data from Aspergillus nidulans was used as a reference to find the orthogroups for the FluG, FlbA-E, SfgA, NsdD, MedA, StuA, VelA-C and VosA regulatory proteins of conidation. The reconciled trees were used to ascertain the orthology-paralogy relationships among the members of the same orthogroup. Protein domains were annotated employing the NCBI web Cd-search tool (Marchler-Bauer & Bryant 2004) against the CDD database. Domain architectures and trees were visualized with Domosaics (Moore et al. 2014).
To construct trees including basal fungi, proteins were first identified from the Mycocosm site. Multiple alignments were then made with Clustal Omega (Sievers et al. 2011), and maximum-likelihood trees were generated using the IQ-Tree server (Trifinopoulos et al. 2016) and drawn in iTOL (Letunic & Bork 2016). Branches were evaluated by 1 000 ultrafast bootstrap replicates and by the SH-aLRT test. Best model selection was carried out by the ModelFinder option included at the IQ-Tree server (Kalyaanamoorthy et al. 2017). Additional blastp and tblastn searches were conducted using the NCBI, JGI-Mycocosm (Grigoriev et al. 2014) and FungiDB (Stajich et al. 2012) databases where necessary.
For AbaA, BrlA and WetA, protein sequences from A. nidulans were used to query their homologues using the HMMER 3.1b2 package (http://www.hmmer.org/). The cut-off E values for homologues of AbaA, BrlA and WetA were set at e−5, e−100 and e−5, respectively. The homologues were aligned by MUSCLE (Edgar 2004) and then submitted to Weblogo (http://weblogo.threeplusone.com/create.cgi) to generate the conserved motifs.
Genetic manipulation of abaA in Monascus ruber
A heterologous gene expression approach was used to determine the effect of abaA expression in Monascus ruber. A. nidulans isolate FGSC A4 (Fungal Genetics Stock Center, USA) was used as the donor of the abaA gene and maintained on PDA slants at 30 °C. Escherichia coli DH 5α and Agrobacterium tumefaciens EHA105 were used for hosting plasmids and were cultivated in LB medium at 28 °C.
Briefly, the abaA gene (AN0422) was amplified from A. nidulans FGSC A4, while the trpC promoter and terminal fragments were cloned from the plasmid pSKH, and the selective marker gene neoR for neomycin resistance was cloned from the plasmid pKN1 (Li et al. 2010). These four DNA fragments were fused by double-joint PCR as illustrated in Supplementary Fig. 12 (Yu et al. 2004). Primers used in this study are listed in Supplementary Table 2. The fused fragment was then introduced into the expression vector pCAMBIA3300 via the vector pMD19-T. The recombinant vector carrying the abaA expression cassette was next introduced into the genome of M. ruber isolate M7 by Agrobacterium tumefaciens-mediated transformation according to Shao et al. (2009). Transformants were selected on potato dextrose agar (PDA) medium containing 15 mg/mL G418 (Sigma-Aldrich, Shanghai, China). Gene integration was confirmed by PCR, cDNA sequencing and Southern blotting. Southern blot assays were performed according to protocols for the DIG-High Prime DNA Labeling & Detection Starter kit I (Roche, Mannheim, Germany). To prepare probes, fragments from the open reading frame of abaA and the selective marker gene neoR were amplified with primer pairs abaA-F/abaA-R, and neo-F/neo-R (Supplementary Table 2), respectively, and then labelled with digoxin after purification with a DNA gel extraction kit (Sangon Biotech, China). In order to verify the relative expression level of abaA in the selected mutants, quantitative real-time RT-PCR was performed using β-actin as a reference gene (Liu et al. 2014). The wild-type and mutants were cultivated at 28 °C on potato dextrose agar (PDA) and an Olympus BH2 compound microscope with differential interference contrast optics was used to take photomicrographs of resulting phenotypes.
Identification and sequencing of the MAT locus of Aspergillus (Emericella) heterothallicus
Isolates 50-3 and 50-5 of A. heterothallicus (=Emericella heterothallica) were obtained from the BDUN culture collection at The University of Nottingham. These were derived from single sporing of the reference isolates WB4982 (MAT-A) and WB5086 (MAT-a), from Kwon & Raper (1967). Isolates were cultivated in malt extract liquid media (20 g malt extract powder, 1 g peptone per L distilled water) at 28 °C, and genomic DNA extracted using a DNeasy Plant Mini Kit (Qiagen) according to manufacturer's instructions.
To characterise the MAT loci, a bridging strategy was used involving PCR with degenerate primers of the internal MAT genes as well as amplification of genes known to be conserved in the external flanking regions of the MAT locus. Initially, fragments of the MAT1-1 and MAT1-2 genes were amplified from A. heterothallicus isolates 50-5 and 50-3, respectively, utilising the degenerate primer sets MAT5-6 and MAT3-4, and MAT5-7 and MAT3-5 respectively [these primers designed for Eurotiomycete fungi; Houbraken & Dyer (2015)] using PCR conditions described by Eagle (2009). Resultant PCR products were gel extracted, ligated into plasmid pTOPO4, then cloned into E. coli prior to DNA sequencing. In parallel, fragments of the SLA2 and APN2 genes [known to flank either side of the MAT locus in many Pezizomyoctina species (Debuchy and Turgeon, 2006, Dyer et al., 2016)] from isolates 50-5 and 50-3 were amplified and sequenced in a similar fashion using SLA2 and APN2 degenerate primer sets designed against conserved sequence found in the genomes of available Aspergillus species (primers used for SLA2 were aaSLA2: AYMGNGARATGGCNGAYYTNGARG and SLA2R: CRTANSDNGGNSWNGCRTTYTG; for APN2 primers used were aaAPN2: CARMGNAARGAYYTNMGNGAYGAYATG and APN2R: GGRTANCCNCCNAYYTGNYKNTC), using PCR conditions described by Eagle (2009). This allowed the subsequent design of species-specific non-degenerate primers for each of the resulting MAT1-1, MAT1-2, SLA2 and APN2 gene fragments. Primers were designed from the SLA2 and APN2 fragments to be orientated inwards towards the MAT locus, and pairs of outward primers were designed from the MAT1-1 and MAT1-2 fragments [see Eagle (2009) for specific details]. This allowed production of a SLA2 to MAT amplicon, and a separate MAT to APN2 amplicon (amplifying outwards from either the MAT1-1-1 or MAT1-2-1 fragments). The resulting products were sequenced by chromosome walking (Eagle 2009). Resulting sequence was interrogated by PSORT II (http://psort.nibb.ac.jp/) and TFSITESCAN (http://www.ifti.org/cgi-bin/ifti/Tfsitescan.pl) programs for the presence of nuclear-targeting and promoter-region motifs.
Identification and sequencing of the MAT loci from Aspergillus (Neosartorya) fennelliae and Aspergillus pseudoglaucus
Isolates 54-1 (CBS 410.89, MATA) and 54-2 (CBS 411.89, MATa) of A. (Neosartorya) fennelliae were obtained from the BDUN culture collection at The University of Nottingham. Both were originally isolated from Marine Sludge in Japan, 1981 (Takada & Udagawa 1985). Similar procedures were used to identify the MAT locus as described above for A. heterothallicus. A bridging strategy was used based on initial degenerate PCR of fragments of the MAT, SLA2 and APN2 genes. This allowed the design of species-specific primers, which were used to amplify SLA2 to MAT and separate MAT to APN2 amplicons, allowing chromosome walking of the entire MAT region. Full experimental details are provided in Eagle (2009).
For studies of A. pseudoglaucus (=Eurotium repens) two isolates were obtained from the BDUN culture collection (University of Nottingham), namely 51-1 (origin unknown) and 51-2 (CBS 529.65) originally isolated in 1965 from Prunus domestica in France (Peterson 2008). Attempts were made to identify MAT loci as described above for A. heterothallicus and A. fennelliae using the bridging strategy with degenerate PCR of fragments of the MAT, SLA2 and APN2 genes. However, it was also necessary to use thermal asymmetric interlaced (TAIL) PCR in combination with the use of the MAT degenerate primers to get sufficient MAT locus sequence (Arie et al. 1997). Successive rounds of TAIL-PCR were performed with degenerate PCR primers to extend the region around the MAT gene fragment (Liu & Whittier 1995). Sequence data was pooled from isolates 51-1 and 51-2 to ensure consistency. See Eagle (2009) for full experimental details.
Sexual biology of Aspergillus clavatus: mating-type diagnostic assay
Twenty isolates of A. clavatus from worldwide locations were obtained from the BDUN collection at the University of Nottingham (isolates 65-1 to 65-20; Supplementary Table 3). Isolates were maintained on Aspergillus complete agar or liquid media (ACM) (Paoletti et al. 2005) at 28 °C, and genomic DNA extracted using a DNeasy Plant Mini Kit (Qiagen) according to manufacturer's instructions.
To study the potential for sexual reproduction in A. clavatus it was first necessary to elucidate the mating types of these isolates in order that directed crosses could be set up. Putative MAT1-1-1 gene sequence data was obtained from The Broad Institute A. clavatus genome screening project (http://www.broad.mit.edu/tools/data/seq.html) (gene locus ID: ACLA_034110) and specific MAT1-1-1 primers were designed from within this sequence to detect the presence of isolates of the MAT1-1 genotype (primers AclM1F: CAGTTGTTCGTAGCAGACGGGG, and AclM1R: CGGTGGAGTATGCTTTGGCGAGG). In parallel, attempts were made to amplify a fragment of the MAT1-2-1 gene sequence using degenerate primers MAT5-7 and MAT3-5 (Houbraken & Dyer 2015) utilising PCR conditions described by Eagle (2009). The resulting putative MAT1-2-1 amplicon from isolate 65-13 was cloned and sequenced. The same bridging strategy as used in A. heterothallicus (see above), involving chromosome walking in from the SLA2 and APN2 flanking genes, was then used to clone and sequence the entire MAT1-2 region of A. clavatus from isolate 65-13 [see Eagle (2009) for specific details; GenBank accession MH401197]. This allowed the design of MAT1-2-1 specific primers within the MAT1-2 idiomorph to detect the presence of isolates of the MAT1-2 genotype (primers AclM2F: ATCAAGGCTCTTCGTGTCATGC, and AclM2R: ATGCTTCTTCTTCATATCTTCTGCC).
The resulting MAT primer sets were used in PCR as diagnostic tools to determine mating type in a screen of genomic DNA from the remaining isolates of A. clavatus. Amplifications were performed using 25 μL reaction volumes containing 2.5 μL 10X PCR Buffer (containing 20 mM MgCl2), 0.2 μL (25 mM each) dNTPs, 2.5 μL (10 μM) of the respective MAT forward and reverse primers, 0.2 μL FastStartTaq Polymerase (Roche, UK), ∼50 ng genomic DNA, and ultrapure water to a final volume of 25 μL. PCR was performed on a Techne Genius thermal cycler, using an initial denaturation step of 94 °C for 5 min; 35 cycles consisting of 1 min at 94 °C, 1 min at 60 °C for MAT1-1 and 55 °C for MAT1-2, and 1 min at 72 °C; followed by a final extension step at 72 °C for 5 min (all steps were performed at a ramp rate of 70 °C/min). Resultant PCR products were resolved on 1.5 % agarose gels and visualized by ethidium bromide staining. The hypothesis of a 1:1 ratio of mating types in the worldwide sample population and ascospore progeny was tested using χ2 and contingency χ2 tests (Fisher 1938).
Sexual biology of Aspergillus clavatus: crossing and progeny analysis
Crosses were then set up in 9 cm Petri dishes between six MAT1-1 strains and three MAT1-2 strains of A. clavatus which were inoculated in all possible pair wise combinations (n = 18), following the barrage crossing procedures of O'Gorman et al. (2009). All crosses were set up on oatmeal agar (Robert et al. 2007; pinhead, Odlums, Ireland) in triplicate, sealed with Nescofilm® and incubated at 25 °C, 28 °C or 30 °C in the dark. The crosses were examined periodically for the presence of cleistothecia for up to five months, using a Nikon-SMZ-2B dissection microscope.
Attempts were then made to isolate ascospore progeny from putative mature cleistothecia. The fruit bodies were cleaned by rolling on 4 % tap water agar to remove adhering conidia as described by Todd et al. (2007). Intact cleistothecia were then added to 500 μL of 0.05 % (v/v) Tween 80 and heat treated at 69 °C for 10 min to inactivate any adhering conidia, with the assumption that the peridium of the cleistothecia served as a barrier to protect the ascospores to some extent (higher temperatures and longer periods were found to kill the ascospores as well; data not shown). The cleistothecia were centrifuged, the supernatant discarded and then 50 μL of 0.05 % (v/v) Tween 80 was added and cleistothecia ruptured by squashing with a needle tip (Todd et al. 2007). The solution was then brought up to 500 μL by addition of 0.05 % Tween 80 (v/v) and vortex-mixed for 1 min to release the ascospores. One hundred μL of a 5 × 105 ascospore mL−1 suspension was then spread inoculated onto ACM plates (Paoletti et al. 2005), which were incubated at 37 °C for 14 h. Single spore cultures were established on ACM by transferring individual germinating ascospore with a LaRu lens cutter attached to a Nikon-OPTIPHOT microscope.
The segregation of five genetic markers was then examined in the ascospore offspring using RAPD-PCR fingerprinting as previously described (Murtagh et al., 1999, O'Gorman et al., 2009, Swilaiman et al., 2013). An initial screen of sixteen RAPD primers revealed four (OPC20, 0PT18, UBC90, OPQ6; sequences available on request) that yielded polymorphisms suitable for genotyping. Finally, cleistothecia were examined by scanning electron microscopy as described by Swilaiman et al. (2013).
Results and discussion
Reproduction via the formation of spores is a property seen throughout the fungal kingdom, which presumably arose early on in the evolution of many different lineages. The ability to produce both a tremendous abundance of asexual and/or sexual spores, combined with the possibility of the long-distance dispersal of these propagules, helps account for the ecological success and widespread occurrence of members of the fungal kingdom (Golan & Pringle 2017). The formation of asexual and sexual spores is in a balance controlled by both environmental factors and intracellular signals (Adams et al., 1998, Rodriguez-Romero et al., 2010, Ruger-Herreros et al., 2011, Cánovas et al., 2016, Marcos et al., 2016). It is therefore of both academic and applied significance to understand the genetic controls of asexual and sexual development, with the prospect of exploiting such knowledge to control detrimental species whilst promoting growth of beneficial species.
In terms of filamentous fungi, A. nidulans and Neurospora crassa have been the most widely used models to study developmental processes up to this point. Research with A. nidulans in particular has established paradigms for the genetic regulation of asexual and sexual reproduction (Adams et al., 1998, Braus et al., 2002, Han and Han, 2010, Etxebeste et al., 2010, Dyer and O'Gorman, 2012, Park and Yu, 2012), as well as the aspergilli in general being used to propose hypotheses concerning the evolution of asexuality and sexual breeding systems (Geiser et al., 1996, Geiser et al., 1998, Varga et al., 2000, Galagan et al., 2005, Dyer, 2007, Dyer and O'Gorman, 2012). However, it has become apparent that there can be significant divergence at the genome level even within a single fungal genus (Galagan et al. 2005). Therefore, the present study was undertaken to assess the phylogenetic distribution of the regulatory pathways of asexual reproduction in a broad taxonomic range of fungi, to gain some indication of their prevalence. In parallel some long-standing questions concerning the control and evolution of sexual development in the aspergilli were addressed. Overall it was found that some features seen in A. nidulans indeed appear to be of relevance to a wide biodiversity of fungi. However, some other features are much less conserved, even within the Eurotiomycetes, and some hypotheses about the origins of sex and asexuality in the aspergilli appear to be incorrect, as will now be described.
Bioinformatic analysis of asexual development in Aspergillus
An A. nidulans-centric approach was used to study the phylogenetic distribution and molecular features of known regulators of conidiation from this species. This involved screening for the presence of a series of upstream activators and repressors, central regulators, as well as velvet regulators (Fig. 1B) in 54 fungal species including 16 Aspergillus species, related Ascomycota and more distant Basidiomycota and Mucoromycota (Fig. 2). This analysis complements and builds on the findings presented by de Vries et al. (2017).
Upstream activators and repressors
With respect to conidiation upregulators, the A. nidulans FluG upstream activator protein was found to possess two characteristic domains, a GlnA domain (glutamine synthetase), and a metallo-dependent hydrolase domain, belonging to the amidohydrolase superfamily (Supplementary Fig. 1). Homologues of the fluG orthogroup were found in the majority of Ascomycota, possibly linked to a role in conidiation as seen in A. nidulans. More distant orthologues were also found in the Basidiomycota, although not in the Mucoromycota (Fig. 2A). Phylogenetic analysis further showed that this orthogroup can be divided into two large groups. One of these encompasses species with proteins that possess only the GlnA domain, with the other containing homologues that have both domains described above. The only exceptions were a subtree encompassing four basidiomycetes and the ascomycete Penicilium chrysogenum, in which proteins only contained the metallo-dependent hydrolase domain. These five species possess proteins that are likely to have lost the GlnA domain after the FluG orthologues became separated from the rest of the species in this orthogroup. In some species of the Pezizomycotina fluG has been lost, specifically in Cladosporium, Botrytis, Trichoderma and Magnaporthe. It was already noted by de Vries et al. (2017) that almost half of the Aspergillus species analysed possess two copies of the fluG gene, possibly suggesting more differentiated regulation of development in these species.
Regarding conidiation repressors, the A. nidulans SfgA repressor protein also has two specific domains: a Gal4-type Zn(II)2Cys6 type transcription factor, which consists of two helices organized around the Zn(II)2Cys6 motif, and a fungal transcription factor regulatory middle homology region, which is present in the large family of fungal zinc cluster transcription factors that contain an N-terminal GAL4-like DNA-binding domain (Supplementary Fig. 2). SfgA was found to be present exclusively in the Eurotiomycetes, being conserved in all Aspergillus species as well as being present in Monascus, Histoplasma, and Penicillium, although not in Talaromyces species (Fig. 2B). All the other Pezizomycotina lack homologues of sfgA (Dothideomycetes, Sordariomycetes, Lecanoromycetes etc.). By contrast, the NsdD repressor protein (a GATA-type zinc-finger transcription factor) whilst also being present in all the Eurotiomycetes had a broader distribution in many other Ascomycota (Fig. 2B). Interestingly A. wentii and A. luchuensis have two paralogs of nsdD, which likely appeared independently by gene duplication. Some other members of the Pezizomycotina, such as certain Sordariomycetes and Leotiomycetes, contain shorter copies of nsdD, which are likely homologues of AnnsdD as they cluster together (Supplementary Fig. 3). Indeed, deletion of nsdD orthologues in Fusarium (csm-1) and Botrytis (ltf1) increases conidiation (Schumacher et al., 2014, Niehaus et al., 2017) as has been reported for A. nidulans (Lee et al. 2016).
The absence of the A. nidulans SfgA and NsdD repressors of conidiation in some other Ascomycota indicates that in such groups the induction of conidiation employs a different mechanism than the derepression exerted by the FluG factor as seen in A. nidulans. Alternatively, given that in many of these cases a homologue of fluG is present in the genome, it is possible that FluG derepression occurs by some other mechanism, or even that FluG directly activates elements of a downstream conidiation pathway. Interestingly Fusarium can undergo microconidiation in liquid media under standard growth conditions (López-Berges et al. 2013) and contains a fluG homologue but not an sfgA homologue. Although it could be argued that the absence of sfgA allows fungi to conidiate in liquid media, this is not the case for other Sordariomycetes also lacking sfgA, such as N. crassa, in which induction of conidation requires growth on a solid surface or particular starvation conditions (Berlin & Yanofsky 1985). In this group of organisms, repression of conidiation in P. chrysogenum poses an interesting case, as it lacks a complete homologue of fluG, but it has homologues of both sfgA and nsdD repressors. Analysis by tblastn against all the Penicillium taxon in NCBI revealed that some Penicillium species contain a complete fluG homologue, while some other species contain N-terminal or C-terminal truncated versions (data not shown).
The next set of results of the bioinformatic analyses concerned the remaining members of the fluffy group of genes, which promote asexual conidiation. FlbA is a regulator of the G-protein signalling (Yu et al. 1996). Accordingly, FlbA contains three different domains (Supplementary Fig. 4): two DEP domains that are responsible for mediating intracellular protein targeting and regulation of protein stability in the cell, and a RGS (Regulator of G-protein Signalling) domain that is an essential part of FlbA because it is involved in the cellular signalling events downstream of G-protein coupled receptors (GPCRs). The DEP domain is present in many signalling molecules, including RGS proteins. This pathway signals through a cAMP-PKA, which is broadly distributed in eukaryotes, and therefore it was expected that most fungal species would contain an flbA homologue. Indeed, FlbA was found to be highly conserved, appearing in all of the species included in this study with the exception of the Saccharomycotina, that have lost one of the domains during their evolution and, surprisingly, Cladosporium in which there are no homologues (Fig. 2A). By contrast, N. crassa and F. graminearum contain two copies of flbA. In the phylogenetic tree two subtrees were observed (Supplementary Fig. 4): the first one has species with just two of the three domains, and the second one has members where all three domains are conserved. We assume that this second subtree comprises species with proteins most orthologous to A. nidulans FlbA.
FlbC is a C2H2 zinc finger transcription factor involved in binding directly to the cis-regulatory element of brlA and inducing its expression (Kwon et al. 2010). FlbC is included in a very large orthogroup encompassing other C2H2 zinc finger transcription factors (e.g. BrlA). It was found to be well conserved throughout the Pezizomycotina, appearing in almost all the species studied (Fig. 2A). In this orthogroup, it was possible to further differentiate the orthologues of FlbC from paralogous proteins involved in other biological processes thanks to the domain architecture combined with the clustering pattern (Supplementary Fig. 5). According to this strategy, FlbC is present in all species of Pezizomycotina (except in Xilomycetes) and Mucoromycota, however was absent from the Basidiomycota and some Ascomycota such as the Taphrinomycotina and Saccharomycotina. Deletion of flbC in Aspergillus species and some Sordariomycetes is consistent with a broad role in fungal development. For example, deletion of flbC in Fusarium resulted in reduced conidiation, whilst in N. crassa and M. oryzae flbC mutants showed a reduction in aerial hyphae in addition to reduced conidiation levels (Son et al., 2014a, Son et al., 2014b, Malapi-Wight et al., 2014, Cao et al., 2016, Matheis et al., 2017, Boni et al., 2018). Overexpression of flbC in A. nidulans produced abnormal vesicles-like structures at the tips (Kwon et al. 2010), which suggests a possible role in the blastic development of the conidiophores.
In the other FluG-dependent pathway in A. nidulans, FlbE interacts with FlbB at the fungal tip in a process necessary to activate FlbB (Herrero-Garcia et al. 2015), and then FlbB induces FlbD (Fig. 1). FlbB and FlbD form a heterodimer that activates the expression of brlA in a cooperative way (Garzia et al. 2010). FlbB contains a basic leucine zipper (bZIP) domain of DNA binding (Etxebeste et al. 2008). The bZIP structural motif contains a basic region and a leucine zipper, composed of alpha helices with leucine residues 7 amino acids apart, which stabilize dimerization with a parallel leucine zipper domain. Analysis of the FlbB phylogenetic distribution revealed that it is found exclusively in the Pezizomycotina (Fig. 2A), and a duplication event is evident that divides the tree into two main subtrees (Supplementary Fig. 6). The upper one contains all the orthologues of AnflbB, whilst the lower one contains other bZIP proteins of the Ascomycota, which suggests the presence of paralogous proteins, which may have acquired new functions during evolution.
FlbD has been reported to possess a Myb-like DNA-binding domain (Wieser & Adams 1995). Myb DNA binding domains display extraordinary similarity to SANT domains, which are involved in histone tail binding and remodelling of nucleosomes (Boyer et al. 2004). Our search for domains using the Cd-search tool against the CDD database showed that FlbD has a SANT domain (Supplementary Fig. 7), which opens the possibility that the role of FlbD is to re-model the chromatin at the brlA promoter to allow its expression. The distribution of flbD perfectly matches the distribution of flbB, with the exception of Leptosphaeria (Fig. 2A), which contains two truncated versions clustering together in the Dothideomycetes cluster but with different domain architecture, which points to a different role. Trichoderma has a truncated version containing only the myb-like DNA binding domain and no additional sequence. Interestingly, despite the perfectly matching flbB and flbD distribution, deletion of flbB in Fusarium and N. crassa did not show any phenotype in conidiation (Son et al., 2014a, Son et al., 2014b, Carrillo et al., 2017). On the other hand, homologues of flbD are essential for the development of the conidiophores in Fusarium and Magnaporthe (Kim et al., 2014b, Son et al., 2014a, Dong et al., 2015, Matheis et al., 2017), and for filamentous growth in Candida (Homann et al. 2009), suggesting that FlbD can also operate without forming a heterodimer with FlbB. In Aspergillus, flbD can also function in a flbB-independent manner orchestrating the formation of the external tissue (peridium) of the fruiting body (cleistothecia) during sexual development (Arratia-Quijada et al. 2012). In addition to the activation of brlA expression, which is not present in Sordariomycetes (see below), it was reported that FlbB may also be a key factor in the transition from metulae to phialide in A. nidulans (Etxebeste et al. 2009). Sordariomycetes contain phialides but not metulae, which can explain the lack of phenotype of the flbB mutants. Although the asexual developmental structures of N. crassa are more simple than the Aspergillus ones, N. crassa displays a complex ontogeny with the formation of blasto-arthrospores during macroconidiation (Cole 1986). Microconidiation in N. crassa ressembles more the development of conidiophores in Aspergillus (Springer 1993). The flbD homologue in N. crassa poses another interesting case: it complements the orthlogous mutation of A. nidulans, but deletion in N. crassa does not show any phenotype (Shen et al. 1998) exposing again the differences in the ontogeny between these organisms.
FlbE has no known domains. In general, the distribution of flbE also matches the distribution of flbB and flbD, with the exception of Podospora. However, flbE seems to be absent from the Leotiomycetes, Orbiliomycetes and Pezizomycetes examined (Fig. 2A). Further analysis showed that Botrytis has a homologue with low homology in the N-terminal part, and Cladonia has two putative copies (one of them shorter). The absence of flbE in some taxa suggest that in these cases, FlbB must be activated in a different way than in A. nidulans, whether this still happens at the tip or not remains unknown.
Regarding the developmental modifiers in A. nidulans, StuA contains a basic helix-loop-helix (bHLH)-like structure of the APSES domain (Dutton et al. 1997). Members of this family participate in developmental processes and cell cycle progression. A StuA homologue was present in all the Pezizomycotina and possible orthologues were detected in the Mucoromycota, which also showed an expansion of the number of copies. However, StuA was generally absent in the Basidiomycota, except for possible retention in Ustilago (Fig. 2B and Supplementary Fig. 8). The domain architecture is very diverse in this orthogroup. Some members have a Sec23_BS domain (sandwich domain) characteristic of SNARE proteins. Some others contain PAT1 domain (topoisomerase II-associated protein), required for accurate chromosomal transmission in yeast. In the Saccharomycotina two stuA homologues of the APSES family were found. In yeast these homologues (PHD1 and SOK2) are involved in pseudohyphal growth, a process with some similarities to the formation of sterigmata cells in the aspergilli, and in Candida EFG1 is involved in hyphal growth and the white-phase cell type (Stoldt et al. 1997). The fission yeast Schizosaccharomyces pombe has two APSES proteins involved in cell cycle (Zhu et al. 1997), which are non-ortholgous to the developmental APSES regulators of other fungi developing more complex structures. Although the deletion of stuA homologues results in a decreased conidiation in Fusarium, Magnaporthe, Aspergillus, Talaromyces and Neurospora, the morphological defects are different. In Magnaporthe and Fusarium stuA seems to be involved in the development of the conidiophore. Macroconidia are produced from intercalarly phialides, rather than from the conidiophore in Fusarium. No difference in morphology was observed between the mutant and the wild type in Magnaporthe (Ohara and Tsuge, 2004, Nishimura et al., 2009). In Aspergillus and Talaromyces stuA mutants showed shorter stalks and absence of metulae and phialides. In this case, a few conidia arise directly from the stalk (Miller et al., 1992, Borneman et al., 2002). Borneman et al. (1992) suggested that StuA controls developmental processes requiring budding, which is in agreement with the non-filamentous phenotype of the deletion mutant of Ustilago (García-Pedrajas et al. 2010) and matches the blastic development of the conidiophores in these fungi. In the case of N. crassa, the deletion mutant was not characterized in depth with respect to conidiation. The mutant showed a stunted appearance, conidiating close to the agar surface (Aramayo et al. 1996). If StuA truly orchestrates blastic development, then residual conidiation of N. crassa could arise during the secondary arthric development of macroconidia. A deeper characterization of the mutant phenotype is required.
The final developmental modifier MedA, showed a taxonomic distribution similar to stuA (Fig. 2B). The medA homologues of the Pezizomycotina seemed to form a distinct cluster (Supplementary Fig. 9), which does not clarify whether homologues in the Zygomycotina and some Basidiomycotina have a similar role to that seen in A. nidulans. Furthermore, medA mutants show different phenotypes depending on the species. In all cases, the levels of conidiation are affected. Whereas in A. nidulans, deletion of medA produces conidiophores with multiple layers of sterigmata cells (metulae and phialides), in A. fumigatus the medusoid aspect was not observed, although mutants still produced a few conidia (Gems and Clutterbuck, 1994, Gravelat et al., 2010). In Fusarium and Magnaporthe, the deletion of medA produced a switch to acropetal conidiation with aberrant conidiophores (Lau and Hamer, 1998, Ohara et al., 2004). In N. crassa deletion of the medA homologue, acon-3, blocked the budding process resulting in major hyphal constrictions (Springer 1993). In contrast, in U. maydis medA mutants grew normally by budding but were incapable of forming conjugation tubes and filamentation (Chacko & Gold 2012). Taken together, it appears that medA homologues contribute to coordinate the switch between filamentous elongation and budding division, but have contrasting roles depending on the species. In agreement with this, monomorphic yeasts (such as S. cerevisiae and S. pombe) lack a medA homologue whereas fungi belonging to other taxonomic groups, that are capable of developing complex reproductive structures, contain an orthologue.
Central regulatory pathway
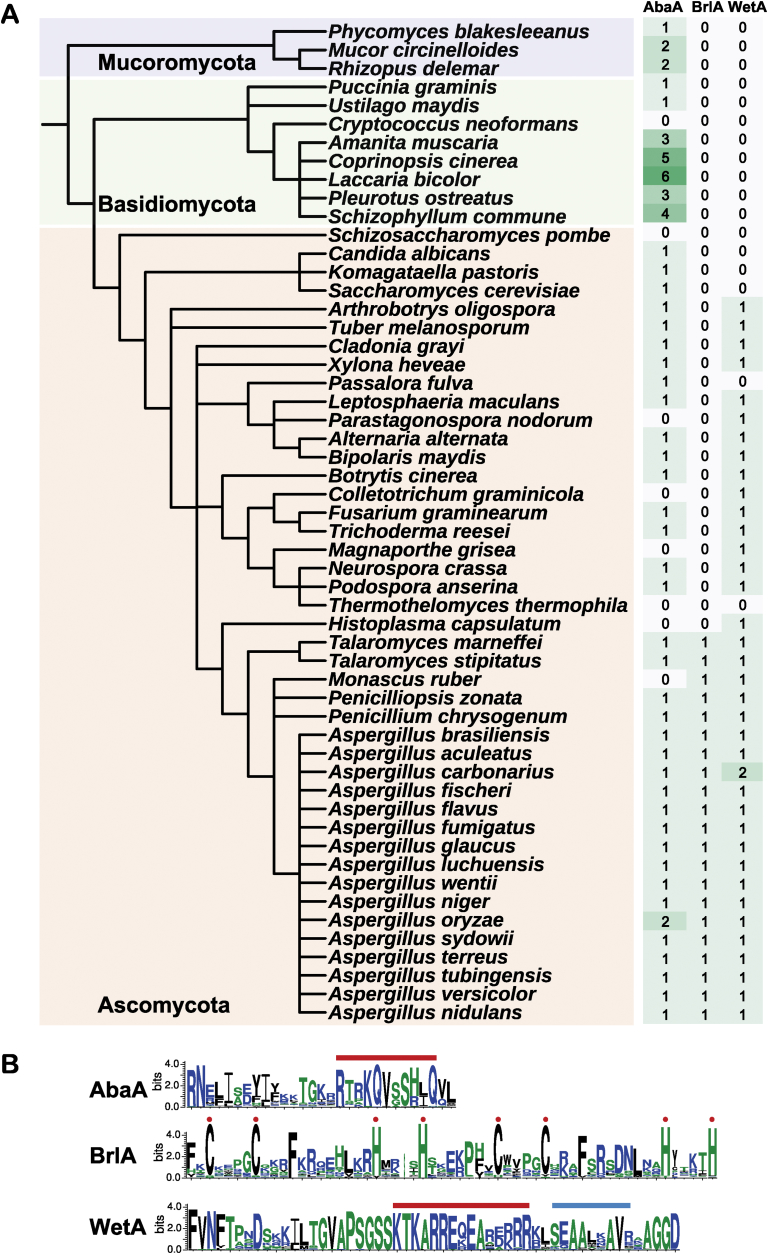
BrlA, AbaA and WetA have been identified as the central regulators for asexual development in A. nidulans (Adams et al., 1998, Park and Yu, 2012). These transcription factors regulate mRNA expression of genes associated with initiation, elongation, and termination of conidiation (Park & Yu 2012). The bioinformatic analysis reported in de Vries et al. (2017) looked especially at the occurrence of the central regulatory pathway (brlA → abaA → wetA) in the Eurotiomycete genomes under investigation. It was found that whereas only one or two elements of the pathway were present in the Ascomycota in general, that all elements of the pathway were present as a central conserved feature in all Aspergillus, Penicillium and Talaromyces species examined. This bioinformatic analysis was extended in the present study to include further outgroup species and also an examination of motifs present in the central regulatory proteins.
It was again found that the central regulatory genes brlA, abaA and wetA are highly conserved in Aspergillus species, and also that conserved DNA binding motifs are present in these central regulatory proteins (Fig. 3A). For example, BrlA orthologues were found to be highly conserved in Aspergillus and Penicillium species and other Eurotiomycetes. However, BrlA was absent from all other Ascomycota, Basidiomycota and Mucoromycota (Fig. 3A). This suggests that BrlA plays a role in the initiation of conidiation specifically in Eurotiales fungi. BrlA orthologues contain a fungal specific C2H2 domain for DNA binding activity (Adams et al. 1988). By contrast, AbaA orthologues could be found in most fungi including both filamentous and yeast-like members of the Ascomycota, suggesting that AbaA is not only involved in conidiophore development but also might have other general functions for fungal morphogenesis. Interestingly, several Basidiomycota such as Schizophyllum commune, Laccaria bicolor, and Coprinopsis cinerea were found to contain more than one AbaA orthologue, whereas A. oryzae was the only member of the aspergilli to contain two AbaA homologues. It was also confirmed that Monascus ruber, a close relative of the aspergilli, lacks an AbaA homologue correlating with a change in conidiation morphology (Hawksworth and Pitt, 1983, Wong and Chien, 1986). AbaA orthologues were found to contain a TEA domain with a DNA binding motif (Burglin, 1991, Andrianopoulos and Timberlake, 1994). TEA domains contain three alpha-helices, two helices with possible DNA binding activity being in the N-terminus of the domain and whose sequences are quite diverse in fungi. The remaining helix is highly conserved in most fungi, and is thought to be a nuclear localisation signal (Fig. 3B, with the conserved RTRKQVSSHLQ sequence shown by a red bar). WetA orthologues were also present throughout the Pezizomycotina. However, Saccharomycetes, including Candida albicans, Pichia pastoris, and Saccharomyces cerevisiae did not contain WetA orthologues (Fig. 3A). WetA orthologues were found to contain a conserved ESC1/WetA-related domain containing a putative 16 amino acid nuclear localization signal and a 9 amino acid transcription activation domain (shown by red and blue bars, respectively, in Fig. 3B) (Marshall and Timberlake, 1991, Son et al., 2014a, Son et al., 2014b, Wu et al., 2017).
Fig. 3.
Orthogroup containing the BrlA, AbaA, and WetA proteins. (A) Distribution of the BrlA, AbaA, and WetA proteins for asexual sporulation across different fungal taxa. (B) Sequence logos of the DNA binding motifs of AbaA, BrlA, and WetA from the fungi examined. Red bars indicate nuclear localization signals (NLS) and the blue bar is the transcription activation domain. Amino acids indicated with red spots are associated with DNA binding activity.
Thus, the central regulatory pathway from A. nidulans appears to be a defining feature of the aspergilli as a whole and it appears likely that the pathway functions in a similar fashion throughout the genus. AbaA and WetA elements of the pathway are also more widely present in the fungal kingdom where it can be speculated that they are also involved in developmental processes such as sporulation, although this awaits experimental confirmation.
Velvet regulatory proteins: phylogenetic distribution and expansion/contraction of the velvet family
Although the velvet proteins have been mainly characterized in Aspergillus, they were found to be present across several different fungal taxa (Fig. 2C). In the aspergilli, all species included in this study contained one copy of veA, velB, velC and vosA, with the exception of A. flavus and A. oryzae, which contain a duplication of vosA (Supplementary Figs 10 & 11). It is rather interesting that T. marneffei, T. stipitatus and P. zonata also have duplications of vosA. There are two possible explanations: an early duplication of vosA in the Eurotiomycetes followed by lost of one of the paralogues in those species that only contain one copy. The second possibility is that independent duplications have led to the vosA paralogues found in these species. This second possibility appears to be more parsimonious due to the following observations. The duplication of vosA seen in A. flavus and A. oryzae is not present in the closely related species A. terreus, which suggests that the duplication occurred after the separation of the A. terreus and A. flavus/A. oryzae clades. The presence of two copies in A. bombycis and A. nomius, which form a monophyletic group together with A. flavus and A. oryzae, but not in A. brevijanus and A. terreus confirms this hypothesis. Indeed, the velC genes from A. flavus and A. oryzae appear to be separated from the rest of the aspergilli velC homologues (Supplementary Fig. 11). Both T. marneffei and T. stipitatus contain vosA paralogues that cluster according to the species, which points to a duplication event occurring independently after the separation of both species. This is further supported by observations of single vosA copies in the other three Talaromyces species available at the Mycocosm site. In particular T. aculeatus is in the same monophyletic group with T. marneffei but not with T. stipitatus, supporting this hypothesis. Taking all these observations above together, it suggests that for unknown reasons vosA has a higher tendency for gene expansions than the other velvet proteins in the Eurotiomycetes. Indeed, using the blastp search tool against the Eurotiomycetes database at the JGI website, we found two independent duplications of veA in Penicillium, no duplications of velB, five independent duplications of velC (two in Aspergillus and three in Penicillium) and 6-7 independent duplications of vosA (two in Aspergillus, one in Penicilliopsis, two in Talaromyces, and 1-2 in Paecilomyces).
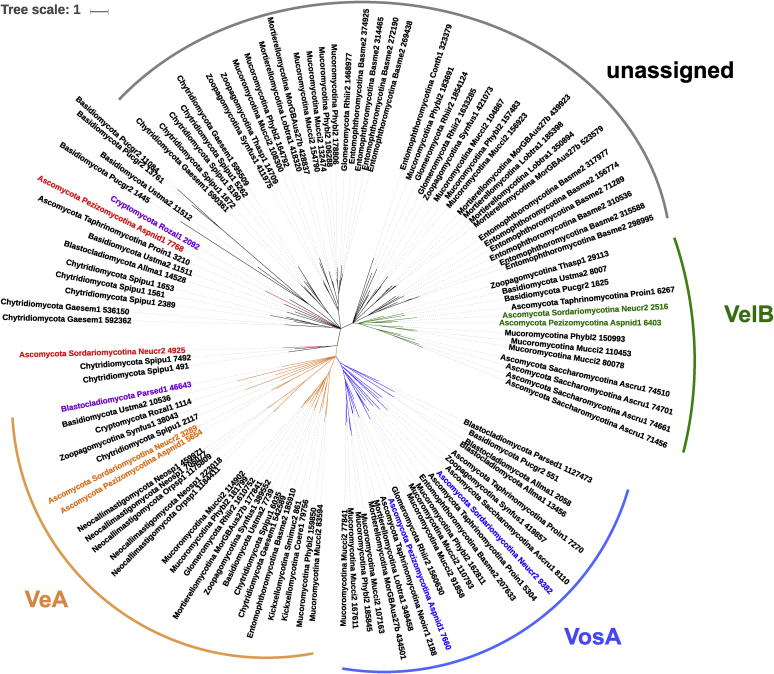
Velvet proteins are specific to fungi (Bayram & Braus 2012) and seem to be widely distributed in this kingdom as they can be found in all the Eurotiomycete species included in this study (Fig. 2C). In order to study in further detail the distribution and evolution of the velvet proteins, we also included early divergent fungi in the analysis, using the velvet domain of AnVeA as a bait to search for homologues in Mycocosm (Grigoriev et al. 2014), and selected all the homologues found from two random species of each fungal phyla (except for the Ascomycota, where the model fungi A. nidulans and N. crassa were purposefully selected and those in which only one species is available in the Mycocosm database) (Fig. 4). Our initial searches and further interrogation using FungiDB (Stajich et al. 2012) could not identify velvet homologues in the 20 species belonging to six different genera of the oomycetes deposited in the databases. Similarly, no homologues could be found in the six species belonging to four different genera of the Microsporidia. However, two velvet proteins were found in the only species of Cryptomycota available on the JGI database, suggesting that either Microsporidia lost their velvet genes or the Cryptomycota have acquired these genes. The two homologues in Cryptomycota are short proteins of 239 and 247 amino acids displaying low similarity between each other (29 % identity and 46 % positives in 205 amino acids according to the blast search), and both contain the velvet domain encompassing most of the protein length. These two copies lie in separate clades in the tree and show a basal location in the branches in agreement with the presumed evolutionary history of the Cryptomycota. One of the clades contains the Cryptomycota velvet protein 1114 and the veA and vosA homologues of A. nidulans and N. crassa (Fig. 4). Homologues in the vosA clade appeared relatively early (in Blastocladiomycota) but seem to be absent in many basal phyla. The vosA and veA clades form a monophyletic group suggesting that vosA may have evolved from veA. Indeed the domain structure of VeA and VosA shares the N-terminal localization of the velvet domain, which is different to the domain organisation seen in VelB and VelC (Bayram & Braus, 2012). The clade containing the other Cryptomycota velvet protein (2092) is not well resolved in the tree and contain subtrees with non-characterized velvet homologues corresponding to basal fungi and basidiomycetes, and another subtree with the velB homologues of A. nidulans and N. crassa. Homologues in the velB clade appear later in Zoopagomycotina. The velC-like homologues encompass a non-monophyletic group of genes that are not well resolved in the tree. Supplementary Fig. 11 also shows a paraphyletic group of the velC-like homologues in the 54 fungal species under analysis, in which the homologues in the Mucoromycota, Basidiomycota and the rest of the Pezizomycotina that do not belong to the Eurotiomycetes form a separate group. A. nidulans velC is located in the base of the so-called unassigned velvets and the velB clade, which makes it difficult to draw conclusions. Available data do not help either to predict a general function for them. For example, deletion of velC does not have any observable phenotype in N. crassa (manuscript in preparation), but it shows a decrease in conidiation, and affects appressoria and plant penetration in M. oryzae (Kim et al. 2014a). By contrast, deletion of velC in A. nidulans produced increased conidiation and reduced number of cleistothecia (Park et al. 2014). In the absence of more genomic sequences of the early divergent fungi of the Cryptomycota and Blastocladiomycota phyla, and homologues in the Microsporidia, it seems that the Cryptomycota homologue in this clade could be the closest form to the origin of the velB/C homologues. The tree shows an increasing expansion of the velvet family from Cryptomycota up to the Mucoromycota and then a contraction in the Basidiomycota and Ascomycota.
Fig. 4.
Phylogenetic tree of the velvet proteins in fungi. The proteins of A. nidulans and N. crassa are coloured as follows: VeA, orange; VelB, green; VelC, red; VosA, blue. The branches of the clades containing those homologues are coloured accordingly. Branches and proteins from the basal fungus of the Cryptomycota phyla are coloured in purple. Black branches represent clades not assigned to any of the above homologues. Branches with bootstrap values lower than 0.8 have been collapsed and are not resolved in the tree.
Overview of asexual development
Taking the results of the bioinformatic analyses above as a whole, a few key observations can be made. Firstly, the fact that many components of the asexual developmental pathway of A. nidulans are absent from members of the Saccharomycotina supports the hypothesis that the difference in their cellular complexity is due in part to the increased diversity in the sporulation machinery seen in the Pezizomycotina (Lengeler et al. 2000). Thus, many species in the Saccharomycotina are unicellular microorganisms, incapable of developing complex multicellular structures (such as conidiophores) or are only able to develop rudimentary ones (e.g. pseudohyphal growth of S. cerevisiae) (Gancedo, 2001, Sudbery, 2011). Upstream regulators of conidiation in the aspergilli are also missing from the Basidiomycotina, many of which undergo only sexual reproduction as part of their life cycle. Secondly, the model derived for asexual development in A. nidulans seems generally applicable to the aspergilli and most Eurotiomycete species, based at least on conservation of the regulatory proteins. Whereas in other members of the Pezizomycotina, homologues of the Aspergillus regulators of conidiation seem to be generally conserved, but perform somewhat different biological roles to accomplish the diverse ontogeny observed in this fungal group. Finally, based on present knowledge it appears that the activation of sporulation seems to be more complex in the aspergilli than in some other taxonomic groupings of the Pezizomycotina, where one or more repressors of conidiation seem to be absent. Considering the evolution of asexual reproduction in the aspergilli, at least two possibilities seem possible. There might have been an acquisition of increased genetic complexity leading to the extant developmental program seen in the aspergilli, and/or the existence of convergent but different genetic strategies to control the onset of sporulation in other taxa. Linked to this, Monascus provides a particularly interesting example as it was found to contain all the upstream genetic regulatory machinery, including the conidiation suppressors and activators of A. nidulans (Fig. 2). It also has homologues for brlA and wetA of the central developmental pathway (Fig. 1, Fig. 3). However, critically it lacks the middle genetic element abaA, which is responsible for the differentiation of the phialides (Sewall et al. 1990). This led to the following section of experimental work.
Genetic manipulation of abaA in Monascus ruber
Members of the genus Monascus are used in the production of Asian foodstuffs and are phylogentically very closely related to the aspergilli (Chen et al., 2015, Chen et al., 2017a, Chen et al., 2017b). However, Monascus has a distinct morphology regarding the development of asexual conidia. Asexual spores are produced either direct from hyphae or produced laterally on short pedicels either singly or in short chains (Hawksworth and Pitt, 1983, Wong and Chien, 1986). The bioinformatic analyses of de Vries et al. (2017) found that the genome of M. ruber contains all the standard genetic regulatory machinery for conidial production seen in the aspergilli. However, it lacks abaA from the central developmental pathway, which is responsible for the differentiation of the phialides in A. nidulans (Sewall et al. 1990). The conidiophore in M. ruber M7 can be likened to a single string of an abacus on the vesicle, whereas that in A. nidulans resembles several strings of an abacus emerging from a swollen vesicle and the phialides i.e. the conidiophore of M. ruber differs in that it lacks the production of a swollen vesicle and the metulae and phialides seen in A. nidulans. It was therefore tempting to speculate that the presence or absence of the abaA gene might be a significant contributory factor to the difference in conidiation form between Monascus and the aspergilli. We therefore examined whether heterologous expression of A. nidulans abaA in M. ruber might lead to a change in conidiation form, perhaps similar to that seen elsewhere in the Eurotiales.
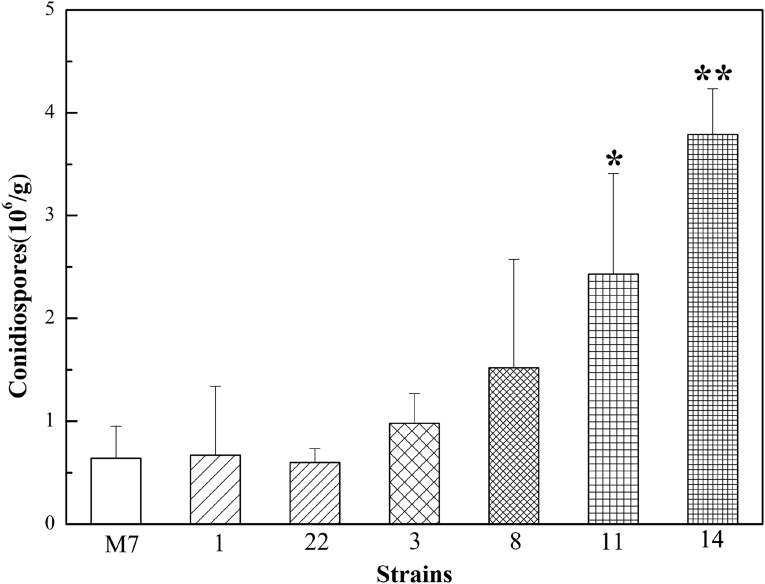
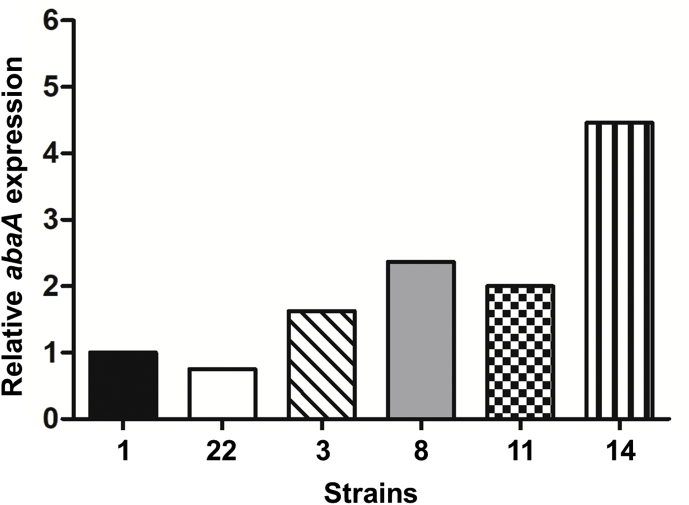
A total of 21 M. ruber transformant strains were obtained in which expression of A. nidulans abaA was confirmed by PCR, cDNA sequencing and Southern blotting (Supplementary Figs 13 & 14). Nine were found to have one copy, ten possessed two copies, and two contained three copies of abaA. Among these, two strains (1 and 22) with one copy of abaA, two strains (3 and 8) with two copies, and two strains (11 and 14) with three gene copies were selected for further investigation. In order to verify the relative expression level of abaA in the selected mutants, quantitative real-time RT-PCR was performed. Results showed that the relative expression level was positively correlated with abaA gene copy number (Supplementary Fig. 15). The conidial morphology was then examined. This revealed that among the six abaA expression strains, most conidiophores were similar to the M7 parent, with no obvious change in micro-morphology or colony macro-morphology, although exceptionally a small number of conidiophores were observed in which one to three conidia were born in two-three-way branches at the top of vesicles (Fig. 5; Supplementary Fig. 16). By contrast, conidial counts showed that production of conidia was significantly increased in some mutants, which was positively correlated with abaA gene copy number, compared to the parental WT (Fig. 6). There was also evidence of earlier germination rates and increased resistance of spores to external stressors in the abaA expression strains, as well as changes in the proteome as a result of abaA expression (Supplementary Figs 17–19).
Fig. 5.
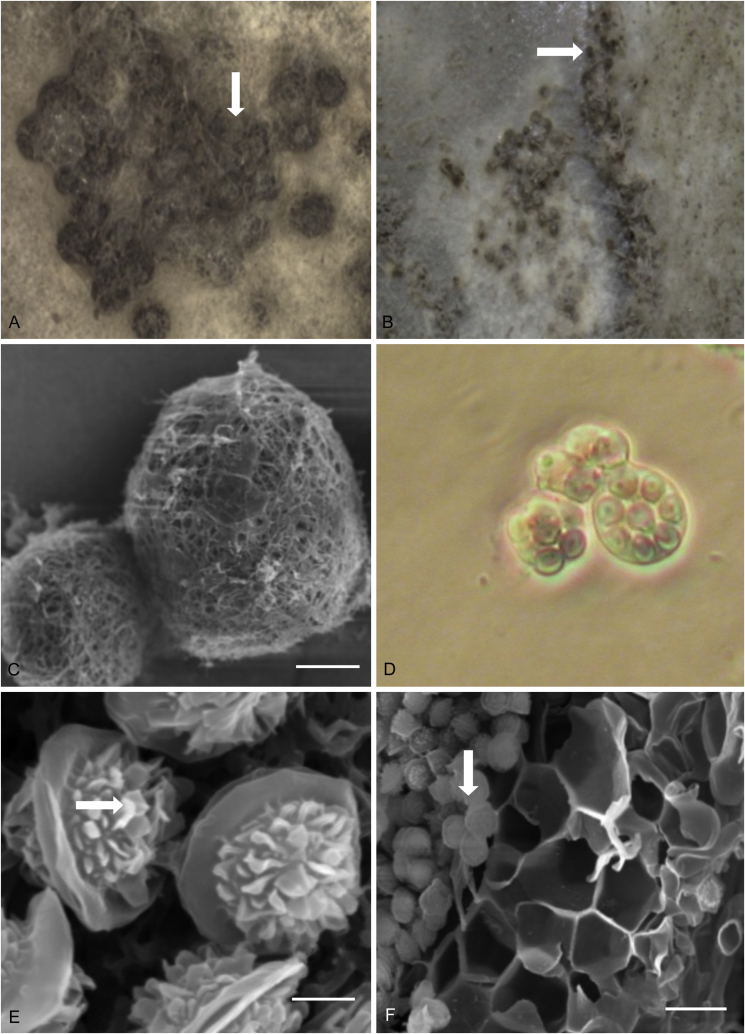
Conidial morphology of wide-type M. ruber M7 (two images) and transformants (1, 22, 3, 8, 11, 14) (three images) expressing one to three copies of the A. nidulans abaA gene. Conidiophores are circled in red, with magnified images shown by the inset arrow. Strains were incubated on PDA (potato dextrose agar) medium at 28 °C, with pre-sterilized glass microscope oblique cutting in the medium. After 4 d, coverslips were taken to observe the conidia morphology under the microscope. Scale bars as indicated.
Fig. 6.
Comparison of conidia production between M. ruber isolate M7 and transformant strains expressing the A. nidulans abaA gene. Strains were cultivated on PDA plates for 10 d before harvesting and counting of conidia. Error bars represent SD. ANOVA analysis of conidial counts was performed, with statistically significant differences between M. ruber isolate M7 and the transformant strains indicated: * represents p < 0.05 and ** represents p < 0.001.
In conclusion, the heterologous expression of abaA in M. ruber had some effect on conidial formation, but it failed to lead to a branching conidiophore form as seen in Aspergillus or Penicillium. Given that AbaA is present in many other members of the Pezizomycotina (Fig. 3) it seems that most likely that gene loss has occurred in the ancestor of Monascus that diverged from an ancestor of the aspergilli, and that Monascus species have then adapted the regulation of the central pathway accordingly. Further evidence for this hypothesis is that the Monascus genome includes brlA, which otherwise only has a narrow distribution in the Eurotiales.
Evolution of sexual breeding systems in Aspergillus
There has been longstanding interest in the evolution and control of sexual reproduction in the fungal kingdom since the earliest reports of different sexes and self-fertility in fungi by Blakeslee in the early 1900s, who introduced the terms homothallism and heterothallism (Houbraken & Dyer 2015). Since then the study of fungal sex has been used to gain insights into the evolution of sex and transitions between self-fertility and cross-fertility that occur throughout the eukaryotic tree of life (Lee et al., 2010, Heitman et al., 2013, Heitman, 2015).
Given that both homothallic and heterothallic breeding systems are widespread in the fungal kingdom, one particular question that has arisen and long-been debated in fungal biology is whether homothallism or heterothallism might be the ancestral sexual state (e.g. Whitehouse, 1949, Metzenberg and Glass, 1990). This is both of fundamental interest, but also has practical ramifications for the exploitation of fungal sex for breeding purposes (Ashton & Dyer 2016). It has been argued that given the long time scales and vast evolutionary distances separating extant species from common evolutionary ancestors, that at best any features of present day sex will be derived. Despite this, it is suggested that the original form of sexual reproduction may have been unisexual (unifactoral), with sexes superimposed as a later feature (Nieuwenhuis et al., 2013, Heitman, 2015). In practice then, models for the evolution of sexual breeding system might be at best, and most reliably, applied with any certainty to related groups of extant taxa. It is also noteworthy that investigations into the evolution of sex in fungi have been greatly assisted over the past 20 years by the molecular identification of mating-type loci, which have been found to be responsible for transitions in modes of sexual reproduction (Heitman et al., 2013, Dyer et al., 2016).
In the case of the Pezizomycotina, different models for the evolution of sexual breeding systems were proposed in the 1990s. The fact that the vast majority of known Aspergillus sexual species are homothallic, combined with phylogenetic reconstruction analysis, led Geiser et al., 1996, Geiser et al., 1998 to propose that this group was derived from a homothallic ancestor. This contrasted with evidence from Cochliobolus species that evolution of homothallism from a heterothallic ancestral strategy was more likely (Yun et al. 1999). This was based on the observation that whereas heterothallic species from the genus exhibited a consistent, conserved arrangement of mating-type genes at the MAT locus, that homothallic species instead had a variable arrangement of MAT genes both in terms of gene arrangement, order and orientation. It was therefore argued that the most parsimonious explanation was that homothallic species arose independently from heterothallic ancestors sharing a common MAT locus structure, accounting for the subsequent variation in homothallic MAT locus arrangement but consistent heterothallic MAT arrangement (Yun et al. 1999). There was also further evidence of sequential MAT gene insertions conferring homothallism in some species. It was later suggested that heterothallism is also the most likely ancestral mating state of members of the genus Stemphylium, which is closely related to Cochliobolus (Inderbitzin et al. 2005). It was hypothesized that homothallic members had arisen by an inversion and fusion event of an ancestral heterothallic MAT loci. In parallel it has been suggested that the ancestor of all extant ascomycete yeast species may have had a heterothallic mating strategy (Butler 2007).
Therefore the genus Aspergillus seemed to be the exception in having arisen from a homothallic ancestor. This apparent anomaly was investigated in the bioinformatic analysis of de Vries et al. (2017), where it was found that all of the presumed asexual species were found to contain either a MAT1-1-1 or MAT1-2-1 mating-type gene, consistent with the presence of either MAT1-1 or MAT1-2 idiomorphs. Adjacent gene synteny was also conserved across all species, again consistent with heterothallism (Dyer et al. 2016). This indicated that heterothallism might be widespread in the aspergilli, bringing into question the supposed homothallic origins of the genus. In the present study, MAT loci were therefore experimentally cloned from a further series of representative heterothallic and homothallic Aspergillus species, to determine whether observations of MAT locus structure could provide a more definitive insight following the approach of Yun et al. (1999).
Identification of MAT loci from A. heterothallicus, A. fennelliae and A. pseudoglaucus
In the case of the heterothallic A. heterothallicus, putative MAT-1-1, MAT1-2-1, SLA2 and APN2 gene fragments were successfully amplified using PCR with degenerate primers. Utilising the bridging strategy, it was then possible to amplify an entire MAT1-1 idiomorph region from isolate 50-5 containing a putative MAT1-1-1 gene, flanked by the SLA2 and APN2 genes (Fig. 7). Sequence analysis of the region revealed the presence of a 1 139 bp open reading frame (ORF), including one putative intron, which was predicted to encode a 362 amino acid MAT1-1-1 protein with a characteristic alpha-box domain (GenBank accession MH401192). Analysis of the putative MAT1-1 protein revealed no clear nuclear targeting signals. Similarly, the bridging strategy was also used to amplify an entire MAT1-2 idiomorph region from isolate 50-3 containing a putative MAT1-2 gene, again flanked by the SLA2 and APN2 genes (Fig. 7). Sequence analysis of the region revealed the presence of a 1 075 bp ORF including two introns, which was predicted to encode a 321 amino acid MAT1-2 protein (GenBank accession MH401191). Analysis of the putative MAT1-2 protein revealed three nuclear targeting signals (KKKH at position 182, RKRR at position 202 and PSERKRR at position 199) upstream of the start site. Possession of nuclear targeting sites is consistent with a role of the MAT1-2-1 gene as a transcriptional activator (Dyer et al. 2016). Further analysis of the MAT1-2 idiomorph region of isolate 50-3 also revealed, intriguingly, the presence of an additional putative ORF which shared high homology with the recently described MAT1-2-4 gene identified from A. fumigatus (Yu et al. 2017). The A. heterothallicus MAT1-2-4 gene was located between the SLA2 and MAT1-2-1 gene (Fig. 7) and comprised a 771 bp ORF (including one putative intron), which was predicted to encode a 242 amino acid MAT1-2-4 protein (GenBank accession MH401191). Analysis of the MAT1-2-4 protein revealed no clear targeting signals, only a transcriptional activator TATA box sequence 35 bp upstream of the start site.
Fig. 7.
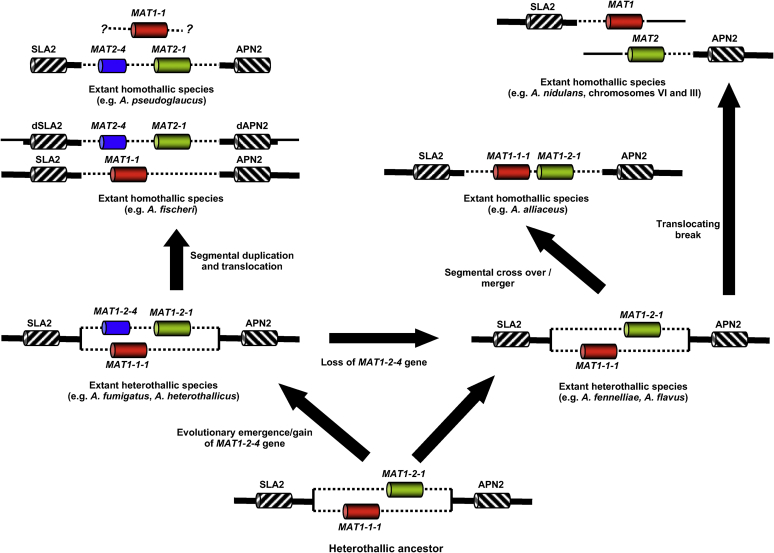
Model proposed to explain the evolution of MAT loci and breeding systems in Aspergillus species from a heterothallic ancestor (not to scale). Adapted in part from Dyer (2007). Mating-type genes are shown in colour: MAT1-1-1 family α-domain in red, MAT1-2-1 HMG family in green, and MAT1-2-4 family in blue. Note that gene nomenclature varies between heterothallic and homothallic species due to presence of idiomorphs only in the former species. Flanking genes (SLA2 and APN2) are shown by diagonal hatching. Dotted lines indicate idiomorph region; heavy bold lines indicate conserved sequence flanking the idiomorph region; suffix ‘d’ indicates disabled pseudogene (Rydholm et al. 2007). Note that the illustration does not show all genes present in the flanking regions (e.g. an APC gene is also present in some species, but syntenic order varies according to species). Furthermore, only limited sequence is available from the A. pseudoglaucus MAT1-1 gene region, as indicated by question marks.
The bridging strategy, using PCR with degenerate primers of MAT-1-1, MAT1-2-1, SLA2 and APN2 and consequent chromosome walking, was also used successfully to amplify entire MAT1-1 and MAT1-2 idiomorph regions from heterothallic A. fennelliae isolates 54-1 and 54-2, respectively. These MAT loci were again flanked by the SLA2 and APN2 genes (Fig. 7). Sequence analysis of the MAT1-1 idiomorph revealed the presence of a 1 160 bp ORF containing one putative intron, which was predicted to encode a 369 amino acid MAT1-1-1 protein (GenBank accession MH401193). Analysis of the putative MAT1-1-1 protein revealed one nuclear localisation sequence (KKKP at position 82), consistent with a role for the MAT1-1-1 gene as a transcriptional activator (Dyer et al. 2016). Analysis of the MAT1-2 idiomorph revealed a 1 072 bp ORF, containing two putative introns, which was predicted to encode a 322 amino acid MAT1-2-1 protein (GenBank accession MH401194). Analysis of the putative MAT1-2-1 protein revealed three nuclear localisation signals (KKKH at position 183, RKRR at position 203 and PSERKRR at position 200). However, unlike A. heterothallicus no MAT1-2-4 gene was found in the region adjoining the MAT1-2-1 gene.
Finally, the bridging strategy was used successfully to identify a 9 437 bp SLA2 to APN2 region from isolates 51-1 and 51-2 of the homothallic A. pseudoglaucus (Fig. 7). This region was found to contain a 1 078 bp MAT1-2-1 gene homologue, which contained two putative introns and was predicted to encode a 321 amino acid MAT1-2-1 protein (an alternative possible ATG start site was also detected 7 amino acids inwards of the proposed MAT1-2-1 start site) (GenBank accession MH401195). Analysis of the putative MAT1-2-1 protein revealed three nuclear targeting signals (KKKH at position 183, KKRR at position 203 and PYEKKRR at position 200 upstream of the start site). Further analysis of the MAT region also revealed the presence of a putative MAT1-2-4 gene homologue, containing one putative intron and which was predicted to encode a 242 amino acid MAT1-2-4 protein (Fig. 7), as reported by Yu et al. (2017). However, no evidence of a MAT1-1-1 gene was found in the SLA2 to APN2 region. Despite this, a 151 bp fragment of a putative MAT1-1-1 family gene was successfully amplified from A. pseudoglaucus by PCR with degenerate primers MAT5-6 and MAT3-4. TAIL-PCR was therefore used to chromosome walk upstream and downstream of this fragment. In total, 1 598 bp of sequence from the MAT region was obtained, which was found to include a 1 125 bp putative MAT1-1-1 family gene, which contained one putative intron and was predicted to encode a 356 amino acid MAT1-1-1 family protein (GenBank accession MH401196). Analysis of the putative MAT1-1-1 family protein revealed three nuclear targeting signals (KKRR at position 83, KRRR at position 84 and RRRP at position 85) upstream of the putative start site. There was no obvious sequence homology to the previous SLA2-APN2 A. pseudoglaucus MAT gene region. Given the presence of two apparently independent MAT loci, these regions were therefore named MAT1 (containing the alpha domain encoding gene) and MAT2 (containing the HMG-domain encoding gene) to recognise their separate locations, consistent with the nomenclature of A. nidulans and A. (Neosartorya) fischeri as recommended by G. Turgeon (Fig. 7) (Turgeon and Yoder, 2000, Paoletti et al., 2007, Rydholm et al., 2007). As a result the MAT1-1-1 alpha domain family gene was named simply MAT1-1 (as there were no alternative idiomorphs in this species), the MAT1-2-1 HMG domain family gene was named MAT2-1, and the novel MAT1-2-4 family gene was named MAT2-4 (Fig. 7) for consistency with previous work (Turgeon and Yoder, 2000, Paoletti et al., 2007, Rydholm et al., 2007, Yu et al., 2017). For further background see Wilken et al. (2017), who have recently proposed updated nomenclature for MAT genes.
RNA expression studies were also undertaken with all of the MAT genes identified from A. heterothallicus, A. fennelliae and A. pseudoglaucus. All of the genes were found to be expressed under the conditions assayed, except for the MAT1-2-4 genes of A. heterothallicus and A. pseudoglaucus (Eagle 2009).
Implications of MAT loci structure for evolution of sex in the aspergilli
Results of the present study provided clear evidence that the heterothallic A. heterothallicus and A. fennelliae shared the same general genomic arrangement of MAT loci as seen previously in the heterothallic A. fumigatus and the asexual aspergilli studied by de Vries et al. (2017) (Fig. 7). The term ‘proto-heterothallic’ has been suggested to be used for such latter species where evidence of heterothallism is present, but a sexual cycle has yet to be demonstrated (Houbraken & Dyer 2015). By contrast, results from the homothallic A. pseudoglaucus added further evidence of a variety of MAT gene arrangements seen in homothallic Aspergillus species (Fig. 7). For example, there is evidence of a translocating break leading to the arrangement of MAT loci in A. nidulans (Galagan et al., 2005, Paoletti et al., 2007), both alpha- and HMG-domains at the same single MAT locus in A. (Petromyces) alliaceus (Ramirez-Prado et al. 2008), and a localised MAT region duplication and then translocation in A. fischeri (Rydholm et al. 2007). The precise situation in A. pseudoglaucus has yet to be determined as only limited sequence could be cloned adjacent to the MAT1-1 gene, but there would appear to be two independent MAT loci present. Following the logic of Yun et al. (1999), it can therefore be strongly argued that it is most likely that the common ancestor of the aspergilli exhibited a heterothallic breeding species. This is on the basis of the general consistency of the heterothallic MAT locus arrangement in the aspergilli, but divergence in the homothallic MAT arrangement (Fig. 7). Thus, it can be envisaged that new Aspergillus species arose as sub groups, containing both MAT1-1 and MAT1-2 isolates, which gradually diverged from each other. Within such groups there might then be occasional evolutionary selection for homothallism (e.g. Murtagh et al. 2000) and the different forms of homothallic MAT loci would then arise as a result of spontaneous mutation, accounting for the inconsistency in their organisation.
One caveat to this conclusion is the recent discovery of the MAT1-2-4 gene in the MAT1-2 idiomorph of a diverse taxonomic range of 10 Aspergillus species including A. fumigatus and A. pseudoglaucus (Yu et al. 2017), and now A. heterothallicus as well (Fig. 7). The gene was shown to be functional in A. fumigatus where gene deletion led to an inability to mate (Yu et al. 2017). However, this gene has not so far been detected or described from most other aspergilli (including A. nidulans) and was absent from the MAT1-2 idiomorphs of A. fennelliae and A. clavatus sequenced in the present study. Given the taxonomic divergence between A. fumigatus, A. pseudoglaucus, A. heterothallicus and other species where the gene has been detected [e.g. A. versicolor and A. carbonarius; Yu et al. (2017)] it might be expected that MAT1-2-4 gene was a conserved ancestral feature of the MAT loci of the aspergilli. So, one possibility is that there have been multiple independent losses of this gene in the evolutionary history of the group (Fig. 7). A number of MAT genes specific to certain groups of the Pezizomycotina have now been described (Wilken et al. 2017). A further caveat is that a fragment of the MAT1-2 gene was found bordering the MAT1-1 idiomorph of A. fumigatus (Paoletti et al. 2005) and the related A. lentulus (Swilaiman et al. 2013), which could indicate evolution from a homothallic ancestor containing both MAT1-1 and MAT1-2 genes (Galagan et al. 2005). However, bioinformatic analysis of the MAT regions of many other aspergilli indicates this to be an unusual occurrence thereby not discrediting the hypothesis of a common heterothallic ancestor. One final consideration is that the apparent predominance of homothallism in the aspergilli is due to the considerable bias in the numbers of homothallic species with Emericella and Eurotium sexual states that have been described [see Table 1 of Dyer & O'Gorman (2012)]. If these are excluded and asexual species included, then there is a bias instead towards heterothallism.
Table 1.
Number of cleistothecia produced by crosses of A. clavatus on oatmeal agar medium at 25 °C in the dark after 10 weeks.
| Crosses | Number of cleistothecia |
|||
|---|---|---|---|---|
|
MAT1-2 | ||||
| 65-16 | 65-19 | 65-20 | ||
| MAT1-1 | 65-2 | + | + | − |
| 65-7 | − | + | − | |
| 65-8 | + | − | − | |
| 56-10 | + | − | − | |
| 65-14 | +++++ | + | − | |
| 65-18 | ++++ | + | − | |
Ratings indicate the mean number of cleistothecia produced from three replicate crosses on Oatmeal agar in 9 cm diameter Petri dishes: -none; +,1–19; ++, 20–39; ++++, 60–79; +++++, 80–100.
Asexuality in the aspergilli and sexual reproduction in A. clavatus
Despite the many supposed benefits of sexual reproduction, approximately 20 % of all fungal species are only known to reproduce by asexual means (Hawksworth et al., 1995, Taylor et al., 1999, Dyer and Kück, 2017). The genus Aspergillus is particularly well known for the predominance of asexual species. Based on the presence of meiotic and mitotic taxa in a series of different Aspergillus phylogenetic clades, Geiser et al. (1996) suggested that asexual fungi are recent derivatives from older meiotic lineages. However, although there might be short-term benefits there would also be long-term costs and Geiser et al. (1996) suggested that the asexual lineages would be more susceptible to extinction. Thus, the aspergilli have been seen to provide a model for the evolution of asexuality in fungi. However, there have been a number of breakthroughs over the past decade indicating that asexuality might not after all be dominant in the genus Aspergillus. Based on results of population genetic analyses, the presence of sex-related genes (as detected in genome sequencing projects), the presence of isolates of complementary mating type, and the induction of sexual reproduction in certain high-profile ‘asexual’ species, it has been argued that a ‘sexual revolution’ is occurring in the genus Aspergillus and the closely related genus Penicillium (Dyer & O'Gorman 2011). As a result, the prevalence of asexuality in the genus is being questioned.
To investigate whether asexual species might have a cryptic sexual cycle, de Vries et al. (2017) investigated whether ‘sex-related’ genes involved with mating processes were present and functionally expressed in supposed ‘asexual’ aspergilli. All of the presumed asexual species examined were found to contain either a MAT1-1 or MAT1-2 idiomorph as well as genes encoding putative pheromone receptors and a pheromone precursor. Furthermore, when the species were grown under conditions conducive to sexual reproduction in the aspergilli it was found that the mating-type, pheromone precursor and receptor genes were expressed in all of the asexual species in the same way as known sexual species. These results suggested the possibility of inducing the sexual cycle in species of applied importance.
This work was extended in the present study by assessing whether it was possible to induce sexual reproduction in the supposed asexual species A. clavatus (Varga et al. 2007). The MAT PCR diagnostic using primer pairs AclM1F with AclM1R, or AclM2F with AclM2R successfully amplified putative MAT gene fragments from all 20 worldwide isolates of A. clavatus. Amplicons of the predicted 244 bp size for MAT1-1 genotypes and 388 bp for the MAT1-2 genotypes were produced in different isolates (Supplementary Fig. 20) indicating a heterothallic breeding system. The overall mating-type distribution did not deviate significantly from a 1:1 ratio [(45 % MAT1-1) n = 9, 55 % MAT1-2 n = 11; Χ2 = 0.80; n = 20; P = 0.654, (p > 0.05)]. When isolates were grouped according to geographic origin there was also no significant difference in the MAT distribution. Based on the equal distribution of mating types, it can be assumed that these populations previously or currently are propagating sexually in their original habitats (Dyer & O'Gorman 2012).
Nine isolates of A. clavatus (six MAT1-1 and three MAT1-2) from different geographic origins were then crossed in all possible pairings under a range of different temperatures on oat meal agar, which had previously been used to induce the sexual cycles of A. fumigatus and A. lentulus (O'Gorman et al., 2009, Swilaiman et al., 2013). Significantly, after four weeks of incubation cleistothecia were observed in three crosses at temperatures between at 25 °C to 30 °C; all contained asci and ascospores when crushed. The cleistothecia formed along the barrage zones between isolates of opposite mating types (Fig. 8). Cleistothecia were superficial, subglobose to ovoid (315−[513]−692 μm), hard, yellowish-brown saffron colour (fawn), uniloculate, nonostiolate covered by dense aerial hyphae, maturing gradually from the centre outward after 4 wk; covered by dense aerial hyphae stromatal peridium. Asci were 8-spored irregularly disposed globose to subglobose, and ascospores hyaline, lenticular (6.0−[6.5]−7.0 μm), variously sculptured, with two equatorial crests. The cleistothecia were similar to those seen in Aspergillus acanthosporus in which the species produce hard and sclerotioid, fawn nonostiolate, unilocular stromata that take 3–4 wk to mature and which at maturity contain hyaline ascospores that have two equatorial ridges (Udagawa & Uchiyama 2002).
Fig 8.
A, B. Sexual reproductive structures in A. clavatus. Paired cultures of isolates 65-14 × 65-16 on oatmeal agar showing formation of fawn to dark brown cleistothecia (arrowed) along the barrage zones following four weeks incubation at 25 °C. C. SEM micrograph of a cleistothecium showing the interwoven hyphae that form the peridial wall. Scale bar = 100 μm. D. A photomicrograph of 8-spored asci. E. SEM micrograph of lenticular ascospores (white arrow) Scale bar = 1 μm. F. Close-up of the peridium of interwoven hyphae with group of ascospores, (white arrow). Scale bar = 10 μm.
Cleistothecia continued to develop in other crosses such that after 10 wk- of incubation cleistothecia were formed in 9 of the attempted 18 crosses (Table 1). Greatest fertility, in terms of number of cleistothecia, was observed in cultures incubated at 25 °C. This was slightly below the optimum temperatures previously described for A. lentulus and A. fumigatus of 28 °C and 30 °C, respectively (O'Gorman et al., 2009, Swilaiman et al., 2013). No additional cleistothecia were observed in any of the crosses at any of the three incubation temperatures when cultures were incubated for a further five months. A few features of the crossing data were noteworthy. Firstly, where cleistothecia formed there was a large variation in isolate fertility depending on the mating partners, with isolate 65-16 (MAT1-2) generally producing the highest number of cleistothecia whilst one isolate (from India) was sterile with all mating partners (Table 1). Secondly, there were relatively few cleistothecia produced overall, with 78 % of the nine fertile crosses producing less than 20 cleistothecia per 9 cm Petri dish, and no crosses producing more than 100 cleistothecia under the incubation conditions. Finally, some crosses were more flexible in their temperature requirement for crossing than others. For example, isolates 65-2, 65-8, 65-10, and 65-14 produced cleistothecia at both 25 and 28 °C, whereas isolates 65-7 and 65-18 were fertile only at 25 °C.
An analysis of recombination was then conducted to confirm that meiosis had taken place, using ascospore-derived progeny from the representative cross 65-14 × 65-16. There was clear evidence of recombination of genetic markers as a result of the sexual cycle. Analysis of the progeny using the PCR mating-type diagnostic revealed a near 1:1 segregation ratio of mating type [MAT1-1:MAT1-2 = 6:6; χ2 = 0.33; p = 0.563, (p > 0.05)]. Moreover, the RAPD analysis showed that the majority of these progeny had recombinant genotypes based on four RAPD-PCR markers. Combining the MAT and the RAPD data revealed that 83 % of the progeny had unique genotypes (Table 2). A representative gel of segregation patterns of the RAPD markers is shown in Supplementary Fig. 21. Note that loss of RAPD markers was observed in some offspring as described elsewhere (Dyer et al. 1996).
Table 2.
Genotypes1 in the parental isolates and 12 ascospore progeny of a cross between A. clavatus isolates 65-14 × 65-16.
| Isolate | Mating type | RAPD band2 |
Genotype3 |
|||
|---|---|---|---|---|---|---|
| OPC 20 | OPT 18 | UBC 90 | OPQ6 | |||
| 65-14 | MAT1-1 | − | − | + | + | P1 |
| 65-16 | MATI-2 | + | + | − | + | P2 |
| 14-16-2 | MAT1-2 | + | + | − | − | A |
| 14-16-4 | MAT1-1 | + | − | + | + | B |
| 14-16-5 | MAT1-1 | − | + | + | − | C |
| 14-16-7 | MAT1-2 | + | + | − | − | D |
| 14-16-8 | MAT1-2 | − | + | + | + | E |
| 14-16-9 | MAT1-2 | − | − | + | + | F |
| 14-16-10 | MAT1-1 | + | − | − | − | G |
| 14-16-11 | MAT1-2 | + | + | − | − | A |
| 14-16-13 | MAT1-1 | + | − | − | + | H |
| 14-16-14 | MAT1-1 | − | − | + | + | P1 |
| 14-16-18 | MAT1-1 | − | − | − | + | J |
| 14-16-19 | MAT1-1 | − | − | + | + | P1 |
| P-value(2- tailed)4 | 1.00 | 0.072 | 1.00 | 0.56 | ||
| Contingency χ25,6 | 0.771 (1) | |||||
Genotypic characterisation based on mating type and RAPD-PCR bands.
RAPD-PCR bands amplified using Operon primers, OMT1 or R108 . ‘+’ and ‘−’ denotes presence or absence, respectively, of particular amplicons.
The genotype of each progeny isolate, defined by unique combinations of mating-type and RAPD markers as distinct from the parental isolates (designated P1 and P2), is identified by a different letter of the alphabet.
Fisher's exact test for deviation from the null hypothesis of independent assortment of mating-type and RAPD markers in the progeny (i.e. a 1:1:1:1 MAT1-1+:MAT1-1-:MAT1-2+:MAT1-2- ratio for each RAPD marker). Fisher's exact test was used instead of the χ2 test because the expected frequencies were <5.
To test for deviation from the null hypothesis of independent assortment of mating-type and RAPD markers in the progeny (i.e. an overall 1:1:1:1 MAT1-1+ :MAT1-1-:MAT1-2+:MAT1-2-ratio for the sum of the RAPD markers).
Number in parenthesis indicates the degree of freedom.
The discovery of sexual reproduction in A. clavatus is significant because it provides further evidence that sexual reproduction might be possible in supposedly ‘asexual’ aspergilli and beyond if partners of compatible mating type and the correct environmental conditions can be identified. Therefore, the original argument of the apparent evolution of asexuality in fungi based on the prevalence of asexuality in the aspergilli (Geiser et al. 1996) has been superseded given new evidence about the possibilities for sexual reproduction in these asexual aspergilli from recent (Dyer and O'Gorman, 2011, Dyer and O'Gorman, 2012) and the present studies. Indeed, the discovery of sexual reproduction in A. clavatus is consistent with phylogenetic work demonstrating that the known sexual species A. acanthosporus clusters together with A. clavatus, which hinted at the possibility of sexual reproduction in the latter species (Peterson 2000). In addition, one species in the section Clavati, Aspergillus ingratus, has been reported to produce saffron-coloured sclerotia when incubated in the dark, representing a possible stage of sexual development (Yaguchi et al. 1993). It is noted under the ‘one fungus, one name’ convention (Hawksworth et al. 2011) that no new Neocarpenteles or other name is now presented for A. clavatus, instead the original A. clavatus epithet is applied to the holomorph.
Conclusions
Members of the genus Aspergillus will no doubt continue to be used as model fungi for a variety of reasons such as their ease of growth under laboratory conditions, the availability of classical genetic and molecular resources, and the economic and biotechnological importance of many of the species (Bennett 2010). Overall results of the present study indicate that results gained with A. nidulans can provide insights into asexual and sexual developmental processes certainly within the aspergilli, and also to the broader fungi kingdom to some extent. However, the growing appreciation of genome diversity in fungi indicates that caution must be exercised before making assumptions based simply on studies in A. nidulans. Also it cannot be assumed that just because homologues of genes are present in different taxa that they have the same functional/mechanistic action. This normally requires some experimental validation. There is also one final irony from the present study. Although some previous theories about the evolution of sexual reproduction in the aspergilli now appear to be incorrect and not applicable to fungi in general, new discoveries about the potential for sexual reproduction in asexual species in the aspergilli now provide a novel model for fungi in general. Therefore Aspergillus has arguably reclaimed its role in the vanguard of fungal biology in this instance.
Acknowledgments
WC was supported by the National Natural Science Foundation of China (No. 31601446). The work by HSP was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIP: No. 2016010945). The work at UW was primarily supported by the Intelligent Synthetic Biology Center of Global Frontier Projects (2015M3A6A8065838). PSD and CEE thank the Biotechnology and Biological Sciences Research Council (UK) for funding. SSS was supported by a Government of Iraq PhD scholarship. The work by DC was supported by the Spanish Ministry of Economy and Competitiveness (MINECO, grant number BIO2015-67148-R). PSD also thanks Jos Houbraken and Rob Samson (CBS/Westerdijk Fungal Biodiversity Institute, The Netherlands) for the donation of A. pseudoglaucus 51-2, and Arun Balajee (CDC, USA) for cultures of A. clavatus. GG thanks David Emms (Department of Plant Sciences, Oxford University) and Santiago Melchor (CSIRC, University of Granada) for help to set up and run Orthofinder in the supercomputing cluster HPC Alhambra.
Footnotes
Peer review under responsibility of Westerdijk Fungal Biodiversity Institute.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.simyco.2018.10.002.
Contributor Information
D. Cánovas, Email: davidc@us.es.
P.S. Dyer, Email: paul.dyer@nottingham.ac.uk.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. s1.
References
- Adams T.H., Boylan M.T., Timberlake W.E. brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell. 1988;54:353–362. doi: 10.1016/0092-8674(88)90198-5. [DOI] [PubMed] [Google Scholar]
- Adams T.H., Wieser J.K., Yu J.H. Asexual sporulation in Aspergillus nidulans. Microbiology and Molecular Biology Reviews. 1998;62:35–54. doi: 10.1128/mmbr.62.1.35-54.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed Y.L., Gerke J., Park H.S. The velvet family of fungal regulators contains a DNA-binding domain structurally similar to NF-κB. PLoS Biology. 2013;11 doi: 10.1371/journal.pbio.1001750. e1001750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrianopoulos A., Timberlake W.E. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Molecular Cell Biology. 1994;14:2503–2515. doi: 10.1128/mcb.14.4.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramayo R., Peleg Y., Addison R. Asm-1+, a Neurospora crassa gene related to transcriptional regulators of fungal development. Genetics. 1996;144:991–1003. doi: 10.1093/genetics/144.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arie T., Christiansen S., Yoder O. Efficient cloning of ascomycete mating-type genes by PCR amplification of the conserved MAT HMG box. Fungal Genetics and Biology. 1997;21:118–130. [PubMed] [Google Scholar]
- Arratia-Quijada J., Sánchez O., Scazzocchio C. FlbD, a Myb transcription factor of A. nidulans, is uniquely involved in both asexual and sexual differentiation. Eukaryotic Cell. 2012;11:1132–1142. doi: 10.1128/EC.00101-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashton G.D., Dyer P.S. Sexual development in fungi and its uses in gene expression systems. In: Schmoll M., Dattenböck C., editors. Gene expression systems of fungi: Applications and advancements. Springer International Publishing; Switzerland: 2016. pp. 335–350. [Google Scholar]
- Bayram O., Braus G.H. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiology Reviews. 2012;36:1–24. doi: 10.1111/j.1574-6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- Bennett J.W. An overview of the genus Aspergillus. In: Machida M., Gomi K., editors. Aspergillus molecular biology and genomics. Caister Academic Press; Norfolk, UK: 2010. pp. 1–17. [Google Scholar]
- Bergel F., Morrison A., Moss A. An antibacterial substance from Aspergillus clavatus. Journal of the Chemical Society. 1944;44:415–421. [Google Scholar]
- Berlin V., Yanofsky C. Isolation and characterization of genes differentially expressed during conidiation of Neurospora crassa. Molecular Cell Biology. 1985;5:849–855. doi: 10.1128/mcb.5.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boni A.C., Ambrósio D.L., Cupertino F.B. Neurospora crassa developmental control mediated by the FLB-3 transcription factor. Fungal Biology. 2018;122:570–582. doi: 10.1016/j.funbio.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Borneman A.R., Hynes M.J., Andrianopoulos A. A basic helix-loop-helix protein with similarity to the fungal morphological regulators, Phd1p, Efg1p and StuA, controls conidiation but not dimorphic growth in Penicillium marneffei. Molecular Microbiology. 2002;44:621–631. doi: 10.1046/j.1365-2958.2002.02906.x. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Latek R.R., Peterson C.L. The SANT domain: a unique histone-tail-binding module? Nature Reviews Molecular Cell Biology. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- Braus G.H., Krappman S., Eckert S.E. Sexual development in ascomycetes. Fruit body formation of Aspergillus nidulans. In: Osiewacz H.D., editor. Molecular biology of fungal development. Marcel Dekker; New York, USA: 2002. pp. 215–244. [Google Scholar]
- Burglin T.R. The TEA domain: a novel, highly conserved DNA-binding motif. Cell. 1991;66:11–12. doi: 10.1016/0092-8674(91)90132-i. [DOI] [PubMed] [Google Scholar]
- Busby T.M., Miller K.Y., Miller B.L. Suppression and enhancement of the Aspergillus nidulans medusa mutation by altered dosage of the bristle and stunted genes. Genetics. 1996;143:155–163. doi: 10.1093/genetics/143.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler G. The Evolution of MAT: the ascomycetes. In: Heitman J., Kronstad J.W., Taylor J.W., Casselton L.A., editors. Sex in fungi: Molecular determination and evolutionary implications. ASM Press; Washington, USA: 2007. p. 3-18. [Google Scholar]
- Cánovas D., Marcos A.T., Gacek A. The histone acetyltransferase GcnE (GCN5). plays a central role in the regulation of Aspergillus asexual development. Genetics. 2014;197:1175–1189. doi: 10.1534/genetics.114.165688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cánovas D., Marcos J.F., Marcos A.T. Nitric oxide in fungi: is there NO light at the end of the tunnel? Current Genetics. 2016;62:513–518. doi: 10.1007/s00294-016-0574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Huang P., Zhang L. Characterization of 47 Cys2 -His2 zinc finger proteins required for the development and pathogenicity of the rice blast fungus Magnaporthe oryzae. New Phytologist. 2016;211:1035–1051. doi: 10.1111/nph.13948. [DOI] [PubMed] [Google Scholar]
- Carrillo A.J., Schacht P., Cabrera I.E. Functional profiling of transcription factor genes in Neurospora crassa. G3 (Bethesda) 2017;7:2945–2956. doi: 10.1534/g3.117.043331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacko N., Gold S. Deletion of the Ustilago maydis ortholog of the Aspergillus sporulation regulator medA affects mating and virulence through pheromone response. Fungal Genetics and Biology. 2012;49:426–432. doi: 10.1016/j.fgb.2012.04.002. [DOI] [PubMed] [Google Scholar]
- Chen W., Chen R., Liu Q. Orange, red, yellow: biosynthesis of azaphilone pigments in Monascus fungi. Chemical Science. 2017;8:4917–4925. doi: 10.1039/c7sc00475c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., He Y., Zhou Y. Edible filamentous fungi from the species Monascus: early traditional fermentations, modern molecular biology, and future genomics. Comprehensive Reviews in Food Science and Food Safety. 2015;14:555–567. [Google Scholar]
- Chen A.J., Hubka V., Frisvad J.C. Polyphasic taxonomy of Aspergillus section Aspergillus (formerly Eurotium), and its occurrence in indoor environments and food. Studies in Mycology. 2017;88:37–135. doi: 10.1016/j.simyco.2017.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole G.T. Models of cell differentiation in conidial fungi. Microbiology Reviews. 1986;50:95–132. doi: 10.1128/mr.50.2.95-132.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debuchy R., Turgeon B.G. Mating-type structure, evolution, and function in euascomycetes. In: Kües U., Fischer R., editors. The mycota I: growth, differentiation and sexuality. Springer; Berlin, Germany: 2006. pp. 293–323. [Google Scholar]
- Dong Y., Zhao Q., Liu X. MoMyb1 is required for asexual development and tissue-specific infection in the rice blast fungus Magnaporthe oryzae. BMC Microbiology. 2015;15:37. doi: 10.1186/s12866-015-0375-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton J.R., Johns S., Miller B.L. StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO Journal. 1997;16:5710–5721. doi: 10.1093/emboj/16.18.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer P.S. Sexual reproduction and significance of MAT in the aspergilli. In: Heitman J., Kronstad J.W., Taylor J.W., Casselton L.A., editors. Sex in fungi: Molecular determination and evolutionary principles. ASM Press; Washington, USA: 2007. pp. 123–142. [Google Scholar]
- Dyer P.S., Inderbitzin P., Debuchy R. Mating-type structure, function, regulation and evolution in the Pezizomycotina. In: Wendland J., editor. Growth, differentiation and sexuality, the Mycota I. (3rd ed) Springer International Publishing; Switzerland: 2016. pp. 351–385. [Google Scholar]
- Dyer P.S., Kück U. Sex and the imperfect fungi. Microbiology Spectrum. 2017;5 doi: 10.1128/microbiolspec.funk-0043-2017. FUNK-0043-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer P.S., Nicholson P., Lucas J.A. Tapesia acuformis as a causal agent of eyespot disease of cereals and evidence for a heterothallic mating system using molecular markers. Mycological Research. 1996;100:1219–1226. [Google Scholar]
- Dyer P.S., O'Gorman C.M. A fungal sexual revolution: Aspergillus and Penicillium show the way. Current Opinion in Microbiology. 2011;14:649–654. doi: 10.1016/j.mib.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Dyer P.S., O'Gorman C.M. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiology Reviews. 2012;36:165–192. doi: 10.1111/j.1574-6976.2011.00308.x. [DOI] [PubMed] [Google Scholar]
- Eagle C.E. School of Biology, University of Nottingham; UK: 2009. Mating-type genes and sexual potential in the ascomycete genera Aspergillus and Penicillium. Ph. D. dissertation. [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biology. 2015;16:157. doi: 10.1186/s13059-015-0721-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxebeste O., Garzia A., Espeso E.A. Aspergillus nidulans asexual development: making the most of cellular modules. Trends in Microbiology. 2010;18:569–576. doi: 10.1016/j.tim.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Etxebeste O., Ni M., Garzia A. Basic-zipper-type transcription factor FlbB controls asexual development in Aspergillus nidulans. Eukaryotic Cell. 2008;7:38–48. doi: 10.1128/EC.00207-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxebeste O., Herrero-García E., Araújo-Bazán L. The bZIP-type transcription factor FlbB regulates distinct morphogenetic stages of colony formation in Aspergillus nidulans. Molecular Microbiology. 2009;73:775–789. doi: 10.1111/j.1365-2958.2009.06804.x. [DOI] [PubMed] [Google Scholar]
- Fisher R.A. 7th ed. Oliver & Boyd; Edinburgh, Scotland: 1938. Statistical methods for research workers. [Google Scholar]
- Galagan J.E., Calvo S.E., Cuomo C. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- Gancedo J.M. Control of pseudohyphae formation in Saccharomyces cerevisiae. FEMS Microbiol Reviews. 2001;25:107–123. doi: 10.1111/j.1574-6976.2001.tb00573.x. [DOI] [PubMed] [Google Scholar]
- García-Pedrajas M.D., Baeza-Montañez L., Gold S.E. Regulation of Ustilago maydis dimorphism, sporulation, and pathogenic development by a transcription factor with a highly conserved APSES domain. Molecular Plant Microbe Interactions. 2010;23:211–222. doi: 10.1094/MPMI-23-2-0211. [DOI] [PubMed] [Google Scholar]
- Garzia A., Etxebeste O., Herrero-Garcia E. The concerted action of bZip and cMyb transcription factors FlbB and FlbD induces brlA expression and asexual development in Aspergillus nidulans. Molecular Microbiology. 2010;75:1314–1324. doi: 10.1111/j.1365-2958.2010.07063.x. [DOI] [PubMed] [Google Scholar]
- Garzia A., Etxebeste O., Rodriguez-Romero J. Transcriptional changes in the transition from vegetative cells to asexual development in the model fungus Aspergillus nidulans. Eukaryotic Cell. 2013;12:311–321. doi: 10.1128/EC.00274-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser D., Frisvad J., Taylor J. Evolutionary relationships in Aspergillus section Fumigati inferred from partial β−tubulin and hydrophobin DNA sequences. Mycologia. 1998;90:831–845. [Google Scholar]
- Geiser D., Timberlake W., Arnold M. Loss of meiosis in Aspergillus. Molecular Biology and Evolution. 1996;13:809–817. doi: 10.1093/oxfordjournals.molbev.a025641. [DOI] [PubMed] [Google Scholar]
- Gems D.H., Clutterbuck A.J. Enhancers of conidiation mutants in Aspergillus nidulans. Genetics. 1994;137:79–85. doi: 10.1093/genetics/137.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan J.J., Pringle A. Long-distance dispersal of fungi. Microbiology Spectrum. 2017;5 doi: 10.1128/microbiolspec.funk-0047-2016. FUNK-0047-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravelat F.N., Ejzykowicz D.E., Chiang L.Y. Aspergillus fumigatus MedA governs adherence, host cell interactions and virulence. Cellular Microbiology. 2010;12:473–488. doi: 10.1111/j.1462-5822.2009.01408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I.V., Nikitin R., Haridas S. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Research. 2014;42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobe V., Krappmann S. The asexual pathogen Aspergillus fumigatus expresses functional determinants of Aspergillus nidulans sexual development. Eukaryotic Cell. 2008;7:1724–1732. doi: 10.1128/EC.00157-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K.H., Han D.M. Genetics and genomics of sexual development of Aspergillus nidulans. In: Machida M., Gomi K., editors. Aspergillus molecular biology and genomics. Caister Academic Press; Norfolk, UK: 2010. pp. 115–137. [Google Scholar]
- Han M.V., Zmasek C.M. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth D.L., Crous P.W., Redhead S.A. The Amsterdam declaration on fungal nomenclature. IMA Fungus. 2011;2:105–112. doi: 10.5598/imafungus.2011.02.01.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth D., Kirk P., Sutton B. CAB International; Oxon, UK: 1995. Ainsworth and Bisby's Dictionary of the fungi. [Google Scholar]
- Hawksworth D.L., Pitt J.I. A new taxonomy for Monoascus species based on cultural and microscopical characters. Australian Journal of Botany. 1983;31:51–61. [Google Scholar]
- Heitman J. Evolution of sexual reproduction: A view from the fungal kingdom supports an evolutionary epoch with sex before sexes. Fungal Biology Reviews. 2015;29:108–117. doi: 10.1016/j.fbr.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Sun S., James T.Y. Evolution of fungal sexual reproduction. Mycologia. 2013;105:1–27. doi: 10.3852/12-253. [DOI] [PubMed] [Google Scholar]
- Herrero-Garcia E., Perez-de-Nanclares-Arregi E., Cortese M.S. Tip-to-nucleus migration dynamics of the asexual development regulator FlbB in vegetative cells. Molecular Microbiology. 2015;98:607–624. doi: 10.1111/mmi.13156. [DOI] [PubMed] [Google Scholar]
- Homann O.R., Dea J., Noble S.M. A phenotypic profile of the Candida albicans regulatory network. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000783. e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn B.W., Moore G.G., Carbone I. Sexual reproduction in Aspergillus flavus. Mycologia. 2009;101:423–429. doi: 10.3852/09-011. [DOI] [PubMed] [Google Scholar]
- Horn B.W., Ramirez-Prado J.H., Carbone I. Sexual reproduction and recombination in the aflatoxin-producing fungus Aspergillus parasiticus. Fungal Genetics and Biology. 2009;46:169–175. doi: 10.1016/j.fgb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Houbraken J., Dyer P.S. Induction of the sexual cycle in filamentous ascomycetes. In: van den Berg M.A., Maruthachalam K., editors. Genetic transformation systems in fungi, Volume 2. Fungal Biology. Springer International Publishing; Switzerland: 2015. pp. 23–46. [Google Scholar]
- Inderbitzin P., Harkness J., Turgeon B.G. Lateral transfer of mating systems in Stemphylium. Proceedings of the National Academy of Sciences USA. 2005;102:11390–11395. doi: 10.1073/pnas.0501918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Han J.H., Kim K.S. Comparative functional analysis of the velvet gene family reveals unique roles in fungal development and pathogenicity in Magnaporthe oryzae. Fungal Genetics and Biology. 2014;66:33–43. doi: 10.1016/j.fgb.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Kim Y., Kim H., Son H. MYT3, a Myb-like transcription factor, affects fungal development and pathogenicity of Fusarium graminearum. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094359. e94359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon N.J., Garzia A., Espeso E.A. FlbC is a putative nuclear C2H2 transcription factor regulating development in Aspergillus nidulans. Molecular Microbiology. 2010;77:1203–1219. doi: 10.1111/j.1365-2958.2010.07282.x. [DOI] [PubMed] [Google Scholar]
- Krijgsheld P., Bleichrodt R., van Veluw G.J. Development in Aspergillus. Studies in Mycology. 2013;74:1–29. doi: 10.3114/sim0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon K., Kim S. A second heterothallic Aspergillus. Mycologia. 1974;66:629-638. [PubMed] [Google Scholar]
- Kwon K., Raper K. Sexuality and cultural characteristics of Aspergillus heterothallicus. American Journal of Botany. 1967;54:36-48. [PubMed] [Google Scholar]
- Lau G.W., Hamer J.E. Acropetal: a genetic locus required for conidiophore architecture and pathogenicity in the rice blast fungus. Fungal Genetics and Biology. 1998;24:228–239. doi: 10.1006/fgbi.1998.1053. [DOI] [PubMed] [Google Scholar]
- Lee M.K., Kwon N.J., Choi J.M. NsdD is a key repressor of asexual development in Aspergillus nidulans. Genetics. 2014;197:159–173. doi: 10.1534/genetics.114.161430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.K., Kwon N.J., Lee I.S. Negative regulation and developmental competence in Aspergillus. Scientific Reports. 2016;6:28874. doi: 10.1038/srep28874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Ni M., Li W. The evolution of sex: a perspective from the fungal kingdom. Microbiology and Molecular Biology Reviews. 2010;74:298–340. doi: 10.1128/MMBR.00005-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler K.B., Davidson R.C., D'Souza C. Signal transduction cascades regulating fungal development and virulence. Microbiology and Molecular Biology Reviews. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive tree of life (iTOL). v3: an online tool for the display and annotation of phylogenetic and other tres. Nucleic Acids Research. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Shao Y., Li Q. Identification of Mga1, a G-protein alpha-subunit gene involved in regulating citrinin and pigment production in Monascus ruber M7. FEMS Microbiology Letters. 2010;308:108–114. doi: 10.1111/j.1574-6968.2010.01992.x. [DOI] [PubMed] [Google Scholar]
- Liu Y., Whittier R. Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674-681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- Liu Q., Xie N., He Y. MpigE, a gene involved in pigment biosynthesis in Monascus ruber M7. Applied Microbiology and Biotechnology. 2014;98:285–296. doi: 10.1007/s00253-013-5289-8. [DOI] [PubMed] [Google Scholar]
- López-Berges M.S., Hera C., Sulyok M. The velvet complex governs mycotoxin production and virulence of Fusarium oxysporum on plant and mammalian hosts. Molecular Microbiology. 2013;87:49–65. doi: 10.1111/mmi.12082. [DOI] [PubMed] [Google Scholar]
- Malapi-Wight M., Kim J.E., Shim W.B. The N-terminus region of the putative C2H2 transcription factor Ada1 harbors a species-specific activation motif that regulates asexual reproduction in Fusarium verticillioides. Fungal Genetics and Biology. 2014;62:25–33. doi: 10.1016/j.fgb.2013.10.008. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bryant S.H. CD-Search: protein domain annotations on the fly. Nucleic Acids Research. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcos A.T., Ramos M.S., Marcos J.F. Nitric oxide synthesis by nitrate reductase is regulated during development in Aspergillus. Molecular Microbiology. 2016;99:15–33. doi: 10.1111/mmi.13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M.A., Timberlake W.E. Aspergillus nidulans wetA activates spore-specific gene expression. Molecular Cell Biology. 1991;11:55–62. doi: 10.1128/mcb.11.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheis S., Yemelin A., Scheps D. Functions of the Magnaporthe oryzae Flb3p and Flb4p transcription factors in the regulation of conidiation. Microbiology Research. 2017;196:106–117. doi: 10.1016/j.micres.2016.12.010. [DOI] [PubMed] [Google Scholar]
- Metzenberg R., Glass N. Mating type and mating strategies in Neurospora. Bioessays. 1990;12:53–59. doi: 10.1002/bies.950120202. [DOI] [PubMed] [Google Scholar]
- Miller K.Y., Wu J., Miller B.L. StuA is required for cell pattern formation in Aspergillus. Genes and Development. 1992;6:1770–1782. doi: 10.1101/gad.6.9.1770. [DOI] [PubMed] [Google Scholar]
- Moore A.D., Held A., Terrapon N. DoMosaics: software for domain arrangement visualization and domain-centric analysis of proteins. Bioinformatics. 2014;30:282–283. doi: 10.1093/bioinformatics/btt640. [DOI] [PubMed] [Google Scholar]
- Murtagh G.J., Dyer P.S., Crittenden P.D. Sex and the single lichen. Nature. 2000;404:564. doi: 10.1038/35007142. [DOI] [PubMed] [Google Scholar]
- Murtagh G.J., Dyer P.S., McClure P.C. Use of randomly amplified polymorphic DNA markers as a tool to study variation in lichen-forming fungi. Lichenologist. 1999;31:257–267. [Google Scholar]
- Niehaus E.M., Schumacher J., Burkhardt I. The GATA-type transcription factor Csm1 regulates conidiation and secondary metabolism in Fusarium fujikuroi. Frontiers in Microbiology. 2017;8:1175. doi: 10.3389/fmicb.2017.01175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwenhuis B.P.S., Billiard S., Vuilleumier S. Evolution of uni- and bifactorial sexual compatibility systems in fungi. Heredity. 2013;111:445–455. doi: 10.1038/hdy.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M., Fukada J., Moriwaki A. Mstu1, an APSES transcription factor, is required for appressorium-mediated infection in Magnaporthe grisea. Bioscience Biotechnology and Biochemistry. 2009;73:1779–1786. doi: 10.1271/bbb.90146. [DOI] [PubMed] [Google Scholar]
- O'Gorman C.M., Fuller H.T., Dyer P.S. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009;457:471–474. doi: 10.1038/nature07528. [DOI] [PubMed] [Google Scholar]
- Ohara T., Inoue I., Namiki F. REN1 is required for development of microconidia and macroconidia, but not of chlamydospores, in the plant pathogenic fungus Fusarium oxysporum. Genetics. 2004;166:113–124. doi: 10.1534/genetics.166.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T., Tsuge T. FoSTUA, encoding a basic helix-loop-helix protein, differentially regulates development of three kinds of asexual spores, macroconidia, microconidia, and chlamydospores, in the fungal plant pathogen Fusarium oxysporum. Eukaryotic Cell. 2004;3:1412–1422. doi: 10.1128/EC.3.6.1412-1422.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M., Rydholm C., Schwier E.U. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Current Biology. 2005;15:1242–1248. doi: 10.1016/j.cub.2005.05.045. [DOI] [PubMed] [Google Scholar]
- Paoletti M., Seymour F.A., Alcocer M.J.C. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Current Biology. 2007;17:1384–1389. doi: 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- Park H.S., Nam T.Y., Han K.H. VelC positively controls sexual development in Aspergillus nidulans. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089883. e89883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.S., Yu J.H. Genetic control of asexual sporulation in filamentous fungi. Current Opinions in Microbiology. 2012;15:669–677. doi: 10.1016/j.mib.2012.09.006. [DOI] [PubMed] [Google Scholar]
- Pel H.J., de Winde J.H., Archer D.B. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nature Biotechnology. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- Peterson S.W. Phylogenetic relationships in Aspergillus based on rDNA sequence analysis. In: Samson R.A., Pitt J.I., editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Harwood Academic Publishers; Amsterdam, The Netherlands: 2000. pp. 323–355. [Google Scholar]
- Peterson S.W. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia. 2008;100:205–226. doi: 10.3852/mycologia.100.2.205. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G. The genetics of Aspergillus nidulans. Advances in Genetics. 1953;5:141–238. doi: 10.1016/s0065-2660(08)60408-3. [DOI] [PubMed] [Google Scholar]
- Pyrzak W., Miller K.Y., Miller B.L. The mating type protein Mat1-2 from asexual Aspergillus fumigatus drives sexual reproduction in fertile Aspergillus nidulans. Eukaryotic Cell. 2008;7:1029–1040. doi: 10.1128/EC.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Prado J.H., Moore G.G., Horn B.W. Characterization and population analysis of the mating-type genes in Aspergillus flavus and A. parasiticus. Fungal Genetics and Biology. 2008;45:1292–1299. doi: 10.1016/j.fgb.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Raper K.B., Fennell D.I. The Williams & Wilkins Company; Baltimore, USA: 1965. The genus Aspergillus. [Google Scholar]
- Robert V., Groenewald M., Epping W. Centraalbureau voor Schimmelcultures; Utrecht, The Netherlands: 2007. CBS yeasts database. [Google Scholar]
- Rodriguez-Romero J., Hedtke M., Kastner C. Fungi, hidden in soil or up in the air: light makes a difference. Annual Reviews of Microbiology. 2010;64:585–610. doi: 10.1146/annurev.micro.112408.134000. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Urra A.B., Jimenez C., Nieto M.I. Signaling the induction of sporulation involves the interaction of two secondary metabolites in Aspergillus nidulans. ACS Chemical Biology. 2012;7:599–606. doi: 10.1021/cb200455u. [DOI] [PubMed] [Google Scholar]
- Ruger-Herreros C., Rodriguez-Romero J., Fernandez-Barranco R. Regulation of conidiation by light in Aspergillus nidulans. Genetics. 2011;188:809–822. doi: 10.1534/genetics.111.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydholm C., Dyer P.S., Lutzoni F. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryotic Cell. 2007;6:868–874. doi: 10.1128/EC.00319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson R.A., Varga J. Molecular systematics of Aspergillus and its teleomorphs. In: Machida M., Gomi K., editors. Aspergillus molecular biology and genomics. Caister Academic Press; Norfolk, UK: 2010. pp. 19–40. [Google Scholar]
- Samson R.A., Varga J., Dyer P.S. Morphology and reproductive mode of Aspergillus fumigatus. In: Latgé J.P., Steinbach W.J., editors. Aspergillus fumigatus and Aspergillosis. ASM Press; Washington USA: 2009. pp. 7–13. [Google Scholar]
- Samson R., Visagie C.M., Houbraken J. Phylogeny, identification and nomenclature of the genus Aspergillus. Studies in Mycology. 2014;78:141–173. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J., Simon A., Cohrs K.C. The transcription factor BcLTF1 regulates virulence and light responses in the necrotrophic plant pathogen Botrytis cinerea. PLoS Genetics. 2014;10 doi: 10.1371/journal.pgen.1004040. e1004040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J.A., Guan Y., Yu J.H. FluG-dependent asexual development in Aspergillus nidulans occurs via derepression. Genetics. 2006;172:1535–1544. doi: 10.1534/genetics.105.052258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewall T.C., Mims C.W., Timberlake W.E. abaA controls phialide differentiation in Aspergillus nidulans. Plant Cell. 1990;2:731–739. doi: 10.1105/tpc.2.8.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y.C., Ding Y.D., Zhao Y. Characteristic analysis of transformants in T-DNA mutation library of Monascus ruber. World Journal of Microbiology and Biotechnology. 2009;25:989–995. [Google Scholar]
- Shen W.C., Wieser J., Adams T.H. The Neurospora rca-1 gene complements an Aspergillus flbD sporulation mutant but has no identifiable role in Neurospora sporulation. Genetics. 1998;148:1031–1041. doi: 10.1093/genetics/148.3.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Molecular Systematics and Biology. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H., Kim M.G., Chae S.K. FgFlbD regulates hyphal differentiation required for sexual and asexual reproduction in the ascomycete fungus Fusarium graminearum. Journal of Microbiology. 2014;52:930–939. doi: 10.1007/s12275-014-4384-6. [DOI] [PubMed] [Google Scholar]
- Son H., Kim M.G., Min K. WetA is required for conidiogenesis and conidium maturation in the ascomycete fungus Fusarium graminearum. Eukaryotic Cell. 2014;13:87–98. doi: 10.1128/EC.00220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M.L. Genetic control of fungal differentiation: the three sporulation pathways of Neurospora crassa. Bioessays. 1993;15:365–374. doi: 10.1002/bies.950150602. [DOI] [PubMed] [Google Scholar]
- Stajich J.E., Harris T., Brunk B.P. FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Research. 2012;40:D675–D681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoldt V.R., Sonneborn A., Leuker C.E. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO Journal. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudbery P.E. Growth of Candida albicans hyphae. Nature Reviews Microbiology. 2011;9:737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Takeda M., Tanabe H. A new mycotoxin produced by Aspergillus clavatus. Chemical and Pharmaceutical Bulletin. 1971;19:1786–1788. doi: 10.1248/cpb.19.1786. [DOI] [PubMed] [Google Scholar]
- Swilaiman S.S., O'Gorman C.M., Balajee S.A. Discovery of a sexual cycle in Aspergillus lentulus, a close relative of A. fumigatus. Eukaryotic Cell. 2013;12:962–969. doi: 10.1128/EC.00040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada M., Udagawa S. A new species of heterothallic Neosartorya. Mycotaxon. 1985;24:395–402. [Google Scholar]
- Taylor J.W., Jacobson D.J., Fisher M.C. The evolution of asexual fungi: reproduction, speciation and classification. Annual Review of Phytopathology. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. [DOI] [PubMed] [Google Scholar]
- Todd R.B., Davis M.A., Hynes M.J. Genetic manipulation of Aspergillus nidulans: meiotic progeny for genetic analysis and strain construction. Natural Protocols. 2007;2:811–821. doi: 10.1038/nprot.2007.112. [DOI] [PubMed] [Google Scholar]
- Trifinopoulos J., Nguyen L.T., von Haeseler A. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research. 2016;44:W232–W235. doi: 10.1093/nar/gkw256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon B.G., Yoder O.C. Proposed nomenclature for mating type genes of fila- mentous ascomycetes. Fungal Genetics and Biology. 2000;31:1–5. doi: 10.1006/fgbi.2000.1227. [DOI] [PubMed] [Google Scholar]
- Udagawa S.I., Uchiyama S. Neocarpenteles: A new ascomycete genus to accommodate Hemicarpenteles acanthosporus. Mycoscience. 2002;43:3–6. [Google Scholar]
- Varga J., Due M., Frisvad J.C. Taxonomic revision of Aspergillus section Clavati based on molecular, morphological and physiological data. Studies in Mycology. 2007;59:89–106. doi: 10.3114/sim.2007.59.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga J., Rigo K., Molnar J. Mycotoxin production and evolutionary relationships among species of Aspergillus section Clavati. Antonie Van Leeuwenhoek. 2003;83:191–200. doi: 10.1023/a:1023355707646. [DOI] [PubMed] [Google Scholar]
- Varga J., Vida Z., Tóth B. Phylogenetic analysis of newly described Neosartorya species. Antonie van Leeuwenhoek. 2000;77:235–239. doi: 10.1023/a:1002476205873. [DOI] [PubMed] [Google Scholar]
- de Vries R.P., Riley R., Wiebenga A. Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. Genome Biology. 2017;18:28. doi: 10.1186/s13059-017-1151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada R., Maruyama J.I., Yamaguchi H. Presence and functionality of mating type genes in the supposedly asexual filamentous fungus Aspergillus oryzae. Applied and Environmental Microbiology. 2012;78:2819–2829. doi: 10.1128/AEM.07034-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieser J., Adams T.H. flbD encodes a Myb-like DNA-binding protein that coordinates initiation of Aspergillus nidulans conidiophore development. Genes and Development. 1995;9:491–502. doi: 10.1101/gad.9.4.491. [DOI] [PubMed] [Google Scholar]
- Whitehouse H. Heterothallism and sex in the fungi. Biological Reviews. 1949;24:411–447. doi: 10.1111/j.1469-185x.1949.tb00582.x. [DOI] [PubMed] [Google Scholar]
- Wilken P.M., Steenkamp E.T., Wingfield M.J. Which MAT gene? Pezizomycotina (Ascomycota) mating-type gene nomenclature reconsidered. Fungal Biology Reviews. 2017;31:199–211. [Google Scholar]
- Wong H.C., Chien C.Y. Ultrastructural studies of the conidial anamorphs of Monascus. Mycologia. 1986;78:593–599. [Google Scholar]
- Wu M.Y., Mead M.E., Kim S.C. WetA bridges cellular and chemical development in Aspergillus flavus. PLoS One. 2017;12 doi: 10.1371/journal.pone.0179571. e0179571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Miller B.L. Aspergillus asexual reproduction and sexual reproduction are differentially affected by transcriptional and translational mechanisms regulated stunted gene expression. Molecular Cell Biology. 1997;17:6191–6201. doi: 10.1128/mcb.17.10.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaguchi T., Someya A., Miyadoh S. Aspergillus ingratus, a new species in Aspergillus section Clavati. Transactions of the Mycological Society of Japan. 1993;34:305–310. [Google Scholar]
- Yu J.H. Regulation of development in Aspergillus nidulans and Aspergillus fumigatus. Mycobiology. 2010;38:229–237. doi: 10.4489/MYCO.2010.38.4.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.H., Hamari Z., Han K.H. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genetics and Biology. 2004;41:973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Yu J.H., Wieser J., Adams T.H. The Aspergillus FlbA RGS domain protein antagonizes G protein signaling to block proliferation and allow development. EMBO Journal. 1996;15:5184–5190. [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Amich J., Will C. The novel Aspergillus fumigatus MAT1-2-4 mating-type gene is required for mating and cleistothecia formation. Fungal Biology and Genetics. 2017;108:1–12. doi: 10.1016/j.fgb.2017.09.001. [DOI] [PubMed] [Google Scholar]
- Yun S., Berbee M., Yoder O. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proceedings of the National Academy of Sciences USA. 1999;96:5592–5597. doi: 10.1073/pnas.96.10.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Takeda T., Whitehall S. Functional characterization of the fission yeast start-specific transcription factor Res2. EMBO Journal. 1997;16:1023–1034. doi: 10.1093/emboj/16.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.