Abstract
Sactipeptides are a subclass of ribosomally synthesized and post-translationally modified peptides (RiPPs). They contain a unique thioether bond, referred to as a sactionine linkage, between the sulfur atom of a cysteine residue and the α-carbon of an acceptor residue. These linkages are formed via radical chemistry and are essential for the spermicidal, antifungal, and antibacterial properties of sactipeptides. Enzymes that form these linkages, called sactisynthases, are AdoMet radical enzymes in the SPASM/Twitch subgroup whose structures are incompletely characterized. Here, we present the X-ray crystal structure to 1.29-Å resolution and Mössbauer analysis of SkfB, a sactisynthase from Bacillus subtilis involved in making sporulation killing factor (SKF). We found that SkfB is a modular enzyme with an N-terminal peptide-binding domain comprising a RiPP recognition element (RRE), a middle domain that forms a classic AdoMet radical partial (β/α)6 barrel structure and displays AdoMet bound to the [4Fe-4S] cluster, and a C-terminal region characteristic of the so-called Twitch domain housing an auxiliary iron-sulfur cluster. Notably, both crystallography and Mössbauer analyses suggest that SkfB can bind a [2Fe-2S] cluster at the auxiliary cluster site, which has been observed only once before in a SPASM/Twitch auxiliary cluster site in the structure of another AdoMet radical enzyme, the pyrroloquinoline quinone biosynthesis enzyme PqqE. Taken together, our findings indicate that SkfB from B. subtilis represents a unique enzyme containing several structural features observed in other AdoMet radical enzymes.
Keywords: enzyme catalysis, enzyme structure, metalloenzyme, crystal structure, catalysis, AdoMet radical enzymes, iron sulfur clusters, radical SAM, sactionine linkage, sporulation killing factor
Introduction
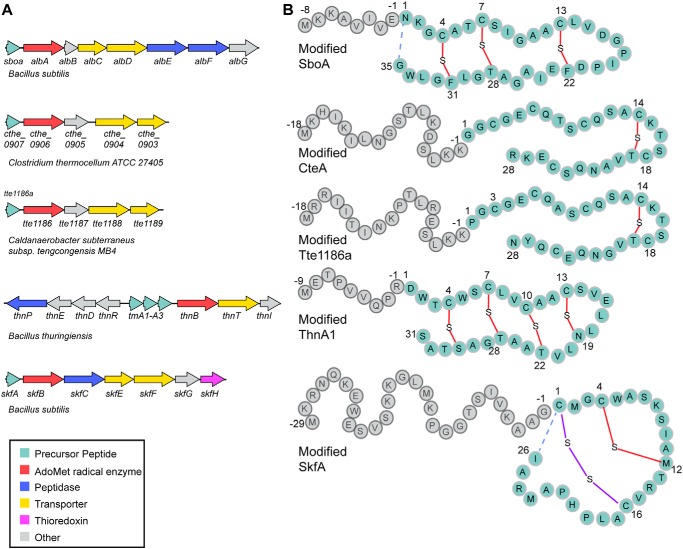
Sulfur to alpha-carbon cross-linked peptides (sactipeptides)5 are a subclass of ribosomally synthesized and post-translationally modified peptide (RiPP) natural products characterized by the presence of one or more thioether bonds between the sulfur atom of a cysteine residue and the α-carbon of an acceptor residue, forming a so-called sactionine linkage (see Fig. 1) (1). To date, six sactipeptides have been identified, and most show chemical and metabolic stability and a narrow spectrum of antimicrobial activity (2–6).
Figure 1.
Sactipeptide biosynthesis. A, gene scheme of several RiPP biosynthetic gene clusters involved in the biosynthesis of sactipeptides. These biosynthetic clusters encode precursor peptides (cyan); AdoMet radical enzymes (red), which are responsible for installing the unique sactionine linkages characteristic of the sactipeptide natural products; peptidases (blue), which are responsible for cleaving the leader peptide; and ABC transporters, which are involved in transporting the sactipeptide out of the producing organism. B, the precursor peptides for each biosynthetic gene cluster in A are shown with the leader peptide sequence, which binds to the biosynthetic enzymes, in gray and the mature peptide, which undergoes post-translational modifications, in cyan. The sactionine linkages formed between the sulfur atom of a cysteine residue and the α-carbon of an acceptor residue are shown in red. Final maturation steps of SboA and SkfA involve an N- to C-terminal macrocyclization (dashed blue lines) step concurrent with a leader peptide cleavage step. Maturation of SkfA also includes disulfide bond formation (purple).
Biosynthesis of sactipeptides commences with a leader peptide–dependent post-translational modification of the precursor peptide core by colocalized radical enzymes, herein called sactisynthases (see Fig. 1A) (1, 2, 7–10). Known sactisynthases include AlbA, SkfB, and presumably CteB, which install a sactionine linkage on precursor peptides SboA, SkfA, and CteA, respectively, during the maturation of the sactipeptides subtilosin A (8), sporulation killing factor (7), and thermocellin (11), respectively (see Fig. 1). These sactisynthases are members of the S-adenosyl-l-methionine (AdoMet) radical enzyme superfamily, which utilize a canonical CX3CXφC motif (where φ is a conserved aromatic amino acid) to coordinate a [4Fe-4S] cluster, thereby named the AdoMet radical cluster (12). AdoMet binds to the site-differentiated iron of the AdoMet radical cluster and is reductively cleaved to produce a highly reactive 5′-deoxyadenosyl radical (5′-dAdo•) species. 5′-dAdo• subsequently abstracts a hydrogen atom from the substrate, forming 5′-deoxyadenosine and a substrate radical, which undergoes further transformation to form the desired product. In the case of the characterized sactisynthases, H-atom abstraction has been shown to occur from the α-carbon of the acceptor residue (8, 13–15).
In addition to a characteristic AdoMet radical core, which houses the active site, sactisynthases are predicted to contain an N-terminal peptide-binding domain called a RiPP recognition element (RRE) (16) and a C-terminal SPASM or Twitch domain (17). RREs are expected to bind the leader sequences of the peptide substrates to secure the substrate to the enzyme, whereas the SPASM or Twitch domains are expected to coordinate additional or auxiliary iron-sulfur clusters (Aux I and Aux II), whose function in sactipeptide generation is not fully established.
SPASM and Twitch domains are not unique to sactisynthases. They are found in several thousand AdoMet radical enzymes that display activities ranging from the dehydrogenation reaction catalyzed by butirosin biosynthetic enzyme BtrN to the amino acid cross-linking reaction catalyzed by the streptide-like biosynthetic enzyme SuiB (Fig. S1). SPASM domains are characterized by the presence of a conserved seven-cysteine motif (CX9–15GX4CgapCX2CX5CX3CgapC) that is used to coordinate two iron-sulfur clusters, Aux I and Aux II (17, 18). The name “SPASM” is derived from the biosynthetic products of the first set of enzymes identified: AlbA, PqqE, anaerobic sulfatase maturating enzyme (anSME), and MftC, which biosynthesize subtilosin A, pyrroloquinoline quinone, anaerobic sulfatase, and mycofactocin, respectively. The so-called Twitch domain is a truncated version of the SPASM domain that binds only one auxiliary cluster (19).
The first set of structures of SPASM and Twitch domain–containing enzymes showed variations in the cysteine ligation of the Aux clusters. anSME (SPASM subclass) and BtrN (Twitch subclass) displayed Aux clusters that were fully ligated, whereas the molybdenum cofactor biosynthetic enzyme MoaA displayed an open coordination site for the binding and positioning of the substrate GTP in the active site (19–21). For anSME and BtrN, auxiliary clusters have been suggested to play a role in oxidizing the radical intermediate species to produce product as neither of these enzymes use their cluster for substrate binding (20). The only structure of a sactisynthase determined to date, CteB, revealed a SPASM domain with two auxiliary clusters and an open coordination site on Aux I (11), the cluster closest to the active site. However, instead of binding the cysteine residue that forms the C–S bond linkage, as predicted for the AlbA system (8), a cysteine residue from the leader sequence of substrate CteA is coordinated to Aux I. The 21-mer CteA fragment utilized in crystallography includes the leader peptide sequence (residues −1 to −18) and the first three residues of the core peptide but did not contain the cysteine involved in sactionine formation (11) (see Fig. 1B). Thus, the role of the Aux I cluster in CteB is not clear at this time.
Here, we present the first view of the sactisynthase SkfB, a Twitch domain–containing family member. SkfB initiates thioether cross-link formation between the α-carbon of Met12 of the precursor peptide SkfA (Fig. 1B) during the maturation of the sactipeptide sporulation killing factor in Bacillus subtilis (1, 7, 13). Although the role of the Aux cluster in SkfB is unknown, it is required for overall activity (7), and recent studies utilizing a radical clock substrate where Met12 is replaced by a cyclopropylglycine residue show that ring opening occurs in a variant where the Aux cluster has been removed (15). The cyclopropylglycine-containing substrate does not undergo thioether cross-link formation. These studies suggest that in SkfB, Aux I does not play any role in the initial H-atom transfer events, but rather in the late stages of the functionalization of the peptide. To further probe the role and nature of Aux I in SkfB, here we investigated SkfB both by crystallography and spectroscopy.
Results
Overall architecture of SkfB is modular
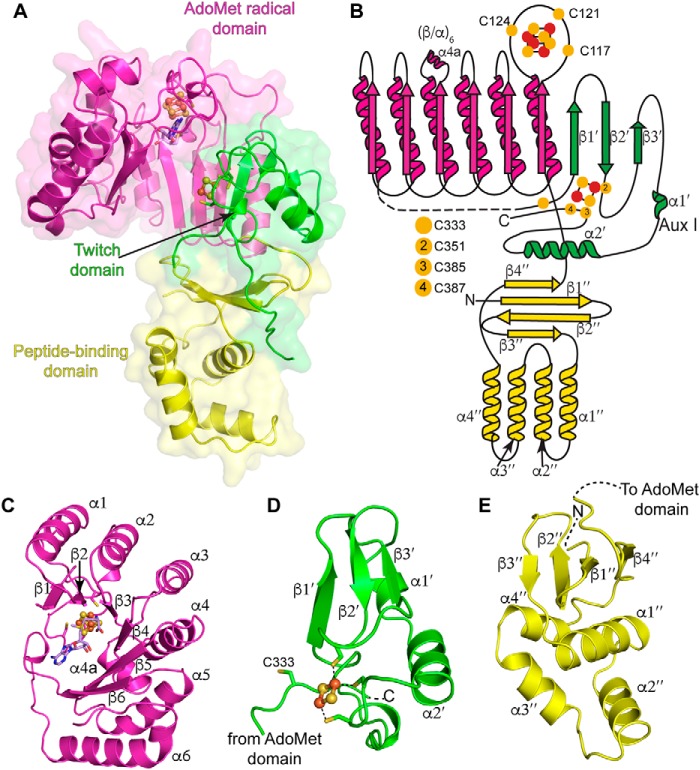
The structure of SkfB from B. subtilis in complex with AdoMet was solved to 1.29-Å resolution using an iron single-wavelength anomalous dispersion technique (Table 1). The final structure contained one molecule in the asymmetric unit with residues 12–322 and 330–401, a molecule of AdoMet bound to a [4Fe-4S] AdoMet radical cluster, and a [2Fe-2S] auxiliary cluster. The SkfB architecture is modular, folding into three distinct domains: an N-terminal peptide-binding domain, an AdoMet radical domain, and a C-terminal Twitch domain (Fig. 2).
Table 1.
Data collection and refinement statistics for SkfB structure
The highest-resolution shell is shown in parentheses. r.m.s., root mean square; asu, asymmetric unit.
| SkfB |
||
|---|---|---|
| Iron peaka | Remote | |
| Data collection | ||
| Beamline | Home Source | APS 24-ID-C |
| Wavelength (Å) | 1.5418 | 0.9792 |
| Space group | C2 | C2 |
| Cell dimensions (Å) | 76.5, 83.1, 61.3 | 76.5, 83.1, 61.3 |
| Resolution (Å) | 50.0–1.97 (2.00–1.97) | 50.0–1.29 (1.34–1.29) |
| Unique reflections | 52,216 | 92,230 |
| Completeness | 97.8 (69.0) | 96.5 (76.2) |
| Redundancy | 3.6 (2.4) | 5.6 (2.6) |
| Rsymb | 0.043 (0.119) | 0.037 (0.499) |
| I/σ(I) | 29.5 (7.7) | 43.6 (1.8) |
| CC1/2d | (0.729) | |
| Model refinement | ||
| Resolution limits (Å) | 50.0–1.29 | |
| Rwork/Rfreec | 14.25/16.85 | |
| No. molecules in asu | 1 | |
| No. atoms | ||
| Protein | 3,226 | |
| [Fe-S] | 12 | |
| AdoMet | 27 | |
| Water | 390 | |
| B-factors (Å2) | ||
| Protein | 20.2 | |
| [Fe-S] | 15.8 | |
| AdoMet | 13.74 | |
| Water | 30.7 | |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | |
| Bond Angles (°) | 0.927 | |
| Rotamer outliers (%) | 0.87 | |
| Ramachandran plot (%) | ||
| Most favored | 98.15 | |
| Additionally allowed | 1.85 | |
| Disallowed | 0.0 | |
a Bijvoet pairs were scaled separately in this data set.
b Rsym = ΣhklΣi |Iihkl − 〈Ihkl〉|/ΣhklΣiIihkl.
c R-factor = Σ(|Fobs| − k|Fcalc|)/Σ(|Fobs|) and Rfree of the R value for a test set of reflections consisting of 7.5% of the diffraction data not used in refinement.
d CC1/2 is Pearson's correlation coefficient.
Figure 2.
Overall architecture of SkfB. A, the partial (β/α)6 TIM barrel of the AdoMet radical domain (magenta) houses the [4Fe-4S] AdoMet radical cluster and an AdoMet molecule (lilac). The C-terminal domain adopts a Twitch domain architecture (green), providing three visible ligands to a [2Fe-2S] auxiliary cluster, Aux I. The N-terminal peptide-binding domain (yellow) displays the architecture of the RRE motif identified in peptide-binding domains of RiPP biosynthetic enzymes. B, topology diagram of SkfB colored by domain with the AdoMet radical domain in magenta, the Twitch domain in green, and the RRE domain in yellow. Yellow circles indicate cysteine residues, and orange and yellow circles represent the iron and sulfur atoms, respectively, of the clusters shown as ball and stick representations. C–E show a closer look at the individual domains of SkfB. C, the active site of SkfB is located within the inner cavity of the AdoMet radical domain and comprises six parallel β-strands. The binding sites of the AdoMet radical cluster and AdoMet (lilac) are located at the top of the partial barrel. D, the C-terminal Twitch domain (green) contains the canonical elements of a SPASM/Twitch fold, a β-hairpin, β1′ and β2′, followed by α2′. In SkfB, a short β-strand, β3′, and α-helix α1′ are found in between β2′ and α2′. E, the N-terminal peptide-binding domain of SkfB (yellow) folds into a three-stranded antiparallel β-sheet (β1″–β3″) and a consecutive helical bundle (α1″–α4″), reminiscent of the winged helix turn helix motif that comprises the RRE. The N-terminal domain ends with a parallel β-strand, β4″ adjacent to β1″. The clusters are shown as ball and stick representations with iron atoms colored orange and sulfur atoms colored yellow.
SkfB adopts a canonical AdoMet radical domain
The central domain of SkfB (residues 106–322) folds into a partial (β/α)6 TIM barrel with six parallel β-strands comprising the inner face of the barrel and six α-helices flanking the outside of the barrel, providing flexibility to the rigid barrel architecture (Fig. 2, A–C). This architecture is characteristic of AdoMet radical enzymes and provides a binding site for cofactors and substrates in a protected cavity that facilitates the corresponding radical mechanisms of members of the superfamily. Herein, we will refer to the partial (β/α)6 TIM barrel architecture as the AdoMet radical domain. Residues from the canonical CX3CXφC motif, Cys117, Cys121, and Cys124, are located on the loop following β1 and ligate an essential [4Fe-4S] cluster, the AdoMet radical cluster, leaving a unique iron for direct coordination of AdoMet (Figs. 2, A–C, and 3A).
Figure 3.
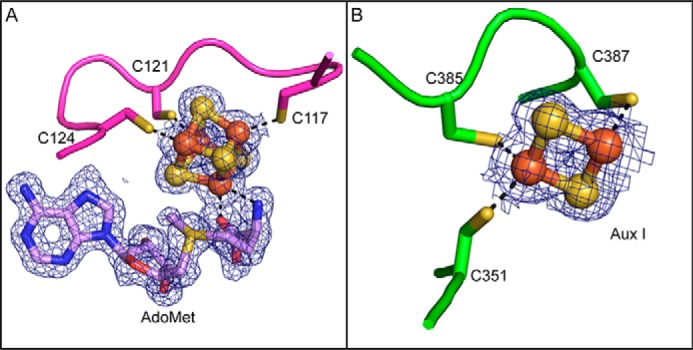
Iron-sulfur clusters and AdoMet density. A, the characteristic CX3CXφC motif, Cys117, Cys121, and Cys124, coordinates the AdoMet radical cluster in SkfB. An AdoMet molecule ligates the site-differentiated iron through a bidentate interaction with the nitrogen of the α-amino moiety and oxygen from the α-carboxyl moiety. B, a [2Fe-2S] cluster is observed in the Aux I site of the Twitch domain, bound by a CX33CXC sequence, Cys351, Cys385, and Cys387. 2Fo − Fc composite omit density is shown in blue and contoured at 1σ.
In the structure of SkfB, the α-amino and α-carboxyl groups of AdoMet are found 2.3 and 2.2 Å, respectively, from the site-differentiated iron of the AdoMet radical cluster. Conserved interactions from the AdoMet radical domain help position AdoMet in a catalytically competent orientation in the active site with all known motifs represented (Fig. 4) (22, 23). In particular, hydrogen bonds from the conserved GGE motif, Gly160, Gly161, and Glu162, and Arg223 orient the α-amino and α-carboxyl moieties of AdoMet. The ribose motif, Ser211, and residue Arg223, located adjacent to helix α4a, serve to position the hydroxyls of the ribose ring in the active site. The adenine moiety of AdoMet is oriented in the active site through hydrophobic interactions from Thr251 of the “GXIXGXXE” (or β5 motif) and from Phe123 of the CX3CXφC motif. Polar backbone interactions from the β6 motif, Leu258, and from the hydrophobic residues of the CX3CXφC motif, Phe123 and Tyr125 (Fig. 4), further help to orient AdoMet in the active site.
Figure 4.
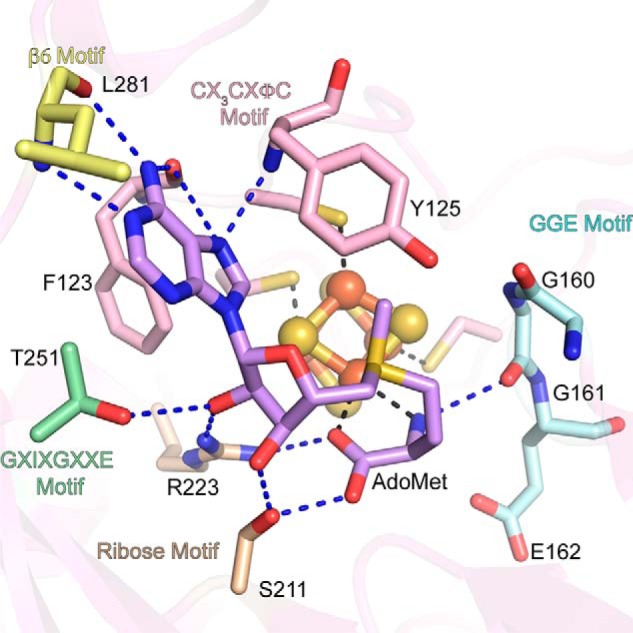
AdoMet-binding motifs are conserved in SkfB. The AdoMet-binding pocket is located within the partial TIM barrel (translucent pink) and includes interactions from the GGE motif, Gly160, Gly161, and Glu162 (cyan); the ribose motif, Ser211 and Arg223 (tan); GXIXGXXE or β5 motif, Thr251 (green); and β6 motif (yellow), Leu281. Arg223 plays a dual role, positioning both the carboxyl group and the ribose ring. Hydrophobic interactions from the CX3CXφC motif Phe123, and the adjacent residue, Tyr125 (pink), orient the adenine ring of AdoMet (lilac) in the active site. The AdoMet radical cluster is shown as a ball and stick representation with iron atoms colored orange and sulfur atoms colored yellow.
The Twitch domain binds a [2Fe-2S] auxiliary cluster
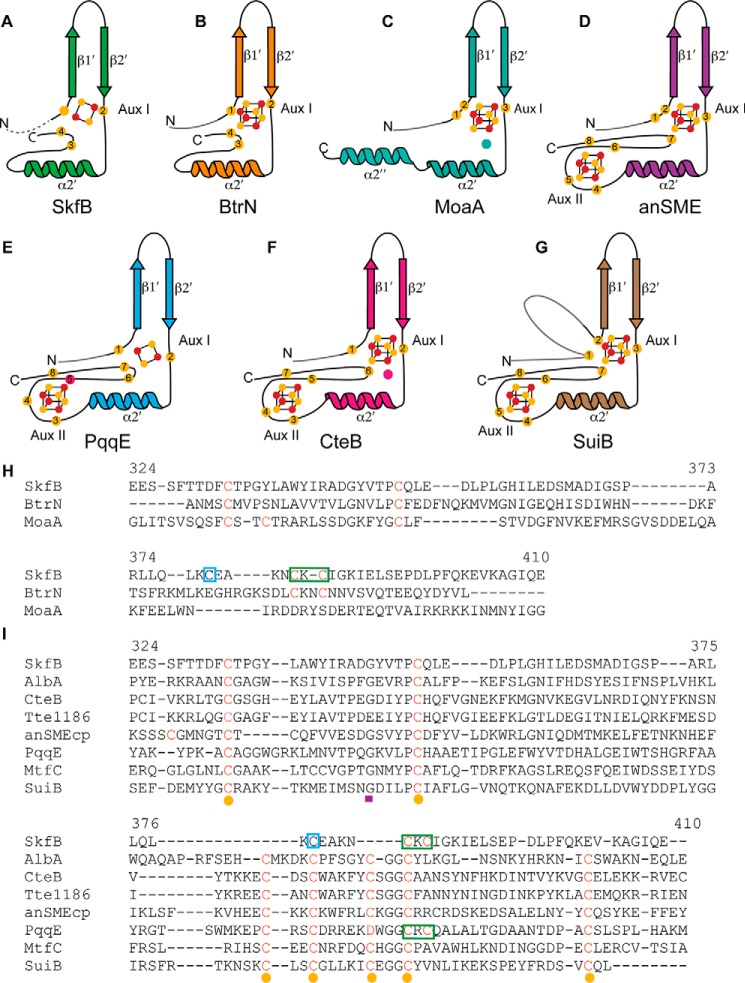
Following helix α6, the SkfB structure wraps back to the N terminus of the AdoMet radical domain and places β1′ of the C-terminal domain adjacent to β1, extending the inner parallel β-sheet by one strand (Fig. 2B). The majority of the linker between the AdoMet radical domain and the C-terminal domain (residues 322–329) is disordered in SkfB. Following the first visible residue of the C-terminal region (Thr330), the structure folds into a three-stranded antiparallel β-sheet, β1′–β3′, followed by two α-helices, a short α1′ and a longer α2′ (Fig. 2, A, B, and D). This C-terminal domain ligates Aux I, which in this structure is a [2Fe-2S] cluster (Fig. 3B). Cluster ligation involves one cysteine from the loop following β2′, Cys351, and two cysteines from the loop following α2′, Cys385 and Cys387 (Fig. 2B). This architecture is as expected for a Twitch domain (Fig. 5, A–D) (17, 19, 20) and is highly similar to the first half of a SPASM domain (Fig. 5, A and E–G), but the identity of the auxiliary cluster as a [2Fe-2S] cluster was a surprise. Although a [2Fe-2S] cluster is observed in the Aux I–binding site of PqqE (Fig. 5E), all other structurally characterized SPASM- or Twitch-containing AdoMet radical enzymes have Aux I clusters that are [4Fe-4S] clusters (Table S1) (11, 19, 20, 24–26).
Figure 5.
Comparison of auxiliary cluster binding by SPASM/Twitch domains. The canonical structural elements of the Twitch (A–C) and SPASM (D–F) domain are shown for each enzyme. A, SkfB (green) binds a [2Fe-2S] cluster using one cysteine following the β-hairpin and two cysteines following α2′, similar to BtrN. SkfB appears to have an open coordination site: although the cysteine corresponding to the cluster ligand before the β-hairpin motif is present, this cysteine is ∼12 Å away from the cluster. B, the Twitch domain of BtrN (orange) provides four cysteine residues to fully ligate a [4Fe-4S] auxiliary cluster. C, MoaA (teal) uses a Twitch domain to bind a [4Fe-4S] cluster with an open coordination site. The unique iron is ligated by substrate, GTP, (teal circle). D, anSME (purple) utilizes a SPASM motif to bind two fully ligated [4Fe-4S] clusters, Aux I and Aux II. E, PqqE (blue) binds two fully ligated clusters, a [2Fe-2S] Aux I cluster and a [4Fe-4S] Aux II cluster, using a unique aspartic acid (red circle) ligand. F, CteB (pink) also binds two [4Fe-4S] auxiliary clusters with an open coordination site on Aux I to which a cysteine residue from the substrate, CteA (pink circle), is bound in the structure. G, the SPASM domain of SuiB ligates two auxiliary clusters. The first cysteine ligand to Aux I is provided by the linker region connecting the SPASM domain to the AdoMet radical domain. The cluster-binding cysteines are shown as yellow circles, and the cluster iron and sulfur atoms are represented as orange and yellow circles, respectively. The linker regions are denoted in gray. H, Twitch domain sequence alignment. Residue numbers are for SkfB. Cysteine ligands to Aux I are indicated in red, yellow spheres represent the first four cysteines of the seven-cysteine motif (CX9–15GX4CgapCX2CX5CX3CgapC), and a purple rectangle denotes the conserved glycine residue. The CXC of SkfB is indicated by a green box, and a blue box designates the cysteine previously proposed to bind the Aux I cluster of SkfB (CX4CXC) (7). I, sequence alignments of SPASM domains with the sequence of SkfB (sequence numbers indicated for SkfB). Cluster-coordinating cysteines are denoted in red, cysteines corresponding to the seven-cysteine motif (CX9–15GX4CgapCX2CX5CX3CgapC) are indicated by yellow circles, and the conserved glycine residue of this motif is denoted by a purple rectangle. Green boxes denote the CXC of SkfB and PqqE, and the blue box indicates the cysteine previously proposed to bind the Aux I cluster of SkfB (CX4CXC) (7).
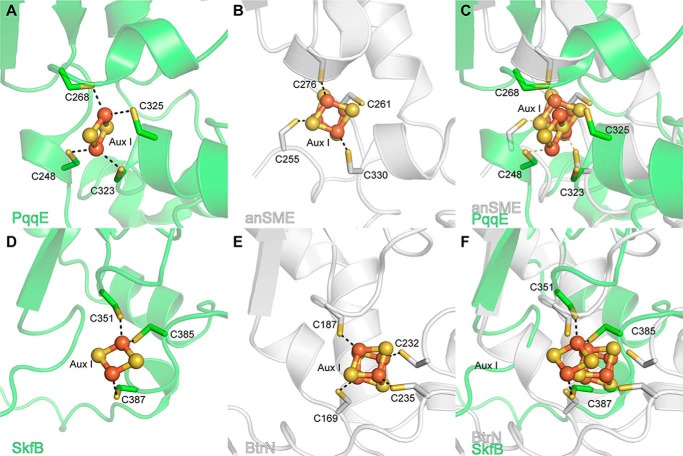
Both SkfB and PqqE utilize a CXC sequence, Cys385 and Cys387 and Cys323 and Cys325, respectively, following helix α2 to ligate the visualized [2Fe-2S] cluster where each cysteine coordinates a different iron of the cluster. A CXC cluster-binding sequence has not been seen in the other SPASM/Twitch enzymes structurally characterized (Figs. 5, H and I, and 6). When the Aux I sites of SkfB and PqqE are overlaid with a [4Fe-4S]-containing Twitch and SPASM enzyme, respectively, the observed [2Fe-2S] cluster superimposes well with Aux I [4Fe-4S] clusters (Fig. 6), and that a [4Fe-4S] cluster should be able to be accommodated at this site as well (Fig. 6).
Figure 6.
Comparison of Aux I iron-sulfur clusters in SPASM/Twitch AdoMet radical enzymes. A, the Aux I site of the PqqE SPASM domain (green) has been shown biochemically to bind either a [4Fe-4S] cluster or a [2Fe-2S] cluster. In the recent structure of PqqE (PDB code 6C8V), the Aux I cluster-binding site is populated by a [2Fe-2S] cluster with two of the ligands provided by a CXC motif, Cys323 and Cys325. B, anSME (PDB code 4K37), shown in white, contains a [4Fe-4S] cluster at the Aux I cluster-binding site ligated by Cys255, Cys261, Cys276, and Cys330. C, when overlaid with anSME (white), the cysteine positions of the PqqE (green) active site appear to be amenable for binding a [4Fe-4S] cluster. D, the Twitch Aux I site of SkfB (green) can bind a [2Fe-2S] cluster with cysteines from the CXC motif, Cys385 and Cys387, binding different irons of the cluster. E, the Twitch Aux I site of BtrN (PDB code 4M7T), shown in white, fully ligates a [4Fe-4S] auxiliary cluster. F, the Aux I site of SkfB (green) overlays well with the Aux I site BtrN (white). To allow for [4Fe-4S] cluster binding, the loop containing residues of the CXC motif of SkfB would have to move slightly. This rearrangement appears possible as this loop corresponds to the C-terminal end of the structure and is highly flexible.
In terms of cluster ligation, the SPASM enzymes anSME, SuiB, and PqqE and the Twitch enzyme BtrN have fully ligated auxiliary clusters (Fig. 5), whereas the Twitch enzyme MoaA and the SPASM enzyme CteB have an open coordination site on Aux I. The current structure of SkfB reveals an open coordination on Aux I, which is surprising as the sequence of SkfB predicts five cysteines in the C-terminal region, four of which are conserved (Fig. 5H). The fourth conserved cysteine, Cys333, was expected to ligate the cluster from its position in the sequence before the β-hairpin in SkfB; coordination of the cluster by a cysteine in this position has been observed in all other characterized SPASM/Twitch family members. However, in this structure, Cys333 is visible about 12 Å away from the cluster on the linker region that connects the AdoMet domain to the Twitch domain (Fig. S2).
Mössbauer spectroscopy
The surprising observation of a [2Fe-2S] cluster in the Aux I site was independently verified by 57Fe Mössbauer spectroscopy. The 4.2 K, 53-mT Mössbauer spectrum of reconstituted SkfB (Fig. 7, vertical bars) can be simulated with two quadrupole doublets with parameters typical of [2Fe-2S]2+ clusters (isomer shift δ = 0.29 mm/s and quadrupole splitting parameter ΔEQ = 0.55 mm/s; 37% of total intensity, dotted line above the data) and [4Fe-4S]2+ clusters (δ = 0.44 mm/s and ΔEQ = 1.18 mm/s; 59% of total intensity, solid line above the data). Together with the 57Fe/SkfB stoichiometry of 5.41 (determined by ICP-MS), these results indicate the presence of 1.0 [2Fe-2S]2+ and 0.8 [4Fe-4S]2+ clusters per polypeptide, consistent with the notion that SkfB harbors one [2Fe-2S] and one [4Fe-4S] cluster. Addition of the cosubstrate, AdoMet, to the sample leads to a noticeable broadening of the high-energy line of the [4Fe-4S]2+ cluster (Fig. S3). The observed perturbation is consistent with binding of AdoMet to the unique iron site of the [4Fe-4S]2+ cluster and can be rationalized with a site-differentiated mixed-valent (Fe2)5+ pair as was observed for other AdoMet radical enzymes previously (27–29).
Figure 7.
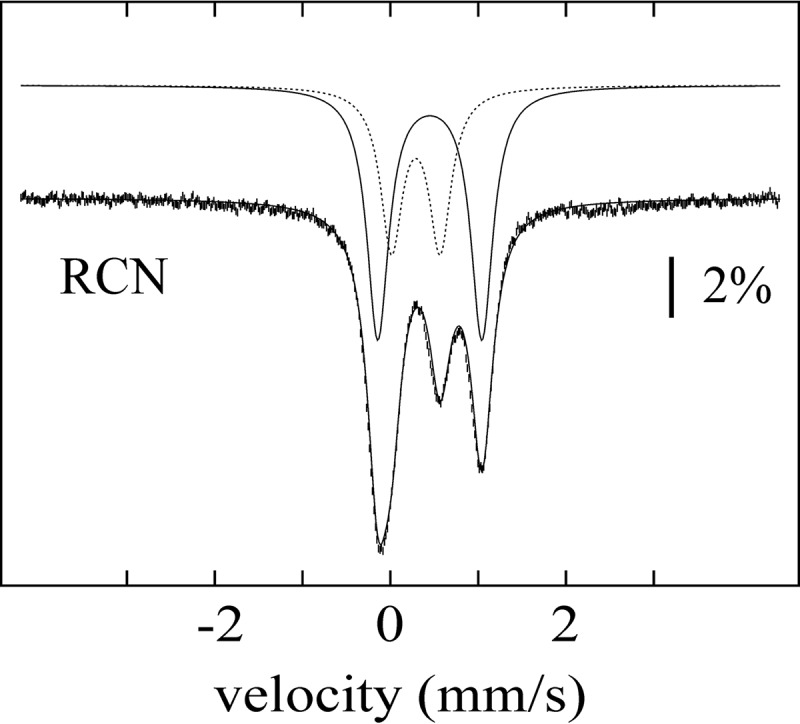
Mössbauer spectrum of SkfB. The spectrum of a sample of SkfB reconstituted with 57Fe (vertical bars) was recorded with the sample kept at 4.2 K in a 53-mT magnetic field applied parallel to the γ-beam. The solid line overlaid with the data is a simulation with two quadrupole doublets using parameters quoted in the text. The individual contributions of the quadrupole doublets associated with the [2Fe-2S]2+ and [4Fe-4S]2+ clusters are shown as dashed and solid lines above the data, respectively. RCN, reconstituted.
We further investigated a reconstituted sample of the SkfB variant in which the three cysteines coordinating the AdoMet radical cluster were substituted with alanine residues (ΔRS SkfB). The Mössbauer spectrum of this variant (Fig. S4) exhibits only one quadrupole doublet with parameters virtually identical to those of the [2Fe-2S]2+ cluster observed in WT SkfB, consistent with notion that the [2Fe-2S]2+ cluster resides in the Aux I site and that the AdoMet radical cluster is a [4Fe-4S]2+ cluster.
The N-terminal peptide-binding domain of SkfB contains a RiPP recognition element motif
The N-terminal domain of SkfB (residues 12–105) sits below the N-terminal region of the AdoMet domain and the C-terminal Twitch domain (Fig. 2, A and B). It folds first into a three-stranded antiparallel β-sheet, β1″–β3″, followed by a helical bundle comprising four consecutive α-helices, α1″–α4″, that stacks against the lower face of the antiparallel β-sheet (Fig. 2E). Following α4″, SkfB folds into a fourth β-strand, β4″, before connecting to the AdoMet radical domain. It is the upper face of the antiparallel β-sheet that forms the interface with the AdoMet radical domain and Twitch domain.
The N-terminal domain architecture of SkfB (Figs. 2E and 8A) is reminiscent of a peptide-binding protein that is involved in the biosynthesis of pyrroloquinoline quinone, PqqD (Fig. 8B), the peptide-binding domains of a nisin biosynthetic enzyme, NisB (Fig. 8C), and a cyanobactin biosynthetic enzyme, LynD (Fig. 8D), an architecture termed the RRE (16, 30–33). The RRE is responsible for binding the substrate precursor peptide in RiPP biosynthetic enzymes and folds into a winged helix-turn-helix motif composed of a three-stranded antiparallel β-sheet and three α-helices. As visualized in NisB and LynD, the leader sequence of the precursor peptide binds to the RRE motif by forming a β-strand, extending the antiparallel β-sheet or the “wing” (Fig. 8, C and D). In SkfB, the antiparallel β-sheet, β1″–β3″, and α1″–α3″ constitute the RRE motif (Figs. 2, B and E, and 8A).
Figure 8.
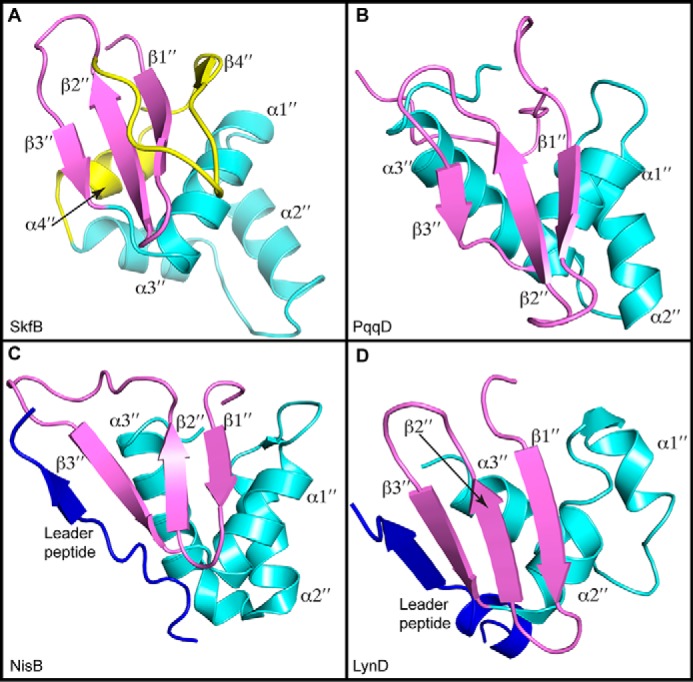
Structural comparisons of RRE domains. A, SkfB exhibits a canonical RRE comprising a three-strand antiparallel β-sheet (β1″–β3″ in purple) and three consecutive α-helices (α1″–α3″ in cyan). Following the RRE motif, SkfB folds into an additional α-helix, α4″, and β-strand, β4″ (yellow). B, the small peptide-binding protein PqqD (PDB code 5SXY) is a standalone RRE domain. C, the RRE domain of NisB (PDB code 4WD9) binds the leader peptide sequence of the peptide substrate NisA (blue) by extending the antiparallel β-sheet or the wing. D, the leader peptide of PatE (blue) also binds to the RRE domain of LynD (PDB code 4V1T) through interactions with the wing.
Structural comparison of AdoMet radical RiPP biosynthetic enzymes
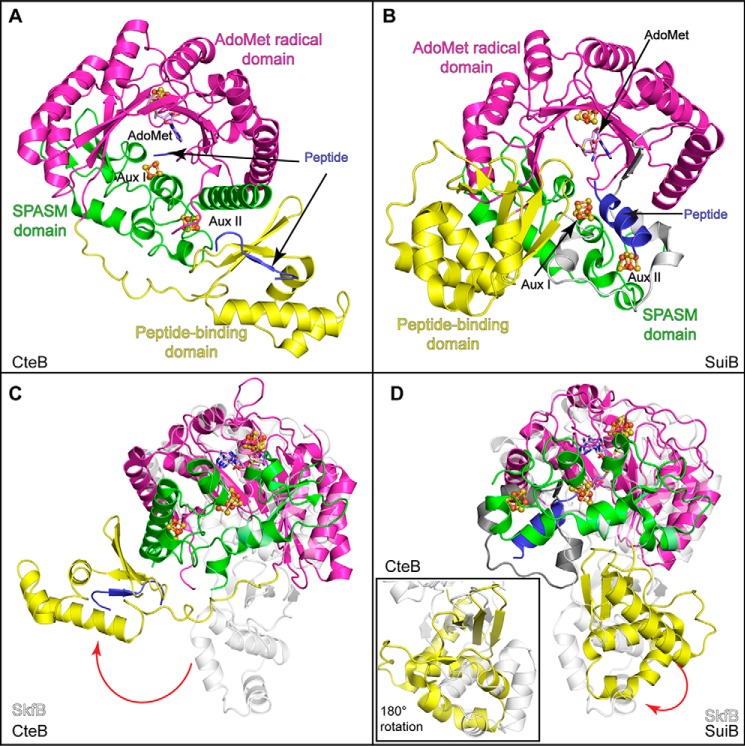
In addition to SkfB, the structures of two other AdoMet radical enzymes involved in the biosynthesis of RiPPs have been determined: the sactisynthase CteB (Fig. 9A) and the enzyme SuiB (Fig. 9B), which is involved in cross-linking two amino acid side chains (Fig. S1). Similar to SkfB, both CteB and SuiB are members of the SPASM/Twitch subfamily and adopt a modular overall architecture comprising three domains: an N-terminal peptide-binding RRE domain, a central AdoMet radical domain, and a C-terminal SPASM domain in place of the C-terminal Twitch domain visualized in SkfB. CteB and SuiB structures contain three [4Fe-4S] clusters, including the AdoMet radical cluster, bound by the canonical CX3CXφC motif, and two auxiliary clusters, Aux I and Aux II, bound by the SPASM domain. In addition, the linkers between the AdoMet radical domain and the SPASM domains were visualized in these structures. In SuiB, this linker region donates the first cysteine ligand to Aux I, resulting in two fully ligated auxiliary clusters (Fig. 5G). In CteB, Aux I has an open coordination site, which is ligated by a cysteine from the N terminus of the peptide substrate CteA (Fig. 5F). Structural comparisons show that the AdoMet radical domain and SPASM domains of CteB and SuiB overlay well with the AdoMet radical and Twitch domains of the SkfB structure (Fig. 9, C and D). The main differences between SkfB and CteB or SuiB are that SkfB has a Twitch rather than SPASM domain, the ligation and nature of the Aux I cluster, and the location of the N-terminal peptide-binding domain.
Figure 9.
Structural comparisons of SkfB with other AdoMet radical enzymes involved in RIPP biosynthesis. A, CteB (PDB code 5WGG) exhibits a trimodular fold composed of a peptide-binding or RRE domain (yellow) followed by an AdoMet-binding domain (magenta), which binds the AdoMet radical cluster and AdoMet (lilac). The C-terminal end of CteB binds two clusters, Aux I and Aux II, using the SPASM domain architecture (green). The leader sequence of CteA (blue) binds to the RRE domain by extending the antiparallel β-sheet. B, SuiB (PDB code 5V1T) demonstrates a similar modular fold to CteB (A). Interestingly, the substrate for SuiB, SuiA (blue), is observed making contacts with the insertion (gray) between the AdoMet radical domain (magenta) and the SPASM domain (green) and not with the peptide-binding domain (yellow). C, the AdoMet radical and Twitch domains of SkfB (white) overlay well with CteB, but the peptide-binding domains are located on opposite sides of the AdoMet radical domain. D, SkfB (white) overlays well with SuiB, and the positions of the N-terminal domains show modest differences. A view of the overlaid N-terminal domains SkfB and SuiB, rotated 180°, is shown in the inset.
The N-terminal domains of both CteB and SuiB adopt typical RRE folds. As aforementioned for NisB and LynD, the leader peptide, CteA, adopts a β-strand conformation and interacts with β3″ of CteB, extending the antiparallel β-sheet of the RRE domain (Fig. 9A). Surprisingly, the leader peptide of SuiA binds to SuiB without interacting with its RRE domain. Instead, SuiA interacts with the AdoMet radical domain, the SPASM domain, and the linker between these two domains (Fig. 9B). There is considerable variability in the placement of the RRE domain with respect to the other domains. CteB has a long flexible linker between α3″ of the RRE domain and β1 of the AdoMet radical domain (Fig. 9A), which could allow flexibility in the positioning of the RRE domain with respect to the rest of the protein structure. In fact, the RRE domain of CteB and the RRE domain of SkfB are observed on the opposite sides of the AdoMet radical domain (Fig. 9C). The placement of the RRE domain of SuiB is more similar to that observed in SkfB (Fig. 9D). Shorter linkers between the RRE domain and the AdoMet radical domain are observed in SuiB (five residues) and SkfB (nine residues) than is seen in CteA (22 residues).
Discussion
Here, we present the structure of an AdoMet radical Twitch enzyme, SkfB, involved in maturation of a sactipeptide natural product. The structure of SkfB allows us to compare and contrast structural features with the previously solved SPASM domain–containing RiPP biosynthetic enzymes, SuiB and CteB, as well as other SPASM and Twitch domain–containing AdoMet radical enzymes for which structures have been determined, anSME, BtrN, MoaA, and PqqE.
In terms of the overall structure, SkfB adopts a modular architecture that resembles SuiB and CteB. The Twitch domain of SkfB overlays well with the SPASM domains of CteB and SuiB but ligates one auxiliary cluster, Aux I, instead of two, a noted difference between SPASM and Twitch enzymes. Surprisingly, the SkfB structure revealed a [2Fe-2S] Aux I cluster ligated by a CX33CXC sequence. This cluster identity was unexpected as the initial biochemical and spectroscopic characterization proposed that SkfB contained a [4Fe-4S] auxiliary cluster ligated by a CX4CXC sequence (7). Moreover, most of the SPASM/Twitch enzymes characterized to date contain [4Fe-4S] auxiliary clusters. However, Mössbauer analysis on the samples prepared in a similar manner as those used for crystallographic analysis also shows that the predominant forms of the clusters in SkfB are a [4Fe-4S] cluster and a [2Fe-2S] with the spectral feature of the [4Fe-4S] cluster perturbed by AdoMet binding. This result is consistent with the AdoMet radical cluster being the one [4Fe-4S] cluster present in this sample.
The recent structure of the SPASM enzyme PqqE revealed a [2Fe-2S] cluster in the Aux I site, which has been reported to be able to bind both [4Fe-4S] and [2Fe-2S] clusters (26, 34, 35). Interestingly, like SkfB, the two-cysteine ligands to Aux I after helix α2′ are provided by a CXC sequence, a motif found primarily in the coordination of [2Fe-2S] clusters (36, 37). Because of the proximity of the cysteine ligands in CXC sequences, both cysteines are expected to ligate the same iron of the [2Fe-2S] clusters. This is not the case in SkfB or PqqE: the CXC sequence ligates a different iron atom of the [2Fe-2S] cluster. This coordination geometry could also allow for [4Fe-4S] cluster binding in SkfB and PqqE with minimal perturbations of the Aux I site as in both cases the site appears to be amenable to rearrangement. The malleability of this site could explain why different cluster identities have been observed in the biochemical and structural data. It is possible that the CXC sequence might be indicative of a physiologically relevant [2Fe-2S] cluster, or it could make a [4Fe-4S] cluster more susceptible to degradation. To understand which species is catalytically relevant, enzyme activity needs to be correlated to cluster content.
The coordination state of the auxiliary clusters of members of the SPASM and Twitch family has long been of debate (11, 17, 19, 20, 38). Although both anSME and BtrN structures reveal fully ligated clusters, an insufficient number of cysteines in the C-terminal sequence of AlbA (seven cysteines to ligate two auxiliary clusters) led to the proposition that AlbA and other sactisynthases may utilize an open coordination site for substrate ligation. It was also hypothesized that these enzymes could utilize a noncysteine ligand as has been seen in LipA (Ser) (39, 40), BioB (Arg) (41), and recently PqqE (Asp) as a ligand to the Aux II cluster (26, 38). In contrast, the C-terminal Twitch domain of SkfB contains five conserved cysteines, four of which correspond with conserved auxiliary cluster ligands in both Twitch and SPASM domains. Therefore, the structures of both SPASM and Twitch sactisynthases were highly anticipated in hope they would shed light on the cluster coordination conundrum.
The first structure of a sactisynthase, CteB, revealed that there is an open coordination site on Aux I of the SPASM domain, whereas Aux II is fully ligated (11). Interestingly, extra density inconsistent with a protein residue was observed near the site-differentiated iron of Aux I and was attributed to a cysteine residue (Cys3 of CteA in Fig. 1B) from the N-terminal fragment of CteA cocrystallized with CteB. The N-terminal fragment used in cocrystallization encompassed the leader peptide sequence (residues −18 to −1) and the first three residues of the core peptide (residues 1–3) of CteA (Fig. 1B and Table S1) but did not include the cysteine involved in thioether bond formation. This cysteine ligation to Aux I could foreshadow binding of the reactive cysteine (Cys14) in the presence of full-length CteA.
The structure of SkfB shows an open coordination site on Aux I. This finding was unexpected as C-terminal sequence alignment of SkfB with SPASM/Twitch enzymes predicts four conserved cysteine ligands (Fig. 5, H and I). Interestingly, the fourth cysteine residue, Cys333, was observed in the structure a distance away from Aux I. Because of the disorder in this region, however, we cannot rule out that Cys333 could serve as a cluster ligand in some enzyme states. A structure of SkfB in which the linker is ordered will be important for making a final determination. Regardless, the structure of SkfB leaves open the possibility that substrate will interact directly with Aux I. Recent studies (15) suggest that Aux I is not essential for binding substrate as hydrogen atom abstraction from substrate can occur without Aux I, indicating that substrate binds and is positioned correctly for the AdoMet radical chemistry in the absence of this cluster. It was also shown that the Aux I is essential for later steps in catalysis, suggesting that interaction between the substrate and Aux I could be important for cyclization, cysteine deprotonation, oxidation of a product-based radical species to generate product, or possibly all three.
The observation that Aux I is not essential for substrate binding (15) is consistent with the presence of a peptide-binding RRE domain as part of the modular structure of SkfB. The three RiPP biosynthetic AdoMet radical enzymes structurally characterized to date all have similar RRE domains, but these domains are present in three different orientations. In the structures of SkfB and SuiB, the RRE domain sits below the AdoMet radical core and SPASM/Twitch domain, whereas the RRE domain of CteB interacts with helix α6 of CteB on the top side of the AdoMet radical core. Mobility of the RRE domain might be important for positioning substrate into the active site, and the structures of SkfB, CteB, and SuiB considered here could represent snapshots of those movements rather than representing enzyme-specific variations.
We predict that SkfA will bind to SkfB using the leader peptide sequence to extend the antiparallel β-sheet of the RRE as seen with CteB. However, the distance between the RRE domain and the active site (indicated open coordination site on Aux I) is short, about 20 Å, compared with 33 Å in CteB. To position the 55-mer SkfA precursor peptide in a catalytically competent orientation, it is possible that SkfA will adopt an ordered secondary structure, the RRE domain of SkfB will undergo a conformational change upon SkfA binding, or a combination of both. How the full-length precursor peptides bind to sactisynthases to allow for sactionine linkage formation and possible conformational changes associated with this interaction still remains to be determined.
Overall, SkfB represents a unique hybrid of features observed in other AdoMet radical enzymes. It has the same domain organization as CteB and SuiB but has a Twitch, not a SPASM, domain. SkfB's Twitch domain is like that of BtrN and MoaA, but its Aux I cluster is most like the Aux I cluster found in PqqE and not in BtrN or MoaA. A sequence similarity network (Fig. 10) shows that SkfB and PqqE are close in sequence space despite one having a Twitch domain and the other having a SPASM domain. Twitch domain–containing enzymes (MoaA, BtrN, and SkfB) do not cluster together nor do SPASM-containing enzymes (anSME, CteB, SuiB, and PqqE). Clustering is not based on function either, with dehydrogenases BtrN and anSME on opposite sides of the Cytoscape diagram (Fig. 10). Thus, it is difficult to predict fold or function based on sequence similarity networks for this AdoMet radical enzyme subfamily. This modular collection of domains seems well-suited to carry out a variety of chemical reactions, and with sections of sequence space left uncharacterized by crystallography, more mix-and-match combinations of domains and cluster types may be left to discover.
Figure 10.
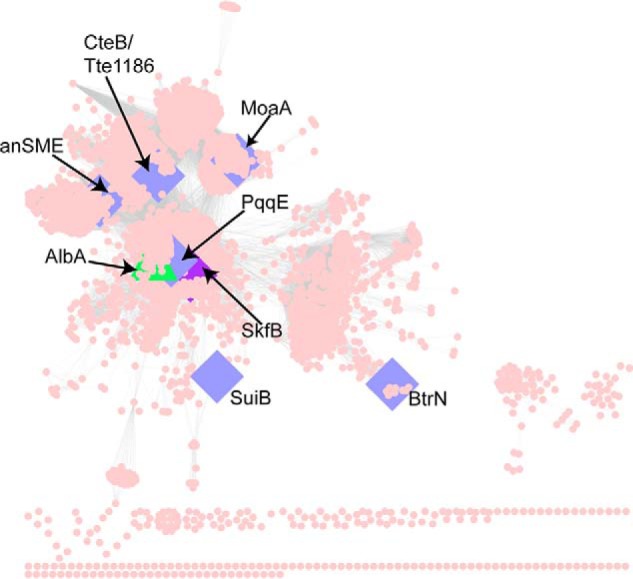
SPASM/Twitch subclass sequence similarity network. The protein similarity network (51) for the SPASM/Twitch subclass is visualized in Cytoscape (52) at a blast probability of 10−20. Sequences were obtained from the Structure Function Linkage Database (53) (http://sfld.rbvi.ucsf.edu/django; Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.), and each node represents sequences that share 50% identity. Nodes corresponding to the previously solved members of the SPASM/Twitch subclass are represented as blue diamonds (11, 19, 20, 24–26), and green indicates family members that have been biochemically characterized but not structurally characterized (8). SkfB, which is structurally characterized here, is shown as a purple diamond.
Experimental procedures
Cloning of SkfB and preparation of constructs to express variants
The skfB gene was previously amplified from the B. subtilis subsp. subtilis strain 168 genome and cloned into a pET28a vector containing a tobacco etch virus cleavable N-terminal His6 tag (13). The ΔRS SkfB construct, which encodes a C117A/C121A/C124A triple variant, was generated by site-directed mutagenesis using the primers listed in Table S2. Standard Sanger sequencing at the University of Michigan DNA Sequencing Core confirmed the sequence of each construct.
Purification of SkfB
WT and ΔRS SkfB were purified and reconstituted as described previously (13).
Crystallization and data collection of SkfB
SkfB was crystallized within an MBraun anaerobic chamber at 21 °C and under a N2 atmosphere using the sitting drop vapor diffusion method. Initial crystals of SkfB were obtained by sparse matrix screening using a mosquito pipetting robot (TTP LabTech). To obtain data-quality crystals, 1.5 μl of SkfB (10 mg/ml, 50 mm PIPES, pH 7.4, 150 mm KCl, 10 mm DTT, and 5 mm AdoMet) was mixed with 0.5 μl of precipitant (0.25 m magnesium formate and 15% PEG 3350) and equilibrated over a 500-μl reservoir. Brown crystals grew within 24 h and were transferred to a Coy Scientific anaerobic chamber (95% argon and 5% H2 atmosphere) for harvesting. To cryoprotect SkfB crystals, the crystals were transferred in four steps into a cryoprotectant solution containing 0.2 m magnesium formate, 20% PEG 3350, and glycerol. Upon each transfer, the glycerol concentration was increased to a final concentration of 20%. Crystals were then flash frozen in liquid nitrogen within the Coy chamber.
An anomalous data set and a native data set of SkfB were collected on the same crystal. The anomalous data set was collected to a resolution of 1.9 Å using an in-house CuKα rotating anode source (Rigaku) with an imaging plate (RAXIS IV, Rigaku). To obtain phasing information, the data set was collected using a pseudo inverse beam in four 90° wedges with 1° increments. Following the home data collection, the crystal was shipped to the Advanced Photon Source (Argonne, IL) where a native data set was collected to a 1.29-Å resolution at beamline 24-ID-C using a Pilatus 6M pixel detector at a temperature of 100 K and a wavelength of 0.9792 Å. All data were processed in HKL 2000.
Structure determination and refinement
The structure of SkfB with AdoMet was solved by an iron single-wavelength anomalous dispersion method in a C2 space group that contained one molecule in the asymmetric unit. Using anomalous data trimmed to 2.5-Å resolution, six iron sites (corresponding to one [4Fe-4S] cluster and one [2Fe-2S] cluster were identified in ShelxD (42) in HKL2MAP and refined in SHARP/autosharp (43, 44). The output experimental maps, with an initial figure of merit of 0.545–2.5-Å resolution, were of sufficient quality to trace the secondary structure of the AdoMet radical partial (β/α)6 TIM barrel domain and the C-terminal Twitch domain using BtrN (Protein Data Bank (PDB) code 4M7T) and MoaA (PDB code 1TV8) as guides. This initial model was used to define the solvent boundary during a subsequent round of solvent flattening in SOLOMON (45) and phase extension to 2.0-Å resolution. Loop regions and side chains with visible density were built into the resulting electron density.
When a near-completed model of SkfB was obtained, containing ∼350 of 410 residues, an AdoMet molecule, and the AdoMet radical cluster, the structure was refined against the full range of the native data set (50.0–1.29-Å resolution) using rigid-body atomic coordinates and atomic displacement (B-factor) refinements in Phenix (46). The resulting Rwork and Rfree were 24.4 and 29.5%, respectively. The model was completed by iterative rounds of model building in Coot (47) and refinement in Phenix (46). Waters were added manually in Coot during final rounds of refinement, and disordered side chains were truncated to the last atom with discernable density. The final model contained residues 12–322 and 330–401, one molecule of AdoMet, a [4Fe-4S] AdoMet radical cluster, and a [2Fe-2S] auxiliary cluster.
Composite omit maps calculated in Phenix (46) were used to validate the final structure, and MolProbity (48) was used to analyze the model geometry. MolProbity indicated that 98.15% of residues were found in the favored region, 1.85% in the allowed region, and 0.0% in the disallowed region of the Ramachandran plot, and ∼99% of residues have favorable rotamers. Figures were generated in PyMOL (49). Crystallography software packages were compiled by SBGrid (50).
Expression, purification, and reconstitution of 57Fe-labeled SkfB for Mössbauer spectroscopy
SkfB variants containing 57Fe were prepared with slight adjustments to the previously published protocol (13). Escherichia coli Rosetta 2 (DE3) cells were cotransformed with pNB529 (SkfB with a tobacco etch virus–cleavable N-terminal His6 tag) and pDB1282 (plasmid containing Azotobacter vinelandii genes iscS, iscU, iscA, hscA, hscB, and fdx for iron-sulfur cluster biogenesis). A starter culture was incubated overnight in LB at 37 °C and 200 rpm. The next day, 15-ml aliquots of starter culture were pelleted by centrifugation (5000 × g) and gently resuspended in 15 ml of a modified M9 minimal medium containing 2% (w/v) glucose. Large-scale growth cultures containing M9 mineral medium (12 × 1.5 liters in 2.8-liter Fernbach flasks) supplemented with ampicillin (0.1 mg/ml), kanamycin (0.034 mg/ml), and 57FeSO4 (25 mm) were inoculated with 15 ml of resuspended starter culture cells. The large cultures were incubated at 37 °C and 175 rpm until the A600 nm reached 0.3, and arabinose and 57FeSO4 were added to final concentrations of 0.07% (w/v) and 0.05 mm, respectively, to induce pDB1282. After the addition of arabinose and 57FeSO4, the cultures were gradually cooled to 18 °C and slowed down to 150 rpm. After the cells reached an A600 nm of 0.6, expression from the skfB gene was induced upon addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 0.1 mm. Cells were allowed to express SkfB at 18 °C for 18 h, harvested by centrifugation at 5000 × g, flash frozen in liquid nitrogen, and stored at −80 °C. SkfB and variant were purified and reconstituted as described previously (13) except that 57FeSO4 was used in the reconstitution.
Mössbauer spectroscopy
Samples were prepared in a Coy Scientific anaerobic chamber. Sample buffer contained 50 mm PIPES, pH 7.4, 150 mm KCl, and 10 mm DTT. Concentrated protein was dispensed into Mössbauer cups and flash frozen with liquid nitrogen in the anaerobic chamber. With samples containing AdoMet, 10 molar eq of enzymatically generated AdoMet was incubated with concentrated SkfB for 20 min prior to flash freezing.
The 57Fe content of protein samples was determined by ICP-MS. SkfB was diluted to 1 μm (based on corrected Bradford concentration) in 1% (v/v) trace-meta-grade nitric acid. Analyses were conducted by the ICP-MS Metals Lab of the Department of Geology and Geophysics at the University of Utah.
Mössbauer spectra were measured on a spectrometer from WEB Research (Edina, MN) with an SVT-400 cryostat from Janis (Wilmington, MA) to maintain the temperature at 4.2 K. A 53-mT magnetic field was applied parallel to the propagation direction of the γ-beam. Isomer shifts are reported with respect to the centroid of the spectrum of α-iron metal at room temperature. Simulations of Mössbauer spectra were carried out with the program WMOSS (SEE Co., Edina, MN).
Author contributions
T. A. J. G., W. M. K., N. A. B., E. J. B., C. K., V. B., and C. L. D. conceptualization; T. A. J. G., W. M. K., N. A. B., E. J. B., C. K., V. B., and C. L. D. investigation; T. A. J. G., V. B., and C. L. D. writing-original draft; T. A. J. G., W. M. K., N. A. B., C. K., V. B., and C. L. D. writing-review and editing; C. K., V. B., and C. L. D. funding acquisition; V. B. and C. L. D. supervision.
Supplementary Material
Acknowledgments
This research used resources of the Advanced Photon Source, a United States Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357 and was conducted at the Northeastern Collaborative Access Team beamlines, which are funded by NIGMS, National Institutes of Health Grant P41 GM103403. The Pilatus 6M detector on 24-ID-C beamline is funded by National Institutes of Health Office of Research Infrastructure Programs High-End Instrumentation Grant S10 RR029205.
This work was supported by a National Science Foundation Graduate Research Fellowship under Grant 1122374 (to T. A. J. G.); NIGMS, National Institutes of Health Grants R35 GM126982 (to C. L. D) and R35 GM 126956, R01 GM72623, and GM 120638 (to V. B.) with Administrative Supplement GM72623 S01 (to V. B.) for the collaboration between V. B. and C. L. D; and Ruth L. Kirschstein Postdoctoral National Research Service Award GM 116353 (to E. J. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4 and Tables S1 and S2.
The atomic coordinates and structure factors (code 6EFN) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- sactipeptide
- sulfur to α-carbon cross-linked peptide
- RiPP
- ribosomally synthesized and post-translationally modified peptide
- AdoMet
- S-adenosyl-l-methionine
- SKF
- sporulation killing factor
- RRE
- RiPP recognition element
- 5′-dAdo•
- 5′-deoxyadenosyl radical
- Aux
- auxiliary iron-sulfur cluster
- anSME
- anaerobic sulfatase maturating enzyme
- mT
- millitesla(s)
- ICP-MS
- inductively coupled plasma mass spectrometry
- TIM
- triose phospahte isomerase.
References
- 1. Flühe L., and Marahiel M. A. (2013) Radical S-adenosylmethionine enzyme catalyzed thioether bond formation in sactipeptide biosynthesis. Curr. Opin. Chem. Biol. 17, 605–612 10.1016/j.cbpa.2013.06.031 [DOI] [PubMed] [Google Scholar]
- 2. Rea M. C., Sit C. S., Clayton E., O'Connor P. M., Whittal R. M., Zheng J., Vederas J. C., Ross R. P., and Hill C. (2010) Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. U.S.A. 107, 9352–9357 10.1073/pnas.0913554107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sit C. S., van Belkum M. J., McKay R. T., Worobo R. W., and Vederas J. C. (2011) The 3D solution structure of thurincin H, a bacteriocin with four sulfur to α-carbon crosslinks. Angew. Chem. Int. Ed. Engl. 50, 8718–8721 10.1002/anie.201102527 [DOI] [PubMed] [Google Scholar]
- 4. Liu W.-T., Yang Y.-L., Xu Y., Lamsa A., Haste N. M., Yang J. Y., Ng J., Gonzalez D., Ellermeier C. D., Straight P. D., Pevzner P. A., Pogliano J., Nizet V., Pogliano K., and Dorrestein P. C. (2010) Imaging mass spectrometry of intraspecies metabolic exchange revealed the cannibalistic factors of Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 107, 16286–16290 10.1073/pnas.1008368107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Datta S., Mori Y., Takagi K., Kawaguchi K., Chen Z.-W., Okajima T., Kuroda S., Ikeda T., Kano K., Tanizawa K., and Mathews F. S. (2001) Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking. Proc. Natl. Acad. Sci. U.S.A. 98, 14268–14273 10.1073/pnas.241429098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Babasaki K., Takao T., Shimonishi Y., and Kurahashi K. (1985) Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: isolation, structural analysis, and biogenesis. J. Biochem. 98, 585–603 10.1093/oxfordjournals.jbchem.a135315 [DOI] [PubMed] [Google Scholar]
- 7. Flühe L., Burghaus O., Wieckowski B. M., Giessen T. W., Linne U., and Marahiel M. A. (2013) Two [4Fe-4S] clusters containing radical SAM enzyme SkfB catalyze thioether bond formation during the maturation of the sporulation killing factor. J. Am. Chem. Soc. 135, 959–962 10.1021/ja310542g [DOI] [PubMed] [Google Scholar]
- 8. Flühe L., Knappe T. A., Gattner M. J., Schäfer A., Burghaus O., Linne U., and Marahiel M. A. (2012) The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A. Nat. Chem. Biol. 8, 350–357 10.1038/nchembio.798 [DOI] [PubMed] [Google Scholar]
- 9. Wieckowski B. M., Hegemann J. D., Mielcarek A., Boss L., Burghaus O., and Marahiel M. A. (2015) The PqqD homologous domain of the radical SAM enzyme ThnB is required for thioether bond formation during thurincin H maturation. FEBS Lett. 589, 1802–1806 10.1016/j.febslet.2015.05.032 [DOI] [PubMed] [Google Scholar]
- 10. Nakai T., Ito H., Kobayashi K., Takahashi Y., Hori H., Tsubaki M., Tanizawa K., and Okajima T. (2015) The radical S-adenosyl-l-methionine enzyme QhpD catalyzes sequential formation of intra-protein sulfur-to-methylene carbon thioether bonds. J. Biol. Chem. 290, 11144–11166 10.1074/jbc.M115.638320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grove T. L., Himes P. M., Hwang S., Yumerefendi H., Bonanno J. B., Kuhlman B., Almo S. C., and Bowers A. A. (2017) Structural insights into thioether bond formation in the biosynthesis of sactipeptides. J. Am. Chem. Soc. 139, 11734–11744 10.1021/jacs.7b01283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sofia H. J., Chen G., Hetzler B. G., Reyes-Spindola J. F., and Miller N. E. (2001) Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29, 1097–1106 10.1093/nar/29.5.1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bruender N. A., and Bandarian V. (2016) SkfB abstracts a hydrogen atom from Cα on SkfA to initiate thioether cross-link formation. Biochemistry 55, 4131–4134 10.1021/acs.biochem.6b00598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bruender N. A., Wilcoxen J., Britt R. D., and Bandarian V. (2016) Biochemical and spectroscopic characterization of a radical S-adenosyl-L-methionine enzyme involved in the formation of a peptide thioether cross-link. Biochemistry 55, 2122–2134 10.1021/acs.biochem.6b00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kincannon W. M., Bruender N. A., and Bandarian V. (2018) A radical clock probe uncouples H-atom abstraction from thioether crosslink formation by the radical SAM enzyme SkfB. Biochemistry 57, 4816–4823 10.1021/acs.biochem.8b00537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burkhart B. J., Hudson G. A., Dunbar K. L., and Mitchell D. A. (2015) A prevalent peptide-binding domain guides ribosomal natural product biosynthesis. Nat. Chem. Biol. 11, 564–570 10.1038/nchembio.1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grell T. A., Goldman P. J., and Drennan C. L. (2015) SPASM and twitch domains in S-adenosylmethionine (SAM) radical enzymes. J. Biol. Chem. 290, 3964–3971 10.1074/jbc.R114.581249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haft D. H., and Basu M. K. (2011) Biological systems discovery in silico: radical S-adenosylmethionine protein families and their target peptides for posttranslational modification. J. Bacteriol. 193, 2745–2755 10.1128/JB.00040-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldman P. J., Grove T. L., Booker S. J., and Drennan C. L. (2013) X-ray analysis of butirosin biosynthetic enzyme BtrN redefines structural motifs for AdoMet radical chemistry. Proc. Natl. Acad. Sci. U.S.A. 110, 15949–15954 10.1073/pnas.1312228110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goldman P. J., Grove T. L., Sites L. A., McLaughlin M. I., Booker S. J., and Drennan C. L. (2013) X-ray structure of an AdoMet radical activase reveals an anaerobic solution for formylglycine posttranslational modification. Proc. Natl. Acad. Sci. U.S.A. 110, 8519–8524 10.1073/pnas.1302417110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hänzelmann P., and Schindelin H. (2006) Binding of 5′-GTP to the C-terminal FeS cluster of the radical S-adenosylmethionine enzyme MoaA provides insights into its mechanism. Proc. Natl. Acad. Sci. U.S.A. 103, 6829–6834 10.1073/pnas.0510711103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dowling D. P., Vey J. L., Croft A. K., and Drennan C. L. (2012) Structural diversity in the AdoMet radical enzyme superfamily. Biochim. Biophys. Acta 1824, 1178–1195 10.1016/j.bbapap.2012.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Vey J. L., and Drennan C. L. (2011) Structural insights into radical generation by the radical SAM superfamily. Chem. Rev. 111, 2487–2506 10.1021/cr9002616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Davis K. M., Schramma K. R., Hansen W. A., Bacik J. P., Khare S. D., Seyedsayamdost M. R., and Ando N. (2017) Structures of the peptide-modifying radical SAM enzyme SuiB elucidate the basis of substrate recognition. Proc. Natl. Acad. Sci. U.S.A. 114, 10420–10425 10.1073/pnas.1703663114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hänzelmann P., and Schindelin H. (2004) Crystal structure of the S-adenosylmethionine-dependent enzyme MoaA and its implications for molybdenum cofactor deficiency in humans. Proc. Natl. Acad. Sci. U.S.A. 101, 12870–12875 10.1073/pnas.0404624101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Barr I., Stich T. A., Gizzi A. S., Grove T. L., Bonanno J. B., Latham J. A., Chung T., Wilmot C. M., Britt R. D., Almo S. C., and Klinman J. P. (2018) X-ray and EPR characterization of the auxiliary Fe–S clusters in the radical SAM enzyme PqqE. Biochemistry 57, 1306–1315 10.1021/acs.biochem.7b01097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lanz N. D., Pandelia M.-E., Kakar E. S., Lee K.-H., Krebs C., and Booker S. J. (2014) Evidence for a catalytically and kinetically competent enzyme–substrate cross-linked intermediate in catalysis by lipoyl synthase. Biochemistry 53, 4557–4572 10.1021/bi500432r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lee K.-H., Saleh L., Anton B. P., Madinger C. L., Benner J. S., Iwig D. F., Roberts R. J., Krebs C., and Booker S. J. (2009) Characterization of RimO, a new member of the methylthiotransferase subclass of the radical SAM superfamily. Biochemistry 48, 10162–10174 10.1021/bi900939w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pandelia M.-E., Lanz N. D., Booker S. J., and Krebs C. (2015) Mössbauer spectroscopy of Fe/S proteins. Biochim. Biophys. Acta 1853, 1395–1405 10.1016/j.bbamcr.2014.12.005 [DOI] [PubMed] [Google Scholar]
- 30. Evans R. L. 3rd, Latham J. A., Xia Y., Klinman J. P., and Wilmot C. M. (2017) Nuclear magnetic resonance structure and binding studies of PqqD, a chaperone required in the biosynthesis of the bacterial dehydrogenase cofactor pyrroloquinoline quinone. Biochemistry 56, 2735–2746 10.1021/acs.biochem.7b00247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsai T. Y., Yang C. Y., Shih H. L., Wang A. H., and Chou S. H. (2009) Xanthomonas campestris PqqD in the pyrroloquinoline quinone biosynthesis operon adopts a novel saddle-like fold that possibly serves as a PQQ carrier. Proteins 76, 1042–1048 10.1002/prot.22461 [DOI] [PubMed] [Google Scholar]
- 32. Koehnke J., Mann G., Bent A. F., Ludewig H., Shirran S., Botting C., Lebl T., Houssen W., Jaspars M., and Naismith J. H. (2015) Structural analysis of leader peptide binding enables leader-free cyanobactin processing. Nat. Chem. Biol. 11, 558–563 10.1038/nchembio.1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ortega M. A., Hao Y., Zhang Q., Walker M. C., van der Donk W. A., and Nair S. K. (2015) Structure and mechanism of the tRNA-dependent lantibiotic dehydratase NisB. Nature 517, 509–512 10.1038/nature13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wecksler S. R., Stoll S., Tran H., Magnusson O. T., Wu S.-P. King D., Britt R. D., and Klinman J. P. (2009) Pyrroloquinoline quinone biogenesis: demonstration that PqqE from Klebsiella pneumoniae is a radical S-adenosyl-l-methionine enzyme. Biochemistry 48, 10151–10161 10.1021/bi900918b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saichana N., Tanizawa K., Ueno H., Pechoušek J., Novák P., and Frébortová J. (2017) Characterization of auxiliary iron–sulfur clusters in a radical S-adenosylmethionine enzyme PqqE from Methylobacterium extorquens AM1. FEBS Open Bio 7, 1864–1879 10.1002/2211-5463.12314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dailey T. A., and Dailey H. A. (2002) Identification of [2Fe-2S] clusters in microbial ferrochelatases. J. Bacteriol. 184, 2460–2464 10.1128/JB.184.9.2460-2464.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paddock M. L., Wiley S. E., Axelrod H. L., Cohen A. E., Roy M., Abresch E. C., Capraro D., Murphy A. N., Nechushtai R., Dixon J. E., and Jennings P. A. (2007) MitoNEET is a uniquely folded 2Fe–2S outer mitochondrial membrane protein stabilized by pioglitazone. Proc. Natl. Acad. Sci. U.S.A. 104, 14342–14347 10.1073/pnas.0707189104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Benjdia A., Guillot A., Lefranc B., Vaudry H., Leprince J., and Berteau O. (2016) Thioether bond formation by SPASM domain radical SAM enzymes: Cα H-atom abstraction in subtilosin A biosynthesis. Chem. Commun. 52, 6249–6252 10.1039/C6CC01317A [DOI] [PubMed] [Google Scholar]
- 39. Harmer J. E., Hiscox M. J., Dinis P. C., Fox S. J., Iliopoulos A., Hussey J. E., Sandy J., Van Beek F. T., Essex J. W., and Roach P. L. (2014) Structures of lipoyl synthase reveal a compact active site for controlling sequential sulfur insertion reactions. Biochem. J. 464, 123–133 10.1042/BJ20140895 [DOI] [PubMed] [Google Scholar]
- 40. McLaughlin M. I., Lanz N. D., Goldman P. J., Lee K.-H., Booker S. J., and Drennan C. L. (2016) Crystallographic snapshots of sulfur insertion by lipoyl synthase. Proc. Natl. Acad. Sci. U.S.A. 113, 9446–9450 10.1073/pnas.1602486113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Berkovitch F., Nicolet Y., Wan J. T., Jarrett J. T., and Drennan C. L. (2004) Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science 303, 76–79 10.1126/science.1088493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sheldrick G. M. (2010) Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr. D Biol. Crystallogr. 66, 479–485 10.1107/S0907444909038360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., and Paciorek W. (2003) Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 10.1107/S0907444903017694 [DOI] [PubMed] [Google Scholar]
- 44. Vonrhein C., Blanc E., Roversi P., and Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol Biol. 364, 215–230 10.1385/1-59745-266-1:215 [DOI] [PubMed] [Google Scholar]
- 45. Abrahams J. P., and Leslie A. G. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52, 30–42 10.1107/S0907444995008754 [DOI] [PubMed] [Google Scholar]
- 46. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 48. Chen V. B., Arendall W. B. 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., and Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schrödinger, LLC, New York [Google Scholar]
- 50. Morin A., Eisenbraun B., Key J., Sanschagrin P. C., Timony M. A., Ottaviano M., and Sliz P. (2013) Cutting edge: collaboration gets the most out of software. Elife 2, e01456 10.7554/eLife.01456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Atkinson H. J., Morris J. H., Ferrin T. E., and Babbitt P. C. (2009) Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS One 4, e4345 10.1371/journal.pone.0004345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smoot M. E., Ono K., Ruscheinski J., Wang P. L., and Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27, 431–432 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Akiva E., Brown S., Almonacid D. E., Barber A. E. 2nd, Custer A. F., Hicks M. A., Huang C. C., Lauck F., Mashiyama S. T., Meng E. C., Mischel D., Morris J. H., Ojha S., Schnoes A. M., Stryke D., et al. (2014) The Structure–Function Linkage Database. Nucleic Acids Res. 42, D521–D530 10.1093/nar/gkt1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.