Abstract
The intracellular deposition of fibrils composed of the microtubule-associated protein Tau is a characteristic feature of Alzheimer's disease (AD) and other fatal neurodegenerative disorders collectively known as tauopathies. Short Tau fibrils spread intracerebrally through transfer between interconnected neurons. Once taken up by a recipient cell, Tau fibrils recruit Tau monomers onto their ends. Based on the number of microtubule-binding repeats, there are two distinct groups of Tau isoforms: three-repeat (3R) Tau and four-repeat (4R) Tau. In AD, all Tau isoforms are deposited, whereas in other tauopathies, only 3R or 4R Tau isoforms are deposited. The molecular basis for these isoform-specific depositions is poorly understood, although conformation-based cross-seeding barriers are key. Here, we used sedimentation assays, EPR spectroscopy, and other structural readouts to better understand the cross-seeding barriers of 4R Tau fibrils. We observed that fibrils formed from truncated Tau (K18), but not full-length Tau (htau40), exhibit a barrier that inhibits 3R Tau recruitment. Investigating an array of differently sized fragments, we found that the Tau C terminus modulates the cross-seeding barrier and that the N terminus plays a synergistic role. Two disease-associated Tau variants, P301S and P301L, also established strong cross-seeding barriers. EPR analysis indicated that fibrils seeded with truncated and mutated Tau, but not htau40, are structurally disordered in the second half of repeat four and onward. These findings suggest that the disorder in this region diminishes the ability of 4R Tau fibrils to recruit 3R Tau monomers, revealing a new mechanism for Tau cross-seeding barriers.
Keywords: Tau protein (Tau), amyloid, conformational change, Alzheimer disease, tauopathy, electron paramagnetic resonance (EPR), fibril, protein aggregation, prion, protein structure, seeding barrier
Introduction
Intracellular deposits of the microtubule associated protein Tau are the pathological hallmark of Alzheimer's disease (AD)2 and other fatal neurodegenerative disorders including progressive supranuclear palsy (PSP), corticobasal degeneration (CBD), and Pick's disease (PID) (1, 2). The majority of tauopathies are sporadic. However, mutations in the Tau gene, MAPT, can cause hereditary forms of disease, known as frontotemporal dementia linked to chromosome 17 (FTDP-17). Some of the mutations cause phenotypes similar to those of the sporadic cases (3). A common characteristic of tauopathies is the successive appearance of Tau pathology in defined brain regions (4–8). There is increasing evidence that this is not a cell-autonomous process, but that instead short Tau fibrils and other aggregates transfer between synaptically connected neurons and then recruit endogenous Tau monomers onto their ends (9, 10). Remarkably, brain homogenates and isolated aggregates from different tauopathies induce disease-characteristic inclusions in cells and mouse brain (11–13), supporting the idea that conformational variants of Tau fibrils result in different strains. The seeded conversion and cell-to-cell spreading of different Tau conformers shares similarities with the propagating principles of prions (14–17).
In the adult human brain, six different Tau isoforms are expressed, ranging in size from 352 to 441 amino acids (18). These isoforms vary by the inclusion or exclusion of one or two inserts in the N terminus and the presence or absence of the second of four semiconserved microtubule-binding repeats in the C terminus. Latter distinction results in three isoforms with three repeats (3R) and another three isoforms with four repeats (4R). The isoforms from each group are expressed at similar levels (19), and all of them are deposited into fibrils in AD (20). On the contrary, in PSP and CBD only 4R Tau is deposited (21, 22); in PID, 3R Tau predominates in the inclusions (23). Tau fibrils are characterized by a cross–β-fold, in which β-strands run perpendicular to the long fibril axis (24) and are arranged parallel, in-register (25). The cryo-EM structure of paired helical and straight filaments from AD (26) reveals a C-shaped cross-β/β-helical core (residues 306–378) made from the third and fourth microtubule-binding repeats and an extra 10 residues in the adjacent C-terminal region. The fully matching core residues between 3R and 4R Tau in these filaments offer an explanation for the co-recruitment of all isoforms (26). The recently solved structure of Tau filaments from PID demonstrates a fold distinct from that observed in AD. The filaments are composed of only 3R Tau isoforms with a core (residues 254–378) of four cross–β-packed stacks, but no β-helical arrangement. The ordered core is larger than that of AD filaments because it also includes residues from the first microtubule-binding repeat. Structural incompatibilities with residues in the second repeat of 4R Tau offer an explanation for the selective recruitment of 3R Tau into these filaments (27). Currently, there are no 4R Tau filaments solved to atomic resolution. Although preferential deposition of 4R Tau is observed in a number of different diseases, it is unknown whether these fibrils vary in conformation. Utilizing the 4R Tau construct K18 (28), which comprises residues 244–372, we identified a seeding barrier that prevents the growth of 3R Tau (29). This barrier was ascribed to conformational heterogeneity in the fibril populations (30). Because the core region of filaments from AD extends six residues beyond the length of K18, the question arises whether residues outside the sequence of K18 influence the seeding properties of Tau fibrils. In this study we evaluated the effects of 4R Tau's N-terminal and C-terminal regions on seeding, assessed the conformational consequences of mutations, and determined structural features in the core of different 4R Tau filaments that prevent 3R Tau growth. The findings have important implications for the spreading of Tau pathology in 4R tauopathies.
Results
htau40 fibrils recruit htau23 monomers
In a first set of experiments, we sought to determine the seeding properties of htau40 (largest isoform, 4R Tau) (18). For this purpose, monomeric htau40 was incubated for 8 days at 37 °C with heparin as an inducer. The fibrils were then sheared for 30 s with a tip sonicator to produce seeds. The breakage of long, unbranched fibrils into short fibrillar species was visualized by negative stain transmission EM (TEM) (Fig. 1, A and B). Tau seeds had an average length of 39 nm (Fig. 1C). The seeds remained in the supernatant when centrifuged at 130,000 × g, different from unsonicated fibrils, which were found in the pellet (Fig. 1D). To test the ability of htau40 seeds to recruit Tau monomers, 10% seeds were mixed with either htau40 or htau23 (shortest isoform, 3R Tau) (18) monomers and allowed to grow for 20–24 h. Fig. 2 provides a schematic depiction of these isoforms. Remarkably not only htau40 but also htau23 was incorporated into the fibrils. Both proteins were exclusively found in the pellets (Fig. 1E). Notice that htau40 seeds appeared in the pellet when incubated with htau23 monomers. We ascribe this characteristic to the extension of the originally short seeds to elongated sedimentable fibrils. The seeding properties of htau40 fibrils were highly reproducible as judged by the combined data of >40 biological replicates for each reaction (Fig. 1F). To examine the kinetics of Tau fibril elongation, we next employed a widely used thioflavin T (ThT) assay that is based on the characteristic fluorescence changes of this dye (31). Conversion of Tau monomers into fibrils results in an increase in ThT fluorescence because of binding of the dye to the fibril core. Both htau23 and htau40 exhibited similar elongation kinetics when added to htau40 seeds (Fig. 1G). The differences in fluorescence intensity are likely due to differences in dye incorporation. When the two Tau monomers were incubated under identical conditions, but in the absence of seeds, no changes in fluorescence intensity were observed (Fig. 1G), suggesting that these proteins remained largely monomeric, in agreement with previous observations (32). The fibrillar nature of aggregates that formed upon seeding was confirmed by TEM (Fig. 1H).
Figure 1.
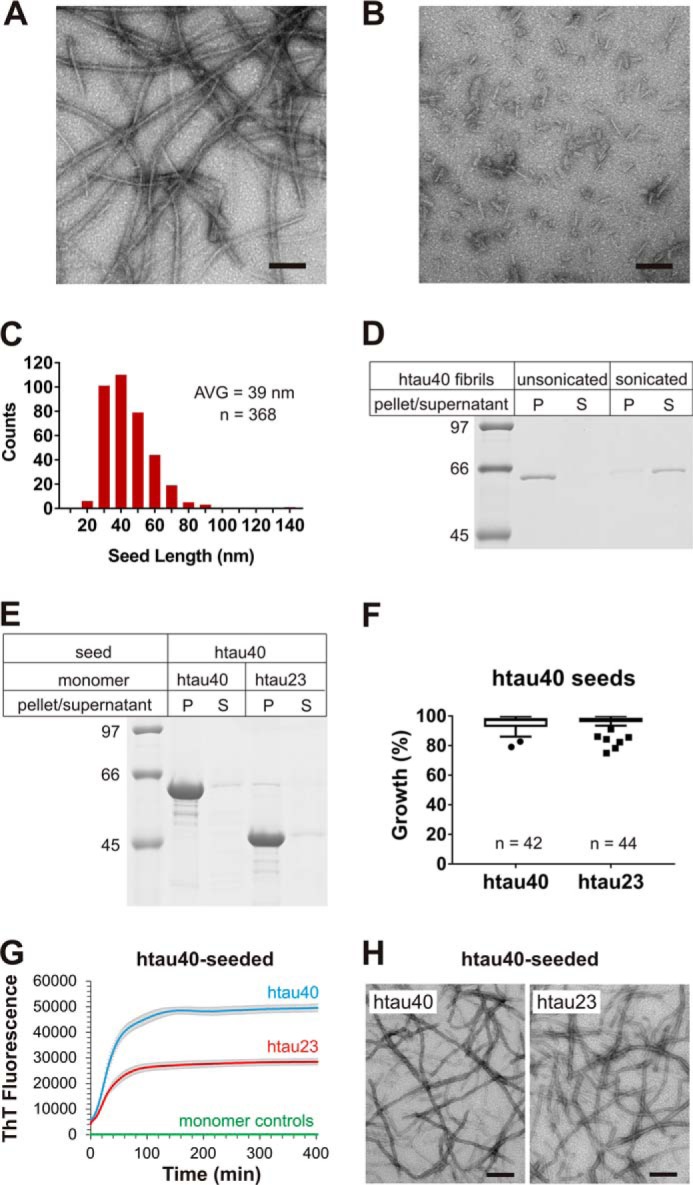
Seeding properties of htau40 fibrils. A and B, htau40 fibrils formed in the presence of heparin were analyzed by EM before (A) and after (B) sonication. Scale bars, 100 nm. C, the size distribution of sonicated fibrils reveals an average (AVG) seed length of 39 nm. D, sonicated and unsonicated fibrils were sedimented by centrifugation and analyzed by SDS–PAGE and Coomassie staining. Equivalent amounts of pellets and supernatants were loaded onto the gel. P, pellet; S, supernatant. htau40 seeds (10% monomer equivalents) were mixed with 10 μm Tau monomers and allowed to grow for 20–24 h at 37 °C. Fibrils were sedimented by ultracentrifugation. E, monomer incorporation was assessed by SDS–PAGE and Coomassie staining. F, quantitative analysis of seeding properties using gel densitometry depicted as box-and-whisker plot (Tukey method). n, number of biological replicates. G, ThT fluorescence measurements of htau40 (blue trace) and htau23 (red trace) growth onto htau40 seeds (10 μm monomers, 5% seeds) in triplicate. The t½ values for these reactions were 27 and 25 min, respectively. Triplicate ThT measurements of htau23 and htau40 monomers in the absence of seeds (overlapping green traces). Error bars represent means ± S.D. H, negative stain EM images of htau40 (left panel) and htau23 (right panel) fibrils after seeding. Scale bars, 200 nm. Collectively, the results demonstrate growth of htau23 and htau40 monomers onto htau40 seeds.
Figure 2.
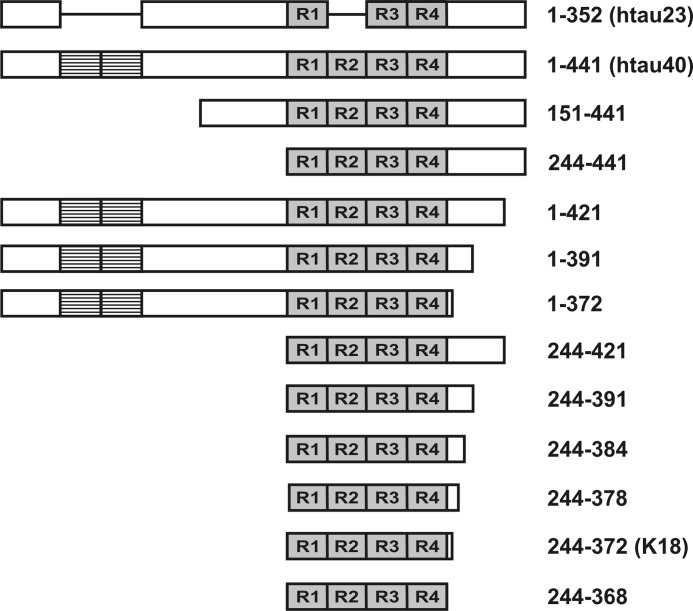
Schematic diagram of htau23, htau40, and truncated variants of htau40. Repeats 1–4 (R1–R4) are shaded. N-terminal inserts are striped. The horizontal bars in htau23 represent regions that are missing in this isoform.
Because previous experiments with K18 fibrils were performed at 25 °C and utilized protein constructs in which the native cysteines were replaced by serines (29), we next asked whether these variations could be responsible for the difference in seeding properties between full-length versus truncated Tau. The aggregation experiments were repeated under new conditions. Sedimentation revealed that neither a change in temperature (Fig. S1A) nor a substitution of cysteines (Fig. S1B) or a simultaneous change in both parameters (Fig. S1C) altered the seeding characteristics of full-length Tau. In all cases htau23 was robustly recruited onto the seeds. Collectively, the results suggest that truncations in 4R Tau alter the fibril structure and thereby affect its cross-seeding properties with 3R Tau.
Effect of truncations at either N or C terminus of htau40 on seeding
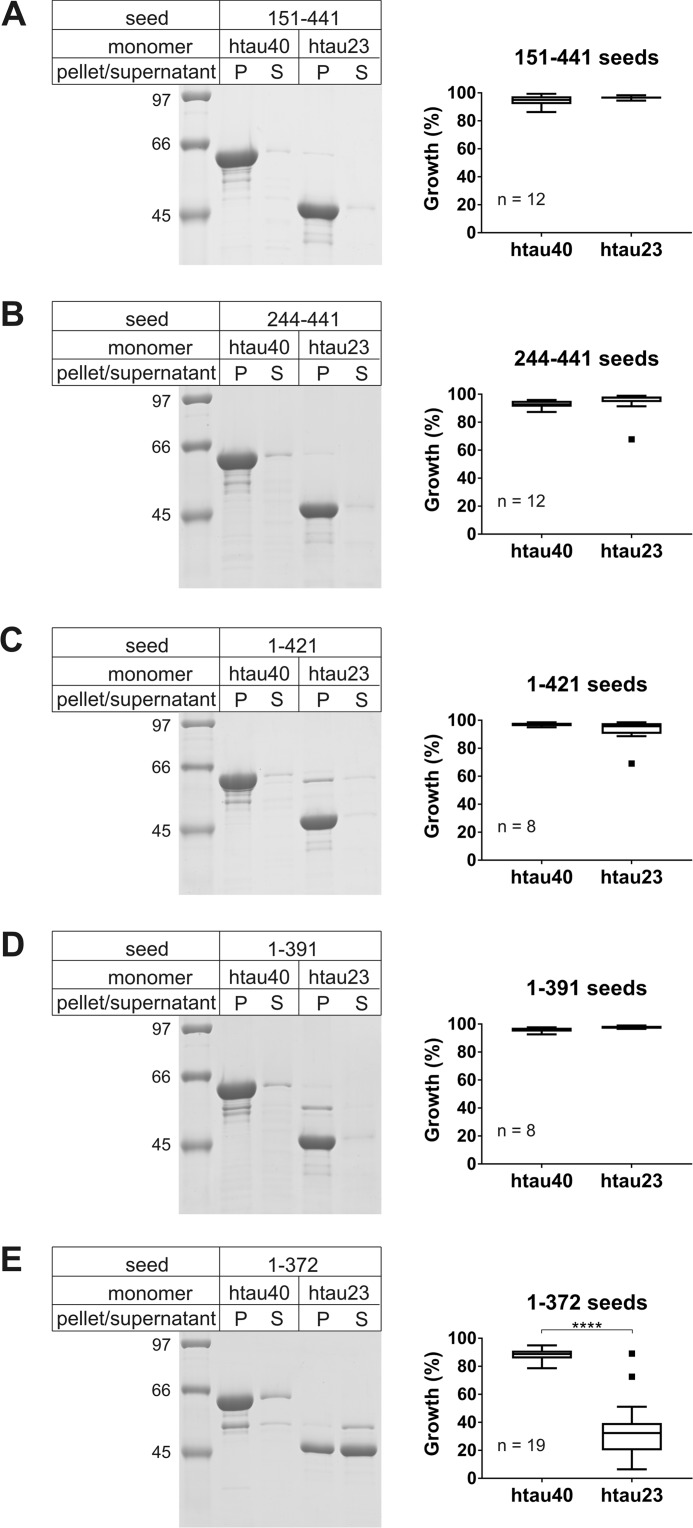
To identify which regions are responsible for altering the seeding properties of 4R Tau, we next generated 11 different Tau variants with truncations at either the N or C terminus or both (Fig. 2). Proteins with truncations at a single end were assessed first. N-terminal truncations included variants 151–441 and 244–441. C-terminal truncations encompassed variants 1–421, 1–391, and 1–372. Fibril formation, seed generation, and growth were carried out as before using short fibrils composed of Tau variants as seeds and full-length Tau monomers for growth. Sedimentation revealed that neither of the two truncations at the N terminus resulted in a barrier that prevented htau23 from growing onto the seeds. In both cases htau23 was found in the fibril containing pellet (Fig. 3, A and B). When the seeding properties of fibrils composed of C-terminally truncated Tau were assessed (Fig. 3, C–E), only fibrils composed of the 1–372 variant showed a significant barrier toward htau23 recruitment (Fig. 3E). Importantly, in all cases recruitment of htau40 was unperturbed. Together, the results suggest that the C-terminal region in Tau affects seeding competency.
Figure 3.
Seeding properties of fibrils composed of N- or C-terminally truncated htau40. After fibril formation and sonication, 10% fibril seeds were combined with 10 μm Tau monomers and allowed to grow for 20–24 h at 37 °C. Fibrils were sedimented by ultracentrifugation. A–E, representative SDS–PAGE gels stained with Coomassie Blue (left panels), and quantitative analysis by densitometry depicted as box-and-whisker plots using the Tukey method (right panels). The horizontal line inside the box represents the median. P, pellet; S, supernatant; n, number of biological replicates. Statistical comparison in E was performed using a paired t test. ****, p ≤ 0.0001. The results demonstrate that only fibrils composed of variant 1–372 exhibit a cross-seeding barrier with htau23.
Effect of combined truncations at the N and C terminus of htau40 on seeding
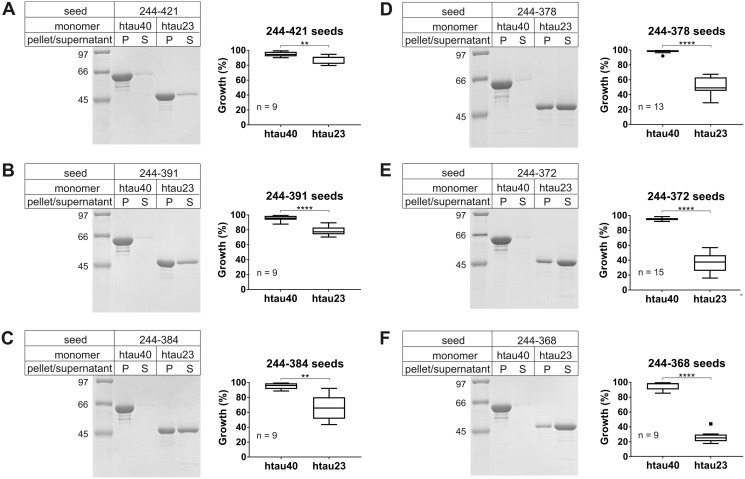
To better understand the structural implications of C-terminal truncations and to test for potential synergism between the N- and C-terminal regions, we next investigated the seeding properties of Tau variants in which the N terminus was removed and the C terminus was systematically shortened. Specifically, fibrils of the following 4R Tau variants were examined: 244–421, 244–391, 244–384, 244–378, 244–372, and 244–368 (Fig. 2). As before, seeding efficiency was assessed by comparing the protein contents in pellets versus supernatants. All seeds effectively recruited htau40. After sedimentation only marginal quantities of protein were detected in the supernatants (Fig. 4). Recruitment of htau23 depended on the type of 4R Tau variant in the seeds, with shorter variants resulting in successively diminished growth. The largest barrier was observed for seeds composed of variant 244–368, which only comprises the repeat region. It is noticeable that any truncation at the C terminus resulted in reduced growth. Even seeds composed of 244–421 and 244–391 showed diminished recruitment of htau23, whereas similar seeds with intact N terminus did not exhibit any barrier (Fig. 3). Combined, the results suggest that the C-terminal region of htau40 affects its cross-seeding properties with htau23 and that the N-terminal region plays a synergistic role.
Figure 4.
Seeding properties of fibrils composed of htau40 truncated at both N and C termini. 10% of fibril seeds were mixed with 10 μm Tau monomers and allowed to grow for 20–24 h at 37 °C. The fibrils were sedimented and analyzed by SDS–PAGE and Coomassie staining. A–F, representative gels (left panels) and densitometric quantification depicted as box-and-whisker plots using the Tukey method (right panels). The horizontal line inside the box represents the median. P, pellet; S, supernatant; n, number of biological replicates. Statistical comparisons were performed using paired t tests. **, p ≤ 0.01; ****, p ≤ 0.0001. The data reveal that successive shortening of the C terminus of htau40 enhances the cross-seeding barrier with htau23.
P301S and P301L induce cross-seeding barrier
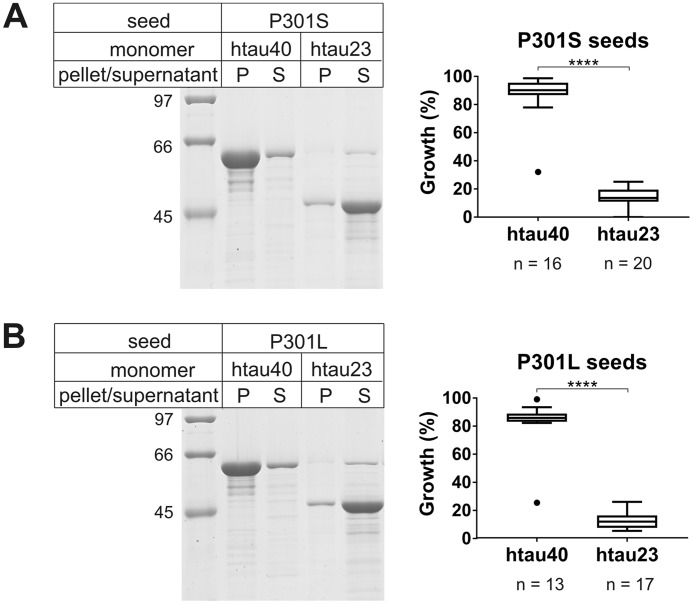
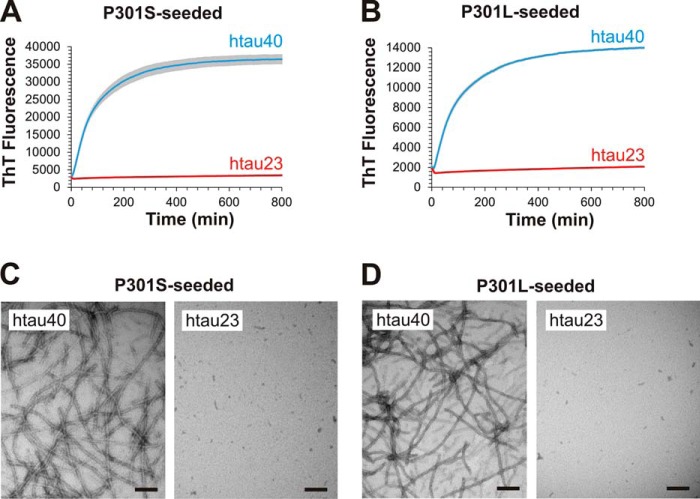
The investigations thus far focused on the effects of truncations on seeding competency. Next, we asked whether familial mutations in htau40 could produce similar seeding barriers as the C-terminal truncations. Of particular interest were the P301S and P301L mutations, because they are known to cause little or no changes in the 3R/4R Tau isoform ratios yet are observed to mimic tauopathies with preferential deposition of 4R Tau (1). P301S and P301L mutants were generated by site-directed mutagenesis using htau40 as a template. The purified protein variants were allowed to form fibrils as described for WT htau40 above and fractured by sonication. Growth of WT htau40 and htau23 monomers onto mutant seeds was assessed using sedimentation. Both P301S and P301L seeds competently recruited htau40 monomers into the fibril. After centrifugation, the majority of htau40 was found in the insoluble pellets (Fig. 5, left panels). The same mutant seeds were unable to recruit htau23. In this case, the majority of protein remained in the supernatant (Fig. 5, left panels). The results were highly reproducible with htau40 exhibiting >80% growth onto both mutant seeds and htau23 monomers exhibiting <15% growth (Fig. 5, right panels).
Figure 5.
Seeding properties of P301S and P301L fibrils. 10% of seeds of P301S and P301L fibrils were mixed with 10 μm Tau monomers and incubated for 20–24 h at 37 °C. After sedimentation, recruitment of Tau monomers was assessed by SDS–PAGE and Coomassie staining. A, representative gel of P301S-seeded reactions (left panel) and densitometric quantification depicted as box-and-whisker plot using the Tukey method (right panel). B, representative gel of P301L-seeded reactions (left panel) and densitometric quantification depicted as box-and-whisker plot using the Tukey method (right panel). P, pellet; S, supernatant; n, number of biological replicates. The horizontal line inside the box represents the median. Statistical comparisons were performed using paired t tests. ****, p ≤ 0.0001. The results indicate that both P301S and P301L fibril seeds are unable to efficiently recruit htau23 monomers.
To assess the kinetics of fibril elongation, we next monitored the changes in ThT fluorescence that occurred when htau23 and htau40 monomers were mixed with either P301S or P301L seeds (Fig. 6, A and B). Consistent with the sedimentation data, only htau40 was recruited onto the seeds, htau23 was not. The t½ values for htau40 growth onto P301S and P301L seeds were 60 and 68 min, respectively. Notably, homotypic recruitment of P301S and P301L monomers onto their respective seeds was even more efficient (>90%) and faster (t½ = 5–9 min) than heterotypic recruitment of htau40 (Fig. S2), in agreement with previous studies that suggested preferential growth of mutant over WT Tau (12, 33–35). To obtain visual confirmation for the recruitment of WT Tau onto mutant seeds, we next collected TEM images of negatively stained samples (Fig. 6, C and D). Whereas htau40 formed long, unbranched fibrils, no fibrils were observed in the case of htau23. Collectively, the data indicate that single substitutions at position 301 are sufficient to produce fibrils with cross-seeding barriers similar to those observed for truncated Tau.
Figure 6.
Fibril growth and EM analysis of P301S- and P301L-seeded reactions. 5% of seeds of P301S and P301L fibrils were mixed with 10 μm Tau monomers. A and B, growth of Tau monomers onto P301S seeds (A) and P301L seeds (B) was monitored by ThT fluorescence at 37 °C (n = 4). Error bars represent means ± S.D. C and D, afterward, all reactions were analyzed by EM. Scale bars, 200 nm. The data reveal that htau23 monomers are unable to grow onto P301S and P301L seeds.
Mutations flanking Pro-301 fail to induce robust cross-seeding barrier
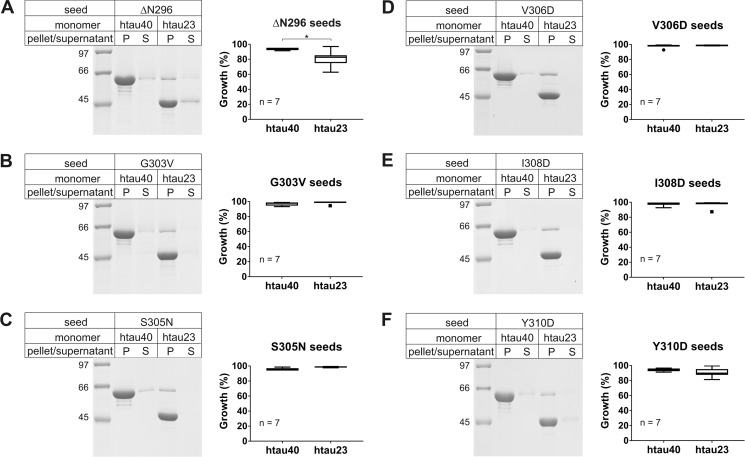
Pro-301 is located in a region of Tau that is important for aggregation. Notably, the PHF6 motif comprising residues 306–311 has been implicated in fibril nucleation (36). To determine whether sequence alterations in the region flanking Pro-301 could alter the seeding specificity of htau40 fibrils, we generated six new htau40 variants. Three of the variants contained familial mutations in the immediate vicinity of Pro-301 (ΔN296, G303V, and S305N). These mutations have been observed to cause 4R tauopathies (37–39). However, they are also known to shift the isoform ratios toward a predominance of 4R Tau (38, 40, 41). The three other variants involved synthetic mutations in PHF6 (V306D, I308D, and Y310D). These mutations were expected to disrupt the packing of β-sheets. All fibrils were formed for 8 days and then fractured as described above. The seeds were mixed with either htau23 or htau40 monomers, incubated overnight, and then sedimented. Surprisingly, only fibrils containing the ΔN296 variant exhibited a small, but significant, decrease in recruitment of htau23 compared with htau40 monomers (Fig. 7A). All other fibril variants efficiently incorporated both htau40 and htau23 monomers (Fig. 7, B–F). Because none of the newly tested htau40 variants produced a cross-seeding barrier comparable with the one observed for P301S and P301L, the data highlight the unique role Pro-301 must play in determining fibril structure.
Figure 7.
Seeding properties of Tau fibrils composed of proteins mutated in the regions immediately flanking Pro-301. 10% of mutant seeds were mixed with Tau monomers, incubated at 37 °C for 20–24 h, and sedimented by ultracentrifugation. Monomer recruitment onto seeds was assessed by SDS–PAGE and Coomassie staining. A–F, representative gels for reactions utilizing different seeds (left panels) and densitometric analysis depicted in box-and-whisker plots using the Tukey method (right panels). The horizontal line inside the box represents the median. P, pellet; S, supernatant; n, number of biological replicates. Statistical comparison in A was performed using a paired t test. *, p ≤ 0.05. The data indicate that only seeds of the ΔN296 mutant exhibited a small barrier for htau23 growth. All other seeds showed unperturbed growth.
The fibril cores of Pro-301 mutants and truncated Tau are distinct from those of htau40
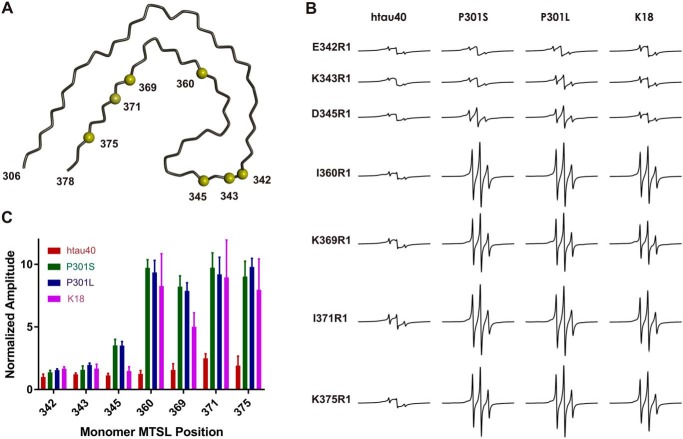
To assess the structural differences between mutant, truncated, and WT Tau fibrils, we next employed site-directed spin labeling in conjunction with continuous wave (CW) EPR spectroscopy. This approach has been successfully used in the past to obtain information on side-chain stacking and local backbone mobility in Tau fibrils (25, 42, 43). The cryo-EM model of paired helical filaments served as a guide for site-directed mutagenesis. In a first step, seven single cysteine mutants of htau40 (at positions 342, 343, 345, 360, 369, 371, and 375) were generated. All cysteines were located at positions that in PHFs point to the aqueous exterior (Fig. 8A). The monomeric Tau mutants were labeled with the paramagnetic nitroxide label MTSL (44) or its nonparamagnetic analogue (45) producing the respective side chains R1 and R1′ (45, 46). Each labeled protein had a high degree of purity as determined by SDS–PAGE and Coomassie staining (Fig. S3A). The EPR spectra of the paramagnetically labeled proteins revealed three sharp lines (Fig. S3B), in agreement with the high mobility and intrinsic disorder of monomeric Tau (47, 48). Next, these proteins were mixed with the preformed seeds of variant Tau fibrils (htau40, P301S, P301L, and K18). After 20–24 h of growth, fibrils were separated from soluble Tau by centrifugation and placed into capillaries for EPR measurements. When grown onto htau40 seeds, all paramagnetically labeled mutants produced single-line EPR spectra with reduced amplitudes (Fig. 8B, left column), characteristic for parallel, in-register arrangement of β-strands, with identical residues in neighboring Tau molecules stacking on top of each other (25). The contributions from mobile Tau in the spectra are less than 2% as judged by spectral subtraction. The loss of hyperfine structure in the EPR spectra can be explained by spin exchange between contacting labels (49). When grown onto mutant (P301S and P301L) and truncated (K18) Tau, only the first three labeled mutants (at positions 342, 343, and 345) produced spectra that were dominated by exchange-narrowed lines (Fig. 8B, center and right columns), indicating that these positions were stacked and part of the structured core. The spectra of all other mutants (at positions 360, 369, 371, and 375) were characterized by three sharp lines, indicating that these positions were highly mobile and outside the structured core. Noticeably, these spectra were similar to those published previously for positions in repeats one and two (42), suggesting that some exchange component, i.e. stacked structure, was still present. The signal amplitude in an EPR spectrum increases with increased mobility (42, 43). An amplitude plot (Fig. 8C) highlights the structural differences between the different fibril variants.
Figure 8.
EPR analysis of fibrils seeded with WT, mutant, and truncated Tau. Seven single cysteine mutants of htau40 were first labeled with the paramagnetic spin label MTSL, then combined with Tau fibril seeds (htau40, P301S, P301L, and K18), and incubated for 20–24 h at 37 °C. A, all cysteines were located at positions that in the paired helical filament point to the aqueous exterior. The depicted model represents the protein backbone of a single Tau layer. In the filament thousands of these layers are stacked on top of each other. The model utilizes PDB accession number 5O3L. Yellow spheres depict Cα carbons at the sites of cysteine substitution. B, representative CW EPR spectra of Tau fibrils collected at X-band. The horizontal row on top signifies the type of seeds that was used. The vertical column on the left identifies the spin labeled htau40 monomer that was grown onto the seed. R1, spin-labeled cysteine. All spectra are normalized to the same number of spins. Scan width, 150 G; modulation, 3 G; incident microwave power, 12 milliwatt. C, plot of the signal amplitudes. The results are represented as the means ± S.D. using three biological replicates. The data highlight differences in the core structures of the fibrils.
The majority of EPR spectra collected thus far was distorted by spin exchange interactions. To obtain spectra free of these distortions, we next repeated the seeding experiments using a mixture of paramagnetically and nonparamagnetically labeled mutants (0.2:0.8 molar ratio). In this case, all spectra of htau40-seeded fibrils exhibited three broad lines with a 68-G separation between outer peaks (Fig. S4A). These spectral features are characteristic for immobilized spin labels (49). Similar spectra were also obtained for the first three sites in fibrils seeded with mutant and truncated Tau (Fig. S4A). The remaining sites in these fibrils produced spectra that, in addition to some immobile components, were dominated by three sharp and narrowly spaced lines with 34-G separation between outer peaks, characteristic for high side chain mobility (49). A plot of the signal amplitudes corroborated these findings (Fig. S4B). Importantly, the plots for spin-diluted and spin-undiluted fibrils revealed the same mobility trends (compare Fig. S4B with Fig. 8C). Combined, the findings indicate that the structured core of htau40 fibrils differs from the structured core of truncated and mutant Tau fibrils by an extension of the fourth repeat and the C-terminally adjacent region.
Pro-301 mutants and htau40 form distinct clouds of fibril conformers
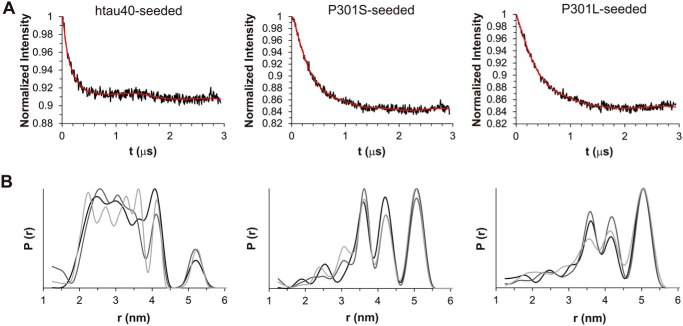
In a last set of experiments we sought to determine whether the structurally distinct fibrils of Pro-301 mutants and htau40 are defined by either single conformers or ensembles of conformers (also known as clouds). To distinguish between these possibilities we employed double electron-electron resonance (DEER) spectroscopy (50, 51), a technique that is widely used to determine distances between paramagnetic centers in proteins (52) and has emerged as a powerful tool for elucidating fibril structure (53–56). Notably, the technique is capable of differentiating fibril conformers within clouds (57–59). In a first step, we generated a double cysteine reporter construct of htau40 (311/328) in which cysteines were introduced at positions 311 and 328. These sites were chosen because they have already been used to report on the fibril structure of truncated Tau (K18) (58). Purified monomers of 311/328 were spin-labeled with MTSL and diluted with a 25-fold molar excess of label-free htau40. The dilution was necessary to ensure that upon incorporation of Tau monomers into the fibril, only intramolecular distances would be measured and not intermolecular distances between labels stacked along the fibril axis. Monomers were mixed with 10% seeds (P301S, P301L, or htau40) and allowed to grow for 24 h at 37 °C. Following sedimentation, the fibrils were subjected to DEER measurements. The dipolar evolution curves of the differently seeded fibrils were collected and then fit by Tikhonov regularization (Fig. 9A). The resulting distance distributions for fibrils seeded with htau40 (Fig. 9B, left panel) were different from those for fibrils seeded with P301S and P301L (Fig. 9B, center and right panels). The former exhibited overall shorter distances, in agreement with the steeper initial drop in intensity in the evolution curve (Fig. 9A, left panel versus center and right panels). Notably, the distributions for the P301S- and P301L-seeded fibrils were similar to the ones observed for K18 fibrils (58). The absence of strong oscillations in the evolution curves corroborates the existence of broad distance distributions (60). Our previous analysis indicated that increased signal-to-noise had only minor effects on these distributions (58). Although the insights on the particular structures are limited, the data allow two important conclusions. First, all three types of fibrils represent clouds of conformers. Second, the clouds of htau40 fibrils are distinct from those of P301S and P301L fibrils.
Figure 9.
DEER analysis of htau40 fibrils seeded with htau40, P301S, or P301L. Spin-labeled monomers of 311/328 were mixed with a 25-fold molar excess of label-free htau40 monomers and grown onto variant seeds. All data are analyzed by Tikhonov regularization. A, representative background corrected dipolar evolution curves shown as black traces with fit lines in red. B, corresponding distance distributions. The differently shaded traces are the results of three biological replicates. Left panels, htau40-seeded reactions. Center panels, P301S-seeded reactions. Right panels, P301L-seeded reactions. The data reveal that all fibrils are clouds of conformers and that the clouds of htau40-seeded fibrils are distinct from those of P301S- and P301L-seeded fibrils.
Discussion
Given the potential of Tau fibrils to spread pathology throughout the brain, it is critical to understand their structures and seeding properties. The present study focused on 4R Tau fibrils. It revealed that htau40 fibrils are fully compatible with 3R Tau monomer recruitment. This stands in marked contrast to fibrils of the frequently used truncated variant, K18, which exhibit a cross-seeding barrier (29, 59, 61). According to our EPR data, a key structural difference between K18 and htau40 fibrils is that the former are disordered in the second half of R4 and the C-terminally adjacent region, whereas the latter are perfectly ordered in the same region. As a consequence, there are fewer residues in K18 fibrils available for stacking of identical residues in 3R Tau monomers. The reduced contact surface of K18 fibrils could explain the cross-seeding barrier, although structural incompatibilities between nonmatching residues in 3R and 4R Tau in other parts of the fibril could contribute as well. It must be noted that K18 lacks residues 373–378 that are part of the AD fibril core (26) and are also structured in the htau40 fibrils investigated herein. Packing of these residues against adjacent β-sheets could help stabilize a C-terminally extended fibril core. We observed that residues beyond position 378, which are not part of the highly ordered core in AD filaments, also influence fibril structure. The majority of these effects occur in synergism with the N terminus. Although the molecular details are not yet resolved, the backfolding of the N terminus onto the third microtubule-binding repeat (62, 63) may play an important role in this.
If preferential deposition in sporadic 4R tauopathies is determined by conformation, why do htau40 fibrils have no cross-seeding barrier? One explanation could be that truncated versions of Tau initiate aggregation. We observe that truncations can alter the cross-seeding properties. Some of the truncations match cleavage sites of proteases that have been implicated in AD and other tauopathies. Caspase 3, for example, cleaves Tau at Asp-421, and fragments of 1–421 have been associated with neurofibrillary tangles in AD (64, 65). Another protease, asparagine endopeptidase, cleaves Tau at Asn-255 and Asn-368. The cleavage products (residues 1–368 and 256–368) were shown to mediate Tau pathology in mice (66). However, little is known about the structure of these fibrils. There are numerous other proteolytic fragments of Tau that have been observed in various biological contexts (67), but the effects on Tau filament structure remain largely unexplored. As pointed out above, we observed that K18-seeded Tau fibrils possessed an increased mobility in the second half of R4 compared with fibrils that were seeded with htau40. These data are in agreement with our previous simulations, which indicated higher flexibility at the C terminus of K18 (30). Notably, upon trypsin digestion, Tau filaments isolated from PSP and CBD produced fragments that terminated at position 369. No such fragments were observed for filaments from AD (68). Here, the shortest fragments extended to position 385. These data suggest that filaments in CBD and PSP are less structured at the C terminus, akin to fibrils of truncated Tau. It is important to point out that fragments of Tau that may be involved in the early stages of aggregation are only one biological means to obtain structurally distinct fibrils. The specific cellular environment, post-translational modifications, and other factors could also play important roles.
Although we demonstrated that self-seeded htau40 fibrils are able to recruit 3R Tau and share an extended core structure with paired helical and straight filaments, our DEER experiments reveal clear structural differences between these fibrils. For one, the distance between the Cα carbons at positions 311 and 328 in AD filaments (4.5 nm) does not agree with the herein observed distance range (2.2–4.2 nm). Importantly, the investigated htau40 fibrils are structurally heterogeneous forming clouds of conformers. The findings are in agreement with recent observations that revealed that fibrils formed from N-terminally truncated 4R Tau (residues 255–441) are also different from AD filaments (56). These structural differences are not completely surprising given that PHFs are compositionally distinct from htau40 filaments. Although filaments in AD and PID have been proposed to have single folds (26, 27), the question of whether there might be alternative conformers, in particular in different brain regions or different subtypes of disease, will need further clarification. Notably, in prion diseases multiple conformers of prion aggregates have been observed in a single brain (69). Furthermore, β-amyloid fibrils, the constituents of extracellular plaques in AD, showed conformational variations between clinical subtypes of AD (70). There are a number of tauopathies including Down's syndrome and chronic traumatic encephalopathy in which fibrils are composed of 3R and 4R Tau. Whether these fibrils are similar to AD fibrils or whether they possess different structures remains to be determined.
To gain further insights into the cross-seeding barriers of Tau fibrils, we investigated the familial mutations P301S and P301L. Both mutations lead to the preferential deposition of 4R Tau isoforms in the brain. We observed that the Pro-301 mutants result in fibrils that are remarkably similar to truncated Tau fibrils in their structural features and seeding properties. The replacement of the single proline results in fibrils with a highly robust cross-seeding barrier. Also, similar to K18 fibrils, residues in the second half of R4 and following are unstructured, leaving an outer surface area with diminished residue-to-residue matching for 3R Tau recruitment. A recent comparison of the atomic models of filaments from AD and PID revealed conserved secondary structure patterns but different turn conformations that changed the cross-β packings (27). Based on these observations, it was proposed that conformational variations in the turn regions could form the basis for structural diversity in Tau fibrils (27). Each microtubule-binding repeat in Tau contains a PGGG motif at its C-terminal end. Three of these motifs (270PGGG273, 332PGGG335, and 364PGGG367) were found in turn regions in PID filaments (27), and two (332PGGG335 and 364PGGG367) were found in AD filaments (26). Although residues 301PGGG304 were not part of the highly ordered core of AD filaments, it is likely that they, too, form a turn that allows residues at the end of R2 to pack against the first two β-strands in R3, as suggested by additional unresolved density in the cryo-EM maps (26). The lack of higher resolution structure was ascribed to the heterotypic stacking of R1 and R2 in these mixed filaments. It is highly likely that the Pro-301 mutations investigated in the present study alter the structural preferences in this potential turn region and thereby change the structural properties of the fibrils. Notably, this would also be consistent with an earlier study that suggested a conformational change in the P301L mutant relative to WT, namely an extension of β-sheet structure (71).
The in vitro studies that were performed here cannot replicate the complex biological environment in the human brain. It is clear that selective pressures acting in specific neuronal populations in particular brain regions may produce clouds of conformers that in one way or another are different from the ones produced in vitro (72, 73). Genetic background could add another layer of complexity that may modulate fibril structure. Despite all this, the work presented herein offers important molecular insights into the cross-seeding barriers of 4R Tau fibrils that are relevant to the selective deposition of 4R Tau isoforms in sporadic and familial 4R tauopathies.
Experimental procedures
Constructs
htau40 WT and htau23 WT were previously cloned into pET-28 as described (43). All point mutations were generated using site-directed mutagenesis following the QuikChange protocol from Stratagene/Agilent Technologies. For EPR experiments the two native cysteines were substituted by serines, and the position of desired MTSL labeling was mutated into a cysteine. The success of all mutagenesis was confirmed by DNA sequencing. All truncated constructs, with the exception of 244–372, which was also previously cloned into pET-28 as described (43), were ordered from Biomatic Corporation as lyophilized pET-28 plasmids containing the genes of interest inserted via the NcoI/XhoI cloning sites. All constructs contained two stop codons preceding the XhoI cloning site to remove the C-terminal His6 tag.
Expression and purification
Plasmids containing the desired inserts were first transformed into Escherichia coli strain BL21(DE3) and then grown on LB (Miller) agar plates. Single colonies were transferred into LB medium (Miller) and agitated for 15–17 h at 37 °C. The cultures were diluted 1:100 with LB medium and again agitated at 37 °C, until optical density reached 0.7–1 at 600 nm. For selection, the growth medium contained kanamycin (50 μg/ml in agar plates and 20 μg/ml in solution) (Gold Biotechnology). Protein expression was induced by addition of 0.5 mm isopropyl β-d-1-thiogalactopyranoside (Gold Biotechnology). Cultures were allowed to shake at 37 °C for another 3.5 h before being pelleted at 3,000 × g and taken up in resuspension buffer (500 mm NaCl, 20 mm PIPES (Research Products International), pH 6.5, 1 mm EDTA (Fisher Scientific), and 50 mm β-mercaptoethanol (Fisher BioReagents). The cells were heated at 80 °C for 20 min and tip-sonicated (Fisher Scientific sonifier 50% power with a 6-mm tip sonifier) on ice for 1 min before being centrifuged at 15,000 × g for 30 min to separate soluble protein from cellular debris. Soluble Tau was precipitated by gently shaking with 55–60% w/v ammonium sulfate (MP Biomedicals) for 3–20 h at 25 °C. Precipitated Tau pellets from a 15,000 × g spin were taken up in H2O with 2 mm DTT (Gold Biotechnology), sonicated for 2 min, syringe-filtered (GxF/GHP 0.45 μm), and loaded onto a cation exchange column (mono S 10/100 GL; GE Healthcare). Proteins were eluted using a linear NaCl gradient (50–1000 mm NaCl, 20 mm PIPES, pH 6.5, 2 mm EDTA), and fractions were pooled based on SDS–PAGE assessment. Pooled ion exchange fractions were loaded onto a Superdex 200 or Superdex 75 (GE Healthcare) gel-filtration column and eluted with 100 mm NaCl, 20 mm Tris (Sigma), pH 7.4, 1 mm EDTA, and 2 mm DTT buffer. Fractions were again assessed using SDS–PAGE and pooled accordingly and then left to precipitate overnight at 4 °C using either an equimolar volume of methanol or a 3-fold volumetric excess of acetone (43), along with 5 mm DTT. Following precipitation pellets were collected with a 15,000 × g spin for 10 min, washed with methanol or acetone, and stored at −80 °C until further use.
Protein solubilization and spin labeling
Methanol or acetone was removed from protein pellets, which were then dissolved in 200 μl of 8 m guanidine hydrochloride (Thermo). Proteins to be used for EPR experiments were at this point labeled by the addition of a 10-fold molar excess of paramagnetic label [1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl]methanethiosulfonate (Toronto Research Chemicals) or the nonparamagnetic analogue [1-acetyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl]methanethiosulfonate (Toronto Research Chemicals) and incubated at 25 °C for 1 h. To remove excess denaturant and unbound label, samples were passed over PD-10 columns (GE Healthcare) and eluted with assembly buffer (100 mm NaCl, 10 mm HEPES (J. T. Baker) and 0.1 mm NaN3 (Fisher Scientific) at pH 7.4). Protein concentration was determined by bicinchoninic acid assay (Pierce), and similar labeling efficiency of all MTSL-labeled proteins was confirmed using CW EPR at 25 °C.
Seed preparation and seeded reactions
Fibrils were formed by incubating a 500 μl mixture of 25 μm Tau, 50 μm heparin (average molecular weight = 4400, Celsus, EN-3225), 0.5 mm tris(2-carboxyethyl)phosphine (TCEP), and assembly buffer for 7–8 days at 37 °C, stirring with a Teflon-coated micro stir bar (5 × 2 mm) at 160 rpm. Fibrils were then subjected to 30 s of sonication with 20% power on a Fisher Scientific sonifier (model 100 with a 2-mm tip) to create fibril fragments or seeds. 10% of seeds (monomer equivalents) were combined with 10 μm Tau monomer, 20 μm heparin, 0.5 mm TCEP, and assembly buffer and let incubate quiescently at 37 °C for 20–24 h. The reactions that included constructs without any cysteines or those to be investigated via EPR did not contain TCEP. After incubation, the samples were centrifuged for 30 min at 130,000 × g. The pellets were separated from supernatants, and the volumes were adjusted using SDS sample buffer. Equal sample amounts were analyzed by SDS–PAGE (12% gels) and Coomassie staining. ImageJ was used on scanned gels to assess the amount of fibril growth for a given reaction by dividing the pixel density of protein bands in either the pellet or supernatant lane by the added pixel density of both. These values (multiplied by 100) are reported in all box-and-whisker plots as a percentage growth. In a reaction with 100% growth, the entire protein density would appear in the pellet lane, and none would appear in the supernatant lane. Box-and-whisker plots were created using GraphPad Prism 7 software, where the Tukey method was used for displaying whiskers and outliers, and statistical comparisons were performed using paired t tests.
Elongation kinetics
After combining 10 μm Tau, 20 μm heparin, 0.5 mm TCEP, 5 μm thioflavin T (Sigma), 5% seeds (monomer equivalents), and assembly buffer, fibril growth was monitored in real time at 37 °C by measuring the fluorescence emission at 480 nm using a BGM Labtech FLUOstar Omega plate reader. Excitation occurred at 440 nm. Prior to the first reading, the reactions were shaken for 2 s at 100 rpm. All readings were taken through the bottom of a Thermo Scientific 96-well optical PolymerBase plate.
CW EPR sample preparation, measurement, and analysis
Fibrils analyzed using CW EPR were formed by allowing a mixture of 30 μm MTSL-labeled monomer (either 100% paramagnetic MTSL-labeled or 20% paramagnetic MTSL-labeled, 80% nonparamagnetic MTSL-labeled monomer), 15 μm heparin, 5% seeds (not labeled with MTSL), and assembly buffer to quiescently incubate at 37 °C for 20–24 h. The htau40 mutants used as monomer had both of the native cysteines mutated into serines and one of the following mutations: E342C, K343C, D345C, I360C, K369C, I371C, or K375C, giving rise to seven different single MTSL-labeled monomer types to be used in these reactions. Following incubation, fibrils were pelleted in a 130,000 × g 30 min spin, supernatant was removed and pelleted fibrils were gently mixed with 10 μl of assembly buffer and loaded into round capillaries (Vitrotubes 0.6-mm inner diameter × 0.84-mm outer diameter). The spectra were depicted as first derivatives and comprised an average of 20 scans collected at room temperature at X-band with 150-G sweep width, 1- or 3-G modulation (as specified), and 12-milliwatt incident microwave power on a Bruker EMX Plus spectrometer fitted with an ER 4119HS resonator. The spectra were normalized to the same number of spins using double integration.
DEER sample preparation, measurement, and analysis
DEER samples were prepared by combining 48 μm cysteine-free non MTSL-labeled Tau, 2 μm Tau double MTSL labeled at positions 311 and 328, 10% seeds, 12.5 μm heparin, and assembly buffer and then incubating quiescently at 37 °C for 24 h. The seeds were assembled as described above, and fibrils indicated as mutant are 100% mutant in that the Tau used in the 48 μm cysteine free, 2 μm MTSL labeled, and seeds all contained the mutation of interest. The fibrils were sedimented in a 30-min 130,000 × g spin, the supernatant was removed, and the pellets were mixed with 10–15 μl of assembly buffer then transferred into quartz Q-band tubes (VITROCOM SFS round cell 1.1-mm inner diameter), flash-frozen, and stored at −80 °C until DEER measurement could be performed. DEER data were collected at Q-band using the four-pulse sequence as described previously (57, 58). A field-sweep spectrum was used to center the operating frequency (ν1), and the pump frequency (ν2) was adjusted to 37 MHz below ν1. For each experiment the gate and τ-0 were optimized and ranged between 64–72 ns and 910–914 ns respectively, and the π/2 pulse at ν1 and the π pulse at ν2, τ1, τ2, and τ3 and τx remained constant at 16, 32, 200, 3000, 100, and 8 ns, respectively. The shot repetition time was optimized for each sample to be 1.2 × T1 (spin-lattice relaxation time), and signal averaging took place for ∼24 h/sample with each scan containing eight-step phase cycling to remove unwanted echoes. The data were collected using a Bruker ELEXSYS E580 spectrometer with an ER 5107D2 dielectric resonator, a 580–400U ELDOR unit as the second microwave source and a 10-W amplifier. Temperature was kept at 80 K using an Oxford CF935 cryostat and liquid nitrogen. The data were analyzed with DEER analysis 2018 (74) using Tikhonov regularization, three-dimensional homogeneous background fit, program-optimized selection utilizing validation tool for zero time, background time, regularization parameter, and phase, whereas cutoff time was kept constant for each sample at 2936 ns.
Transmission electron microscopy
Formvar/carbon-coated 200 mesh copper grids (Electron Microscopy Sciences) were exposed to a 10-μl droplet containing ∼2.5 μm Tau for 1.5 min followed by exposure to a 10-μl droplet of 2% uranyl acetate for 1.5 min. Images were recorded with a FEI Tecnai T12 Biotwin transmission electron microscope at 100 keV equipped with a Gatan CCD camera. Quantification of seed length was done by analyzing micrographs of sonicated seeds using ImageJ software.
Author contributions
H. A. W., G. W., B. M., R. N., and M. M. conceptualization; H. A. W. and M. M. data curation; H. A. W., R.M., and M. M. formal analysis; H. A. W. and M. M. validation; H. A. W. and R.M. investigation; H. A. W. and M. M. visualization; H. A. W. and M. M. writing-review and editing; M. M. resources; M. M. supervision; M. M. funding acquisition; M. M. writing-original draft; M. M. project administration; G. W., B. M., R. N., and M. M. structural interpretation.
Supplementary Material
Acknowledgments
We thank Drs. S. S. Eaton and G. R. Eaton for technical support on the DEER measurements and Dr. M. R. Holden for help in the collection of EM images. This project was also supported by Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health under contract HHSN261200800001E. Further support was provided by the Intramural Research Program of National Institutes of Health, Frederick National Lab, Center for Cancer Research.
This work was supported by National Institutes of Health Grant R21AG051833 (to M. M.). This work was also supported by National Natural Science Foundation of China Grant 11674065 (to G. W.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
- AD
- Alzheimer's disease
- PSP
- progressive supranuclear palsy
- CBD
- corticobasal degeneration
- PID
- Pick's disease
- 3R
- three-repeat
- 4R
- four-repeat
- DEER
- double electron-electron resonance
- TEM
- transmission electron microscopy
- ThT
- thioflavin T
- CW
- continuous wave
- TCEP
- tris(2-carboxyethyl)phosphine
- PHF
- paired helical filament
- MTSL
- [1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl]methanethiosulfonate.
References
- 1. Lee V. M., Goedert M., and Trojanowski J. Q. (2001) Neurodegenerative tauopathies. Annu. Rev. Neurosci. 24, 1121–1159 10.1146/annurev.neuro.24.1.1121 [DOI] [PubMed] [Google Scholar]
- 2. Buée L., Bussière T., Buée-Scherrer V., Delacourte A., and Hof P. R. (2000) Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Brain Res. Rev 33, 95–130 10.1016/S0165-0173(00)00019-9 [DOI] [PubMed] [Google Scholar]
- 3. Ghetti B., Oblak A. L., Boeve B. F., Johnson K. A., Dickerson B. C., and Goedert M. (2015) Frontotemporal dementia caused by microtubule-associated protein Tau gene (MAPT) mutations: a chameleon for neuropathology and neuroimaging. Neuropathol. Appl. Neurobiol. 41, 24–46 10.1111/nan.12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Braak H., and Braak E. (1991) Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 10.1007/BF00308809 [DOI] [PubMed] [Google Scholar]
- 5. Williams D. R., Holton J. L., Strand C., Pittman A., de Silva R., Lees A. J., and Revesz T. (2007) Pathological Tau burden and distribution distinguishes progressive supranuclear palsy-parkinsonism from Richardson's syndrome. Brain 130, 1566–1576 10.1093/brain/awm104 [DOI] [PubMed] [Google Scholar]
- 6. Irwin D. J., Brettschneider J., McMillan C. T., Cooper F., Olm C., Arnold S. E., Van Deerlin V. M., Seeley W. W., Miller B. L., Lee E. B., Lee V. M., Grossman M., and Trojanowski J. Q. (2016) Deep clinical and neuropathological phenotyping of Pick disease. Ann. Neurol. 79, 272–287 10.1002/ana.24559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saito Y., Ruberu N. N., Sawabe M., Arai T., Tanaka N., Kakuta Y., Yamanouchi H., and Murayama S. (2004) Staging of argyrophilic grains: an age-associated tauopathy. J. Neuropathol. Exp. Neurol. 63, 911–918 10.1093/jnen/63.9.911 [DOI] [PubMed] [Google Scholar]
- 8. McKee A. C., Stern R. A., Nowinski C. J., Stein T. D., Alvarez V. E., Daneshvar D. H., Lee H. S., Wojtowicz S. M., Hall G., Baugh C. M., Riley D. O., Kubilus C. A., Cormier K. A., Jacobs M. A., Martin B. R., et al. (2013) The spectrum of disease in chronic traumatic encephalopathy. Brain 136, 43–64 10.1093/brain/aws307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Calignon A., Polydoro M., Suárez-Calvet M., William C., Adamowicz D. H., Kopeikina K. J., Pitstick R., Sahara N., Ashe K. H., Carlson G. A., Spires-Jones T. L., and Hyman B. T. (2012) Propagation of Tau pathology in a model of early Alzheimer's disease. Neuron 73, 685–697 10.1016/j.neuron.2011.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu L., Drouet V., Wu J. W., Witter M. P., Small S. A., Clelland C., and Duff K. (2012) Trans-synaptic spread of Tau pathology in vivo. PLoS One 7, e31302 10.1371/journal.pone.0031302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Clavaguera F., Akatsu H., Fraser G., Crowther R. A., Frank S., Hench J., Probst A., Winkler D. T., Reichwald J., Staufenbiel M., Ghetti B., Goedert M., and Tolnay M. (2013) Brain homogenates from human tauopathies induce Tau inclusions in mouse brain. Proc. Natl. Acad. Sci. U.S.A. 110, 9535–9540 10.1073/pnas.1301175110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sanders D. W., Kaufman S. K., DeVos S. L., Sharma A. M., Mirbaha H., Li A., Barker S. J., Foley A. C., Thorpe J. R., Serpell L. C., Miller T. M., Grinberg L. T., Seeley W. W., and Diamond M. I. (2014) Distinct Tau prion strains propagate in cells and mice and define different tauopathies. Neuron 82, 1271–1288 10.1016/j.neuron.2014.04.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Narasimhan S., Guo J. L., Changolkar L., Stieber A., McBride J. D., Silva L. V., He Z., Zhang B., Gathagan R. J., Trojanowski J. Q., and Lee V. M. Y. (2017) Pathological Tau strains from human brains recapitulate the diversity of tauopathies in nontransgenic mouse brain. J. Neurosci. 37, 11406–11423 10.1523/JNEUROSCI.1230-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goedert M., Eisenberg D. S., and Crowther R. A. (2017) Propagation of Tau aggregates and neurodegeneration. Annu. Rev. Neurosci. 40, 189–210 10.1146/annurev-neuro-072116-031153 [DOI] [PubMed] [Google Scholar]
- 15. Collinge J. (2016) Mammalian prions and their wider relevance in neurodegenerative diseases. Nature 539, 217–226 10.1038/nature20415 [DOI] [PubMed] [Google Scholar]
- 16. Collinge J., and Clarke A. R. (2007) A general model of prion strains and their pathogenicity. Science 318, 930–936 10.1126/science.1138718 [DOI] [PubMed] [Google Scholar]
- 17. Ayers J. I., Giasson B. I., and Borchelt D. R. (2018) Prion-like spreading in tauopathies. Biol. Psychiatry 83, 337–346 10.1016/j.biopsych.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goedert M., Spillantini M. G., Jakes R., Rutherford D., and Crowther R. A. (1989) Multiple isoforms of human microtubule-associated protein Tau: sequences and localization in neurofibrillary tangles of Alzheimer's disease. Neuron 3, 519–526 10.1016/0896-6273(89)90210-9 [DOI] [PubMed] [Google Scholar]
- 19. Goedert M., and Jakes R. (1990) Expression of separate isoforms of human Tau protein: correlation with the Tau pattern in brain and effects on tubulin polymerization. EMBO J. 9, 4225–4230 10.1002/j.1460-2075.1990.tb07870.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goedert M., Spillantini M. G., Cairns N. J., and Crowther R. A. (1992) Tau proteins of Alzheimer paired helical filaments: abnormal phosphorylation of all six brain isoforms. Neuron 8, 159–168 10.1016/0896-6273(92)90117-V [DOI] [PubMed] [Google Scholar]
- 21. Flament S., Delacourte A., Verny M., Hauw J. J., and Javoy-Agid F. (1991) Abnormal Tau proteins in progressive supranuclear palsy: similarities and differences with the neurofibrillary degeneration of the Alzheimer type. Acta Neuropathol. 81, 591–596 10.1007/BF00296367 [DOI] [PubMed] [Google Scholar]
- 22. Ksiezak-Reding H., Morgan K., Mattiace L. A., Davies P., Liu W. K., Yen S. H., Weidenheim K., and Dickson D. W. (1994) Ultrastructure and biochemical composition of paired helical filaments in corticobasal degeneration. Am. J. Pathol 145, 1496–1508 [PMC free article] [PubMed] [Google Scholar]
- 23. Delacourte A., Robitaille Y., Sergeant N., Buée L., Hof P. R., Wattez A., Laroche-Cholette A., Mathieu J., Chagnon P., and Gauvreau D. (1996) Specific pathological Tau protein variants characterize Pick's disease. J. Neuropathol. Exp. Neurol. 55, 159–168 10.1097/00005072-199602000-00004 [DOI] [PubMed] [Google Scholar]
- 24. Berriman J., Serpell L. C., Oberg K. A., Fink A. L., Goedert M., and Crowther R. A. (2003) Tau filaments from human brain and from in vitro assembly of recombinant protein show cross-β structure. Proc. Natl. Acad. Sci. U.S.A. 100, 9034–9038 10.1073/pnas.1530287100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Margittai M., and Langen R. (2004) Template-assisted filament growth by parallel stacking of Tau. Proc. Natl. Acad. Sci. U.S.A. 101, 10278–10283 10.1073/pnas.0401911101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fitzpatrick A. W. P., Falcon B., He S., Murzin A. G., Murshudov G., Garringer H. J., Crowther R. A., Ghetti B., Goedert M., and Scheres S. H. W. (2017) Cryo-EM structures of Tau filaments from Alzheimer's disease. Nature 547, 185–190 10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Falcon B., Zhang W., Murzin A. G., Murshudov G., Garringer H. J., Vidal R., Crowther R. A., Ghetti B., Scheres S. H. W., and Goedert M. (2018) Structures of filaments from Pick's disease reveal a novel Tau protein fold. Nature 561, 137–140 10.1038/s41586-018-0454-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gustke N., Trinczek B., Biernat J., Mandelkow E. M., and Mandelkow E. (1994) Domains of Tau protein and interactions with microtubules. Biochemistry 33, 9511–9522 10.1021/bi00198a017 [DOI] [PubMed] [Google Scholar]
- 29. Dinkel P. D., Siddiqua A., Huynh H., Shah M., and Margittai M. (2011) Variations in filament conformation dictate seeding barrier between three- and four-repeat Tau. Biochemistry 50, 4330–4336 10.1021/bi2004685 [DOI] [PubMed] [Google Scholar]
- 30. Yu X., Luo Y., Dinkel P., Zheng J., Wei G., Margittai M., Nussinov R., and Ma B. (2012) Cross-seeding and conformational selection between three- and four-repeat human Tau proteins. J. Biol. Chem. 287, 14950–14959 10.1074/jbc.M112.340794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Naiki H., Higuchi K., Hosokawa M., and Takeda T. (1989) Fluorometric determination of amyloid fibrils in vitro using the fluorescent dye, thioflavin T1. Anal. Biochem. 177, 244–249 10.1016/0003-2697(89)90046-8 [DOI] [PubMed] [Google Scholar]
- 32. Meyer V., Dinkel P. D., Rickman Hager E., and Margittai M. (2014) Amplification of Tau fibrils from minute quantities of seeds. Biochemistry 53, 5804–5809 10.1021/bi501050g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miyasaka T., Morishima-Kawashima M., Ravid R., Kamphorst W., Nagashima K., and Ihara Y. (2001) Selective deposition of mutant Tau in the FTDP-17 brain affected by the P301L mutation. J. Neuropathol. Exp. Neurol. 60, 872–884 10.1093/jnen/60.9.872 [DOI] [PubMed] [Google Scholar]
- 34. Aoyagi H., Hasegawa M., and Tamaoka A. (2007) Fibrillogenic nuclei composed of P301L mutant Tau induce elongation of P301L Tau but not wild-type Tau. J. Biol. Chem. 282, 20309–20318 10.1074/jbc.M611876200 [DOI] [PubMed] [Google Scholar]
- 35. Strang K. H., Croft C. L., Sorrentino Z. A., Chakrabarty P., Golde T. E., and Giasson B. I. (2018) Distinct differences in prion-like seeding and aggregation between Tau protein variants provide mechanistic insights into tauopathies. J. Biol. Chem. 293, 2408–2421 10.1074/jbc.M117.815357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. von Bergen M., Friedhoff P., Biernat J., Heberle J., Mandelkow E. M., and Mandelkow E. (2000) Assembly of Tau protein into Alzheimer paired helical filaments depends on a local sequence motif (306VQIVYK311) forming β structure. Proc. Natl. Acad. Sci. U.S.A. 97, 5129–5134 10.1073/pnas.97.10.5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pastor P., Pastor E., Carnero C., Vela R., Garcia T., Amer G., Tolosa E., and Oliva R. (2001) Familial atypical progressive supranuclear palsy associated with homozigosity for the delN296 mutation in the Tau gene. Ann. Neurol. 49, 263–267 10.1002/1531-8249(20010201)49:2%3C263::AID-ANA50%3E3.0.CO%3B2-K [DOI] [PubMed] [Google Scholar]
- 38. Ros R., Thobois S., Streichenberger N., Kopp N., Sánchez M. P., Pérez M., Hoenicka J., Avila J., Honnorat J., and de Yébenes J. G. (2005) A new mutation of the Tau gene, G303V, in early-onset familial progressive supranuclear palsy. Arch. Neurol. 62, 1444–1450 10.1001/archneur.62.9.1444 [DOI] [PubMed] [Google Scholar]
- 39. Iijima M., Tabira T., Poorkaj P., Schellenberg G. D., Trojanowski J. Q., Lee V. M., Schmidt M. L., Takahashi K., Nabika T., Matsumoto T., Yamashita Y., Yoshioka S., and Ishino H. (1999) A distinct familial presenile dementia with a novel missense mutation in the Tau gene. Neuroreport 10, 497–501 10.1097/00001756-199902250-00010 [DOI] [PubMed] [Google Scholar]
- 40. Yoshida H., Crowther R. A., and Goedert M. (2002) Functional effects of Tau gene mutations deltaN296 and N296H. J. Neurochem. 80, 548–551 10.1046/j.0022-3042.2001.00729.x [DOI] [PubMed] [Google Scholar]
- 41. Hasegawa M., Smith M. J., Iijima M., Tabira T., and Goedert M. (1999) FTDP-17 mutations N279K and S305N in Tau produce increased splicing of exon 10. FEBS Lett. 443, 93–96 10.1016/S0014-5793(98)01696-2 [DOI] [PubMed] [Google Scholar]
- 42. Margittai M., and Langen R. (2006) Side chain-dependent stacking modulates Tau filament structure. J. Biol. Chem. 281, 37820–37827 10.1074/jbc.M605336200 [DOI] [PubMed] [Google Scholar]
- 43. Siddiqua A., and Margittai M. (2010) Three- and four-repeat Tau coassemble into heterogeneous filaments: an implication for Alzheimer disease. J. Biol. Chem. 285, 37920–37926 10.1074/jbc.M110.185728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Berliner L. J., Grunwald J., Hankovszky H. O., and Hideg K. (1982) A novel reversible thiol-specific spin label: papain active site labeling and inhibition. Anal. Biochem. 119, 450–455 10.1016/0003-2697(82)90612-1 [DOI] [PubMed] [Google Scholar]
- 45. Gross A., Columbus L., Hideg K., Altenbach C., and Hubbell W. L. (1999) Structure of the KcsA potassium channel from Streptomyces lividans: a site-directed spin labeling study of the second transmembrane segment. Biochemistry 38, 10324–10335 10.1021/bi990856k [DOI] [PubMed] [Google Scholar]
- 46. Chen M., Margittai M., Chen J., and Langen R. (2007) Investigation of α-synuclein fibril structure by site-directed spin labeling. J. Biol. Chem. 282, 24970–24979 10.1074/jbc.M700368200 [DOI] [PubMed] [Google Scholar]
- 47. Cleveland D. W., Hwo S. Y., and Kirschner M. W. (1977) Physical and chemical properties of purified Tau factor and the role of Tau in microtubule assembly. J. Mol. Biol. 116, 227–247 10.1016/0022-2836(77)90214-5 [DOI] [PubMed] [Google Scholar]
- 48. Schweers O., Schönbrunn-Hanebeck E., Marx A., and Mandelkow E. (1994) Structural studies of Tau protein and Alzheimer paired helical filaments show no evidence for β-structure. J. Biol. Chem. 269, 24290–24297 [PubMed] [Google Scholar]
- 49. Margittai M., and Langen R. (2008) Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: molecular insights from electron paramagnetic resonance spectroscopy. Q. Rev. Biophys. 41, 265–297 10.1017/S0033583508004733 [DOI] [PubMed] [Google Scholar]
- 50. Milov A. D., Salikhov K. M., and Shchirov M. D. (1981) Apllication of the double resonance method to electron spin echo in a study of the spatial distribution of paramagnetic centers in solids. Sov. Phys. Solid State 23, 565–569 [Google Scholar]
- 51. Jeschke G., Pannier M., and Spiess H. W. (2000) Double electron-electron resonance. Biol. Magn. Reson. 19, 493–512 [Google Scholar]
- 52. Jeschke G. (2012) DEER distance measurements on proteins. Annu. Rev. Phys. Chem. 63, 419–446 10.1146/annurev-physchem-032511-143716 [DOI] [PubMed] [Google Scholar]
- 53. Karyagina I., Becker S., Giller K., Riedel D., Jovin T. M., Griesinger C., and Bennati M. (2011) Electron paramagnetic resonance spectroscopy measures the distance between the external β-strands of folded α-synuclein in amyloid fibrils. Biophys. J. 101, L1–3 10.1016/j.bpj.2011.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bedrood S., Li Y., Isas J. M., Hegde B. G., Baxa U., Haworth I. S., and Langen R. (2012) Fibril structure of human islet amyloid polypeptide. J. Biol. Chem. 287, 5235–5241 10.1074/jbc.M111.327817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pornsuwan S., Giller K., Riedel D., Becker S., Griesinger C., and Bennati M. (2013) Long-range distances in amyloid fibrils of α-synuclein from PELDOR spectroscopy. Angew. Chem. Int. Ed. Engl. 52, 10290–10294 10.1002/anie.201304747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fichou Y., Vigers M., Goring A. K., Eschmann N. A., and Han S. (2018) Heparin-induced Tau filaments are structurally heterogeneous and differ from Alzheimer's disease filaments. Chem. Commun. (Camb.) 54, 4573–4576 10.1039/c8cc01355a ; Correction (2018) Chem. Commun. (Camb.) 54, 8653 [DOI] [PubMed] [Google Scholar]
- 57. Siddiqua A., Luo Y., Meyer V., Swanson M. A., Yu X., Wei G., Zheng J., Eaton G. R., Ma B., Nussinov R., Eaton S. S., and Margittai M. (2012) Conformational basis for asymmetric seeding barrier in filaments of three- and four-repeat Tau. J. Am. Chem. Soc. 134, 10271–10278 10.1021/ja303498q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Meyer V., Dinkel P. D., Luo Y., Yu X., Wei G., Zheng J., Eaton G. R., Ma B., Nussinov R., Eaton S. S., and Margittai M. (2014) Single mutations in Tau modulate the populations of fibril conformers through seed selection. Angew. Chem. Int. Ed. Engl. 53, 1590–1593 10.1002/anie.201308473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Meyer V., Holden M. R., Weismiller H. A., Eaton G. R., Eaton S. S., and Margittai M. (2016) Fracture and growth are competing forces determining the fate of conformers in Tau fibril populations. J. Biol. Chem. 291, 12271–12281 10.1074/jbc.M116.715557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Berliner L. J., Eaton G. R., and Eaton S. S. (2000) Biological Magnetic Resonance: Distance Measurements in Biological Systems by EPR, Kluwer Academic/Plenum Publishers, New York [Google Scholar]
- 61. Dinkel P. D., Holden M. R., Matin N., and Margittai M. (2015) RNA binds to Tau fibrils and sustains template-assisted growth. Biochemistry 54, 4731–4740 10.1021/acs.biochem.5b00453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Carmel G., Mager E. M., Binder L. I., and Kuret J. (1996) The structural basis of monoclonal antibody Alz50's selectivity for Alzheimer's disease pathology. J. Biol. Chem. 271, 32789–32795 10.1074/jbc.271.51.32789 [DOI] [PubMed] [Google Scholar]
- 63. Bibow S., Mukrasch M. D., Chinnathambi S., Biernat J., Griesinger C., Mandelkow E., and Zweckstetter M. (2011) The dynamic structure of filamentous Tau. Angew. Chem. Int. Ed. Engl. 50, 11520–11524 10.1002/anie.201105493 [DOI] [PubMed] [Google Scholar]
- 64. Gamblin T. C., Chen F., Zambrano A., Abraha A., Lagalwar S., Guillozet A. L., Lu M., Fu Y., Garcia-Sierra F., LaPointe N., Miller R., Berry R. W., Binder L. I., and Cryns V. L. (2003) Caspase cleavage of Tau: linking amyloid and neurofibrillary tangles in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 100, 10032–10037 10.1073/pnas.1630428100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rissman R. A., Poon W. W., Blurton-Jones M., Oddo S., Torp R., Vitek M. P., LaFerla F. M., Rohn T. T., and Cotman C. W. (2004) Caspase-cleavage of Tau is an early event in Alzheimer disease tangle pathology. J. Clin. Invest. 114, 121–130 10.1172/JCI200420640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang Z., Song M., Liu X., Kang S. S., Kwon I. S., Duong D. M., Seyfried N. T., Hu W. T., Liu Z., Wang J. Z., Cheng L., Sun Y. E., Yu S. P., Levey A. I., and Ye K. (2014) Cleavage of Tau by asparagine endopeptidase mediates the neurofibrillary pathology in Alzheimer's disease. Nat. Med. 20, 1254–1262 10.1038/nm.3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Quinn J. P., Corbett N. J., Kellett K. A. B., and Hooper N. M. (2018) Tau proteolysis in the pathogenesis of tauopathies: neurotoxic fragments and novel biomarkers. J. Alzheimers Dis. 63, 13–33 10.3233/JAD-170959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Taniguchi-Watanabe S., Arai T., Kametani F., Nonaka T., Masuda-Suzukake M., Tarutani A., Murayama S., Saito Y., Arima K., Yoshida M., Akiyama H., Robinson A., Mann D. M. A., Iwatsubo T., and Hasegawa M. (2016) Biochemical classification of tauopathies by immunoblot, protein sequence and mass spectrometric analyses of sarkosyl-insoluble and trypsin-resistant Tau. Acta Neuropathol. 131, 267–280 10.1007/s00401-015-1503-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Puoti G., Giaccone G., Rossi G., Canciani B., Bugiani O., and Tagliavini F. (1999) Sporadic Creutzfeldt–Jakob disease: co-occurrence of different types of PrP(Sc) in the same brain. Neurology 53, 2173–2176 10.1212/WNL.53.9.2173 [DOI] [PubMed] [Google Scholar]
- 70. Qiang W., Yau W. M., Lu J. X., Collinge J., and Tycko R. (2017) Structural variation in amyloid-β fibrils from Alzheimer's disease clinical subtypes. Nature 541, 217–221 10.1038/nature20814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. von Bergen M., Barghorn S., Li L., Marx A., Biernat J., Mandelkow E. M., and Mandelkow E. (2001) Mutations of Tau protein in frontotemporal dementia promote aggregation of paired helical filaments by enhancing local β-structure. J. Biol. Chem. 276, 48165–48174 10.1074/jbc.M105196200 [DOI] [PubMed] [Google Scholar]
- 72. Falcon B., Cavallini A., Angers R., Glover S., Murray T. K., Barnham L., Jackson S., O'Neill M. J., Isaacs A. M., Hutton M. L., Szekeres P. G., Goedert M., and Bose S. (2015) Conformation determines the seeding potencies of native and recombinant Tau aggregates. J. Biol. Chem. 290, 1049–1065 10.1074/jbc.M114.589309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Guo J. L., Narasimhan S., Changolkar L., He Z., Stieber A., Zhang B., Gathagan R. J., Iba M., McBride J. D., Trojanowski J. Q., and Lee V. M. (2016) Unique pathological Tau conformers from Alzheimer's brains transmit Tau pathology in nontransgenic mice. J. Exp. Med. 213, 2635–2654 10.1084/jem.20160833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Jeschke G., Chechnik V., Ionita P., Godt A., Zimmermann H., Banham J., Timmel C. R., Hilger D., and Jung H. (2006) DEER Analysis 2006: a comprehensive software package for analyzing pulsed ELDOR data. Appl. Magn. Reson. 30, 473–498 10.1007/BF03166213 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.